Abstract
Recent advances and future perspectives of machine learning techniques offer promising applications in medical imaging. Machine learning has the potential to improve different steps of the radiology workflow including order scheduling and triage, clinical decision support systems, detection and interpretation of findings, postprocessing and dose estimation, examination quality control, and radiology reporting. In this article, the authors review examples of current applications of machine learning and artificial intelligence techniques in diagnostic radiology. In addition, the future impact and natural extension of these techniques in radiology practice are discussed.
© RSNA, 2018
Introduction
Recent advances in machine learning offer promise in numerous industries and applications, including medical imaging (1). Within the innovations of data science, machine learning is a class of techniques and area of research that is enabling computers to learn like humans and to extract or classify patterns. Machines may further be able to analyze more data sets and extract features from data that humans may not be able to do (2). Recent research and developments are enabling technologies that hold promise now and in the future for diagnostic imaging (3). In this review article, we will first define what is meant by “machine learning” at broad and granular levels, providing an introduction into how such techniques can be developed and applied to imaging interpretation. Second, we will provide examples of applications of machine learning in diagnostic radiology. Third, we will discuss the key barriers and challenges in clinical application of machine learning techniques. Finally, we will discuss the future direction and natural extension of machine learning in radiology and beyond radiology in medicine.
Fundamentals of Machine Learning
Definition of Machine Learning
Machine learning is a method of data science that provides computers with the ability to learn without being programmed with explicit rules (2). Machine learning enables the creation of algorithms that can learn and make predictions. In contrast to rules-based algorithms, machine learning takes advantage of increased exposure to large and new data sets and has the ability to improve and learn with experience (3,4).
Machine Learning Categories
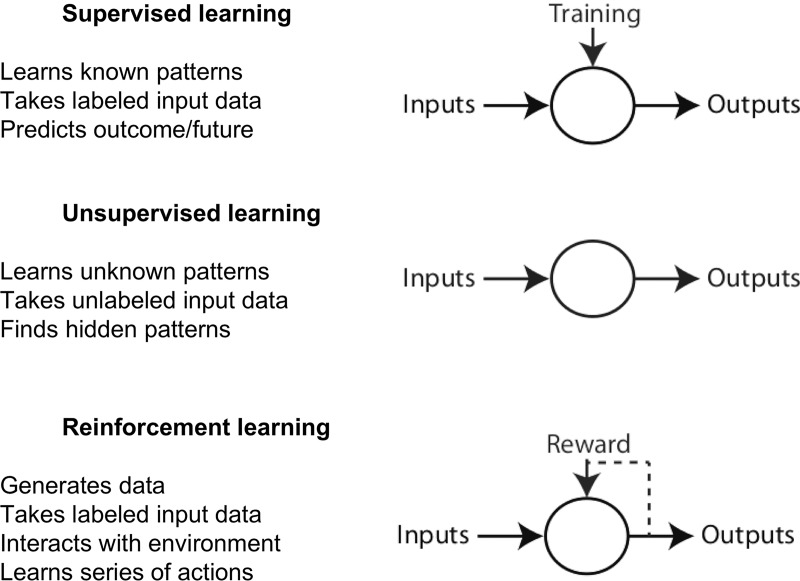
Machine learning tasks are typically classified into three broad categories (5), depending on the type of task: supervised, unsupervised, and reinforcement learning (Fig 1).
Figure 1:
Image shows different categories of machine learning.
In supervised learning, data labels are provided to the algorithm in the training phase (there is supervision in training). The expected outputs are usually labeled by human experts and serve as ground truth for the algorithm. The goal of the algorithm is usually to learn a general rule that maps inputs to outputs. In machine learning, ground truth refers to the data assumed to be true. In unsupervised learning, no data labels are given to the learning algorithm. The goal of the machine learning task is to find the hidden structure in the data and to separate data into clusters or groups. In reinforcement learning, a computer program performs a certain task in a dynamic environment in which it receives feedback in terms of positive and negative reinforcement (such as playing a game against an opponent) (6). Reinforcement learning is learning from the consequences of interactions with an environment without being explicitly taught. Examples of supervised and unsupervised learning techniques are provided in Figure 2. A machine learning paradigm may use a combination of supervised and unsupervised methods with a reinforcement feedback loop.
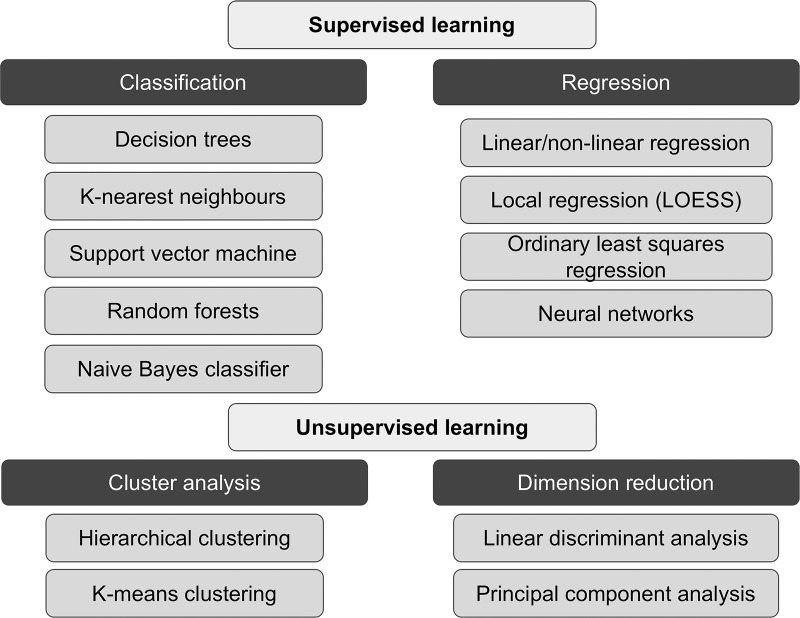
Figure 2:
Image shows summary of supervised and unsupervised learning paradigms and subcategories, with examples in each subcategory.
Artificial Neural Networks
Artificial neural networks (Fig 3) are statistical and mathematical methods that are a subset of machine learning. These networks are inspired by the way biologic nervous systems process information with a large number of highly interconnected processing elements, which are called neurons, nodes, or cells (7). An artificial neural network is structured as one input layer of neurons, one or more “hidden layers,” and one output layer. Each hidden layer is made up of a set of neurons, where each neuron is fully connected to all neurons in the previous layer. The strength of each connection is quantified with its own weight. For the network to yield the correct outputs (eg, correct detection and classification of findings on images), the weights must be set to suitable values, which are estimated through a training process. Learning in artificial neural networks could be supervised, partially supervised, or unsupervised.
