Abstract
Gain of the long arm of chromosome 17 (17q) is a cytogenetic hallmark of high-risk neuroblastoma, yet its contribution to neuroblastoma pathogenesis remains incompletely understood. Combining whole-genome and RNA sequencing of neuroblastomas, we identified the prohibitin (PHB) gene as highly expressed in tumors with 17q gain. High PHB expression correlated with poor prognosis and was associated with loss of gene expression programs promoting neuronal development and differentiation. PHB depletion induced differentiation and apoptosis and slowed cell cycle progression of neuroblastoma cells, at least in part through impaired ERK1/2 activation. Conversely, ectopic expression of PHB was sufficient to increase proliferation of neuroblastoma cells and was associated with suppression of markers associated with neuronal differentiation and favorable neuroblastoma outcome. Thus, PHB is a 17q oncogene in neuroblastoma that promotes tumor cell proliferation and dedifferentiation.
Keywords: Genetics, Oncology
Keywords: Cancer, Oncogenes
Prohibitin is a 17q oncogene that contributes to de-differentiation of neuroblastoma cells and represents a potential target for therapy.
Introduction
Neuroblastoma is a childhood tumor that arises from developing neural crest cells of the sympathetic nervous system and is characterized by a broad range of clinical behaviors (1). Although low-risk disease is readily treated with standard chemotherapy regimens or surgery and some tumors regress spontaneously, relapses are common, and fewer than half of all patients with high-risk disease survive longer than 5 years after diagnosis (2, 3). The genomes of primary neuroblastoma tumors are characterized by a relative paucity of somatic mutations, making identification of tumor-specific genetic targets a challenge (4). Gain of the long arm of chromosome 17 (17q) has long been recognized as the most common cytogenetic abnormality of high-risk neuroblastoma and is present in over 60% of tumors (5). Gain of 17q21 extending to 17qter correlates with poor prognosis, though the causes and consequences of 17q21-qter segment gain remain largely elusive (5, 6). One gene in this region with several known roles in cancer pathogenesis is prohibitin (PHB) (7). PHB and its homolog, PHB2, comprise the PHB family of proteins and may function as monomers, as homo- and heterodimers, or as a hetero-oligomeric ring complex in mitochondria (8). High expression of PHB is observed in a variety of cancers, including non–small cell lung cancer, pancreatic ductal adenocarcinoma, and gallbladder tumors (9–11). Moreover, cancer cells expressing high levels of PHB show an enhanced capacity for migration and invasion, cellular qualities associated with metastatic disease (12, 13). Notably, it was recently reported that PHB is required for dictating the directionality of invasive colorectal cancer cells (14). Although PHB has been extensively characterized in adult tumors, its role in pediatric cancers remains undefined. Because patients with disseminated disease have poor outcomes among neuroblastoma cases (1, 15), the identification and targeting of factors involved in migration, invasion, and metastasis are urgent.
Furthermore, recent whole-genome sequencing of paired diagnostic and relapse neuroblastomas identified mutations leading to activation of the RAS/MAPK pathway as a frequent genetic feature of relapse and high-risk tumors (16). Among these mutations are inactivating mutations in the tumor suppressor neurofibromin 1, a repressor of RAS activity, as well as activating mutations in neuroblastoma RAS (NRAS) and the receptor tyrosine kinase anaplastic lymphoma kinase (ALK) (16, 17). PHB has been reported to contribute to RAS/MAPK signaling by facilitating the activation of c-RAF by RAS in HeLa and CL1-0 cells (18). Several studies have sought to target PHB as a means of reducing signaling through the MAPK pathway to inhibit the migratory capacity or growth of T cell leukemia, pancreatic cancer, gallbladder cancer, and lung cancer cells (10, 11, 19, 20). Here, we present evidence that PHB contributes to features of high-risk neuroblastoma tumors with 17q gain, such as enhanced proliferation, and promotes a state of cellular dedifferentiation. PHB-mediated activation of ERK in neuroblastoma cells may represent a therapeutic target for patients with tumors harboring activating RAS/MAPK pathway mutations.
Results
PHB is highly expressed in neuroblastomas with 17q gain, and its expression is associated with loss of neural differentiation gene expression.
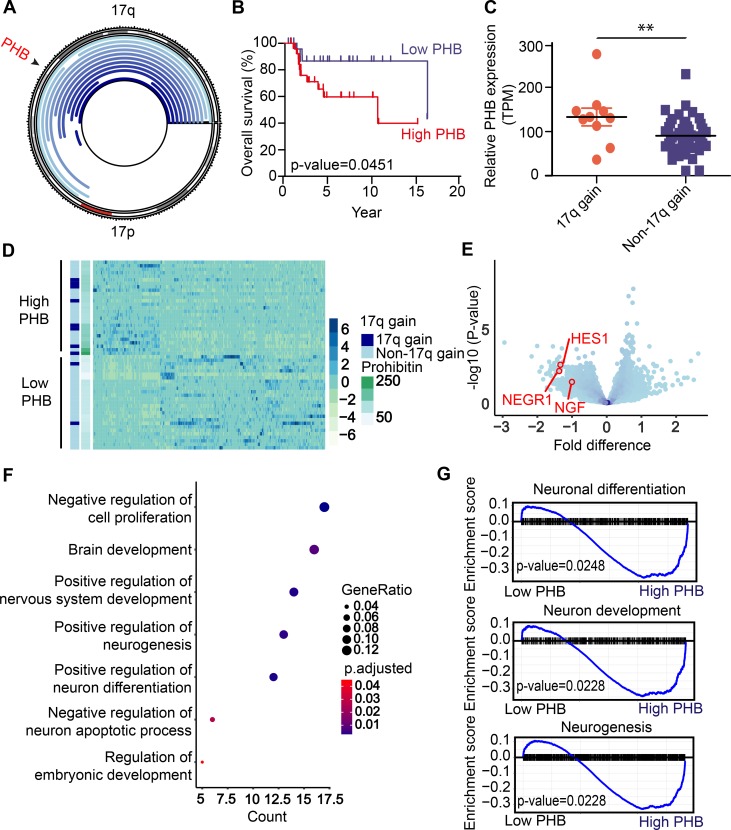
Although 17q-21qter gain has long been recognized as a feature of high-risk neuroblastoma, its contribution to neuroblastoma pathogenesis has largely remained elusive. Based on prior studies that link the 17q gene PHB to breast, gallbladder, and pancreatic cancers, we hypothesized that PHB might contribute to the effects of 17q gain in neuroblastoma (7, 10, 11). To test whether PHB is gained and expressed highly in 17q-gained neuroblastoma, we analyzed previously acquired whole-genome sequencing data of 56 pairs of neuroblastoma and normal lymphocyte controls to analyze copy number variations affecting chromosome 17 (21). Consistent with previous reports in breast cancer (22, 23), PHB was gained in the majority of neuroblastomas with copy number gains on 17q (Figure 1A). Moreover, in agreement with prior reports (5, 6), patients whose tumors had 17q gain had significantly poorer overall survival than patients with no or whole 17 chromosome gain (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127130DS1). High-throughput mRNA sequencing of the same tumors revealed that patients with elevated tumor PHB expression had significantly worse survival than those with lower expression (Figure 1B). In addition, expression of PHB was significantly elevated in tumors with 17q gain compared with tumors with no or whole 17 chromosome gain (Figure 1C). Publicly available neuroblastoma data sets corroborated the correlation between high PHB expression and poor prognosis and the association between 17q gain and elevated PHB transcript levels (Supplemental Figure 1, B and C).
Figure 1. Chromosome 17q gain in neuroblastoma is associated with high PHB copy number and expression and correlates with adverse patient prognosis.
