Abstract
Monoterpenes (C10 isoprenoids) are a structurally diverse group of natural compounds that are attractive to industry as flavours and fragrances. Monoterpenes are produced from a single linear substrate, geranyl diphosphate, by a group of enzymes called the monoterpene cyclases/synthases (mTC/Ss) that catalyse high-energy cyclisation reactions involving unstable carbocation intermediates. Efforts towards producing monoterpenes via biocatalysis or metabolic engineering often result in the formation of multiple products due to the nature of the highly branched reaction mechanism of mTC/Ss. Rational engineering of mTC/Ss is hampered by the lack of correlation between the active site sequence and cyclisation type. We used available mutagenesis data to show that amino acids involved in product outcome are clustered and spatially conserved within the mTC/S family. Consensus sequences for three such plasticity regions were introduced in different mTC/S with increasingly complex cyclisation cascades, including the model enzyme limonene synthase (LimS). In all three mTC/S studied, mutations in the first two regions mostly give rise to products that result from premature quenching of the linalyl or α-terpinyl cations, suggesting that both plasticity regions are involved in the formation and stabilisation of cations early in the reaction cascade. A LimS variant with mutations in the second region (S454G, C457V, M458I), produced mainly more complex bicyclic products. QM/MM MD simulations reveal that the second cyclisation is not due to compression of the C2-C7 distance in the α-terpinyl cation, but is the result of an increased distance between C8 of the α-terpinyl cation and two putative bases (W324, H579) located on the other side of the active site, preventing early termination by deprotonation. Such insights into the impact of mutations can only be obtained using integrated experimental and computational approaches, and will aid the design of altered mTC/S activities towards clean monoterpenoid products.
Keywords: Monoterpenoids, Monoterpene synthase, Functional plasticity, Limonene synthase, Site-directed mutagenesis, QM/MM MD simulations, Synthetic biology
Introduction
Terpenoids are a large and diverse group of natural products with more than 80,000 identified compounds in the Dictionary of Natural Compounds (http://dnp.chemnetbase.com). Most terpenoids are commonly found in plants and their biological roles range from inter-species communication, to intracellular signalling, and defence against predatory species1. All terpenoids are produced from the universal linear C5 isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are combined by prenyl transferases to generate pyrophosphate substrates of varying lengths. The latter are then used by terpene synthases/cyclases to produce monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20) or larger terpene skeletons. Monoterpenoids are of considerable industrial interest; they are, for example, used as pharmaceuticals, fragrances, antiseptics, and biofuels2. Previously, we have developed a flexible platform for the production of diverse monoterpene hydrocarbon scaffolds, through the ‘plug-and-play’ insertion of varying plant monoterpene cyclases/synthases (mTC/S) into an engineered E. coli strain containing a bacterial/yeast hybrid mevalonate (MVA) pathway3. Most mTC/S introduced into our platform produced a mixture of monoterpenoids but the production of ‘clean’ or pure products is desirable for commercial applications. Rational engineering of mTC/S enzymes is, however, hampered by the lack of a clear correlation between the active site sequence and cyclisation type, given that closely related enzymes may produce different product profiles4–5.
All mTC/S enzymes catalyse high-energy cyclisation reactions involving unstable carbocation intermediates originating from a single linear substrate (geranyl diphosphate; GPP). The reaction is initiated by metal-dependent ionisation of GPP; the resulting carbocation can then react along several channels to form linear, monocyclic or bicyclic monoterpene hydrocarbons. After the initial and highly conserved ionisation step, the enzyme effectively provides a productive template for the often complex cyclisation cascade, in which the formation of the α-terpinyl cation (TPC, generated as a result of C1-C6 cyclisation) is the first intermediate for all mono- and bi-cyclic products (Figure 1). The reaction cascade is ultimately quenched by deprotonation or water capture6. Because each carbocation intermediate can undergo a range of cyclizations and isomerisations before the reaction terminates, many mTC/S enzymes are capable of producing multiple products. Crystal structures have been reported for several plant mTC/S enzymes, including a bornyl diphosphate synthase from Salvia officinalis7, two limonene synthases from Mentha spicata8 and Citrus sinensis9–10, a 1,8-cineole synthase from Salvia fruticosa11, and a γ-terpinene synthase from Thymus vulgaris12. In addition, two bacterial mTC/S structures have recently been reported, a linalool synthase and 1,8-cineole synthase from Streptomyces clavuligerus13. The plant enzymes invariantly consist of two domains: a C-terminal α-helical catalytic domain exhibiting a class I terpene cyclase fold, and an N-terminal α-barrel domain with an as yet unknown function. Though the overall sequence conservation is low, the structure of the α-helical fold is highly conserved (Figure 2). The active site has two conserved motifs, the aspartate rich DDXXD motif and the NSE/DTE motif, required for the binding of the three catalytically essential Mg2+ or Mn2+ ions. The bacterial enzymes lack the N-terminal domain, and are more similar to previously characterized sesquiterpene synthases.
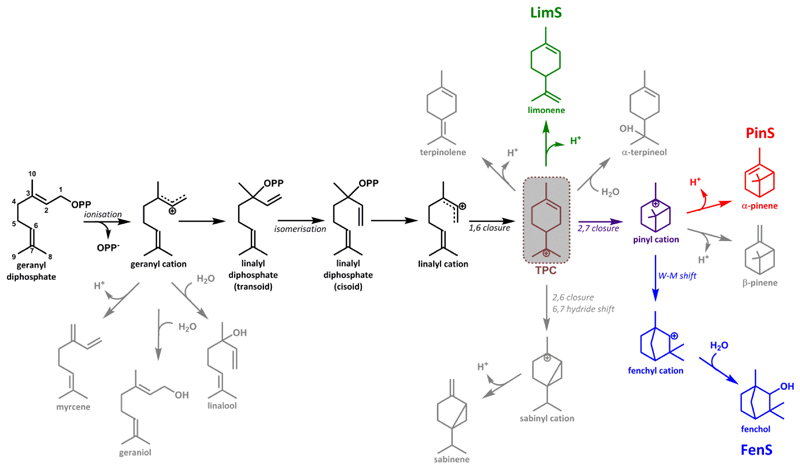
Figure 1. Proposed mechanism for the formation of limonene, pinene, fenchol and common by-products.
All mTC/S reactions commonly start with the ionization of the geranyl diphosphate (GPP) substrate resulting in the geranyl cation, which subsequently can undergo a wide range of cyclizations and rearrangements before the reaction is terminated by deprotonation or water attack. The formation of all cyclic products requires the isomerization of the geranyl cation to the linalyl cation via linalyl diphosphate. The latter can cyclize to yield the α-terpinyl cation (TPC, grey box). The main reaction cascades of LimS, PinS, and FenS following the formation of TPC are indicated in green, red and blue respectively. Premature quenching of intermediates may result in the formation of various by-products (grey). Carbon atom numbering of intermediates refers to that for the GPP substrate.
