Abstract
Bovine respiratory disease (BRD) is economically significant, and influenza D virus (IDV) is commonly identified in cattle with BRD. Mannheimia haemolytica (MHA) is an opportunistic bacterial contributor to BRD; surveillance data suggest that MHA and IDV co-infection occurs in cattle. The objective of this study was to evaluate the synergistic pathogenesis in cattle co-infected with IDV and MHA. Sixteen dairy calves were randomly assigned to four groups of four calves. The IDV+MHA+ group received D/bovine/C00046N/Mississippi/2014 (D/46N) intranasally at 0 days post-inoculation (DPI) and Mannheimia haemolytica D153 (MHA D153) intratracheally at 5 DPI. The IDV+MHA− group received only D/46N at 0 DPI; the IDV−MHA+ group received only MHA D153 at 5 DPI; and the IDV−MHA− group received neither agent. Clinical scores were calculated twice daily. At 10 DPI, IDV+MHA+, IDV−MHA+, and IDV−MHA− calves were euthanized and evaluated for pathologic lesions. The IDV+ groups seroconverted to IDV by 10 DPI. Clinical scores were higher in IDV+ groups than IDV− groups on 2–5 DPI (p = 0.001). After MHA challenge on 5 DPI, clinical scores (6–10 DPI) were slightly lower in IDV+MHA+ group than IDV−MHA+ group (p < 0.05) but not significantly different between MHA+ groups and MHA− groups. The average gross pathology score was higher for IDV−MHA+ group than groups IDV−MHA− and IDV+MHA+; however, no significant differences were identified among groups. Under the conditions of this study, infection with IDV before MHA enhance neither clinical disease nor lung pathology, relative to calves infected with MHA alone.
Keywords: Pathogenesis, influenza D virus, Mannheimia haemolytica, bovine respiratory disease, cattle
Introduction
Bovine respiratory disease (BRD) is one of the most economically significant diseases of the cattle industry. The pathogenesis is known to be multifactorial, with stress and a primary viral infection compromising host immunity to allow for a severe secondary bacterial infection of the lower respiratory tract, which may lead to pneumonia or death of the animal. A secondary bacterial infection develops because of immune suppression caused by the primary viral infection or other factors. Antibiotics and vaccines are used to limit infection. However, neither strategy completely reduces severity of BRD. Both bacteria (e.g. Mannheimia haemolytica [MHA], Pasteurella multocida, Histophilus somni, and Mycoplasma bovis) and viruses (e.g. bovine viral diarrhea virus, infectious bovine rhinotracheitis virus, bovine respiratory syncytial virus, and parainfluenza type-3 virus) are considered important BRD pathogens. Among these pathogens, MHA is one of the most commonly identified bacterial pathogens, and the clinical signs caused by MHA infection can last for several days post-challenge (Grissett et al., 2015).
In 2011, a novel influenza virus thought to be a new subtype of influenza C virus was isolated from swine in Oklahoma (Hause et al., 2013). Later, the virus was shown to be santigenically and genetically distinct from influenza C virus and was designated influenza D virus, or IDV (Hause et al., 2014). Serosurveillance suggested that domestic animals (i.e. swine, cattle, camelids, and small ruminants) throughout North America, Asia, Africa, and South America (Chiapponi et al., 2016; Ducatez et al., 2015; Ferguson et al., 2015; Foni et al., 2017; Foni et al., 2016; Hause et al., 2014; Hause et al., 2013; Horimoto et al., 2016; Jiang et al., 2014; Luo et al., 2017; Murakami et al., 2016; Quast et al., 2015; Sreenivasan et al., 2015) and wild animals (i.e. feral swine) have been exposed to IDV (Ferguson et al., 2018), and IDV has been recovered from both cattle and domestic swine (Ferguson et al., 2015; Hause et al., 2013). Among these species, surveillance data showed that IDV is enzootic in cattle, and cattle are proposed to be the natural reservoir of IDV (Ferguson et al., 2015; Hause et al., 2013). This has been supported by challenge studies in cattle showing that IDV causes a mild respiratory disease and can be transmitted effectively among cattle by in-pen contact (Ferguson et al., 2016). Previous studies have shown that, while IDV can be isolated in healthy cattle, it is more commonly isolated from sick cattle (Ferguson et al., 2015). Metagenomic analysis has shown that IDV is a common isolate in cattle diagnosed with BRD (Ng et al., 2015). However, the role of IDV in the pathogenesis of BRD is not well defined.
The goal of this study was to assess the potential role of IDV in development of BRD when IDV infection precedes infection with MHA, a known BRD bacterial agent shown to be present in cattle infected with IDV (Ducatez et al., 2015). A secondary objective was to assess the potential risk of viremia associated with IDV infection. Our results showed that, MHA infection following IDV infection or IDV infection alone was not adequate to lead to development of BRD, and that IDV does have the potential to cause transient viremia in experimentally inoculated cattle.
Results
Clinical scores rose in cattle infected with IDV and/or MHA.
To test the hypothesis that the severity of respiratory disease will increase after MHA challenge when IDV infection is present, 16 dairy calves were randomly assigned to 4 groups: groups IDV+MHA+ and IDV+MHA− received D/Bovine/C00046N/Mississippi/2014 (abbreviated as D/46N) intranasally at 0 DPI, as described elsewhere (Ferguson et al., 2016). Groups IDV+MHA+ and IDV−MHA+ received Mannheimia haemolytica D153 (abbreviated as MHA D153) intratracheally at 5 DPI, as described elsewhere (Lhermie et al., 2016; Riffault et al., 2010). Group IDV−MHA− received neither pathogen (Fig. 1A). All cattle used in this study were confirmed to be seronegative to the testing IDV and with low serological titers against both leukotoxin and whole cell (MHA bacterium) (Table 1). Clinical signs were evaluated twice a day, and clinical scores were quantified by eight different clinical measurements associated with BRD (e.g. temperature, respiratory rates, attitude, nasal discharge, ocular discharge, spontaneous cough, induced cough, and lung sounds).