(A) Plot of chromosome 17 with regions of copy number gain (shown in blue) as detected in 56 patient samples of tumor-normal paired whole-genome sequencing. PHB locus indicated (black arrowhead). (B) Kaplan-Meier curve displaying overall survival for patients with low versus high PHB mRNA expression measured using RNA-Seq. P value calculated by log-rank test. (C) PHB expression in patients with 17q versus whole 17 or no gain. **P < 0.05, 2-tailed t test. (D) Heatmap of the significantly differentially expressed genes in patients with high versus low PHB expression. Statistical significance assessed with the Mann-Whitney U test. (E) Volcano plot showing differentially expressed genes in low- versus high-PHB groups. Neuronal differentiation genes NEGR1, HES1, and NGF marked (red circles). (F) GO analysis showing most GO categories significantly enriched in differentially expressed genes in low versus high PHB–expressing tumors. P values calculated with Fisher’s exact test. (G) GSEA indicating enrichment and upregulation of genes involved in neuronal differentiation, neuron development, and neurogenesis in patient tumors expressing PHB at low levels. P values calculated with Fisher’s exact test.
Given the pleiotropic nature of PHB cellular function, including contributions to mitochondrial function, signal transduction, and gene expression regulation (18, 24), we hypothesized that differences in PHB expression would be associated with diverse changes in gene expression programs. To determine gene expression changes associated with high levels of tumor PHB, we assessed differences in gene expression between tumors with low and high PHB transcript levels. Gene expression differed significantly between the 2 groups (Figure 1D), and several genes implicated in neuronal differentiation and favorable prognosis, such as neuronal growth regulator 1 (NEGR1), nerve growth factor (NGF), and hairy and enhancer of split 1 (HES1), were upregulated in patients with low PHB (refs. 25–27 and Figure 1E). Gene ontology (GO) analysis revealed significant differential gene expression in programs governing neuron differentiation and neurogenesis (Figure 1F). Moreover, gene set enrichment analysis (GSEA) demonstrated enrichment and upregulation of gene sets involved in neuronal differentiation and development in tumors with low PHB expression (Figure 1G). Thus, PHB is commonly gained and highly expressed in neuroblastomas with 17q gain, and elevated PHB expression is associated with gene expression programs that promote the growth and dedifferentiation of neuroblastoma cells.
PHB is expressed in neuroblastoma cell lines and is required for cell survival and cell cycle progression.
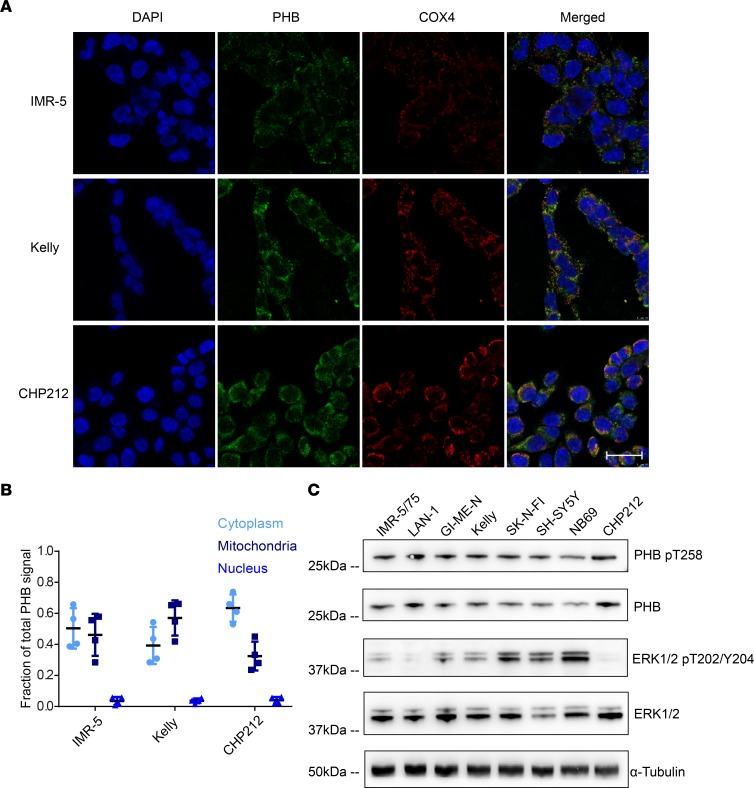
PHB is reported to localize to several subcellular compartments to carry out its diverse functions, including the nucleus, mitochondria, and plasma membrane (18, 24, 28). To further delineate the role of PHB in neuroblastoma cells, we assessed its subcellular localization by immunofluorescence. PHB was found to localize primarily in the cytoplasm and mitochondria, as indicated by colocalization with cytochrome c oxidase subunit 4 (COX4) (29), while little PHB was detected in the nucleus, as indicated by colocalization with DAPI (Figure 2, A and B). Site-specific phosphorylation of PHB has been implicated in regulating its function (30). Phosphorylation of PHB at threonine 258 has been found to contribute to activation of c-RAF by RAS. In addition, increased levels of PHB T258 phosphorylation can augment signaling through the ERK pathway and increase the migratory capabilities of cancer cells (12, 18). PHB T258 phosphorylation was detected in a panel of 8 neuroblastoma cell lines (Figure 2C; see complete unedited blots in the supplemental material), though levels of phosphorylated PHB T258 did not correlate with levels of phosphorylated ERK1/2 (Supplemental Figure 2), suggesting that this modified PHB participates in MAPK pathway–independent functions. Indeed, PHB T258 phosphorylation is also reported to facilitate the targeting of PHB to mitochondria (31). The detection of PHB T258 phosphorylation in neuroblastoma cells nonetheless presents the possibility that PHB may augment c-RAF activation in these cells. Moreover, ERK and its activated phosphorylated form was detected in these cells, consistent with basal activation of ERK signaling in neuroblastoma cell lines (Figure 2C). This led us to hypothesize that PHB may contribute to ERK activation in neuroblastoma cells.
Figure 2. Prohibitin is expressed in neuroblastoma cell lines, has diverse subcellular localization, and is posttranslationally modified to influence RAS/MAPK signaling.
(A) Immunofluorescence showing subcellular localization of PHB (shown in green). Mitochondrial staining indicated by COX4 (shown in red). Nuclear staining indicated by DAPI (shown in blue). Scale bar: 50 μm. (B) Fraction of PHB signal quantified in each compartment. Data represent 4 technical replicates. (C) Western blot analysis demonstrating detection of PHB phosphorylated at T258, as well as ERK1/2 phosphorylated at T202/Y204.
To test this hypothesis, we depleted PHB using shRNA interference in neuroblastoma cell lines. We used 3 independent shRNAs and observed depletion of PHB using Western blot analysis as compared with a control shRNA targeting GFP (Figure 3A). Depletion of PHB was associated with reduced phosphorylation of ERK1/2 at threonine 202 and tyrosine 204 (T202/Y204) in IMR-5/75 and NB01 cells by as much as 77% and 80%, respectively, with no significant effects on total levels of ERK1/2 proteins (Figure 3A and Supplemental Figure 3). PHB knockdown in Kelly cells was not associated with reduction of levels of activated ERK, suggesting that the requirement of PHB for ERK activation is cell type specific (Supplemental Figure 4A). PHB knockdown was not associated with changes in phosphorylation of c-RAF serine 338, a residue associated with c-RAF activation by RAS (refs. 32, 33, Figure 3A, and Supplemental Figure 3). Given that phosphorylation of other residues is important for c-RAF activation, notably tyrosine 341, threonine 491, and serine 494, the lack of reduction of serine 338 phosphorylation does not preclude impaired c-RAF activation after PHB knockdown. In addition, levels of PHB2 were found to decrease following knockdown, and we therefore cannot exclude that loss of PHB2 contributes to the observed phenotypes.
Figure 3. PHB knockdown impairs ERK activation, reduces cell viability, slows cell cycle progression, and induces apoptosis in neuroblastoma cells.
(A) Western blot analysis displaying quantities of phosphorylated ERK1/2 at T202/Y204 in IMR-5/75 cells. ERK1/2 results are representative of 3 independent experiments. (B) IMR-5/75 cell proliferation following PHB knockdown as measured with the RTCA iCelligence system. Each condition was tested in duplicate. (C) FACS plots showing IMR-5/75 cell cycle distribution following PHB knockdown. (D) Quantification of cells in S phase after PHB knockdown. Data represent mean ± SD. *P < 0.001, 2-tailed t test. (E) Cell viability of IMR-5/75 cells expressing ectopic PHB-V5 following transduction of an shRNA against the 3′ UTR of PHB. Viability measured with the CellTiter-Glo luminescent viability assay. Data represent mean ± SEM. *P < 0.001, 2-tailed t test. (F) Representative FACS plots of TUNEL-stained IMR-5/75 neuroblastoma cells after PHB knockdown. (G) Quantification of TUNEL-positive cells after PHB knockdown. Data represent mean ± SD. *P < 0.001, 2-tailed t test. Three technical replicates shown. Bonferroni’s correction was applied to account for multiple comparisons.