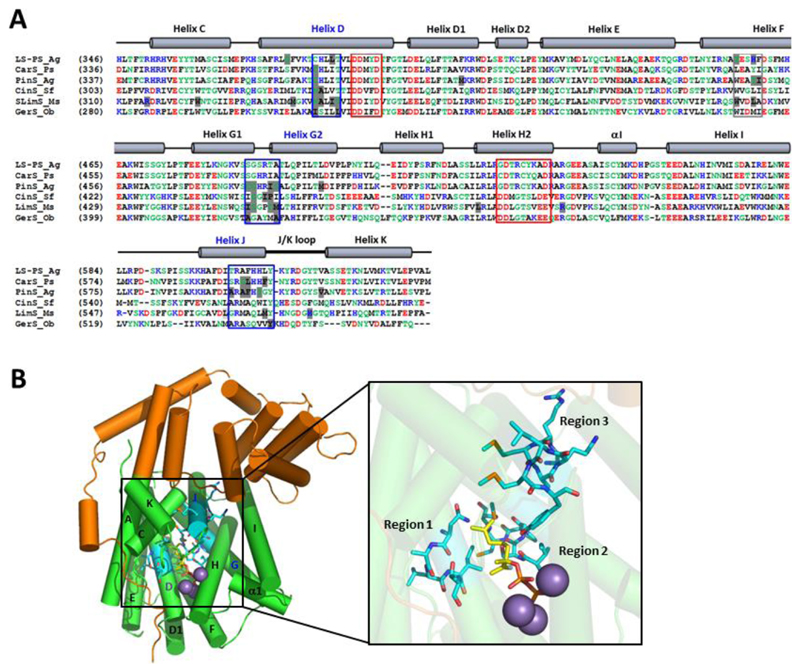
Figure 2. Identification and location of clustered plasticity residues in the plant mTC/S family.
(A) Multiple sequence alignment of the C-terminal domains of mTC/S enzymes that have been mutated to variants with altered product profiles. Non-polar residues are in black, polar residues in green, positively charged residues in blue, and negatively charged residues are in red. The conserved DDxxD and NSE/DTE motifs are indicated with red boxes. Mutated amino acid residues identified from literature studies that resulted in altered product profiles are highlighted in light grey. The three plasticity regions identified and investigated in this study are marked with blue boxes. A potential fourth plasticity region not investigated in this study is marked with a grey box. LS-PS_Ag is (–)-limonene/(–)-α-pinene synthase from Abies grandis24; CarS_Ps is (+)-3-carene synthase from Picea sitchensis26; PinS_Ag is (–)-pinene synthase from Abies grandis23; CinS_Sf is 1,8-cineole synthase from Salvia fruticosa11; LimS_Ms is (–)-(4S)-limonene synthase from Mentha spicata L.27–28; GerS_Ob is geraniol synthase from Ocimum basilicum48. The secondary structure of terpene synthases/cyclases is indicated above the alignment49. (B) The three identified plasticity regions were mapped onto the structure of LimS (pdb 2ONG)8. The N-terminal domain is in orange, the C-terminal terpene cyclase domain is in green, the substrate analogue in yellow, and the Mn2+ ions in purple. The three plasticity regions are highlighted in cyan.
Bornyl diphosphate synthase (BPPS), the only monoterpene with the diphosphate (PPi) group incorporated in the product, has been the focus of much research experimentally and computationally14. Conversion of GPP by BPPS yields ~75% (+)-bornyl diphosphate, with (+)-camphene, (+)-α-pinene, (±)-limonene and terpinolene formed as side products15–16. Computational studies in the gas phase and in BPPS have shown that the free energy surface for the reaction is highly complex17–19. The enzyme actually destabilises some of the reaction intermediates on the pathway, particularly in the early stages of the reaction to prevent the formation of side-products. Simulations by Major and Weitman18 have shown that a combination of electrostatics and dynamics is involved in directing the product outcome in BPPS. Recently, Major20 has investigated the role of electrostatic control in terpene cyclases further, showing that the electrostatic interactions due to PPi and D351/D100 (in BPPS/trichodiene synthase, respectively) raise the barrier to C-C bond formation in the first cyclisation to form TPC.
It is generally believed that the active site pocket places steric constraints on the reaction intermediates and as such determines the product outcome21–22. Several mutational studies on various mTC/S enzymes revealed that just a few amino acid substitutions are enough to sufficiently reshape the active site and change the product outcome11, 23–28, demonstrating a high level of plasticity, in this group of enzymes. This ability to drive change via a small number of mutations allows for rapid product diversification, and explains why closely related mTC/S can produce different product spectra. For example, a reciprocal single amino acid substitution is responsible for interconversion of carene synthase and sabinene synthase activity in two related mTC/S enzymes from Picea sitchensis26. Limonene synthase (LimS) from Mentha spicata can be converted into a phellandrene synthase via the introduction of three amino acid substitutions. The presence of a polar Asn residue at position 345 is implicated in the formation of limonene, mutation to a non-polar residue is thought to allow TPC to undergo further cyclizations and/or hydride shifts leading to the formation of pinene or phellandrene28. The 1,8-cineole synthase from S. fruticosa was converted to a sabinene synthase by introducing just a single Asn to Ile amino acid change11. Interestingly, the Asn residue identified to be essential for water activation in cineole synthase (Asn338) is equivalent to the Asn residue essential for limonene formation in LimS. Further literature research revealed that this amino acid position is implicated in product outcome in several other mTC/S including a pinene synthase from Abies grandis23 (Cys372), two sabinene hydrate synthases from Thymus vulgaris25 (Asn350 and Ile346, respectively), and a fungal cineole synthase29 (Asn136). The apparent spatial conservation of functional plasticity at this amino acid position prompted us to look at other possible correlations between identified plasticity residues in the different mTC/S enzymes in more detail.
In this study, we integrated classic site-directed mutagenesis with synthetic biology methods and molecular dynamics (MD) simulations using combined quantum mechanics/molecular mechanics (QM/MM) to investigate the influence of active site amino acid composition on product outcome. We show that amino acids involved in product outcome are clustered and spatially conserved within the mTC/S family, and gained unique insights into the impact of mutations in these plasticity regions. Mutation to a consensus sequence in one of the identified plasticity regions resulted in a variant with a drastically altered product profile, which can be explained through simulation. These results will help guide future engineering efforts of this important group of enzymes towards the production of clean monoterpenoid products.
Experimental section
Bacterial strains, plasmids and media
All E. coli strains were routinely grown in Lysogeny Broth (LB) or on LB agar plates including antibiotic supplements as appropriate (carbenicillin, 100 μg ml-1; kanamycin, 50 μg mL-1). Cloning, mutagenesis and plasmid propagation was performed using E. coli Stellar cells (Takara Bio), in vivo production of monoterpenoids was performed using E. coli DH5α cells (New England Biolabs), and recombinant protein production was performed using E. coli Arctic Express (DE3) cells (Agilent).
Synthetic genes encoding native FenS, LimS and PinS, without plastidial signal sequences were codon optimised for expression in E. coli and sub-cloned into the NcoI-XhoI restriction sites of pETM-11 fused to a Tobacco Etch Virus (TEV) protease cleavable N-terminal His6-tag (GeneArt, Life Techonologies)3. The resulting pseudo-mature constructs, truncated at the RRX8W motif30–31, are shown in Table S1 of the Supporting Information. For in vivo production of monoterpenoids the genes were re-cloned into a BglBrick vector32 containing a geranyl diphosphate synthase (GPPS) from Abies grandis (pBb-GPPS-mTC/S series) and co-transformed with pVMA, containing the bacterial/yeast hybrid MVA pathway, as described previously3. A list of all plasmids used in this study is shown in Table S2 of the Supporting Information.
