Abstract
Although cardiovascular disease (CVD) is the leading cause of morbimortality worldwide, promising new drug candidates are lacking. We compared the arterial high-resolution proteome of patients with advanced versus early-stage CVD to predict, from a library of small bioactive molecules, drug candidates able to reverse this disease signature. Of the approximately 4000 identified proteins, 100 proteins were upregulated and 52 were downregulated in advanced-stage CVD. Arachidonyl trifluoromethyl ketone (AACOCF3), a cytosolic phospholipase A2 (cPLA2) inhibitor was predicted as the top drug able to reverse the advanced-stage CVD signature. Vascular cPLA2 expression was increased in patients with advanced-stage CVD. Treatment with AACOCF3 significantly reduced vascular calcification in a cholecalciferol-overload mouse model and inhibited osteoinductive signaling in vivo and in vitro in human aortic smooth muscle cells. In conclusion, using a systems biology approach, we have identified a potentially new compound that prevented typical vascular calcification in CVD in vivo. Apart from the clear effect of this approach in CVD, such strategy should also be able to generate novel drug candidates in other complex diseases.
Keywords: Vascular Biology
Keywords: Atherosclerosis, Cardiovascular disease
A cytosolic phospholipase A2 inhibitor that prevents typical vascular calcification in cardiovascular disease in a mouse model was identified using a systems biology approach.
Introduction
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide, accounting for about 32% of all global deaths (1, 2). CVD results from multifactorial causative mechanisms and constitutes a high economic burden on society and the health care system. Typically, CVD, including coronary artery disease, stroke, or peripheral artery disease, involves endothelial dysfunction resulting from a vascular injury and is characterized by progressive arterial thickening, leading ultimately to thrombosis and vascular occlusion. This atherosclerotic process starts with early lipid deposition in the inner layers of the artery walls and progresses to late complications, such as fibrosis and calcification. While calcification of arterial intima is associated with plaque rupture and myocardial infarction, calcification of arterial media leads to vessel stiffening and increased pulse-wave velocity and reduces cardiac perfusion, with the consequence of ventricular hypertrophy and heart failure (3, 4).
Although significant advancements have been made in understanding the pathophysiology of CVD (5), there is still progress to be made to unravel the detailed molecular complexity of the disease and to transform this information into new drug candidates. The extreme complexity and multifactorial character of CVD has prompted researchers to shift from investigating already well-known candidate pathways or single molecules to unbiased omics-based studies (6). Omics allows the study of the entire “system” simultaneously, leading to a better representation of the complex molecular mechanism of CVD (7) and, thus, ultimately enabling the major aim of identifying truly novel CVD targets. This has resulted in a number of recent omics studies in the field identifying potential biomarkers of CVD (8) but not necessarily resulting in novel drugs yet. The latter could be attributed to the fact that potential targets need to be transformed into drugs, which often takes a substantial amount of time and effort, and/or that most studies employed genomic and transcriptomic strategies, which not necessarily translate to changes in proteins, the major drug targets (9).
As a response to the ever-increasing omics data and in an effort to target their optimal use for drug discovery, a number of initiatives have evolved to create a shortcut associating omics molecular signatures to drugs and small compounds. The Connectivity Map (CMap) is the most advanced example of this type of approach (10), where 564 gene expression profiles from cultured human cells treated with 164 bioactive small molecules were generated and associated via pattern-mining software. This resource was later updated to include associated signatures to 1,309 compounds (https://portals.broadinstitute.org/cmap/). Attempts have been made to use CMap in CVD but were limited to mouse models (11) or in vitro studies (12) or focused on human stem cells (13). Furthermore, none of these studies tested the therapeutic validity of the selected small CMap molecules.
To our knowledge this is the first effort to identify novel drugs in CVD using drug repurposing based on human molecular signatures. We generated a high-resolution human proteome signature of advanced-stage CVD using peripheral artery samples that allowed the identification of a potentially novel drug candidate, arachidonyl trifluoromethyl ketone (AACOCF3), a cytosolic phospholipase A2 (cPLA2) inhibitor. In vivo and in vitro investigations showed that AACOCF3 exerts a potent inhibitory action on arterial calcification.
Results
CVD patient clinical data used in vessel proteome analysis.
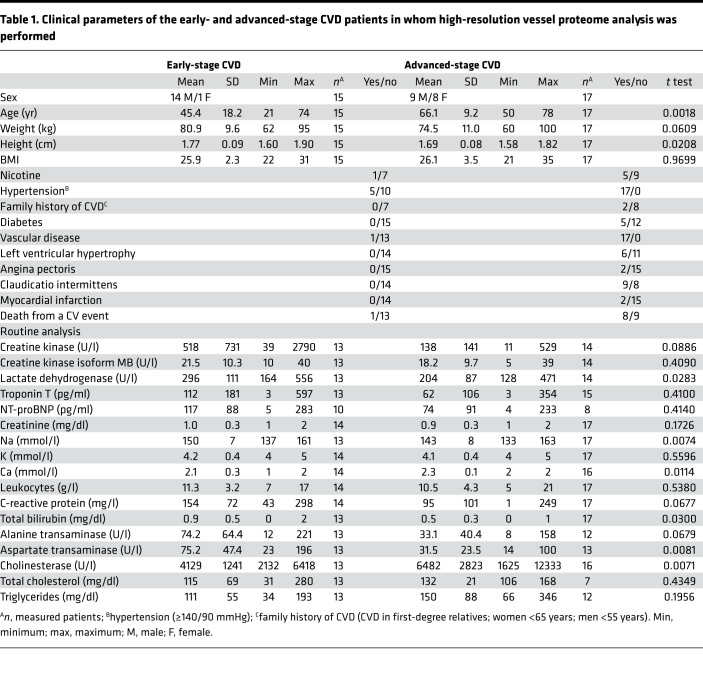
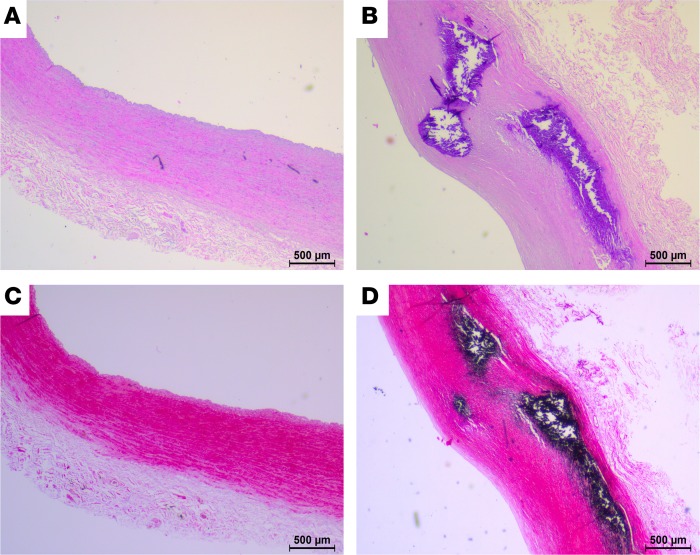
For vessel proteome analysis of CVD vessels, common iliac, external iliac, and superficial femoral arteries from 15 patients with early-stage CVD and 17 patients with advanced-stage CVD were employed (Table 1). Advanced-stage CVD patients had clear CVD phenotypes (vascular disease, left ventricular hypertrophy, angina pectoris, intermittent claudication, myocardial infraction). These cardiovascular phenotypes were not found in early-stage CVD patients (Table 1). H&E staining (Figure 1, A and C) and von Kossa staining (Figure 1, B and D) confirmed intima media thickening and calcification in arteries from the advanced-stage CVD group compared with early-stage CVD patients. Advanced-stage CVD patients were older than early-stage CVD patients. The female/male ratio was also different between the groups, with almost exclusively females in the early-stage CVD group. The advanced-stage CVD group had a higher proportion of patients with hypertension, diabetes, and nicotine use. The medication use of the 2 groups was very diverse, with no clear distinguishable features between the groups (Supplemental Table 3; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125638DS1).
Table 1. Clinical parameters of the early- and advanced-stage CVD patients in whom high-resolution vessel proteome analysis was performed.
Figure 1. Histology of human arteries.