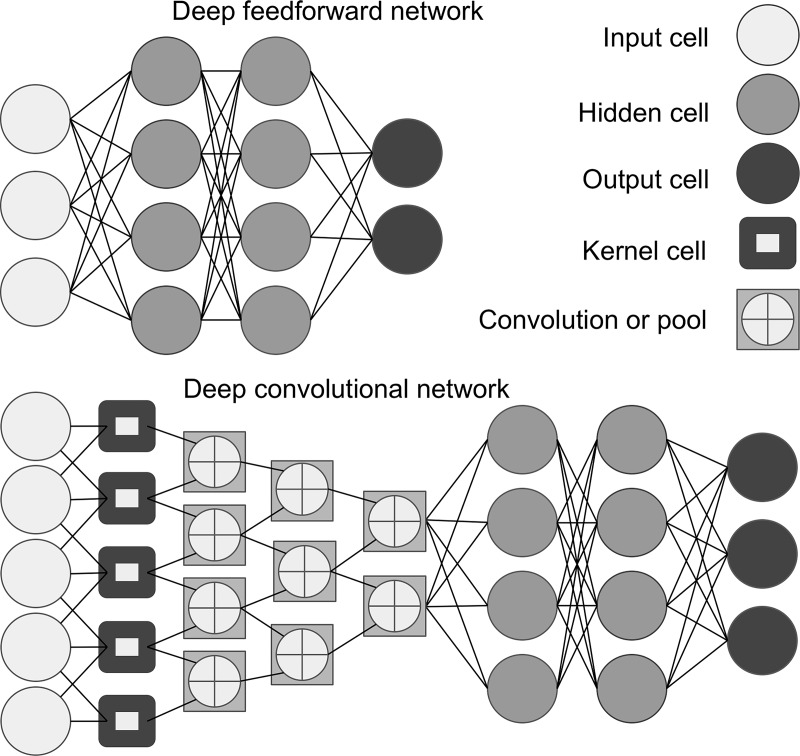
Figure 3:
Image shows artificial neural network, an interconnected group of processing elements similar to network of neurons in the brain. Each processing element is called a cell (also called neuron or node). Multiple hidden layers with nodes allow for multiple mathematical calculations to generate outputs. Deep learning is an artificial neural network algorithm that contains more than one hidden layer. Feedforward neural network (top panel) is the simplest type of artificial neural network. In this network, information moves in only one direction (forward) from input nodes, through hidden nodes, and to output nodes. Convolutional neural network (bottom panel) is a type of feedforward artificial neural network built from multiple hidden layers including convolutional layers, pooling layers, fully connected layers, and normalization layers. Convolution layer is comprised of filter elements known as kernels.
Deep Learning and Convolutional Neural Networks
Deep learning (also known as deep structured learning, hierarchical learning, or deep machine learning) is a subset of artificial neural network algorithms that contain more than one hidden layer (typically many more, and thus are “deep”). In other words, deep learning algorithms are based on a set of algorithms that attempt to model high-level abstractions in data (8). A typical application of machine learning, a use case, is object recognition on images. An example of object recognition with deep learning and details on the analysis performed in different layers of a neural network is available online (9).
Deep learning models can be categorized as typical (or normal) networks that take vector form (one-dimensional) inputs (ie, nonstructured input) or convolutional neural networks (CNNs) that takes two-dimensional or three-dimensional shaped inputs (ie, structured input). Given the configural information among neighboring pixels or voxels on images (structured input), CNNs have gained great interest in medical image analysis, particularly for feature extraction from images (10).
Convolution is a mathematical operation that has applications in finding patterns in signals or filtering signals. CNNs are formed by a stack of an input, an output layer, as well as multiple hidden layers that filter (convolve) the inputs to get the useful information (Fig 3). The hidden layers of a CNN typically consist of convolutional layers, pooling layers, fully connected layers, and normalization layers.
The main actors on CNNs are the convolution layers, which are comprised of filter elements known as kernels (11). The pooling (or downsampling) layer is used to reduce the spatial dimensions to gain computational performance and also to reduce the chance of overfitting. CNNs (Fig 4) are currently the most commonly applied machine learning technique in medical imaging (4,11).
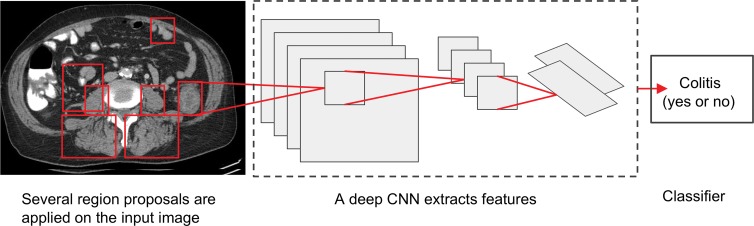
Figure 4:
Image shows proposed deep convolutional neural network (CNN) system for detection of colitis. In the first step, several thousand automated regions are applied on each CT section with an algorithm that finds all possible places where objects can be located (region proposal). For each region proposal, feature extraction and computation are performed by implementation of CNN with multiple hidden layers by using pretrained data sets. In the last step, classifier algorithm (eg, linear support vector machine) could be used for colitis classification.
Transfer Learning
Transfer learning is a machine learning approach that applies knowledge learned from a previous task to a different but related task. Transfer learning allows us to use the already existing labeled data for a new but related task. For example, a CNN pretrained on ImageNet (http://www.image-net.org) for nonmedical image classification and visual recognition has been used for feature extraction and survival prediction of lung tumors on CT scans (12). ImageNet is a large database of images that have been annotated by hand to indicate what objects are pictured. ImageNet Large Scale Visual Recognition Challenge is an annual contest in which software programs compete to correctly detect and classify objects on images. Knowledge gained from nonmedical image analysis is transferable to medical image analysis. Transfer learning creates a promising opportunity for rapid progress of machine learning in different domains.
Machine Learning Data Sets
In general, machine learning techniques are developed by using a train-test system (5). Three primary sets of data for training, testing, and validation are ideally needed. The training data set is used to fit the model. During training, the algorithm learns from examples. The validation set is used to evaluate different model fits on a separate data and to tune the model parameters. Most training approaches tend to overfit training data, meaning that they find relationships that fit the training data set well but do not hold in general. Therefore, successive iterations of training and validation may be performed to optimize the algorithm and avoid overfitting. In the testing set, after a machine learning algorithm is initially developed, the final model fit may then be applied to an independent testing data set to assess the performance, accuracy, and generalizability of the algorithm.
Open-Source Tools for Deep Neural Network Machine Learning
Machine learning algorithms can be deployed with relative simplicity given low cost of software tools and if armed with an appropriate foundation of knowledge (3). There are numerous open-source tools for deep learning (summarized in Table 1). There has been an increasing trend by independent software developers, data scientists, and corporate entities such as Google to democratize machine learning technologies (Fig 5).
Table 1:
Open-Source Tools for Deep Neural Network Machine Learning

Note.—Websites were last accessed on July 26, 2017.
Figure 5:
Image shows that feature extraction and object recognition, qualified by using confidence indicators, is made simple with toolkits such as TensorFlow object detection application programming interface. (Image courtesy of Omid Khalilzadeh, MD, MPH, Massachusetts General Hospital, Boston, Mass.)
