Abstract
The physicochemical properties of nucleic acids are dominated by their highly charged phosphodiester backbone chemistry. The polyelectrolyte structure decouples information content (base sequence) from bulk properties such as solubility and has been proposed as a defining trait of all informational polymers. However, this conjecture has not been tested experimentally. Here, we describe the encoded synthesis of a genetic polymer with an uncharged backbone chemistry: alkyl-phosphonate nucleic acids (phNA), in which the canonical, negatively charged phosphodiester is replaced by an uncharged P-alkyl-phosphonodiester backbone. Using synthetic chemistry and polymerase engineering, we describe the enzymatic, DNA-templated synthesis of P-methyl- and P-ethyl-phNAs, and the directed evolution of specific streptavidin-binding phNA aptamer ligands directly from random-sequence, mixed P-methyl- / P-ethyl-phNA repertoires. Our results establish a first example of the DNA-templated enzymatic synthesis and evolution of an uncharged genetic polymer and provide a foundational methodology for their exploration as a source of novel, functional molecules.
Introduction
All currently known genetic systems in biology are founded on genetic polymers based on the ribofuranose nucleic acids, DNA and RNA. Indeed, several features of nucleic acid chemistry appear to be uniquely suited for seamless genetic information storage and readout. These include the unusual kinetic stability of phosphate esters and diesters1 and the dominant influence of the polyanionic phosphodiester backbone on the physicochemical properties of nucleic acids. This aspect serves to decouple physicochemical properties such as solubility from information content (i.e. nucleotide sequence), as well as to aid information readout by stabilizing a linear, extended conformation through charge repulsion2. Consequently, a “polyelectrolyte theory of the gene” has been proposed, positing that a highly charged backbone chemistry is a defining attribute of all (possible) information storing polymers, i.e. genetic materials3.
In accordance with this concept, multiple variations of the nucleobase and sugar ring congener chemistry have been found to be compatible with genetic function, yielding DNA and RNA bearing novel basepairs4 and synthetic genetic polymers (xeno-nucleic acids, XNAs)5. Some of these can be replicated (synthesized and reverse transcribed) by natural or engineered polymerases and this has enabled the in vitro evolution of novel ligands (aptamers) and catalysts6, 7 as well as the inception of a semi-synthetic organism with an expanded genetic alphabet8. However, all systems reported to date are nonetheless reliant on a polyanionic phosphodiester backbone.
The development of functional genetic polymers based on variant backbone chemistries has generally proven more challenging. However, a few examples have been described including the N3’-P5’ phosphoroamidates (PN) in which the bridging 3’-oxygen is replaced by an amino group, phosphorothioates (PS) and boranophosphates (PB), in which the non-bridging oxygen on the phosphate is replaced by respectively sulfur (αS) or borane (αBH3) (Supplementary Fig. 1) and 3′-2′ phosphonomethyl-threosyl nucleic acid (tPhoNA)9, in which threose replaces ribose and a methyl group is inserted between the phosphorous and the 3‘-oxygen. Unlike many other examples that are only accessible by solid-phase synthesis these can be replicated non-enzymatically (PN)10 or enzymatically by DNA polymerases (PS, PB, tPhoNA)11, 12 and (in some cases) evolved into ligands13. However, all of these polymers retain the polyanionic nature of the canonical diester backbone and therefore conform to the polyelectrolyte hypothesis.
Also consistent with the theory, a variety of polycationic backbone chemistries including guanidinium-, S-methylthiourea and nucleosyl-amino acid linkages (Supplementary Fig. 1) have been synthesized by solid phase methods14. However, these cationic polymers have so far not been shown to be amenable to either non-enzymatic or enzymatic replication and their capacity for genetic function therefore remains to be explored. A more robust challenge to the polyelectrolyte theory would be the elaboration of genetic function in uncharged genetic polymers. However, the design of functional nucleic acid isomorphs with uncharged backbones has proven especially demanding, with even minor modifications showing drastic effects on nucleic acid solubility, duplex conformation and stability, rendering the majority of such backbone chemistries incompatible with information transfer and genetic function15, 16.
Nevertheless, some uncharged oligonucleotide chemistries accessible by solid-phase synthesis have been shown to yield highly specific probes for hybridization and gene silencing, notably P-alkyl-phosphonate nucleic acids (phNA)17, phosphoro-diamidate morpholino oligomers (PMO)18, and peptide nucleic acids (PNA)19. Template-encoded synthesis and replication of these polymers would enable a more thorough exploration of their sequence space and provide a stringent test of their capacity for genetic function. Previous efforts in this area have demonstrated sporadic (template-dependent) incorporation of P-methyl-phosphonate dTTP by enzymatic synthesis20, 21 as well as non-enzymatic template-dependent synthesis strategies based on reductive amination of PNA-pentamers enabling encoded synthesis and a model selection22. However, the potential of these chemistries for genetic function and evolution has not been fully explored.
Here we report the discovery of engineered polymerase variants using in silico model building and in vitro evolution that enable the synthesis and reverse transcription of uncharged P-alkyl-phosphonate nucleic acids (phNAs) at full substitution. We also demonstrate that uncharged phNA genetic polymers can support Darwinian evolution by the de novo selection of specific streptavidin-binding aptamers directly from random-sequence phNA repertoires. Together, our results demonstrate that an uncharged genetic polymer not found in nature has the capacity for genetic function, and establish the first synthetic platform for its encoded synthesis and evolution.
Results
In order to explore strategies for the encoded synthesis and evolution of genetic polymers with uncharged backbone chemistries by polymerase engineering we chose to revisit P-alkyl-phosphonate nucleic acids (phNA) as substrates20, 21. In phNAs the negatively charged, non-bridging oxygen of the canonical phosphodiester linkage is replaced by an uncharged alkyl substituent (specifically a methyl (Met), or ethyl (Et) group) (Fig. 1a, Supplementary Fig. 1). Previous work involving such phNAs in the context of antisense technologies has shown that these chemistries are broadly biocompatible (non-toxic and nuclease resistant) and retain an ability to base pair (with target RNA)17, 23. Furthermore, phNAs combine an uncharged alkyl-phosphonodiester backbone chemistry with close steric and electronic analogy to canonical nucleic acids and therefore provide a tractable challenge for polymerase engineering.
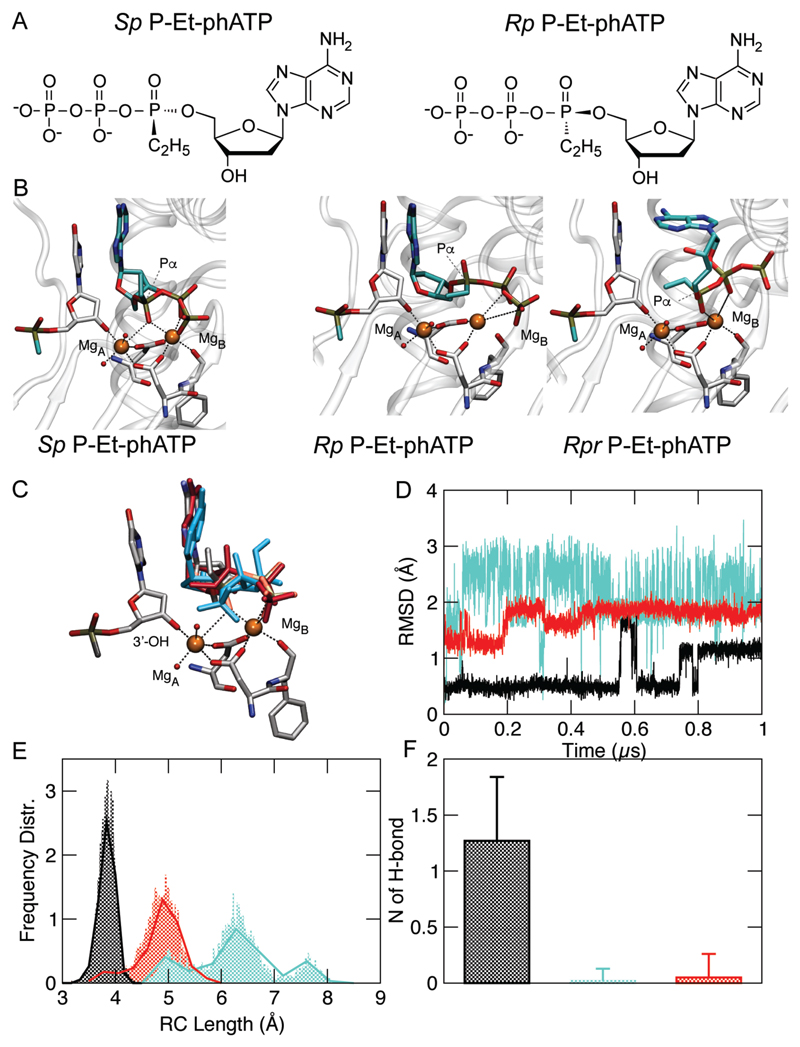
Figure 1. Global and structural modelling of Pol / P-Et-phATP diastereoisomer complexes.