Gene synthesis, cloning and site-directed mutagenesis
Oligonucleotide design for gene synthesis of the C-terminal domain of native and variant α-pinene synthase from Pinus taeda (PinS) was performed using the GeneGenie tool33. The input parameters and DNA output sequence are shown in Table S2. The resulting oligonucleotides (Table S3) were obtained from Integrated DNA Technologies (Ultramer DNA oligonucleotides) and assembled using the SpeedyGenes method34. Two intermediate DNA blocks were synthesized using oligonucleotides 1-8 and 9-18, using the two outermost oligonucleotides (i.e. numbers 1 and 8, and 9 and 18) as primers for the PCR. The remaining inner oligonucleotides (i.e. numbers 2-7 and 10-17) were pooled together in an equimolar mixture as PCR template. The PCR reaction was performed as described34. When synthesizing PinS variant sequences, the variant oligonucleotides (marked with an asterisk (*) in Table S3) were used for the intermediate block synthesis instead of the original native sequence oligonucleotide. Endonuclease digestion for error correction using Surveyor nuclease (Integrated DNA Technologies) was performed on the intermediate DNA blocks as described35. Full-length sequences were synthesised by pooling equal amounts of the purified block digests as PCR template, and the two outermost oligonucleotides (numbers 1 and 18) as primers34. The full-length fragments encoding the C-terminal domain of PinS were cloned into the pGPPSmTC/S27 vector linearized with Vector_IF_Fw and maPSpt_IF_Rv (Table S4) using the InFusion HD cloning kit (Takara Bio) according to the manufacturer’s instructions, resulting in plasmids pBb-PinS_var1, pBb-PinS-var2 and pBb-PinS_var3. Correct DNA synthesis and cloning was confirmed by standard Sanger sequencing.
Consensus sequences were introduced in fenchol synthase from Lavandula viridis (FenS) and limonene synthase from Mentha spicata (LimS) using the QuikChange site-directed mutagenesis method (Stratagene) using plasmids pBbGPPSmTC/S9 and pBbGPPSmTC/S15 as templates, respectively. FenS and LimS variants 3 were generated following two rounds of QuikChange to incorporate all mutations. The QuikChange primers used are shown in Table S4. Correct insertion of mutations in the resulting plasmids pBb-LimSvar1 to 3 and pBb-FenSvar1 to 3 was confirmed by standard Sanger sequencing.
Full-length native and variant mTC/S genes were sub-cloned into pETM-11 for recombinant expression using InFusion cloning. The vector backbone was linearized with primers Vector_IF_Fw/Rv and the mTC/S genes were amplified from their respective pBb vectors using pET_IF_Fw/Rv (Table S4).
Monoterpenoid production in E. coli
For monoterpenoid production, the pGPPSmTC/S plasmids were co-transformed with plasmid pMVA into E. coli DH5α and grown as described3. Briefly, expression strains were inoculated in terrific broth (TB) supplemented with 0.4% glucose in glass screw capped vials, and induced for 72h at 30° C with 50 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 25 nM anhydro-tetracycline. A 20% (v/v) n-nonane layer was added to capture the volatile monoterpenoid products. After induction, the nonane overlay was collected, dried over anhydrous MgSO4 and mixed at a 1:1 ratio with ethyl acetate containing sec-butylbenzene as internal standard. The samples were analysed by GC-MS (vide infra).
Expression and purification of native and variant mTC/S
Recombinant native and variant mTC/S proteins were expressed in E. coli Arctic Express (DE3). Cells were freshly transformed with a pET-mTC/S plasmid, grown in LB medium and induced with 0.1 mM IPTG for 16 h at 12° C according to the manufacturer’s instructions. The recombinant proteins were purified as described previously13. Briefly, the cells were harvested and resuspended in buffer A (25 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT, 4 mM MgCl2 and 5% (v/v) glycerol). The cells were lysed by sonication and the debris was removed by centrifugation. The supernatant was loaded onto a His-Trap column (GE Healthcare) pre-equilibrated with buffer A containing 25 mM imidazole. The column was washed with buffer A containing 25 mM imidazole, and the proteins were eluted by increasing the imidazole concentration to 500 mM. The purified proteins were desalted prior to removal of the His-tag by overnight incubation with TEV protease at 4°C with gentle mixing. The TEV protease was removed by passing the protein mixtures through a His-Trap column, and the proteins collected in the flow-through were concentrated and loaded onto a Hiload Superdex (26/60) S200 column (GE Healthcare) pre-equilibrated with buffer A. Purified protein fractions were concentrated to 8-10 mg/ml, aliquoted and stored at -80°C.
In vitro biotransformations
All biotransformation reactions with purified mTC/S were prepared in triplicate in buffer A supplemented with 4 mM MnCl2 in glass vials. For product profile determination, the reactions contained 2 mM GPP and 20 μM purified mTC/S, and a 20% (v/v) n-nonane layer was added to capture the volatile monoterpenoid products. The vials were incubated at 25°C with gentle shaking for 16 h. After incubation, the nonane overlay was collected, dried over anhydrous MgSO4 and mixed at a 1:1 ratio with ethyl acetate containing sec-butylbenzene as internal standard. The samples were analysed by GC-MS.
GC-MS analysis
Samples were injected onto an Agilent Technologies 7890 BGC equipped with an Agilent Technologies 5977 AMSD. The products were separated on a DB-WAX column (30 m x 0.32 mm i.d., 0.25 μm film thickness, Agilent Technologies). The injector temperature was set at 240°C with a split ratio of 20:1 (1 μl injection). The carrier gas was helium with a flow rate of 1 ml/min and a pressure of 5.1 psi. The following oven program was used: 50°C (1 min hold), ramp to 68°C at 5°C/min (2 min hold), and ramp to 230°C at 25°C/min (2 min hold). The ion source temperature of the mass spectrometer (MS) was set to 230°C and spectra were recorded from m/z 50 to m/z 250. Compound identification was carried out using authentic standards and comparison to reference spectra in the NIST library of MS spectra and fragmentation patterns as described previously3.