Why Now? Convergence of Computing Power and Data
Major advances in processing units based on massive concurrent parallel processing chip architectures for graphics processing, combined with parallel computing approaches (historically available for graphical rendering and gaming), have rapidly accelerated the capability of artificial neural networks by making truly deep neural networks possible. In addition, enterprises are amassing large stores of digital data including medical images, which have already been digital for decades. Furthermore, there has been democratization of many free open-source algorithms available for machine learning such as Caffe, Torch, and TensorFlow (see Table 1). Large amounts of training data are also available, such as generalized ImageNet (13,14).
Why Machine Learning Is Powerful
Fundamentally, machine learning is powerful because it is not “brittle.” A rules-based approach may break when exposed to the real world, because the real world often offers examples that are not captured within the rules that programmer uses to define an algorithm. With machine learning, the system simply uses statistical approximation to respond most appropriately based on its training set, which means that it is flexible. Additionally, machine learning is a powerful tool because it is generic, that is, the same concepts are used for self-driving cars as is used for medical imaging interpretation. Generalizability of machine learning allows for rapid expansion in different fields, including medicine.
Machine Learning versus Artificial Intelligence
Compared with machine learning, artificial intelligence (or machine intelligence) encompasses a broader range of intelligent functions performed by computers such as problem solving, planning, knowledge representation, language processing, or “learning.” Therefore, machine learning is one type of artificial intelligence (15). For example, rule-based algorithms, such as computer-aided diagnosis used for several years in mammography, represent a type of artificial intelligence but not a type of machine learning. Computer-aided diagnosis is, however, a broader term and may incorporate machine learning approaches. By definition, machine learning algorithms improve automatically through experience and are not rule based. Machine learning is becoming more popular at different use cases, and in fact many artificial intelligence applications are currently using machine learning approaches (15).
Applications of Machine Learning in Diagnostic Imaging
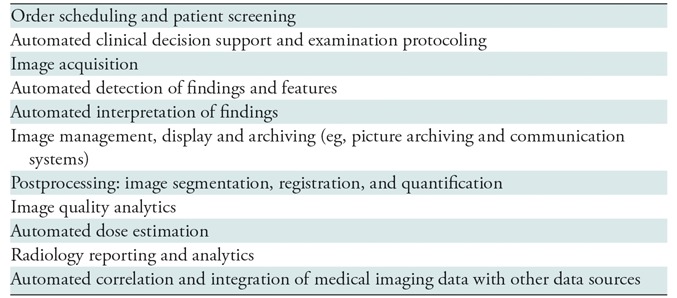
Although most of the literature is focused on the role of machine learning in detection of radiology findings, machine learning also has the potential to improve different steps of radiology workflow (Table 2), as described in the following sections.
Table 2:
Clinical Applications of Machine Learning in Radiology
Order Scheduling and Patient Screening
Intelligent scheduling facilitated by machine learning techniques can optimize patient scheduling and reduce the likelihood of missing care as a result of not attending the medical and radiology appointments. A project led by Dr Efren Flores at Massachusetts General Hospital (Boston, Mass) is using machine learning and predictive analytics for identification of patients who are at high risk for missing radiology care and not attending their appointments (16). The team is developing individualized solutions to reduce the chance of missing care.
In addition, machine learning applications are proposed for patient safety screening (17) or enhancement of safety reports (18), which have the potential for applications in radiology practice (for example, MRI safety screening or administration of contrast material).
Image Acquisition
Machine learning could make imaging systems intelligent. Machine learning–based data processing methods have the potential to decrease imaging time (19). Further, intelligent imaging systems could reduce unnecessary imaging, improve positioning, and help improve characterization of the findings. For example, an intelligent MR imager may recognize a lesion and suggest modifications in the sequence to achieve optimal characterization of the lesion (4).
Automated Detection of Findings
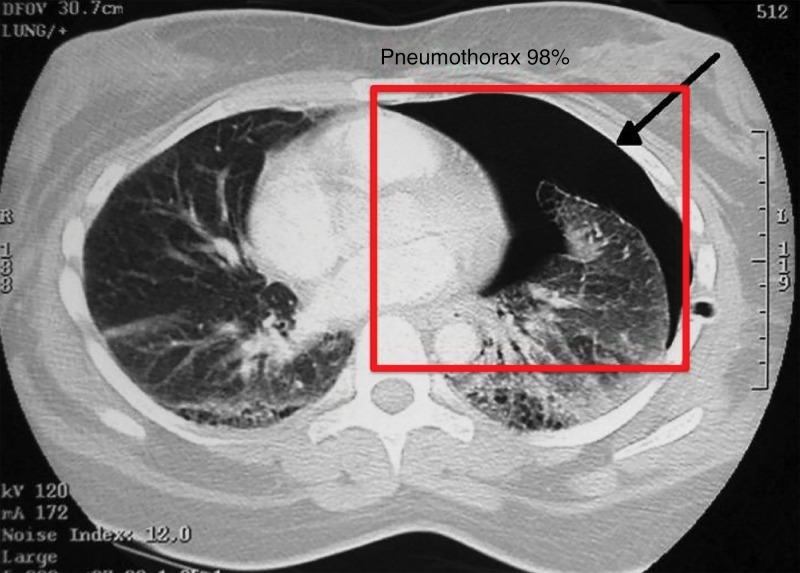
Automated detection of findings within medical images in radiology is an area where machine learning can make an impact immediately. For instance, the extraction of incidental findings such as pulmonary and thyroid nodules (20–22) has been demonstrated to be possible with machine learning techniques. Further machine learning research has also been performed for detection of critical findings such as pneumothorax (Fig 6), fractures, organ laceration, and stroke (23–29).
Figure 6:
Image shows feature extraction example in medical imaging use case. Automated detection of critical findings such as pneumothorax in medical imaging is one application of machine learning. Heads-up display or method of highlighting relevant findings in a picture archiving and communication system or other image viewing system is example of how machine learning can be productized and integrated into radiology workflow.
The algorithms that fall within the categories of computer-aided detection and computer-aided diagnosis have been used for decades (30–32). In mammography, computer-aided diagnosis has shown effectiveness (33). However, there is controversy that computer-aided diagnosis is to some extent ignored by some mammographers and may have limited benefit clinically (34).
Breast cancer screening is one of the first areas where machine learning is expected to be incorporated into radiology practice (35). Several studies have shown the diagnostic value of machine learning techniques in different breast imaging modalities including mammography (36), US (37), MRI (38), and tomosynthesis (39).
Interest has grown in the role of machine learning in detection, classification, and management of pulmonary nodules (40). For example, a deep learning system to classify pulmonary nodules performs within the interobserver variability of experienced human observers (41). Machine learning algorithms have also aided in reduction of false-positive results in detection of pulmonary nodules (20). The recent Kaggle Data Science Bowl saw nearly 10 000 participants compete for $1 million in prize money; competitors achieved high levels of performance in identifying candidates likely to be diagnosed with lung cancer within 1 year (https://www.kaggle.com/c/data-science-bowl-2017). A follow-up challenge has been proposed to bring these models to the clinic (https://www.datasciencebowl.com/totheclinic).