Representative images of H&E staining (A and C) and von Kossa staining (B and D) of iliac artery sections from an early- (A and B) and an advanced-stage (C and D) CVD patient. Black staining in B and D corresponds to calcium deposits. Scale bar: 500 μm.
Arterial protein signature of advanced-stage CVD.
Proteome analysis of the arteries led to the detection of about 4000 proteins in total (2500–3500 proteins per patient, detected in at least 60% of samples). Samples were analyzed in 2 batches, and only proteins that were significantly differentially expressed in 1 batch and displayed the same expression direction in the other batch were selected. This led to the selection of 152 proteins, with 100 and 52 that were upregulated and downregulated in advanced-stage CVD, respectively (Supplemental Table 4). The change in abundance of selected proteins present in the CVD signature (SHMT1, EFEMP1, KDM5D: increase; GUCY1B2: decrease) was confirmed by Western blot analyses of the arteries (Supplemental Figure 1), supporting in general the validity of the CVD vessel proteome analysis.
Pathway analysis with the Reactome database (https://reactome.org/) revealed that these proteins mainly participated in signal transduction (37%); transcription (26%); metabolism (22%); immune system (22%); metabolism of proteins (21%); and developmental biology (18%). Among the 29 proteins involved in metabolism, 14 were related to lipid metabolism, including SREBF1, ATP-binding cassette subfamily A member 2 (ABCA2), and PTB domain–containing engulfment adapter protein 1 (GULP1), involved in cholesterol synthesis and transport. The CVD signature also contains the protein AMBP, involved in calcium crystallization, and EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1), involved in chondrogenesis.
AACOCF3 as a potential drug candidate for CVD treatment.
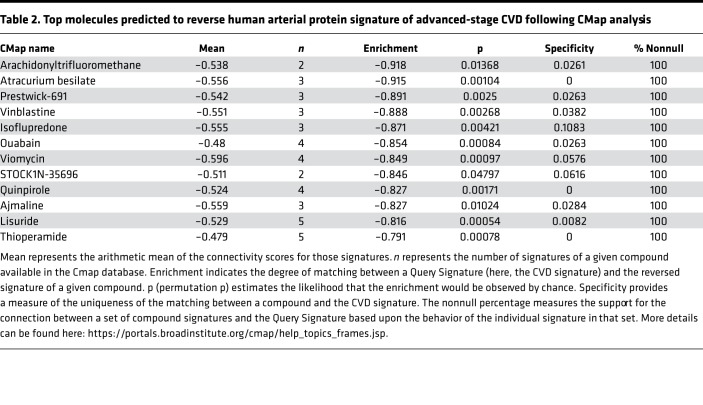
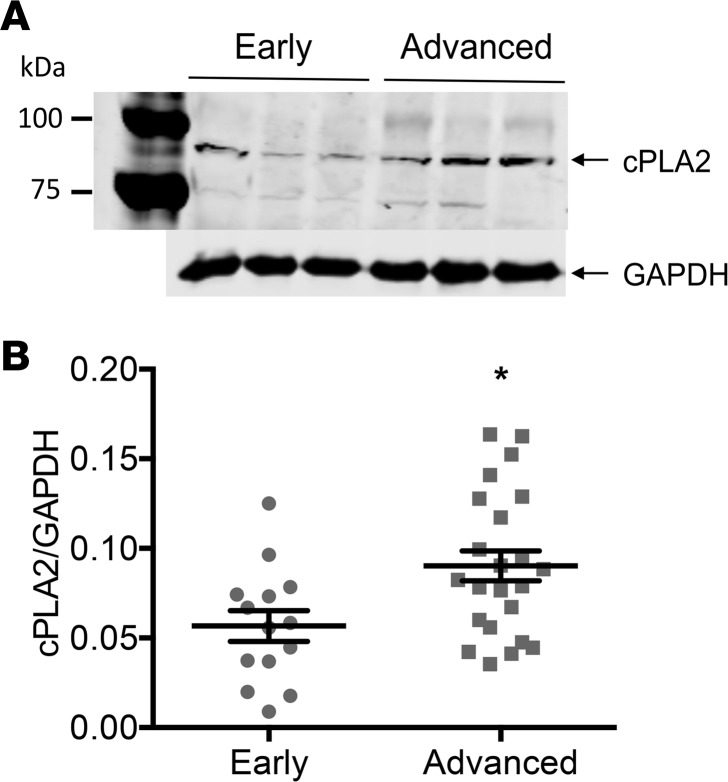
We next used this 152-protein signature to identify potentially new drug candidates for CVD treatment. Using CMap (10), we screened 1309 small molecules able to reverse this CVD signature in silico. Among the molecules of the CMap database, 12 were predicted to significantly reverse the CVD signature (Table 2). Among these 12 molecules, arachidonyltrifluoromethane, also called arachidonic acid trifluoromethyl ketone (AACOCF3), an inhibitor of cPLA2, showed the highest negative enrichment score (Table 2). Immunohistochemistry showed that cPLA2 was present in all vascular areas in early- and late-stage CVD (Supplemental Figure 2A). Semiquantitative analysis showed that cPLA2 was significantly more abundant in the intima of advanced- compared with early-stage CVD arteries, while no significant difference was observed in the tunica media and the tunica externa (Supplemental Figure 2B). Western Blot analysis confirmed that cPLA2 expression was significantly higher (1.4-fold) in arteries from advanced-stage CVD patients compared with early-stage CVD patients (Figure 2).
Table 2. Top molecules predicted to reverse human arterial protein signature of advanced-stage CVD following CMap analysis.
Figure 2. cPLA2 expression was increased in the arteries of advanced- compared with early-stage CVD patients.
(A) Representative cPLA2 and GAPDH Western blots in iliac arteries from early- (n = 3) versus advanced-stage (n = 3) CVD patients. (B) Scatter dot plots (arithmetic mean ± SEM) of cPLA2 protein expression normalized to GAPDH in iliac arteries from early- (n = 14) versus advanced-stage (n = 23) CVD patients. *P < 0.05 vs. early-stage CVD group (unpaired Mann-Whitney test).
AACOCF3 protects against vascular calcification.
Arterial calcifications are observed during medial calcification and late-stage atherosclerosis (4, 14). Vascular smooth muscle cells (VSMCs) are important mediators for these calcific processes (4). Due to important clinical implications, we have focused our interventional approach on the effects of AACOCF3 in VSMCs.
To evaluate the potential protective effects of AACOCF3 on CVD, experiments were performed in the cholecalciferol-overload (cholecalciferol is also known as vitamin D3) mouse vascular calcification model without or with additional treatment with AACOCF3. Cholecalciferol overload significantly increased serum calcium, calcium × phosphorus (CaxPi) product, and FGF23 C-terminal concentrations and reduced serum phosphorus levels to similar levels in both control- and AACOCF3-treated mice (Table 3).
Table 3. Effect of AACOCF3 during cholecalciferol overload.
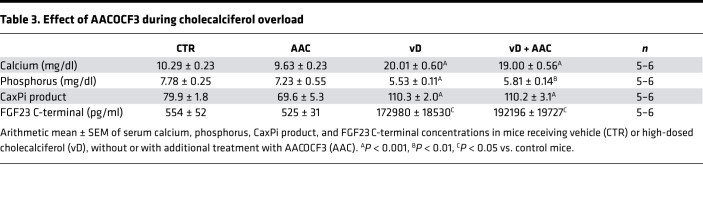
Cholecalciferol overload triggered aortic calcification in mice, an effect significantly attenuated by additional treatment with AACOCF3 (Figure 3A). In control mice, AACOCF3 did not modify aortic calcium deposition. The mRNA level of the gene encoding the most prevalent cPLA2 subtype, cPLA2α (Pla2g4a) (15), was significantly higher in aortic tissue of cholecalciferol-treated mice than in that of control mice (Figure 3B). AACOCF3 treatment alone did not significantly affect aortic Pla2g4a mRNA expression. Western blot analysis showed that cPLA2 protein was clearly present in the aortic tissue, but no significant change was observed after vitamin D or AACOCF3 treatments (Supplemental Figure 3).
Figure 3. AACOCF3 reduced vascular calcification in vivo.