Global and local structural properties of Polymerase / primer complex accommodating the incoming different P-Et-phATP diastereoisomers in the active site of RB69 DNA polymerase26. A) Chemical structure the Sp and Rp P-Et-phATP diastereoisomers. B) Snapshot extracted from our MD simulations displaying the incoming Sp P-Et-phATP and Rp / Rpr P-Et-phATP diastereoisomers in the polymerase active site. Dashed lines indicate Mg-O coordination. Carbons are coloured in white, oxygen in red, nitrogen in blue while phosphorus in maroon. C-F) Color code: black identifies Ethyl-Sp system, red Ethyl-Rp while cyan represents Ethyl-Rpr system. C) View of the active site of Pol / phDNA / P-Et-phATP ternary complex. Incoming phATP extracted from Ethyl-Sp (coloured accordingly with the atom species), Ethyl-Rp (in red) and Ethyl-Rpr (in cyan) systems are overlapped for comparison. (D) RMSD of the incoming P-Et-phATP (heavy atoms only) as a function of time (1µs). (E) Length of the reaction coordinate describing the nascent phosphodiester bond (i.e. the length between -3’O- of the 3’-end nucleotide and the αP of the incoming phATP. (F) Population of Watson-Crick H-bond between the incoming phATP and its complementary base on the DNA template strand.
In order to prepare building blocks for use in polymerase-catalysed phNA synthesis reactions, we first synthesized the P-α-ethyl-phosphonyl-β,γ-diphosphate (P-Et-phNTPs) equivalents of all four deoxyribonucleoside triphosphates (P-Et-phATP, P-Et-phGTP, P-Et-phTTP, P-Et-phCTP), as well as the P-α-methyl-phosphonyl-β,γ-diphosphate (P-Met-phNTPs) analogues of deoxyuridine and deoxycytidine (P-Met-phUTP, P-Met-phCTP), following the method of Dyatkina24 (Supplementary Fig. 2a). phNTP yields after HPLC purification varied between 0.5-10%.
As is the case with the charged phosphorothioate (PS) and boranophosphate (PB) nucleic acids (Supplementary Fig. 1), substitution of the non-bridging oxygen on the (α) phosphorus is stereogenic. Indeed, synthesized P-α-alkyl-phNTP analogues presented a racemic mix of Rp and Sp diastereoisomers, which could be resolved by high-pressure liquid chromatography (HPLC) (Supplementary Fig. 2B). We HPLC purified and tested putative Rp and Sp P-Met– and P-Et-phNTP diastereoisomers for enzymatic incorporation and found that, by analogy to the case with PS-dNTPs (where the stereocenter is created by the S substitution (Supplementary Fig. 1))25, and P-α-methyl-dTTP21, only one of the diastereoisomers was a substrate for incorporation by polymerases. However, while enzymatic incorporation clearly points to selective incorporation of just one of the phNTP diastereoisomers, HPLC mobility shifts provide an insufficient criterion to distinguish which diastereoisomer (Rp or Sp) was incorporated. In order to probe the stereochemistry of the likely substrate we performed structural modelling and extensive classical molecular dynamics (MD) simulations of the high-resolution, polB-family RB69 DNA polymerase primer-template holoenzyme (PDBid 4M3R)26 complex, offering either the Sp or Rp P-ethyl (P-Et) or P-methyl (P-Met) -phATP diastereoisomers as substrates (Fig. 1, Supplementary Fig. 3). Importantly, in order to sample a wider conformational phATP space, additional MD simulations were performed on different structural variants of Met-Rp and Et-Rp substrates that were rotated along the P-Met/P-Et-phNTP ε-dihedral angle (namely Rpr) (Fig. 1b, c).
Our theoretical investigations unequivocally showed that only the Sp phATP substrates were able to productively engage (and remain) within the polymerase holoenzyme active site (thus forming the polymerase-primer-template complex). This is demonstrated by three different and well-established structural/chemical determinants we monitored along the 6 μs of MD trajectory to evaluate the reactivity of the precatalytic complex; namely: i) the root-mean-square deviation (RMSD) of the incoming substrate measuring its stability within polymerase active site27, ii) the length between the -3’O- (-3’OH) nucleophile of the primer 3’-end and the α-Phosphorus of the incoming phATP assessing the formation of a prone-to-react Michaelis-Menten complex28 and iii) the ability to form Watson-Crick (WC) H-bonds to the instructive template base, a condition known to further assist the incorporation of the incoming nucleotide to the growing primer strand29, 30.
While the Sp phATP (in both P-methyl- and P-ethyl configuration) appeared to form a stable complex (lowest RMSD fluctuations, see Fig. 1, Supplementary Fig. 3) with a reaction coordinate of ca. 3.5Å and 1-1.5 WC H-bonds to the template base indicating a productive holoenzyme complex (Fig. 1b-d), both the Rp and Rpr diastereoisomer (in both P-methyl- and P-ethyl configuration) showed formation of a much less stable complex, a significantly extended reaction coordinate (4.5 – 8.0Å) and no WC H-boding to the template base (Fig. 1e-f Supplementary Fig. 3). Hence, these data suggest that the the Rp configuration phNTP substrates are incompatible with catalysis.
Thus, in silico modelling strongly suggests that polymerases will selectively incorporate only the Sp P-alkyl-phNTP diastereoisomers, which preserves the critical interaction of the negatively charged proRp-oxygen with the catalytically essential proximal Mg2+ ion (MgA) in the polymerase active site28, 30. Indeed, this is consistent with previous results that suggested stereospecific incorporation of only the Sp P-Met-phTTP by Terminal transferase21.
While P-Met-phTTPs had previously been shown to be substrates for sporadic incorporation by some polymerases20, 21, we wished to identify enzymatic strategies for phNA synthesis at full substitution, i.e. comprising all four phNTPs. We initiated our screening efforts using all four of the P-ethyl-phNTPs (P-Et-phNTPs: P-Et-phATP, P-Et-phGTP, P-Et-phCTP, P-Et-phTTP), as these had been higher yielding in chemical synthesis than the P-Met-phNTPs. We first screened a range of commercial polymerases, as well as our in-house panel of engineered XNA polymerases and reverse transcriptases8, for their ability to catalyse primer extension by P-Et-phNTPs on a DNA template, using the previously described high-throughput polymerase activity assay (PAA)31. Several commercially available polymerases could extend primers using P-Et-phNTPs, but were incapable of phNA synthesis beyond more than one to two incorporations at full substitution. In contrast, some of the polymerases previously engineered for alternative, charged XNA substrates displayed superior (if weak) phNA synthesis activity at full substitution, as judged by PAA and primer extension assays. In particular, RT521L, a mutant of the polB-family DNA polymerase from Thermococcus gorgonarius (Tgo) (Tgo: V93Q, D141A, E143A, E429G, A485L, I521L, K726R), previously described as a reverse transcriptase for several XNAs (as well as polymerase for α-L-threofuranosyl nucleic acid (TNA))6, showed promising activity. We therefore chose RT521L as the starting point for further engineering toward a more efficient phNA polymerase. By analogy with racemic mixtures of phosphorothioate (PS dNTPs), RT521L catalyzed phNA synthesis directly from the racemic P-α-Et-phNTP mix generated by chemical synthesis, with presumed selective incorporation of the Sp phNTP diastereoisomers (Fig. 1, Supplementary Fig. 3).
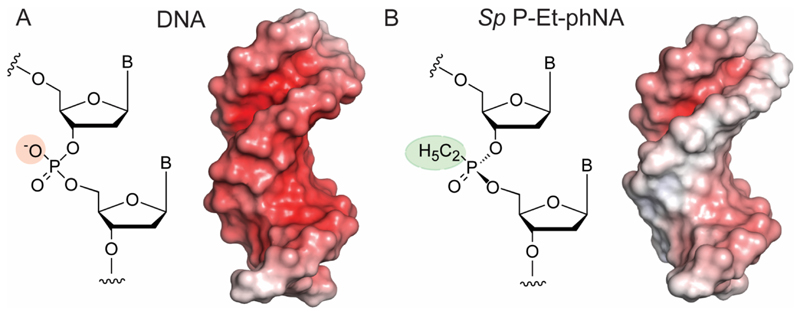
Based on this data, we built a simple model of a Sp P-Et-phNA / DNA heteroduplex (assuming minimal changes to the helical conformation) in order to map the putative interaction surface with the polymerase. Our model suggested a steep reduction in negative surface charge potential (Figure 2) as well as steric deviations from the canonical DNA-DNA homoduplex. These changes from the canonical DNA duplex structure define the challenges for polymerase engineering including steric and ionic incompatibilities both in the context of the polymerase catalytic site as well as primer-template duplex binding surface due to the replacement of the negatively charged oxygen by the bulkier ethyl-groups.
Figure 2. Surface electrostatic potential in DNA x DNA and phNA x DNA duplexes.
A) Chemical structure (left) and solvent-accessible surface area of a canonical DNA homoduplex (DNA x DNA) (Sequence: CGCGAATTCGCG, PDB: 3U2N) (right) surface electrostatic potential calculated by APBS28 B) Chemical structure of Sp P-ethyl-phNA (left) and model of a DNA x Sp P-ethyl-phNA heteroduplex (right) (based on A) with a surface electrostatic potential calculated by APBS showing much reduced negative charge potential (in red) compared to DNA.