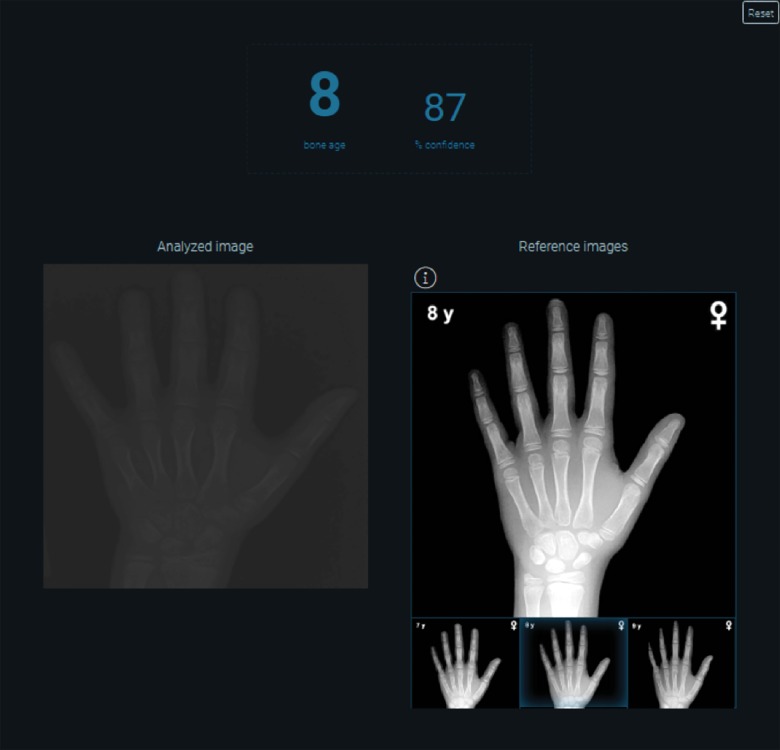
Bone age analysis and automated determination of anatomic age based on medical imaging holds considerable utility for pediatric radiology and endocrinology. Dr. Synho Do and colleagues created an algorithm that accurately characterizes bone age based on inputs of hand radiographs of pediatric patients (Fig 7) (42).
Figure 7:
Image shows automated bone age algorithm based on machine learning techniques. Opportunities exist to automate and help replace more manual workflows, such as use of book-based references.
Other potential use cases of machine learning include line detection (43), prostate cancer detection at MRI (44–46), determination of coronary artery calcium score (47), or detection and segmentation of brain lesions (48,49).
Automated Interpretation of Findings
Interpretation of the detected findings in medical imaging (either normal or abnormal) requires a high level of expert knowledge, experience, and clinical judgment based on each clinical case scenario. A simple example is intra-abdominal free air, which is most likely a normal finding in a postoperative patient but a critical finding in a patient without recent surgery. For a machine to function as an independent image interpreter, extensive acquisition of data-derived knowledge is required (50). Interpretation-based systems have been developed to find life-threatening abnormalities on the images (eg, intracranial hemorrhage), although this system is for prioritizing studies on a worklist as opposed to performing a final read for the study (51).
Several studies have shown that machine learning could improve interpretation of findings as an aid to the radiologist (4). Feature extraction from breast MR images by machine learning could improve interpretation of findings for breast cancer diagnosis (38). A machine-learning method based on radiologic traits (semantic features such as contour, texture, and margin) of the incidental pulmonary nodules has been shown to improve accuracy of cancer prediction and diagnostic interpretation of pulmonary nodules (40).
Automated Clinical Decision Support and Examination Protocoling
Machine learning techniques could further enhance radiology decision support tools (52). It has been suggested that an artificial intelligence simulation framework can approximate optimal human decisions even in complex and uncertain environments (53). Intelligent clinical decision support systems could improve quality of care and imaging efficiency and reduce the likelihood of adverse events or mistakes in examination protocoling (54).
Postprocessing: Image Segmentation, Registration, and Quantification
As more imaging data are becoming available, with the help of machine learning, medical imaging has made considerable progress in postprocessing tasks such as image registration, segmentation, and quantification. An intelligent medical imaging paradigm is data driven and tends to learn clinically useful information from the medical images (55). Extraction of clinically relevant data from medical images requires accurate image registration and segmentation. Several studies have used machine learning approaches for image segmentations, such as segmentation of breast density on mammography (56), body organs (35), or joint and musculoskeletal tissues at MRI (57).
Machine learning could also be used for intermodality image synthesis. For example, estimation of CT images from the corresponding MR images may be possible by using a generative adversarial network (58). The generative adversarial networks are neural networks that use two competing neural network models: a noise generator model that produces noise data and a discriminator model that distinguishes real data from noise. Over the process of training, the discriminator model learns to better distinguish noise from real data. Compared with other neural networks, the generative adversarial networks show superior performance in image generation tasks (59).
Image registration is often used in multimodality overlays such as PET/CT registration and for comparison or subtraction of images. Deep learning could play a considerable role in image registration, in which manual contouring and registration is time consuming and may suffer from inter- or intrarater variability (60). An example is application of unsupervised deep learning for deformable registration of brain MR images (61).
Machine learning could be used for quantitative assessment of three-dimensional structures in cross-sectional imaging (62,63). Wang et al used a CNN-based algorithm to accurately segment adipose tissue volume on CT images (62). Brain MRI anatomic segmentation has also been performed by using deep learning algorithms for delineation and quantitative assessment of brain structures and lesions (63).
Image Quality Analytics
Trained human observers (eg, experienced radiologists) are considered to be the reference for task-based evaluation of medical image quality. However, a long time is required to evaluate a large number of images. To address this problem, machine learning numerical observers (also known as model observers) have been developed as a surrogate for human observers for image quality assessment (64). A model observer can be applied to optimize parameters and evaluate image quality of low-dose CT iterative reconstructions (65).
Automated image quality evaluation using deep learning has been attempted by researchers. For instance, work has been performed in automated image quality evaluation of liver MRI (66).
A recent study (67) suggested that neural network algorithms could be applied for noise reduction from low-dose CT images. Training with an adversarial network improved the ability of the CNN to generate images with a quality similar to that of routine-dose CT (67).
Automated Radiation Dose Estimation
Machine learning could be used for organ-specific classification and organ radiation dose estimation from CT data sets. A recent study (68) revealed accuracy of higher than 96% in organ mapping and organ-specific radiation dose estimation when employing a deep CNN classifier on CT data sets. More work on automated radiation dose estimation has been done in the field of radiation oncology, which has the potential for similar applications in diagnostic radiology (69,70). For example, a machine learning framework for radiation therapy planning for prostate cancer with MRI has shown reduction in dosage to the organs at risk and a boosted dose delivered to the cancerous lesions (71).
Radiology Reporting and Analytics
Machine learning techniques have been widely applied in natural language processing. Data extraction from free-text radiology reports with natural language processing has applications for quality assurance and performance monitoring, as well as large-scale testing of clinical decision support (72). Natural language processing engines may extract findings and organ measurements from narrative radiology reports and categorize extracted measurements (73). This can provide radiologic input data for other machine learning applications that process medical data. Machine learning techniques could also be used to extract terminology from radiology reports for quality improvement and analytics (74). Machine learning and natural language processing algorithms could help track radiologists’ recommendation and reduce the chance of disconnect in communication of follow-up recommendations (75).