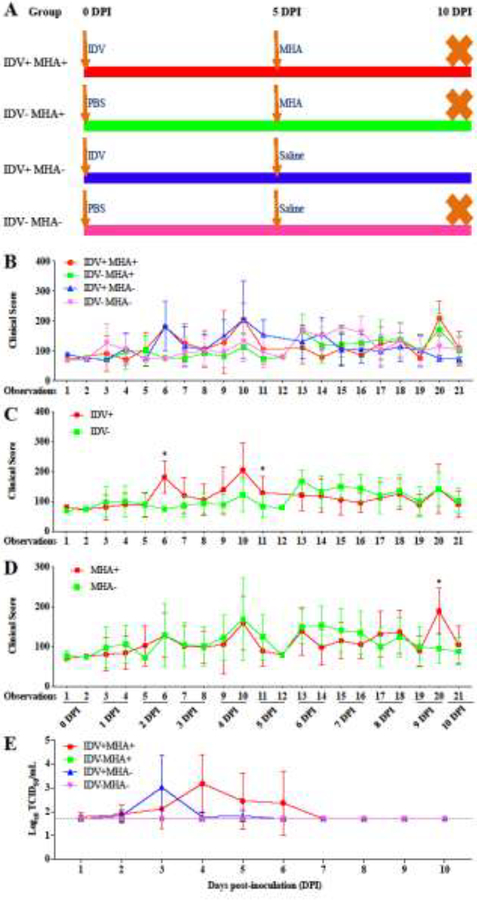
Fig. 1.
(A) Experimental design to evaluate pathogenesis of co-infections of influenza D virus and Mannheimia haemolytica in cattle. The 4–6 month-old calves (n = 16) were randomly assigned to 4 groups: IDV+MHA+ (n = 4), IDV−MHA+ (n = 4), IDV+MHA− (n = 4), and IDV−MHA− (n = 4). The cattle in groups IDV+MHA+ and IDV+MHA− received 107 TCID50 of D/46N intranasally at 0 DPI, and the cattle in groups IDV−MHA+ and IDV−MHA− were inoculated intranasally with sterile PBS at 0 DPI. At 5 DPI, the cattle in groups IDV+MHA+ and IDV−MHA+ received 109 CFU of MHA by intratracheal instillation, and the cattle in groups IDV+MHA− and IDV−MHA− received sterile saline intratracheally. The cattle in groups IDV+MHA+, IDV−MHA+, IDV+MHA−, and IDV−MHA− were in separate isolation stalls in the same building. (B) Dynamics of clinical scores in the experimental and control cattle. Clinical signs were evaluated twice daily (~12 hours interval, 21 observations in total), and clinical scores were quantified by eight different clinical measurements associated with BRD, including body temperature, respiratory rates, attitude, nasal discharge, ocular discharge, spontaneous cough, induced cough, and lung sounds. Clinical scores range from 0 to 755, with higher clinical scores indicating more abnormal clinical signs. (C) Dynamics of clinical scores in calves inoculated with IDV (groups IDV+MHA+ and IDV+MHA−) and calves without IDV exposure (groups IDV−MHA+ and IDV−MHA−). (D) Dynamics of clinical scores in calves inoculated with MHA (groups IDV+MHA+ and IDV−MHA+) and calves without MHA exposure (groups IDV+MHA− and IDV−MHA−). (E) Dynamics of viral shedding in experimental and control cattle. Dashed line indicates detection limit of 1.699 Log10 TCID50/mL. In Figures 1B and 1E, red line = IDV+MHA+, green line = IDV−MHA+, blue line = IDV+MHA−, pink line = IDV−MHA−. A p-value of 0.05 was determined as statistically significant, and the individual observations in Figures 1C and 1D with a statistically significant difference were indicated as stars.
Table 1.
Detection of MHA specific anti-whole cell and anti-leukotoxin antibodies in the sera of 16 calves used in this study.
| Antibody titers against whole cell (ng IgG) | Antibody titers against leukotoxin (ng IgG) | ||||
|---|---|---|---|---|---|
| Group | ID | 0 DPI | 10 DPI | 0 DPI | 10 DPI |
| IDV+MHA+ | B26 | 0.000 | 0.000 | 0.244 | 0.268 |
| B33 | 0.000 | 0.000 | 0.135 | 0.245 | |
| B50 | 0.000 | 0.161 | 0.000 | 0.017 | |
| B55 | 0.000 | NA* | 0.000 | NA | |
| IDV−MHA+ | B28 | 0.188 | 0.713 | 0.056 | 0.043 |
| B30 | 0.266 | 0.151 | 0.071 | 0.083 | |
| B38 | 0.070 | 0.097 | 0.026 | 0.229 | |
| B57 | 0.000 | 0.000 | 0.000 | 0.000 | |
| IDV+MHA− | B27 | 0.279 | NA | 1.026 | NA |
| B31 | 0.667 | 0.935 | 0.134 | 0.095 | |
| B51 | 0.016 | 0.000 | 0.118 | 0.372 | |
| B52 | 0.000 | 0.000 | 0.186 | 0.486 | |
| IDV−MHA− | B29 | 0.160 | 0.100 | 0.410 | 0.000 |
| B32 | 1.387 | 1.408 | 0.134 | 0.565 | |
| B35 | 0.000 | 0.000 | 1.337 | 1.222 | |
| B56 | 0.045 | 0.000 | 1.402 | 0.997 | |
NA, serum were not available for test.
Based on the clinical scoring system used, the range of possible clinical scores was 55 to 755; higher clinical score being associated with more severe clinical signs. Results showed that the 16 experimental calves had actual clinical scores in the range from 57 to 387 (Fig. 1B). From 2 to 5 DPI, the clinical scores for the cattle from the IDV+ groups (IDV+MHA+ and IDV+MHA−), which received IDV intranasally on 0 DPI, were significantly higher than those for the cattle from the other two groups IDV−MHA+ and IDV−MHA−, which received sterile PBS (p = 0.001) (Fig. 1C). The cattle in groups IDV+MHA+ and IDV+MHA− showed minor increases in depression and coughing scores after receiving IDV inoculation, leading to increased clinical scores. The clinical scores were peaked at 4 DPI but diminished thereafter (Fig. 1C). These results are consistent with the results from a prior study (Ferguson et al., 2016).