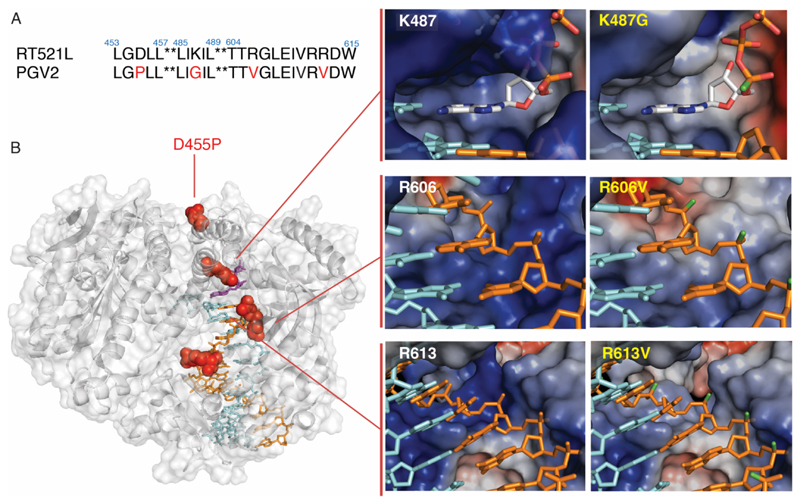
We overlaid this simple P-Et-phNA / DNA heteroduplex model on the primer-template duplex in the tertiary complex structure of the polB family phage RB69 DNA polymerase29, with cross referencing to the secondary complex structures of polB family DNA polymerases from the Pyro- and Thermococcales (Pfu, KOD and 9°N)32, 33 more closely related to Tgo. This approach indicated a list of (putative) unfavourable contacts and potential steric clashes between the P-ethyl-groups (protruding from the P-Et-phNA helix apex) and the polymerase surface (Supplementary Figs. 4, 5), involving several residues (Tgo RT521L: K487, R606, R613, Q665, T667, R668) known to make direct ionic and / or H-bonding interactions with the corresponding negatively charged proSp oxygens on the canonical phosphodiester backbone of a nascent DNA strand.
Next, we prepared and screened polymerase libraries with positional saturation mutagenesis (comprising all 20 amino acids) focused on these residues involved in putative steric and ionic clashes by PAA31, and were able to discover mutations that enhanced phNA synthesis. While none of the mutations at residues Q665, T667 and R668 appeared to yield clear improvements, some mutations at residues K487, R606, R613 (in particular K487G, R606V and to a lesser extent, R613V), both alone and in combination improved the efficiency of phNA synthesis. Combined, they yielded the new phNA polymerase “GV2” (Tgo RT521L: K487G, R606V, R613V) (Fig. 3a-b), which displayed enhanced synthesis of fully-modified P-Et-phNA oligomers on a range of (favourable templates) (Supplementary Fig. 6).
Figure 3. Polymerase mutations enabling phNA synthesis.
A) Sequence alignment of RT521L and phNA polymerase PGV2, mutated residues are highlighted in red. B) PGV2 mutations (red) mapped onto the structure of the ternary complex of the closely related KOD polymerase (PDB 5OMF) (left panel); surface structure and electrostatic surface potential changes upon mutation is shown for three mutations: K487G (top right), R606V (middle right) and R613V (bottom right panel) with nascent strand (orange), with or without Sp-P-alkyl substitution (green), and template strand (cyan).
All three residues mutated in GV2 encode positively charged amino acids and are highly conserved in the Thermococcales polB family polymerases. K487 in particular is conserved universally across the entire polB family, with close analogues apparent in the structures of every known polymerase family, where the residue is often substituted for arginine. Indeed, importance and role of this strictly conserved residue in polymerase catalysis, has been the focus of extensive bioinformatics, physicochemical and structural studies29, 34. High-resolution structures of the catalytic steps of DNA synthesis (in human Pol η) underline the key role for the cationic head group of arginine (pol η: R61, analogous to K487) in charge neutralization during the catalytic step35. Replacement with a less bulky side chain (glycine in GV2) may serve to both remove steric clashes of the lysine side-chain with the bulky ethyl group on the incoming P-Et-phNTP substrate, as well as reduce the unfavourable energetics of the unpaired positive charge of the K487 amino group in the weakly polar transition state of phNTP incorporation.
Similarly, the R606 residue is well conserved in the Pyro- and Thermococcales polB-family polymerases, (substituted by a lysine in most mesophilic polB homologs) and its mutation to smaller, uncharged valine (R606V) in GV2 may eliminate potential steric and ionic clashes with the newly incorporated ethyl-phosphonate nucleotide at the n+1 position of the nascent strand (Fig. 3b, Supplementary Fig. 4, 5). Although the analogous R613V mutation did not yield an improvement by itself, in combination with K487G and R606V, P-Et-phNA synthesis was enhanced, presumably due to removal of steric and / or ionic incompatibilities once the phNA nascent strand reaches the n+3 position. (Fig. 3b). Thus, in all three cases, mutation of large cationic amino acids to smaller, uncharged, nonpolar amino acid side chains improved phNA synthesis. The mutations likely reduce the steric and ionic clashes with the ethyl groups on the incoming P-α-Et-phNTP and nascent P-Et-phNA strand, eliminating the energetic penalty of unsatisfied ionic bonds, while potentially providing novel hydrophobic and van der Waal’s interactions with the nonpolar nascent strand P-Et-phNA backbone. This suggests that the key changes to the polymerase that enabled enzymatic phNA synthesis required both a reduction of the steric bulk as well as local positive charge surface potential in the primer-template binding interface (Fig. 3).
Having removed all apparent suboptimal contacts identified in our simple model, we sought to further improve phNA polymerase activity by in vitro evolution using compartmentalized self-tagging (CST)36. Starting from both the original RT521L and the GV2 triple mutant frameworks, we created four different polymerase libraries for CST selections, each composed of ~2x109 polymerase variants. Mutations were focused on different polymerase submotifs located in the fingers and thumb domains previously shown to be important for substrate recognition or primer-template binding. After two rounds of CST, one of the libraries (motif 5), targeting residues L453-K468 (Supplementary Fig. 7), yielded a range of polymerases with a capacity for phNA synthesis, although none superior to GV2.
The best performing polymerase from this library derived from the parental polymerase RT521L and comprised the mutations RT521L: E398K, R379K M467L, D455P, I449T and K557E. Introduction of the K487G, R606V, R613V triple mutation into this polymerase did not augment its phNA synthetase activity. However, the reverse introduction of just the D455P mutation into the GV2 background, yielding polymerase PGV2 (RT521L: D455P, K487G, R606V, R613V)(Fig. 3), substantially improved phNA synthesis, enabling full-length synthesis of 57 nucleotide (nt) phNA oligomers on three different favourable, pyrimidine-rich templates (Supplementary Fig. 6). However, the synthesis of fully substituted P-Et-phNAs on mixed-sequence templates with a balanced proportion of purines and pyrimidines remained challenging. Dissecting properties of less favourable templates revealed a clear bias against the incorporation of pyrimidine P-Et-phNTPs, in particular P-Et-phTTP (Supplementary Fig. 6), whereby sequences devoid of, or low in, pyrimidines (and especially phT) were synthesized with much higher efficiency.
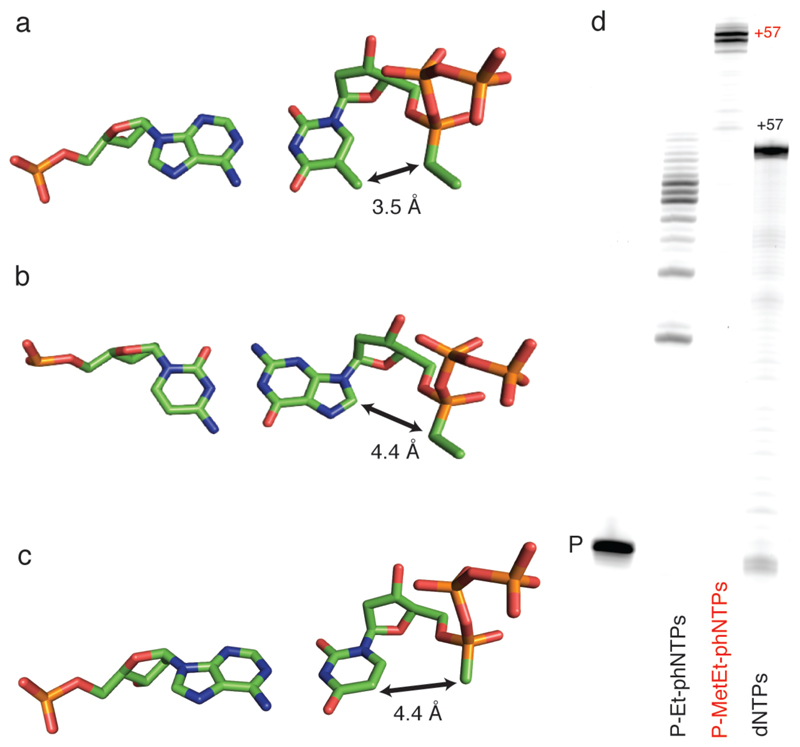
To better understand this bias, we examined the likely conformation of both pyrimidine and purine Sp P-Et-phNTPs bound in the homologous RB69 polymerase active site26, assuming close conformational and structural analogy to dNTPs as revealed in RB69 tertiary complex structures (Fig. 4). Inspection of this simple model indicated a potential intramolecular steric clash in the dA: Sp P-Et-phTTP pair between the ethyl-group on the α-phosphorus and the C5-methyl group of thymidine in the catalytically poised nucleotide conformation28 (Fig. 4a). As this close contact is absent in the phNTP purine pairs (e.g. the dC: Sp P-Et-phGTP pair) (Fig. 4b), we hypothesized that this might be a potential cause for the inefficient incorporation of P-Et-phTTP. In order to relieve the steric incompatibility (Fig. 4a) the P-Et-phTTP nucleotide would be forced to deviate from the canonical catalytically poised triphosphate conformation and adopt a non-optimal alignment between the primer 3’-OH and the Sp P-α-ethyl-phosphonate group with a likely negative impact on phNTP incorporation.
