Abstract
Protein unfolding thermodynamic parameters are conventionally extracted from equilibrium thermal and chemical denaturation experiments. Despite decades of work, the degree of structure and the compactness of denatured states populated in these experiments are still open questions. Here, building on previous works, we show that thermally and chemically denatured protein states are distinct from the viewpoint of far-ultraviolet circular dichroism experiments that report on the local conformational status of peptide bonds. The differences identified are independent of protein length, structural class, or experimental conditions, highlighting the presence of two sub-ensembles within the denatured states. The sub-ensembles, UT and UD for thermally induced and denaturant-induced unfolded states, respectively, can exclusively exchange populations as a function of temperature at high chemical denaturant concentrations. Empirical analysis suggests that chemically denatured states are ∼50% more expanded than the thermally denatured chains of the same protein. Our observations hint that the strength of protein backbone−backbone interactions and identity versus backbone−solvent/co-solvent interactions determine the conformational distributions. We discuss the implications for protein folding mechanisms, the heterogeneity in relaxation rates, and folding diffusion coefficients.
Denatured protein states are perhaps the most challenging to experimentally characterize given the vast conformational space that is sampled and the extreme conditions employed to populate them. While it is well established that a folded protein can be denatured by different perturbations (temperature, chemical denaturants, pH, force, pressure, and mutations), there has always been a question about the nature of ensembles that are populated.1–4 This has been further renewed due to the explosion of research in intrinsically disordered proteins (IDPs), their functional significance, their dimensions, and the advantages and limitations of experimental techniques used to characterize them.5–7
Of particular interest are the differences in conformational preferences of thermally and chemically denatured protein states. The simplest spectroscopic technique used to study denaturation is far-ultraviolet (far-UV) circular dichroism (hereafter termed CD) that interrogates the local conformational preference of peptide bonds.8 Earlier studies have pointed to differences in the magnitude of the CD signal between thermally and chemically denatured states on specific proteins.9–20 Here, we extend these works by collecting CD data from the literature, show that the differences observed previously are robust to structure−sequence changes, and explain their implications.
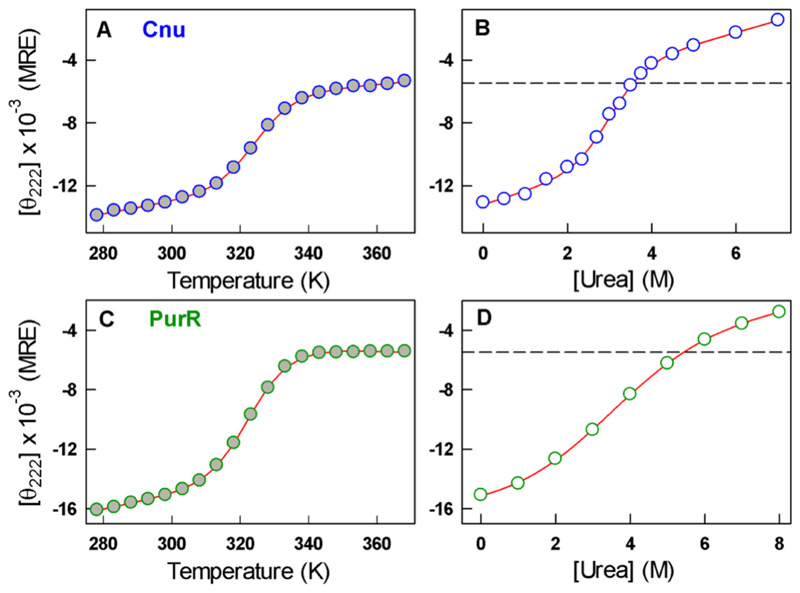
To illustrate, Figure 1A plots the thermal denaturation data of Cnu, a bacterial nanosensor of environmental conditions.21,22 The unfolding curve is apparently two-state-like with a thermally denatured ensemble exhibiting a CD signal of −5000 deg cm2 dmol−1 [mean residue ellipticity (MRE) units] at 222 nm. A urea-induced unfolding experiment at a constant temperature, on the other hand, reveals a similar two-state-like transition but with a denatured state displaying a CD signal of −2000 deg cm2 dmol−1 (Figure 1B). As an additional example, we show the thermal and chemical unfolding curve of PurR, a member of the LacI DNA-binding domain family. Clear differences in the CD signals of the denatured states are also apparent in these data (Figure 1C,D), consistent with earlier works.9–20
Figure 1.