(A) Scatter dot plots (arithmetic mean ± SEM) (n = 5–6, μg/mg protein) of calcium content in aortic tissue of mice receiving vehicle (CTR) or high-dosed cholecalciferol (vD), without or with additional AACOCF3 (AAC) treatment. (B) Scatter dot plots (arithmetic mean ± SEM) (n = 5–6; arbitrary units [a.u.]) of Pla2g4a relative mRNA expression in aortic tissue of mice receiving vehicle or high-dosed cholecalciferol, without or with additional AACOCF3 treatment. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control mice; †P < 0.05 vs. vD-treated mice (1-way ANOVA with Tukey-HSD post-hoc test).
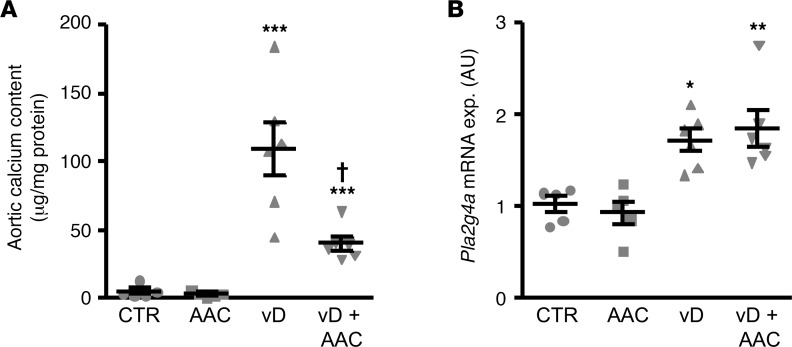
These effects were paralleled by inhibition of aortic osteogenic/chondrogenic signaling by AACOCF3. As shown in Figure 4, cholecalciferol overload significantly upregulated mRNA expression of osteogenic transcription factors Msx2 and Cbfa1 and of osteogenic enzyme tissue-nonspecific alkaline phosphatase (Alpl), markers of increased osteogenic/chondrogenic transdifferentiation in the aortic tissue, effects again significantly ameliorated by AACOCF3 treatment. Thus, AACOCF3 inhibited aortic osteoinductive signaling and vascular calcification in the cholecalciferol-overload mouse model.
Figure 4. AACOCF3 ameliorated aortic osteogenic/chondrogenic signaling during cholecalciferol overload–induced calcification in vivo.
Scatter dot plots (arithmetic mean ± SEM) (n = 5–6; arbitrary units [a.u.]) of Msx2 (A), Cbfa1 (B), and Alpl (C) relative mRNA expression in aortic tissue of mice receiving vehicle (CTR) or high-dosed cholecalciferol (vD), without or with additional treatment with AACOCF3 (AAC). *P < 0.05, **P < 0.01 vs. control mice; †P < 0.05 vs. vD-treated mice (Steel-Dwass method for A and 1-way ANOVA with Games-Howell post-hoc test for B and C).
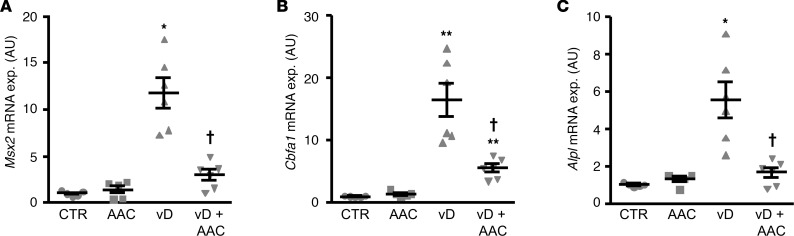
To investigate possible mechanisms of the protective effects of AACOCF3 on vascular calcification, further experiments were performed in primary human aortic smooth muscle cells (HAoSMCs) during high-phosphate conditions in the presence or absence of AACOCF3. As shown by alizarin red staining and quantification of calcium deposition, treatment with calcification medium increased calcification of HAoSMCs that was significantly blunted by treatment with AACOCF3 (Figure 5, A and B). Similarly, AACOCF3 significantly suppressed phosphate-induced MSX2, CBFA1, and ALPL mRNA expression (Figure 5, C–E) as well as ALPL activity (Figure 5F) in HAoSMCs. In control conditions, AACOCF3 did not significantly affect mineralization but tended to reduce CBFA1 mRNA levels (P = 0.202) and significantly downregulated MSX2 and ALPL mRNA expression in HAoSMCs (Figure 5, A–F). Thus, AACOCF3 suppressed phosphate-induced osteogenic/chondrogenic transdifferentiation and calcification of HAoSMCs in vitro.
Figure 5. AACOCF3 suppressed phosphate-induced calcification and osteogenic/chondrogenic signaling in HAoSMCs.
Representative images showing alizarin red staining (A, n = 3) and scatter dot plots (arithmetic mean ± SEM) of calcium content (B, n = 4, μg/mg protein) in HAoSMCs following treatment with control (CTR) or with calcification medium (Calc.), without or with additional treatment with 10 μM AACOCF3 (AAC). The calcified areas are shown as red staining. (C–G) Scatter dot plots (arithmetic mean ± SEM) of MSX2 (C), CBFA1 (D), and ALPL (E) relative mRNA expression (n = 6; arbitrary units [a.u.]) and ALPL activity (F, n = 4, U/mg protein) in HAoSMCs as well as arachidonic acid (AA) levels (G, n = 4; ng/mg protein) in the cell culture medium of HAoSMCs following treatment with control or with β-glycerophosphate (Pi), without or with additional treatment with 10 μM AACOCF3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control-treated HAoSMCs; †P < 0.05, ††P < 0.01, †††P < 0.001 vs. HAoSMCs treated with Calc./Pi alone (1-way ANOVA with Games-Howell post-hoc test for B, C, and F or with Tukey-HSD post-hoc test for D, E, and G).
Moreover, phosphate triggered the release of arachidonic acid (AA) in the cell culture medium of HAoSMCs (Figure 5G). Treatment with AACOCF3 significantly inhibited both basal and Pi-induced release of AA, the product of PLA2 activity.
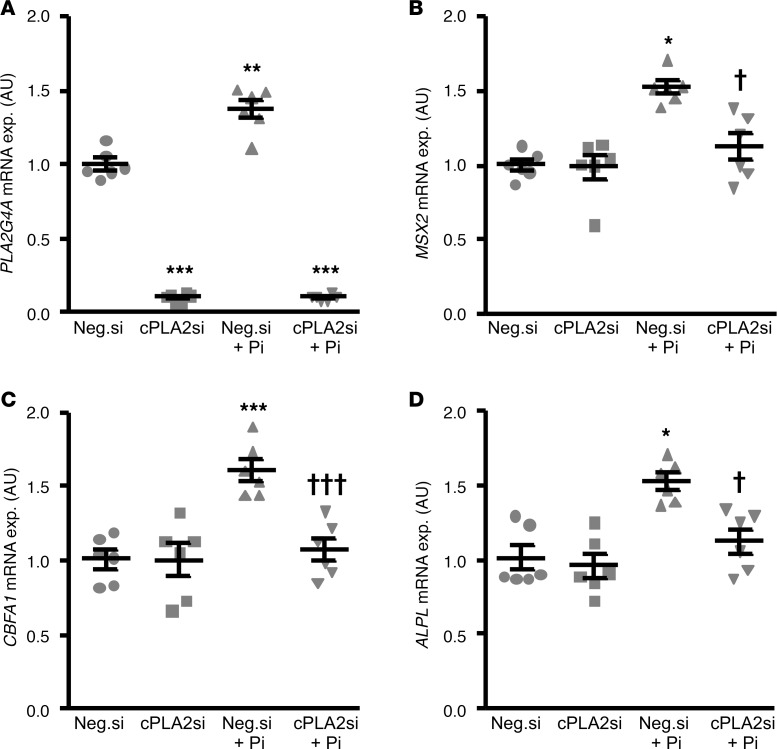
To confirm the involvement of cPLA2 in vascular calcification, the endogenous expression of cPLA2 in HAoSMCs was suppressed by silencing of the PLA2G4A gene using siRNA. As a result, transfection with cPLA2 siRNA significantly suppressed PLA2G4A mRNA levels in HAoSMCs, as compared with negative control-silenced HAoSMCs (Figure 6A). Phosphate treatment upregulated PLA2G4A mRNA expression in negative control-silenced HAoSMCs (Figure 6A). Furthermore, phosphate significantly increased MSX2, CBFA1, and ALPL mRNA expression in negative control siRNA–transfected HAoSMCs, effects significantly blunted in cPLA2 siRNA silenced HAoSMCs (Figure 6, B–D). Taken together, the protective effects of AACOCF3 on vascular calcification may involve, at least in part, inhibition of cPLA2-mediated osteoinductive signaling.