Figure 4. Structural context of Sp P-alkyl-phNTP incorporation.
A) Structure of a model of the polymerase active site based on the ternary complex structures of RB69 polymerase (PDB: 3CFP, PDB 4FJH). The estimated distance between the C5-methyl and P-ethyl groups (assuming no conformational adjustments) is shown for A) Sp P-Et-phTTP : dA pair; for B) between C5 and P-methyl group of the Sp P-Met-phUTP: dA pair and compared C) between C8 and P-ethyl group for the Sp P-Et-phGTP : dC pair. Note that P-Met-phU : dA shows similar distance as P-Et-phG:dC pair. D. PAGE of primer extension on an unbiased template TempN (Supplementary Table 5) showing synthesis of using P-Et-phNTPs with multiple pausing (mostly corresponding to template dApdA) or full-length synthesis of using P-MetEt-phNTP mix by PGV2 (P: unextended primer). Note the reduced electrophoretic mobility of full-length phNA (57 nt) compared to DNA control (57 nt).
Indeed, detailed high-resolution studies of the active site and catalytic trajectory of dNTP incorporation have shown the exquisite sensitivity of polymerase catalysis to even minor deviations from the ideal geometry and inter-atomic distances35. To test this hypothesis, we synthesized the sterically less conflicted α-methyl-phosphonate analogues of dUTP and dCTP, in which both the steric bulk on the α-phosphorus (by replacing the ethyl- with a methyl-group) and at the nucleobase by replacing thymine with the equivalent uracil (i.e. removing the C5 methyl group) (Fig. 4c). Indeed, both P-Met-phUTP and P-Met-phCTP proved to be superior substrates, consistent with our structural interpretations. We therefore switched to a blend of purine ethyl- and pyrimidine methyl-phosphonate triphosphates as substrates (P-Et-phATP, P-Et-phGTP, P-Met-phUTP, P-Met-phCTP). This blend allowed the efficient, encoded synthesis of phNA oligomers (57 nt) with balanced nucleotide composition (Fig. 4d). For clarity, we will refer to these mixed P-Met-/ P-Et-phNA substituted genetic polymers as “P-MetEt-phNAs”.
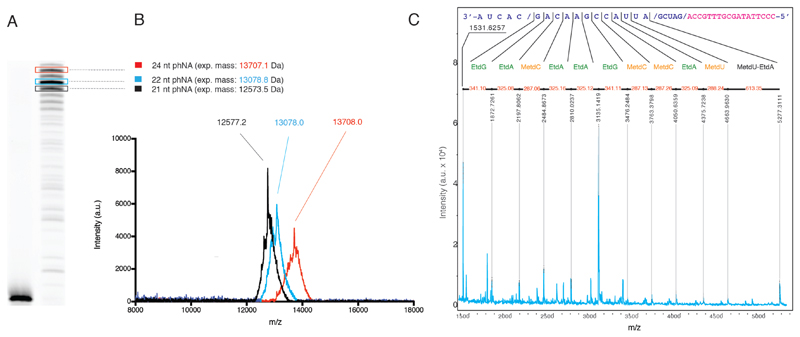
Wholesale replacement of the polyanionic backbone of DNA with a near-isosteric but uncharged, alkyl-substituted analogue in phNA would be expected to alter a polymer’s bulk physicochemical properties. Indeed, mixed sequence P-MetEt-phNA strands (despite covalent linkage to DNA sequences used to prime synthesis) exhibited substantially altered (i.e. retarded) mobility in polyacrylamide gel electrophoresis compared to equivalent DNA strands (Fig. 4d) (as observed previously for short P-Met-dT oligomers20), presumably due to the reduced charge to mass ratio. However, electrophoretic mobility shifts can occur as a result of many effects, including the possibility that P-alkyl substituents lead to an increased hydrodynamic radius of phNA compared with DNA. To conclusively verify DNA-templated synthesis of a defined phNA sequence by the PGV2 polymerase we analyzed an enzymatically synthesized P-MetEt-phNA strand (comprising the full-length product (n) as well as early termination products (n-2 and n-3) using MALDI-TOF (Matrix Assisted Laser Desorption/Ionization - Time-of-Flight) mass-spectrometry (MS) (Fig. 5a-b). Expected masses of phNA oligomers were observed. We also sought to determine a partial phNA sequence using MS-fragmentation. The divergent masses of P-Et-phNA purines and P-Met-phNA pyrimidines were clearly observed and allowed sequence determination, confirming templated synthesis and genetic information transfer from DNA to P-MetEt-phNA (Fig. 5C).
Figure 5. Mass-spectrometry analysis of phNA synthesis.
a) P-MetEt-phNA primer extension products on template TempN25, Supplementary Table 6) including full-length synthesis (red), n-2 (cyan) and n-3 (black) products are PAGE-purified for characterization for mass-spectrometry (MS). b), MALDI-TOF MS of each purified phNA oligomer excised from PAGE showing expected and measured masses; a.u., arbitrary units; m/z, mass-to-charge ratio. c) Fragmentation analysis using MALDI-ISD allows direct sequencing of part of the phNA oligomer confirming sequence and clearly resolving purine P-Et- and pyrimidine P-Met phNA nucleotides.
However, sequencing by MS fragmentation at best provides partial sequence information and a crude measure of the fidelity of information transfer. Therefore, we next sought to establish whether P-MetEt-phNA oligomers could be reverse transcribed and the encoded genetic information be transferred back to cDNA allowing standard sequencing. This would require a phNA-reverse transcriptase (RT) capable of synthesizing complementary DNA from a phNA template as previously achieved for a number of XNAs6, 37. This proved a challenging proposition not only due to the weakened interaction of the uncharged, sterically bulky phNA template with the polymerase as well as the known poor stability of Sp phNA-DNA heteroduplexes, which substantially reduce melting temperatures17, 38, 39. Earlier work on the S-isomer of glycerol nucleic acids (S-GNA), which pairs only weakly with DNA, had shown that the polymerase active site can stabilize a weak heteroduplex (DNA x S-GNA) to enable templated synthesis40. However, we failed to detect either binding or extension of DNA primers on enzymatically synthesized P-MetEt-phNA templates. We therefore resorted to poly-dA tailing of an enzymatically synthesized 72-nt P-MetEt-phNA oligomer strand. Although Terminal transferase (TdT) had previously been described to be able to utilize P-Met-phdTTPs as substrates21, it proved inefficient at synthesizing a poly-dA tail on a phNA oligomer. However, the engineered Tgo 6G12 polymerase, previously used to poly-dA tail HNA (hexitol nucleic acid)6 also allowed efficient poly-dA tailing of phNA. This allowed stable primer binding and oligo-dT-primed reverse transcription by the engineered RT521L reverse transcriptase. Although phNA reverse transcription remained inefficient (Supplementary Fig. 8), the resulting cDNA product could be amplified by RT-PCR, cloned and sequenced, revealing synthesis of the correct sequence with an aggregate fidelity (comprising both P-MetEt-phNA synthesis and reverse transcription) of 5.1 x 10-3 (Supplementary Table 1), at the lower end of previously studied XNAs systems6. Further studies developing more efficient phNA reverse transcription may be needed to provide a more comprehensive picture of phNA replication fidelity.