Automated Data Integration and Analysis
In electronic medical records, a heterogeneous pool of data from various data sources exist (called multiview data). There is a high throughput of data from different sources including medical histories and progress notes, laboratory results, pathology reports and images, radiology reports and images, genomics, and safety reports into medical record systems. The availability of these data provides unprecedented opportunities for data mining but also raises challenges for integration of heterogeneous data sources (for example, imaging data vs textual data) (76). Various machine learning techniques such as kernel methods, matrix factorization models, and network-based fusion methods could be applied for data integration and analysis (77).
Key Barriers and Challenges
Collection of high-quality ground truth data, development of generalizable and diagnostically accurate techniques, and workflow integration are the key challenges facing adoption of machine learning in radiology practice.
Machine Learning Performance: Large Data Sets Typically Required but Not Always
Peter Norvig of Google demonstrated that large volumes of data may overcome deficiencies in machine learning algorithms (78). Narrow-scope machine learning algorithms may not require large amounts of training data, but instead may require high-quality ground truth training data. In medical imaging analysis, as with other kinds of machine learning, the amount of data that is required varies largely on the task to perform. For example, segmentation tasks may only require a small set data. On the other hand, performance classification tasks (eg, classifying a liver lesion as malignant vs benign) may require many more label examples, which may also be largely dependent on the number of classifiers to distinguish between (79).
Confounders in source data may result in possible failures of machine learning algorithms. Rare findings or features may also be possible weaknesses due to lack of large volume of a particular feature for neural networks, which are therefore vulnerable to inaccuracies (43).
Variance and bias are issues that may result in poor performance of a machine learning algorithm. Bias is erroneous assumptions in the algorithm that can result in missing the associations (underfitting). High variance can cause an algorithm to learn the data too well and start fitting random noise (overfitting). An optimal model is not only accurate in representation of the training data, but also generalizable to unseen data. An overfitted algorithm overreacts to minor variations from the training data. Therefore, the algorithm performs well on the training data and poorly with the new data. Overfitting is a major challenge in machine learning, particularly when a model is excessively complex (80).
Ground Truth Annotation
Extensive ground truth annotation is often required for proper training of machine learning algorithms. Multiple technology companies and academic research projects rely on trained radiologists to annotate what is considered ground truth on radiology reports and images (64). Extensive labor costs, time, and resources are required for endeavors to be properly implemented. Also, the validation process must be highly robust, otherwise the algorithm could be subject to overfitting to a particular subclassification of data (80).
Defining Standards
Appropriate development of artificial intelligence tools necessitates defining standardized use cases and annotation tools. These use cases will need to be consistent with clinical practice, as well as regulatory, legal, and ethical issues that accompany artificial intelligence in medical imaging. The clinical panels of the American College of Radiology Data Science Institute, in conjunction with other medical specialty societies, are defining these standardized use cases (https://www.acr.org/Advocacy/Informatics/Data-Science-Institute).
In addition, a standard approach could make image annotations interoperable between different information technology systems and software applications that communicate and exchange data. The National Cancer Institute's Annotation and Image Markup model offers a possible standard approach to annotation of images and image features (81).
Regulation and Workflow Integration
Machine learning–based algorithms are not currently well integrated into picture archiving and communication system workstations. Many systems incorporate and require a separate workstation or network node for sending images for analysis. Within the ecosystem of the radiology informatics value chain, more work is needed to better incorporate novel machine learning technologies. Standards may need to be set for interoperability of machine learning algorithms with existing systems. Vendors and researchers alike must aim to create platforms that will allow for continuous learning and upgrades of machine learning algorithms. Machine learning algorithms need to be updated continuously based on possible changes in the model through exposure to more data.
An important step toward integration of machine learning in the clinical setting is approval from the U.S. Food and Drug Administration (FDA). Before clinical use, machine learning applications should submit specific information about the algorithm development and clinical validation to the FDA. Clinical validation studies should show sufficient agreement with human experts. The FDA is facing challenges in regulating this software and is currently developing appropriate regulatory pathways for machine learning applications. For example, human expert validation is challenging for machine learning algorithms designed to find associations in data that have eluded the human eye. Another example is algorithms that continue to learn in the hands of users and perform better over time. This is challenging because the FDA needs assurance that the performance will improve consistently and will not decline. The FDA may need different regulatory approaches for software that functions like a “black box” does and just provide the clinical advice or software, which allow health care professionals to review independently the basis for the recommendations (82).
Deciphering the Black Box of Artificial Intelligence
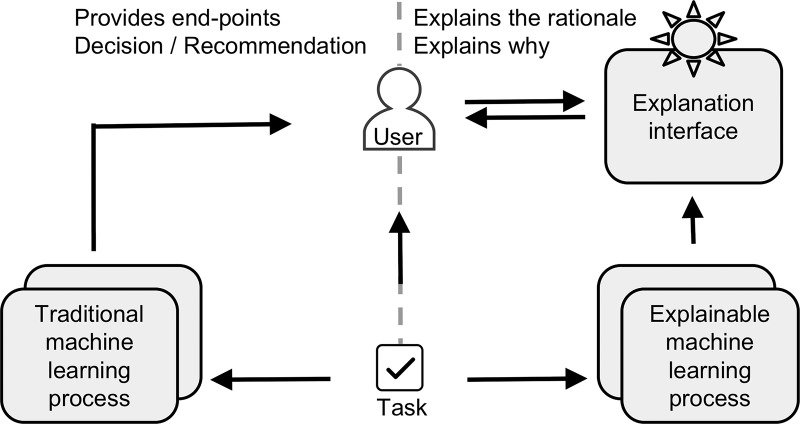
By its very nature, machine learning develops complex and high-dimensional functions that cannot be explained in simple terms. This makes interpretability of machine learning one of the main challenges for its acceptance in the areas where identifying the underlying causes and logic is important (such as health care). Currently, there is limited visibility into deconstructing drivers of a machine’s decision when learning is unsupervised. Work such as the Explainable Artificial Intelligence program by the Defense Advanced Research Projects Agency is being performed such that artificial intelligence and machine learning–based algorithms can be better understood in how these models reach their conclusions (https://www.darpa.mil/program/explainable-artificial-intelligence) (Fig 8).
Figure 8:
Image compares traditional (left panel) and explainable (right panel) artificial intelligence systems. Defense Advanced Research Projects Agency is developing explainable artificial intelligence systems that will have ability to explain their rationale through explanation interface. This approach will enable human users to more effectively manage and more confidently trust artificial intelligent systems.
Visual saliency is the perceptual quality that makes some items stand out to our vision from their neighbors and immediately grab our attention. Visual saliency maps could highlight areas within images that have grabbed the attention of human observers to perform a classification task. The saliency maps could provide “explicability” for the machine learning models and improve the accuracy for detection of findings (83).
