Abstract
PRRX1 is a homeodomain transcriptional factor, which has two isoforms, PRXX1A and PRRX1B. The PRRX1 isoforms have been demonstrated to be important in pancreatic cancer, especially in the regulation of epithelial-to-mesenchymal transition (EMT) in Pancreatic Ductal Adenocarcinoma (PDAC) and of mesenchymal-to-epithelial transition (MET) in liver metastasis. In order to determine the functional underpinnings of PRRX1 and its isoforms, we have unraveled a new interplay between PRRX1 and the FOXM1 transcriptional factors. Our detailed biochemical analysis reveals the direct physical interaction between PRRX1 and FOXM1 proteins that requires the PRRX1A/B 200-222/217 amino acid (aa) region and the FOXM1 Forkhead domain. Additionally, we demonstrate the cooperation between PRRX1 and FOXM1 in the regulation of FOXM1-dependent transcriptional activity. Moreover, we establish FOXM1 as a critical downstream target of PRRX1 in pancreatic cancer cells. We demonstrate a novel role for PRRX1 in the regulation of genes involved in DNA repair pathways. Indeed, we show that expression of PRRX1 isoforms may limit the induction of DNA damage in pancreatic cancer cells. Finally, we demonstrate that targeting FOXM1 with the small molecule inhibitor FDI6 suppress pancreatic cancer cell proliferation and induces their apoptotic cell death. FDI6 sensitizes pancreatic cancer cells to Etoposide and Gemcitabine induced apoptosis. Our data provide new insights into PRRX1’s involvement in regulating DNA damage and provide evidence of a possible PRRX1-FOXM1 axis that is critical for PDAC cells.
Keywords: PRRX1A, PRRX1B, FOXM1, pancreatic cancer, DNA damage
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is predicted to become the second leading cause of cancer-related death in the United States by 2030 and remains the cancer with the lowest 5-year relative survival rate (8%) (1,2). This can be explained in part by the lack of early detection methods, proclivity for metastasis and the underlying chemoresistance of PDAC cells. Recent advances in Genetically Engineered Mouse Models (GEMMs) of pancreatic cancer, such as the lineage-labeled Pdx1-cre;LSL-KrasG12D/+;p53fl/+;R26YFP (abbreviated as “KPCY”) mouse model, reveal that epithelial-to-mesenchymal transition (EMT) and dissemination of pancreatic preneoplastic cells precedes PDAC tumor formation (3). Pancreatic epithelial cell plasticity plays an important role during PDAC initiation, progression, and metastasis. Thus, specific genes and pathways that play functional roles along this continuum are of paramount importance to provide new mechanistic insights and also offer new therapeutic perspectives.
We undertook previously an unbiased approach to identify genes involved in pancreatic epithelial cell plasticity during pancreatic ductal development, acinar-to-ductal metaplasia (ADM) and pancreatic intraepithelial neoplasia (PanIN) (4). This analysis revealed that the Paired-Related homeobox 1 (Prrx1) transcription factor (TF) was the most up-regulated TF shared amongst these three processes (4). The alternative splicing of the PRRX1 gene give rise to two isoforms: PRRX1A (245 amino acids) and PRRX1B (217 amino acids) (5,6). Our work established that PRRX1A regulates EMT in primary PDAC and PRRX1B promotes the reverse process, mesenchymal-to-epithelial transition (MET), in liver metastasis (7). This isoform-specific regulation of EMT-MET axis builds upon previous findings, that PRRX1 induces EMT in breast (8), colorectal (9,10) and gastric cancer (11). Furthermore, loss (or low levels) of PRRX1 is required for MET and metastatic colonization in breast and colorectal cancer (8,10). It is possible that competition occurs de novo between PRRX1A and PRRX1B to drive EMT or MET respectively, in the context of tumor progression. Therefore, the study of the underlying mechanisms regulating PRRX1 isoform switching and the identification of possible co-factors involved will provide useful new insights into PRRX1 biology.
Forkhead Box M1 (FOXM1) is a member of the Forkhead box family of TF that shares a winged-helix DNA-binding domain (12). FOXM1 is upregulated in a variety of solid tumors and is well renown within the FOX genes for its crucial role in regulating the cell cycle. In the context of pancreatic cancer, FOXM1 is overexpressed and has been shown to regulate migration, invasion, and EMT (13–15).
In this study, we have identified FOXM1 as a critical downstream target of PRRX1 and also a novel binding partner of PRRX1. We observe cooperation between FOXM1 and PRRX1 in the induction of FOXM1-dependent transcriptional activity. Additionally, we describe a new role for PRRX1 in the DNA damage response (DDR) via the induction of key DNA repair genes. Our results reveal that expression of PRRX1 may limit the induction of DNA damage. Furthermore, we demonstrate that FOXM1 inhibition via FDI6, a small molecule inhibitor of FOXM1 DNA binding (16), increases DNA damage, reduces PDAC cell growth and induces their apoptosis. Finally, treatment with FDI6 also promotes induction of apoptosis by Gemcitabine, part of a chemotherapy regimen for PDAC in the clinical setting, suggesting FOXM1 as a potential target to improve survival of pancreatic cancer patients.
