Abstract
1,3-Butadiene (BD) is an environmental and occupational toxicant classified as a human carcinogen. BD is metabolically activated by cytochrome P450 monooxygenases to 3,4-epoxy-1-butene (EB), which alkylates DNA to form a range of nucleobase adducts. Among these, the most abundant are the hydrolytically labile N7-guanine adducts such as N7-(2-hydroxy-3-buten-1-yl)-guanine (N7-EB-dG). We now report that N7-EB-dG can be converted to the corresponding ring open N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxy-3-buten-1-yl)-formamidopyrimidine (EB-Fapy-dG) adducts. EB-Fapy-dG lesions were detected in EB-treated calf thymus DNA and in EB-treated mammalian cells using quantitative isotope dilution nanoLC-ESI-MS/MS. EB-Fapy-dG adduct formation in EB-treated calf thymus DNA was concentration dependent and was greatly accelerated at an increased pH. EB-FAPy-dG adduct amounts were 2-fold higher in base excision repair-deficient NEIL1−/− mouse embryonic fibroblasts (MEF) as compared to isogenic controls (NEIL1+/+), suggesting that this lesion may be a substrate for NEIL1. Furthermore, NEIL1−/− cells were sensitized to EB treatment as compared to NEIL1+/+ fibroblasts. Overall, our results indicate that ring-opened EB-FAPy-dG adducts form under physiological conditions, prompting future studies to determine their contributions to genotoxicity and mutagenicity of 1,3-butadiene.
Graphical Abstract
Introduction
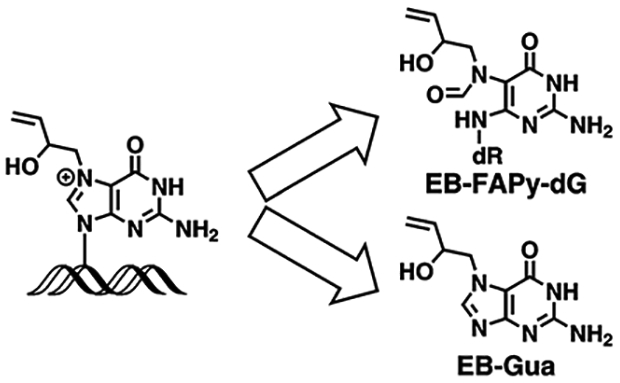
1,3-Butadiene (BD) is a recognized human and animal carcinogen present in cigarette smoke,1 automobile exhaust,2 and wood burning fumes.3 BD is also an important industrial chemical broadly used in polymer and rubber production, with 519,400 workers exposed to BD in the United States.4 BD is metabolically activated by cytochrome P450 monooxygenases to three inherently reactive epoxides: 3,4-epoxy-1-butene (EB), 1,2,3,4-diepoxybutane (DEB), and 1,2-dihydroxy-3,4-epoxybutane (EBD) (Figure S-1).5 These electrophilic species can enter cell nuclei and react with nucleophilic bases of DNA to form covalent nucleobase adducts.6 DNA adducts are thought to be responsible for the carcinogenicity of BD, as they can lead to DNA polymerase errors during replication.7–12 A number of BD-DNA adducts have been identified, including N-7-(2-hydroxy-3-buten-1-yl)-guanine (EB-Gua I),13, 14 N-7-(1-hydroxy-3-buten-2-yl) guanine (EB-Gua II),13, 14 N7-(2,3,4-trihydroxybut-1-yl)-guanine (N7-THBG),15, 16 N1-(2,3,4-trihydroxybut-1-yl)-adenine (N1-THB-Ade),17 1,4-bis-(guan-7-yl)-2,3-butanediol (bis-N7G-BD)18,19 and 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2′-deoxyadenosine (1,N6-HMHP-dA) (Figure S-1).20, 21 Among these, N7-guanine adducts (EB-Gua and N7-THBG) are by far the most abundant and have been detected in BD-treated laboratory animals,15, 18, 22 occupationally exposed workers,16 and current smokers.16
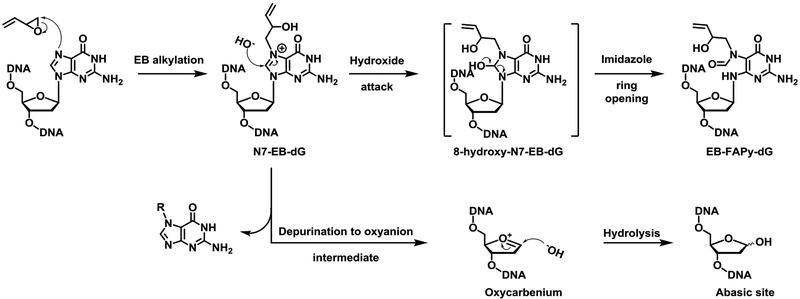
Despite their predominant formation in cells and tissues, N7-guanine adducts of BD may not be the most biologically relevant because they retain the ability to pair with cytosine and are hydrolytically labile because of the intrinsic destabilization of the glycosydic bond when the N7 position of guanine is alkylated.23–26 Spontaneous depurination of N7-THBG and EB-Gua releases free base adducts from the DNA backbone and leads to the formation of abasic sites (Scheme 1).25, 26 Alternatively, the C8 position of EB-Gua could be attacked by hydroxide anions, leading to imidazole ring opening and the formation of N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (EB-Fapy-dG) adducts (Scheme 1).27, 28
Scheme 1.
EB-FAPy-dG formation from dG following exposure to EB.
Imidazole ring scission upon FAPy adduct formation changes the molecular shape of the parent nucleobase, potentially modifying its base pairing preferences and leading to mispairing during DNA replication.29, 30 Specifically, keto-enol tautomerization of FAPy-dG adducts can lead to favorable base pairing to adenosine.31, 32 FAPy adducts can be recognized by DNA repair enzymes and excised from the DNA backbone.33–38 Unsubstituted (FAPy-dG) and alkyl-FAPy adducts (N5-alkyl-FAPy-dG) are known substrates for the base excision repair (BER) enzymes 8-oxoguanine glycosylase (OGG1),36, 37 endonuclease III-like protein (NTHL1),36, 38 and endonuclease 8-like proteins I – III (NEIL1, NEIL2, and NEIL3 respectively).36, 38
The most studied example of the N5-alkyl-FAPy-dG adducts are those formed upon exposure to aflatoxin B1 (AFB1), a potent liver carcinogen produced by the common soil fungus Aspergillus flavus.39, 40 Dietary exposure to AFB1 is recognized as a major risk factor for hepatocellular carcinoma.41–43 Metabolic activation of AFB1 yields AFB1-exo-8,9-epoxide,44 which alkylates DNA to form trans-8,9-dihydro-8-(N7-guanyl)-9-hydroxy-aflatoxin B1 (AFB1-N7-dG), and the corresponding ring open AFB1-β-FAPy adducts.45 AFB1-β-FAPy-dG adducts have been shown to induce high levels of G→ T transversion mutations in Escherichia coli, nearly 6 times more frequent than G→ T transversions caused by the cationic AFB1-N7-dG adduct.46 AFB1-FAPy-dG’s ability to induce G → T transversion mutations is consistent with G→ T mutations in codon 249 of the p53 tumor suppressor gene observed in AFB1-treated human hepatocytes cultures.47, 48 In addition, AFB1-induced G → T mutations in the ras oncogene contribute to liver tumor progression.49 Recently, the bulky AFB1-FAPy-dG adducts have been shown to be excised by NEIL1.50
In addition to AFB1-β-FAPy, several other N5-alkyl-FAPy-dG adducts have been detected in vivo. This includes Me-FAPy-dG and Me-FAPy-dA induced by methylating agents and FAPy adducts of antitumor nitrogen mustards (NM).27, 29, 51, 52 When Me-FAPy-bearing M13mp18 plasmid vectors were introduced in E. Coli bacteria, they blocked DNA replication and increased mutational frequency following SOS induction.29 Gruppi et al detected NM-FAPy-dG adduct and FAPy-dG-NM-dG cross-links in human mammary tumor (MDA-MB-321) cells treated with bis-(2-chloroethyl)-ethylamine (NM).52 Overall, previous studies indicate that N5-alkyl-FAPy-dG adducts can form in mammalian cells treated with alkylating agents, although their prevalence is often unknown, and their biological significance is not fully understood.
We hypothesized that by analogy with other N7-alkylguanines, BD-induced N7-EB-dG adducts can form the corresponding EB-Fapy-dG lesions in the presence of hydroxide anions. Authentic standards of 3,4-epoxy-1-butene (EB)-derived guanine Fapy adducts (EB-Fapy-dG) were prepared for the first time and structurally characterized by NMR and mass spectrometry. Using isotope dilution liquid chromatography-tandem mass spectrometry methods developed in our laboratory, we investigated concentration dependence for EB-Fapy-dG adduct formation in CT DNA and quantified these novel adducts in NEIL deficient and proficient MEF cells treated with EB.
Experimental
Note: 3,4-Epoxy-1-butene (EB) is a known carcinogen and must be handled with adequate safety precautions.
LC-MS grade water, methanol and acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA). Racemic 3,4-epoxy-1-butene ((±)-EB), N,N-dimethylformamide dimethyl acetal, dimethoxytrityl chloride, trifluoroethanol, dichloroacetic acid, and calf thymus DNA were obtained from Sigma-Aldrich (Millwaukee, WI/St. Louis, MO). 2’-Deoxyguanosine was obtained from Carbosynth (San Diego, Ca). Hypersep Hypercarb SPE cartridges (100 mg/3 mL) were obtained from Thermo Scientific (Waltham, MA). Omega Nanosep 10K filters were obtained from PALL Life Science (Port Washington, NY). All other chemicals and solvents were purchased from Sigma-Aldrich (Millwaukee, WI; St. Louis, MO) unless specified otherwise. Isotopically labeled 15N3-dG was provided by Dr. Rajat Subhra Das and Professor Ashis Basu (University of Connecticut).
Cell culture of wild type mouse embryonic fibroblast (MEF) cells and NEIL1 isogenic knockouts
Wild type MEF (NEIL1+/+) and isogenic NEIL1−/− MEF cells were a gift from Prof. Robert Turesky’s laboratory (University of Minnesota), used with permission from Prof. R. Stephen Lloyd (Oregon Health and Science University).53 Both cell lines were seeded at a density of 1 × 106 cells in DMEM supplemented with 10% FBS. Cells were cultured in an incubator humidified atmosphere of 5% carbon dioxide, 95% air at 37 °C.
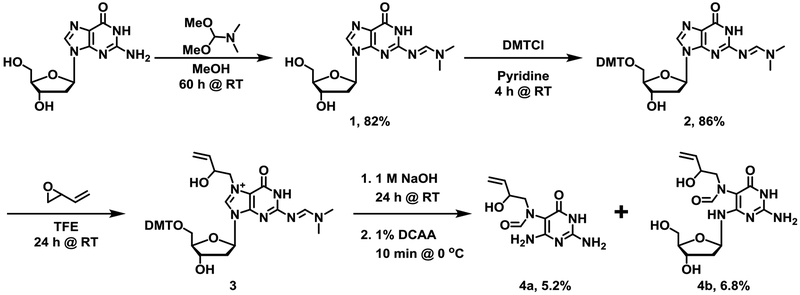
Synthesis of N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (EB-FAPy-dG, Scheme 2)
Scheme 2.