Given previous reports detailing the sensitivity of neuroblastoma cells to MEK inhibition and linking PHB to cellular survival (34, 35), we hypothesized that PHB expression may contribute to neuroblastoma cell survival. Indeed, the viability of IMR-5/75, SH-SY5Y, GI-ME-N, and NB69 (Supplemental Figure 4, B and C) cells was dramatically reduced following shRNA-mediated PHB knockdown. In addition, PHB knockdown impaired the proliferation of IMR-5/75 cells but not the proliferation of retinal pigment epithelial (RPE) cells, indicating that PHB is a neuroblastoma-specific dependency and is not required for the growth of untransformed cells (Figure 3B and Supplemental Figure 4D). Flow cytometric analysis revealed that knockdown led to accumulation of cells in the G1 phase of the cell cycle and reduced the number of cells in the S and G2 phases (Figure 3, C and D, and Supplemental Figure 4, E and F), a result consistent with a reduced proliferative phenotype and with the upregulation of antiproliferative gene expression programs. Importantly, expression of a recombinant PHB cDNA conjugated to a V5 epitope tag (PHB-V5) lacking the 3′ UTR sequence of PHB was able to significantly rescue the reduced viability of IMR-5/75 cells upon knockdown with a 3′ UTR–targeting shRNA (Figure 3E). Thus, loss of cell viability upon knockdown was mostly due to reduction in PHB transcript levels. Taken together, these results highlight the requirement of PHB for ERK activation as well as the survival and proliferation of neuroblastoma cells, the latter of which may in part be attributed to slowed cell cycle progression.
Prohibitin depletion leads to apoptosis and differentiation of neuroblastoma cells.
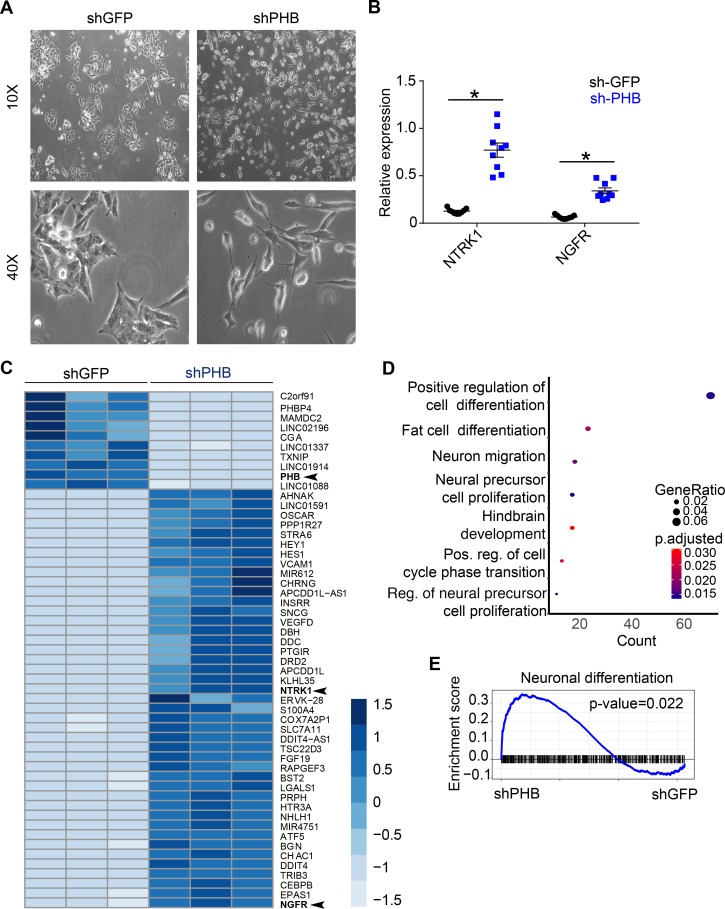
Studies of PHB in mitochondria have delineated a role for the protein in regulating the mitochondrial apoptotic pathway and have demonstrated that pharmacological disruption of PHB leads to apoptosis (28, 36). Given these results, we hypothesized that shRNA-mediated knockdown of PHB might induce apoptosis of neuroblastoma cells. In line with our hypothesis, Western blot analysis revealed increased levels of cleaved caspase-3 upon PHB knockdown but not in controls (Figure 3A). Moreover, flow cytometric analysis detected the formation of nicked DNA ends, an established marker of apoptosis (37), following knockdown (Figure 3, F and G, and Supplemental Figure 4G). In addition to this evidence of apoptosis, neuroblastoma cells displayed marked morphological changes following knockdown, namely the formation of neurites (Figure 4A and Supplemental Figure 4H), a change that has been linked to the differentiation of neuroblastoma cells upon exposure to all-trans-retinoic acid (ATRA) (38). Indeed, significant morphological changes associated with depletion of PHB have been reported in epithelial and pancreatic cancer cells (10, 18). This differentiation phenotype was further supported by the increased expression of neurotrophic receptor tyrosine kinase 1 (NTRK1) and nerve growth factor receptor (NGFR) (Figure 4B), markers associated with neuronal differentiation and favorable patient outcome (23, 39). These morphological and molecular signs of differentiation are in line with analysis of RNA-Seq of patient tumors, through which we found upregulation of prodifferentiation gene expression programs in tumors with low PHB expression (Figure 1G). To assess whether similar changes in gene expression could be observed in cells depleted of PHB, we performed RNA-Seq of cells after PHB knockdown. Gene expression after knockdown differed significantly compared with controls (Figure 4C). Notably, NTRK1 and NGFR were among the most differentially upregulated genes in PHB-knockdown cells, findings we validated by qRT-PCR (Figure 4B). The most significantly different GO category between the 2 groups was positive regulation of cell differentiation (Figure 4D), and GSEA analysis revealed that neural differentiation gene expression programs were upregulated in PHB-knockdown cells while cell cycle regulation was downregulated (Figure 4E and Supplemental Figure 4I). These results suggest that PHB contributes to the regulation of neuroblastoma cell differentiation and that high expression of PHB promotes tumor dedifferentiation.
Figure 4. PHB knockdown promotes differentiation of neuroblastoma cells.
(A) Photomicrographs of Kelly cells after transduction with shRNAs against either PHB or GFP. Photomicrographs representative of 3 independent samples. (B) NTRK1 and NGFR mRNA expression measured by quantitative reverse transcription PCR (qRT-PCR) after PHB knockdown. Data represent mean ± SEM. n = 3, *P < 0.001, 2-tailed t test. (C) Heatmap showing significantly differentially expressed genes in IMR-5/75 after PHB knockdown. PHB, NTRK1, and NGFR are labeled in bold and denoted by arrowheads. n = 3, and P < 0.001. (D) GO analysis of differentially expressed genes after PHB knockdown. n = 3, Fisher’s exact test. (E) GSEA plot of neuronal differentiation after PHB knockdown. n = 3, Fisher’s exact test. Stated n values indicate number of biological replicates.
Ectopic expression of prohibitin promotes proliferation and dedifferentiation of neuroblastoma cells.