QM/MM MD simulations
Models of wild type (WT) LimS and mutants were built from the 2.7 Å resolution crystal structure of LimS with 2-fluorogeranyl diphosphate (PDB id 2ONG)8. The protonation states of titratable residues were estimated using PropKa3.136–37. All Asp and Glu residues were predicted to be negatively charged and Lys and Arg residues positively charged at pH 7. All His residues were predicted to be singly protonated and visual inspection of their hydrogen bonding environment showed that protonation should be on NE1 in all cases. The enzyme was solvated using a box of TIP3P38 water molecules (with a minimum buffer or 13 Å around the protein) using the solvate plugin of the VMD package39. Twenty three sodium ions were added to neutralize the system using the autoionize plugin of VMD39. The CHARMM27 forcefield40 was used to describe the protein. As the aim of this work is not to simulate the entire multi-step reaction mechanism, models were built of the system after ionisation to the α-terpinyl cation (TPC) and inorganic diphosphate (PPi), as this is the crucial species in the terpene cyclase cascade. Partial atomic charges were assigned for TPC and PPi consistent with the CHARMM27 forcefield. The position of TPC and PPi substrate were based on the position of the fluorinated GFPP analogue resolved in the crystal structure. Due to the minimal differences in the structure of the inhibitor and substrate (F vs H), the position in the crystal structure was considered a suitable starting point for the simulations. It has been suggested that many terpene cyclase/synthase structures contain substrates bound in unreactive conformations14, 41; however structures containing the larger and more flexible FPP building block for sesquiterpenes are more prevalent than monoterpenes. The parameter set developed by Allner et al.42 was used to describe the three Mg2+ ions. The setup of the model consisted of: (i) minimisation of the positions of the hydrogen atoms (all heavy atoms fixed); (ii) minimisation of the solvent (with all protein heavy atoms fixed); (iii) energy minimisation of the entire system with positional restraints of 5 kcal mol–1 Å–2 applied to all Cα atoms; (iv) canonical ensemble (NVT) thermalisation to 300 K over 20 ps with positional restraints of 5 kcal mol–1 Å–2 on Cα atoms; (v) thermal equilibration at 300K for 100 ps with positional restraints of 5 kcal mol–1 Å–2 on Cα atoms; (vi) 140 ps of NPT equilibration with decreasing restraints on the Cα atoms; (vi) 2 ns production simulation. Ten sets of isothermal-isobaric ensemble (NPT) MD simulations were performed at 300 K for each enzyme, repeating steps (iv)-(vi) to obtain 10 models with different initial conditions. QM/MM MD simulations at the SCC-DFTB level were carried out on 4 CPUs using the sander code43 of AMBER1644. SHAKE restraints were also applied to the hydrogen atoms of the QM region to prevent unwanted proton transfers occurring, as the SCC-DFTB method can underestimate the barrier to proton transfer45. Langevin dynamics was used for temperature control (collision frequency of 5 ps–1 for equilibration and 2 ps–1 for production), and pressure was controlled by coupling to an external bath (AMBER16 default settings) for NPT conditions. For analysis, structures were aligned based on positions of the heavy atoms of active site residues, i.e. residues within 5 Å of TPC (residues 315, 348, 349, 352, 427, 445, 453, 454, 458, 492, 493, 496, and 573). Average linkage hierarchical clustering was carried out using the CPPTRAJ utility of AmberTools 1644 to identify representative structures of the ternary complex over the course of the simulations. Five clusters were generated based on RMS to the position of PPi without any further fitting of structures to highlight any differences in the position of TPC. The simulations were analysed to find key distances between the cation and active site residues and histograms generated using a bin size of 0.1 Å and normalisation of the population to 1.
Results and Discussion
Identification and location of clustered plasticity residues in the plant mTC/S family
A multiple sequence alignment of mTC/S enzymes that have been mutated to produce altered product profiles revealed that amino acids involved in product outcome are clustered and spatially conserved throughout the plant mTC/S enzyme family (Figure 2, panel A). Active site residues interacting with the PPi moiety of GPP are highly conserved, but residues interacting with the carbon-chain moiety of GPP are variable. The mutational hotspots identified in the alignment are amongst the variable residues, and can be primarily found in three helices lining the active site of mTC/S enzymes (Figure 2, panel B). The first region is located in helix D, at the bottom of the active site cavity, and includes Asn345 in LimS and Asn388 in CinS, both these residues are thought to play an essential role in the formation of their respective products limonene27–28 and 1,8-cineole11. The second region is located on helices G1 and G2 and the loop connecting the two. Residues in this region have been implicated in stabilization of the carbocation intermediates through the presence of local partial charges in the catalytic pocket11. The third identified plasticity region is located on helix J, mutations in this region primarily affect bicyclic mTC/S23–24, 26. Amino acids in the third region are not amongst the residues directly interacting with substrate. A possible fourth cluster can be identified on helix F (grey box in Figure 2, panel A), the effect of mutating this region was not investigated in this study.
A second multiple sequence alignment comprising all 36 mTC/S enzymes in our mTC/S library3, yielded the amino acid occurrence at each position as well as the average, or consensus sequence for each of the three regions (see Table S5 in the Supporting Information). Looking at the family as a whole, there are very few positively charged residues present in these regions, and there is an almost complete absence of negatively charged residues; only one aspartic acid is present in region 3 of a terpinolene synthase from sweet basil. An arginine residue in region 3 is strictly conserved among all 36 enzymes in the alignment. Interestingly, mTC/S enzymes from pine trees contain His residues in all three regions; these enzymes produce mainly bicyclic monoterpenoids such as pinene, 3-carene, sabinene and camphene. In general, however, all three regions contain mainly polar and non-polar residues. This is in line with the notion that the active sites of terpene synthases are rich in relatively inert amino acids that do not react directly with the reaction intermediates, and that the shape of the active site ultimately determines the product outcome22, 46–47.
We introduced the consensus sequences of the three plasticity regions (Figure 3A, Table S5) into three mTC/S enzymes from different plant species with increasingly complex cyclisation cascades (see Figure 1). LimS from Mentha spicata has the simplest cyclisation cascade. Following the formation of TPC, the first intermediate for all cyclic products, the reaction is immediately terminated by deprotonation at C8 resulting in the monocyclic olefin (–)-(4S)-limonene as the main product (96%)8, 27, 50. The mechanism of (–)-α-pinene synthase from Pinus taeda (PinS) involves a second cyclisation of TPC (2,7-ring closure) followed by deprotonation, yielding the bicyclic products α-pinene (85%) and β-pinene (11%) as the main products23, 51. The final enzyme used in this study is a fenchol synthase from Lavandula viridis (FenS), and has the most complex cyclisation cascade of the three enzymes. In FenS the pinyl cation undergoes a Wagner-Meerwein rearrangement, yielding the fenchyl cation, before the reaction is terminated by water capture, resulting in the bicyclic monoterpene alcohol fenchol as the main product (65%)52–53.
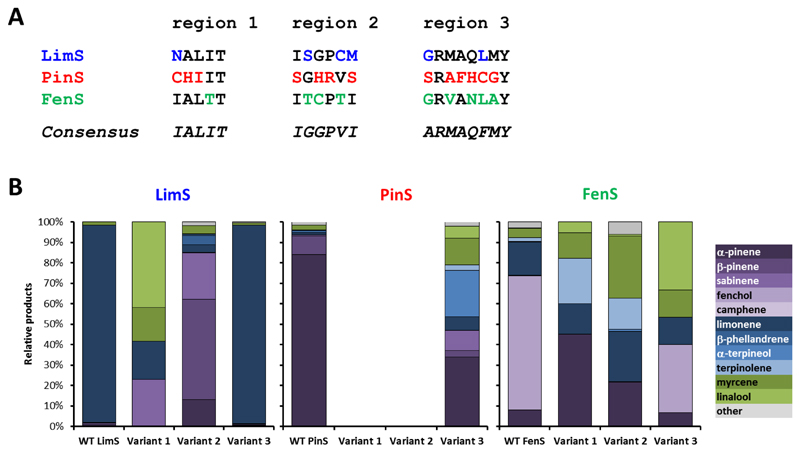
Figure 3. Effect of plasticity region consensus sequences on the product profiles of different plant mTC/S.
(A) Native vs consensus sequences of the targeted enzymes. Each residue targeted by mutagenesis is marked in colour. (B) Relative product profiles achieved upon insertion of the variant enzymes into our E. coli monoterpenoid production strain. Bicyclic monoterpenoids are shaded in purple, monocyclic monoterpenoids in blue and linear monoterpenoids in green. Geraniol and derivatives are omitted from the comparison, as they are (mainly) produced by endogenous E. coli activity3, 54. Only VAR3-PinS 3 produces geraniol in significant amounts as was demonstrated in vitro (see Figure S4 in the Supporting Information). A full break-down of the product profiles can be found in the Supporting Information (Figures S1-S3, and Table S7).