Chemical synthesis of EB-FAPy-dG starting from double protected 2’-deoxyguanosine.
(E)-N’-(9-((2R,4S,5R)-4-Hydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-6-oxo-6,9-dihydro-1H-purin-2-yl)-N,N-dimethylformimidamide (2’-dG-N2-N,N-dimethylformimidamide) (1)
2’-Deoxyguanosine (1 g, 3.70 mmol) was dissolved in dry methanol (9.5 mL) under argon, followed by the dropwise addition of N,N-dimethylformamide dimethyl acetal (2 mL, 14.8 mmol) over 5 minutes. The resulting suspension was stirred at room temperature for 60 h. The precipitate was collected by vacuum filtration, and the solid was washed with cold methanol to yield a white solid (980 mg, 81.7% yield). ESI+-MS/MS (1): 323.0 [M + H]+ → 207.1 [M – dR + H]+. 1H-NMR (500 MHz, DMSO-d6): 11.32 (s, N1-H, 1H), 8.56 (s, C8-H, 1H), 8.04 (s, N=CH-N(Me)2, 1H), 6.25 (t, J = 7 Hz, C’1-H, 1H), 5.30 (d, J = 3 Hz, C’3-OH, 1H), 4.94 (t, J = 5 Hz, C’5-OH, 1H), 4.42 – 4.34 (m, C’3-H, 1H), 3.89 – 3.79 (m, C’4-H, 1H), 3.59 – 3.51 (m, C’5-H, 2H), 3.16 (s, N-(CH3)2, 3H), 3.04 (s, N-(CH3)2, 3H), 2.64 – 2.64 (qt, J = 7 Hz, C’2-H, 1H), 2.28 – 2.19 (ddd, J = 14, 6, 3 Hz, C’2-H, 1H). 13C-NMR (400 MHz, DMSO-d6): 158.46, 158.06, 157.74, 150.10, 137.12, 120.16, 88.20, 83.24, 71.39, 62.29, 41.12, 35.12.
(E)-N’-(9-((2R,4S,5R)-5-((Bis(4-methoxyphenyl)(phenyl)methoxy)methyl)-4-hydroxytetrahydrofuran-2-yl)-6-oxo-6,9-dihydro-1H-purin-2-yl)-N,N-dimethylformimidamide (2)
Protected dG (compound 1 in Scheme 2) (958 mg, 2.97 mmol) was dissolved in anhydrous pyridine (20 mL) and 300 mg of 4,4-dimethoxytrityl chloride (DMTCl) was added. Three aliquots of 300 mg DMTCl were added to the solution every 15 minutes (1.2 g total, 2.97 mmol), and the solution was allowed to stir at room temperature for 4 h. The solution was concentrated in vacuo to yield a brownish red oil, which was purified by silica column chromatography using methanol in dichloromethane to yield pure compound 2 (Scheme 2) as a white solid (1.6 g, 86%). ESI+-MS/MS (2): 625.0 [M + H]+ → 207.1 [M – dR + H]+ and 303.1 [DMT+]. 1H-NMR (500 MHz, CDCl3): 8.93 (s, N1-H, 1H), 8.59 (s, C8-H, 1H), 7.72 (s, N=CH-N(Me)2, 1H), 7.42 (d, J = 8 Hz, DMT-H, 2H), 7.36 – 7.26 (m, DMT-H, 8H), 6.82 (d, J = 8, DMT-H, 4H), 6.40 (t, J = 6 Hz, C’1-H, 1H), 4.68 – 4.58 (m, C’4-H,1H), 4.18 – 4.11 (m, C’3-H, 1H), 3.80 (s, DMT-OCH3, 6H), 3.47 – 3.42 (m, C’5-H, 1H), 3.34 – 3.29 (m, C’5-H, 1H), 3.14 (s, N-(CH3)2, 3H), 3.08 (s, N-(CH3)2, 3H), 2.76 (s, C’3-OH, 1H), 2.62 – 2.54 (q, J = 5 Hz, C’2-H, 1H), 2.58 – 2.49 (m, C’2-H, 1H).
N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (4)
Protected dG (compound 2) (1 g, 1.6 mmol) was dissolved in 8.17 mL trifluoroethanol, and 1.93 mL of 3,4-epoxy-1-butene (24 mmol, 10 equiv.) was added dropwise over 15 min. The solution was allowed to stir for 24 h at room temperature and concentrated in vacuo to yield a white gum. Intermediate 3 was isolated by silica column purification (eluent methanol in dichloromethane with 1% triethylamine) to yield an off-white solid (150 mg, 13.5% yield). ESI+-MS/MS (3): 695 [M + H]+ → 277.2 [M – dR + H]+, 303.1 [DMT+], and 207.1 [M – dR – EB + H]+.
Compound 3 (150 mg, 0.216 mmol) was dissolved in 100 μL trifluoroethanol and 100 μL 1 M NaOH, and the solution was stirred for 16 h at room temperature. The pH was adjusted to 7.0 using 5% HCl, and the solution was concentrated under vacuum to yield an off-white solid. The resulting solid was chilled in an ice bath (0 °C) for 15 min and dissolved in 250 μL of 1% dichloroacetic acid in dichloromethane. The solution was stirred for 10 min and immediately neutralized to pH 7.0 using triethylamine. The solution was concentrated in vacuo to yield a reddish yellow solid.
The resulting crude product was resuspended in 1 mL of water and purified by semi preparative RP-HPLC using Zorbax XDB C18 column (250 mm × 10 mm, 5 μ, Agilent Technologies, Palo Alto, CA) with a gradient of water and acetonitrile at a flow rate of 1 mL/min. Solvent composition was changed from 0 to 3% B over the course of 5 min, 3 to 5% B over the course of 5 min, 5 to 10% B over the course of 10 min, 10 to 75% over the course of 5 min, 75 to 0% over the course of 3 min, and held at 0% B over 7 min. A set of peaks eluting at 9.5, 10.7, and 11.8 min corresponded to the EB-FAPy nucleobase (4a, Scheme 2), and peaks eluting at 13.7, 14.1, and 14.8 min corresponded to the desired EB-FAPy nucleoside (4b, Scheme 2) (Figures S-2 and S-3). The collected HPLC fractions of interest were concentrated in vacuo and characterized by ESI+-MS/MS, 1H-NMR, and HSQC. 15N3-EB-Fapy adducts were prepared analogously starting with 5 mg of 15N3–dG to be used as internal standards for mass spectrometry.
N-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-N-(2-hydroxybut-3-en-1-yl)-formamido-pyrimidine (4a): ESI+-MS/MS (4a): 240.1 [M + H]+ → 222.2 [M – H2O + H]+ and 194.1 [M – H2O – CO + H]+. 1H-NMR (600 MHz, DMSO-d6): 10.34 (s, N1-H, 1H), 8.10 (s, HC=O, 0.5 H), 7.79 (s, HC=O, 0.5 H), 6.47 (s, N2H2, 2H), 6.34 (s, C4-NH2, 2H), 6.13 (s, OH, 1H), 5.85 – 5.75 (m, CH=CH2, 1H), 5.30 – 5.18 (dd, J = 17, 6 Hz, CH=CH2, 1H), 5.15 – 5.00 (dd, J = 12, 5 Hz, CH=CH2, 1H), 4.79 – 4.69 (q, J = 7 Hz, CH-OH, 1H), 3.55 – 3.47 (m, CH2, 2H).
N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (4b): ESI+-MS/MS: 356.1 [M + H]+ → 240.1 [M – dR + H]+, 222.2 [M – dR – H2O + H]+ and 194.1 [M – dR – H2O – CO + H]+. 1H-NMR (600 MHz, DMSO-d6): 10.34 (s, N1-H, 1H), 7.90 (s, HC=O, 0.25 H), 7.83 (s, HC=O, 0.25 H), 7.77 (s, HC=O, 0.5 H), 6.45 (s, N2H2, 2H), 6.35 (s, C4-NH, 1H), 6.0 (s, C’5-OH, 0.5 H) 5.85 – 5.75 (m, CH=CH2, 1H), 5.80 (s, C’5-OH, 0.5 H), 5.75 (s, C’3-OH, 0.5 H), 5.64 (s, C’3-OH, 0.5 H), 5.30 – 5.18 (m, J = 17, 6 Hz, CH=CH2, 1H), 5.15 – 5.00 (m, J = 12, 5 Hz, CH=CH2, 1H), 4.77 – 4.70 (q, J = 8 Hz, CH-OH, 1H), 4.28 – 4.22 (m, C’1-H, 1H), 4.08 – 3.98 (m, C’3-H, 1H), 3.73 – 3.64 (m, CH2, 2H), 3.50 (m, C’4-H, 1H) 3.20 (dd, J = 13, 10 Hz, C’5-H, 1H), 3.12 – 3.04 (dd, J = 18, 3 Hz, C’5-H, 1H), 2.92 – 2.86 (dd, J = 13, 3 Hz, C’2-H, 1H), 2.87 – 2.81 (dd, J = 13, 9 Hz, C’2-H, 1H)
UV-Vis characterization of EB-FAPy and its molar concentration measurements (Figure S-4):
UV spectrophotometry measurements for EB-Fapy base (4b in Scheme 2) were conducted in solutions of 0.1N HCl (pH: 1.0), water (pH: 7.0) and 0.1N NaOH (pH: 12.0) for the determination of absorption maxima. Molar concentrations of EB-Fapy standard solutions were determined by UV spectrophotometry using the molar extinction coefficient (ε) of 12,589.2 M-1 cm−1 at 266 nm at neutral pH.54
Treatment of Calf-Thymus DNA with 3,4-epoxy-1-butene to determine dose-dependent increase in EB-FAPy-dG
To investigate dose-dependent formation of EB-FAPy-dG adducts at high pH, calf thymus DNA (1 mg/mL in 10 mM Tris-HCl buffer, pH 7.5) was treated with 10 μM, 50 μM, 100 μM, 250 μM, 500 μM, 1 mM, 5 mM, or 10 mM EB at 37 °C for 24 h. The unreacted EB was extracted with diethyl ether (3× at 2 mL), and alkylated DNA was precipitated with cold ethanol. The recovered DNA was washed with 70% EtOH and 100% EtOH. DNA was incubated 1 N NaOH (pH 12) for 1 h at room temperature to induce EB-FAPy-dG adduct formation. The solution was neutralized with 0.1M HCl, followed by ethanol precipitation of DNA as described above. DNA was then reconstituted in 200 μL 10 mM tris, pH 7.0 in milli-Q water, and its concentration was estimated by nanodrop UV spectrophotometer measurement (Thermo Scientific, Waltham, MA). DNA was accurately quantified via HPLC analysis of dG in enzymatic digests and processed for nanoLC-ESI+-MS/MS analysis as described below.
To investigate dose-dependent formation of EB-FAPy-dG under physiological conditions, CT DNA was treated with 500 μM, 1 mM, 5 mM, or 10 mM EB as described above. Precipitated and washed DNA was then reconstituted in 500 μL 10 mM Tris-HCl buffer, pH 7.0 and incubated at 37 °C for 72 h. DNA was subsequently processed for nanoLC-ESI+-MS/MS analysis as described below.