Few abnormal clinical signs were observed from 5 to 10 DPI in the cattle from treatment groups IDV+MHA+, IDV−MHA+, and IDV+MHA−; however, for reasons that are unknown, the cattle in the control group IDV−MHA− showed minor increases in clinical scores at 6 and 7 DPI (Fig. 1B). From 6 to 10 DPI, the clinical scores for the cattle, which received both IDV and MHA (i.e. IDV+MHA+), were slightly lower than those received MHA only (i.e. IDV−MHA+) (Fig. 1B) (p = 0.028). On the other hand, the clinical scores for the cattle from the MHA+ groups (i.e., IDV+MHA+ and IDV−MHA+), which received MHA intratracheally at 5 DPI, were not significantly different than those for the cattle from the MHA− groups (i.e. IDV+MHA− and IDV−MHA−), which received saline at 5 DPI (Fig. 1D) (p = 0.169).
Examined separately, clinical scores for cattle receiving MHA only trended to increase after 5 DPI, whereas scores for cattle receiving both IDV and MHA appeared to generally trend to decrease after 5 DPI. Clinical signs in groups receiving MHA after 5 DPI were very mild and consisted of mild, sporadic depression and coughing.
MHA anti-whole cell IgG and MHA anti-leukotoxin antibodies were also assessed between groups using the Kruskal-Wallis test at 0 DPI and 10 DPI. No differences were found between groups in either MHA anti-whole cell IgG at 0 DPI or 10 DPI, or MHA anti-leukotoxin antibody levels at 10 DPI. A near significant difference in MHA anti-leukotoxin antibody levels at Day 0 was observed between groups (p = 0.057). This near significance was due to serendipitously higher levels of MHA anti-leukotoxin antibody in groups not receiving MHA challenge (IDV−MHA− and IDV+MHA−). Those groups receiving MHA challenge (IDV+MHA+ and IDV−MHA+) had similar MHA anti-leukotoxin antibody concentrations (p = 0.991).
Experimental cattle were confirmed with IDV infection by viral titration and seroconversion.
Viral titration showed that cattle in groups IDV+MHA+ and IDV+MHA−, which received IDV inoculation at 0 DPI, began to shed IDV at 1 or 2 DPI, peaked at 3 or 4 DPI up to 4.699 Log10 TCID50/mL, and lasted until 5 or 6 DPI (Fig. 1E). The virus shedding pattern was consistent with those results from a prior study (Ferguson et al., 2016). No virus was shed from calves in groups that did not receive IDV. The sera collected from all 16 calves at 10 DPI were subjected to hemagglutination inhibition (HI) assays, and results showed that all eight cattle which received IDV inoculation (groups IDV+MHA+ and IDV+MHA−) seroconverted, with a maximum HI title of 1:320 (Table 2).
Table 2.
Serological responses in cattle using HI assay against D/bovine/C00046N/Mississippi/2014.
| Group | ID | Days post-inoculation (DPI)a | |||
|---|---|---|---|---|---|
| 0 | 2 | 5 | 10 | ||
| IDV+ MHA+ | 26 | <1:10 | <1:10 | <1:10 | 1:320 |
| 33 | <1:10 | <1:10 | <1:10b | 1:213.33 | |
| 50 | <1:10 | <1:10 | <1:10 | 1:160 | |
| 55 | <1:10 | <1:10b | <1:10 | 1:13.33 | |
| IDV− MHA+ | 28 | <1:10 | <1:10 | <1:10 | <1:10 |
| 30 | <1:10 | <1:10 | <1:10 | <1:10 | |
| 38 | <1:10 | <1:10 | <1:10 | <1:10 | |
| 57 | <1:10 | <1:10 | <1:10 | <1:10 | |
| IDV+ MHA− | 27 | <1:10 | <1:10 | <1:10b | 1:213.33 |
| 31 | <1:10 | <1:10 | <1:10 | 1:266.67 | |
| 51 | <1:10 | <1:10 | <1:10 | 1:106.67 | |
| 52 | <1:10 | <1:10 | <1:10 | 1:80 | |
| IDV− MHA− | 29 | <1:10 | <1:10 | <1:10 | <1:10 |
| 32 | <1:10 | <1:10 | <1:10 | <1:10 | |
| 35 | <1:10 | <1:10 | <110 | <1:10 | |
| 56 | <1:10 | <1:10 | <1:10 | <1:10 | |
The cattle inoculated with MHA after IDV did not significantly increase either virus titer or the length of viral shedding. At 10 DPI, all four calves from group IDV+MHA+, which received both IDV and MHA, were euthanized. Results from viral titration demonstrated the presence of IDV in 9 out of 15 tissue sections including the nasal turbinate (viral titer range: 1.930–2.199 Log10TCID50/mL), soft palate (viral titer range: 2.199–4.032 Log10TCID50/mL), trachea (viral titer range: 1.930–3.699 Log10TCID50/mL), bronchi (viral titer range: 2.137–2.532 Log10TCID50/mL), and lung (viral titer range: 2.032–2.366 Log10TCID50/mL) (Table 3).
Table 3.
Titration of viral loads in respiratory tissues collected from cattle in group IDV+MHA+ that received both IDV and MHA.
| Cattle | Viral titers in respiratory tissue (Log10 TCID50/mL)a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TUR1 | TUR2 | SP | UT | MT | LT | BR | RCR | RCCD | RMD | RA | LCR | LCCD | LCD | RCD | |
| B26 | 1.930 | 2.199 | 2.366 | NDc | ND | ND | 2.199 | ND | ND | 2.199 | ND | ND | ND | ND | ND |
| B33 | 2.199 | 2.199 | 4.032 | 1.930 | 2.366 | ND | 2.532 | ND | ND | 2.199 | ND | ND | ND | ND | ND |
| B50 | 2.032 | 2.199 | 2.199 | ND | 2.199 | ND | 2.137 | ND | ND | 2.032 | ND | ND | ND | ND | ND |
| B55 | 2.199 | 2.199 | N/Ab | 2.199 | 2.699 | 3.699 | 2.366 | ND | 2.366 | ND | ND | ND | ND | ND | ND |
Abbreviations: Nasal turbinate (TUR1), Ethmoid turbinate (TUR2), Soft palate (SP), Upper trachea (UT), Middle trachea (MT), Lower trachea (LT), Bronchus (BR), Right Cranial (RCR), Right cranial caudal (RCCD), Right Middle (RMD), Right accessory (RA), Right caudal (RCD), Left cranial (LCR), Left cranial caudal (LCCD), Left caudal (LCD);
N/A: Sample is not available;
ND: not detected with a detection limit of 1.699 Log10 TCID50/mL.