To assess the effects of high PHB expression on neuroblastoma, we ectopically expressed PHB-V5 in IMR-5/75 and SH-SY5Y neuroblastoma cell lines. Western blot analysis confirmed stable expression of PHB-V5 in these cells (Figure 5A). Expression of PHB-V5 was sufficient to increase phosphorylation of c-RAF at serine 338 in IMR-5/75 cells but not in NB01 or SH-SY5Y cells. PHB-V5 expression was not sufficient to increase phosphorylation of ERK1/2 at T202/Y204 in any of the three cell lines tested (Supplemental Figure 5). Given that expression of ectopic PHB targeted to the plasma membrane has been reported to increase ERK activation (12), this result suggests that PHB-V5 is not completely targeted to the plasma membrane and likely also localizes to other compartments, such as the mitochondrion. Ectopic PHB expression dramatically increased the rate of IMR-5/75 cell proliferation (Figure 5B), a finding consistent with a role for PHB in cell cycle regulation (Supplemental Figure 4I). Because previous studies have implicated PHB in cell migration and invasion (10–12, 14, 18, 20), we assessed the ability of ectopic PHB expression to promote a migratory phenotype in neuroblastoma. PHB-V5–expressing cells displayed enhanced migration relative to controls as assessed by the scratch assay (Figure 5C and Supplemental Figure 6C). Given that PHB-V5 increased the proliferation of neuroblastoma cells, we cannot exclude the effects of increased cell numbers on the apparent increase in migration we observed. In addition, SH-SY5Y cells expressing PHB-V5 displayed significant morphological alterations and grew in mounds characteristic of the undifferentiated state of this cell line (refs. 40, 41, Figure 5D, and Supplemental Figure 6A). A similar morphological and growth phenotype was observed in NB01 cells (Supplemental Figure 6B). This dedifferentiated phenotype was further substantiated by downregulation of NTRK1 and NGFR upon PHB-V5 expression (Figure 5E and Supplemental Figure 6D). Thus, PHB is able to promote proliferation and dedifferentiation of neuroblastoma cells.
Figure 5. Ectopic expression of PHB promotes proliferation, migration, and dedifferentiation of neuroblastoma cells.
(A) Western blot analysis of cells after stable expression of PHB-V5 compared with empty vector control cells. Contrast of blots was enhanced equally for clarity. (B) Proliferation of IMR-5/75 cells stably expressing PHB-V5 compared with empty vector control cells as measured with the RTCA iCelligence system. Each condition was tested in triplicate. (C) Migration of IMR-5/75 and SH-SY5Y cells stably expressing PHB-V5 compared with empty vector control cells as measured by scratch assay. Data represent mean ± SD. n = 3, and **P < 0.05 for IMR-5/75; n = 1, and P > 0.05 for SH-SY5Y; 2-tailed t test. (D) Number of mounds formed by SH-SY5Y cells stably expressing PHB-V5 compared with empty vector control cells. Data represent mean ± SD. n = 3, and **P < 0.001; 2-tailed t test. (E) Expression of NTRK1 and NGFR measured by qRT-PCR. Data represent mean ± SD. ***P < 0.0003, and **P < 0.005; 2-tailed t test. Stated n values represent number of biological replicates.
Pharmacological targeting of PHB is sufficient to promote differentiation and reduce ERK activation of neuroblastoma cell lines in vitro and in a patient-derived ALK-mutant neuroblastoma xenograft in vivo.
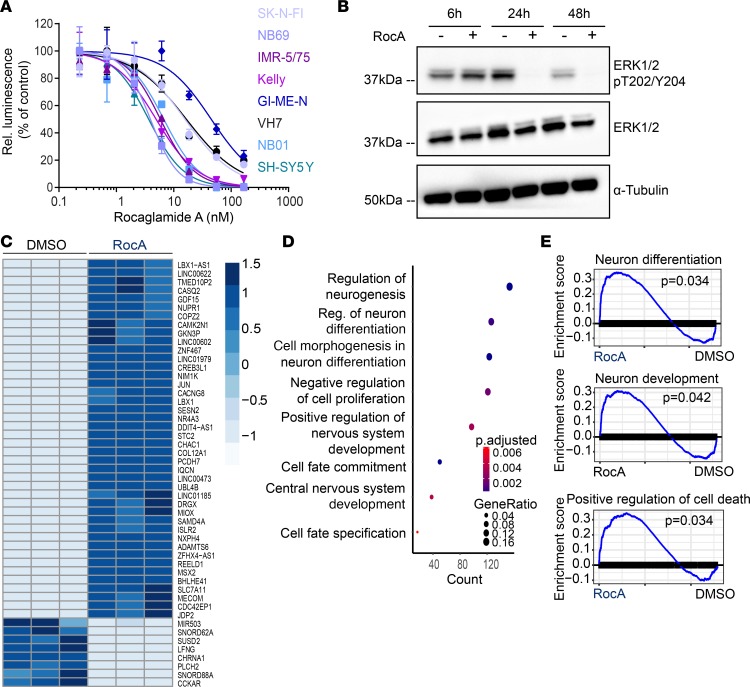
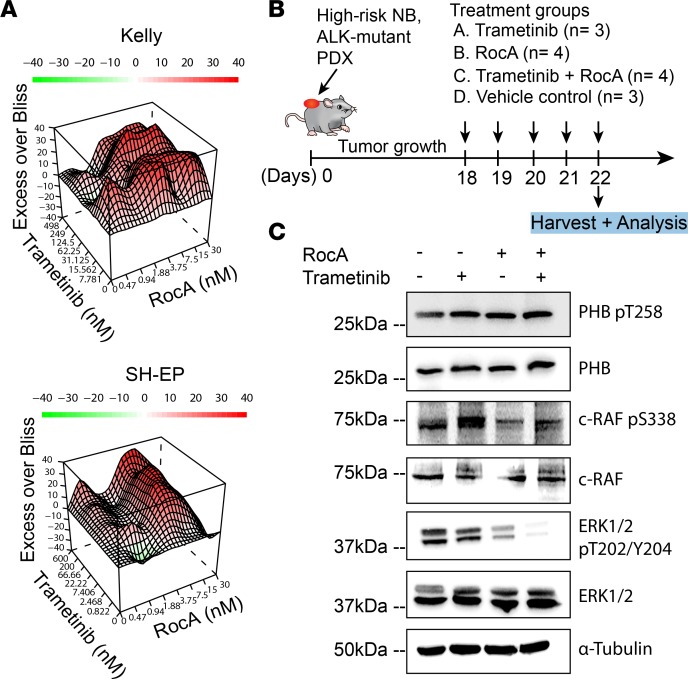
Given our findings that PHB overexpression was associated with several malignant features of neuroblastoma, including proliferation and dedifferentiation, we asked whether pharmacological disruption of PHB might help counter these processes. Rocaglamide A (RocA) can bind to PHB and has been reported to inhibit ERK activation and the growth and migration of a variety of cancer cells, including pancreatic cancer, non–small cell lung cancer, T cell leukemia, and melanoma (10, 19, 20, 42). Although RocA has been reported to bind other molecules in addition to PHB, such as the translation factor eukaryotic initiation factor 4A (EIF4A) (43), and to inhibit autophagy in non–small cell lung cancer cells (44), we reasoned that its effects on PHB might prove useful in assessing the feasibility of inhibiting ERK activation in neuroblastoma in the absence of a more specific PHB inhibitor. All neuroblastoma cell lines tested exhibited greater sensitivity to RocA treatment than RPE cells, and all neuroblastoma cell lines except for GI-ME-N exhibited greater sensitivity than VH7 fibroblast cells, suggesting that RocA exhibits tumor-specific antiproliferative effects (Figure 6A and Supplemental Figure 7, A and B). In addition, RocA treatment led to reduced ERK phosphorylation in IMR-5/75 and GI-ME-N cells 24 and 48 hours after treatment and to a more modest reduction in phosphorylated ERK in NB69 cells 48 hours after treatment (Figure 6B and Supplemental Figures 8 and 9). RocA treatment did not reduce phosphorylated ERK at any tested time point in SH-SY5Y cells (Supplemental Figures 8 and 9). In addition, RocA treatment reduced c-RAF phosphorylation in IMR-5/75 and NB69 cells 24 hours after treatment (Supplemental Figure 10, A and B). The sensitivity of SH-SY5Y cells to RocA in the absence of a detectable reduction in ERK activation likely reflects a MAPK-independent effect of RocA. To test whether RocA induces similar gene expression changes as genetic PHB depletion, we performed RNA-Seq on IMR-5/75 cells following RocA treatment. As with our findings in patient tumors with low PHB expression and in PHB-depleted cells, there were significant changes in gene expression between cells treated with RocA and controls (Figure 6C). Differentially expressed GO terms included regulation of neurogenesis, cell fate commitment, and cell morphogenesis involved in neuron differentiation (Figure 6D). GSEA revealed enrichment and upregulation of gene sets governing differentiation, development, and cell death in cells treated with RocA (Figure 6E). Because of recent interest in MEK inhibition as a therapeutic strategy for neuroblastomas with MAPK pathway mutations (35, 45), and our data indicating that PHB is required for ERK activation, we hypothesized that RocA and trametinib, an MEK inhibitor previously studied in neuroblastoma (16), could exhibit synergistic antineuroblastoma effects. Treatment of 2 ALK-mutant cell lines, Kelly and SH-EP, with RocA and trametinib revealed strong synergy between the compounds as assessed using excess over Bliss analysis (Figure 7A). Because ALK is known to activate ERK in cancers including neuroblastoma, this finding suggests that pharmacological targeting of PHB is a promising therapeutic avenue for MAPK-dependent neuroblastoma (46, 47). The ability of RocA to inhibit ERK and synergize with trametinib in vitro led us to test this approach in mice harboring ALK-mutant neuroblastoma patient–derived xenografts (Figure 7B). RocA treatment was effective in reducing levels of phosphorylated ERK (Figure 7C). Intriguingly, RocA treatment was as effective as trametinib at doses used in previous reports in reducing ERK phosphorylation (Figure 7C and Supplemental Figure 11). Consistent with their synergistic effects in vitro, combining RocA with trametinib led to loss of detectable ERK activation (Figure 7C). Because of mouse weight loss, perhaps owing to RocA effects on protein translation (Supplemental Figure 12A), in vivo treatment was halted after monitoring tumor growth for 4 days (Supplemental Figure 12B). In summary, we find that PHB is a compelling target for blocking ERK activation in MAPK-dependent and ALK-mutant neuroblastomas.
