Abstract
Background: Infiltrating lobular carcinoma (ILC) is the second most common histologicaI subtype of breast cancer, accounting for 10% of all cases. ILC has a characteristic genomic profile. ILC shows a high frequency of cadherin 1 (CDH1) mutations, along with loss of phosphatase and tensin homolog (PTEN), activation of alpha serine/threonine kinase (AKT), and mutations in T-box transcription factor (TBX3) and forkhead box protein A1 (FOXA1). We suspected that another gene, von Willebrand factor (VWF), might also be part of the profile, since coagulation tests reveal significant differences in patients with breast cancer. Materials and Methods: For newly-diagnosed breast cancer, the association between VWF and histology in the GDC Breast Cancer dataset in The Cancer Genome Atlas (TCGA) was evaluated. The following were used to access and analyze the data: Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/); Xena browser (https://xenabrowser.net); cBioportal (http://cbioportal.org); Oncomine (https://oncomine.org); and Prediction Analysis of Microarray 50 (PAM50). Results: Patients with ILC had higher VWF RNA expression than patients with infiltrating ductal carcinoma and other histology. The difference of expression was present to the same degree in both pre-menopausal and post-menopausal patients. Nine alterations in VWF and PTEN were significantly co-occurrent. Considering all histologies in 843 samples, Tukey’s honest significant difference post hoc test showed that VWF RNA expression of the normal subtype was significantly greater than that of the other subtypes (p<0.001). Conclusion: Our finding of significantly higher VWF RNA expression in the PAM50 normal (non-basal-like) breast cancer subtype suggests that VWF protein measurement might complement or corroborate PAM50 results. VWF and PAM50 results both suggesting a low risk of recurrence might make the decision whether to give chemotherapy easier, especially if VWF protein were an independent predictor.
Keywords: Breast cancer, survival, von Willebrand factor, The Cancer Genome Atlas, coagulation
Infiltrating lobular carcinoma (ILC) is the second most common histological subtype of breast cancer, accounting for 10% of all cases. ILC differs from infiltrating ductal carcinoma (IDC) in its pathological features and responsiveness to systemic therapy. ILC derives a distinct benefit from systemic therapy compared to IDC (1).
ILC has a characteristic genomic profile, exhibiting a high frequency of cadherin 1 (CDH1) mutation, along with loss of phosphatase and tensin homolog (PTEN), activation of alpha serine/threonine kinase (AKT), and mutations in T-box transcription factor (TBX3) and forkhead box protein A1 (FOXA1) (2,3). The genomic profile of lobular carcinoma in situ is similar (4).
We suspected that another gene, von Willebrand factor (VWF), might be part of the profile, since coagulation tests reveal significant differences in patients with breast cancer (5,6). One report documents VWF up-regulation in ILC and suggests an effect on cell adhesion (7).
VWF, the largest human plasma protein, is a multimeric glycoprotein that mediates platelet adhesion to both the subendothelial matrix and endothelial surfaces and acts as a carrier for coagulation factor VIII in the circulation (6). Besides its essential role in hemostasis, VWF has an effect on tumors, mainly by inhibiting angiogenesis and apoptosis (6). Increased expression of VWF gene is associated with poorer survival in primary lower-grade gliomas (8).
In the current study, we used The Cancer Genome Atlas (TCGA) to investigate whether VWF might be part of the genetic profile of ILC.
Materials and Methods
For newly diagnosed breast cancer, this study evaluated the association between VWF gene expression and histology in the GDC Breast Cancer dataset in TCGA (9). The following were used to access and analyze the data: Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/); Xena browser (https://xenabrowser.net) (10); cBioportal (http://cbioportal.org) (11); Oncomine (https:// oncomine.org) (12); and Prediction Analysis of Microarray 50 (PAM50), a Food and Drug Administration-approved tooI for intrinsic subtyping of breast cancer using quantitative reverse transcriptase-polymerase chain reaction that assays the expression of 50 genes. VWF Data from these assays were sourced from Oncomine. Comparisons were made only for VWF mRNA. In the case of the PAM50, we did not look at data from any of the 50 genes included in the test; we looked only at whether the PAM50 result was reported as normal or abnormal.