The variant mTC/S enzymes harbouring consensus plasticity sequences in either region 1 (VAR1), region 2 (VAR2) or region 3 (VAR3) were expressed in our E. coli monoterpene dual-plasmid production strain for the rapid generation of full product profiles without the need for expensive GPP substrate and laborious protein purification. Variant strains were grown in two-phase shake flask cultures using glucose as carbon source and n-nonane as the organic phase to trap the volatile monoterpene products. The products were identified and quantified by GC-MS analysis as described previously3. Most variant mTC/S enzymes were active in our platform and produced product profiles that are different from the native enzymes, with the exception of VAR1-PinS (C373I, H374A, I375L) and VAR2-PinS (S481I, H483G, R484P, S486I), which did not produce any detectable monoterpenoids, and VAR3-LimS (G566A, L571F) which has a product profile nearly identical to WT-LimS (see Figure 3, panel B, and Table S6 in the Supporting Information). Interestingly, most of the other variants produced product profiles rich in products that are the result of premature quenching of the carbocations. VAR1-FenS (T344I) and VAR2-FenS (T450G, C451G, T453V) no longer produce fenchol. Fenchol is formed via water attack of the fenchyl cation52–53, however both variants produce mostly products that result from premature quenching of the linalyl cation or TPC, suggesting that both plasticity regions are involved in the formation and stabilisation of cations in the reaction cascade.
A notable exception is VAR2-LimS (S454G, C457V, M458I), which produces mainly more complex bi-cyclic products (49% β-pinene, 23% sabinene and 13% α-pinene). The replacement of three polar residues in this variant by non-polar residues thus prevents quenching of the reaction by deprotonation and the subsequent formation of limonene. It has previously been proposed that polar and aromatic side-chains can stabilize TPC long enough for deprotonation to occur, in effect preventing its migration to more complex products22, 28.
The role of the third plasticity region is unclear, as only the bicyclic mTC/Ss PinS and FenS were affected by mutations in this area. Both VAR3-PinS (S595A, A597M, F598M, H599Q, C600F, G601M) and VAR3-FenS (G561A, V563M, N565Q, L566F, A567M) have more subtle changes to their product profiles, i.e. changes in product ratios. Interestingly, most previously reported mutations in this area resulted in altered product profiles of bicyclic mTC/S enzymes in a similar fashion. The F597W mutation in a β-pinene synthase of Grand Fir flipped the α/β-pinene ratio23. Similarly, the presence of a Leu or Phe at position 596 promotes the formation of carene and sabinene respectively in carene or sabinene synthase of Sitka Spruce, with neighbouring residues providing a synergistic effect. In fact, the position of residue 596 in the third plasticity region, equivalent to PinS Gly601 and FenS Ala567, is sufficiently close to the C4 carbon of TPC to facilitate stabilisation and promote further migration26. Based on these results, the third plasticity region is most likely important in carbocation stabilisation in the later stages of more complex reaction cascades, fine-tuning the product outcomes of bicyclic mTC/S.
QM/MM MD simulations of LimS variants
Plant mTC/Ss undergo significant structural rearrangement during the reaction cycle55 and there are few crystal structures available of mTC/S in the closed form with the Mg2+ ions and substrate analogue resolved. LimS is one such example and is a useful model to gain insight into the impact of mutations in the identified regions of plasticity. As TPC is a critical intermediate in the formation of both mono and bicyclic products, we have built models of WT-LimS and the three variants described above at the stage in the reaction where TPC and PPi are already formed. Ten 2 ns SCC-DFTB/CHARMM27 MD simulations treating TPC by QM were carried out for each model system and structures analysed to identify important interactions between TPC and the enzyme. Deprotonation of the C8 methyl group must occur for the final stage in the production of (–)-(4S)-limonene and despite much experimental effort, the nature of the catalytic base remains unanswered. Residues H579 and W324 were identified as important for activity in the work of Srividya et al.27; however, H579 can be exchanged for other non-basic amino acids with only minor reduction in activity. Srividya et al. also postulate that W324 could act as the base, with protonation occurring at Cγ, but this is unlikely (see Supporting Information). Xu et al.56 have recently identified Y573 as important in the cyclisation mechanism in LimS, showing that both the hydroxyl group and the aromatic ring are important for function. It has also been suggested that the PPi moiety released by the ionisation step may play a further role in the reaction by acting as the catalytic base57–61.
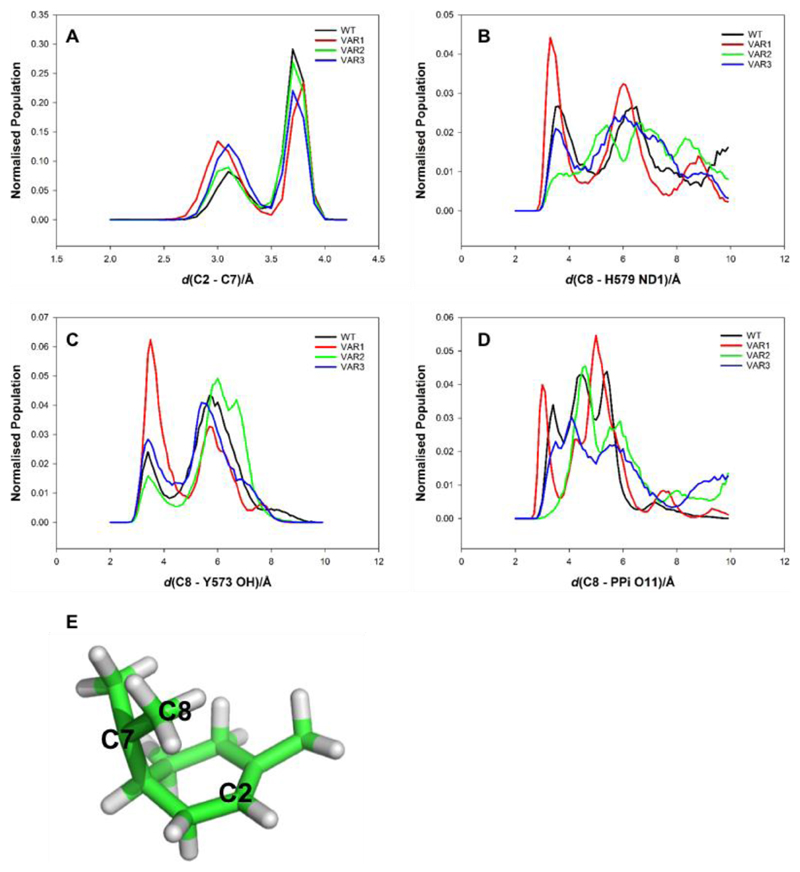
Figure 4 shows histograms of some important distances between TPC and surrounding residues in the active site of LimS. The data presented here is a combination of all 10 simulations and histograms showing the data for the individual simulations can be found in the Supporting Information. As mentioned above, VAR2-LimS is interesting as it produced mainly more complex bicyclic products. One simple explanation for the preference of VAR2-LimS for bicyclic products could be that the mutations change the shape of the active site forcing the atoms involved in the formation of the second ring (C2-C7) closer together. However, all the LimS variants show a very similar distribution of C2-C7 distances (Figure 4 panel A), with two main peaks (at 3 Å and at 3.7 Å).