RESULTS
PRRX1 positively regulates FOXM1 expression and activity
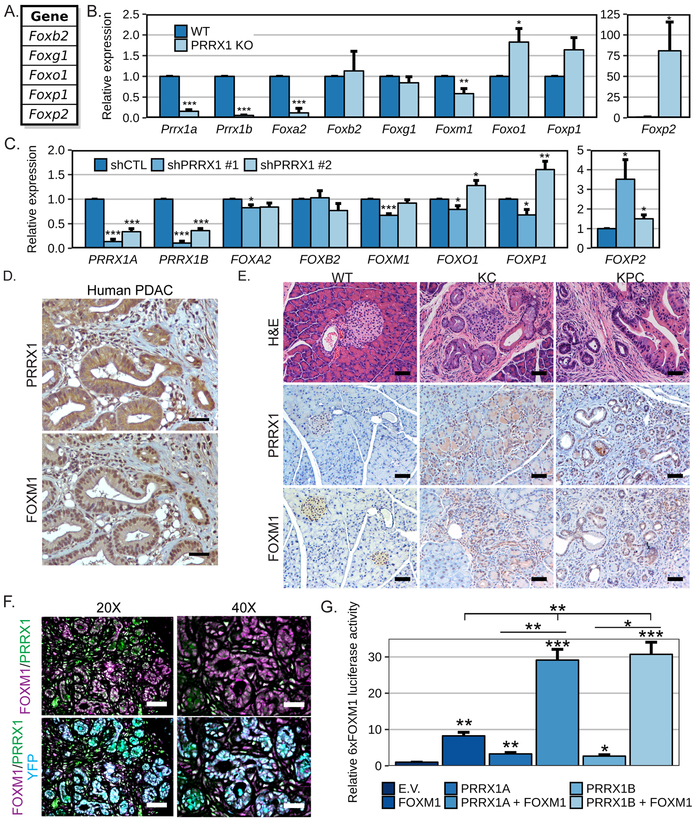
Previous ChIP-seq results from our group identified shared and specific target genes of PRRX1 isoforms in pancreatic tumor cells (4). Interestingly, PRRX1B was found bound to a set of genes from the Forkhead family of transcription factors (unpublished observations, Fig. 1A), suggesting PRRX1 may be a central regulator of FOX TFs gene expression. Therefore, we analyzed the expression of a panel of selected FOX genes in mouse embryonic fibroblasts (MEFs) established from wild type (WT) or global Prrx1 knockout (KO) mice. Prrx1KO MEF cells showed significantly lower expression of Foxa2 and Foxm1 compared to WT MEF cells (Fig.1B). By contrast, Foxo1 and Foxp2 expression were significantly higher in Prrx1KO compared to WT MEFs, while a trend for increased Foxp1 was also observed (Fig.1B). These results were validated in PANC1 human pancreatic tumor cells in which PRRX1 expression was knocked down by shRNAs. PANC1 cells expressing an shRNA targeting both isoforms of PRRX1 reduced levels of PRRX1A by 86% (shPRRX1 #1) or 66% (shPRRX1 #2) and of PRRX1B by 89% (shPRRX1 #1) or 64% (shPRRX1 #2) (Fig.1C). Similar to MEFs, loss of PRRX1 expression significantly reduced expression of FOXA2 and FOXM1, and increased FOXP2 in PANC1 cells (Fig.1C). Knockdown of PRRX1 expression in PANC1 also regulated expression of FOXO1 and FOXP1, although conflicting results were observed (Fig.1C).
Figure 1. PRRX1 and FOXM1 in PDAC.
A. List of genes identified in Prrx1b ChIP-sequencing (7). The relative mRNA expression levels of PRRX1 isoforms and the indicated FOX genes were measured by qPCR B. in MEF established from wild type (WT) or PRRX1 KO mice (n=4-5) and C. in PANC1 cells stably expressing either a non-targeting shRNA (shCTL) or shRNA targeting PRRX1 (n=5). D. Consecutive sections from a human PDAC patient tissue stained for PRRX1 and FOXM1 by IHC. A representative 20X field is shown for each with a 50μm scale bar. E-F. Mouse pancreatic tissue from a wild type (WT; 18w; normal), Pdx1-Cre;KrasG12D/+ (KC; 41w; PanIN) or Pdx1-Cre;KrasG12D/+;p53R175H/+;R26YFP (KPCY; 16w; PDAC) were analyzed. E. Morphology was assessed by H&E staining and the expression of PRRX1 and FOXM1 were analyzed by IHC. A representative 20X field is shown for each with a 50μm scale bar. F. Immunofluorescence staining of tissue from KPCY mice for FOXM1 (Magenta), PRRX1 (Green), YFP (Cyan) and DAPI (Blue) staining are shown. Representative 20X and 40X fields are shown for the merged channels with 50 or 25μm scale bars respectively. The single channel images are shown in Supplementary Fig.1B. G. PANC1 cells were transiently transfected with the 6xFOXM1 luciferase and the SV40-renilla reporters with or without the indicated PRRX1 or FOXM1 constructs. The luciferase and renilla activities were measured 48 hrs. post-transfection. The 6xFoxm1 luciferase activity was normalized to the renilla and is shown relative to empty vector (E.V.) control cells (n=3 independent experiments; *=p<0.05, **=p<0.01 and ***=p<0.001).