Gene expression is reported as fragments per kilobase of transcript per million mapped reads upper quartile (fpkm-uq), which is an RNA-Seq-based method for expression normalization (13).
Simple statistics were calculated to identify patterns of mutual exclusivity or co-occurrence. For each pair of query genes (e.g. VWF and PTEN), an odds ratio (OR) was calculated (Equation 1) that indicates the likelihood that the events in the two genes are mutually exclusive or co-occurrent across the selected cases
OR=(A×D)/(B×C) (Eq. 1)
where A was the number of cases in which both genes were altered; B was the number of cases in which VWF but not PTEN was altered; C was the number of cases in which PTEN but not VWF was altered; and D was the number of cases in which neither gene was altered. Each pair was then assigned to one of three categories indicative of a tendency toward mutual exclusivity, co-occurrence, or of no association, respectively. Only Fisher’s exact test was performed to determine whether the identified relationship was significant for a gene pair. The test is used to support co-occurrence when the number of tumors with alterations in both genes is significantly higher than expected by chance. Likewise, it suggests mutual exclusivity when the number of tumors with alterations in both genes is significantly lower (11).
Results
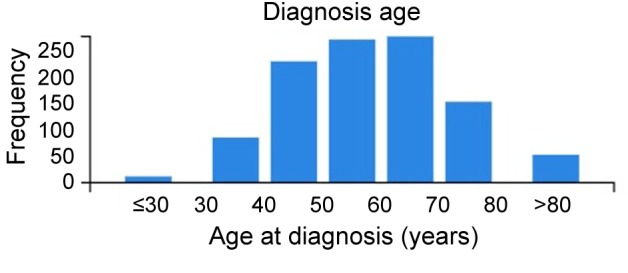
We analyzed the Breast Invasive Carcinoma (TCGA) cohort of 1,113 samples from women. The age distribution of patients is shown in Figure 1. Race information: 69.3% were White, 16.8% were Black, 5.5% Asian, 0.1% American Indian, 8.3% unclassified. Ethnicity information: 80% were non-Hispanic, 4% were Hispanic, 16% were unclassified (unclassified can refer to race or non-Hispanic, Hispanic classifications). Out of the tumors that were staged, 26% were stage T1, 58% stage T2, 12% stage T3, and 4% stage T4 (one tumor had no staging).
Figure 1. Distribution of age at diagnosis of women patients in the GDC The Cancer Genome Atlas breast cancer cohort.
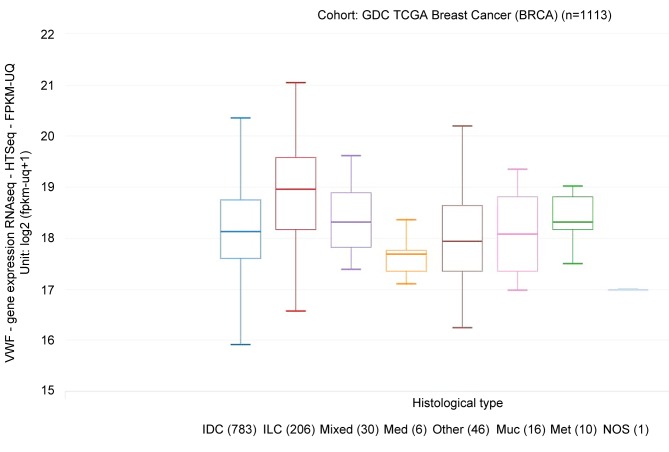
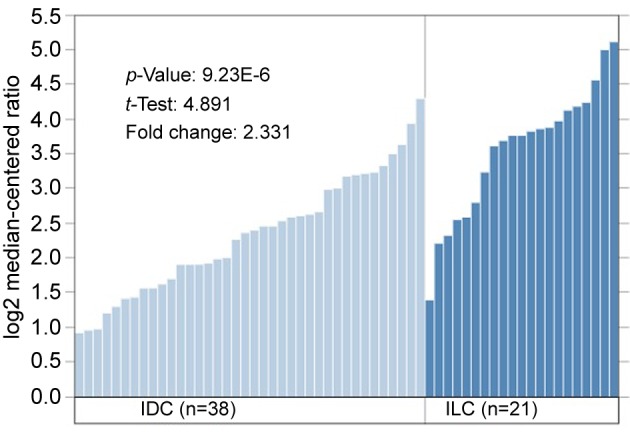
VWF mRNA expression was significantly related to histology. Patients with ILC had higher VWF mRNA expression than patients with IDC and other histology (Figure 2). The difference of VWF mRNA expression was present to the same degree in both pre-menopausal and post-menopausal breast cancer. Data from Zhao et al. (7) show a 2.331-fold change (ILC/IDC) in VWF mRNA expression that is significant (data shown in Figure 3; these data confirm the analysis in Figure 2).