Figure 4. Histograms of key interatomic distances from QM/MM MD simulations of the terpinyl cation (TPC) in WT and LimS variants.
(A) Histogram of the C2 - C7 distance involved in the second cyclisation to form bicyclic products. (B) Histogram of the heavy atom separation (C8 - H579 ND1) that would be involved in the formation of limonene if H579 is the base. (C) Histogram of the heavy atom separation (C8 - Y573 OH) that would be involved in the formation of limonene if Y573 is the base. (D) Histogram of the heavy atom separation (C8 - PPi O11) an indication of the distances that would be involved in the formation of limonene if the inorganic diphosphate PPi group acts as the base. (E) Structure of TPC showing atoms C2 and C7 involved in the second cyclisation and C8 the site of deprotonation.
Because the preference of VAR2-LimS for the formation of bicyclic products cannot be explained by compression of TPC, we must consider the role of the base in the deprotonation step. In order to deprotonate C8, the atoms involved in this C− H…X interaction must approach to distances similar to those of a conventional hydrogen bond, usually defined by a heavy atom separation of <3.5 Å62. There are many examples of histidine residues acting as catalytic bases in enzymatic reactions63, making H579 a strong candidate for the catalytic base in LimS. H579 was predicted to be singly protonated on NE2 in analysis by PropKa64, leaving ND1 available to accept a proton. Figure 4 panel B shows a histogram of the distance between C8 and ND1 of H579. Simulations of WT-LimS, VAR1-LimS and VAR3-LimS all show that C8 and ND1 of H579 can approach each other in line with a possible proton transfer, whereas they do not in VAR2-LimS. (The histogram of the distance in Figure 4B shows a peak in the histogram at around 3.5 Å for WT-LimS, VAR1-LimS and VAR3-LimS, whereas the VAR2-LimS model does not have a peak until 5.3 Å.) The histogram of the distance between C8 and Y573 OH during simulation shows a small peak in the population at ~3.5 Å and then a second maximum at ~5.7 Å for all variants (Figure 4 panel C). However, the population of the first maximum in the VAR2-LimS data is significantly smaller than that observed for the other models. After ionisation, PPi remains in close proximity to TPC, bound by the Mg2+ ions. Figure 4 panel D shows the histograms for the C8 – PPi O11 distance. There is a maximum in the population at ~3.5 Å for WT, VAR1-LimS and VAR3-LimS, but there is no significant population for this distance until ~4.5 Å in the simulations of VAR2-LimS. Major has shown that the PPi moiety has a strong electrostatic influence on the reaction in members of the other terpene cyclase family20. The relative positioning of PPi and TPC will be important in guiding the product outcome, whether or not PPi actively participates as the base in the reaction. D496, part of the DTE motif binding the Mg2+ ions, was observed to interact directly with TPC in some structures, but the population of any reactive conformations is low for all models, particularly WT-LimS and VAR2-LimS (see Supporting Information). The equivalent D496 residue in BPPS20 was shown to play only a minor role in the electrostatic control of the reaction.
The simulations show that there is very good agreement between the data obtained for WT-LimS and VAR3-LimS which show very similar product distributions, producing mainly limonene. The data for the VAR1-LimS simulations also has significant overlap with that obtained for WT-LimS, but the population of structures showing shorter distances to the candidates for the catalytic base is higher for VAR1-LimS, indicating that reaction termination prior to cyclisation is more likely. VAR1-LimS was found to produce predominantly linear products, but can produce mono and bicyclic products if early termination of the reaction is avoided. The data for VAR2-LimS differs from all the other models, with C8 generally positioned further from any of the putative bases. If the deprotonation reaction cannot occur, the cascade is not terminated at this point, allowing further cyclisation and rearrangements to occur and more complex products to form. This is consistent with the product distribution of VAR2-LimS forming ~10% monocyclic products (limonene and phellandrene) and ~85% bicyclic products.
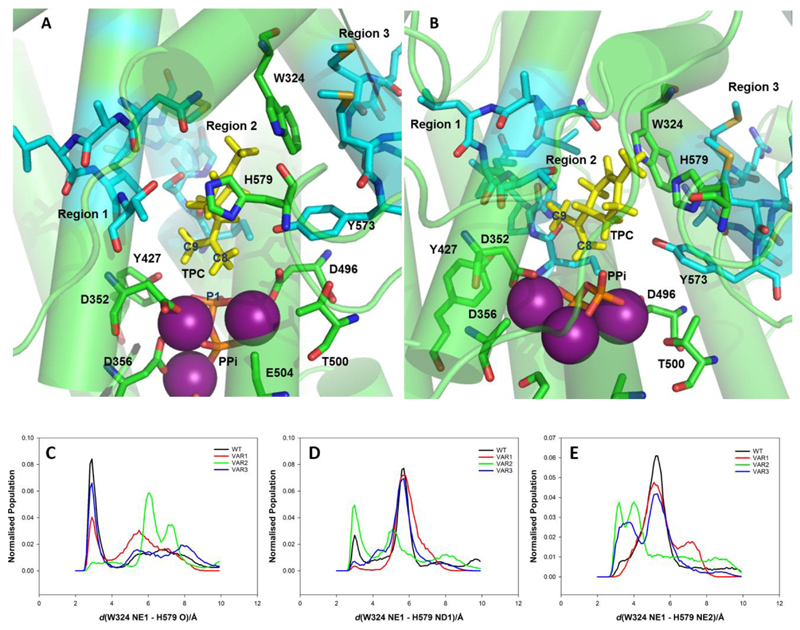
Figure 5 shows structures from cluster analysis of the MD simulations for WT-LimS (panel A) and VAR2-LimS (panel B), where H579 adopts a different position relative to W324 in the VAR2-LimS model. Figure 5 panel C shows the histogram of the distance between the sidechain of W324 and the backbone carbonyl of H579. There is a peak in the population at ~2.9 Å in the histograms for WT-LimS, VAR1-LimS and VAR3-LimS, consistent with the formation of a hydrogen bond in the majority of the simulations. However, this hydrogen bond interaction is almost entirely absent in VAR2-LimS, with the atoms involved separated by more than 6 Å for the majority of the simulation time. Instead, the simulations of VAR2-LimS show an interaction between the sidechains of W324 and H579 (Figure 5 panels D and E show the distance between W324 NE1 and ND1 or NE2 of H579, respectively). This interaction leads to the greater separation of TPC and H579 as described above. Mutation in region 2 of LimS thus impacts on the interactions between the reacting species and W324 and H579, the two residues identified as important for activity in LimS by Srividya et al. that are located on the opposite side of the active site cavity from the site of mutation.
Figure 5. The model of (A) WT-LimS and (B) VAR2-LimS with the terpinyl cation (TPC) and inorganic phosphate (PPi), a critical stage in determining product outcome in the terpene cyclase casade.