Radiologist’ Job Perspective and Medicolegal Issues
The performance of machine learning systems for clinical diagnosis and decision making need to be monitored. Physicians take the ownership of the medical diagnoses and treatments delivered to patients (84). In case of medical errors, the manufacturer and developers of the machine learning systems may not be accountable, given that by definition, the computers are learning and relearning based on data sets in ways unknown to the developers. Clinical advice provided by the machine learning software may need to be reviewed by an expert health care professional, who may or may not approve the recommendation provided by the software. Preferably, the basis for the recommendations should also be provided to the health care professional and reviewed (85). For the foreseeable future, machine learning is not expected to replace the radiologists. Instead, these techniques are expected to aid the radiologists, enhance the radiology workflow, and improve the radiologists’ diagnostic accuracy. The machine learning systems may help to identify patterns and associations that may normally evade human eyes. Currently, many artificial intelligence systems are being developed on fairly obvious tasks that are of little challenge to the human. The artificial intelligence systems could add more value if efforts are focused on tasks that are challenging for radiologists.
Future Directions of Machine Learning in Radiology and Beyond Radiology in Medicine
Availability of a large amount of electronic medical record data allows for creation of an interdisciplinary data pool. Machine learning extracts knowledge from this big data and produces outputs that could be used for individual outcome prediction analysis and clinical decision making. This could make way for personalized medicine (or precision medicine), in which individual variability in genes, environment, and lifestyle factors for each person is taken into account for disease prevention, treatment, and prognostication.
Interdisciplinary Collaborations and Precision Medicine
Machine learning models have rapidly improved in the past few years. Google recently demonstrated a multimodel machine learning approach, which provides a template for the development of future systems that are more accurate and more broadly applicable to different disciplines, including medicine (86). Diagnostic imaging may be one of the first medical disciplines subject to the application of machine learning algorithms, but other fields such as pathology, cardiology, dermatology, and gastroenterology also have potential use cases (87,88).
Machine learning approaches to the interrogation of a wide spectrum of such data (sociodemographic, imaging, clinical, laboratory, and genetic) has the potential to further personalize health care, far beyond what would be possible through imaging applications alone (89). Precision medicine require the use of novel computational techniques to harness the vast amounts of data required to discover individualized disease factors and treatment decisions (87,90).
Radiomics is a process designed to extract a large number of quantitative features from radiology images (91). Radiomics is an emerging field for machine learning that allows for conversion of radiologic images into mineable high-dimensional data. For instance, Zhang et al (87) evaluated over 970 radiomics features extracted from MR images by using machine learning methods and correlated with features to predict local and distant treatment failure of advanced nasopharyngeal carcinoma.
Predictive Analytics
Prediction of treatment response and prognosis are areas where machine learning may hold promise. Early phases of this work have recently begun. For instance, brain tumor response to therapy can be estimated accurately with machine learning (92). Oakden-Rayner et al (93) used features within chest CTs to predict longevity of patients through detecting features indicative of overall individual health within those CTs.
Extension to the Electronic Medical Record and Other Nonimaging Clinical Data
In the future, imaging data will be linked more readily to nonimaging data in electronic medical records and other large data sets. Deep learning, when applied to electronic medical record data, can help derive patient representations that may lead to clinical predictions and augmentation of clinical decision support systems (94). Miotto et al evaluated medical records from over 700 000 patients with an unsupervised deep learning representation titled “deep patient.” They found broadly predictive characteristics for various health states (95).
In conclusion, with the current fast pace in development of machine learning techniques, and deep learning in particular, there is prospect for a more widespread clinical adoption of machine learning in radiology practice. Machine learning and artificial intelligence are not expected to replace the radiologists in the foreseeable future. These techniques can potentially facilitate radiology workflow, increase radiologist productivity, improve detection and interpretation of findings, reduce the chance of error, and enhance patient care and satisfaction.
Summary
Machine learning has the potential to improve different steps of the radiology workflow.
Essentials
■ Machine learning comprises a set of statistical tools that can allow computers to perform tasks by learning from examples without being explicitly programmed. Artificial neural networks are a subset of machine learning inspired by the human brain neuronal network. Deep learning is a subset of artificial neural network algorithms that contain more than one hidden processing layer.
■ Machine learning has the potential to improve different steps of the radiology workflow including order scheduling and patient screening, clinical decision support systems, detection and interpretation of findings, postprocessing and dose estimation, and radiology reporting.
■ Collection of high-quality ground truth data, development of generalizable and diagnostically accurate techniques, and workflow integration are key challenges for the creation and adoption of machine learning models in radiology practice.
■ Machine learning has the potential to personalize health care further. Availability of large electronic medical record data allows for creation of interdisciplinary big data sets that could be used for individual outcome prediction analysis and clinical decision making.
■ For the foreseeable future, widespread application of machine learning algorithms in diagnostic radiology is not expected to reduce the need for radiologists. Instead, these techniques are expected to improve radiology workflow, increase radiologist productivity, and enhance patient care and satisfaction.
Current address: Department of Radiology, Mount Sinai Health System, Icahn School of Medicine at Mount Sinai, New York, NY.
Disclosures of Conflicts of Interest: G.C. disclosed no relevant relationships. O.K. disclosed no relevant relationships. M.M. disclosed no relevant relationships. S.D. disclosed no relevant relationships. A.E.S. disclosed no relevant relationships. O.S.P. disclosed no relevant relationships. J.R.G. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: held stock/stock options in Montage Healthcare Solutions. Other relationships: disclosed no relevant relationships. P.V.P. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: has grants/grants pending with Medical Imaging and Technology Alliance. Other relationships: disclosed no relevant relationships. J.A.B. disclosed no relevant relationships. K.J.D. disclosed no relevant relationships.
Abbreviation
- CNN
- convolutional neural network
References
- 1.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. RadioGraphics 2017;37(2):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16(5):933–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science 2015;349(6245):255–260. [DOI] [PubMed] [Google Scholar]
- 4.Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 2017;208(4):754–760. [DOI] [PubMed] [Google Scholar]
- 5.Deo RC. Machine learning in medicine. Circulation 2015;132(20):1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge, Mass: MIT Press, 2012. [Google Scholar]
- 7.Dayhoff JE, DeLeo JM. Artificial neural networks: opening the black box. Cancer 2001;91(8 Suppl):1615–1635. [DOI] [PubMed] [Google Scholar]
- 8.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 9.Geitgey A. Machine learning is fun! Part 3: deep learning and convolutional neural networks. Medium. https://medium.com/@ageitgey/machine-learning-is-fun-part-3-deep-learning-and-convolutional-neural-networks-f40359318721. Published June 13, 2016. Accessed November 12, 2017.