IDV infection caused transient viremia in cattle.
Among the IDV-inoculated animals, viremia was detected in calf B55 (in group IDV+MHA+) at 2 DPI (titer of 2.95 Log10[Copies of Virus]/0.2 mL) and in calves B33 (in group IDV+MHA+) and B27 (in group IDV+MHA−) at 5 DPI (titers of 3.85 and 4.80 Log10[Copies of Virus]/0.2 mL, respectively). The viruses in calves B55 and B27 were further confirmed by MDCK cell based viral titration with titers of 3.032 and 2.699 Log10 TCID50/mL, respectively (Table 4).
Table 4.
Titration of viral loads in the positive sera collected from the cattle inoculated with IDV.
| IDa (Group) | Days post-inoculation (DPI) | Log10 (Copies of virus) / 0.2 mL | Log10 TCID50/mL |
|---|---|---|---|
| B27 (IDV+MHA−) | 5 | 3.98 | 2.699 |
| B33 (IDV+MHA+) | 5 | 4.80 | NDb |
| B55 (IDV+MHA+) | 2 | 2.95 | 3.032 |
Note:
Sera from all eight calves inoculated with IDV were tested, and only the results from three positive sera samples are shown;
ND, not detected with a detection limit of 1.699 Log10 TCID50/mL.
Gross lung lesions were more severe in calves infected with MHA alone than in the negative control group but were not different between calves infected with both IDV and MHA and calves in negative control group.
Our prior study showed that IDV caused tracheal inflammation with multifocal areas of epithelial neutrophil infiltration and mild epithelial attenuation (Ferguson et al., 2016). To test the hypothesis that the severity of disease would increase after challenge with MHA when an IDV infection is present, we compared the gross lesions among cattle infected with MHA alone (group IDV−MHA+, n = 4), those with IDV and MHA (group IDV+MHA+, n = 4), and those with neither IDV nor MHA (group IDV−MHA−, n = 4). Gross lung lesions were scored 0–3 based on the percentage of lung affected (0: no gross lesions; 1: <25% of lobe affected; 2: 25–75% of lobe affected; 3: >75% of lobe affected). All lung lobes were scored (right cranial, right cranial caudal, right middle, right caudal, left cranial, left cranial caudal, and left caudal) and the scores were added together to result in a gross lesion score (ranging from 0–8, with a possible maximum score of 21).
Among the four cattle inoculated with MHA alone (group IDV−MHA+), three cattle showed varying degrees of MHA-associated pathogenesis whereas one did not show any gross lesions (Table 5). Multifocal red areas were observed across multiple lung tissues in all three affected cattle (Fig. 2B). The average lesion score was 3 and the highest lesion score was 8 (Table 5).
Table 5.
Gross lesion scores in the lung tissues from the cattle received MHA D153 and in negative control cattle.
| Group | ID | Lung tissues of cattlea | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RCR | RCCD | RMD | RCD | LCR | LCCD | LCD | Total Gross Lesion Score | ||
| IDV+MHA+ | B26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B33 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | |
| B50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| B55 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 4 | |
| 1.50(±2.34)b | |||||||||
| IDV−MHA+ | B28 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| B30 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | |
| B38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| B57 | 2 | 3 | 0 | 0 | 1 | 1 | 1 | 8 | |
| 3.00±(2.64) | |||||||||
| IDV−MHA− | B29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| B35 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | |
| B56 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| 0.75(±0.95) | |||||||||
Abbreviations: Right Cranial (RCR), Right cranial caudal (RCCD), Right Middle (RMD), Right accessory (RA), Right caudal (RCD), Left cranial (LCR), Left cranial caudal (LCCD), Left caudal (LCD);
average ± standard deviation of Gross Lesion Score/cattle for each group.
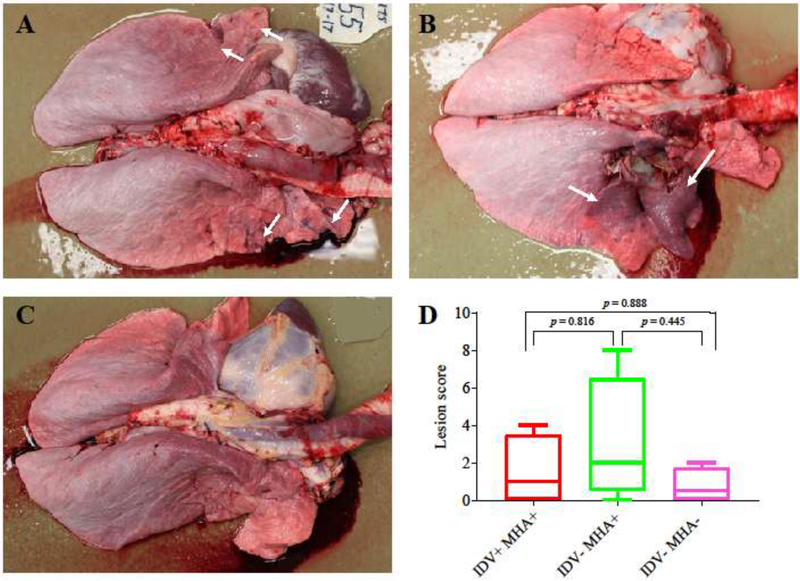
Fig. 2.
Gross pathology in the experimental and control cattle. Multifocal red areas (white arrows) were observed in the lungs of cattle from groups IDV+MHA+ (A) and IDV−MHA+ (B), but not in the lung of cattle from group IDV−MHA− (C); (D) Gross lung lesions were scored 0–3 based on the percentage of lung affected (0: no gross lesions; 1: <25% of lobe affected; 2: 25–75% of lobe affected; 3: >75% of lobe affected). All lung lobes were scored (right cranial, right cranial caudal, right middle, right caudal, left cranial, left cranial caudal, and left caudal) and the scores were added together to result in a gross lesion score (ranging from 0–8, with a possible maximum score of 21). The higher the gross pathology score, the more serious the pathogenesis. Box and whisker plots depicting gross pathology scores for IDV+MHA+, IDV−MHA+, and IDV−MHA− groups. A p-value of 0.05 was determined as statistically significant.