Enzymatic hydrolysis of DNA to 2’-deoxynucleosides and sample preparation for nanoLC-ESI+-MS/MS
DNA (100 μg) was enzymatically digested in the presence of nuclease P1 (5 U) in 50 μL 10 mM tris-HCl/5 mM ZnCl2 (pH 5.5) for 18 h. The pH of the solution was raised to 7.0 by the addition of 1 μL of 1 M ammonium bicarbonate, followed by incubation with 120 mU phosphodiesterase I, 105 mU phosphodiesterase II, 35 U DNase, and 22 U alkaline phosphatase, in the presence of 15 mM MgCl2 at 37 °C for 18 h to yield 2’-deoxynucleosides.
Enzymatic DNA digests were spiked with 15N3-EB-FAPy-dG internal standard (1 pmol) and purified by solid-phase extraction on Hypercarb Hypersep SPE cartridges (100 mg/3 mL). SPE cartridges were conditioned using 3 mL of methanol, followed by 3 mL of water. DNA samples (200 μL) were loaded under neutral pH, washed with 3 mL of water and 10 % methanol, and then eluted with 70 % methanol in water. SPE fractions were dried under vacuum and reconstituted in 16 μL of 5 mM ammonium formate, pH 4.5 for nanoLC-ESI+-MS/MS analysis as described below.
pH dependence for EB-FAPy-dG and EB-Gua II formation in EB-treated CT DNA
To investigate pH dependence for EB-Fapy-dG and EB-Gua II adduct formation, calf thymus DNA (1 mg/mL in 10 mM Tris-HCl buffer, pH 7.5) was treated with 5 mM EB at 37 °C for 24 h (in duplicate). Following extraction of unreacted EB with diethyl ether (3 × 2 mL), DNA was precipitated with cold ethanol. The recovered DNA was washed with 70% ethanol and re-suspended in 10 mM Tris buffer (200 μL) adjusted to pH 7.5, 9.0, 10.0, 11.0, or 12.0. DNA was incubated for 1 h, precipitated with cold ethanol, and reconstituted in 10 mM Tris buffer, pH 7.0 (500 μL). Samples were incubated at 37 °C for 72 h to allow for EB-FAPy-dG formation, followed by heating at 70 °C for 1 h to release any remaining EB-Gua as a free base. The DNA backbone was precipitated using 100% EtOH and washed as described above, while the supernatant was concentrated under vacuum. Each sample was reconstituted in 200 μL of water, spiked with 15N5-EB-Gua II internal standard (200 fmol),14 and subjected to SPE enrichment using Hypercarb SPE cartridges (100 mg, Thermo).
To quantify EB-Fapy-dG adducts, the precipitated, partially depurinated DNA was reconstituted in 10 mM Tris, pH 7.5 buffer (200 μL). DNA aliquots (100 μg) were taken and subjected to enzymatic digestion and solid phase extraction on Hypersep Hypercarb cartridges as described above. EB-Gua II and EB-Fapy-dG adduct numbers were expressed per 106 normal nucleotides, and the data were plotted together as a function of pH.
Cytotoxicity of EB in mouse embryonic fibroblasts (MEF) NEIL1+/+ and NEIL−/− cells
Wild type and NEIL−/− MEF cells53 were plated in 100 μL of Dulbecco’s Modified Eagle’s Medium containing 10% FBS at a density of 1 × 104 cells/well. Cells were permitted to adhere in humidified atmosphere of 5% carbon dioxide, 95% air, at 37 °C for 24 h. Cells were treated with 0 – 20 mM EB in 100 μL of serum-free medium for 3 h at 37 °C. Following treatment, cell media was replaced with 200 μL of fresh FBS-containing media, and the cells were maintained for an additional 48 h at 37 °C. Cell viability was established using an Alamar Blue assay55 using a Synergy HI Microplate reader (BioTek, Winooski, VT).
EB-Fapy-dG and EB-Gua II adduct formation in EB-treated MEF cells
Wild type and NEIL−/− MEF cells were grown in Dulbecco’s modified Eagle’s media (DMEM) supplemented with 10% fetal bovine serum. Cells were cultured in a humidified atmosphere of 5% carbon dioxide, 95% air, at 37 °C in 15 cm dishes until at least 70% confluency. Adhered HT1080 cells (1 × 107) were treated with 500 μM EB (in triplicate) for 24 h at 37 °C. Following treatment, EB containing media was discarded, and the cells were washed twice with ice-cold phosphate-buffered saline (PBS) to remove EB. Cells were harvested by trypsin treatment, and DNA was extracted as reported previously using Qiagen Cell Lysis solution.16 DNA concentrations were determined by dG analysis as described below. Extracted DNA (50 μg) was subjected to enzymatic digestion and SPE enrichment for EB-FAPy-dG and EB-Gua II analysis as described above.
nanoLC-ESI+-MS/MS analysis of EB-Fapy-dG
Quantitative analyses of EB-FAPy-dG were conducted by nanoLC-ESI+-MS/MS using a TSQ Quantiva mass spectrometer (Thermo Scientific, Waltham, MA) interfaced with a Dionex UltiMate 3000 RSLC nanoHPLC system (Thermo Scientific, Waltham, MA). Samples were loaded onto a pulled-tip fused silica column with a 100 μm inner diameter packed in-house with 15 cm of 5 μm Synergy Hydro RP resin (Phenomenex, Torrence, Ca) that served as both a resolving column and as a nanospray ionization emitter. HPLC solvents were comprised of 5 mM ammonium formate, pH 4.5 (solvent A) and 90/10 acetonitrile/solvent A (solvent B). Solvent composition was kept at 2% solvent B over 6 min, followed by an increase to 20% over 10 min, further to 80% over 2 min, held at 80% B over 2 min, then decreased to 2% over 2 min, and finally re-equilibrated at 2% B for 11 min. HPLC flow rate was kept at 1.0 μL/min for the first 6 min of the run, then reduced to 0.3 μL/min, followed by final equilibration for 11 min at 1 μL/min. Under these conditions, EB-FAPy and its internal standard (15N3–EB-FAPy) eluted at ~ 13.8 min. Electrospray ionization was achieved at a spray voltage of 2600 V and a capillary temperature of 350 °C. Collision induced dissociation was performed with Ar as a collision gas (1.0 mTorr) at a collision energy of 10 V. MS/MS parameters were optimized for maximum response during infusion of a standard solution of EB-FAPy.
nanoLC-ESI+-MS/MS analysis of EB-FAPy-dG was performed in the selected reaction monitoring mode by following the neutral loss of dR and dR + water from protonated molecules of the analyte (m/z 356.1 [M + H]+ → 240.1 [M + H − dR]+ and m/z 356.1 [M + H]+ → 222.1 [M + H – dR – H2O]+, respectively). The corresponding mass transitions corresponding to 15N3-labeled internal standard were m/z 359.1 [M + H]+ → 243.1 [M + H − dR]+ and m/z 359.1 [M + H]+ → 225.1 [M + H – dR – H2O]+). Analyte concentrations were determined using the relative response ratios calculated from HPLC-ESI+-MS/MS peak areas in extracted ion chromatograms corresponding to EB-FAPy-dR and its internal standard.
nanoLC-ESI+-MS/MS analysis of EB-Gua II
NanoLC-ESI+-MS/MS analyses of EB-Gua II were conducted using a TSQ Quantiva mass spectrometer (Thermo Scientific, Waltham, MA) interfaced with a Dionex UltiMate 3000 RSLC nanoHPLC system (Thermo Scientific, Waltham, MA). Samples were loaded onto a pulled-tip fused silica column with a 100 μm inner diameter packed in-house with 15 cm of 5 μm Synergy Hydro RP resin (Phenomenex, Torrence, Ca) that served both as a resolving column and as a nanospray ionization emitter. HPLC solvents included (A) 0.01% acetic acid in LC-MS grade water and (B) 0.02% acetic acid in LC-MS grade acetonitrile. Solvent composition was kept at 2% solvent B over 8 min, followed by an increase to 30% over 9 min, further to 80% over 1 min, held at 80% B over 10 min, then decreased to 2% over 1 min, and finally re-equilibrated at 2% B for 8 min. The flow rate was 0.7 μL/min for the first 7.5 min of the run, then reduced to 0.3 μL/min, until the final equilibration for 11 min at 0.7 μL/min. Sample injection volume was 4 μL. Under these conditions, both analyte its internal standard eluted as a complex peak around 16.3 min.
Electrospray ionization was achieved at a spray voltage of 3100 V and a capillary temperature of 350 °C. Collision induced dissociation was performed with Ar as a collision gas (1.5 mTorr). MS tuning was performed upon infusion of standard solutions to obtain maximum sensitivity. HPLC-ESI+-MS/MS analysis was performed in the selected reaction monitoring (SRM) mode using 15N5 labeled internal standard (15N5-EB-Gua II). SRM transitions included a neutral loss of 1-hydroxy-3-buten-2-yl from protonated molecules of EB-Gua II m/z 222.1 (M + H)+ → 152.1 (M – C4H8O + H)+ with a collision energy (CE) of 15 V and a neutral loss of 1-hydroxy-3-buten-2-yl and ammonia (m/z 222.1 (M + H)+ → m/z 135.0 (M – C4H8O – NH3 + H)+) with a CE of 29 V as observed previously.13, 14 Similarly, SRM transitions for 15N5-EB-Gua II included m/z 227.1 (15N5-M + H)+ → 157.0 (15N5-M – C4H8O + H)+, CE = 17 V and m/z 227.1 (15N5-M + H)+ → 139.0 (15N5-M – C4H8O – 15NH3 + H)+, CE = 32 V.14 Quantitative analyses were conducted using SRM transitions m/z 222.1 (M + H)+ → m/z 152.1 (M – C4H8O + H)+ for EB-Gua II and m/z 227.1 (15N5-M + H)+ → 157.0 (15N5-M – C4H8O + H)+ for its internal standard.
DNA Quantitation by HPLC-UV Analysis of dG
To quantify DNA extracted from cells and to detect any potential RNA contamination, 5 μg aliquots of DNA were taken and digested to 2′-deoxynucleosides in the presence of phosphodiesterase I (120 mU), phosphodiesterase II (105 mU), DNase (35 U) and alkaline phosphatase (22 U) in 20 μL 10 mM Tris-HCl/15 mM ZnCl2 (pH 7.0) for 18 h at 37 °C. Quantitative analysis of dG was conducted by HPLC-UV using an Agilent Technologies 1100 HPLC system equipped with a diode array UV detector and an autosampler. Samples were loaded onto an Atlantis T3 C18 column (2.1 × 150 mm, 5 μm, from Waters Corporation, Milford, MA) eluted with a gradient of 5 mM ammonium formate, pH 4.0 (A) and methanol (B). Solvent composition was changed linearly from 3 to 30% B over 15 min, further increased to 80% B over 3 min, held at 80% B for 1 min, and returned to 3% B over 2 min, where it was kept for the final 8 min of the HPLC run. UV absorbance was monitored at 260 nm. With this method, dG eluted as a sharp peak at 11.7 min (Figure S-5). dG amounts were calculated from HPLC peak areas using a calibration curve constructed by injecting known amounts of dG.
