Abstract
Summary
Methods for antibody structure prediction rely on sequence homology to experimentally determined structures. Resulting models may be accurate but are often stereochemically strained, limiting their usefulness in modeling and design workflows. We present the AbPredict 2 web-server, which instead of using sequence homology, conducts a Monte Carlo-based search for low-energy combinations of backbone conformations to yield accurate and unstrained antibody structures.
Availability & implementation
We introduce several important improvements over the previous AbPredict implementation: (1) backbones and sidechains are now modeled using ideal bond lengths and angles, substantially reducing stereochemical strain, (2) sampling of the rigid-body orientation at the light-heavy chain interface is improved, increasing model accuracy, and (3) runtime is reduced 20-fold without compromising accuracy, enabling the implementation of AbPredict 2 as a fully automated web-server (http://abpredict.weizmann.ac.il). Accurate and unstrained antibody model structures may in some cases obviate the need for experimental structures in antibody optimization workflows.
Introduction
Antibodies are the main soluble component of the mammalian immune system and have the ability to bind a virtually boundless repertoire of molecules with exquisite specificity and affinity(Janeway, 2005). The antibody variable fragment (Fv) comprises two chains, light and heavy, in each of which three hypervariable complementarity-determining regions (CDRs) encode most of the interactions with ligands. The CDRs rest on a conserved framework, which provides structural stability. Structural diversity in the Fv is observed primarily in the CDRs, which vary in length, backbone conformation, and amino acid sequence. The rigid-body orientation (RBO) of the light relative to the heavy chain is another important degree of freedom that affects the conformation of the antigen-binding site(Chailyan, Marcatili, & Tramontano, 2011). Owing to their versatility, antibodies are by far the most important class of protein used in biomedical research(Hattori et al., 2010; Nogi et al., 2008),(Lawson, 2012) and clinical applications(Li & Zhu, 2010; Reichert & Valge-Archer, 2007; Vezina, Cotreau, Han, & Gupta, 2017).
Structure determination is often an essential step in optimizing antibody affinity, specificity, and stability; but structure determination is time-consuming and may fail, particularly with antibodies that are not stable enough to be produced to homogeneity in large quantities. Structure prediction may bypass this obstacle, providing a way to design improved antibody variants directly from sequence, but both high accuracy and stereochemical quality are essential prerequisites for reliable design. Most antibody structure-prediction methods use sequence homology to natural antibody structures to select backbones for modeling. Automated web-servers, such as RosettaAntibody(Sircar, Kim, & Gray, 2009), PIGS(Marcatili, Rosi, & Tramontano, 2008), and SAbPred(Dunbar et al., 2016), start by identifying the segments corresponding to the CDRs and framework in the query sequence; template structures from the Protein Data Bank (PDB) are then chosen for each segment by sequence identity and the resulting structure model is refined by energy minimization. Previous assessments of antibody structure-prediction methods noted that although models were accurate, stereochemical strain was often high(Almagro et al., 2014).
By contrast with sequence-homology based methods, we recently described AbPredict, which uses Rosetta to search for combinations of backbone conformations observed in natural antibody structures(Norn, Lapidoth, & Fleishman, 2017). In a benchmark, we found that in some cases AbPredict improved accuracy in conformationally diverse segments, including HCDR2 and 3, relative to homology-based methods. For example, one of our benchmark targets (PDB entry: 4KMT) is identical in sequence to the HCDR3 of another antibody, but the root-mean-square deviation (rmsd) between these two HCDR3 backbones is 2.7 Å. Instead of selecting the sequence-identical but conformationally distant HCDR3 backbone, AbPredict selected as template an HCDR3 which exhibited <1 Å rmsd despite having ≤10% sequence identity to 4KMT(Norn et al., 2017). Furthermore, unlike many other methods(Weitzner, Kuroda, Marze, Xu, & Gray, 2014)-17, AbPredict does not require expert rules or post-filtering to produce accurate and unstrained models. These results encouraged us to further improve model accuracy, stereochemical quality, and runtime efficiency enabling the method’s implementation as a web-server (http://abpredict.weizmann.ac.il).