The model is based on the 2.7 Å resolution crystal structure of LimS (pdb 2ONG)8. The C-terminal terpene cyclase domain is in green, TPC is shown in yellow, and the Mg2+ ions in purple and PPi in CPK colours. The three plasticity regions are highlighted in cyan. Limonene is formed by the deprotonation of the C8 methyl group. The catalytic base has yet to be identified, but residues H579, W32427 and Y57356 have been identified as important for activity in previous studies. These putative bases are more distant from C8 in VAR2-LimS, allowing the second cyclisation to occur. (C) Histogram of the distance between the backbone carbonyl of H579 and the imidazole ring of W324 which form a hydrogen bond (W324 NE1 – H579 O) in some simulations. (D) Histogram of the distance between the sidechain of H579 and the imidazole ring of W324 which form a hydrogen bond (W324 NE1 – H579 NE2) in some simulations. (E) Histogram of the distance between the sidechain of H579 and the imidazole ring of W324 which form a hydrogen bond (W324 NE1 – H579 ND1) in some simulations.
Conclusions
In conclusion, here we described the identification of three clustered plasticity regions located in identical positions in helices around the active site pocket of plant mTC/S enzymes. We generated ‘consensus’ sequences for these mutational hotspots and introduced these in three different plant mTC/S backbones with increasingly complex cyclisation cascades. The variant enzymes were expressed and tested in our E. coli monoterpene production strain to rapidly obtain full product profiles. Using this systematic mutagenesis approach we found that the different identified areas are involved in different stages in the cyclisation cascade. Furthermore, we have performed QM/MM MD simulations of models of WT-LimS and the constructed variants with the α-terpinyl cation to aid our understanding of how these mutations alter the product distribution. The nature of the catalytic base for the final deprotonation step in the formation of limonene remains unclear and our simulations show that H579, Y573 and the pyrophosphate (PPi) released by the ionisation of the substrate can all adopt conformations consistent with reaction in WT-LimS, VAR1-LimS and VAR3-LimS, but not VAR2-LimS. The site of deprotonation of TPC is further from any of the possible base candidates in VAR2-LimS, which presumably allows the reaction cascade to continue. The mutations in VAR2-LimS do not lead to any compression of the C2-C7 distance favouring the second cyclisation, but rather alter the position of W324 and H579 which are residues important for the activity of LimS27. Such insights into the impact of mutations can only be obtained from simulation and is a key benefit of adopting a multidisciplinary approach to investigate enzyme catalysis. The results obtained in this study will help provide a general basis to guide rational engineering of members of the plant monoterpene synthase family, and is an important step towards the predictable and tuneable engineering of mTCSs for efficient production of desired terpenoids.
Supplementary Material
Additional figures and tables as described in the text. This information is available free of charge on the ACS Publications website.
Acknowledgments
This work was funded by the U.K. Biotechnology and Biological Sciences Research Council (BBSRC; BB/M000354/1, BB/M017702/1 and BB/L027593/1). M.W.v.d.K. is a BBSRC David Phillips Fellow (BB/M026280/1). N.S.S. was a Royal Society Wolfson Merit Award holder and an Engineering and Physical Sciences Research Council (EPSRC; EP/J020192/1) Established Career Fellow.
References
- 1.Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- 2.Beller HR, Lee TS, Katz L. Natural products as biofuels and bio-based chemicals: fatty acids and isoprenoids. Nat Prod Rep. 2015;32:1508–1526. doi: 10.1039/c5np00068h. [DOI] [PubMed] [Google Scholar]
- 3.Leferink NGH, Jervis AJ, Zebec Z, Toogood HS, Hay S, Takano E, Scrutton NS. A ‘plug and play’ platform for the production of diverse monoterpene hydrocarbon scaffolds in Escherichia coli. ChemistrySelect. 2016;1:1893–1896. doi: 10.1002/slct.201600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–29. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 5.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–42. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 6.Degenhardt J, Köllner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–37. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 7.Whittington DA, Wise ML, Urbansky M, Coates RM, Croteau RB, Christianson DW. Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc Natl Acad Sci U S A. 2002;99:15375–80. doi: 10.1073/pnas.232591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyatt DC, Youn B, Zhao Y, Santhamma B, Coates RM, Croteau RB, Kang C. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci U S A. 2007;104:5360–5. doi: 10.1073/pnas.0700915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar RP, Morehouse BR, Matos JO, Malik K, Lin H, Krauss IJ, Oprian DD. Structural characterization of early Michaelis complexes in the reaction catalyzed by (+)-limonene synthase from Citrus sinensis using fluorinated substrate analogues. Biochemistry. 2017;56:1716–1725. doi: 10.1021/acs.biochem.7b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morehouse BR, Kumar RP, Matos JO, Olsen SN, Entova S, Oprian DD. Functional and structural characterization of a (+)-limonene synthase from Citrus sinensis. Biochemistry. 2017;56:1706–1715. doi: 10.1021/acs.biochem.7b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampranis SC, Ioannidis D, Purvis A, Mahrez W, Ninga E, Katerelos NA, Anssour S, Dunwell JM, Degenhardt J, Makris AM, Goodenough PW, et al. Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell. 2007;19:1994–2005. doi: 10.1105/tpc.106.047779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolph K, Parthier C, Egerer-Sieber C, Geiger D, Muller YA, Kreis W, Muller-Uri F. Expression, crystallization and structure elucidation of gamma-terpinene synthase from Thymus vulgaris. Acta Crystallogr F Struct Biol Commun. 2016;72:16–23. doi: 10.1107/S2053230X15023043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karuppiah V, Ranaghan K, Leferink NGH, Johannissen LO, Shanmugam M, Ni Cheallaigh A, Bennett N, Kearsey L, Takano E, Gardiner J, Van der Kamp M, et al. Structural basis of catalysis in the bacterial monoterpene synthases linalool synthase and 1,8-cineole synthase. ACS Catal. 2017;7:6268–6282. doi: 10.1021/acscatal.7b01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Major DT, Freud Y, Weitman M. Catalytic control in terpenoid cyclases: multiscale modeling of thermodynamic, kinetic, and dynamic effects. Curr Opin Chem Biol. 2014;21:25–33. doi: 10.1016/j.cbpa.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Wise ML, Pyun H-J, Helms G, Assink B, Coates RM, Croteau RB. Stereochemical disposition of the geminal dimethyl groups in the enzymatic cyclization of geranyl diphosphate to (+)-bornyl diphosphate by recombinant (+)-bornyl diphosphate synthase from Salvia officinalis. Tetrahedron. 2001;57:5327–5334. [Google Scholar]
- 16.Wise ML, Savage TJ, Katahira E, Croteau R. Monoterpene synthases from common sage (Salvia officinalis) cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J Biol Chem. 1998;273:14891–9. doi: 10.1074/jbc.273.24.14891. [DOI] [PubMed] [Google Scholar]
- 17.Hong YJ, Tantillo DJ. Quantum chemical dissection of the classic terpinyl/pinyl/bornyl/camphyl cation conundrum-the role of pyrophosphate in manipulating pathways to monoterpenes. Org Biomol Chem. 2010;8:4589–4600. doi: 10.1039/c0ob00167h. [DOI] [PubMed] [Google Scholar]
- 18.Major DT, Weitman M. Electrostatically guided dynamics--the root of fidelity in a promiscuous terpene synthase? J Am Chem Soc. 2012;134:19454–62. doi: 10.1021/ja308295p. [DOI] [PubMed] [Google Scholar]
- 19.Weitman M, Major DT. Challenges posed to bornyl diphosphate synthase: diverging reaction mechanisms in monoterpenes. J Am Chem Soc. 2010;132:6349–6360. doi: 10.1021/ja910134x. [DOI] [PubMed] [Google Scholar]