Thermal (filled circles, panels A and C) and chemical unfolding (empty circles and at 298 K, panels B and D) curves of Cnu at pH 5.0 (blue) and PurR at pH 7.0 (green) monitored by far-UV CD at 222 nm and reported in MRE units (degrees square centimeters per decimole). Dashed lines in panels B and D represent the highest-temperature far-UV CD signal. Red curves are fits to two-state models.
The differences reported above could have multiple origins: errors in concentration measurement, effect of aromatics,23 and even nontrivial intrinsic sequence effects arising from preferential interaction with the chemical denaturants.24,25 To identify potential reasons for this deviation, we generated a database of far-UV CD data from thermal and chemical denaturation experiments that are reported in absolute units (MRE units) (see the Supporting Information). CD signals reported in absolute units are independent of protein concentration, protein length, and slight differences in sample path length and thus serve as a common ground for comparison across varied proteins and research laboratories. The database consists of 31 proteins of predominantly the α-helical or α/β structural type, with protein lengths spanning 29−150 residues. A majority of the data are at pH 7 and 298 K and include both urea-based (∼47%) and GuHCl-based (∼53%) denaturation.
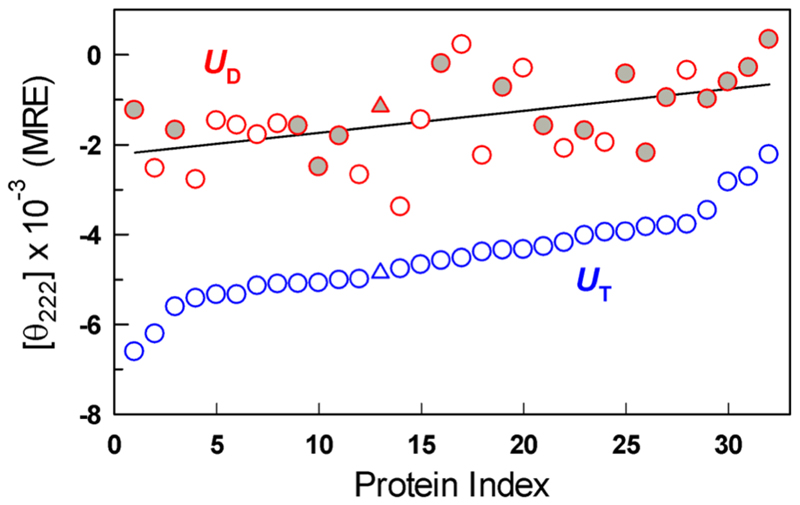
A comparison of the far-UV CD signal at 222 nm for all the proteins between the highest temperature (in the absence of denaturant) and highest denaturant concentration (at a fixed temperature) reveals a consistent difference between the two, as noted in Figure 1, with a mean absolute difference of ∼3100 deg cm2 dmol−1 (Figure 2). This suggests that thermally and chemically denatured protein states are fundamentally different and that the differences cannot be ascribed to protein type, structural class, nature of the denaturant, or temperature. The CD spectral shapes are distinct and reversible (see Figures S1 and S2 for select proteins), indicating that the differences cannot be attributed to errors in the estimation of concentration. Because far-UV CD is sensitive to the peptide bond conformations and the extent of alignment of amide transition dipoles (specifically that of the amide carbonyl at 222 nm), these differences can be taken as strong evidence that the two perturbants (chemical denaturants and temperature) contribute to different conformational sub-ensembles within the denatured state.
Figure 2.
Far-UV CD signal at 222 nm of thermally (UT) and chemically denatured states (UD at ∼298 K) for the protein database used in this work (Table S1). Filled and empty red circles represent signals of proteins denatured at high concentrations of GuHCl and urea, respectively. The triangle represents the signal at 230 nm for protein index 13.
Can the sub-ensembles, UT and UD for thermally and chemically denatured states, respectively, exclusively exchange populations within the denatured state? Thermal perturbations of urea or GuHCl-denatured states of folded proteins reveal a continuous and reversible increase in the intensity of the CD signal at 222 nm (Figure 3A and Figures S3 and S4).10,11,13–15,17–20 Even IDPs exhibit a tendency where the signal at 222 nm displays a negative slope with temperature (Figure 3B).26–29 Interestingly, the CD signal range and slopes (34.7 ± 9.4 deg cm2 dmol−1 K−1) are very similar despite the large differences in sequence. The CD signals at high temperatures and in the absence of denaturant (UT) agree very well with the CD signals at high denaturant concentration and high temperatures (UD, High T) with a mean absolute difference of just ∼550 deg cm2 dmol−1 (Figure 3C) for a small database of proteins (Table S2). This agreement is evidence that the two sub-ensembles can indeed exclusively exchange populations with the temperature effect trumping the denaturant-induced conformational distribution.