Figure 2. von Willebrand factor (VWF) mRNA expression according to histology. Patients with infiltrating lobular carcinoma (ILC) had higher VWF mRNA expression than patients with infiltrating ductal carcinoma (IDC) and other histologies. Tukey’s honest significant difference post hoc test: ILC vs.: IDC, p<0.001; medullary carcinoma (Med.), p=0.038; and mucinous carcinoma (Muc.), p=0.004. Excluding ILC, VWF mRNA expression in IDC did not significantly differ from that of any other histology. Met: Metaplastic carcinoma; NOS: infiltrating carcinoma, not otherwise specified.
Figure 3. Data from Zhao et al. (24) show a 2.331-fold increase of von Willebrand factor (VWF) in infiltrating lobular carcinoma (ILC) compared to infiltrating ductal carcinoma (IDC). To show the fold change, microarray data was log2-transformed to generate a normal distribution of the data, a requirement for most statistical tests and a commonly used scale for microarray data. Oncomine displays log2, median centered values on the y-axis of bar charts, as above (https://oncomine.org).
Tukey’s honest significant difference post hoc test confirmed that VWF mRNA expression in ILC was significantly increased over that of IDC (p<0.001), medullary carcinoma (p=0.038), and mucinous carcinoma (p=0.004). Excluding ILC, VWF mRNA expression in IDC was not significantly different from that of any other histology.
Because of the relationship between ABO blood group and serum VWF levels (14), we performed univariate analysis of variance, general linear model using VWF mRNA expression as the dependent variable, histological type as the fixed factor, and ABO mRNA expression as covariate. Variability of VWF mRNA expression was significant (p<0.001) and independent of ABO mRNA expression (p<0.001).
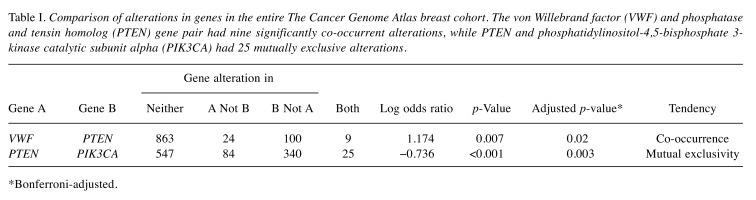
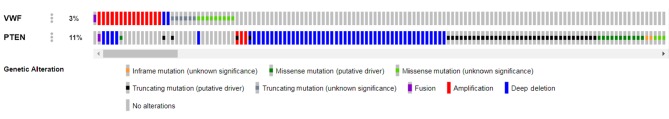
PTEN mutations are characteristic of ILC (2,3). Nine alterations in VWF and PTEN were significantly co-occurrent (p=0.007; Table I and Figure 4). In contrast, 25 alterations in PTEN and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) were mutually exclusive.
Table I. Comparison of alterations in genes in the entire The Cancer Genome Atlas breast cohort. The von Willebrand factor (VWF) and phosphatase and tensin homolog (PTEN) gene pair had nine significantly co-occurrent alterations, while PTEN and phosphatidylinositol-4,5-bisphosphate 3- kinase catalytic subunit alpha (PIK3CA) had 25 mutually exclusive alterations.
*Bonferroni-adjusted.