- 20.Major DT. Electrostatic control of chemistry in terpene cyclases. ACS Catal. 2017;7:5461–5465. [Google Scholar]
- 21.Schrepfer P, Buettner A, Goerner C, Hertel M, van Rijn J, Wallrapp F, Eisenreich W, Sieber V, Kourist R, Brück T. Identification of amino acid networks governing catalysis in the closed complex of class I terpene synthases. Proc Natl Acad Sci U S A. 2016;113:E958–E967. doi: 10.1073/pnas.1519680113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christianson DW. Structural and chemical biology of terpenoid cyclases. Chem Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyatt DC, Croteau R. Mutational analysis of a monoterpene synthase reaction: altered catalysis through directed mutagenesis of (−)-pinene synthase from Abies grandis. Arch Biochem Biophys. 2005;439:222–33. doi: 10.1016/j.abb.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Katoh S, Hyatt D, Croteau R. Altering product outcome in Abies grandis(−)-limonene synthase and (−)-limonene/(−)-alpha-pinene synthase by domain swapping and directed mutagenesis. Arch Biochem Biophys. 2004;425:65–76. doi: 10.1016/j.abb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Krause ST, Kollner TG, Asbach J, Degenhardt J. Stereochemical mechanism of two sabinene hydrate synthases forming antipodal monoterpenes in thyme (Thymus vulgaris) Arch Biochem Biophys. 2013;529:112–21. doi: 10.1016/j.abb.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Roach CR, Hall DE, Zerbe P, Bohlmann J. Plasticity and evolution of (+)-3-carene synthase and (−)-sabinene synthase functions of a sitka spruce monoterpene synthase gene family associated with weevil resistance. J Biol Chem. 2014;289:23859–69. doi: 10.1074/jbc.M114.571703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srividya N, Davis EM, Croteau RB, Lange BM. Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc Natl Acad Sci U S A. 2015;112:3332–7. doi: 10.1073/pnas.1501203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Ai Y, Wang J, Zhang Y, Yang D. Converting S-limonene synthase to pinene or phellandrene synthases reveals the plasticity of the active site. Phytochemistry. 2017;137:34–41. doi: 10.1016/j.phytochem.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Shaw JJ, Berbasova T, Sasaki T, Jefferson-George K, Spakowicz DJ, Dunican BF, Portero CE, Narvaez-Trujillo A, Strobel SA. Identification of a fungal 1,8-cineole synthase from Hypoxylon sp. with common specificity determinants to the plant synthases. J Biol Chem. 2015;290:8511–8526. doi: 10.1074/jbc.M114.636159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci U S A. 1998;95:4126–33. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams DC, McGarvey DJ, Katahira EJ, Croteau R. Truncation of limonene synthase preprotein provides a fully active 'pseudomature' form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry. 1998;37:12213–20. doi: 10.1021/bi980854k. [DOI] [PubMed] [Google Scholar]
- 32.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. Journal of Biological Engineering. 2011;5:1–14. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swainston N, Currin A, Day PJ, Kell DB. GeneGenie: optimized oligomer design for directed evolution. Nucleic Acids Res. 2014;42:W395–400. doi: 10.1093/nar/gku336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Currin A, Swainston N, Day PJ, Kell DB. SpeedyGenes: an improved gene synthesis method for the efficient production of error-corrected, synthetic protein libraries for directed evolution. Protein Eng Des Sel. 2014;27:273–80. doi: 10.1093/protein/gzu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saaem I, Ma S, Quan J, Tian J. Error correction of microchip synthesized genes using Surveyor nuclease. Nucleic Acids Res. 2012;40:e23. doi: 10.1093/nar/gkr887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 37.Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien TE, Bertolani SJ, Tantillo DJ, Siegel JB. Mechanistically informed predictions of binding modes for carbocation intermediates of a sesquiterpene synthase reaction. Chem Sci. 2016;7:4009–4015. doi: 10.1039/c6sc00635c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allnér O, Nilsson L, Villa A. Magnesium ion–water coordination and exchange in biomolecular simulations. J Chem Theory Comput. 2012;8:1493–1502. doi: 10.1021/ct3000734. [DOI] [PubMed] [Google Scholar]
- 43.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Case DA, Betz RM, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, et al. AMBER 2016. University of California; San Francisco: 2016. [Google Scholar]
- 45.Elstner M. The SCC-DFTB method and its application to biological systems. Theor Chem Acc. 2006;116:316–325. [Google Scholar]
- 46.Greenhagen BT, O'Maille PE, Noel JP, Chappell J. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc Natl Acad Sci U S A. 2006;103:9826–31. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 48.Fischer MJC, Meyer S, Claudel P, Steyer D, Bergdoll M, Hugueney P. Determination of amino-acidic positions important for Ocimum basilicum geraniol synthase activity. Adv Biosci Biotechnol. 2013;4:242–249. [Google Scholar]
- 49.Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277:1815–20. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 50.Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R. 4S-limonene synthase from the oil glands of spearmint (Mentha spicata). cDNA isolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. J Biol Chem. 1993;268:23016–24. [PubMed] [Google Scholar]
- 51.Phillips MA, Wildung MR, Williams DC, Hyatt DC, Croteau R. cDNA isolation, functional expression, and characterization of (+)-α-pinene synthase and (−)-α-pinene synthase from loblolly pine (Pinus taeda): stereocontrol in pinene biosynthesis. Arch Biochem Biophys. 2003;411:267–76. doi: 10.1016/s0003-9861(02)00746-4. [DOI] [PubMed] [Google Scholar]
- 52.Croteau R, Miyazaki JH, Wheeler CJ. Monoterpene biosynthesis: Mechanistic evaluation of the geranyl pyrophosphate:(−)-endo-fenchol cyclase from fennel (Foeniculum vulgare) Arch Biochem Biophys. 1989;269:507–516. doi: 10.1016/0003-9861(89)90134-3. [DOI] [PubMed] [Google Scholar]
- 53.Croteau R, Shaskus J, Cane DE, Saito A, Chang C. Enzymic cyclization of [1-18O]geranyl pyrophosphate to l-endo-fenchol. J Am Chem Soc. 1984;106:1142–1143. [Google Scholar]
- 54.Zhou J, Wang C, Yoon S-H, Jang H-J, Choi E-S, Kim S-W. Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. J Biotechnol. 2014;169:42–50. doi: 10.1016/j.jbiotec.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 55.van der Kamp MW, Sirirak J, Zurek J, Allemann RK, Mulholland AJ. Conformational change and ligand binding in the aristolochene synthase catalytic cycle. Biochemistry. 2013;52:8094–105. doi: 10.1021/bi400898k. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Xu J, Ai Y, Farid RA, Tong L, Yang D. Mutational analysis and dynamic simulation of S-limonene synthase reveal the importance of Y573: Insight into the cyclization mechanism in monoterpene synthases. Arch Biochem Biophys. 2018;638:27–34. doi: 10.1016/j.abb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Gao Y, Honzatko RB, Peters RJ. Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat Prod Rep. 2012;29:1153–75. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J Biol Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 59.Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol. 2011;7:431. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2010;469:116. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seemann M, Zhai G, de Kraker J-W, Paschall CM, Christianson DW, Cane DE. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 62.Hadfield AT, Mulholland AJ. Active-site dynamics of ASADH—A bacterial biosynthetic enzyme. Int J Quantum Chem. 1999;73:137–146. [Google Scholar]
- 63.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins: Struct, Funct, Bioinf. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.