Figure 6. RocA treatment impairs ERK activation and promotes differentiation of neuroblastoma cells in vitro.
(A) Dose-response curve of cells treated with RocA as measured with CellTiter-Glo luminescent viability assay 72 hours after treatment. Data represent 3 technical replicates. (B) Western blot analysis of IMR-5/75 cells 6, 24, and 48 hours after treatment with 50 nM RocA compared with DMSO-treated cells. Data represent a single experiment. (C) Heatmap showing differentially expressed genes in IMR-5/75 cells 24 hours following treatment with RocA (50 nM) compared with DMSO-treated cells. n = 3. (D) GO analysis of differentially expressed genes in IMR-5/75 cells 24 hours following treatment with RocA (50 nM) compared with DMSO-treated cells. n = 3, Fisher’s exact test. (E) GSEA analysis in IMR-5/75 cells 24 hours following treatment with RocA (50 nM) compared with DMSO-treated cells. n = 3, Fisher’s exact test. Stated n values indicate number of biological replicates.
Figure 7. RocA treatment impairs ERK activation in an ALK-mutant, patient-derived xenograft in vivo.
(A) Heatmaps showing excess over Bliss synergy scores for combination treatment with RocA and trametinib in neuroblastoma cells. Values greater than 1 (shown in red) denote synergistic combinations while values less than 1 (shown in green) denote antagonistic combinations. Data represent 3 technical triplicates. (B) Dosing schedule of RocA, trametinib, and vehicle controls for patient-derived xenograft treatment. (C) Western blot analysis of patient-derived xenografts of mice treated with RocA, trametinib, or RocA and trametinib combination treatment compared with vehicle control–treated mice.
Discussion
Here, we have found that the PHB gene is highly expressed in neuroblastomas harboring gains on 17q and that elevated PHB expression correlates with poor prognosis. High PHB expression in tumors was associated with the suppression of genes controlling the differentiation of neural cells. Depletion of PHB promoted differentiation, cell cycle arrest, and apoptosis and reduced ERK activation in 2 of 3 neuroblastoma cell lines tested. Overexpression of PHB was sufficient to enhance proliferation and dedifferentiation. Further, pharmacological targeting of PHB was lethal to neuroblastoma cells, promoted differentiation, and inhibited ERK activation in vitro and in an ALK-mutant neuroblastoma xenograft in vivo, suggesting that PHB represents a promising therapeutic target in high-risk, MAPK-dependent neuroblastomas. Given these findings, further research is warranted to fully substantiate the role of PHB in MAPK pathway activation in neuroblastoma.
The relatively low mutational burden of neuroblastomas has inspired efforts to identify nonmutational events driving disease pathogenesis, including gene and chromosomal copy number variations, such as 17q gain. Our finding that elevated PHB in neuroblastomas was associated with 17q gain and dedifferentiation of neuroblastoma, with our experimental evidence linking PHB to a dedifferentiated phenotype in vitro, suggests a connection between PHB, 17q gain, and unfavorable, undifferentiated neuroblastoma (48). With the understanding that neuroblastoma arises from embryonic neural crest cells that fail to terminally differentiate (49), much effort has been exerted in identifying therapies to induce neuroblastoma differentiation. ATRA and 13-cis-retinoic acid are 2 clinically important compounds used to treat neuroblastoma by inducing differentiation (50–52). However, retinoid therapy is often ineffective in treating high-risk cases, and patients may become resistant to retinoids because of amplification of the MYCN protooncogene (50, 53), necessitating the identification of new therapeutic targets to promote differentiation. Our finding that depletion of PHB induced morphological changes similar to ATRA treatment and promoted prodifferentiation gene expression programs suggests that PHB is one such candidate target for therapeutic neuroblastoma differentiation (38). These findings are in line with a previous report detailing loss of PHB protein levels following ATRA-induced differentiation of SK-N-SH neuroblastoma cells (54). Future studies should explore the mechanism by which PHB suppresses differentiation in neuroblastoma to better exploit its therapeutic potential.
Furthermore, the ability to sequence complete genomes of healthy and diseased tissues has led to the development of rational therapies tailored to target disease-specific mutations. Recent sequencing efforts of relapsed neuroblastoma have inspired the development of therapies targeting the RAS/MAPK pathway, mutations in which are enriched in relapsed disease (16, 35, 45). The results of our RocA and MEK inhibitor treatment of an ALK-mutant neuroblastoma patient-derived xenograft present PHB as a compelling therapeutic target for blocking oncogenic signaling in vivo. Although combination of RocA with trametinib treatment resulted in some mouse toxicity, an effect that might be attributed to additional effects of RocA, such as inhibition of protein translation through binding EIF4A (43), it is conceivable that molecules with greater specificity for targeting PHB might prove effective in blocking oncogenic signaling while having a more favorable toxicity profile. Interestingly, it has been reported that inhibition of EIF4A with the small molecule FL3 synergizes with trametinib treatment in melanoma cells (55). Further research is therefore needed to establish whether the observed synergism between RocA and trametinib is due to effects on PHB, EIF4A, or both proteins.
Although it is plausible that loss of PHB at the plasma membrane contributes to the upregulation of prodifferentiation gene expression, increased apoptosis, and impaired cell cycle progression, we cannot exclude that loss of PHB in other subcellular compartments contributes to these phenotypes. Although PHB T258 phosphorylation was abundantly detected in neuroblastoma cell lines, a lack of positive correlation between PHB phosphorylated T258 levels and phosphorylated ERK levels implies that a substantial quantity of this modified PHB is involved in MAPK pathway–independent functions. Indeed, it has been demonstrated that phosphorylation of T258 can induce translocation of PHB from the cytoplasm to mitochondria, where it promotes proliferation in bladder cancer cells (31). Furthermore, because our immunofluorescence analysis demonstrated significant quantities of PHB in the mitochondria of neuroblastoma cells, it is highly likely that mitochondrial PHB plays a significant role in neuroblastoma physiology. PHB has been shown to regulate the proteolytic processing of mitochondrial dynamin-like GTPase (OPA1), a key mediator of the mitochondrial apoptotic pathway and regulator of mitochondrial morphology. Loss of PHB promotes the processing of OPA1 into its short isoform, rendering cells sensitive to apoptotic stimuli by enabling the release of cytochrome c and impairing mitochondrial fusion (28, 56). It is likely that the increased apoptosis we observed following PHB knockdown was due to enhanced processing of OPA1 into its short isoform. In addition, mitochondrial morphology and fission-fusion dynamics have been implicated as determinants of cellular differentiation states. Interestingly, increased mitochondrial fission is reported to be associated with proliferation and invasiveness of cancer cells and dedifferentiation of stem cells (57, 58). Because increased PHB in mitochondria is predicted to suppress fission, it is possible that the enhanced proliferation and dedifferentiation of neuroblastoma cells expressing high levels of PHB are due to nonmitochondrial functions of the protein. Further research is necessary to determine whether PHB appreciably alters fission-fusion dynamics in neuroblastoma and whether these dynamics are associated with the aforementioned phenotypes.