Figure 6. Silencing of cPLA2 inhibited phosphate-induced osteogenic/chondrogenic signaling in HAoSMCs.
Scatter dot plots (arithmetic mean ± SEM) (n = 6, arbitrary units [a.u.]) of PLA2G4A (A), MSX2 (B), CBFA1 (C), and ALPL (D) relative mRNA expression in HAoSMCs following silencing with negative control siRNA (Neg.si) or cPLA2 siRNA (cPLA2si), without or with additional treatment with β-glycerophosphate (Pi). *P < 0.05, **P < 0.01, ***P < 0.001 vs. Neg.si-silenced HAoSMCs; †P < 0.05, †††P < 0.001 vs. Neg.si-silenced and Pi-treated HAoSMCs (1-way ANOVA with Games-Howell post-hoc test for A or with Tukey-HSD post-hoc test for C and Steel-Dwass method for B and D).
AACOCF3 treatment of ApoE–/– mice.
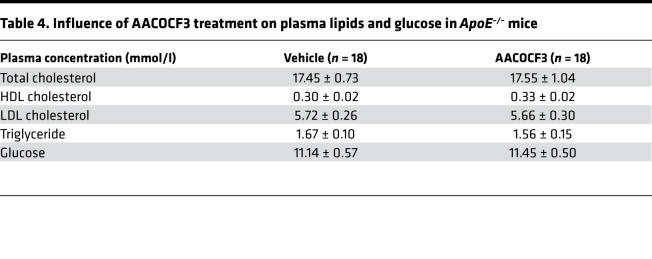
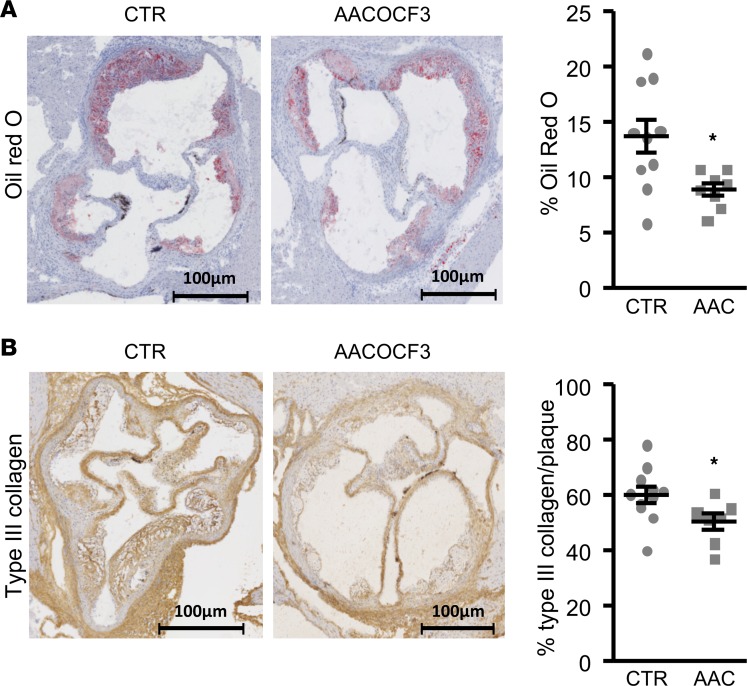
AACOCF3 was tested in another preclinical model of CVD, ApoE–/– mice known to develop atherosclerotic lesions. In our conditions, ApoE–/– mice fed a high-cholesterol diet displayed dyslipidemia (Table 4, vehicle) and lipid deposition (Figure 7A) and increased type III collagen expression (Figure 7B) in the aortic sinus, in agreement with previous reports (16). Ten weeks of treatment with AACOCF3 did not significantly modify plasma lipids and glucose levels (Table 4). In contrast, analysis of the aortic root showed that AACOCF3 had no influence on the total plaque area, but it significantly inhibited lipid deposition in the plaques (–35%; Figure 7A) compared with vehicle-treated mice. In parallel, AACOCF3 significantly reduced type III collagen plaque expression (–16%; Figure 7B) but had no significant influence on total collagen accumulation, as assessed by Sirius red staining (Supplemental Figure 4). In parallel, AACOCF3 did not changed αSMA (myofibroblast marker) and significantly increased MOMA2 (macrophage marker) staining (Supplemental Figure 4). Finally, AACOCF3 did not significantly change the expression of calcification gene markers (Alpl and Cbfa1) in aortic tissue (Supplemental Figure 5). Collectively, the experiments using the ApoE–/– mouse model showed that, while AACOCF3 reduces lipid accumulation, it has no effect on atherosclerotic lesions.
Table 4. Influence of AACOCF3 treatment on plasma lipids and glucose in ApoE–/– mice.
Figure 7. Influence of AACOCF3 treatment in ApoE–/– mice.
Representative images and scatter dot plots (arithmetic mean ± SEM) of quantification (percentage) of lipid deposits by oil red O staining (A, n = 8–10) and of type III collagen by immunostaining (B, n = 7–11) in aortic sinus cryosections of 6-week-old ApoE–/– male mice fed a high-cholesterol diet and simultaneously treated with AACOCF3 (AAC, 10 mg/kg) or vehicle (CTR) for 10 weeks. *P < 0.05 vs. control mice (unpaired 2-tailed t test). Scale bar: 100 μm.
Discussion
In the present study, we explored the possibility of omics-based disease signature correction by small bioactive compounds to identify and validate in vivo CVD drugs. Applying this approach to the high-resolution human arterial protein signature of advanced-stage CVD, we identified among >1000 compounds the cPLA2 inhibitor AACOCF3 that reversed this CVD signature. AACOCF3 is a cell-permeant trifluoromethyl ketone analog of AA (17) that behaves as a slow- and tight-binding inhibitor of cPLA2 (18–21). A review of the literature showed that cPLA2 inhibition has not been used in CVD management yet, suggesting AACOCF3 as a possible novel drug candidate for CVD treatment. Our preclinical analysis of the cholecalciferol-overload mouse model demonstrates that AACOCF3 was efficient in vivo at attenuating vascular calcification, one of the major features of CVD. To our knowledge, our work demonstrates for the first time the protective action of AACOCF3 against vascular injury.
Functional analysis of the proteins present in the human CVD vessel signature highlighted enrichment of proteins associated with cholesterol and lipid metabolism. Moreover, we also detected an increase in EFEMP1, also called fibulin-3, a protein involved in the regulation of osteogenic/chondrogenic transdifferentiation of VSMCs (22). Because vascular calcification is preferentially associated with advanced-stage CVD, we focused on the mechanisms of AACOCF3 in this process. Vascular calcification is mediated mainly by osteogenic/chondrogenic reprogramming of VSMCs in response to various triggers, especially hyperphosphatemia (14, 23). In liver tissue, excessive phosphate intake in rats is associated with increased AA concentrations (24). The osteogenic/chondrogenic transdifferentiated VSMCs are characterized by increased expression and activity of osteogenic and chondrogenic transcription factors as well as of osteogenic enzymes, such as tissue nonspecific ALPL, leading to active mineralization of the vascular tissue (22, 25–27). We show here that AACOCF3 treatment attenuates vascular osteogenic/chondrogenic transdifferentiation and calcification in vitro and in the cholecalciferol-overload mouse model in vivo. Moreover, vascular cPLA2 expression is increased during calcification conditions, both in vitro and in vivo. Accordingly, osteogenic/chondrogenic transdifferentiation of VSMCs during elevated phosphate levels in vitro is blunted by silencing of cPLA2, and, thus, cPLA2 knockdown mimics the protective effects of AACOCF3. These observations strongly suggest that the in vivo protective effect of AACOCF3 may result, at least partially, from its direct action on VSMCs through inhibition of cPLA2-dependent osteoinductive signaling.