Aptamers are synthetic ligands composed of single-stranded (ss) nucleic acids that can be selected to bind target molecules with high affinity and specificity and have typically been evolved using polyanionic backbones either composed of RNA or DNA, or analogues such as 2’O-methyl RNA (2’Ome-RNA)41, or more recently from XNA backbones such as Fluoroarabino nucleic acids (FANA), hexitol nucleic acids (HNA) and TNA6, 42, 43, 44, although an uncharged PNA version of a DNA anti-thrombin aptamer has been described45. The capacity for phNA synthesis developed herein now opened the opportunity to explore whether uncharged phNA backbones could support not only information storage but also Darwinian evolution by the de novo selection of uncharged phNA aptamers directly from random-sequence P-MetEt-phNA repertoires. Due to the relative inefficiency of phNA reverse transcription, we chose to adapt a previously described ‘DNA display’ strategy43, 46 (Supplementary Fig. 9), involving display of a ss-phNA attached to the encoding double-stranded DNA, for phNA aptamer selection. We reasoned that this approach would also allow potential solubility issues of the uncharged phNA oligomers to be decoupled from the aptamer discovery process and enable high stringency selection conditions due to the higher sensitivity of PCR compared to RT-PCR.
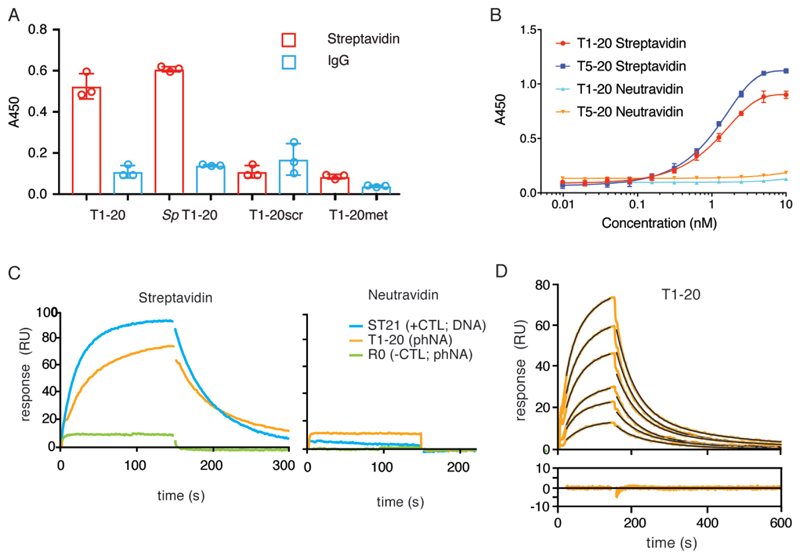
Starting from a P-MetEt-phNA library containing a 25 nt random sequence (N25) displayed on a 5’-tethered DNA duplex region (Supplementary Fig. 9), we performed 5 rounds of selection against immobilized streptavidin (SA), a well-characterised aptamer protein target47. Next-generation sequencing of selection pools revealed the emergence of a dominant sequence (Supplementary Table 2). We chose to characterize this dominant clone (“T1”) as well as another sequence that was less strongly enriched (“T5”). Both molecules bound strongly to SA as judged by an Enzyme-Linked Oligonucleotide Assay (ELONA) (Supplementary Fig. 10a-b). In order to dissect the contributions of different molecular domains to SA binding, we investigated a range of variants to the phNA aptamer structure. Aptamer variants (T1-20 and T5-20) in which the last 20 bases of the phNA sequence (corresponding to the conserved primer binding site) at the 3’ end were trimmed, showed a similar specific SA binding as judged by ELONA (Fig 6a, Supplementary Fig. 10b), with no more than background binding to control proteins IgG or neutravidin (an unrelated biotin binding protein derived from G. gallus avidin) (Fig. 6b). Likewise, scrambled control T1-20scr (and T1scr) showed no binding to SA (or IgG) (Fig. 6a, Supplementary Fig. 10b) indicating that the precise T1-20 phNA sequence is required for SA binding.
Figure 6. Characterisation of phNA aptamers.
A) ELONA assay of different aptamer clones for binding to Streptavidin (SA)(red) or a control protein (immunoglobulin (IgG) (cyan)) comprising phNA aptamers T1-20 (Supplementary Fig. 10), Sp T1-20 (T1-20 synthesized from purified Sp phNTP mix (Supplementary Figs 2, 12), T1-20scr (sequence scrambled T1-20) and T1-20met (T1-20 synthesized from all P-Met-phNTPs) B) Concentration dependent binding (ELONA) of T1-20 / T5-20 aptamers to SA (red / blue) and Neutravidin (NA) (orange / cyan). High-lighting the specificity of the selected clones. C, SPR sensogram showing binding of DNA anti-SA aptamer ST2147 (cyan) and phNA aptamers T1-20 (orange), R0 (green) to SA (left panel) and NA (right panel). D, SPR sensograms with residuals of the curves fit for T1-20 aptamer. Summary of aptamer binding and kinetics determined from SPR (Supplementary Fig. 11, Supplementary Table 3).
ELONA titration suggested binding affinities in the low nanomolar range but in order to obtain more precise measurements of dissociation constants (KD), we examined kinetics of binding to SA by surface plasmon resonance (SPR). These measurements suggested affinities for SA in the mid nanomolar range with the trimmed phNA aptamers T1-20 and T5-20 showing tighter binding for T1-20: KD = 90-240nM; T5-20: KD = 50-80nM (Fig. 6c-d, Supplementary Fig. 11, Supplementary Table 3) broadly comparable to the previously described DNA aptamer ST21 (KD = 40nM)47.
We sought to further map out the SA binding motif by synthesizing a range of variants of the T1 phNA aptamer (Fig. 6a, Supplementary Fig. 10b-c). First, we examined the effect of the phNA backbone alkyl substituents on aptamer binding. To this end we synthesized T1 and T1-20 variants with an alternative phNA backbone composition replacing the mixed P-MetEt-phNA backbone of the selected aptamer with an all P-Met- (T1met, T1-20met) or all P-Et-phNA backbone (T1et). In both cases SA binding was reduced to near background levels as judged by ELONA (Fig. 6a, Supplementary Fig. 10b) suggesting that not only the sequence, but the precise arrangement of methyl and ethyl substituents on the phNA backbone is critical for the formation of the SA binding motif.
In the DNA-display approach to aptamer selection we used, the phNA aptamer motif remains attached to a piece of double-stranded (ds) DNA encoding its sequence. In order to determine the potential contribution of the dsDNA tail to aptamer binding, we prepared a range of aptamer T1 aptamer variants in which one of the DNA strands (the primer strand) was replaced by a 2’-Omethyl-RNA (forming a DNA x 2’OmeRNA heteroduplex) (T1Ome), as well as variants with a chimeric RNA x DNA primer strand (retaining just the final five DNA nucleotides) (5T1) or an all-DNA primer strand (1T1) with just a single dT-FAM (6-carboxyfluorescein) residue positioned at the junction. The 5T1 allowed removal of the RNA portions using RNase digestion (and dissociation of the complementary DNA strand) yielding 5T1x, which comprises the all phNA aptamer domain retaining a ssDNA tail of just dN5 deoxinucleotides. In the 1T1 aptamer nuclease digestion with Bal31(removing both DNA primer and complementary sequence) yielded 1T1x, comprising the T1 phNA aptamer domain with just a single 5’-FAM-dTp residue. While T1Ome, 5T1 (and 1T1) showed essentially unchanged SA binding (Supplementary Fig. 10b-c), indicating that the chemical nature of the ds tail (DNA x DNA, DNA x 2’OmeRNA, DNA x RNA) does not influence SA binding. However, binding of 5T1x was reduced, suggesting potential interference by a single-stranded pentanucleotide DNA tail. In contrast, both 1T1 and 1T1x retained comparable and near-full SA binding activity (Supplementary Fig. 10c). All together, these results suggest that SA binding is mediated by the phNA segment, with both phNA sequence and the nature and sequence of P-alkyl substituents critical for binding, while the dsDNA portion makes little or no contribution to the functional aptamer motif (Fig. 6; Supplementary Fig. 10).
Finally, both binding of both T1 and T5 phNA aptamers to SA could be abolished by addition of biotin (Supplementary Fig. 10c), again underlining aptamer specificity and suggesting that the T1 and T5 phNA aptamers bind at (or close to) the biotin binding pocket, which is known to be rich in hydrophobic residues48 or alternatively may interact with the flexible binding loop, which undergoes a substantial conformational rearrangement upon biotin binding49.
Discussion
All natural nucleic acid substrates are polyanionic. Consequently, both the active site and primer-template interaction surface of DNA (and RNA) polymerases from all polymerase families are highly adapted to these highly negatively charged substrates and comprise numerous cationic amino acids, which form a near-continuous positively charged surface with a strong cationic charge potential. Furthermore, both cationic (Lys, Arg) and hydrophilic (Ser, Thr, Asn, Gln) amino acids mediate key ionic and hydrogen-bonding interactions with the incoming substrate, transition state and nascent strand duplex (Fig. 2, 3). While we and others have previously succeeded in engineering polymerases for the synthesis and reverse transcription of a range of xeno nucleic acids (XNAs)50 not found in nature, all of these retain the canonical phosphodiester backbone of DNA and RNA. Enzymatic synthesis and reverse transcription of nucleic acids with a modified backbone chemistry at full substitution has only been reported for phosphorothioate (PS) DNA11, an essentially isosteric and isoelectronic analogue of DNA that retains full negative backbone charge. Thus, despite a nearly isosteric structural configuration at the nucleotide level, incorporation of phNTP substrates (comprising reduced negative charge compared to dNTPs) as well as synthesis of uncharged phNA backbones represent a new category of challenge for polymerase engineering. Indeed, the wholesale replacement of the negatively charged phosphodiester with an uncharged P-alkyl-phosphonodiester backbone in the nascent strand required multiple mutations in the nascent-strand interaction surface to support phNA synthesis (Fig. 3).