Figure 4. Diagrammatic representation of nine significantly co-occurring alterations in von Willebrand factor (VWF) and phosphatase and tensin homolog (PTEN). VWF and PTEN were altered in 3% and 11%, respectively, of 996 sequenced cases (cBioportal.org).
VWF is part of a network of neighboring genes, including PTEN, PIK3CA, and erythroblastic leukemia viral oncogene homolog 2 (ERB2), implicated in the genesis of ILC (Figure 5).
Figure 5. von Willebrand factor (VWF) and its network of neighboring genes, including phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) and erythroblastic leukemia viral oncogene homolog 2 (ERB2), implicated in the genesis of infiltrating lobular carcinoma. An arrow indicates a directed interaction and a line an undirected interaction. Green arrow: Control of phosphorylation; blue arrow: control of state of change; brown lines indicate targeting by a drug (cBioportal.org). FGF3,4,19: Fibroblast growth factor 3,4,19; YWHAZ: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta; FGFR1: fibroblast growth factor receptor 1; PTK2: protein tyrosine kinase 2; TP53: tumor protein 53; COL14A1: collagen type XIV alpha 1 chain; ACN2: actinin alpha 2.
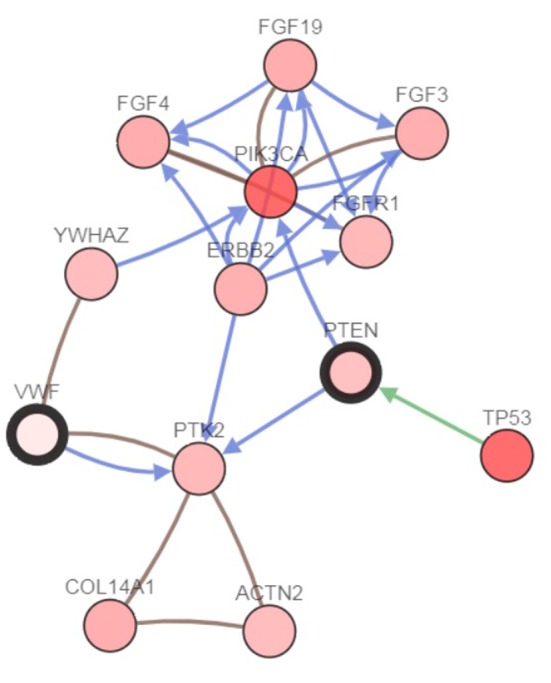
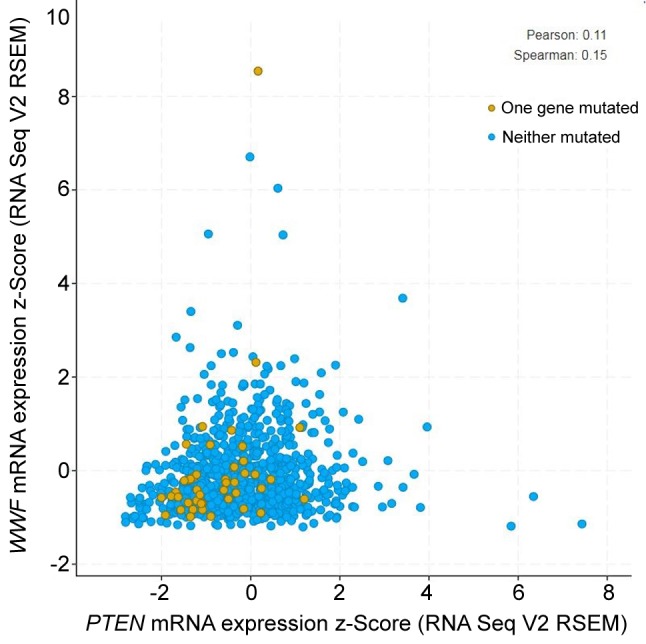
Figure 6 shows the correlation of VWF and PTEN mRNA expression in 963 breast cancer cases. The positive correlation was significant (p=0.000627); although weak, the correlation is still important.
Figure 6. Correlation of mRNA expression of von Willebrand factor (VWF) with that of phosphatase and tensin homolog (PTEN) in 963 breast cancer cases (p=0.000627).