Implementation
With the intent of developing a fully automated web-server for non-expert users, AbPredict 2 only requires a fasta-formatted sequence of the query antibody. Starting from the query sequence, it uses HMMer(Eddy, 1998) to identify the sequences corresponding to the heavy and light variable domains and determine the type of light chain (λ or κ). Atomistic modeling starts with a random combination of four backbone fragments (two fragments for the Vl and Vh, which comprise CDRs 1 and 2 and the light and heavy chain framework regions, respectively, and two fragments for LCDR3 and HCDR3); it then threads the query sequence on the backbone fragments and performs a simulated annealing Monte-Carlo search over all conformational degrees of freedom. In each conformational move, the method randomly samples from precomputed databases a backbone conformation belonging to the Vl, Vh, LCDR3, HCDR3 or the RBO, followed by combinatorial sidechain packing and sidechain and backbone minimization to reduce stereochemical strain(Lapidoth et al., 2015). At the end of each trajectory, the lowest-energy structure sampled during the trajectory is selected. We introduced two improvements relative to the previous implementation of AbPredict. The previous AbPredict implementation used bond lengths and angles that were derived from a template antibody to model all query antibodies, leading to bias towards the template and to stereochemical strain in the models. By contrast, AbPrdict 2 models all backbone and sidechain bond lengths and angles with ideal values, substantially reducing stereochemical strain. Thus, the resulting structures produced by AbPredict 2 are more relaxed and therefore more suitable for downstream modelling, such as molecular-dynamics simulations or sequence design. Second, AbPredict 2 samples the RBO from a database of experimentally observed RBOs generated by superimposing all of the Fvs in the PDB on a common reference frame, thereby improving modeling accuracy. The server’s precomputed RBO and conformation databases are automatically updated as new antibody structures are deposited in the PDB, allowing for continued improvement in model quality as the PDB grows. Scripts, documentation, and instructions for executing AbPredict 2 independent of the web server are available in http://www.rosettacommons.org. Expert users may use this standalone version of AbPredict 2, which provides control over all modelling parameters, such as number of trajectories.
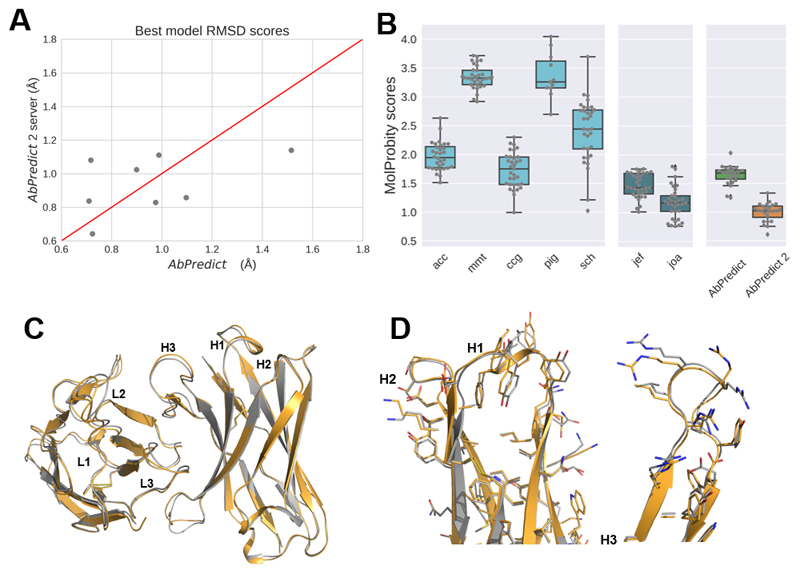
We analyzed the accuracy and stereochemical strain in models produced for eight query antibodies that were part of the AMA-II blind benchmark of antibody structure modeling methods(Almagro et al., 2014). For fair comparison, we eliminated from the conformation databases any structures deposited after the start of the AMA-II experiment (February, 2014). All the top ranked models produced by AbPredict 2 deviated by <1.2 Å rmsd over backbone-carbonyl atoms (Fig. 1A). Furthermore, according to MolProbity(Davis, Murray, Richardson, & Richardson, 2004), the top models exhibited stereochemical quality that is expected of structures at resolutions <1.2 Å (Fig. 1B) — an improvement even relative to methods that used MolProbity as a model-selection criterion(Shirai et al., 2014; Weitzner et al., 2014). Hence, model structures from AbPredict 2 are at the level of accuracy of AbPredict and exhibit substantially improved stereochemical quality relative to the previous version of AbPredict and all methods that participated in the AMA-II benchmark. These improvements may in some cases allow reliable design based on AbPredict 2 models without requiring experimental structures(Baran et al., 2017).
Figure 1. High accuracy and stereochemical quality in AbPredict 2 model structures.
(A) Comparison of models produced using AbPredict and the AbPredict 2 web-server for eight antibodies from the AMA-II benchmark, demonstrating similar model accuracy(Almagro et al., 2014). (B) Comparison of the MolProbity scores of AbPredict versus AbPredict 2 and all methods examined in the AMA-II benchmark(Almagro et al., 2014), demonstrating improvement in the stereochemical quality of the model structures in AbPredict 2 relative to all others. Note that joa(Shirai et al., 2014) and jef(Shirai et al., 2014; Weitzner et al., 2014) use MolProbity scores as a model-selection criterion. MolProbity scores represent the crystallographic resolution at which structures with similar stereochemical quality are observed (lower scores are stereochemically better)(Chen et al., 2010). The MolProbity score assesses sidechain and backbone dihedral outliers and steric overlaps. (C & D) Comparison of top anti-DNA Fv A52 model from the AbPredict 2 server (gold) and the crystal structure (PDB entry: 4M61, gray). Carbonyl rmsd: 0.6 Å, MolProbity score: 0.9.