In the dual challenge group (with IDV and MHA), similar to those cattle in group IDV−MHA+, three calves showed varying degrees of MHA-associated pathogenesis whereas one did not show any gross lesions (Table 5). The average lesion score was 1.5, and the highest lesion score was 4 (Table 5).
No major lesions were observed in the four calves in the control group although small red depressed areas were found in the lung tissues of two experimental cattle. The average lesion score was 0.75 and the highest lesion score was 2 (Table 5).
Statistical analyses showed that there was no significant difference between the gross lesion scores among the cattle inoculated with MHA alone (group IDV−MHA+) and those inoculated with IDV and MHA (group IDV+MHA+) (p = 0.816), between cattle inoculated with IDV and MHA (group IDV+MHA+) and those in the negative control group (group IDV−MHA−) (p = 0.888), and between cattle inoculated with MHA alone (group IDV−MHA+) and those in the negative control group (group IDV−MHA−) (p = 0.445) (Fig. 2D).
To confirm that the gross lesions in lung tissues of cattle challenged with MHA were indeed caused by MHA, we determined the copy number of MHA in 12 selected lung tissues with gross lesion scores > 0 (Table 5) by quantitative PCR (qPCR) with MHA specific primers and probes. Results showed that 3 out of 12 tested lung tissues were MHA positive: the right cranial tissues of cattle B28 (gross lesion score of 1) with a bacterial load of 6.01 Log10 Copies/50 mg tissue; the right cranial tissues of B57 (gross lesion score of 2) had a bacterial load of 6.85 Log10 Copies/50 mg tissue, and that the right cranial caudal of B57 (gross lesion score of 3) had a bacterial load of 6.00 Log10 Copies/50 mg tissue (Table 6). Surprisingly, all four tissues tested in the dual infection group (IDV+MHA+) were MHA negative but had gross lesion scores consistent with infection, as determined by the pathologist who was blinded to treatment status (Table 5). All of the tested tissues from the negative control group (IDV−MHA−) were MHA negative.
Table 6.
Quantification of MHA in 12 cattle lung tissues with gross lesion score >0 (Table 5).
| Group | Cattle ID | Lung tissue of cattlea | Gross Lesion Score | Logio Copies/ 50 mg |
|---|---|---|---|---|
| IDV+/MHA+ | B33 | LCR | 1 | NDb |
| LCD | 1 | ND | ||
| B55 | RCR | 1 | ND | |
| LCR | 1 | ND | ||
| IDV−/MHA+ | B28 | RCR | 1 | 6.01 |
| LCCD | 1 | ND | ||
| B30 | RCD | 1 | ND | |
| B57 | RCR | 2 | 6.85 | |
| RCCD | 3 | 6.00 | ||
| LCR | 1 | ND | ||
| IDV−/MHA− | B35 | RCR | 1 | ND |
| RCCD | 1 | ND |
Abbreviation: Right Cranial (RCR), Right cranial caudal (RCCD), Right caudal (RCD), Left cranial (LCR), Left cranial caudal (LCCD), Left caudal (LCD);
ND: MHA was not detected with a detection limit of 2 × 104 Copies/50 mg tissue by qPCR.
Discussion
BRD is a leading cause of economic loss in the United States beef cattle industry every year. In this study, we evaluated the synergistic effects of MHA and IDV, two pathogens commonly found in cattle with BRD. Our results showed that, under the conditions of this study, IDV infection prior to MHA did not result in an increase in clinical disease or in lung pathology.
Mannheimia haemolytica is an opportunistic pathogen present in the nasopharynx and tonsils of cattle, although the virulence of this bacterium is strain and serotype dependent. Usually the serotypes 2 and 4 MHA are nonpathogenic whereas serotype 1 and 6 are pathogenic (Singh et al., 2011; Tatum et al., 1998). In healthy cattle, MHA are present as a part of normal microbial flora in these tissues with nonpathogenic MHA predominating, but pathogenic strains of MHA can become predominant in the upper respiratory tract and further transmit to and cause colonization in the trachea and lungs if the microenvironmental status is changed due to stress or infections by other agents (Briggs and Frank, 1992). Laboratory duplication of disease consistent with MHA infection has been accomplished for some strains of MHA (Lhermie et al., 2016). In this study, we used Mannheimia haemolytica D153, which was recovered from pneumonic calf lung and belongs to serotype 1 of MHA (Hauglund et al., 2013). A dose of 109 colony forming units (CFU) of MHA D153 resulted in mild gross lung lesions in the cattle challenged intratracheally by MHA D153 alone. However, none of the four cattle inoculated with MHA D153 showed overt signs of clinical BRD, and one animal had no lesions at necropsy suggesting that infection was mild or inoculation was not sufficient. The three animals with lesions had limited areas of consolidation, indicating mild effects of MHA. Nevertheless, through MHA specific qPCR, two out of four calves in the IDV−MHA+ group, which had gross lesions in their lungs, were confirmed to have MHA colonization in their lungs whereas the cattle in negative control (IDV−MHA−) group were MHA negative. In a prior study (Tatum et al., 1998), the wild type MHA D153 caused more lung pathogenesis in cattle including one death, although gross lung lesions observed were similar to those in our study; the average clinical score in the previous report was 4 (out of a possible 10) whereas our study had a clinical score of 3. Of note, serological titers against leukotoxin and those against whole cell (MHA bacterium) were low in all cattle used in this study (Table 1), and the serological titers for most cattle did not increase at 10 DPI (5 days post inoculation of MHA), indicating that most cattle used for MHA challenges had no or a low level of exposure to MHA before the MHA challenge. In the prior study (Tatum et al., 1998), 3–4 week-old Holstein calves were used whereas in this study, 4–6 month-old Holstein and Jersey calves were used. The age of the cattle could have impacted virulence. Nevertheless, our study did confirm the MHA D153 challenge was successful, although virulence was mild.