There exist multiple PLA2 isoforms classified as secreted (sPLA2) and cytosolic (cPLA2) that have different structures and various physiological functions. PLA2G4A, i.e., cPLA2, is a calcium-dependent cPLA2. Some secreted isoforms, such as the lipoprotein-associated PLA2 (Lp-PLA2) or sPLA2-X or sPLA2-V (28), were shown as causative in atherosclerosis, and some of their inhibitors were in clinical trials but did not reach the market (29). Expression of cPLA2 has been previously found in arteries from atherosclerotic patients, mainly localized in the intimal regions with an inflammatory infiltrate consisting of macrophages and smooth muscle cells (30). Our results confirm a localization of cPLA2 in the intima, but the clinical relevance of this observation remains to be established because immunolocalization was not assessed directly within atherosclerotic plaques. Moreover, a PLA2G4A gene variant was associated with a decreased risk of coronary artery disease, although the functional consequence of the mutation remains still elusive (31). Our results are in agreement with those observations, since we found that cPLA2 was indeed present in arteries and its expression was increased in advanced-stage CVD. So far, to our knowledge the use of a cPLA2 inhibitor as a possible drug candidate for CVD treatment has never been proposed.
A first limitation of our study is that we have deliberately compared patients with advanced vascular disease and early vascular disease. While these features were confirmed by histology, the initial screen was based on presence or absence of risk factors, and, hence, differences in age, blood pressure, and other cardiovascular risk factors between the group are present. We cannot rule out that elements of the proteomic data are driven by these risk factors rather than specifically by the vascular phenotype. However, by selecting patients with multiple risk factors and advanced disease, we believe that our findings represent a molecular signature of advanced arteriosclerosis, rather than a single risk factor. A further limitation might be that the CVD signature was established from peripheral arteries (iliac and femoral) and cannot necessarily be extrapolated to a CVD signature in other arterial locations. However, our results show that AACOCF3 treatment was efficient in the aorta of the animal models, suggesting that its protective action is not limited to peripheral arteries and that, apparently, the molecular mechanisms overlap between these different vessel compartments/segments. We are also aware that the specificity of AACOCF3 for cPLA2 is likely not exclusive. Indeed, it has been reported that AACOCF3 was able to inhibit cyclooxygenases 1 and 2 (32). However, our results show that the effect of AACOCF3 could be reproduced by cPLA2 knockdown in vitro.
In conclusion, using a systems biology approach to optimally describe the molecular mechanisms of human advanced-stage CVD, we identified a potentially new compound that reversed typical vascular lesions in CVD in vivo. Apart from the clear effect of this approach in CVD, such a strategy should be able to generate novel targets in other complex diseases.
Methods
Arterial sample acquisition and CVD grading method.
18 common iliac arteries, 15 superficial femoral arteries, and 2 external iliac arteries were collected from 15 early- and 17 advanced-stage CVD patients, with the following specific distribution: early-stage CVD (11 common iliac arteries, 2 external iliac arteries, 4 superficial femoral arteries) and advanced-stage CVD (7 common iliac arteries, 11 superficial femoral arteries). All advanced-stage CVD patients had clear CVD phenotypes (vascular disease, left ventricular hypertrophy, angina pectoris, claudication intermittens, myocardial infraction). These cardiovascular phenotypes were not found early-stage CVD patients exhibits (Table 1). Following harvesting, the vascular tissues were snap frozen in liquid nitrogen and stored at –80°C until preparation for proteomics analysis. Blood and urine samples were also collected from the same patients for biochemical analysis.
Sample preparation and LC-MS/MS analysis.
Approximately 10–20 mg (net weight) of tissue specimen was homogenized in lysis buffer consisting of 0.1 M Tris-HCl, pH 7.6, supplemented with 4% SDS and 0.1 M DTE. Homogenization was performed with the bullet blender homogenizer (Next Advance) (33). Ten micrograms of each protein sample were analyzed in SDS-PAGE (5% stacking, 12% separating). Electrophoresis was stopped when samples just entered the separating gel. Gels were fixed with 30% methanol, 10% acetic acid for 30 minutes, followed by 3 washes with water (5 minutes each), and stained with Coomassie colloidal blue overnight. Each band was excised from the gel and further sliced into small pieces (1–2 mm). Gel pieces were destained with 40% acetonitrile, 50 mM NH4HCO3 and then reduced with 10 mM DTE in 100 mM NH4HCO3 for 20 minutes at room temperature. After reduction, samples were alkylated with 54 mM iodoacetamide in 100 mM NH4HCO3 for 20 minutes at room temperature in the dark. Samples were then washed with 100 mM NH4HCO3 for 20 minutes at room temperature, followed by another wash with 40% acetonitrile, 50 mM NH4HCO3 for 20 minutes at room temperature and a final wash with ultrapure water under the same conditions. Gel pieces were dried in a centrifugal vacuum concentrator and trypsinized overnight in the dark at room temperature by adding 600 ng trypsin per sample (trypsin stock solution: 10 ng/μl in 10 mM NH4HCO3, pH 8.5). Peptides were extracted after incubation with 50 mM NH4HCO3 for 15 minutes at room temperature, followed by 2 incubations with 10% formic acid, acetonitrile (1:1) for 15 minutes at room temperature. Peptides were eluted in a final volume of 600 μl and filtered with PVDF filters (Merck Millipore) before dried in a centrifugal vacuum concentrator. Dried peptides were reconstituted in mobile phase A buffer (0.1% formic acid, pH 3.5) and processed with LC-MS/MS analysis.
2.5 μg of protein digest was injected from each sample into a Dionex Ultimate 3000 RSLS nano flow system configured with a Dionex 0.1 × 20 mm 5 μm C18 nano trap column. The mobile phase was 2% ACN/0.1% formic acid with a flow rate of 5 μl/min. The analytical column was an Acclaim PepMap C18 nano column, 75 μm × 50 cm, 2 μm 100 Å, at a flow rate of 300 nl/min. The trap and nano flow column were maintained at 35°C. The samples were eluted with a gradient starting at 1% B for 5 minutes, rising to 5% B at 10 minutes, then to 25% B at 360 minutes, and 65% B at 480 minutes. Mobile phase A was 0.1% formic acid, while mobile phase B was 80% ACN/0.1% formic acid. The column was washed and reequilibrated prior to each sample injection. The eluent was ionized using a Proxeon nano spray ESI source operating in positive ion mode. For mass spectrometry analysis, an Orbitrap LTQ Velos (Thermo Finnigan) was operated in MS/MS mode, scanning from 380 to 2,000 m/z. Ionization voltage was 2.6 kV, and the capillary temperature was 275°C. The resolution of ions in MS1 was 60,000 and 7,500 for higher-energy collisional dissociation (HCD) MS2. The top 20 multiply charged ions were selected from each scan for MS/MS analysis using HCD at 35% collision energy. Dynamic exclusion was enabled with a repeat count of 1, exclusion duration of 30 seconds.
MS data processing and quantification.
Raw files were analyzed with Proteome Discoverer 1.4 software package (Thermo Finnigan), using the Sequest search engine and the Uniprot human (Homo sapiens) reviewed database, downloaded on May 30, 2016. The search was performed using carbamidomethylation of cysteine as static and oxidation of methionine as dynamic modifications. Two missed cleavage sites, a precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.05 Da were allowed. FDR validation was based on q values: target FDR (strict): 0.01, target FDR (relaxed): 0.05. Quantification analysis was performed at the peptide level, with the clustering approach as previously described (34), with some minor modifications. Peptides were clustered using the “window” of mass (±5 ppm) and retention time (±15 minutes). Only peptides present in 60% of the samples in at least 1 group were further processed for quantification and statistical analysis (Mann-Whitney). The differentially expressed proteins selected for further analysis were considered those with a fold change of ≥1.2 or ≤0.83 in both batches and a P value of ≤0.05 in at least 1 batch.
CMap analysis.
Each upregulated and downregulated protein ID from the CVD signature was converted into gene ID and were analyzed with the bioinformatic tool CMap (build 02). CMap is a collection of genome-wide transcriptional expression signatures (6,100 treatment vs. vehicle control pairs) from cultured human cells (MCF7, PC3, and HL60) treated with 1,309 bioactive small-chemical molecules at varying concentrations and analyzed by simple pattern-matching algorithms (10).