- 10.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017;19(1):221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wernick MN, Yang Y, Brankov JG, Yourganov G, Strother SC. Machine learning in medical imaging. IEEE Signal Process Mag 2010;27(4):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul R, Hawkins SH, Balagurunathan Y, et al. Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma. Tomography 2016;2(4):388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: a large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009. [Google Scholar]
- 14.Li FF, Deng J, Li K. ImageNet: constructing a large-scale image database. J Vis 2010;9(8):1037. [Google Scholar]
- 15.Neapolitan RE, Jiang X. Contemporary artificial intelligence. Boca Raton, Fla: CRC, 2012. [Google Scholar]
- 16.Glover M, 4th, Daye D, Khalilzadeh O, et al. Socioeconomic and demographic predictors of missed opportunities to provide advanced imaging services. J Am Coll Radiol 2017;14(11):1403–1411. [DOI] [PubMed] [Google Scholar]
- 17.Marella WM, Sparnon E, Finley E. Screening electronic health record-related patient safety reports using machine learning. J Patient Saf 2017;13(1):31–36. [DOI] [PubMed] [Google Scholar]
- 18.Fong A, Howe JL, Adams KT, Ratwani RM. Using active learning to identify health information technology related patient safety events. Appl Clin Inform 2017;8(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golkov V, Dosovitskiy A, Sperl JI, et al. Q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE Trans Med Imaging 2016;35(5):1344–1351. [DOI] [PubMed] [Google Scholar]
- 20.Setio AA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 2016;35(5):1160–1169. [DOI] [PubMed] [Google Scholar]
- 21.Zeng JY, Ye HH, Yang SX, et al. Clinical application of a novel computer-aided detection system based on three-dimensional CT images on pulmonary nodule. Int J Clin Exp Med 2015;8(9):16077–16082. [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Y, Paul AK, Kim N, et al. Computer-aided diagnosis for classifying benign versus malignant thyroid nodules based on ultrasound images: a comparison with radiologist-based assessments. Med Phys 2016;43(1):554–567. [DOI] [PubMed] [Google Scholar]
- 23.Reza Soroushmehr SM, Davuluri P, Molaei S, et al. Spleen segmentation and assessment in CT images for traumatic abdominal injuries. J Med Syst 2015;39(9):87. [DOI] [PubMed] [Google Scholar]
- 24.Pustina D, Coslett HB, Turkeltaub PE, Tustison N, Schwartz MF, Avants B. Automated segmentation of chronic stroke lesions using LINDA: lesion identification with neighborhood data analysis. Hum Brain Mapp 2016;37(4):1405–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier O, Schröder C, Forkert ND, Martinetz T, Handels H. Classifiers for ischemic stroke lesion segmentation: a comparison study. PLoS One 2015;10(12):e0145118. [Published correction appears in PLoS One 2016;11(2):e0149828.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Xie Y, Reeves AP. Automated 3D closed surface segmentation: application to vertebral body segmentation in CT images. Int J CARS 2016;11(5):789–801. [DOI] [PubMed] [Google Scholar]
- 27.Do S, Salvaggio K, Gupta S, Kalra M, Ali NU, Pien H. Automated quantification of pneumothorax in CT. Comput Math Methods Med 2012;2012:736320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herweh C, Ringleb PA, Rauch G, et al. Performance of e-ASPECTS software in comparison to that of stroke physicians on assessing CT scans of acute ischemic stroke patients. Int J Stroke 2016;11(4):438–445. [DOI] [PubMed] [Google Scholar]
- 29.Burns JE, Yao J, Muñoz H, Summers RM. Automated detection, localization, and classification of traumatic vertebral body fractures in the thoracic and lumbar spine at CT. Radiology 2016;278(1):64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi R, Kajikawa Y. Computer-aided diagnosis: a survey with bibliometric analysis. Int J Med Inform 2017;101:58–67. [DOI] [PubMed] [Google Scholar]
- 31.van Ginneken B. Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning. Radiol Phys Technol 2017;10(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suhail Z, Sarwar M, Murtaza K. Automatic detection of abnormalities in mammograms. BMC Med Imaging 2015;15(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dromain C, Boyer B, Ferré R, Canale S, Delaloge S, Balleyguier C. Computed-aided diagnosis (CAD) in the detection of breast cancer. Eur J Radiol 2013;82(3):417–423. [DOI] [PubMed] [Google Scholar]
- 34.Berlin L. Archive or discard computer-aided detection markings: two schools of thought. J Am Coll Radiol 2015;12(11):1134–1135. [DOI] [PubMed] [Google Scholar]
- 35.Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a random forest algorithm: a feasibility study. Phys Med Biol 2016;61(17):6553–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging; (Bellingham: ) 2016;3(3):034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu P, Lee WM, Roubidoux MA, Yuan J, Wang X, Carson PL. Automated 3D ultrasound image segmentation to aid breast cancer image interpretation. Ultrasonics 2016;65:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bickelhaupt S, Paech D, Kickingereder P, et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging 2017;46(2):604–616. [DOI] [PubMed] [Google Scholar]
- 39.Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys 2016;43(12):6654–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Balagurunathan Y, Atwater T, et al. Radiological image traits predictive of cancer status in pulmonary nodules. Clin Cancer Res 2017;23(6):1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging 2017;30(4):427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakhani P. Deep convolutional neural networks for endotracheal tube position and x-ray image classification: challenges and opportunities. J Digit Imaging 2017;30(4):460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le MH, Chen J, Wang L, et al. Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Phys Med Biol 2017;62(16):6497–6514. [DOI] [PubMed] [Google Scholar]
- 45.Turkbey B, Fotin SV, Huang RJ, et al. Fully automated prostate segmentation on MRI: comparison with manual segmentation methods and specimen volumes. AJR Am J Roentgenol 2013;201(5):W720–W729. [DOI] [PubMed] [Google Scholar]
- 46.Kwak JT, Xu S, Wood BJ, et al. Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Med Phys 2015;42(5):2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuhbaeck A, Otaki Y, Achenbach S, et al. Coronary calcium scoring from contrast coronary CT angiography using a semiautomated standardized method. J Cardiovasc Comput Tomogr 2015;9(5):446–453. [DOI] [PubMed] [Google Scholar]
- 48.García-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 2013;17(1):1–18. [DOI] [PubMed] [Google Scholar]
- 49.Mortazavi D, Kouzani AZ, Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: a review. Neuroradiology 2012;54(4):299–320. [DOI] [PubMed] [Google Scholar]
- 50.Velikova M, Lucas PJ, Samulski M, Karssemeijer N. On the interplay of machine learning and background knowledge in image interpretation by Bayesian networks. Artif Intell Med 2013;57(1):73–86. [DOI] [PubMed] [Google Scholar]
- 51.vRad and MetaMind collaborate on deep learning powered workflows to help radiologists accelerate identification of life-threatening abnormalities. PRWeb. http://www.prweb.com/releases/2015/06/prweb12790975.htm. Published June 16, 2015. Accessed July 22, 2017.