We also improved the computational efficiency relative to the previous implementation of AbPredict. AbPredict 2 performs 50 simulated annealing Monte Carlo steps and a total of 500 trajectories (instead of 150 and 3,000, respectively, in AbPredict), resulting in 20-fold reduction in runtime. Depending on server load, total runtime from query submission to structure models is under an hour. The 500 resulting models are clustered by carbonyl rmsd and the lowest-energy structures from the top-three clusters are presented as the predicted models using JSmol(Hanson, Prilusky, Renjian, Nakane, & Sussman, 2013). The three models are also provided as PDB-formatted structures for download. Exemplary models produced by AbPredict 2 are shown in Figure 1C-D.
Acknowledgements
This research was supported by a European Research Council Starting Grant [335439], the Israel Science Foundation through its Center for Research Excellence in Structural Cell Biology and its joint India-Israel Research Program [1775/12; 2281/15], and a research grant from Anne Christopoulos and Carolyn Hewitt.
References
- Almagro JC, Teplyakov A, Luo J, Sweet RW, Kodangattil S, Hernandez-Guzman F, Gilliland GL. Second antibody modeling assessment (AMA-II) Proteins. 2014;82(8):1553–1562. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- Baran D, Pszolla MG, Lapidoth GD, Norn C, Dym O, Unger T, et al. Fleishman SJ. Principles for computational design of binding antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(41):10900–10905. doi: 10.1073/pnas.1707171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailyan A, Marcatili P, Tramontano A. The association of heavy and light chain variable domains in antibodies: implications for antigen specificity. The FEBS Journal. 2011;278(16):2858–2866. doi: 10.1111/j.1742-4658.2011.08207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Research. 2004;32(Web Server issue):W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J, Krawczyk K, Leem J, Marks C, Nowak J, Regep C, et al. Deane CM. SAbPred: a structure-based antibody prediction server. Nucleic Acids Research. 2016;44(W1):W474–W478. doi: 10.1093/nar/gkw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Hanson RM, Prilusky J, Renjian Z, Nakane T, Sussman JL. JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied toProteopedia. Israel Journal of Chemistry. 2013;53(3-4):207–216. [Google Scholar]
- Hattori T, Umetsu M, Nakanishi T, Togashi T, Yokoo N, Abe H, et al. Kumagai I. High affinity anti-inorganic material antibody generation by integrating graft and evolution technologies: potential of antibodies as biointerface molecules. The Journal of Biological Chemistry. 2010;285(10):7784–7793. doi: 10.1074/jbc.M109.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C. Immunobiology: The Immune System In Health And Disease. Harcourt Health Sciences. 2005 [Google Scholar]
- Lapidoth GD, Baran D, Pszolla GM, Norn C, Alon A, Tyka MD, Fleishman SJ. AbDesign: An algorithm for combinatorial backbone design guided by natural conformations and sequences. Proteins. 2015;83(8):1385–1406. doi: 10.1002/prot.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ADG. Antibody-enabled small-molecule drug discovery. Nature Reviews. Drug Discovery. 2012;11(7):519–525. doi: 10.1038/nrd3756. [DOI] [PubMed] [Google Scholar]
- Li J, Zhu Z. Research and development of next generation of antibody-based therapeutics. Acta Pharmacologica Sinica. 2010;31(9):1198–1207. doi: 10.1038/aps.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcatili P, Rosi A, Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24(17):1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- Nogi T, Sangawa T, Tabata S, Nagae M, Tamura-Kawakami K, Beppu A, et al. Takagi J. Novel affinity tag system using structurally defined antibody-tag interaction: application to single-step protein purification. Protein Science: A Publication of the Protein Society. 2008;17(12):2120–2126. doi: 10.1110/ps.038299.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norn CH, Lapidoth G, Fleishman SJ. High-accuracy modeling of antibody structures by a search for minimum-energy recombination of backbone fragments. Proteins. 2017;85(1):30–38. doi: 10.1002/prot.25185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nature Reviews. Drug Discovery. 2007;6(5):349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- Shirai H, Ikeda K, Yamashita K, Tsuchiya Y, Sarmiento J, Liang S, et al. Nakamura H. High-resolution modeling of antibody structures by a combination of bioinformatics, expert knowledge, and molecular simulations. Proteins: Structure, Function, and Bioinformatics. 2014;82(8):1624–1635. doi: 10.1002/prot.24591. [DOI] [PubMed] [Google Scholar]
- Sircar A, Kim ET, Gray JJ. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Research. 2009;37(Web Server issue):W474–W479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina HE, Cotreau M, Han TH, Gupta M. Antibody-Drug Conjugates as Cancer Therapeutics: Past, Present, and Future. Journal of Clinical Pharmacology. 2017;57(Suppl 10):S11–S25. doi: 10.1002/jcph.981. [DOI] [PubMed] [Google Scholar]
- Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3.0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins. 2014;82(8):1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]