Our observation that PHB knockdown resulted in accumulation of cells in the G1 phase of the cell cycle with concurrent decreases in G2/M and S phases suggests that PHB plays a role in the G1/S transition in neuroblastoma cells. Cyclin D1 is capable of shortening the G1 phase and promoting the transition to S phase in response to mitogenic signaling through the MAPK pathway (59, 60). It possible that the apparent G1 arrest we observe in IMR-5/75 cells after PHB knockdown is due to impaired ERK activation in these cells. Indeed, previous reports have noted that both siRNA-mediated depletion of PHB and RocA treatment results in G1 phase arrest and reduced cyclin D1 levels in Jurkat cells and in non–small cell lung cancer cells (19, 20).
Finally, although the results of our scratch assays suggest a role for PHB in neuroblastoma cell migration and add to the increasing evidence implicating the gene in cancer cell metastasis (10–12, 14, 20), the confounding effects of increased proliferation after PHB-V5 expression necessitate further research to define PHB as a promoter of neuroblastoma cell migration. The poor outcomes of patients with metastatic neuroblastoma make the identification of new targets, such as PHB, extremely important (1, 15), and future studies are warranted to assess whether PHB is indeed a viable target for suppressing this highly malignant process.
Methods
Reagents.
Oligonucleotide primers were obtained from Eurofins Genomics. Please see Supplemental Table 2 for a complete list of oligonucleotide primers used in this study. Sequences of NTRK1 and NGFR qRT-PCR primers were obtained from Dzieran et al. (39). HPRT1 qRT-PCR primers were a gift from Cornelia Eckert (Charité-Universitätsmedizin Berlin, Berlin, Germany). Please see Supplemental Table 3 for a complete list of plasmid vectors used in this study. pLX304 was a gift from David Root (Addgene plasmid 25890) (61). Please see Supplemental Table 4 for a complete list of antibodies used for Western blotting and immunofluorescence.
Cell culture.
Human tumor cell lines were obtained from the American Type Culture Collection. The identity of all cell lines was verified by short tandem repeat genotyping (Genetica DNA Laboratories). The absence of Mycoplasma sp. contamination was determined using a Lonza MycoAlert system. Neuroblastoma cell lines were cultured in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with penicillin, streptomycin, and 10% FCS (Thermo Fisher Scientific). RPE cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with penicillin, streptomycin, and 10% FCS. To assess the number of viable cells, cells were trypsinized, resuspended in medium, and sedimented at 500 g for 5 minutes. Cells were then resuspended in medium, mixed in a 1:1 ratio with 0.02% trypan blue (Thermo Fisher Scientific), and counted with a Bio-Rad TC20 cell counter.
Lentivirus production and cell transduction.
Lentivirus production was carried out as previously described (62). In brief, HEK293T cells were transfected with TransIT-LT1 (Mirus) in a 2:1:1 ratio of the lentiviral vector and psPAX2 and pMD2.G packaging plasmids (Addgene), according to the manufacturer’s instructions. Viral supernatant was collected 48 and 72 hours after transfection. Supernatant was pooled, filtered, and stored at –80°C. Neuroblastoma and RPE cells were transduced with virus particles in the presence of 8 μg/ml hexadimethrine bromide (Macherey-Nagel). Cells were transduced for 1 day in antibiotic-free medium and then grown in full medium for 1 day. Neuroblastoma cells were then selected for 2 days with puromycin hydrochloride (2 μg/ml) while RPE cells were selected for 2 days with 5 μg/ml puromycin.
Cell viability assays.
Viability assays on cells were carried out 2 days following selection. Viability assays on cells treated with RocA (Active Biochem) and trametinib (Absource Diagnostics) were performed 72 hours after treatment. We seeded 500 cells per well in white, flat-bottom, 96-well plates (Corning). Cell viability was measured using CellTiter-Glo Luminescent Reagent according to the manufacturer’s protocol (Promega). Luminescence was measured on a Promega GlowMax-Multi Detection System.
Western blotting.
Whole-cell protein lysates were prepared by lysing cells in 15 mM HEPES, 150 mM NaCl, 10 mM EGTA, and 2% (v/v) Triton X-100 supplemented with cOmplete (Roche) and PhosStop (Roche) phosphatase inhibitors. Protein concentrations were assessed by BCA assay (Santa Cruz Biotechnology). For 10 minutes, 20 μg of protein was denatured in Laemmli buffer at 95°C. Samples were run on NuPage 10% polyacrylamide, 1 mm Bis-Tris Protein Gels (Thermo Fisher Scientific) and transferred to PVDF membranes (Roche). Membranes were blocked with 10% dry milk or 10% BSA (Carl Roth) in TBS with 0.1% (v/v) Tween-20 (Carl Roth). Membranes were probed with primary antibodies overnight at 4°C and then with secondary antibodies conjugated to horseradish peroxidase for 1 hour at room temperature (Supplemental Table 4). Mouse anti-prohibitin (E-5, sc-377037, Santa Cruz Biotechnology) was used for detection of prohibitin by Western blot. Chemiluminescent detection of proteins was carried out using Immunocruz Western blotting luminol reagent (Santa Cruz Biotechnology) and the Fusion FX7 imaging system (Vilber Lourmat). Densitometry was performed using ImageJ (NIH).
Immunofluorescence and colocalization analysis.
Immunofluorescence analysis was carried out with assistance by the Molecular Cytology Core Facility at Memorial Sloan Kettering Cancer Center. Cells were plated on a 4-well, glass millicell EZ slide (MilliporeSigma) and fixed in 4% paraformaldehyde. Slides were stained using a Ventana XT automated immunohistochemical staining machine. Permeabilization was carried out according to the Ventana XT manufacturer’s protocol. Cells were blocked in 10% NGS with 2% BSA in PBS for 30 minutes. Cells were incubated with primary antibodies for 5 hours and then incubated with secondary antibodies for 32 minutes. Cells were washed with PBS before imaging. Rabbit anti-prohibitin (sc-28259, Santa Cruz Biotechnology) was used for detection of prohibitin by immunofluorescence and was used at a concentration of 2.5 μg/ml. COX4 antibody (Cell Signaling Technology, 4844) was used at a concentration of 1.2 μg/ml. Confocal images were acquired using a Leica TCS SP5 II (Leica Microsystems). Colocalization analysis was performed using ImageJ (NIH). First, the total coverage area of PHB, COX4, and DAPI was quantified. Next, PHB images were merged with COX4 and DAPI images separately to determine the area of overlap. The threshold tool was used to identify and quantitate the area of overlap above the nonoverlapped signal.
qRT-PCR.
qRT-PCR was performed using 50 ng of template DNA and 0.5 μM primers (Supplemental Table 2) with SYBER Green PCR Master Mix (Thermo Fisher Scientific) in FrameStar 96-well plates (4titude). Reactions were run and monitored on a StepOnePlus Real-Time PCR System, and Ct values were calculated with StepOne Plus software (Thermo Fisher Scientific).
iCelligence.
Ten thousand cells were seeded in each well of E-Plate L8 disposable plates (ACEA Biosciences) and used on the RTCA iCelligence system (ACEA Biosciences) for reading of impedance measurements. Each condition was monitored in duplicate. Cells transduced with PHB-V5 or empty vector were monitored for 96 hours. Cells transduced with shPHB 1–3 or shGFP (Dharmacon Inc.) were monitored for 200 hours.
Flow cytometry–based assays.
One million cells were seeded in 10-cm dishes 24 hours before transduction with shRNA vectors (Addgene). Cells were fixed and stained 72 hours following transduction. Cell cycle progression was studied using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific). Double-strand break detection was performed by TUNEL using the APO-BrdU TUNEL Assay Kit (Thermo Fisher Scientific). Staining was performed according to the manufacturer’s protocol. Cells were analyzed following staining on a BD LSR Fortessa flow cytometer (BD Biosciences).