Mixed ILC/IDC breast cancers have distinct histopatho-logical characteristics and are often diagnosed at a late stage, although survival does not vary significantly from ILC and IDC (15). In the TCGA cohort, 30 cases were recorded as having a mixed histology; however, TCGA does not specify what the mixture was.
VWF mRNA expression in ILC was not related to T-stage (p=0.2) nor the presence of metastases (p=0.97). Nor was VWF mRNA expression in IDC affected by T-stage or the presence of metastases (p=0.55). VWF RNA expression, copy number, or mutations had no effect on survival of patients with IDC or ILC.
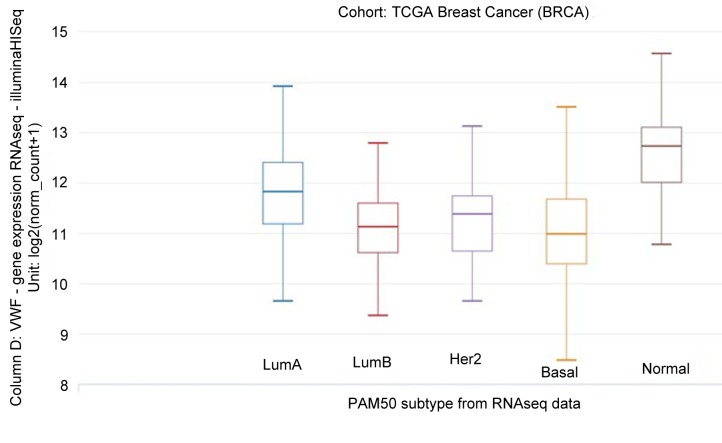
VWF mRNA expression was related to PAM50 subtype (Figure 7). Considering all histologies in 843 samples, Tukey’s honest significant difference post hoc test showed that VWF mRNA expression of the normal subtype was significantly greater than that of the other subtypes (p<0.001).
Figure 7. VWF mRNA expression according to the Prosigna Breast Cancer Prognostic Gene Signature Assay (PAM50) subtype in 840 samples, all histologies. Tukey’s honest significant difference post hoc test showed that VWF mRNA expression of the normal subtype was significantly greater than that of the other subtypes (p<0.001). LumA: Luminal A; LumB: luminal B; HER2: human epidermal growth factor receptor 2.
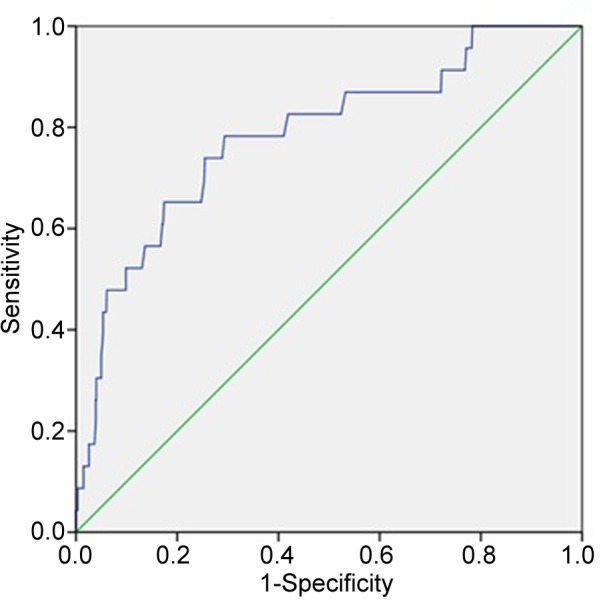
A receiver operating characteristic curve was constructed with the data used in Figure 7 to determine the optimum cut-off for VWF mRNA expression above which the PAM50 subtype was probably normal and below which it was probably abnormal (Figure 8). Using a cut-off of 12 fpkm-uq, sensitivity was 45%, specificity was 99% positive predictive value=78%, negative predictive value=97%, and accuracy=97% (p<0.001).