Figure 3.
(A) Transitions within the unfolded ensemble observed by far-UV CD in our lab for the folded proteins Cnu at pH 5 and 8, 7 M urea (blue and green), PDD F43W at pH 3, 7 M GuHCl (red) and PurR at pH 7, and 8 M urea (magenta). (B) Transitions within the IDP ensembles (in the absence of denaturant) of RVCaB (blue), α-synuclein (magenta), the phosphodiesterase γ-subunit (cyan), Caldesmon (black), a disordered peptide C-pep (green), and the IDP CytR at 7 M urea as a reference (red). The latter two data sets were generated in our lab. (C) Comparison of far-UV CD signals of folded proteins at high temperatures (UT, in the absence of denaturant) and under high-denaturant, high-temperature conditions (UD, High T).
Numerous studies highlight that denatured states of folded proteins and IDPs expand with an increase in denaturant concentration while collapsing to compact globules at higher temperatures in the absence of denaturants.28–37 Thus, it is likely that the thermally and chemically denatured states (Figure 2) represent the expanded and collapsed conformations, respectively, that are often identified in simulations38,39 and have been reported in prior small angle X-ray scattering and CD studies.9,12,16,17 From a purely structural view of the CD signals and employing the Uversky−Fink empirical relation between the relative CD signals and hydrodynamic volumes characterized by Stokes radii (RS),40 we arrive at a simple expression for the relative dimensions of UT and UD given the CD signals in Figure 2 (Table S1):
This relation signifies that chemically denatured states at 298 K are on average ∼50% more expanded than thermally denatured states (in the absence of denaturant), potentially due to specific interactions of the denaturant with the protein backbone.41–43 The caricature of UD employed here (as a single sub-ensemble) is an approximation, as the dimensions of chemically denatured states themselves change with denaturant in a continuous30,31,36,44 or cooperative manner.19
What specific structural features in the denatured ensemble could contribute to the observed differences? CD experiments point to an enhancement of the propensity of left-handed polyproline II-like local conformations (PPII) in homopolymeric peptides, denatured proteins, and IDPs with increasing urea and GuHCl concentrations.45–49 Temperature, on the other hand, has an opposite effect on the PPII conformational bias.50 However, a comparison of far-UV CD spectra from select proteins studied in our lab does not show the signature appearance of a peak in the CD spectrum between 220 and 230 nm (Figure S1), though the trends follow earlier observations.50 NMR experiments clearly demonstrate that an increase in temperature does not promote helical structure acquisition in IDPs or disordered proteins.28 Interestingly, the temperature dependence of the CD signal of the urea-unfolded state of LacI DBD is similar to that of the short fragments generated using extensive digestion of the protein by proteinase K under native conditions.13 These experiments raise questions about employing a structural view of CD signals at high temperatures and high denaturant concentrations. Resolving these conflicts, with experiments and simulations, could provide a better understanding of the denatured state characteristics.
From a folding mechanistic perspective, the two sub-ensembles could exchange populations in a direction orthogonal to the reaction coordinate for folding (Figure 4A). In such a scenario, kinetic experiments starting from UD will be characterized by a first step in which the unfolded population equilibrates with UT, after which the folding reaction proceeds. Second, UD and UT could equilibrate with each other and the folded state (three-state triangular mechanism), while a third scenario is one in which folding proceeds strictly from either of the ensembles depending on the perturbation. Temperature-jump (T-jump) experiments on the villin headpiece reveal an order of magnitude difference in relaxation rates when the folding reaction is initiated from UT or UD, with the latter being slower,51 the only such evidence available to date.
Figure 4.
(A) Folding free energy surface with multiple minima corresponding to denatured (UD and UT) and folded (N) states at non-zero denaturant concentrations. Coordinate 2 could represent any conformational feature that distinguishes the two denatured state sub-ensembles. (B) One-dimensional (1D) free energy profiles under stabilizing (blue) and destabilizing (red) conditions. The arrows represent the downhill-like conformational redistribution that occurs on switching to conditions favorable for folding. Panel B is not a projection of panel A onto a 1D coordinate.