Mound formation assays.
Five hundred thousand cells transduced with either PHB-V5 or empty vector were seeded in each well of a 6-well plate and cultured for 24 hours. Mounds were then counted on an Axio Observer light microscope (Carl Zeiss). Mounds were defined as distinct clusters of cells observed to be growing on top of one another at distinct foci (i.e., not growing as an adherent monolayer).
Scratch assay.
Twenty thousand cells transduced with either PHB-V5 or empty vector were seeded in both chambers of a 2-chambered removable silicone insert (Ibidi) placed in each well of a 6-well plate. After 24 hours of culture in serum-deprived medium, the inserts were removed to create a scratch and to mark the 0-hour time point. Scratch images were acquired with an Axio Observer light microscope (Carl Zeiss). Following 24 and 48 hours of incubation, scratch images were again acquired. The images were processed and the size of the scratches was quantified using the ImageJ (NIH) image processing program.
RNA-Seq.
RNA-Seq of patient samples was carried out as described in Peifer et al., from which the data were obtained (21). Total RNA was isolated from cells 72 hours after shRNA-mediated PHB knockdown or 24 hours after 50 nM RocA treatment. Libraries were sequenced on HiSeq 2000 v4 instruments with 2× 125-bp paired-end reads (Illumina). Libraries were mapped with STAR to GRCh38 using the Gencode v27 annotation (63). Gene abundance was estimated using featureCounts in STAR, counting only alignments with both mates mapped and allowing for fractional counting of multi-mapping and multi-overlapping reads (63).
Bioinformatic analysis.
Differential expression analyses were performed with R software (version 3.4.0) and the dedicated package DESeq2 (64). Genes were filtered out below a threshold of 2.5 reads per million reads. Normalization of counts among different samples was performed by the trimmed mean of M values method. The exactTest function was used to detect differential expression. GSEA on GO terms and Molecular Signatures Database gene set collections was done with the clusterProfiler R package using the overrepresentation test with the hypergeometric model to assess whether the number of selected genes associated with a GO term was larger than expected (65).
Patient-derived xenograft treatment.
Tumor fragments from an ALK-mutant tumor (see Supplemental Table 1 for clinical characteristics of patient-derived xenograft) were transplanted into NMRI (nu/nu) mice (Experimental Pharmacology and Oncology, Berlin, Germany). Tumor growth was monitored with caliper measurements. Mice were randomized into 4 groups to receive trametinib (3 mg/kg, oral, n = 3), RocA (2.5 mg/kg, intraperitoneal injection, n = 4), trametinib with RocA (3 mg/kg; 2.5 mg/kg, oral; intraperitoneal injection, n = 4), or vehicle (oral; intraperitoneal injection, n = 3). Trametinib was administered once daily and RocA was administered once every 2 days. Mice were sacrificed by cervical dislocation 22 days after transplantation or when body weight loss exceeded 10% of baseline.
Availability of data and materials.
Analyzed data are openly available at the Zenodo digital repository (doi:10.5281/zenodo.2567654). Whole-genome sequencing data of neuroblastoma tumors are deposited at the European Genome-phenome Archive (https://ega-archive.org/) under accession number EGAS00001308 (21).
Statistics.
All data analysis was performed using R software (version 3.4.0) or GraphPad Prism 7. Graphs were created using GraphPad Prism 7. Plotted data represent mean ± SD unless otherwise indicated. Differential gene expression analyses were performed using Wald’s test and corrected for multiple testing using the method of Benjamini and Hochberg unless otherwise indicated (64, 66). Genes with P values less than 0.001 were considered significantly differentially expressed.
Study approval.
Written and informed consent was obtained for the use of the patient-derived xenograft sample in accordance with Charité-Universitätsmedizin Berlin guidelines. All in vivo patient-derived xenograft experiments were compliant with animal welfare guidelines defined by the Charité-Universitätsmedizin Berlin.
Author contributions
ICM contributed to the study design and collection and interpretation of the data. YB performed in vitro differentiation and migration assays, Western immunoblotting analysis, RNA-Seq, and whole-genome sequencing data analysis. JT analyzed tumor genome-sequencing data. FK performed RNA-Seq data analysis. MVO performed immunofluorescence analysis, vector design, and subcloning. HDG performed experiments and analyzed data. JR contributed to mouse xenograft study design. AK, AE, and JHS contributed to study design. ICM and AGH wrote the manuscript, to which all authors contributed.
Supplementary Material
Acknowledgments
We thank Natalie Timme, Youjia Han, Rocío Chamorro Gonzalez, Gerard Arrey Tané, and members of the Schulte lab for helpful discussions of the project. AGH is supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) — 398299703, the Berliner Krebsgesellschaft e.V., and the Berlin Institute of Health Charité Clinician Scientist Program. This work was supported by NIH R01 CA214812, NIH P30 CA008748, Cycle for Survival, the Burroughs Wellcome Fund, the Josie Robertson Investigator Program, the Rita Allen Foundation, and the St. Baldrick’s Arceci Innovation Award. AK is a Damon Runyon-Richard Lumsden Foundation Clinical Investigator. We are deeply grateful to the patients whose participation has made this study possible.
Version 1. 04/18/2019
In-Press Preview
Version 2. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: AK is a consultant for Novartis.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e127130. https://doi.org/10.1172/jci.insight.127130.
Contributor Information
Ian C. MacArthur, Email: imacarth@mail.einstein.yu.edu.
Yi Bei, Email: yi.bei@charite.de.
Michael V. Ortiz, Email: ortizm2@mskcc.org.
Joern Toedling, Email: joern.toedling@charite.de.
Filippos Klironomos, Email: filippos.klironomos@charite.de.
Jana Rolff, Email: j.rolff@epo-berlin.com.
Angelika Eggert, Email: angelika.eggert@charite.de.
Johannes H. Schulte, Email: johannes.schulte@charite.de.
Alex Kentsis, Email: kentsisa@mskcc.org.