Preparation of AACOCF3 solutions.
AACOCF3 was supplied by Cayman Chemical (item 62120). For in vivo treatments, the initial solution of AACOCF3 (50 mg/5 ml ethanol) was concentrated to 250 mg/ml and 20 μl was diluted in 4 ml of a 0.5% methylcellulose (Fisher Scientific, product 10246190) in water. That solution was used to treat the animals. For in vitro treatment with AACOCF3 (Cayman Chemical), 10 mM stock solution in ethanol, was diluted to a final concentration of 10 μM in cell culture medium. Equal amounts of vehicle were used as control.
Mouse model of vascular calcification.
Female C57BL/6 mice were treated with AACOCF3 (10 mg/kg) or its vehicle (methylcellulose 0.5% containing 0.5% M ethanol) via gavage and were fed a calcium-reduced diet (Altromin, C1031). Subsequently, mice were injected subcutaneously with 400,000 IU/kg BW of cholecalciferol (MilliporeSigma) or vehicle for 3 days (35, 36). After 6 days of treatment, blood was collected by retroorbital puncture under isoflurane inhalation anaesthesia. The mice were immediately sacrificed by cervical dislocation, and aortic tissues were collected and snap frozen for further experiments.
Mouse model of atherosclerosis.
ApoE–/– mice (37, 38) (6-week-old males) (Charles River) were fed a high-cholesterol diet (1.25% cholesterol, 6% fat, and 2% sucrose [Ssniff, Germany diet, TD.96335 mod. – 1.25% cholesterol]) and simultaneously treated (gavage with 200 μl; 5 days a week) with AACOCF3 (10 mg/kg) or its vehicle (methylcellulose 0.5% containing 0.5% M ethanol) for 10 weeks. The mice were sacrificed by pentobarbital injection, and the heart aortic sinus was collected and snap frozen in liquid nitrogen for further analysis. Blood was collected 1 week before sacrifice by intracaudal puncture in heparinized tubes. Ice-cold blood was centrifuged for 5 minutes at 1500 g, and upper plasma phase was collected and immediately frozen at –80°C.
Biochemical analysis.
The plasma concentrations of phosphorus and calcium were measured by the QuantiChrom Phosphate assay kit and QuantiChrom Calcium assay kit, respectively (BioAssay Systems). An ELISA kit was used to determine plasma concentrations of FGF23 C-terminal (Immutopics), according to the manufacturer’s instructions. Blood glucose concentration was measured using a glucometer (Roche Diagnostic). Plasma concentrations of cholesterol and triglyceride were measured by autoanalyzer (Horiba Medical).
Immunohistology.
For human, histologic paraffin sections of iliac arteries samples were cut at 2 μm, mounted on charged slides, and stained with H&E and von Kossa for pathological evaluation. cPLA2 staining was performed on 1–2 μm paraffin sections of A. ilia communis samples of histopathologically classified early (n = 15) and advanced (n = 24) patients, using a 1:50 dilution of the rabbit anti-human PLA2G4A antibody (ProSci Incorporated). Heat-induced epitope retrieval was performed at pH 8. For detection, the EnVision+ System-HRP Labelled Polymer Anti-Rabbit kit (Dako) was used. Nuclei were counterstained with hematoxylin, and sections were coverslipped with gelatin. The expression levels of cPLA2 were evaluated per tissue section (vessel cross section), distinguishing between the different vessel layers (tunica intima, tunica media, and tunica externa). The abundance of cPLA2 was scored as follows: 0.5 for no expression, 1 for weak expression, 2 for moderate expression, and 3 for strong expression. Images were acquired using the AxioImager Z1 microscope (Carl Zeiss MicroImaging Inc.). All evaluations were performed in a blinded manner.
For aortic sinuses of ApoE–/– mice, lipid accumulation and collagen expression were analyzed on serial cross-cryosections of the heart as previously described (39). For lipid analysis, cryosections were post-fixed with formaldehyde and stained with oil red O and counterstained with hematoxylin. For collagen, cryosections were post-fixed with methanol/acetone and exposed to a type III collagen primary antibody (Acris, BP8014) and revealed with an anti-rabbit HRP secondary antibody (DAKO, 4010). Samples were imaged with a Nikon Y-FL microscope equipped with a computer-based imaging system. Staining was expressed as the percentage of total sinus area. Reported intensities used for further statistical analysis correspond to the mean value of 5 sections per animal.
Western blot.
Arteria iliaca communis samples of histopathologically classified control (n = 15) and disease (n = 24) patients were homogenized in lysis buffer (Invitrogen) supplemented with proteinase inhibitors (Roche). Next, 30 μg protein was loaded on 10% SDS-polyacrylamide gels. EFEMP1, GUCY1B2, KDM5D, cPLA2, and SHMT1 were detected with a rabbit anti-human EFEMP1 antibody (1:1000, Abcam), rabbit anti-human GUCY1B2 antibody (1:500, Thermo Fisher Scientific), rabbit anti-human KDM5D (Jarid 1D, 1:5000, Novus Biologicals), rabbit anti-human PLA2G4A antibody (1:500, ProSci Incorporated), and rabbit anti-human SHMT1 (1 250, MilliporeSigma), respectively, followed by a goat anti-rabbit IR dye 680 secondary antibody (1:7500, LI-COR Biosciences). All samples were normalized toward GAPDH (1:2000, BPS Bioscience) as internal control. Visualization and quantitative analysis were performed on Odyssey (LI-COR Biosciences) via Odyssey V3.0 software (LI-COR Biosciences).
Cell culture of primary HAoSMCs.
Primary HAoSMCs (Thermo Fisher Scientific) were cultured in Waymouth’s MB 752/1 medium and Ham’s F-12 nutrient mixture in a 1:1 ratio (Thermo Fisher Scientific) containing 10% FBS (Thermo Fisher Scientific) and 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific) (40, 41). HAoSMCs were used in all experiments from passages 4 to 11, and each experiment was performed in at least 2 different batches of HAoSMCs at different passages, depending on the availability of the cells. HAoSMCs were treated for 24 hours (qRT-PCR, AA release), 7 days (ALPL assay), or 11 days (calcification) with 2 mM β-glycerophosphate (MilliporeSigma) (25, 36, 40, 42, 43) or with 10 μM AACOCF3. Equal amounts of vehicle were used as control. For the mineralization analysis and alizarin red staining, treatment with 10 mM β-glycerophosphate and 1.5 mM CaCl2 (MilliporeSigma) for 11 days was used as calcification media (44). Fresh media with agents were added every 2–3 days. HAoSMCs were transfected with 10 nM cPLA2 siRNA (Thermo Fisher Scientific, s10592) or with 10 nM negative control siRNA (Thermo Fisher Scientific, 4390843) using siPORT amine transfection agent (Thermo Fisher Scientific) according to the manufacturer’s protocol. The cells were used 48 hours (qRT-PCR) after silencing, and silencing efficiency was determined by quantitative RT-PCR.
Analysis of calcification.
To quantify vascular calcification, abdominal aortic tissues were incubated overnight at 37°C in 0.6 M HCl. HAoSMCs were incubated in 0.6 M HCl for 24 hours at 4°C. Calcium content in the supernatant was determined by using QuantiChrom Calcium assay kit (BioAssay Systems) according to the manufacturer’s protocol. Tissues and HAoSMCs were lysed with 0.1 M NaOH/ 0.1% SDS and protein concentration was determined by the Bradford assay (Bio-Rad). The results are shown normalized to total protein concentration (45). To visualize calcification, HAoSMCs were fixed with 4% paraformaldehyde and stained with 2% alizarin red (pH 4.5). The calcified areas are shown as red staining (36).
ALPL activity assay.
ALPL activity in HAoSMCs was determined by using the ALP colorimetric assay kit (Abcam) according to the manufacturer’s protocol (22, 41). The results are shown normalized to total protein concentration measured by the Bradford assay (Bio-Rad).
AA release.
Medium AA levels were determined by using an AA ELISA kit (Elabscience) according to the manufacturer’s instructions. The results are shown normalized to total protein concentration measured by the Bradford assay (Bio-Rad).
RNA isolation and quantitative RT-PCR.