Our prior study clearly demonstrated that IDV can cause tracheal inflammation with multifocal areas of epithelial neutrophil infiltration and mild epithelial attenuation (Ferguson et al., 2016). Both serological data and viral shedding data confirmed that the cattle were successfully infected with IDV in this study. Of interest, this study suggests that IDV infection can lead to viremia in cattle, as was shown in swine infected with IDV (Ferguson et al., 2018). Another study showed that IDV was present in animal serum samples during IDV surveillance (Zhai et al., 2017). Soft palate tissue has been identified as a significant site of infection for a number of organisms including IDV infection in cattle (Ferguson et al., 2018), influenza A virus infections in ferrets (Lakdawala et al., 2015), and other pathogens, including both viruses (e.g., porcine reproductive and respiratory syndrome virus and porcine circovirus-2) and bacteria (e.g., Streptococcus porcinus, Streptococcus dysgalactiae, Staphylococcus aureus, Staphylococcus hyicus, Streptococcus suis, Yersinia enterocolitica, Salmonella spp., and Listeria monocytogenes) (Kernaghan et al., 2012). Thus, it is likely that IDV could enter the blood stream through soft palate in cattle because of the rich distribution of lymphoid tissue in soft palate.
In this study, we hypothesized that tracheal inflammation and epithelial injury from prior IDV infection could facilitate colonization of MHA. Based on prior work, we challenged cattle with MHA at 5 DPI, when histologic tracheal damage was expected to be present (Ferguson et al., 2016). After MHA challenge on 5 DPI, on 6–10 DPI, clinical scores were a slightly lower in the IDV+MHA+ group than the IDV−MHA+ group (p < 0.05). However, cattle infected with both MHA and IDV did not show more severe pathogenesis than those cattle infected with either MHA or IDV alone, indicating no increase in severity of disease caused by Mannheimia with prior IDV infection. Through MHA specific qPCR, two tested calves in the IDV−MHA+ group, which had gross lesions in their lungs, were confirmed to have MHA colonization in their lungs. Thus, IDV appears to compromise the host sufficiently to lead to minor increases in the MHA induced clinical signs but not lung pathology.
Limitations of this study include the small animal number per group (n = 4), although power analysis suggested that four were enough to provide 80% power to identify a 2-fold difference in disease pathogenesis (e.g. gross lesion score, clinical score). The day for MHA challenge was chosen based the results from a prior study that demonstrated tracheal inflammation at both day 4 and 6 post IDV inoculation (Ferguson et al., 2016). Nevertheless, consistent with this prior study (Ferguson et al., 2016), viral shedding in nasal swabs from the cattle inoculated with IDV peaked on 3 or 4 DPI (Fig. 1C), and therefore an earlier administration of MHA may be more optimal. In addition, the cattle used in this study were healthy and may not represent cattle in the field; these cattle were housed indoors under BSL-2 conditions and had limited space to exhibit subtle behavioral changes associated with illness. Furthermore, challenge with MHA by mid-tracheal inoculation bypassed a significant portion of the respiratory tract affected by IDV and may have reduced the opportunity to observe MHA−IDV synergism. The MHA challenge reported here was intentionally mild, in effort to observe subtle synergism, if any existed, and not to overwhelm the host with a severe MHA challenge. Finally, the prior study suggested IDV did not cause any significant lung lesions in the cattle (Ferguson et al., 2016), and thus, the cattle in the IDV+MHA− group were not euthanized and subjected to gross lesion analyses; this presents a potential limitation of this study due to the possible variations across individual experiments.
In summary, although IDV can cause mild respiratory signs and tracheal inflammation in cattle, under the conditions of this study, IDV infection did not enhance the MHA induced clinical signs or lung pathology. Dose, route, and timing optimization of MHA and IDV co-infection could potentially increase synergism of BRD signs and virulence. However, the pathogenesis of BRD is multifactorial, and, thus, it will be necessary to understand the synergistic effects of mixed infections of IDV and other pathogens, especially those co-present in the respiratory tracts of sick cattle, such as bovine adenovirus 3 and bovine rhinitis A virus, and their roles in development of BRD (Ng et al., 2015). Given this complexity, elucidation of the role of IDV in BRD may require an approach that assesses multiple agents simultaneously in a large, naturally exposed population.
Materials and Methods
Viruses and Bacteria.
The influenza D virus strains D/bovine/Mississippi/C00013N/2014 (abbreviated as D/13N) and D/Mississippi/C00046N/2014 (abbreviated as D/46N) were isolated from sick cattle in Mississippi (Ferguson et al., 2015). The bacterial strain D153 of Mannheimia haemolytica (MHA) was obtained from the National Animal Disease Center, US Department of Agriculture. The 16s rRNA gene of this bacterial strain was sequenced to confirm the identity of the isolate before it was used for challenge. The IDVs were propagated in Human Rectal Tumor cells (HRT-18G, ATCC) in Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific, Asheville, NC, USA) supplemented with 1 μg/mL of TPCK-trypsin (Gibco, New York, USA) at 37°C under 5% CO2. Viral titers were determined by TCID50 in HRT-18G cells. MHA was incubated in Brain Heart Infusion (BHI) broth at 37°C. Prior to challenge, MHA was washed and suspended in physiological saline (0.9% NaCl).
RNA and DNA extraction.
Viral RNA was extracted by using the GeneJET Viral DNA and RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Genomic DNA was extracted from bacterial culture and 50 mg of cattle lung tissue using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions.
Hemagglutination (HA) and HI assay.
HA and HI assays were performed as previously described (Ferguson et al., 2015). Briefly, serum samples were treated 1:3 with receptor-destroying enzyme (Denka Seiken Co., Tokyo, Japan) at 37°C for a minimum of 18 hours, followed by heat inactivation at 55°C for 30 min. Inactivated serum was diluted to a final concentration of 1:10 with 1× PBS. The HA and HI assays were conducted with 0.5% turkey red blood cells at 4°C against a testing IDV; samples with an HI titer ≥1:40 were considered IDV-seropositive. All sera were tested against influenza D/13N and D/46N.
Quantitative PCR (qPCR) and quantitative reverse transcription PCR (qRT-PCR).
qRT-PCR was used to determine the copy numbers of IDVs in tissues as previously described (Ferguson et al., 2016). Briefly, qRT-PCR was performed in triplicate by using TaqMan Fast Virus 1-step Master Mix (Life Technology, Carlsbad, CA) with the PB1 specific primer set: Forward 5’-GCTGTTTGCAAGTTGATGGG-3’; Reverse 5’-TGAAAGCAGGTAACTCCAAGG-3’; and FAM-Probe 5’-TTCAGGCAAGCACCCGTAGGATT-3’. Viral copies in samples were determined with the standard curve, which was generated by the plasmid containing the target gene segment (PB1 of IDV).