NanoLC-ESI+-MS/MS method validation
Calf thymus DNA (CT DNA, 100 μg, in triplicate) was spiked with increasing amounts of EB-FAPy-dG (40–2000 fmol) and 15N3-EB-FAPy-dG (1 pmol, internal standard). Each sample was analyzed as described above. The linearity of our nanoLC-ESI+-MS/MS methodology was determined by plotting the observed analyte/internal standard peak area ratios versus the expected peak area ratios. The method limit of detection was defined as analyte signal at least 3 times the height of the background. Accuracy and precision of the method were calculated for three samples (1000 fmol, high point; 400 fmol, middle point; 100 fmol, low point). Accuracy was determined by subtracting the average observed peak area ratio (analyte/internal standard) from the expected peak area ratio, followed by dividing the difference by the expected peak area ratio multiplied by 100. Accuracy was expressed as the % difference. Precision was calculated by dividing the standard deviation between samples by the mean observed peak area multiplied by 100, expressed as % coefficient of variation (%CV). Validation results are presented in the Results section and shown in the Supporting Information.
Results
Synthesis and structural characterization of EB-Fapy-dG
In our initial efforts to synthesize authentic standards of EB-Fapy-dG adducts, 2’-deoxyguanosine (dG) was reacted with excess (±)-EB at pH 7.2, followed by treatment with 1N NaOH to induce imidazole ring opening as shown in Scheme 1. Although ESI+-MS/MS analyses of reaction mixtures revealed the presence of small amounts of EB-Fapy-dG (M = 356.1), the product could not be purified due to the low adduct yields and the complexity of the reaction mixtures (results not shown).
To improve EB-Fapy-dG yields and to minimize anomerization reactions,28 doubly protected dG (compound 2 in Scheme 2) was employed as a starting material for reaction with EB. A similar approach was previously used by Rizzo et al in the synthesis of Me-FAPy-dG.56 Following reaction of 2 with EB to generate N7-substituted intermediate 3, the reaction mixtures were incubated in the presence of 1N NaOH to induce imidazole ring opening. Following deprotection steps, EB-FAPy-dG (4b in Scheme 2) was isolated by semi-preparative HPLC (Figure S-2). All major HPLC peaks were collected and subjected to HPLC-ESI+-MS/MS analysis. We found that HPLC fractions collected at 13.7, 14.1, and 14.8 min contained EB-FAPy nucleoside (M = 356.1) (comp. 4b in Scheme 2). HPLC peaks eluting at 9.5, 10.7, and 11.8 min (Figure S-3) contained EB-FAPy free base (comp. 4a in Scheme 2, M = 239.1). The isotopically labeled analog, 15N3- EB-FAPy-dG, was synthesized analogously using 15N3-dG as a starting material.
HPLC purified EB-FAPy base and EB-FAPy-dG were initially characterized by UV spectrophotometry (Figure S-4) and ESI+ MS/MS (Figures S-6–S-8). At neutral pH, UV λ max was determined to be 266 nm. When solution pH was adjusted to pH 1.0 with 1 M HCl, λ max shifted to 264 nm. Conversely, when solution pH was adjusted to pH 12.0 using 1 M NaOH, λ max moved to 274 nm. Overall, UV absorption profiles of the novel adduct (Figure S-4) matched the published spectra of other alkyl-FAPy-dG adducts.57
The main ESI+ MS/MS fragmentation pathways for EB-FAPy base (M = 239.1) were the neutral loss of water (m/z 240.1 [M + H]+ → 222.1 [M – H2O + H]+) and a loss of both water and carbon monoxide from the protonated molecules of the adduct and (m/z 240.1 [M + H]+ → 194.1) [M – H2O - CO + H]+ (Figure S-6). ESI+ MS/MS fragmentation of EB-FAPy-dG was characterized by the neutral loss of sugar, a loss of sugar and water, and a loss of sugar, water, and carbon monoxide from the protonated parent molecules (m/z 356.1 [M + H]+ → 240.1 [M + H − dR]+, 222.1 [M + H – dR – H2O]+, and 194.1 [M – dR – H2O – CO + H]+ (Figure S-7). ESI+ MS/MS spectrum of 15N3-labeled EB-FAPy-dG (m/z 356.1 [M + H]+) contained similar fragments at 243.1 [M + H − dR]+, 225.1 [M + H – dR – H2O]+, and 197.1 [M – dR – H2O – CO + H]+ (Figure S-8).
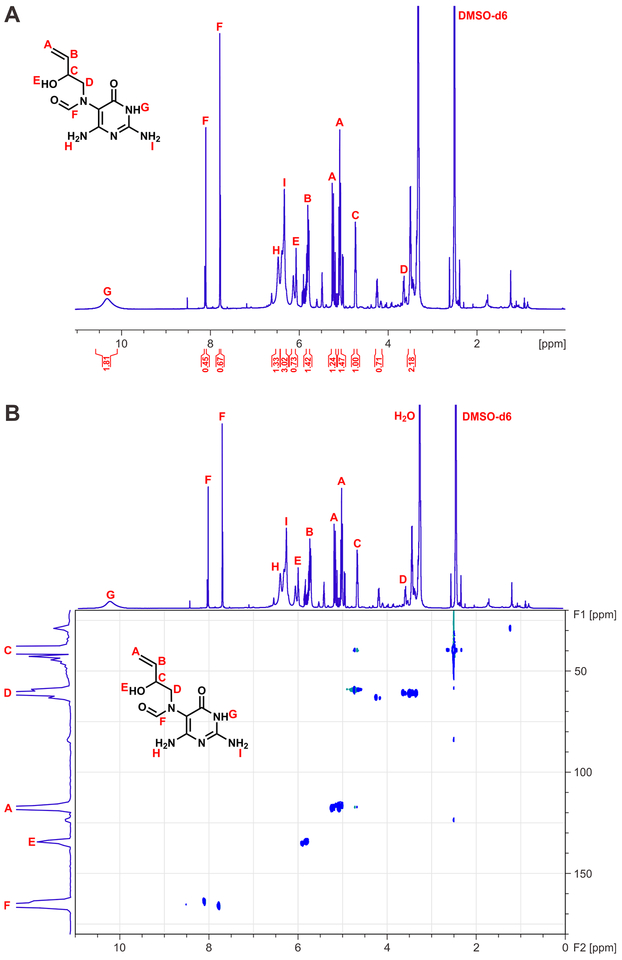
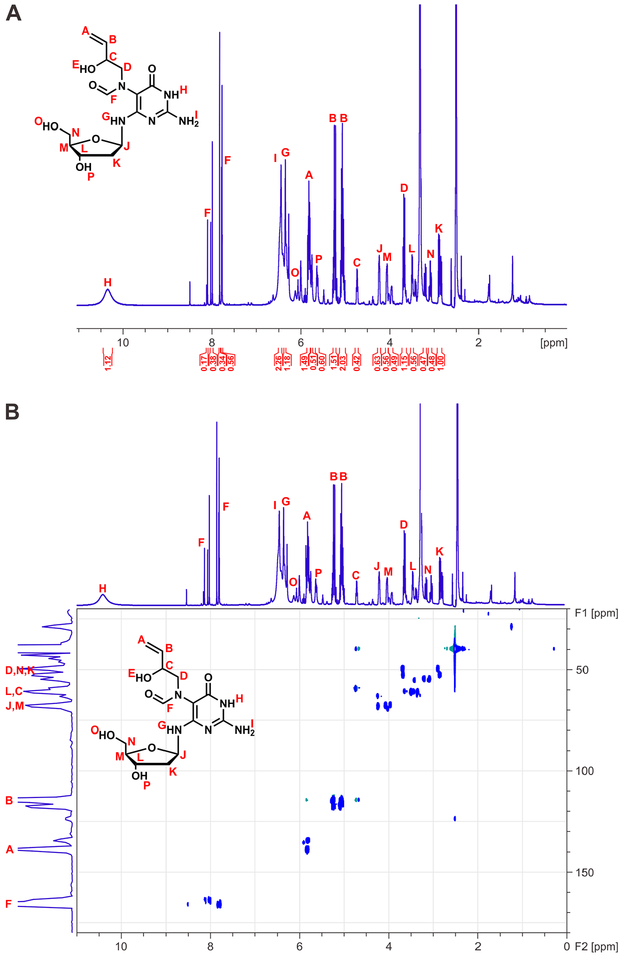
EB-FAPy base and EB-FAPy nucleoside (Compounds 4A and 4B in Scheme 2) were further structurally characterized by 1H-NMR and 1H-13C HSQC using 600 MHz NMR. 1H-NMR confirmed the presence of the 2-hydroxybut-3-en-1-yl group by the observation of geminal alkene protons at 5.22 and 5.08 ppm, an internal alkene at 5.82 ppm, a quartet at 4.26 ppm (2-position of substituent), and a multiplet at 3.51 ppm (Figure 1A). Further analysis of the EB-FAPy nucleobase by 1H-13C HSQC revealed 1H signals at 5.80, 5.22, and 5.08 ppm, which correlated to 13C signals at 117.6 and 134.5, confirming the presence of a terminal alkene in the adduct structure (Figure 1B). Furthermore, the 1H-quartet at 4.74 ppm and a multiplet at 3.51 ppm correlated with carbon atoms observed at 63.2 and 60.9 ppm, respectively, as expected for the 2-hydroxybut-3-en-1-yl substituent (Figure 1B). The presence of formamido group in adduct structure was confirmed by the presence of 1H signals at 7.78 and 8.10 ppm, which correlated to 13C signals at 164.2 and 166.2 ppm in the HSQC analysis (Figure 1B).
Figure 1.
H-NMR (A) and HSQC spectra of synthetic EB-FAPy base (B) (4a in Scheme 2)
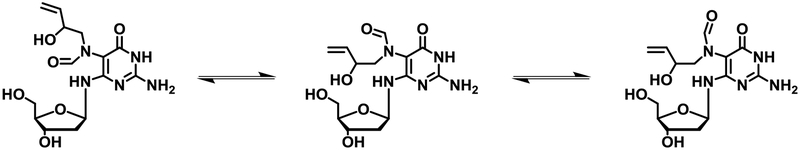
Our observation of two major formamide signals in the NMR spectra is consistent with the EB-FAPy molecule existing as a mixture of E/Z formamide isomers caused by sterically hindered rotation about the C5-N5-bond (Scheme 3).56, 58, 59 Additional formamide peaks can be attributed to the presence of rotamers caused by the slow rotation around the N5-CO bond respectively (Scheme 3).56, 58–60 N1H, exocyclic amines at C2 and C4, and the alcohol OH were observed at 10.34, 6.47, 6.34, and 6.13 ppm, respectively (Figure 1A).
Scheme 3.
Proposed structures of E/Z formamide isomers of EB-FAPy nucleoside.