Total RNA was isolated from aortic tissue and HAoSMCs by using Trizol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions (46–48). Reverse transcription of total RNA was performed using oligo(dT)12–18 primers (Thermo Fisher Scientific) and SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). Quantitative RT-PCR was performed with the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and iQ Sybr Green Supermix (Bio-Rad) according to the manufacturer’s instructions. The mouse and human primer sequences are given in Supplemental Tables 1 and 2, respectively. The specificity of the PCR products was confirmed by analysis of the melting curves. All PCRs were performed in duplicate, and relative mRNA expression was calculated by the 2–ΔΔCt method using GAPDH as housekeeping gene normalized to the control group.
Statistics.
Proteomics statistical analysis was based on the Mann-Whitney test performed using R package. For both batches, protein changes between early-stage and advanced-stage CVD with P ≤ 0.05 were considered as statistically significant. For other statistical analysis, data are shown as scatter dot plots, with arithmetic mean ± SEM; n indicates the number of independent experiments performed at different passages of the cells or the number of mice examined, respectively. Normality was tested with the Shapiro-Wilk test. Nonnormal data sets were transformed (log, reciprocal, or sqrt) prior to statistical testing to provide normality according to the Shapiro-Wilk test. Statistical testing was performed by 1-way ANOVA followed by Tukey test for homoscedastic data or Games-Howell test for heteroscedastic data. Nonnormal data were tested by the Steel-Dwass method. Two groups were compared by unpaired 2-tailed t test or Mann-Whitney test. P < 0.05 was considered statistically significant (22).
Study approval.
The study has been conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent has been provided by each patient or their representative. Human studies were reviewed and approved by the local ethics committee (Ethikkommission - Medizinische Universität Graz, Graz, Austria; approval 26–355 ex 13/14). All animal experiments were conducted according to the recommendations of the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) as well as the German law for the welfare of animals and were approved by local authorities (ethics committee for animal experiment, G32217; Berlin, Germany). The project was approved by the local (Inserm/UPS US006 CREFRE) and national ethics committees (ethics committee for animal experiment, CEEA122; Toulouse, France; approval 02604.02).
Author contributions
JPS, JK, JLB, AV, CD, JV, IA, HM, BP, FL, and DVL designed the research; MM, SVL, VL, AL, TTDL, JV, IA, BB, NS, GF, CD, and WM performed the experiments; JPS, JK, JSSB, JLB, AV, CD, JV, IA, HM, BP, and FL analyzed and interpreted the data; and JSSB, JPS, JK, JLB, AV, CD, JV, IA, HM, BP, FL, and SN wrote the manuscript. All authors gave final approval of the version to be published.
Supplementary Material
Acknowledgments
This work was supported by the European Union Seventh Framework Program (FP7/2007-2013–603288-SysVasc, to JPS, VL, SVL, CD, FL, BP, JLP, HM, JSSB, JV, AV, and JL), the European Union ERA CVD JTC2017 PROACT (ANR-17-ECVD-0006 to JK, GF, CD, JSSB, and JPS; 01KL1805 via the Federal Ministry of Education and Research to HM), INSERM, the “Fondation pour la Recherche Médicale” (grant DEQ20170336759 to JK, GF, CD, JSSB and JPS), the British Heart Foundation Centre of Research Excellence Award (RE/13/5/30177), the Deutsche Forschungsgemeinschaft (AL2054/1-1, VO2259/2-1), the Berlin Institute of Health, and the Else Kröner-Fresenius-Stiftung to TTDL, JV, IA, BB, and NS. Immunopathological analysis of cPLA2 was provided by the iPATH.Berlin – Core Unit Immunopathology for Experimental Models of the Charité – Universitätsmedizin Berlin (Berlin, Germany).
Version 1. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e125638. https://doi.org/10.1172/jci.insight.125638.
Contributor Information
Manousos Makridakis, Email: mmakrid@bioacademy.gr.
Sophie Van Linthout, Email: sophie.van-linthout@charite.de.
Vasiliki Lygirou, Email: vlygirou@hotmail.com.
Ioana Alesutan, Email: ioana.alesutan@charite.de.
Beate Boehme, Email: beate.boehme@charite.de.
Nadeshda Schelski, Email: sibnade4ka31@zedat.fu-berlin.de.
Dirk Von Lewinski, Email: dirk.von-lewinski@medunigraz.at.
William Mullen, Email: William.Mullen@glasgow.ac.uk.
Christian Delles, Email: Christian.Delles@glasgow.ac.uk.
Guylène Feuillet, Email: guylene.feuillet@inserm.fr.
Florian Lang, Email: florian.lang@uni-tuebingen.de.
Burkert Pieske, Email: burkert.pieske@charite.de.
Jean-Loup Bascands, Email: jean-loup.bascands@inserm.fr.
Harald Mischak, Email: mischak@mosaiques-diagnostics.com.
Jean-Sebastien Saulnier-Blache, Email: Jean-Sebastien.Saulnier-Blache@inserm.fr.
Jakob Voelkl, Email: jakob.voelkl@charite.de.
Antonia Vlahou, Email: vlahoua@bioacademy.gr.
Julie Klein, Email: julie.klein@inserm.fr.
References
- 1.Roth GA, et al. Demographic and epidemiologic drivers of global cardiovascular mortality. N Engl J Med. 2015;372(14):1333–1341. doi: 10.1056/NEJMoa1406656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin EJ, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;135(10):e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albanese I, Khan K, Barratt B, Al-Kindi H, Schwertani A. Atherosclerotic Calcification: Wnt Is the Hint. J Am Heart Assoc. 2018;7(4):e007356. doi: 10.1161/JAHA.117.007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durham AL, Speer MY, Scatena M, Giachelli CM, Shanahan CM. Role of smooth muscle cells in vascular calcification: implications in atherosclerosis and arterial stiffness. Cardiovasc Res. 2018;114(4):590–600. doi: 10.1093/cvr/cvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17(11):1410–1422. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 6.Rüegg C, Tissot JD, Farmer P, Mariotti A. Omics meets hypothesis-driven research. Partnership for innovative discoveries in vascular biology and angiogenesis. Thromb Haemost. 2008;100(5):738–746. doi: 10.1160/TH08-06-0348. [DOI] [PubMed] [Google Scholar]
- 7.Dominiczak AF, et al. Systems biology to battle vascular disease. Nephrol Dial Transplant. 2010;25(4):1019–1022. doi: 10.1093/ndt/gfq025. [DOI] [PubMed] [Google Scholar]
- 8.Vernon ST, Hansen T, Kott KA, Yang JY, O’Sullivan JF, Figtree GA. Utilizing state-of-the-art “omics” technology and bioinformatics to identify new biological mechanisms and biomarkers for coronary artery disease. Microcirculation. 2019;26(2):e12488. doi: 10.1111/micc.12488. [DOI] [PubMed] [Google Scholar]
- 9.Mokou M, Lygirou V, Vlahou A, Mischak H. Proteomics in cardiovascular disease: recent progress and clinical implication and implementation. Expert Rev Proteomics. 2017;14(2):117–136. doi: 10.1080/14789450.2017.1274653. [DOI] [PubMed] [Google Scholar]
- 10.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 11.Xi D, Zhao J, Zhao M, Fu W, Guo Z, Chen H. Identification of Gene Expression Changes in the Aorta of ApoE Null Mice Fed a High-Fat Diet. Genes (Basel) 2017;8(10):E289. doi: 10.3390/genes8100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang CH, et al. Identify potential drugs for cardiovascular diseases caused by stress-induced genes in vascular smooth muscle cells. PeerJ. 2016;4:e2478. doi: 10.7717/peerj.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, et al. Gene expression profiling of CD133-positive cells in coronary artery disease. Mol Med Rep. 2015;12(5):7512–7516. doi: 10.3892/mmr.2015.4359. [DOI] [PubMed] [Google Scholar]