If the sub-ensembles are populated along the reaction coordinate to folding, then the rates could again be different if the folding is initiated from UT or UD (Figure 4B). In such cases, even proteins with smaller folding thermodynamic barriers could display slow or multiple relaxation rates, as the dynamic term (i.e., the folding diffusion coefficient) itself could be quite slow due to the initial pre-equilibration between the two sub-ensembles apart from the expected rough landscape. This is more likely in proteins rich in α-helices whose structural features are dominated by local interactions and as shown experimentally and theoretically in Barstar52,53 and bACBP.54,55
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00089.
Tables S1 and S2, Figures S1–S4, and additional references (PDF)
Funding
This work was supported by the BIRAC-SRISTI GYTI award (2018/009) to A.N. The instrumentation used in this work was supported by Wellcome Trust/DBT India Alliance Intermediate Fellowship IA/I/15/1/501837 to A.N.N.
Abbreviations
- CD
circular dichroism
- PPII
polyproline II
- MRE
mean residue ellipticity
- RS
Stokes radius
- IDP
intrinsically disordered protein
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: An isolated three-stranded antiparallel beta-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- (3).McCarney ER, Kohn JE, Plaxco KW. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit Rev Biochem Mol Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- (4).Bowler BE. Residual structure in unfolded proteins. Curr Opin Struct Biol. 2012;22:4–13. doi: 10.1016/j.sbi.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Haran G. How, when and why proteins collapse: the relation to folding. Curr Opin Struct Biol. 2012;22:14–20. doi: 10.1016/j.sbi.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Schuler B, Soranno A, Hofmann H, Nettels D. Single-Molecule FRET Spectroscopy and the Polymer Physics of Unfolded and Intrinsically Disordered Proteins. Annu Rev Biophys. 2016;45:207–231. doi: 10.1146/annurev-biophys-062215-010915. [DOI] [PubMed] [Google Scholar]
- (7).Holehouse AS, Pappu RV. Collapse Transitions of Proteins and the Interplay Among Backbone, Sidechain, and Solvent Interactions. Annu Rev Biophys. 2018;47:19–39. doi: 10.1146/annurev-biophys-070317-032838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Manning MC, Woody RW. Theoretical CD studies of polypeptide helices: examination of important electronic and geometric factors. Biopolymers. 1991;31:569–586. doi: 10.1002/bip.360310511. [DOI] [PubMed] [Google Scholar]
- (9).Sosnick TR, Trewhella J. Denatured states of ribonuclease A have compact dimensions and residual secondary structure. Biochemistry. 1992;31:8329–8335. doi: 10.1021/bi00150a029. [DOI] [PubMed] [Google Scholar]
- (10).Agashe VR, Udgaonkar JB. Thermodynamics of Denaturation of Barstar: Evidence for Cold Denaturation and Evaluation of the Interaction with Guanidine Hydrochloride. Biochemistry. 1995;34:3286–3299. doi: 10.1021/bi00010a019. [DOI] [PubMed] [Google Scholar]
- (11).Kuhlman B, Raleigh DP. Global analysis of the thermal and chemical denaturation of the N-terminal domain of the ribosomal protein L9 in H2O and D2O. Determination of the thermodynamic parameters, deltaH(o), deltaS(o), and deltaC(o)p and evaluation of solvent isotope effects. Protein Sci. 1998;7:2405–2412. doi: 10.1002/pro.5560071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Farruggia B, Pico GA. Thermodynamic features of the chemical and thermal denaturations of human serum albumin. Int J Biol Macromol. 1999;26:317–323. doi: 10.1016/s0141-8130(99)00054-9. [DOI] [PubMed] [Google Scholar]
- (13).Felitsky DJ, Record MT. Thermal and urea-induced unfolding of the marginally stable lac repressor DNA-binding domain: A model system for analysis of solute effects on protein processes. Biochemistry. 2003;42:2202–2217. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- (14).Oliva FY, Muñoz V. A simple thermodynamic test to discriminate between two’state and downhill folding. J Am Chem Soc. 2004;126:8596–8597. doi: 10.1021/ja048309w. [DOI] [PubMed] [Google Scholar]
- (15).Fung A, Li P, Godoy-Ruiz R, Sanchez-Ruiz JM, Muñoz V. Expanding the realm of ultrafast protein folding: gpW, a midsize natural single-domain with alpha+beta topology that folds downhill. J Am Chem Soc. 2008;130:7489–7495. doi: 10.1021/ja801401a. [DOI] [PubMed] [Google Scholar]