1H-NMR and 1H-13C HSQC analysis of EB-FAPy nucleoside confirmed its structure as N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (Figure 2A). NMR peaks corresponding to 2’-deoxyribose protons were observed at 4.25, 4.03, 3.50, 3.20, 3.08, 2.89, and 2.84 ppm. As compared to the NMR spectrum of the free EB-FAPy base, EB-FAPy nucleoside exhibited a small downfield shift of the protons corresponding to 2-hydroxybut-3-en-1-yl substituent at 4.74 ppm and 3.68 ppm (Figure 2A). Furthermore, 1H signals corresponding to the formamide group were observed at 7.90, 7.83, and 7.77 ppm (Figure 2A). 13C chemical shifts corresponding to the 2-hydroxybut-3-en-1-yl substituent observed HSQC spectra were similar to the ones for the free base (Figure 2B, 1B). As was the case for NMR spectra of EB-FAPy base (Figure 1A), multiple proton signals corresponding to the formamide group were observed due to the presence of E/Z formamide isomers and rotamers of the adduct (Scheme 3, Figure 2A).56, 58, 59
Figure 2.
H-NMR (A) and HSQC spectra of EB-FAPy-dG (4b in Scheme 2).
Collectively, our mass spectrometry and NMR spectroscopy results for synthetic EB-FAPy adduct are consistent with the structure of N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (EB-Fapy-dG, 4b in Scheme 3).
Development and validation of nanoLC-ESI+-MS/MS method for EB-FAPy-dG
Authentic standards of EB-FAPy-dG and its 15N3- labeled analog (15N3- EB-FAPy-dG) were used to develop an isotope dilution nanoLC-ESI+-MS/MS method to allow for its accurate quantitation in EB-treated DNA and in human cells exposed to EB. In our approach, DNA (50 μg) was fortified with known amounts of 15N3- EB-FAPy-dG internal standard, followed by enzymatic hydrolysis to release EB-FAPy-dG nucleoside. EB-FAPy-dG and its internal standard were enriched by solid phase extraction on Carbograph SPE cartridges. EB-FAPy-dG is a very polar molecule (calculated logP = – 1.498) and does not retain well on most SPE columns. We found that among SPE cartridges tested (Isolute ENV+ (50 mg/1 mL, Biotage, Charlotte, NC), Sep-pack C18 (50 mg/1 mL, Waters Corp., Millford, MA), Strata X polymeric C18 (30 mg/ 1 mL, Phenomenex, Torrance, CA)) graphitized carbon based packing (Carbograph, Grace or Hypersep Hypercarb, ThermoFisher Scientific) was the only one that allowed for good retention of EB-FAPy-dG, leading to good SPE recovery (90.3%) and the removal of impurities that interfered with nanoLC-ESI+-MS/MS analysis.
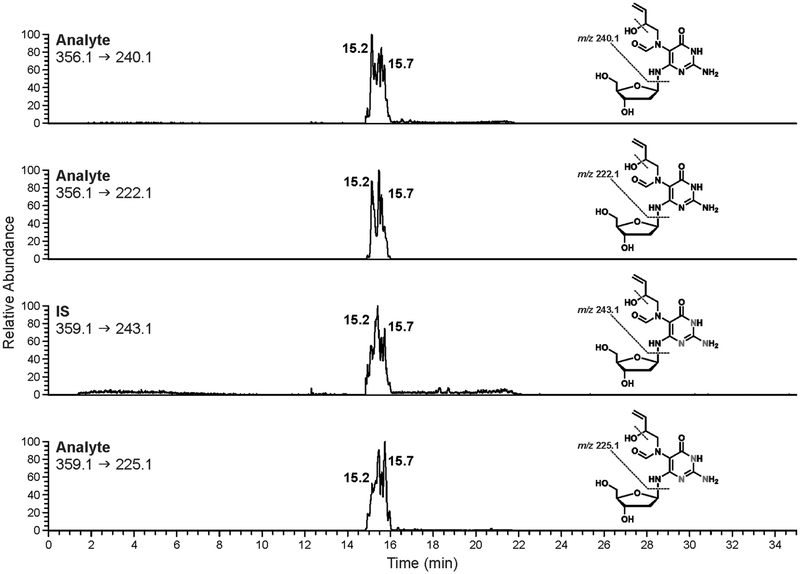
SPE fractions containing EB-FAPy-dG and its internal standard were concentrated under vacuum and re-constituted for isotope dilution nanoLC-ESI+-MS/MS analysis. We chose nanoLC because it allows for the best sensitivity in mass-spectrometry based approaches for DNA adduct detection.22 EB-FAPy-dG does not retain well on standard C18 packing materials. Furthermore, it exhibits a complex HPLC peak shape due to the presence of multiple E/Z formamide isomers/rotamers in its structure (Scheme 3). A number of HPLC stationary phases (Synergi Max-RP (Phenomenex, Torrance, CA), Hypersil Gold aq (Thermo Scientific, Waltham, MA), Zorbax SB-C18 (Agilent Technologies, Santa Clara, CA) and solvent systems (0.05% acetic acid, 0.1 % formic acid, 15 mM ammonium acetate (pH 6.8) with methanol or acetonitrile) were tested. The best HPLC retention and peak shape were achieved with Synergi Hydro RP packing material (Phenomenex, Waltham, MA) (Figure 3). When using a gradient of acetonitrile in 5 mM ammonium formate, pH 4.5, EB-FAPy-dG eluted as a complex peak at 14.5–16 min (Figure 3) as a result of multiple rotamers/atroisomers in its structure (Scheme 3). Our attempts to focus all EB-FAPy-dG isomers into a sharper HPLC peak were unsuccessful as they compromised analyte retention and reduced the method’s sensitivity.
Figure 3.
NanoLC-ESI+-MS/MS traces of EB-FAPy-dG (200 fmol on column) and its 15N3-internal standard (200 fmol on column).
As noted above, ESI MS/MS spectrum of EB-FAPy-dG (m/z 356.1, Figure S-7) is characterized by two major fragments corresponding to a neutral loss of deoxyribose sugar (m/z 240.1) and both dR and a molecule of water (m/z 222.1). 15N3-EB-Fapy-dG internal standard exhibited analogous signals at m/z 243.1 and 225.1, respectively (Figure S-8). Therefore, our quantitative nanoLC-ESI+-MS/MS methodology is based on selected reaction monitoring of mass transitions m/z 356.1 → 240.1 (analyte) and m/z 359.1 → 243.1 (internal standard). Secondary (qualifier) SRM transitions (m/z 356.1 → 222.1 (analyte) and m/z 359.1 → 225.1 (internal standard) were used to confirm signal identities. Mass spectrometer parameters were optimized for maximum sensitivity upon infusion of synthesized standards.
The newly developed nanoLC-ESI+-MS/MS method was validated by analyzing CT DNA spiked with known amounts of the analyte (40 – 2000 fmol) and its internal standard (1 pmol). nanoLC-ESI+-MS/MS limit of detection (LOD) was calculated as 1.2 EB-FAPy-dG per 107 nucleotides, while the limit of quantification was 3.1 EB-FAPy-dG per 107 nucleotides (Figure S-9). The accuracy (% difference) of the method was calculated as 97.7%, 91.7%, and 97.8% for the high, middle, and low validation points. The precision of the nanoLC-ESI+-MS/MS method (% CV) was calculated as 0.3, 3.9, and 3.1 % CV for the high, middle, and low validation points.
EB-Fapy-dG adduct detection and quantitation in CT DNA
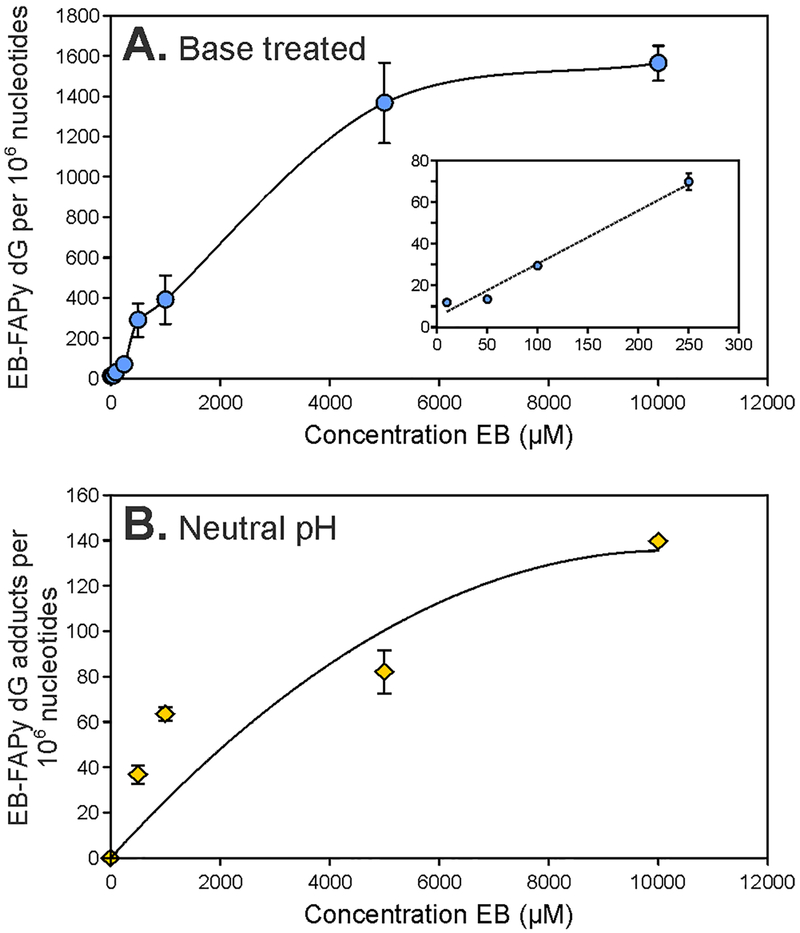
To determine whether EB-Fapy-dG adducts can be formed in DNA, calf thymus DNA was treated with increasing concentrations of EB. Aliquots of treated DNA were taken and either analyzed directly or treated with NaOH to induce imidazole ring opening. The newly developed isotope dilution nanoLC-ESI+-MS/MS methodology was employed to allow for accurate quantification of EB-Fapy-dG. Samples treated with NaOH exhibited a concentration dependent formation of EB-Fapy-dG, with adduct numbers ranging from 1.2 to 156.3 EB-FAPy-dG adducts per 105 nucleotides, depending on EB concentration. EB-FAPy-dG adduct numbers increased linearly between 10 μM and 5 mM EB, followed by a plateau (Figure 4A). EB-FAPy-dG were also detected in samples kept at physiological pH without base treatment, although the amounts were 10-fold lower (36.8 ± 2.4, 63.6 ± 4.0, 82.0 ± 2.9, and 139.6 ± 9.5 adducts per 106 nucleotides in DNA treated with 500 μM, 1 mM, 5 mM, and 10 mM EB, respectively, second-order polynomial equation, R2 = 0.98) (Figure 4B, Figure S-10).
Figure 4.
Concentration dependent formation of EB-Fapy-dG adducts in CT DNA treated with increasing concentrations of EB, followed by treatment with base (1N NaOH for 1 h) (A) and concentration dependent formation of EB-FAPy-dG adducts in CT DNA treated with increasing concentrations of EB and incubated at 37 °C at neutral pH for 3 days.