Figure 8. Receiver operating characteristic curve of the data used in Figure 7 in order to determine the optimum cut-off for von Willebrand factor (VWF) mRNA expression above which the Prosigna Breast Cancer Prognostic Gene Signature Assay (PAM50) subtype was probably normal and below which it was probably abnormal determine. Area under curve=0.784 (95% confidence intervaI=0.682-0.887), standard error=0.052, p<0.001. Using a cut-off of 12 sensitivity was 45%, specificity was 99%. positive predictive value=78%, negative predictive value=97% accuracy 97%.
Discussion
Armand Trousseau first reported the relationship of malignant tumors and coagulation in 1865. Trousseau diagnosed the syndrome in himself 2 years later, dying soon afterward of gastric cancer. Recent findings suggest that genetic pathways within tumor cells might trigger thrombotic phenomena, thereby worsening prognosis (16).
Von Willebrand disease (vWD) is the most common hereditary coagulation disorder in humans, and results from a deficiency in the quality or quantity of VWF. vWD presents with bleeding diathesis: easy bruising, frequent nosebleeds, and bleeding gums. Women may experience menorrhagia and hemorrhage during parturition (17).
Some studies have found that VWF might promote cancer dissemination (6). Tumor cells actively interact with coagulation cascade factors such as VWF, thereby enhancing their metastatic capacity (6). A survey of 92 patients with vWD from 54 Italian hemophilia treatment centers revealed that many cancers occur in people with vWD (106 carcinomas) (18). In addition, cancer-cell-derived VWF mediates gastric cancer metastasis, and VWF may be a new therapeutic target (19). Other studies indicate that VWF might protect against tumor dissemination [reviewed in (6)]. In patients with breast cancer, elevated serum VWF is associated with advanced stage and tumor progression (20).
As noted above, patients with ILC had higher VWF mRNA expression than patients with IDC or other histology. Additional evidence of the involvement of VWF in ILC is the relationship of VWF to PTEN. PTEN is part of the ILC genomic profile (2,3). PTEN and VWF were found to share nine co-occurrent alterations in breast cancer, and mRNA expression of the two genes significantly correlated.
The PTEN gene codes for an enzyme that acts as a tumor suppressor. The PTEN enzyme is a tyrosine phosphatase: it modifies other proteins and lipids by removing phosphate groups. PTEN tyrosine phosphatase signals cells to stop dividing and to undergo apoptosis. The PTEN tyrosine phosphatase helps control cell migration, the adhesion of cells to surrounding tissues, and angiogenesis. Additionally, PTEN tyrosine phosphatase maintains the stability of a cell’s genetic information. All of these functions prevent uncontrolled cell growth and the formation of tumors (21). Loss of expression of the PTEN tyrosine phosphatase is associated with poor outcome in breast cancer (21).
Biological processes or pathways in cancer are often deregulated through different genes or by multiple different mechanisms. But cancer gene mutations usually do not occur at random. Mutations of certain cancer genes tend to co-occur, indicating that they may work in tandem to drive tumor formation and development (11). This appears to be the case with the nine co-occurring alterations of VWF and PTEN in ILC.
In contrast, mutations of other genes appear in a mutually exclusive fashion, suggesting that two genes may have highly similar downstream components. The concept of mutual exclusivity can be exploited to identify previously unknown mechanisms that contribute to oncogenesis and cancer progression. In mutual exclusivity, events in genes associated with a specific cancer tend to be mutually exclusive across a set of tumors – that is, each tumor is likely to have only one of the genetic events (11). The 25 mutually exclusive mutations in PTEN and PIK3CA are an example of mutual exclusivity presented here (22).
In a cohort of women with breast cancer with all histologies, recurrence and death of women with basal-like subtype was significantly higher than for those with normal (non-basal-like) subtype (23). GDC TCGA data analyzed here demonstrated the same relationship of basal-like subtype to early recurrence.
PAM50 intrinsic subtypes are not equally distributed across breast cancer histologies (24), and VWF is not one of the genes analyzed in PAM50, although VWF RNA data we analyzed here is part of the GDC TCGA breast cancer cohort. Our finding of significantly higher VWF mRNA expression in those with normal (non-basal-like) subtype suggests that serum VWF measurement might complement or corroborate PAM50 results. VWF and PAM50 results, both suggesting low risk of recurrence, might make the decision whether to give chemotherapy easier, especially if VWF was an independent predictor.