- 14.Lang F, et al. Therapeutic Interference With Vascular Calcification-Lessons From Klotho-Hypomorphic Mice and Beyond. Front Endocrinol (Lausanne) 2018;9:207. doi: 10.3389/fendo.2018.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie CC. Cytosolic phospholipase A2: physiological function and role in disease. J Lipid Res. 2015;56(8):1386–1402. doi: 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Getz GS, Reardon CA. Animal models of atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(5):1104–1115. doi: 10.1161/ATVBAHA.111.237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimble LA, Street IP, Perrier H, Tremblay NM, Weech PK, Bernstein MA. NMR structural studies of the tight complex between a trifluoromethyl ketone inhibitor and the 85-kDa human phospholipase A2. Biochemistry. 1993;32(47):12560–12565. doi: 10.1021/bi00210a002. [DOI] [PubMed] [Google Scholar]
- 18.Riendeau D, et al. Arachidonyl trifluoromethyl ketone, a potent inhibitor of 85-kDa phospholipase A2, blocks production of arachidonate and 12-hydroxyeicosatetraenoic acid by calcium ionophore-challenged platelets. J Biol Chem. 1994;269(22):15619–15624. [PubMed] [Google Scholar]
- 19.Bartoli F, Lin HK, Ghomashchi F, Gelb MH, Jain MK, Apitz-Castro R. Tight binding inhibitors of 85-kDa phospholipase A2 but not 14-kDa phospholipase A2 inhibit release of free arachidonate in thrombin-stimulated human platelets. J Biol Chem. 1994;269(22):15625–15630. [PubMed] [Google Scholar]
- 20.Gelb MH, Jain MK, Berg OG. Inhibition of phospholipase A2. FASEB J. 1994;8(12):916–924. doi: 10.1096/fasebj.8.12.8088457. [DOI] [PubMed] [Google Scholar]
- 21.Street IP, et al. Slow- and tight-binding inhibitors of the 85-kDa human phospholipase A2. Biochemistry. 1993;32(23):5935–5940. doi: 10.1021/bi00074a003. [DOI] [PubMed] [Google Scholar]
- 22.Luong TTD, et al. Fibulin-3 Attenuates Phosphate-Induced Vascular Smooth Muscle Cell Calcification by Inhibition of Oxidative Stress. Cell Physiol Biochem. 2018;46(4):1305–1316. doi: 10.1159/000489144. [DOI] [PubMed] [Google Scholar]
- 23.Voelkl J, et al. Zinc Inhibits Phosphate-Induced Vascular Calcification through TNFAIP3-Mediated Suppression of NF- J Am Soc Nephrol. 2018;29(6):1636–1648. doi: 10.1681/ASN.2017050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun S, et al. A High Phosphorus Diet Affects Lipid Metabolism in Rat Liver: A DNA Microarray Analysis. PLoS ONE. 2016;11(5):e0155386. doi: 10.1371/journal.pone.0155386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steitz SA, et al. Smooth muscle cell phenotypic transition associated with calcification: upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ Res. 2001;89(12):1147–1154. doi: 10.1161/hh2401.101070. [DOI] [PubMed] [Google Scholar]
- 26.Lanzer P, et al. Medial vascular calcification revisited: review and perspectives. Eur Heart J. 2014;35(23):1515–1525. doi: 10.1093/eurheartj/ehu163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voelkl J, et al. SGK1 induces vascular smooth muscle cell calcification through NF-κB signaling. J Clin Invest. 2018;128(7):3024–3040. doi: 10.1172/JCI96477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenson RS, Hurt-Camejo E. Phospholipase A2 enzymes and the risk of atherosclerosis. Eur Heart J. 2012;33(23):2899–2909. doi: 10.1093/eurheartj/ehs148. [DOI] [PubMed] [Google Scholar]
- 29.Kokotou MG, Limnios D, Nikolaou A, Psarra A, Kokotos G. Inhibitors of phospholipase A. Expert Opin Ther Pat. 2017;27(2):217–225. doi: 10.1080/13543776.2017.1246540. [DOI] [PubMed] [Google Scholar]
- 30.Elinder LS, Dumitrescu A, Larsson P, Hedin U, Frostegård J, Claesson HE. Expression of phospholipase A2 isoforms in human normal and atherosclerotic arterial wall. Arterioscler Thromb Vasc Biol. 1997;17(10):2257–2263. doi: 10.1161/01.ATV.17.10.2257. [DOI] [PubMed] [Google Scholar]
- 31.Hartiala J, et al. Genetic contribution of the leukotriene pathway to coronary artery disease. Hum Genet. 2011;129(6):617–627. doi: 10.1007/s00439-011-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leis HJ, Windischhofer W. Inhibition of cyclooxygenases 1 and 2 by the phospholipase-blocker, arachidonyl trifluoromethyl ketone. Br J Pharmacol. 2008;155(5):731–737. doi: 10.1038/bjp.2008.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makridakis M, Vlahou A. GeLC-MS: A Sample Preparation Method for Proteomics Analysis of Minimal Amount of Tissue. Methods Mol Biol. 2018;1788:165–175. doi: 10.1007/7651_2017_76. [DOI] [PubMed] [Google Scholar]
- 34.Lygirou V, et al. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J Transl Med. 2018;16(1):104. doi: 10.1186/s12967-018-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han MS, et al. Functional cooperation between vitamin D receptor and Runx2 in vitamin D-induced vascular calcification. PLoS ONE. 2013;8(12):e83584. doi: 10.1371/journal.pone.0083584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alesutan I, et al. Augmentation of phosphate-induced osteo-/chondrogenic transformation of vascular smooth muscle cells by homoarginine. Cardiovasc Res. 2016;110(3):408–418. doi: 10.1093/cvr/cvw062. [DOI] [PubMed] [Google Scholar]
- 37.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258(5081):468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 38.Plump AS, et al. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71(2):343–353. doi: 10.1016/0092-8674(92)90362-G. [DOI] [PubMed] [Google Scholar]
- 39.Lesnik P, Haskell CA, Charo IF. Decreased atherosclerosis in CX3CR1–/– mice reveals a role for fractalkine in atherogenesis. J Clin Invest. 2003;111(3):333–340. doi: 10.1172/JCI15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alesutan I, Musculus K, Castor T, Alzoubi K, Voelkl J, Lang F. Inhibition of Phosphate-Induced Vascular Smooth Muscle Cell Osteo-/Chondrogenic Signaling and Calcification by Bafilomycin A1 and Methylamine. Kidney Blood Press Res. 2015;40(5):490–499. doi: 10.1159/000368524. [DOI] [PubMed] [Google Scholar]
- 41.Alesutan I, et al. Involvement Of Vascular Aldosterone Synthase In Phosphate-Induced Osteogenic Transformation Of Vascular Smooth Muscle Cells. Sci Rep. 2017;7(1):2059. doi: 10.1038/s41598-017-01882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao XB, et al. MiR-133a modulates osteogenic differentiation of vascular smooth muscle cells. Endocrinology. 2013;154(9):3344–3352. doi: 10.1210/en.2012-2236. [DOI] [PubMed] [Google Scholar]
- 43.Qiao W, Chen L, Zhang M. MicroRNA-205 regulates the calcification and osteoblastic differentiation of vascular smooth muscle cells. Cell Physiol Biochem. 2014;33(6):1945–1953. doi: 10.1159/000362971. [DOI] [PubMed] [Google Scholar]
- 44.Villa-Bellosta R, Millan A, Sorribas V. Role of calcium-phosphate deposition in vascular smooth muscle cell calcification. Am J Physiol, Cell Physiol. 2011;300(1):C210–C220. doi: 10.1152/ajpcell.00229.2010. [DOI] [PubMed] [Google Scholar]
- 45.Alesutan I, et al. Inhibition of osteo/chondrogenic transformation of vascular smooth muscle cells by MgCl2 via calcium-sensing receptor. J Hypertens. 2017;35(3):523–532. doi: 10.1097/HJH.0000000000001202. [DOI] [PubMed] [Google Scholar]
- 46.Voelkl J, et al. Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice. J Clin Invest. 2013;123(2):812–822. doi: 10.1172/JCI64093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voelkl J, et al. Up-regulation of hepatic alpha-2-HS-glycoprotein transcription by testosterone via androgen receptor activation. Cell Physiol Biochem. 2014;33(6):1911–1920. doi: 10.1159/000362968. [DOI] [PubMed] [Google Scholar]
- 48.Voelkl J, et al. AMP-activated protein kinase α1-sensitive activation of AP-1 in cardiomyocytes. J Mol Cell Cardiol. 2016;97:36–43. doi: 10.1016/j.yjmcc.2016.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.