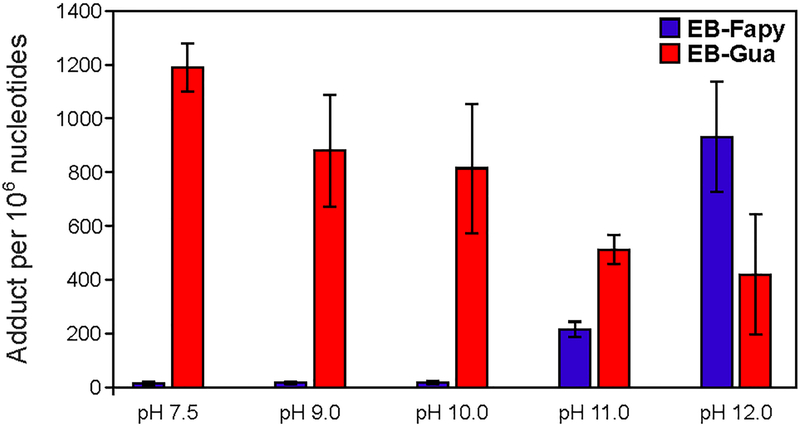
To establish pH requirements for EB-FAPy formation following treatment with EB, and to determine what fraction of N7-EB-dG adducts converts to EB-FAPy (Scheme 1), CT DNA was treated with 5 mM EB, followed by incubation at increasing pH (pH 7.5, 8.0, 9.0, 10.0, 11.0, 12.0) to induce EB-FAPy formation. For comparison, EB-Gua adducts were quantified in the same samples using the previously reported nanoLC-ESI+-MS/MS methodology.13–15 EB-Fapy-dG adducts were detected in all samples, with levels measured at 13.9 ± 6.4, 17.4 ± 4.2, 18.2 ± 7.0, 215.1 ± 29.1, and 931.4 ± 204.3 adducts per 106 nucleotides at pH 7.5, 9.0, 10.0, 11.0, and 12.0 (blue bars in Figure 5). EB-Gua levels in the same samples were measured at 1190.2 ± 89.3, 879.4 ± 207.6, 813.8 ± 240.4, 512.3 ± 53.3, and 420.6 ± 223.5 adducts per 106 nucleotides at pH 7.5, 9.0, 10.0, 11.0, and 12.0 respectively (red bars in Figure 5). Overall, EB-Gua adducts revealed an inverse relationship with EB-FAPy-dG, consistent with N7-EB-dG serving as a precursor for EB-Fapy-dG formation (Scheme 1).
Figure 5.
pH dependent formation of EB-FAPy-dG and EB-Gua II adducts in CT DNA treated with 5 mM EB.
EB-FAPy-dG formation in NEIL1+/+ and NEIL1−/− MEF cells treated with EB
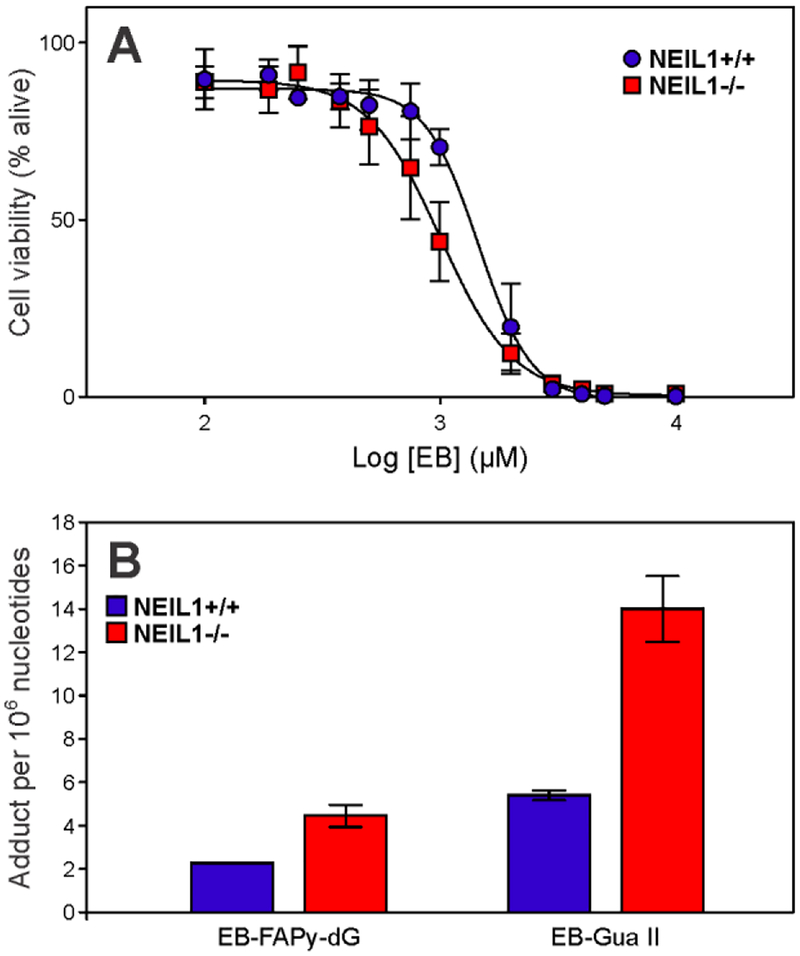
To investigate potential formation of EB-FAPy-dG in living cells treated with EB, mouse embryonic fibroblasts (MEF) were employed. Both NEIL1−/− and wild type cells were included. To determine the toxicity of EB in wild type and NEIL1−/− MEF cells, they were treated with increasing amounts of EB from 0 to 20,000 μM for 3 h, and cell viability after 48 h was determined using the Alamar Blue assay.55 NEIL1+/+ cells were found to be 1.5-fold less sensitive to EB (IC50 = 1,456 ± 126 μM, R2 = 0.98) as compared to NEIL1−/− cells (IC50 = 1,006 ± 108 μM, R2 = 0.97) (Figure 6A).
Figure 6.
Toxicity (A) and DNA adduct formation (B) in NEIL1+/+ and NEIL1−/− MEF cells treated with EB (500 μM)
We next investigated EB-Fapy-dG and EB-Gua II adduct formation in NEIL1+/+ and NEIL1−/− MEF cells treated with 500 μM of EB for 24 h. Following enzymatic digestion to 2’-deoxynucleosides and adduct enrichment, both EB-FAPy-dG and EB-Gua II were quantified by nanoLC-ESI+-MS/MS (Figure 6B). EB-treated NEIL1−/− cells had nearly 2-fold greater numbers of EB-FAPy-dG adducts (4.45 ± 0.51 EB-FAPy-dG per 106 nucleotides) as compared to NEIL1+/+ cells (2.27 ± 0.032 EB-FAPy-dG per 106 nucleotides, p-value < 0.01, Figure 6B). Similarly, NEIL1−/− cells had nearly 3-fold greater numbers of EB-Gua II adducts (14.01 ± 1.52 EB-Gua II per 106 nucleotides) as compared to NEIL1+/+ cells (5.35 ± 0.22 EB-Gua II per 106 nucleotides, p-value < 0.01, (Figure 6B)).
Discussion
1,3-Butadiene (BD) is a known carcinogen present in cigarette smoke,1 automobile exhaust,2 urban air, and utilized in many industrial processes.4 Previous studies revealed that the reactive epoxide metabolites of BD (EB, DEB, and EDB) are capable of alkylating nucleophilic positions of DNA to form a range of covalent BD-DNA adduct.6, 22 Although cationic N7-guanine adducts (N7-THBG and EB-Gua) are the most abundant,7–12 they are unlikely to be responsible for the mutagenic and genotoxic effects of BD because they do not disrupt Watson-Crick base pairing23, 24 and are spontaneously released form DNA to form apurinic sites.61–63 Alternatively, N7-alkyl-dG adducts can undergo base-catalyzed imidazole ring opening to yield hydrolytically stable and strongly genotoxic N5-alkyl-Fapy-dG adducts. However, to our knowledge, there are no previous reports of BD-derived FAPy adducts in vitro or in vivo, and it is unknown whether these adducts contribute to the observed mutagenic and genotoxic effects of BD.
In the present study, authentic N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxybut-3-en-1-yl)formamidopyrimidine (EB-Fapy-dG) was prepared for the first time by a synthetic strategy starting with 5’-protected dG (Scheme 2).56, 64 EB-FAPy free base and EB-FAPy-dG were characterized by UV-vis, ESI+-MS/MS, 1H-NMR, and 1H-13C HSQC (Figures S-4, 2, 3).
NMR analysis of purified EB-FAPy revealed this adduct exists in equilibrium between several related structures (Scheme 3 and Figures 1 and 2). Previous studies with AFB1-FAPy-dG and Me-FAPy-dG have shown the C5-N5 bond is capable of restricted rotation to form E/Z formamide isomers, while the formyl N-CO bond is capable of rotation as well to yield stable rotamers.56, 58, 59 We hypothesize that similar isomers exist for EB-FAPy-dG (Scheme 3). Additional possible isomers include β- and α-anomers and regioisomers formed following ring opening of the deoxyribose sugar and re-closure of the deoxyribose sugar involving the 5’-hydroxyl group to yield pyranose nucleosides.58, 65 However, previous studies reported that trityl protection of the 5’-OH prevents sugar anomerization and pyranose/furanose conversion in AFB1-FAPy-dG and Me-FAPy-dG containing DNA.28, 56, 59 In our synthetic scheme (Scheme 2), 5’-OH trityl protected dG was used, and we observed no evidence for EB-FAPy-dG isomerization to the corresponding pyranose species.
EB-FAPy-dG adduct formation was greatly accelerated at elevated pH (Figure 5). EB-FAPy-dG adduct numbers in CT DNA treated with 5 mM EB at pH 7.5 – 10.0 were below 20 adducts per 106 nucleotides, but increased to 215.1 ± 29.1, and 931.4 ± 204.3 adducts per 106 nucleotides at pH 11.0 and 12.0 respectively (Figure 5). This can be explained by a competition between two alternative fates of the hydrolytically unstable N7-EB-dG adducts (Scheme 1). Under physiological conditions, the preferred pathway involves N-glycosidic bond cleavage to form a free base EB-Gua adduct and an abasic site (Figure 5). At high pH, EB-FAPy-dG can undergo imidazole ring opening to form EB-FAPy-dG (Scheme 1, Figure 5).
If not repaired, EB-FAPy-dG adducts are expected to contribute to mutagenicity and genotoxicity of BD. Other alkyl-FAPy adducts adducts have been shown to inhibit DNA replication,29, 66–68 specifically blocking the extension step of lesion bypass.68 Me-FAPy-dG adducts block eukaryotic DNA polymeraeses α, β, and hPol δ/ PCNA,69 but are bypassed by translesion synthesis polymerases hPol η, κ, and hRev1/Pol ζ.69 All three TLS polymerases primarily insert the correct base (C) opposite Me-FAPy-dG, but hPol η and κ misinsert T, G, and A opposite the lesion and produce −1 deletion products.69 Recently, Minko et al reported that the α-anomer of nitrogen mustard-FAPy-dG adducts causes targeted G → T transversions in primate COS7 cells.70
Our cytotoxicity experiments show that NEIL1−/− MEF cells are 1.5-fold more sensitive to EB than NEIL1+/+ MEF cells, and therefore EB-FAPy-dG could potentially be a substrate for NEIL1 enzyme (Figure 6A). NEIL1 is a BER bifunctional DNA glycosylase enzyme that recognizes a range of adducts including thymine glycol, FAPy-G, FAPy-A, 8-oxoguanine, 5-hydroxyuracil, dihydroxyuracil, spiroaminohydantioins, guanidinohydantoins, and AFB1-FAPy-dG.50, 71, 72 Furthermore, both EB-FAPy-dG and EB-Gua II adducts were more abundant in NEIL1−/− treated with EB as compared to repair proficient NEIL1+/+ cells (Figure 6B). Taken together, these results suggest that the NEIL1 may participate in EB-FAPy-dG and EB-Gua II repair via the base excision repair mechanism. In addition to NEIL1, other BER enzymes such as formamidopyrimidine DNA glycosylase (FPG) and E. Coli endonuclease IV33, 34 may contribute to EB-FAPy-dG adduct repair. Indeed, our preliminary data indicate that EB-FAPy can be cleaved from the DNA backbone by bacterial FPG (Figure S-11).