Conflicts of Interest
None declared.
Authors’ Contributions
All contributed equally in data collection, data analysis, manuscript writing, and revision.
References
- 1.Barroso-Sousa R, Metzger-Filho O. Differences between invasive lobular and invasive ductal carcinoma of the breast: results and therapeutic implications. Ther Adv Med Oncol. 2016;8:261–266. doi: 10.1177/1758834016644156. PMID: 27482285. DOI: 10.1177_1758834016644156 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GM, Factor RE, Collins LC, Allison KH, Chen YY, Jensen K, Johnson NB, Oesterreich S, Mills GB, Cherniack AD, Robertson G, Benz C, Sander C, Laird PW, Hoadley KA, King TA, Perou CM. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. PMID: 26451490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, Fumagalli D, Brown D, Rothe F, Vincent D, Kheddoumi N, Rouas G, Majjaj S, Brohee S, Van LP, Maisonneuve P, Salgado R, Van BT, Lambrechts D, Bose R, Metzger O, Galant C, Bertucci F, Piccart-Gebhart M, Viale G, Biganzoli E, Campbell PJ, Sotiriou C. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34:1872–1881. doi: 10.1200/JCO.2015.64.0334. PMID: 26926684. DOI: 10.1200/JCO.2015.64.0334. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Schizas M, Geyer FC, Selenica P, Piscuoglio S, Sakr RA, Ng CKY, Carniello JVS, Towers R, Giri DD, de Andrade VP, Papanastasiou AD, Viale A, Harris RS, Solit DB, Weigelt B, Reis-Filho JS, King TA. Lobular carcinomas in situ display intralesion genetic heterogeneity and clonal evolution in the progression to invasive lobular carcinoma. Clin Cancer Res. 2019;25:674. doi: 10.1158/1078-0432.CCR-18-1103. PMID: 30185420. DOI: 10.1158/1078-0432.CCR-18-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tas F, Kilic L, Duranyildiz D. Coagulation tests show significant differences in patients with breast cancer. Tumour Biol. 2014;35:5985–5992. doi: 10.1007/s13277-014-1793-4. PMID: 24596033. DOI: 10.1007/s13277-014-1793-4. [DOI] [PubMed] [Google Scholar]
- 6.Franchini M, Frattini F, Crestani S, Bonfanti C, Lippi G. von Willebrand factor and cancer: A renewed interest. Thromb Res. 2013;131:290–292. doi: 10.1016/j.thromres.2013.01.015. PMID: 23394808. DOI: 10.1016/j.thromres.2013. 01.015. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. PMID: 15034139. DOI: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrer S, Rheinstein PH, Rosenzweig KE. Increased expression of von Willebrand factor gene is associated with poorer survival in primary lower grade glioma. Glioma. 2018;1:132–135. doi: 10.4103/glioma.glioma_17_18. PMID: 30272052. DOI: 10.4103/glioma.glioma_17_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, Rao A, Schultz A, Li X, Sumazin P, Williams C, Mestdagh P, Gunaratne PH, Yau C, Bowlby R, Robertson AG, Tiezzi DG, Wang C, Cherniack AD, Godwin AK, Kuderer NM, Rader JS, Zuna RE, Sood AK, Lazar AJ, Ojesina AI, Adebamowo C, Adebamowo SN, Baggerly KA, Chen TW, Chiu HS, Lefever S, Liu L, MacKenzie K, Orsulic S, Roszik J, Shelley CS, Song Q, Vellano CP, Wentzensen N, Weinstein JN, Mills GB, Levine DA, Akbani R. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690–705. doi: 10.1016/j.ccell.2018.03.014. PMID: 29622464. DOI: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman M, Craft B, Brooks AN, Zhu J, Haussler D. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv 326470. 2018 [Google Scholar]