Our engineering strategy involved the reconfiguration of key ionic interactions by substituting highly conserved lysine and arginine residues for smaller uncharged and hydrophobic amino acids (glycine, valine) in order to reduce the positive surface charge potential and steric bulk of the polymerase nascent strand interaction interface (Fig. 3), followed by further improvements through in vitro evolution. The resulting engineered polymerase PGV2 (RT521L8: D455P, K487G, R606V, R613V) enabled the general template-directed synthesis of mixed P-Met- / P-Et-phNA (Fig. 4) as well as all-P-Et-phNA (albeit on a more restricted set of sequences) (Supplementary Fig. 6) and opens up the sequence space of uncharged P-alkyl-phNAs for exploration as shown here by the in vitro evolution of the first phNA aptamers (Fig. 6, Supplementary Fig. 10).
We observed an improvement in the efficiency of phNA synthesis with an all P-Met-phNTP blend (Supplementary Fig. 12a) suggesting that even in the purine P-Et-phNTPs there remain steric incompatibilities in the active site and / or the nascent strand -polymerase interaction. Indeed, comparison of the MD simulations of the ternary P-Met-phATP- vs. P-Et-phATP polymerase holoenzyme complex (Fig.1, Supplementary Fig. 2) suggests a more cognate, catalytically-poised complex for the P-Met-phATP. This is evident both from the slightly shorter reaction coordinate and the more favourable placement of the Pα between the two catalytic metal ions, allowing coordination of both MgA and MgB by the non-bridging oxygen. Finally, the internuclear distance between MgA-MgB is closer to a ready-to-react Michaelis-Menten complex (± 3.8 Å), a critical parameter to define a well-ordered precatalytic complex28.
Steady-state kinetics of nucleotide incorporation of dNTPs vs P-Et- / P-Met-phNTPs (Supplementary Table 5) suggests the loss of the aforementioned interaction leads to an increase in Km of up to 100-fold in both the “wild-type” RT521L and the engineered PGV2 polymerase, resulting in inefficient utilization of phNTPs. On the other hand, kcat (and Vmax) values remain essentially identical, suggesting that a fractional negative charge likely is retained on the phNTP Rp oxygen (due to polarity of the phosphorus – oxygen double bond) and may be sufficient to allow productive interaction with the active site MgA and MgB ions and enable the catalytic step. This would imply that a full negative charge may not be required for the polymerase catalytic step, boding well for the development of enzymatic synthesis strategies for other uncharged (or charge-reduced) backbones.
Currently, engineered polymerases are available for only for P-Met- and P-Et- (and mixed P-Met-, P-Et-) phNA synthesis, but other substituents should in principle be accessible to enzymatic synthesis, including the near isosteric and isoelectronic Perfluoro-methyl and ethyl substituents as well as substituted alkyls. Bulkier alky or other substituents would likely require further engineering of both the polymerase active site and nascent strand interaction surface due to steric incompatibilities. Nevertheless, polymerase engineering should allow extension of P-substituents to a wider range of larger, branched or unsaturated hydrocarbon chains, which would afford opportunities for post-synthetic modification51 as well as increasingly hydrophobic characteristics. Indeed, P-MetEt-phNA bulk physicochemical properties already diverge substantially from canonical nucleic acids (as well as previously explored XNAs). The transition to an uncharged hydrophobic backbone in phNAs significantly reduces polarity and hence solubility in aqueous solvents. However, for phNA synthesis and also characterization it is desirable for aqueous solubility to be maintained, at least to some degree, as neither polymerases nor standard molecular biology assays (e.g. ELONA) are currently able to function in non-aqueous solvents. We have exploited the fact that aqueous solubility of phNAs can be readily modulated by conjugation to DNA (or charged XNA) oligonucleotides (e.g. derived from primers and/or templates). Indeed, while the anti-streptavidin phNA aptamers T1 and T5 (Fig. 6, Supplementary Fig. 10) with a mixed P-Met /-Et-phNA backbone retained full solubility in aqueous buffers while attached to a 20-mer DNA duplex. Aqueous solubility was reduced when the duplex was shortened or removed. However, a DNA pentanucleotide (5’-FAM-dNp5) (equivalent to a six negative charges, 5T1x), and even as little as a single 5’-FAM-dTp residue (equivalent to just two negative charges (1T1x), was sufficient to maintain low micromolar solubility in aqueous solution in the presence of a detergent (Tween20), as well as SA binding as judged by ELONA (Supplementary Fig. 10c).
Substitution of one of the non-bridging oxygens at the Pα phosphorus with an uncharged alkyl group creates a new stereocenter and both P-Met- and P-Et-phNTPs are synthesized as a racemic mixture of roughly equal proportions of the Rp - and Sp diastereoisomers. Upon separation by HPLC, only one of the diastereoisomers is incorporated by the polymerase (Supplementary Fig. 2) by the polymerase as would be expected based on previous observation with phosphorothioates (PS-dNTPs)25 and boranophosphates (PB-dNTPs)12, which bear an analogous stereochemical configuration as well as early data on incorporation of P-Met-phTTP by terminal transferase (TdT)21, which identified the Sp diastereoisomer as the polymerase substrate. Indeed, our own data on HPLC mobility (for the P-Met-phNTPs) as well as modelling and MD simulation of the P-Met-phATP and the P-Et-phATP polymerase holoenzyme complex very strongly indicate that only the Sp configuration is compatible with polymerase incorporation (Fig. 1, Supplementary Fig. 3). Furthermore, as established previously the hybridization of Sp-phNA with DNA is weakened compared to Rp -phNA-DNA heteroduplexes, which show similar stability to DNA homoduplexes17, 38, 39. We determined the thermal stability of a partial DNA x Sp -phNA duplex (synthesized by PGV2) and compared it to the equivalent DNA duplex and indeed found only very weak stabilization by the Sp-phNA x DNA interaction (ca. 0.15 °C / base pair (bp) vs. 0.58°C / bp for DNA)(Supplementary Fig. 13, Supplementary Table 4). Taken together, our data strongly suggests that only Sp phNTPs are the substrates for polymerases. Furthermore, again by analogy with PS-dNTPs, the “wrong” diastereoisomer is a sufficiently poor substrate that it does not efficiently compete and inhibit polymerase activity. Consequently, a racemic substrate mixture yields essentially identical product yields (Supplementary Fig. 12b) as a diastereochemically pure phNTP mix. Finally, another clear indication of polymerase stereoselectivity from a racemic mix is that the T1-20 phNA aptamer shows similarly strong binding (and specificity) when synthesized from a stereopure (Sp T1-20) as from a racemic phNTP mix (Fig. 6a).
The template-directed synthesis, reverse transcription and evolution of P-Met- / P-Et-phosphonate nucleic acid polymers (phNAs) described herein clearly demonstrates that genetic information storage, propagation and evolution are compatible with and can be performed by an uncharged backbone chemistry. However, in many ways the phNA “genetic system” remains beholden to a polyelectrolyte template (and / or appendix). For example, phNA replication requires both a DNA template for synthesis and yields a DNA product strand in reverse transcription and therefore effectively proceeds via a polyanionic intermediate (phNA -> DNA -> phNA). Furthermore, at least in the limited sequence space explored by phNA aptamers some charged groups are required, even though just one to two negative charges appear sufficient to retain solubility and binding activity (Supplementary Fig. 10c). Therefore, while our results indicate that a charged backbone (polyelectrolyte) chemistry may not be a strict prerequisite for genetic information storage, propagation and evolution, it is currently still required at intermediate stages of the genetic cycle.
The enzymatic synthesis of phNA oligomers provides a platform for the isolation of phNA aptamer ligands, with the future possibility of phNA catalyst (phNAzyme) discovery, which may allow the expansion of nucleic acid catalysis into non-aqueous solvents. Future advances should allow for a generalization of this approach and extension to phNA isomorphs and potentially other uncharged or charge neutral genetic polymers. By providing access to the sequence space of increasingly diverse genetic polymers with non-canonical uncharged backbone chemistries, these approaches should expand the synthetic genetics toolbox and enhance the potential for the discovery of novel ligands, catalysts and nanostructures with physicochemical and pharmacological properties divergent from those of DNA and RNA. Such studies should also allow for a deeper investigation into the chemical parameters most critical for genetic function.
Materials and Methods
Chemical Synthesis of α-P-Alkyl-2’-deoxynucleoside Triphosphate analogs
The synthesis of phNTPs was carried out using a method analogous to that of Dyatkina21, with a few modifications. For the synthesis of P-α-ethyl-phosphonate dNTPs we used Ethylphosphonic dichloride as the alkylphosphonylating reagent, whereas Methylphosphonic dichloride was used for the pilot-scale synthesis of P-α-methyl-dUTP and P-α-methyl-dCTP. For the majority of our large-scale synthesis of phNA, we used custom-synthesized phNTPs from Jena (P-α-methyl-dCTP) or GeneACT (P-α-methyl-dUTP and P-α-ethyl-dATP).