- 52.Bal M, Amasyali MF, Sever H, Kose G, Demirhan A. Performance evaluation of the machine learning algorithms used in inference mechanism of a medical decision support system. Sci World J 2014;2014:137896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett CC, Hauser K. Artificial intelligence framework for simulating clinical decision-making: a Markov decision process approach. Artif Intell Med 2013;57(1):9–19. [DOI] [PubMed] [Google Scholar]
- 54.Kohli M, Dreyer KJ, Geis JR. Rethinking radiology informatics. AJR Am J Roentgenol 2015;204(4):716–720. [DOI] [PubMed] [Google Scholar]
- 55.Rueckert D, Glocker B, Kainz B. Learning clinically useful information from images: past, present and future. Med Image Anal 2016;33:13–18. [DOI] [PubMed] [Google Scholar]
- 56.Kallenberg M, Petersen K, Nielsen M, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging 2016;35(5):1322–1331. [DOI] [PubMed] [Google Scholar]
- 57.Pedoia V, Majumdar S, Link TM. Segmentation of joint and musculoskeletal tissue in the study of arthritis. MAGMA 2016;29(2):207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie D, Trullo R, Petitjean C, Ruan S, Shen D. Medical image synthesis with context-aware generative adversarial networks. http://arxiv.org/abs/1612.05362. Published December 16, 2016. Accessed July 1, 2017. [DOI] [PMC free article] [PubMed]
- 59.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. http://arxiv.org/abs/1406.2661. Published June 10, 2014. Accessed October 29, 2017.
- 60.Ghose S, Holloway L, Lim K, et al. A review of segmentation and deformable registration methods applied to adaptive cervical cancer radiation therapy treatment planning. Artif Intell Med 2015;64(2):75–87. [DOI] [PubMed] [Google Scholar]
- 61.Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D. Unsupervised deep feature learning for deformable registration of MR brain images. Med Image Comput Comput Assist Interv 2013;16(Pt 2):649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Qiu Y, Thai T, Moore K, Liu H, Zheng B. A two-step convolutional neural network based computer-aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput Methods Programs Biomed 2017;144:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017;30(4):449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalayeh MM, Marin T, Brankov JG. Generalization evaluation of machine learning numerical observers for image quality assessment. IEEE Trans Nucl Sci 2013;60(3):1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eck BL, Fahmi R, Brown KM, et al. Computational and human observer image quality evaluation of low dose, knowledge-based CT iterative reconstruction. Med Phys 2015;42(10):6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esses SJ, Lu X, Zhao T, et al. Automated image quality evaluation of T2 -weighted liver MRI utilizing deep learning architecture. J Magn Reson Imaging 2017 Jun 3. [Epub ahead of print] [DOI] [PubMed]
- 67.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging 2017;36(12):2536–2545. [DOI] [PubMed] [Google Scholar]
- 68.Cho J, Lee E, Lee H, et al. Machine learning powered automatic organ classification for patient specific organ dose estimation. SIIM 2017 Scientific Session. Massachusetts General Hospital, Harvard Medical School, 2017. [Google Scholar]
- 69.Coates J, Souhami L, El Naqa I. Big data analytics for prostate radiotherapy. Front Oncol 2016;6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valdes G, Solberg TD, Heskel M, Ungar L, Simone CB, 2nd. Using machine learning to predict radiation pneumonitis in patients with stage I non-small cell lung cancer treated with stereotactic body radiation therapy. Phys Med Biol 2016;61(16):6105–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shiradkar R, Podder TK, Algohary A, Viswanath S, Ellis RJ, Madabhushi A. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol 2016;11(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai T, Giannopoulos AA, Yu S, et al. Natural language processing technologies in radiology research and clinical applications. RadioGraphics 2016;36(1):176–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sevenster M, Buurman J, Liu P, Peters JF, Chang PJ. Natural language processing techniques for extracting and categorizing finding measurements in narrative radiology reports. Appl Clin Inform 2015;6(3):600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassanpour S, Langlotz CP, Amrhein TJ, Befera NT, Lungren MP. Performance of a machine learning classifier of knee MRI reports in two large academic radiology practices: a tool to estimate diagnostic yield. AJR Am J Roentgenol 2017;208(4):750–753. [DOI] [PubMed] [Google Scholar]
- 75.Oliveira L, Tellis R, Qian Y, Trovato K, Mankovich G. Follow-up recommendation detection on radiology reports with incidental pulmonary nodules. Stud Health Technol Inform 2015;216:1028. [PubMed] [Google Scholar]
- 76.Greene CS, Tan J, Ung M, Moore JH, Cheng C. Big data bioinformatics. J Cell Physiol 2014;229(12):1896–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Wu FX, Ngom A. A review on machine learning principles for multi-view biological data integration. Brief Bioinform 2016 Dec 22 [Epub ahead of print]. [DOI] [PubMed]
- 78.Russell S, Norvig P. Artificial intelligence: a modern approach, global edition. 3rd ed. Harlow, England: Pearson Education, 2016. [Google Scholar]
- 79.Figueroa RL, Zeng-Treitler Q, Kandula S, Ngo LH. Predicting sample size required for classification performance. BMC Med Inform Decis Mak 2012;12(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18(4):570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mongkolwat P, Kleper V, Talbot S, Rubin D. The National Cancer Informatics Program (NCIP) Annotation and Image Markup (AIM) Foundation model. J Digit Imaging 2014;27(6):692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosenblatt M, Boutin MM, Nussbaum SR. Innovation in medicine and device development, regulatory review, and use of clinical advances. JAMA 2016;316(16):1671–1672. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Borji A, Jay Kuo CC, Itti L. Learning a combined model of visual saliency for fixation prediction. IEEE Trans Image Process 2016;25(4):1566–1579. [DOI] [PubMed] [Google Scholar]
- 84.Stamm JA, Korzick KA, Beech K, Wood KE. Medical malpractice: reform for today’s patients and clinicians. Am J Med 2016;129(1):20–25. [DOI] [PubMed] [Google Scholar]
- 85.Altman RB. Artificial intelligence (AI) systems for interpreting complex medical datasets. Clin Pharmacol Ther 2017;101(5):585–586. [DOI] [PubMed] [Google Scholar]
- 86.Kaiser L, Gomez AN, Shazeer N, et al. One model to learn them all. http://arxiv.org/abs/1706.05137. Published June 16, 2017. Accessed June 25, 2017.
- 87.Zhang B, He X, Ouyang F, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett 2017;403:21–27. [DOI] [PubMed] [Google Scholar]
- 88.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen JH, Asch SM. Machine learning and prediction in medicine: beyond the peak of inflated expectations. N Engl J Med 2017;376(26):2507–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 2017;69(21):2657–2664. [DOI] [PubMed] [Google Scholar]
- 91.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nie D, Zhang H, Adeli E, Liu L, Shen D. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. Med Image Comput Comput Assist Interv 2016;9901:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oakden-Rayner L, Carneiro G, Bessen T, Nascimento JC, Bradley AP, Palmer LJ. Precision radiology: predicting longevity using feature engineering and deep learning methods in a radiomics framework. Sci Rep 2017;7(1):1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tran T, Kavuluru R. Predicting mental conditions based on “history of present illness” in psychiatric notes with deep neural networks. J Biomed Inform 2017;75S:S138–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016;6(1):26094. [DOI] [PMC free article] [PubMed] [Google Scholar]