- 11.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. PMID: 23550210. DOI: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pander A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. PMID: 15068665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahriyari L. Effect of normalization methods on the performance of supervised learning algorithms applied to HTSeq-FPKM-UQ data sets: 7SK RNA expression as a predictor of survival in patients with colon adenocarcinoma. Brief Bioinform, 2017 doi: 10.1093/bib/bbx153. PMID: 29112707. DOI: 10.1093/bib/bbx153. [DOI] [PubMed] [Google Scholar]
- 14.Franchini M, Capra F, Targher G, Montagnana M, Lippi G. Relationship between ABO blood group and von Willebrand factor levels: From biology to clinical implications. Thromb J. 2007;5:14. doi: 10.1186/1477-9560-5-14. PMID: 17894864. DOI: 10.1186/1477-9560-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zengel B, Yararbas U, Duran A, Uslu A, Eliyatkin N, Demirkiran MA, Cengiz F, Simsek C, Postaci H, Vardar E, Durusoy R. Comparison of the clinicopathological features of invasive ductal, invasive lobular, and mixed (invasive ductal + invasive lobular) carcinoma of the breast. Breast Cancer. 2015;22:374–381. doi: 10.1007/s12282-013-0489-8. PMID: 23925582. DOI: 10.1007/s12282-013-0489-8. [DOI] [PubMed] [Google Scholar]
- 16.Varki A. Trousseau’s syndrome: Multiple definitions and multiple mechanisms. Blood. 2007;110:1723–1729. doi: 10.1182/blood-2006-10-053736. PMID: 17496204. DOI: 10.1182/blood-2006-10-053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler JE. Von Willebrand disease type 1: A diagnosis in search of a disease. Blood. 2003;101:2089–2093. doi: 10.1182/blood-2002-09-2892. PMID: 12411289. DOI: 10.1182/blood-2002-09-2892. [DOI] [PubMed] [Google Scholar]
- 18.Franchini M, Di PC, Santoro C, Castaman G, Siboni SM, Zanon E, Linari S, Gresele P, Pasca S, Coppola A, Santoro R, Napolitano M, Ranalli P, Tagliaferri A. Cancers in patients with von Willebrand disease: A diseasefrom the Italian Association of Haemophilia Centres. Semin Thromb Hemost. 2016;42:36–41. doi: 10.1055/s-0035-1564844. PMID: 26595151. DOI: 10.1055/s-0035-1564844. [DOI] [PubMed] [Google Scholar]
- 19.Yang AJ, Wang M, Wang Y, Cai W, Li Q, Zhao TT, Zhang LH, Houck K, Chen X, Jin YL, Mu JY, Dong JF, Li M. Cancer cell-derived von Willebrand factor enhanced metastasis of gastric adenocarcinoma. Oncogenesis. 2018;7:12. doi: 10.1038/s41389-017-0023-5. PMID: 29362409. DOI: 10.1038/s41389-017-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohsig LM, Damin DC, Stefani SD, Castro CG Jr., Roisenberg I, Schwartsmann G. von Willebrand factor antigen levels in plasma of patients with malignant breast disease. Braz J Med Biol Res. 2001;34:1125–1129. doi: 10.1590/s0100-879x2001000900004. PMID: 11514835. [DOI] [PubMed] [Google Scholar]
- 21.Depowski PL, Rosenthal SI, Ross JS. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod Path. 2001;14:672. doi: 10.1038/modpathol.3880371. PMID: 11454999. DOI: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 22.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. PMID: 15805248. DOI: 10.1158/ 0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 23.Gulbahce HE, Bernard PS, Weltzien EK, Factor RE, Kushi LH, Caan BJ, Sweeney C. Differences in molecular features of triple-negative breast cancers based on the age at diagnosis. Cancer Res. 2018;124(24):4676–4684. doi: 10.1002/cncr.31776. PMID: 30311638. DOI: 10.1002/cncr.31776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao YG, Xiao AZ, Park HI, Newcomer RG, Yan M, Man YG, Heffelfinger SC, Sang QX. Endometase/matrilysin-2 in human breast ductal carcinoma in situ and its inhibition by tissue inhibitors of metalloproteinases-2 and -4: A putative role in the initiation of breast cancer invasion. Cancer Res. 2004;64:590–598. doi: 10.1158/0008-5472.can-03-1932. [DOI] [PubMed] [Google Scholar]