Primer Extension Assays
Typical primer extension reaction mixes were composed of the following: 100μM nucleoside triphosphate and analogues thereof; 1μM primer; 2μM template; 0.5 mg /mL polymerase 1x Thermopol buffer (NEB) supplemented by 4mM MgSO4. Typical primer extension experiments were carried out using Peltier thermal cyclers and PCR plates or tubes. Primers were annealed to templates by incubation at 95° C for 1 min, then slowly cooling to 4°C. Typical primer extensions were carried out for five cycles of 1 hour at 50°C followed by 15min at 65°C.
Polymerase Structure-Guided mutagenesis and directed evolution
Site-directed mutagenesis, directed evolution and screening of polymerase mutants was carried out using established methods as detailed by Pinheiro, Cozens et al.3–5, with the use of phNTPs during screening for polymerase activity and selection experiments being the only significant change. Detailed protocols are available in the Supplementary Materials and Methods.
Mass Spectrometry of polymerase-synthesized phNA
We synthesized phNA from a DNA primer oligonucleotide “FD” (5’-CCCCTTATTAGCGAAAGCCA-3’) using a template encoding for 25 phNA incorporations TSD (5’-ATGCTGTATCGCGTATGCATCGCATCTGGCAAACGCTAATAAG-GGG-3’). Three different products of the primer extension were PAGE purified on a denaturing 8M Urea gel, excised from the gel, electroeluted, isopropanol-precipitated and desalted by dialysis using a 2kDa cut-off membrane (Thermo). Oligo samples, 0.6 μl in water, were spotted onto a MALDI target followed by 0.6 μl of 3-hydroxypicolinic acid. Some oligo samples were vacuum dried, resuspended in 25 μl, 0.1 M TEAA (triethylammonium acetate) and desalted using a zip-tip C18 (Merck Millipore, USA). The zip-tip C18 was washed 3× 10 μl of 0.1 M TEAA and then 3× 10 μl water. Next, the oligo was eluted directly onto a MALDI target with 5 μl of 3-hydroxypicolinic acid. All mass spectrometric measurements were carried out in positive ion mode on an Ultraflex III TOF-TOF instrument (Bruker Daltonik).
MALDI-ISD experiments
30μl -70 μl of the given samples were concentrated in a Speed Vac (Savant) to less than 10ul and centrifuged to settle some pink precipitates. The supernatant were removed and 1 μl of 50% MeCN was added to re-solubilise the precipitates. 0.6 μl of each sample was spotted onto a MALDI target followed by 0.6 μl of 3-hydroxypicolinic acid and air-dried. The dried sample spots were washed with 1 μl of water, air-dried and further 0.6 μl of matrix was added. All ISD experiments were carried out in positive ion mode on an Ultraflex III TOF-TOF instrument (Bruker Daltonik,). The spectra were processed using Flex Analysis v3.3 (Bruker Daltonik) and annotated manually.
Reverse Transcription of phNA
The scheme is summarized in Supplementary Fig. 8. phNA was transcribed from DNA template TnD (5’-CCCTCCTTCTTCCTCTTCCCCTCACGATGCTGCGACTCAGATA CAGCACTTAGCCACGTAGTGCTGTTCGGTACATCGATCTGGCAAACGCTAATCAGGGG CTATTATTAATT-3’) using the 6-FAM labelled NAPfd DNA primer (5’ -FAM-CAGTATCGACAAAGGACCCCTTATTAGCGTTTGCCA-3’). Transcription products were PAGE-purified and isopropanol-precipitated before polydeoxyadenylation using 6G12 polymerase (1μgmL-1) and 1mM dATP for 2hr at 65°C. Poly(dA)-phNA oligomers were purified by isopropanol precipitation, then reverse-transcribed using the biotinylated bL3polyT primer (5’- Biotin-TEG-CAGGAAACAGCTATGACAAAT(24) -3’), RT521L polymerase (0.5μgmL-1), 1mM dNTPs at 50°C for 1 hr, 55°C 1hr, 60°C 1hr and 65°C 1hr. The RT products were bound to M1 Streptavidin-coated magnetic beads (Invitrogen), washed with bead-washing buffer (10mM Tris-HCl, 200mM NaCl, 0.1% Tween-20) and the washed beads were preserved in elution buffer (10mM Tris-HCl, 0.1%Tween-20). Beads were used directly in PCR reactions using the NAP and L3 primers (5’-CAGTATCGACAAAGGA-3’, 5’-CAGGAAACAGCTATGACAA-3’). Semi-outnesting PCR was carried out using the NAP/Ts7 primer pair (NAP and Ts7 5’-CCCTCCTTCTTCCTCTTCCC-3’). PCR products were cloned using the TOPO-TA cloning kit (Invitrogen) and cloned products were transformed into 10β cells (NEB), and individual colonies were sequenced by Sanger sequencing.
Selection of phNA aptamers against Streptavidin
The selection scheme is summarized in Supplementary Fig. 9. Detailed protocols for the selection are described in the Supplementary Materials and Methods.
ELONA streptavidin binding assays
Selected and control phNA aptamer sequences were synthesized from a FITC-labelled DNA primer (or 2’O-methyl primer) and a DNA template and PAGE-purified as described previously. For binding assays the DNA portion of the phNA aptamer was hybridized to its complementary sequence at a 1:1 ratio. All sequences were hybridized and assayed in phosphate-buffered saline (PBS) and 0.1% Tween. Aptamer clones were bound to Streptavidin wells (or Neutravidin, or Goat IgG controls) for 1 hour, then unbound aptamers were washed three times with buffer, and an anti-FITC Fab-HRP (Roche) was bound at 0.15 units mL-1, and unbound Fab was removed. After washing the wells three times with wash buffer, the assay was developed using Ultra-TMB ELISA reagent (Thermo). Peroxidase reactions were stopped with 1M H2SO4 before absorbance measurements (450 nm).
Surface Plasmon Resonance with phNA aptamers
Surface Plasmon Resonance (SPR) measurements were made using a BIAcore 2000 instrument (GE Healthcare UK Ltd., UK) at a flow rate of 20 μl min-1 in running buffer (140 mM NaCl, 2.5 mM KCl, 1.6 mM KH2PO4, 15 mM Na2HPO4, 0.02% Tween-20, pH 7.5) at 20°C. For all experiments, immobilized streptavidin (S4762-.5MG, Sigma) or neutravidin (Pierce) surfaces (~4000 or 20,000 RU per flow cell respectively) were prepared using an amine coupling kit (GE Life Sciences UK Ltd, UK) with CM4 sensor chips (GE Life Sciences UK Ltd, UK) in 5 mM sodium acetate, pH 5.5, then equilibrated overnight in running buffer. DNA or phNA aptamers in running buffer were injected for 150 s at a series of concentrations (500 nM, 250 nM, 125 nM, 62.5 nM and 31.3 nM) and dissociation monitored for 1,200 s. Binding data was double-referenced by subtracting data from a blank flow cell as well as molar equivalent injections of “R0”. Data were fitted to a parallel association with biphasic dissociation model by non-linear least squares.
Supplementary Material
Acknowledgments
This work was supported by a Trinity College Cambridge fellowship (S.A-F.) and by the Medical Research Council (S.A-F. A.T., B.T.P, S.P-C., P.H., program no. MC_U105178804) and NICHD/ NIH Intramural Research Program (A.V. & R.W.) and a European Molecular Biology Organization (EMBO) Long-Term Fellowship (VG, ALTF 103-2018).
Footnotes
Contributions
S.A-F. and P.H. conceived and designed the experiments. S.A-F. performed all experiments except SPR measurements (A.T, B.T.P.), Mass spectrometry (S.P-C) and steady-state kinetics (A.V., R.W.) and Modelling and MD simulations (V.G., M. O.). All authors discussed the results, and jointly wrote the manuscript.