To determine the copy number of MHA in cattle lung tissues by qPCR, TaqMan Fast Virus 1-step Master Mix (Life Technology, Carlsbad, CA) and MHA specific primers and probe sets (Forward 5’-AGCAGCGACTACTCGTGTTGGT-3’; Reverse 5’-AAGACTAAAATCGGATAGCCTGAAACG -3’; and FAM-Probe 5’- AGGCTGGGCGTGGTTAGTATTAGAAGAGGG -3’) were used. Genomic DNA from fresh prepared MHA D153 was extracted and used to build a MHA specific standard curve. The genomic DNA from the lung tissues were used as templates in qPCR to test the presence of MHA, and the copies of MHA were determined using the MHA D153 specified standard curve.
Determination of TCID50 and CFU.
For viral titration, the TCID50 was determined in HRT-18G cells using supernatants of nasal swabs and tissues as previously described (Ferguson et al., 2016). For bacterial titration, the CFU was determined using BHI broth agar plates.
Animal experiments.
The 4–6 months old male calves (n = 16) included 8 Holsteins and 8 Jersey breed, were confirmed to be IDV seronegative against two genetic variants D/13N and D/46N using the HI assay. After completion of the study serum samples collected before and after challenge were tested for serum anti-leukotoxin and anti-whole cell (bacterium) antibodies both on the day of challenge and 10 DPI as described elsewhere (Confer et al., 2009). Calves were blocked by breed and randomly assigned to 4 groups: IDV+MHA+ (n = 4), IDV−MHA+ (n = 4), IDV+MHA− (n = 4), and IDV−MHA− (n = 4). The cattle in groups IDV+MHA+ and IDV+MHA− received 107 TCID50 of D/46N intranasally (in a volume of 10 mL) at 0 DPI, and the cattle in groups IDV−MHA+ and IDV−MHA− were inoculated with 10 mL sterile PBS at 0 DPI. At 5 DPI, the cattle in groups IDV+MHA+ and IDV−MHA+ received 109 CFU of MHA (in volume of 30 mL) by intratracheal instillation (Lhermie et al., 2016), and the cattle in groups IDV+MHA− and IDV−MHA− received 30 mL sterile saline, the same used to re-suspend MHA. Prior to intratracheal instillation, calves were sedated with 0.01 mg/kg butorphanol, 0.02 mg/kg xylazine, and 0.04 mg/kg ketamine given together by intravenous injection. The four calves in each experimental group were housed in separate isolation stalls with separate air handling, and personnel wore personal protective equipment (disposable coveralls, gloves, boot covers, and N95 masks) to prevent cross contamination.
At 10 DPI, 12 cattle in groups IDV+MHA+, IDV−MHA+ and IDV−MHA− were euthanized, and tissue sections across the respiratory tract were collected for pathogenesis analyses, including turbinates [Nasal turbinate (TUR1), Ethmoid turbinate (TUR2)], Soft palate (SP), trachea [Upper trachea (UT), Middle trachea (MT), Lower trachea (LT)], Bronchus (BR), and lung [Right Cranial (RCR), Right cranial caudal (RCCD), Right Middle (RMD), Right accessory (RA), Right caudal (RCD), Left cranial (LCR), Left cranial caudal (LCCD), Left caudal (LCD)]. A total of 12 lung tissues with a gross lesion score >0 were selected for bacteria titration using MHA specific qPCR. Because the pathogenesis of IDV infection was characterized in a prior study (Ferguson et al., 2016), and because the calves in group IDV+MHA− developed only mild clinical signs, the calves in this group were not euthanized.
Clinical observation and scoring.
Clinical signs were evaluated twice daily by a veterinarian, and clinical scores were quantified using a previously described clinical scoring system (Miao et al., 2004). The scoring system includes assessment of rectal temperature, respiratory rate and character, ocular and nasal secretions, and presence of cough or difficulty breathing. Lung fields were auscultated for the presence of large airway sounds consistent with pulmonary consolidation, crackles, wheezes, or absence of sounds.
Gross lesions score:
Gross lung lesions were scored 0–3 based on the percentage of lung affected (0: no gross lesions; 1: <25% of lobe affected; 2: 25–75% of lobe affected; 3: >75% of lobe affected). All lung lobes were scored (right cranial, right cranial caudal, right middle, right caudal, left cranial, left cranial caudal, and left caudal) and the scores were added together to result in a gross lesion score.
Statistical analyses.
Because data could not be assumed to be normally distributed, nonparametric methods were used for statistical analysis. The Friedman test was used to compare clinical scores between groups, accounting for repeated measures over time. When the Friedman test was significant, groups were compared at individual time points using Exact Wilcoxon Two-Sample tests. This approach was used to test three research hypotheses. The first hypothesis was that clinical scores in IDV challenged animals were different than in non-IDV challenged animals. This hypothesis was assessed using IDV challenged animals (IDV+MHA+ plus IDV+MHA−) and IDV non-challenged (IDV−MHA− plus IDV−MHA+) between study day 2 through 5 (observations 5 through 11). The second hypothesis was that clinical scores in MHA challenged animals were different than scores in MHA non-challenged animals. This hypothesis was assessed using MHA challenged (IDV+MHA+ plus IDV−MHA+) and non-challenged groups (IDV+MHA− plus IDV−MHA−) between study days 6–10 (observations 14 through 21). The third hypothesis was that, among animals challenged with MHA, prior challenge with IDV resulted in different clinical scores than animals not receiving prior challenge with IDV. This hypothesis was tested by comparing IDV+MHA+ to IDV−MHA+) between study days 6–10 (observations 14 through 21). Gross lesion scores were compared between three groups (IDV+MHA−, IDV+MHA+ and IDV−MHA−) using the Kruskal-Wallis test. A p-value of 0.05 was determined statistically significant for all analyses. SAS 9.4 was used for statistical calculations.
Biosafety and animal handling.
All laboratory and animal experiments were conducted under BSL-2 conditions, with investigators wearing appropriate protective equipment, and in compliance with protocols approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Mississippi State University.