Conclusions
In conclusion, we investigated the formation of ring-opened EB-FAPy-dG adducts in DNA and MEF cells treated with 3,4-epoxy-1-butene, a carcinogenic metabolite of 1,3-butadiene. Authentic standards of EB-FAPy-dG and 15N3- EB-FAPy-dG were prepared and structurally characterized by mass spectrometry and NMR. A sensitive isotope dilution nanoLC-ESI+-MS/MS methodology was developed for the detection and quantitation of EB-FAPy-dG in vitro and in cell cultures. EB-Fapy adducts formed in a concentration dependent manner in EB-treated CT DNA. EB-FAPy-dG was also detected in EB-treated cells, with higher adduct numbers observed in cells deficient in NEIL1 enzyme. We conclude that the EB-Fapy-dG adducts form under physiological conditions and may contribute to genotoxicity and mutagenicity of BD.
Supplementary Material
Supplmental Figure 1, Metabolism of 1,3-Butadiene and structures of characterized DNA adducts. SI Figure 2, Representative HPLC trace for purification of EB-FAPy-dG (4b in Scheme 2). SI Figure 3, Representative HPLC-UV trace for purification of EB-FAPy base. SI Figure 4, UV absorption spectra of EB-Fapy base in solutions of 0.1N HCl (pH: 1.0), water (pH: 7.0) and 0.1N NaOH (pH: 12.0). λ max, neutral pH : 274 nm, λ max, acidic pH(1.0): 266 nm, λ max, basic pH(13.0):264 nm. SI Figure 5, HPLC-UV analysis of dG in enzymatic digests of DNA isolated from MEF NEIL1+/+ cells. SI Figure 6, MS/MS fragmentation patterns of synthetic EB-FAPy free base. SI Figure 7, MS/MS fragmentation patterns of synthetic EB-FAPy-dG. SI Figure 8, ESI+-MS/MS spectrum of 15N3-EB-FAPy-dG (internal standard for mass spectrometry). SI Figure 9, Validation of nanoLC-ESI+-MS/MS assay for EB-FAPy-dG. SI Figure 10, Representative nanoLC-ESI-MS/MS trace for EB-FAPy-dG in calf thymus DNA treated with 5 mM EB and no base treatment. SI Figure 11, EB-FAPy base release from the DNA backbone by bacterial FPG and acid hydrolysis (0.1 N HCl at 70 °C for 1 h).
Funding information
This work was supported by the U.S. National Cancer Institute [grant number R01 ES-100670].
Abbreviations
- 1,N6-HMHP-dA
1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2′-deoxyadenosine
- AFB1
aflatoxin B1
- AFB1-N7-dG
trans-8,9-dihydro-8-(N7-guanyl)-9-hydroxy-aflatoxin B1
- BD
1,3-Butadiene
- BER
base excision repair
- bis-N7G-BD
1,4-bis-(guan-7-yl)-2,3-butanediol
- CE
collision energy
- CT DNA
Calf thymus DNA
- DEB
1,2,3,4-diepoxybutane
- dG
2’-deoxyguanosine
- DMEM
Dulbecco’s modified Eagle’s media
- DMTCl
4,4-dimethoxytrityl chloride
- EB
3,4-epoxy-1-butene
- EBD
1,2-dihydroxy-3,4-epoxybutane
- EB-Fapy-dG
N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-(2-hydroxy-3-buten-1-yl)-formamidopyrimidine
- EB-Gua I
N-7-(2-hydroxy-3-buten-1-yl)-guanine
- EB-Gua II
N-7-(1-hydroxy-3-buten-2-yl) guanine
- MEF
mouse embryonic fibroblasts
- N1-THB-Ade
N1-(2,3,4-trihydroxybut-1-yl)-adenine
- N7-EB-dG
N7-(2-hydroxy-3-buten-1-yl)-guanine
- N7-THBG
N7-(2,3,4-trihydroxybut-1-yl)-guanine
- NM
bis-(2-chloroethyl)-ethylamine
- OGG1
8-oxoguanine glycosylase
- PBS
phosphate-buffered saline
- SRM
selected reaction monitoring
References
- (1).Hecht SS (2003) Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer 3, 733–744. [DOI] [PubMed] [Google Scholar]
- (2).Pelz N, Dempster NM, and Shore PR (1990) Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J. Chromatogr. Sci 28, 230–235. [DOI] [PubMed] [Google Scholar]
- (3).Gustafson P, Barregard L, Strandberg B, and Sallsten G (2007) The impact of domestic wood burning on personal, indoor and outdoor levels of 1,3-butadiene, benzene, formaldehyde and acetaldehyde. J. Environ. Monit 9, 23–32. [DOI] [PubMed] [Google Scholar]
- (4).Cheng H, Sathiakumar N, Graff J, Matthews R, and Delzell E (2007) 1,3-Butadiene and leukemia among synthetic rubber industry workers: exposure-response relationships. Chem. Biol. Interact 166, 15–24. [DOI] [PubMed] [Google Scholar]
- (5).Duescher RJ, and Elfarra AA (1994) Human liver microsomes are efficient catalysts of 1,3-butadiene oxidation: evidence for major roles by cytochromes P450 2A6 and 2E1. Arch. Biochem. Biophys 311, 342–349. [DOI] [PubMed] [Google Scholar]
- (6).Blair IA, Oe T, Kambouris S, and Chaudhary AK (2000) 1,3-butadiene: cancer, mutations, and adducts. Part IV: Molecular dosimetry of 1,3-butadiene. Res. Rep. Health Eff. Inst, 151–190. [PubMed] [Google Scholar]
- (7).Kotapati S, Wickramaratne S, Esades A, Boldry EJ, Quirk DD, Pence MG, Guengerich FP, and Tretyakova NY (2015) Polymerase bypass of N6-deoxyadenosine adducts derived from epoxide metabolites of 1,3-butadiene. Chem. Res. Toxicol 28, 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Carmical JR, Nechev LV, Harris CM, Harris TM, and Lloyd RS (2000) Mutagenic potential of adenine N6 adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ. Mol. Mutagen 35, 48–56. [PubMed] [Google Scholar]
- (9).Carmical JR, Zhang M, Nechev L, Harris CM, Harris TM, and Lloyd RS (2000) Mutagenic potential of guanine N2 adducts of butadiene mono- and diolepoxide. Chem. Res. Toxicol 13, 18–25. [DOI] [PubMed] [Google Scholar]
- (10).Carmical JR, Zhang M, Nechev L, Harris CM, Harris TM, and Lloyd RS (2000) Mutagenic potential of guanine N2 adducts of butadiene mono- and diolepoxide volume 13, number 1, january 2000, pp 18–25. Chem. Res. Toxicol 13, 430. [DOI] [PubMed] [Google Scholar]
- (11).Steen AM, Meyer KG, and Recio L (1997) Analysis of hprt mutations occurring in human TK6 lymphoblastoid cells following exposure to 1,2,3,4-diepoxybutane. Mutagenesis 12, 61–67. [DOI] [PubMed] [Google Scholar]
- (12).Seo KY, Jelinsky SA, and Loechler EL (2000) Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res 463, 215–246. [DOI] [PubMed] [Google Scholar]
- (13).Tretyakova N, Lin Y, Sangaiah R, Upton PB, and Swenberg JA (1997) Identification and quantitation of DNA adducts from calf thymus DNA exposed to 3,4-epoxy-1-butene. Carcinogenesis 18, 137–147. [DOI] [PubMed] [Google Scholar]
- (14).Tretyakova NY, Chiang SY, Walker VE, and Swenberg JA (1998) Quantitative analysis of 1,3-butadiene-induced DNA adducts in vivo and in vitro using liquid chromatography electrospray ionization tandem mass spectrometry. J. Mass Spectrom 33, 363–376. [DOI] [PubMed] [Google Scholar]
- (15).Koc H, Tretyakova NY, Walker VE, Henderson RF, and Swenberg JA (1999) Molecular dosimetry of N-7 guanine adduct formation in mice and rats exposed to 1,3-butadiene. Chem. Res. Toxicol 12, 566–574. [DOI] [PubMed] [Google Scholar]
- (16).Sangaraju D, Villalta P, Goggin M, Agunsoye MO, Campbell C, and Tretyakova N (2013) Capillary HPLC-accurate mass MS/MS quantitation of N7-(2,3,4-trihydroxybut-1-yl)-guanine adducts of 1,3-butadiene in human leukocyte DNA. Chem. Res. Toxicol 26, 1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhao C, Vodicka P, RJ S, and Hemminki K (2000) Human DNA adducts of 1,3-butadiene, an important environmental carcinogen. Carcinogenesis 21, 107–111. [DOI] [PubMed] [Google Scholar]
- (18).Goggin M, Swenberg JA, Walker VE, and Tretyakova N (2009) Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res 69, 2479–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Goggin M, Anderson C, Park S, Swenberg J, Walker V, and Tretyakova N (2008) Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem. Res. Toxicol 21, 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, and Tretyakova N (2010) Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem. Res. Toxicol 23, 118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Goggin M, Seneviratne U, Swenberg JA, Walker VE, and Tretyakova N (2010) Column switching HPLC-ESI+-MS/MS methods for quantitative analysis of exocyclic dA adducts in the DNA of laboratory animals exposed to 1,3-butadiene. Chem. Res. Toxicol 23, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sangaraju D, Villalta PW, Wickramaratne S, Swenberg J, and Tretyakova N (2014) NanoLC/ESI+ HRMS3 quantitation of DNA adducts induced by 1,3-butadiene. J. Am. Soc. Mass Spectrom 25, 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Watson JD, and Crick FH (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738. [DOI] [PubMed] [Google Scholar]
- (24).Boysen G, Pachkowski BF, Nakamura J, and Swenberg JA (2009) The formation and biological significance of N7-guanine adducts. Mutat. Res 678, 76–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gates KS (2009) An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol 22, 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gates KS, Nooner T, and Dutta S (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol 17, 839–856. [DOI] [PubMed] [Google Scholar]
- (27).Tudek B (2003) Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol 36, 12–19. [DOI] [PubMed] [Google Scholar]
- (28).Pujari SS, and Tretyakova N (2017) Chemical biology of N5-substituted formamidopyrimidine DNA adducts. Chem Res Toxicol 30, 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tudek B, Boiteux S, and Laval J (1992) Biological properties of imidazole ring-opened N7-methylguanine in M13mp18 phage DNA. Nucleic Acids Res 20, 3079–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Graziewicz MA, Zastawny TH, Olinski R, Speina E, Siedlecki J, and Tudek B (2000) Fapyadenine is a moderately efficient chain terminator for prokaryotic DNA polymerases. Free Radic. Biol. Med 28, 75–83. [DOI] [PubMed] [Google Scholar]
- (31).Jena NR, Mark AE, and Mishra PC (2014) Does tautomerization of FapyG influence its mutagenicity? Chemphyschem 15, 1779–1784. [DOI] [PubMed] [Google Scholar]
- (32).Gehrke TH, Lischke U, Gasteiger KL, Schneider S, Arnold S, Muller HC, Stephenson DS, Zipse H, and Carell T (2013) Unexpected non-Hoogsteen-based mutagenicity mechanism of FaPy-DNA lesions. Nat Chem Biol 9, 455–461. [DOI] [PubMed] [Google Scholar]