- (16).Wang Q, Christiansen A, Samiotakis A, Wittung-Stafshede P, Cheung MS. Comparison of chemical and thermal protein denaturation by combination of computational and experimental approaches. II. J Chem Phys. 2011;135 doi: 10.1063/1.3656692. 175102. [DOI] [PubMed] [Google Scholar]
- (17).Tischer A, Auton M. Urea-temperature phase diagrams capture the thermodynamics of denatured state expansion that accompany protein unfolding. Protein Sci. 2013;22:1147–1160. doi: 10.1002/pro.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Naganathan AN, Muñoz V. Thermodynamics of Downhill Folding: Multi-Probe Analysis of PDD, a Protein that Folds Over a Marginal Free Energy Barrier. J Phys Chem B. 2014;118:8982–8994. doi: 10.1021/jp504261g. [DOI] [PubMed] [Google Scholar]
- (19).Singh R, Hassan MI, Islam A, Ahmad F. Cooperative Unfolding of Residual Structure in Heat Denatured Proteins by Urea and Guanidinium Chloride. PLoS One. 2015;10:e0128740. doi: 10.1371/journal.pone.0128740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zaidi S, Haque MA, Ubaid-Ullah S, Prakash A, Hassan MI, Islam A, Batra JK, Ahmad F. Denatured states of yeast cytochrome c induced by heat and guanidinium chloride are structurally and thermodynamically different. J Biomol Struct Dyn. 2017;35:1420–1435. doi: 10.1080/07391102.2016.1185039. [DOI] [PubMed] [Google Scholar]
- (21).Narayan A, Campos LA, Bhatia S, Fushman D, Naganathan AN. Graded structural polymorphism in a bacterial thermosensor protein. J Am Chem Soc. 2017;139:792–802. doi: 10.1021/jacs.6b10608. [DOI] [PubMed] [Google Scholar]
- (22).Narayan A, Naganathan AN. Tuning the Continuum of Structural States in the Native Ensemble of a Regulatory Protein. J Phys Chem Lett. 2017;8:1683–1687. doi: 10.1021/acs.jpclett.7b00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Aromatic side-chain contribution to farultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- (24).Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- (25).Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- (26).Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- (27).Ishijima J, Nagasaki N, Maeshima M, Miyano M. RVCaB, a calcium-binding protein in radish vacuoles, is predominantly an unstructured protein with a polyproline type II helix. J Biochem. 2007;142:201–211. doi: 10.1093/jb/mvm130. [DOI] [PubMed] [Google Scholar]
- (28).Kjaergaard M, Norholm AB, Hendus-Altenburger R, Pedersen SF, Poulsen FM, Kragelund BB. Temperature-dependent structural changes in intrinsically disordered proteins: formation of alpha-helices or loss of polyproline II? Protein Sci. 2010;19:1555–1564. doi: 10.1002/pro.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Munshi S, Gopi S, Subramanian S, Campos LA, Naganathan AN. Protein plasticity driven by disorder and collapse governs the heterogeneous binding of CytR to DNA. Nucleic Acids Res. 2018;46:4044–4053. doi: 10.1093/nar/gky176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sinha KK, Udgaonkar JB. Dependence of the Size of the Initially Collapsed Form During the Refolding of Barstar on Denaturant Concentration: Evidence for a Continuous Transition. J Mol Biol. 2005;353:704–718. doi: 10.1016/j.jmb.2005.08.056. [DOI] [PubMed] [Google Scholar]
- (31).Sherman E, Haran G. Coil-globule transition in the denatured state of a small protein. Proc Natl Acad Sci U S A. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ziv G, Thirumalai D, Haran G. Collapse transition in proteins. Phys Chem Chem Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nettels D, Muller-Spath S, Kuster F, Hofmann H, Haenni D, Ruegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, et al. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc Natl Acad Sci U S A. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Teufel DP, Johnson CM, Lum JK, Neuweiler H. Backbone-driven collapse in unfolded protein chains. J Mol Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- (35).Ciasca G, Campi G, Battisti A, Rea G, Rodio M, Papi M, Pernot P, Tenenbaum A, Bianconi A. Continuous thermal collapse of the intrinsically disordered protein tau is driven by its entropic flexible domain. Langmuir. 2012;28:13405–13410. doi: 10.1021/la302628y. [DOI] [PubMed] [Google Scholar]