Acknowledgments
We would like to thank Dr. Anthony Confer and Marie Montelongo from Oklahoma State University for quantifying anti-MHA whole cell and anti-MHA leukotoxin antibodies in the sera of the cattle used in this study, and we thank Dr. Patricia Cox for her editorial assistance. We are also grateful for critical discussion from Dr. Mariette Ducatez and Dr. Gilles Meyer. In addition, we would like to thank Michelle Banes for her assistance in propagating bacteria used in the animal experiments, and to thank Hui Wang, Lucas Ferguson, Charles Provine, Will Crosby, Brigitte Martin, Lei Zhong, Kaitlyn Waters, Minhui Guan, and Kaijun Jiang for their assistance with this study. Caitlyn Outlaw was supported by the National Institute of Health (grant # 2T35OD010432–16).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Briggs RE, Frank GH, 1992. Increased elastase activity in nasal mucus associated with nasal colonization by Pasteurella haemolytica in infectious bovine rhinotracheitis virus-infected calves. Am J Vet Res 53, 631–635. [PubMed] [Google Scholar]
- Chiapponi C, Faccini S, De Mattia A, Baioni L, Barbieri I, Rosignoli C, Nigrelli A, Foni E, 2016. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg Infect Dis 22, 352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confer AW, Ayalew S, Montelongo M, Step DL, Wray JH, Hansen RD, Panciera RJ, 2009. Immunity of cattle following vaccination with a Mannheimia haemolytica chimeric PlpE-LKT (SAC89) protein. Vaccine 27, 1771–1776. [DOI] [PubMed] [Google Scholar]
- Ducatez MF, Pelletier C, Meyer G, 2015. Influenza D virus in cattle, France, 2011–2014. Emerg Infect Dis 21, 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, Genova S, Webby R, Wan XF, 2015. Influenza D virus infection in Mississippi beef cattle. Virology 486, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Luo K, Olivier AK, Cunningham FL, Blackmon S, Hanson-Dorr K, Sun H, Baroch J, Lutman MW, Quade B, Epperson W, Webby R, DeLiberto TJ, Wan XF, 2018. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg Infect Dis 24, 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, Barton K, McCuan K, Webby RJ, Wan XF, 2016. Pathogenesis of Influenza D Virus in Cattle. J Virol 90, 5636–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foni E, Chiapponi C, Baioni L, Zanni I, Merenda M, Rosignoli C, Kyriakis CS, Luini MV, Mandola ML, Bolzoni L, Nigrelli AD, Faccini S, 2017. Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci Rep 7, 11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foni E, Chiapponi C, Faccini S, Baioni L, Barbieri I, Rosignoli C, Merenda M, Zanni I, Manfredi R, Sandri G 2016. The circulation of Influenza D in pig breeding farms of Northern Italy. In: Atti della Società Italiana di Patologia ed Allevamento dei Suini, XLII Meeting Annuale, Montichiari, Italia, 10–11 Marzo 2016, 151–155. [Google Scholar]
- Grissett GP, White BJ, Larson RL, 2015. Structured literature review of responses of cattle to viral and bacterial pathogens causing bovine respiratory disease complex. J Vet Intern Med 29, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauglund MJ, Tatum FM, Bayles DO, Maheswaran SK, Briggs RE, 2013. Genome Sequences of Mannheimia haemolytica Serotype A1 Strains D153 and D193 from Bovine Pneumonia. Genome Announc 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F, 2014. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio 5, e00031–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS pathogens 9, e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Hiono T, Mekata H, Odagiri T, Lei Z, Kobayashi T, Norimine J, Inoshima Y, Hikono H, Murakami K, Sato R, Murakami H, Sakaguchi M, Ishii K, Ando T, Otomaru K, Ozawa M, Sakoda Y, Murakami S, 2016. Nationwide Distribution of Bovine Influenza D Virus Infection in Japan. PLoS One 11, e0163828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM, 2014. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 49, 493–496. [DOI] [PubMed] [Google Scholar]
- Kernaghan S, Bujold AR, MacInnes JI, 2012. The microbiome of the soft palate of swine. Animal Health Research Reviews 13, 110–120. [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K, 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhermie G, Ferran AA, Assie S, Cassard H, El Garch F, Schneider M, Woerhle F, Pacalin D, Delverdier M, Bousquet-Melou A, Meyer G, 2016. Impact of Timing and Dosage of a Fluoroquinolone Treatment on the Microbiological, Pathological, and Clinical Outcomes of Calves Challenged with Mannheimia haemolytica. Front Microbiol 7, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Ferguson L, Smith DR, Woolums AR, Epperson WB, Wan XF, 2017. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 501, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Woolums AR, Zarlenga DS, Brown CC, Brown JC Jr., Williams SM, Scott MA, 2004. Effects of a single intranasal dose of modified-live bovine respiratory syncytial virus vaccine on cytokine messenger RNA expression following viral challenge in calves. Am J Vet Res 65, 725–733. [DOI] [PubMed] [Google Scholar]
- Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, Nishihara M, Hause B, Horimoto T, 2016. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg Infect Dis 22, 1517–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E, 2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 89, 5340–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, Miller G, Lauer D, Voss S, Pollock S, Cunha CW, Christopher-Hennings J, Nelson E, Li F, 2015. Serological evidence for the presence of influenza D virus in small ruminants. Vet Microbiol 180, 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffault S, Meyer G, Deplanche M, Dubuquoy C, Durand G, Soulestin M, Castagne N, Bernard J, Bernardet P, Dubosclard V, Bernex F, Petit-Camurdan A, Deville S, Schwartz-Cornil I, Eleouet JF, 2010. A new subunit vaccine based on nucleoprotein nanoparticles confers partial clinical and virological protection in calves against bovine respiratory syncytial virus. Vaccine 28, 3722–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Ritchey JW, Confer AW, 2011. Mannheimia haemolytica: bacterial-host interactions in bovine pneumonia. Vet Pathol 48, 338–348. [DOI] [PubMed] [Google Scholar]
- Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, Pillatzki A, Nelson E, Wang D, Kaushik RS, Li F, 2015. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J Virol 89, 11990–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum FM, Briggs RE, Sreevatsan SS, Zehr ES, Ling Hsuan S, Whiteley LO, Ames TR, Maheswaran SK, 1998. Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolytica serotype 1: characterization and virulence. Microb Pathog 24, 37–46. [DOI] [PubMed] [Google Scholar]
- Zhai SL, Zhang H, Chen SN, Zhou X, Lin T, Liu R, Lv DH, Wen XH, Wei WK, Wang D, Li F, 2017. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg Infect Dis 23, 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]