- (33).Chetsanga CJ, and Lindahl T (1979) Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res 6, 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Christov PP, Banerjee S, Stone MP, and Rizzo CJ (2010) Selective incision of the alpha-N-methyl-formamidopyrimidine anomer by Escherichia coli endonuclease IV. J Nucleic Acids 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tudek B, van Zeeland AA, Kusmierek JT, and Laval J (1998) Activity of Escherichia coli DNA-glycosylases on DNA damaged by methylating and ethylating agents and influence of 3-substituted adenine derivatives. Mutat. Res 407, 169–176. [DOI] [PubMed] [Google Scholar]
- (36).Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, and Bohr VA (2005) Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem 280, 40544–40551. [DOI] [PubMed] [Google Scholar]
- (37).Dherin C, Radicella JP, Dizdaroglu M, and Boiteux S (1999) Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res 27, 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, et al. (2009) Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 8, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Klich MA (2007) Aspergillus flavus: the major producer of aflatoxin. Mol Plant Pathol 8, 713–722. [DOI] [PubMed] [Google Scholar]
- (40).Schindler AF, Palmer JG, and Eisenberg WV (1967) Aflatoxin production by Aspergillus flavus as related to various temperatures. Appl Microbiol 15, 1006–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang JS, Huang T, Su J, Liang F, Wei Z, Liang Y, Luo H, Kuang SY, Qian GS, et al. (2001) Hepatocellular carcinoma and aflatoxin exposure in Zhuqing Village, Fusui County, People’s Republic of China. Cancer Epidemiol. Biomarkers Prev 10, 143–146. [PubMed] [Google Scholar]
- (42).Li FQ, Yoshizawa T, Kawamura O, Luo XY, and Li YW (2001) Aflatoxins and fumonisins in corn from the high-incidence area for human hepatocellular carcinoma in Guangxi, China. J Agric Food Chem 49, 4122–4126. [DOI] [PubMed] [Google Scholar]
- (43).Ding X, Wu L, Li P, Zhang Z, Zhou H, Bai Y, Chen X, and Jiang J (2015) Risk assessment on dietary exposure to aflatoxin B1 in post-harvest peanuts in the Yangtze River Ecological Region. Toxins (Basel) 7, 4157–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Shimada T, and Guengerich FP (1989) Evidence for cytochrome P-450NF, the nifedipine oxidase, being the principal enzyme involved in the bioactivation of aflatoxins in human liver. Proc. Natl. Acad. Sci. U. S. A 86, 462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Hertzog PJ, Smith JR, and Garner RC (1982) Characterisation of the imidazole ring-opened forms of trans-8,9-dihydro-8,9-dihydro-8-(7-guanyl)9-hydroxy aflatoxin B1. Carcinogenesis 3, 723–725. [DOI] [PubMed] [Google Scholar]
- (46).Bailey EA, Iyer RS, Stone MP, Harris TM, and Essigmann JM (1996) Mutational properties of the primary aflatoxin B1-DNA adduct. Proc. Natl. Acad. Sci. U. S. A 93, 1535–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, and Harris CC (1991) Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 350, 427–428. [DOI] [PubMed] [Google Scholar]
- (48).Bressac B, Kew M, Wands J, and Ozturk M (1991) Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350, 429–431. [DOI] [PubMed] [Google Scholar]
- (49).McMahon G, Davis EF, Huber LJ, Kim Y, and Wogan GN (1990) Characterization of c-Ki-ras and N-ras oncogenes in aflatoxin B1-induced rat liver tumors. Proc. Natl. Acad. Sci. U. S. A 87, 1104–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Vartanian V, Minko IG, Chawanthayatham S, Egner PA, Lin YC, Earley LF, Makar R, Eng JR, Camp MT, et al. (2017) NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc Natl Acad Sci U S A 114, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Graziewicz MA, Zastawny TH, Olinski R, and Tudek B (1999) SOS-dependent A-->G transitions induced by hydroxyl radical generating system hypoxanthine/xanthine oxidase/Fe3+/EDTA are accompanied by the increase of Fapy-adenine content in M13 mp18 phage DNA. Mutat. Res 434, 41–52. [DOI] [PubMed] [Google Scholar]
- (52).Gruppi F, Hejazi L, Christov PP, Krishnamachari S, Turesky RJ, and Rizzo CJ (2015) Characterization of nitrogen mustard formamidopyrimidine adduct formation of bis(2-chloroethyl)ethylamine with calf thymus DNA and a human mammary cancer cell line. Chem Res Toxicol 28, 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, et al. (2006) The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A 103, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Cavalieri LF, Bendich A, and . (1948) Ultraviolet absorption spectra of purines, pyrimidines and triazolopyrimidines. J. Am. Chem. Soc 70, 3875–3880. [DOI] [PubMed] [Google Scholar]
- (55).O’Brien J, Wilson I, Orton T, and Pognan F (2000) Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem 267, 5421–5426. [DOI] [PubMed] [Google Scholar]
- (56).Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, and Rizzo CJ (2008) Site-specific synthesis and characterization of oligonucleotides containing an N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylfor mamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem Res Toxicol 21, 2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Singer B, and Grunberger D (1983) Molecular biology of mutagens and carcinogens. pp. Plenum Press, New York. [Google Scholar]
- (58).Tomasz M, Lipman R, Lee MS, Verdine GL, and Nakanishi K (1987) Reaction of acid-activated mitomycin C with calf thymus DNA and model guanines: elucidation of the base-catalyzed degradation of N7-alkylguanine nucleosides. Biochemistry 26, 2010–2027. [DOI] [PubMed] [Google Scholar]
- (59).Brown KL, Deng JZ, Iyer RS, Iyer LG, Voehler MW, Stone MP, Harris CM, and Harris TM (2006) Unraveling the aflatoxin-FAPY conundrum: structural basis for differential replicative processing of isomeric forms of the formamidopyrimidine-type DNA adduct of aflatoxin B1. J. Am. Chem. Soc 128, 15188–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Boiteux S, Belleney J, Roques BP, and Laval J (1984) Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res 12, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Rusyn I, Asakura S, Li Y, Kosyk O, Koc H, Nakamura J, Upton PB, and Swenberg JA (2005) Effects of ethylene oxide and ethylene inhalation on DNA adducts, apurinic/apyrimidinic sites and expression of base excision DNA repair genes in rat brain, spleen, and liver. DNA Repair (Amst) 4, 1099–1110. [DOI] [PubMed] [Google Scholar]
- (62).Rios-Blanco MN, Faller TH, Nakamura J, Kessler W, Kreuzer PE, Ranasinghe A, Filser JG, and Swenberg JA (2000) Quantitation of DNA and hemoglobin adducts and apurinic/apyrimidinic sites in tissues of F344 rats exposed to propylene oxide by inhalation. Carcinogenesis 21, 2011–2018. [DOI] [PubMed] [Google Scholar]
- (63).O’Brien PJ, and Ellenberger T (2004) Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem 279, 9750–9757. [DOI] [PubMed] [Google Scholar]
- (64).Christov PP, Son KJ, and Rizzo CJ (2014) Synthesis and characterization of oligonucleotides containing a nitrogen mustard formamidopyrimidine monoadduct of deoxyguanosine. Chem Res Toxicol 27, 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Berger M, and Cadet J (1985) Isolation and characterization of the radiation-induced degradation products of 2’-deoxyguanosine in oxygen-free aqueous-solutions. Zeitschrift Fur Naturforschung Section B-a Journal of Chemical Sciences 40, 1519–1531. [Google Scholar]
- (66).Boiteux S, and Laval J (1983) Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochem Biophys Res Commun 110, 552–558. [DOI] [PubMed] [Google Scholar]
- (67).O’Connor TR, Boiteux S, and Laval J (1988) Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res 16, 5879–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Asagoshi K, Terato H, Ohyama Y, and Ide H (2002) Effects of a guanine-derived formamidopyrimidine lesion on DNA replication: translesion DNA synthesis, nucleotide insertion, and extension kinetics. J. Biol. Chem 277, 14589–14597. [DOI] [PubMed] [Google Scholar]
- (69).Christov PP, Yamanaka K, Choi JY, Takata K, Wood RD, Guengerich FP, Lloyd RS, and Rizzo CJ (2012) Replication of the 2,6-diamino-4-hydroxy-N5-(methyl)-formamidopyrimidine (MeFapy-dGuo) adduct by eukaryotic DNA polymerases. Chem. Res. Toxicol 25, 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Minko IG, Rizzo CJ, and Lloyd RS (2017) Mutagenic potential of nitrogen mustard-induced formamidopyrimidine DNA adduct: contribution of the non-canonical alpha-anomer. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, and Mitra S (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U. S. A 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, and Izumi T (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem 277, 30417–30420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplmental Figure 1, Metabolism of 1,3-Butadiene and structures of characterized DNA adducts. SI Figure 2, Representative HPLC trace for purification of EB-FAPy-dG (4b in Scheme 2). SI Figure 3, Representative HPLC-UV trace for purification of EB-FAPy base. SI Figure 4, UV absorption spectra of EB-Fapy base in solutions of 0.1N HCl (pH: 1.0), water (pH: 7.0) and 0.1N NaOH (pH: 12.0). λ max, neutral pH : 274 nm, λ max, acidic pH(1.0): 266 nm, λ max, basic pH(13.0):264 nm. SI Figure 5, HPLC-UV analysis of dG in enzymatic digests of DNA isolated from MEF NEIL1+/+ cells. SI Figure 6, MS/MS fragmentation patterns of synthetic EB-FAPy free base. SI Figure 7, MS/MS fragmentation patterns of synthetic EB-FAPy-dG. SI Figure 8, ESI+-MS/MS spectrum of 15N3-EB-FAPy-dG (internal standard for mass spectrometry). SI Figure 9, Validation of nanoLC-ESI+-MS/MS assay for EB-FAPy-dG. SI Figure 10, Representative nanoLC-ESI-MS/MS trace for EB-FAPy-dG in calf thymus DNA treated with 5 mM EB and no base treatment. SI Figure 11, EB-FAPy base release from the DNA backbone by bacterial FPG and acid hydrolysis (0.1 N HCl at 70 °C for 1 h).