Anton G. Henssen, Email: henssenlab@gmail.com.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mahapatra S, Challagundla KB. StatPearls. Cancer, Neuroblastoma. [Updated 2019 Mar 8]. In: StatPearls [Internet]. Treasure Island, Florida, USA: StatPearls Publishing; 2019. https://www.ncbi.nlm.nih.gov/books/NBK448111/
- 3.Hero B, et al. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J Clin Oncol. 2008;26(9):1504–1510. doi: 10.1200/JCO.2007.12.3349. [DOI] [PubMed] [Google Scholar]
- 4.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bown N, et al. 17q gain in neuroblastoma predicts adverse clinical outcome. U.K. Cancer Cytogenetics Group and the U.K. Children’s Cancer Study Group. Med Pediatr Oncol. 2001;36(1):14–19. doi: 10.1002/1096-911X(20010101)36:1<14::AID-MPO1005>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Bown N, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340(25):1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, et al. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 1992;52(6):1643–1646. [PubMed] [Google Scholar]
- 8.Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J Cell Mol Med. 2006;10(2):353–363. doi: 10.1111/j.1582-4934.2006.tb00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X, et al. Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc Natl Acad Sci U S A. 2015;112(25):7779–7784. doi: 10.1073/pnas.1505629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan Z, He Y, Alattar M, Chen Z, He F. Targeting the prohibitin scaffold-CRAF kinase interaction in RAS-ERK-driven pancreatic ductal adenocarcinoma. Mol Cancer. 2014;13:38. doi: 10.1186/1476-4598-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, et al. Prohibitin overexpression predicts poor prognosis and promotes cell proliferation and invasion through ERK pathway activation in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:68. doi: 10.1186/s13046-016-0346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu CF, et al. Raf activation by Ras and promotion of cellular metastasis require phosphorylation of prohibitin in the raft domain of the plasma membrane. Oncogene. 2013;32(6):777–787. doi: 10.1038/onc.2012.86. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Ma LL, et al. Prohibitin, relocated to the front ends, can control the migration directionality of colorectal cancer cells. Oncotarget. 2017;8(44):76340–76356. doi: 10.18632/oncotarget.19394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 16.Eleveld TF, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47(8):864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hölzel M, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142(2):218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajalingam K, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol. 2005;7(8):837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 19.Polier G, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem Biol. 2012;19(9):1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Yurugi H, et al. Targeting prohibitins with chemical ligands inhibits KRAS-mediated lung tumours. Oncogene. 2017;36(33):4778–4789. doi: 10.1038/onc.2017.93. [DOI] [PubMed] [Google Scholar]
- 21.Peifer M, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(7575):700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orsetti B, et al. Genomic and expression profiling of chromosome 17 in breast cancer reveals complex patterns of alterations and novel candidate genes. Cancer Res. 2004;64(18):6453–6460. doi: 10.1158/0008-5472.CAN-04-0756. [DOI] [PubMed] [Google Scholar]
- 23.Westermark UK, Wilhelm M, Frenzel A, Henriksson MA. The MYCN oncogene and differentiation in neuroblastoma. Semin Cancer Biol. 2011;21(4):256–266. doi: 10.1016/j.semcancer.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21(55):8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 25.Takita J, et al. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer Sci. 2011;102(9):1645–1650. doi: 10.1111/j.1349-7006.2011.01995.x. [DOI] [PubMed] [Google Scholar]
- 26.Grynfeld A, Påhlman S, Axelson H. Induced neuroblastoma cell differentiation, associated with transient HES-1 activity and reduced HASH-1 expression, is inhibited by Notch1. Int J Cancer. 2000;88(3):401–410. doi: 10.1002/1097-0215(20001101)88:3<401::AID-IJC12>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawara A, Brodeur GM. Role of neurotrophins and their receptors in human neuroblastomas: a primary culture study. Eur J Cancer. 1997;33(12):2050–2053. doi: 10.1016/S0959-8049(97)00280-3. [DOI] [PubMed] [Google Scholar]
- 28.Sato S, et al. Marine natural product aurilide activates the OPA1-mediated apoptosis by binding to prohibitin. Chem Biol. 2011;18(1):131–139. doi: 10.1016/j.chembiol.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Lithgow T. Targeting of proteins to mitochondria. FEBS Lett. 2000;476(1–2):22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- 30.Ande SR, Xu YXZ, Mishra S. Prohibitin: a potential therapeutic target in tyrosine kinase signaling. Signal Transduct Target Ther. 2017;2:17059. doi: 10.1038/sigtrans.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, et al. Akt phosphorylates Prohibitin 1 to mediate its mitochondrial localization and promote proliferation of bladder cancer cells. Cell Death Dis. 2015;6:e1660. doi: 10.1038/cddis.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terrell EM, Morrison DK. Ras-mediated activation of the Raf family kinases. Cold Spring Harb Perspect Med. 2019;9(1):a033746. doi: 10.1101/cshperspect.a033746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong H, Lee J, Guan KL. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 2001;20(14):3716–3727. doi: 10.1093/emboj/20.14.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhury I, Thompson WE, Thomas K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J Cell Physiol. 2014;229(8):998–1004. doi: 10.1002/jcp.24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart LS, et al. Preclinical therapeutic synergy of MEK1/2 and CDK4/6 inhibition in neuroblastoma. Clin Cancer Res. 2017;23(7):1785–1796. doi: 10.1158/1078-0432.CCR-16-1131. [DOI] [PubMed] [Google Scholar]
- 36.Tortelli TC, et al. Accumulation of prohibitin is a common cellular response to different stressing stimuli and protects melanoma cells from ER stress and chemotherapy-induced cell death. Oncotarget. 2017;8(26):43114–43129. doi: 10.18632/oncotarget.17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyrylkova K, Kyryachenko S, Leid M, Kioussi C. Detection of apoptosis by TUNEL assay. Methods Mol Biol. 2012;887:41–47. doi: 10.1007/978-1-61779-860-3_5. [DOI] [PubMed] [Google Scholar]
- 38.Nagai J, Yazawa T, Okudela K, Kigasawa H, Kitamura H, Osaka H. Retinoic acid induces neuroblastoma cell death by inhibiting proteasomal degradation of retinoic acid receptor α. Cancer Res. 2004;64(21):7910–7917. doi: 10.1158/0008-5472.CAN-04-1178. [DOI] [PubMed] [Google Scholar]
- 39.Dzieran J, et al. MYCN-amplified neuroblastoma maintains an aggressive and undifferentiated phenotype by deregulation of estrogen and NGF signaling. Proc Natl Acad Sci U S A. 2018;115(6):E1229–E1238. doi: 10.1073/pnas.1710901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shipley MM, Mangold CA, Szpara ML. Differentiation of the SH-SY5Y human neuroblastoma cell line. J Vis Exp. 2016;(108):53193. doi: 10.3791/53193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doudican NA, Orlow SJ. Inhibition of the CRAF/prohibitin interaction reverses CRAF-dependent resistance to vemurafenib. Oncogene. 2017;36(3):423–428. doi: 10.1038/onc.2016.214. [DOI] [PubMed] [Google Scholar]
- 43.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534(7608):558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao C, et al. Rocaglamide enhances NK cell-mediated killing of non-small cell lung cancer cells by inhibiting autophagy. Autophagy. 2018;14(10):1831–1844. doi: 10.1080/15548627.2018.1489946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodfield SE, Zhang L, Scorsone KA, Liu Y, Zage PE. Binimetinib inhibits MEK and is effective against neuroblastoma tumor cells with low NF1 expression. BMC Cancer. 2016;16:172. doi: 10.1186/s12885-016-2199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8(1):11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Delisle L, et al. Activated ALK signals through the ERK-ETV5-RET pathway to drive neuroblastoma oncogenesis. Oncogene. 2018;37(11):1417–1429. doi: 10.1038/s41388-017-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang LL, et al. Neuroblastoma of undifferentiated subtype, prognostic significance of prominent nucleolar formation, and MYC/MYCN protein expression: a report from the Children’s Oncology Group. Cancer. 2013;119(20):3718–3726. doi: 10.1002/cncr.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross RA, Spengler BA, Domènech C, Porubcin M, Rettig WJ, Biedler JL. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995;6(4):449–456. [PubMed] [Google Scholar]
- 50.Reynolds CP, Matthay KK, Villablanca JG, Maurer BJ. Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003;197(1–2):185–192. doi: 10.1016/s0304-3835(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 51.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med. 1999;341(16):1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 52.Øra I, Eggert A. Progress in treatment and risk stratification of neuroblastoma: impact on future clinical and basic research. Semin Cancer Biol. 2011;21(4):217–228. doi: 10.1016/j.semcancer.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Finklestein JZ, et al. 13-cis-retinoic acid (NSC 122758) in the treatment of children with metastatic neuroblastoma unresponsive to conventional chemotherapy: report from the Childrens Cancer Study Group. Med Pediatr Oncol. 1992;20(4):307–311. doi: 10.1002/mpo.2950200407. [DOI] [PubMed] [Google Scholar]
- 54.Li QF, et al. Localization of prohibitin in the nuclear matrix and alteration of its expression during differentiation of human neuroblastoma SK-N-SH cells induced by retinoic acid. Cell Mol Neurobiol. 2011;31(2):203–211. doi: 10.1007/s10571-010-9608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malka-Mahieu H, et al. Synergistic effects of eIF4A and MEK inhibitors on proliferation of NRAS-mutant melanoma cell lines. Cell Cycle. 2016;15(18):2405–2409. doi: 10.1080/15384101.2016.1208862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. 2009;1793(1):27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuyàs E, et al. Mitostemness. Cell Cycle. 2018;17(8):918–926. doi: 10.1080/15384101.2018.1467679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albanese C, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270(40):23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 60.Jiang W, et al. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene. 1993;8(12):3447–3457. [PubMed] [Google Scholar]
- 61.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8(8):659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henssen AG, et al. PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat Genet. 2017;49(7):1005–1014. doi: 10.1038/ng.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analyzed data are openly available at the Zenodo digital repository (doi:10.5281/zenodo.2567654). Whole-genome sequencing data of neuroblastoma tumors are deposited at the European Genome-phenome Archive (https://ega-archive.org/) under accession number EGAS00001308 (21).