- (36).Borgia A, Zheng W, Buholzer K, Borgia MB, Schuler A, Hofmann H, Soranno A, Nettels D, Gast K, Grishaev A, Best RB, et al. Consistent View of Polypeptide Chain Expansion in Chemical Denaturants from Multiple Experimental Methods. J Am Chem Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Stenzoski NE, Luan B, Holehouse AS, Raleigh DP. The Unfolded State of the C-Terminal Domain of L9 Expands at Low but Not at Elevated Temperatures. Biophys J. 2018;115:655–663. doi: 10.1016/j.bpj.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Maity H, Reddy G. Folding of Protein L with Implications for Collapse in the Denatured State Ensemble. J Am Chem Soc. 2016;138:2609–2616. doi: 10.1021/jacs.5b11300. [DOI] [PubMed] [Google Scholar]
- (39).Reddy G, Thirumalai D. Collapse Precedes Folding in Denaturant-Dependent Assembly of Ubiquitin. J Phys Chem B. 2017;121:995–1009. doi: 10.1021/acs.jpcb.6b13100. [DOI] [PubMed] [Google Scholar]
- (40).Uversky VN, Fink AL. The chicken-egg scenario of protein folding revisited. FEBS Lett. 2002;515:79–83. doi: 10.1016/s0014-5793(02)02441-9. [DOI] [PubMed] [Google Scholar]
- (41).O’Brien EP, Dima RI, Brooks B, Thirumalai D. Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: Lessons for protein denaturation mechanism. J Am Chem Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- (42).England JL, Haran G. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu Rev Phys Chem. 2011;62:257–277. doi: 10.1146/annurev-physchem-032210-103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Canchi DR, Garcia AE. Backbone and side-chain contributions in protein denaturation by urea. Biophys J. 2011;100:1526–1533. doi: 10.1016/j.bpj.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci U S A. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Shi Z, Woody RW, Kallenbach NR. Is polyproline II a major backbone conformation in unfolded proteins? Adv Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- (46).Chellgren BW, Creamer TP. Short sequences of non-proline residues can adopt the polyproline II helical conformation. Biochemistry. 2004;43:5864–5869. doi: 10.1021/bi049922v. [DOI] [PubMed] [Google Scholar]
- (47).Whittington SJ, Chellgren BW, Hermann VM, Creamer TP. Urea promotes polyproline II helix formation: implications for protein denatured states. Biochemistry. 2005;44:6269–6275. doi: 10.1021/bi050124u. [DOI] [PubMed] [Google Scholar]
- (48).Tiffany ML, Krimm S. New chain conformations of poly(glutamic acid) and polylysine. Biopolymers. 1968;6:1379–1382. doi: 10.1002/bip.1968.360060911. [DOI] [PubMed] [Google Scholar]
- (49).Rucker AL, Pager CT, Campbell MN, Qualls JE, Creamer TP. Host-guest scale of left-handed polyproline II helix formation. Proteins: Struct, Funct, Genet. 2003;53:68–75. doi: 10.1002/prot.10477. [DOI] [PubMed] [Google Scholar]
- (50).Elam WA, Schrank TP, Campagnolo AJ, Hilser VJ. Temperature and urea have opposing impacts on polyproline II conformational bias. Biochemistry. 2013;52:949–958. doi: 10.1021/bi301435p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhu L, Ghosh K, King M, Cellmer T, Bakajin O, Lapidus LJ. Evidence of Multiple Folding Pathways for the Villin Headpiece Subdomain. J Phys Chem B. 2011;115:12632–12637. doi: 10.1021/jp206238y. [DOI] [PubMed] [Google Scholar]
- (52).Sarkar SS, Udgaonkar JB, Krishnamoorthy G. Unfolding of a Small Protein Proceeds via Dry and Wet Globules and a Solvated Transition State. Biophys J. 2013;105:2392–2402. doi: 10.1016/j.bpj.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Naganathan AN, Sanchez-Ruiz JM, Munshi S, Suresh S. Are Protein Folding Intermediates the Evolutionary Consequence of Functional Constraints? J Phys Chem B. 2015;119:1323–1333. doi: 10.1021/jp510342m. [DOI] [PubMed] [Google Scholar]
- (54).Voelz VA, Jager M, Yao SH, Chen YJ, Zhu L, Waldauer SA, Bowman GR, Friedrichs M, Bakajin O, Lapidus LJ, Weiss S, et al. Slow Unfolded-State Structuring in Acyl-CoA Binding Protein Folding Revealed by Simulation and Experiment. J Am Chem Soc. 2012;134:12565–12577. doi: 10.1021/ja302528z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Munshi S, Naganathan AN. Imprints of function on the folding landscape: functional role for an intermediate in a conserved eukaryotic binding protein. Phys Chem Chem Phys. 2015;17:11042–11052. doi: 10.1039/c4cp06102k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.