References
- 1.Westheimer FH. Why nature chose phosphates. Science. 1987;235(4793):1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 2.Benner SA. Understanding Nucleic Acids Using Synthetic Chemistry. Acc Chem Res. 2004;37(10) doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 3.Benner SA, Hutter D. Phosphates, DNA, and the search for nonterrean life: a second generation model for genetic molecules. Bioorg Chem. 2002;30(1):62–80. doi: 10.1006/bioo.2001.1232. [DOI] [PubMed] [Google Scholar]
- 4.Malyshev DA, Romesberg FE. The Expanded Genetic Alphabet. Angew Chem Int Ed. 2015;54(41):11930–11944. doi: 10.1002/anie.201502890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr Op Chem Biol. 2012;16(3–4):245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- 6.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336(6079):341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AI, Pinheiro VB, Smola MJ, Morgunov AS, Peak-Chew S, Cozens C, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518(7539):427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509(7500):385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Cozens C, Jaziri F, Rozenski J, Marechal A, Dumbre S, et al. Phosphonomethyl Oligonucleotides as Backbone-Modified Artificial Genetic Polymers. J Am Chem Soc. 2018;140(21):6690–6699. doi: 10.1021/jacs.8b03447. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SL, Blain JC, Zielinska D, Gryaznov SM, Szostak JW. Fast and accurate nonenzymatic copying of an RNA-like synthetic genetic polymer. Proc Natl Acad Sci USA. 2013;110(44):17732–17737. doi: 10.1073/pnas.1312329110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, et al. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nature biotech. 2004;22(6):755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 12.Shaw BR, Dobrikov M, Wang X, Wan J, He K, Lin JL, et al. Reading, writing, and modulating genetic information with boranophosphate mimics of nucleotides, DNA, and RNA. Ann N Y Acad Sci. 2003;1002:12–29. doi: 10.1196/annals.1281.004. [DOI] [PubMed] [Google Scholar]
- 13.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37(47):16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 14.Meng M, Ducho C. Oligonucleotide analogues with cationic backbone linkages. Beilstein J Org Chem. 2018;14:1293–1308. doi: 10.3762/bjoc.14.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen PE. DNA analogues with nonphosphodiester backbones. Annu Rev Biophys Biomol Struct. 1995;24:167–183. doi: 10.1146/annurev.bb.24.060195.001123. [DOI] [PubMed] [Google Scholar]
- 16.Steinbeck C, Richert C. The role of ionic backbones in RNA structure: An unusually stable non-Watson-Crick duplex of a nonionic analog in an apolar medium. J Am Chem Soc. 1998;120(45):11576–11580. [Google Scholar]
- 17.Micklefield J. Backbone modification of nucleic acids: synthesis, structure and therapeutic applications. Curr Med Chem. 2001;8(10):1157–1179. doi: 10.2174/0929867013372391. [DOI] [PubMed] [Google Scholar]
- 18.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489(1):141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr Issues Mol Biol. 1999;1(1–2) [PubMed] [Google Scholar]
- 20.Dineva MA, Chakurov S, Bratovanova EK, Devedjiev I, Petkov DD. Complete template-directed enzymatic synthesis of a potential antisense DNA containing 42 methylphosphonodiester bonds. Bioorgan Med Chem. 1993;1(6):411–414. doi: 10.1016/s0968-0896(00)82151-3. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi H, Endo T, Kaji A. Enzymic synthesis of oligonucleotides containing methylphosphonate internucleotide linkages. Biochemistry. 1990;29(37):8747–8753. doi: 10.1021/bi00489a035. [DOI] [PubMed] [Google Scholar]
- 22.Brudno Y, Birnbaum ME, Kleiner RE, Liu DR. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat Chem Biol. 2010;6(2):148–155. doi: 10.1038/nchembio.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami A, Blake KR, Miller PS. Characterization of sequence-specific oligodeoxyribonucleoside methylphosphonates and their interaction with rabbit globin mRNA. Biochemistry. 1985;24(15):4041–4046. doi: 10.1021/bi00336a036. [DOI] [PubMed] [Google Scholar]
- 24.Arzumanov AA, Dyatkina NB. An Alternative Route for Preparation of Alpha-Methylphosphonyl-Beta,Gamma-Diphosphates of Thymidine Derivatives. Nucleos Nucleot. 1994;13(5):1031–1037. [Google Scholar]
- 25.Burgers PMJ, Eckstein F. Stereochemistry of Internucleotide Bond Formation by Polynucleotide Phosphorylase from Micrococcus-Luteus. Biochemistry. 1979;18(3):450–454. doi: 10.1021/bi00570a010. [DOI] [PubMed] [Google Scholar]
- 26.Xia S, Konigsberg WH. Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant. Protein Sci. 2014;23(4):508–513. doi: 10.1002/pro.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genna V, Gaspari R, Dal Peraro M, De Vivo M. Cooperative motion of a key positively charged residue and metal ions for DNA replication catalyzed by human DNA Polymerase-eta. Nucleic Acids Res. 2016;44(6):2827–2836. doi: 10.1093/nar/gkw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genna V, Donati E, De Vivo M. The Catalytic Mechanism of DNA and RNA Polymerases. ACS Catal. 2018;8(12):11103–11118. [Google Scholar]
- 29.Genna V, Carloni P, De Vivo M. A Strategically Located Arg/Lys Residue Promotes Correct Base Paring During Nucleic Acid Biosynthesis in Polymerases. J Am Chem Soc. 2018;140(9):3312–3321. doi: 10.1021/jacs.7b12446. [DOI] [PubMed] [Google Scholar]
- 30.Genna V, Colombo M, De Vivo M, Marcia M. Second-Shell Basic Residues Expand the Two-Metal-Ion Architecture of DNA and RNA Processing Enzymes. Structure. 2018;26(1):40. doi: 10.1016/j.str.2017.11.008. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cozens C, Pinheiro VB, Vaisman A, Woodgate R, Holliger P. A short adaptive path from DNA to RNA polymerases. Proc Natl Acad Sci USA. 2012;109(21):8067–8072. doi: 10.1073/pnas.1120964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wynne SA, Pinheiro VB, Holliger P, Leslie AG. Structures of an apo and a binary complex of an evolved archeal B family DNA polymerase capable of synthesising highly Cy-dye labelled DNA. PloS One. 2013;8(8):e70892. doi: 10.1371/journal.pone.0070892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergen K, Betz K, Welte W, Diederichs K, Marx A. Structures of KOD and 9 degrees N DNA polymerases complexed with primer template duplex. Chembiochem. 2013;14(9):1058–1062. doi: 10.1002/cbic.201300175. [DOI] [PubMed] [Google Scholar]
- 34.Genna V, Vidossich P, Ippoliti E, Carloni P, De Vivo M. A Self-Activated Mechanism for Nucleic Acid Polymerization Catalyzed by DNA/RNA Polymerases. J Am Chem Soc. 2016;138(44):14592–14598. doi: 10.1021/jacs.6b05475. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487(7406):196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro VB, Loakes D, Holliger P. Synthetic polymers and their potential as genetic materials. BioEssays. 2013;35(2):113–122. doi: 10.1002/bies.201200135. [DOI] [PubMed] [Google Scholar]
- 37.Dunn MR, Chaput JC. Reverse Transcription of Threose Nucleic Acid by a Naturally Occurring DNA Polymerase. Chembiochem. 2016;17(19):1804–1808. doi: 10.1002/cbic.201600338. [DOI] [PubMed] [Google Scholar]
- 38.Thiviyanathan V, Vyazovkina KV, Gozansky EK, Bichenchova E, Abramova TV, Luxon BA, Lebedev A, Gorenstein DG. Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry. Biochemistry. 2002;41(3):827–238. doi: 10.1021/bi011551k. [DOI] [PubMed] [Google Scholar]
- 39.Vyazovkina EV, Savchenko EV, Lokhov SG, Engels JW, Wickstrom E, Lebedev AV. Synthesis of specific diastereomers of a DNA methylphosphonate heptamer, d (CpCpApApApCpA), and stability of base pairing with the normal DNA octamer d (TpGpTpTpTpGpGpC) Nucleic Acids Res. 1994;22(12):2404–2409. doi: 10.1093/nar/22.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai CH, Chen J, Szostak JW. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc Natl Acad Sci USA. 2007;104(37):14598–14603. doi: 10.1073/pnas.0704211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol. 2005;12(1):25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Alves Ferreira-Bravo I, Cozens C, Holliger P, DeStefano JJ. Selection of 2߰-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015;43(20):9587–9599. doi: 10.1093/nar/gkv1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nature Chem. 2012;4(3):183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 44.Rangel AE, Chen Z, Ayele TM, Heemstra JM. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 2018;46(16):8057–8068. doi: 10.1093/nar/gky667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee EJ, Lim HK, Cho YS, Hah SS. Peptide nucleic acids are an additional class of aptamers. RSc Adv. 2013;3(17):5828–5831. [Google Scholar]
- 46.Ichida JK, Zou K, Horhota A, Yu B, McLaughlin LW, Szostak JW. An in vitro selection system for TNA. J Am Chem Soc. 2005;127(9):2802–2803. doi: 10.1021/ja045364w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bing T, Yang XJ, Mei HC, Cao ZH, Shangguan DH. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorgan Med Chem. 2010;18(5):1798–1805. doi: 10.1016/j.bmc.2010.01.054. [DOI] [PubMed] [Google Scholar]
- 48.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Structural Origins of High-Affinity Biotin Binding to Streptavidin. Science. 1989;243(4887):85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 49.Freitag S, LeTrong I, Klumb L, Stayton PS, Stenkamp RE. Structural studies of the streptavidin binding loop. Protein Sci. 1997;6(6):1157–1166. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houlihan G, Arangundy-Franklin S, Holliger P. Engineering and application of polymerases for synthetic genetics. Curr Opin Biotechnol. 2017;48:168–179. doi: 10.1016/j.copbio.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Krishna H, Caruthers MH. Alkynyl phosphonate DNA: a versatile “click”able backbone for DNA-based biological applications. J Am Chem Soc. 2012;134(28):11618–11631. doi: 10.1021/ja3026714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.