These results demonstrate a possible link between PRRX1 and FOX genes in MEFs and PDAC cells, and thus we analyzed PRRX1 and FOXM1 expression in pancreatic tissues from human PDAC patient and mouse models of pancreatic cancer. Immunohistochemistry (IHC) of human PDAC tissue demonstrated that both PRRX1 and FOXM1 are highly expressed in PDAC (Fig. 1D). Furthermore, a more detailed analysis of IHC from mouse tissue revealed that both PRRX1 and FOXM1 are increased early on during transformation, in pancreatic intraepithelial neoplasia (PanIN) lesions harvested from Pdx1-Cre;LSL-KrasG12D/+ (KC) (17) and Pdx1-Cre;LSL-KrasG12D/+;p53R175H/+;R26YFP (KPCY) (3,18) mice (Fig.1E). Both PRRX1 and FOXM1 remained elevated in PDAC tumor tissue from KPCY mice compared to WT controls (Fig.1E).
Strikingly, co-immunofluorescence staining of FOXM1 and PRRX1 in mouse PDAC tissues revealed co-localization of both transcription factors, suggesting they might cooperate in the regulation of target genes (Fig.1F). In order to determine if FOXM1 and PRRX1 cooperatively regulate FOXM1-dependent transcriptional activity, we used the 6xFOXM1-luciferase reporter containing 6 copies of the FOXM1 DNA binding site (19). Transfection of PANC1 cells with a construct encoding FOXM1 increased the activity of the 6xFOXM1-luciferase reporter construct (Fig.1G). Interestingly, the exogenous expression of PRRX1A or PRRX1B alone was sufficient to induce activity of the FOXM1 reporter (Fig.1G). Surprisingly, the activity of the 6xFOXM1-luciferase reporter was dramatically increased by co-transfection of FOXM1 construct with a plasmid encoding either PRRX1A or PRRX1B (Fig.1G). Knockdown of PRRX1 in PANC1 cells also reduced expression of the canonical FOXM1 transcriptional targets CDC25B and CCNB1 (Supplementary Fig. 2A). Furthermore, we observed that co-expression of FOXM1 and PRRX1 cooperatively activated the known PRRX1 target gene, Tenascin-C (Supplementary Fig.2B), suggesting that FOXM1 may also help to stimulate canonical PRRX1-mediated transcriptional networks.
PRRX1 and FOXM1 physically interact
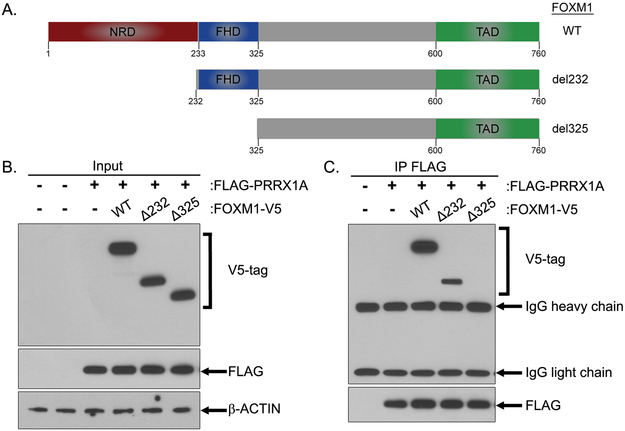
Since our results revealed potential cooperation between FOXM1 and PRRX1, we next tested if they can physically interact, and if so, to delineate which domains may be involved. To that end, we generated N-terminal deletion mutants of FOXM1 lacking the N-terminal repressor domain (NRD) or both the NRD and Forkhead domain (FHD) (Fig.2A). The different FOXM1 constructs were transiently co-expressed with a FLAG-tagged WT PRRX1A in HEK293T cells (Fig.2B). Next, we immunoprecipitated the FLAG-tagged PRRX1A and observed that it binds WT FOXM1 and FOXM1Δ232, but not to the FOXM1Δ325 deletion mutant (Fig.2C). These results demonstrate that the Forkhead domain of FOXM1, whose loss is unique to FOXM1Δ325, is necessary for its binding to PRRX1.
Figure 2. FOXM1 interacts with PRRX1 through its Forkhead domain (FHD).
A. Schematic representation of the C-terminal V5-tagged FOXM1 wild type and deletion mutant constructs. NRD (N-terminal repressor domain), FHD (ForkHead domain) and TAD (Transactivation Domain). B-C. HEK293T cells were transiently transfected with the FLAG-tagged PRRX1A with either the FOXM1 constructs shown in (A) or the empty vector. The cells were lysed at 48 hours post-transfection. B. Western Blot analysis of the indicated protein expression levels in the protein lysates (Input). C. The FLAG-PRRX1A was immunoprecipitated using a FLAG antibody and co-immunoprecipitation of FOXM1 constructs was analyzed by Western Blot.
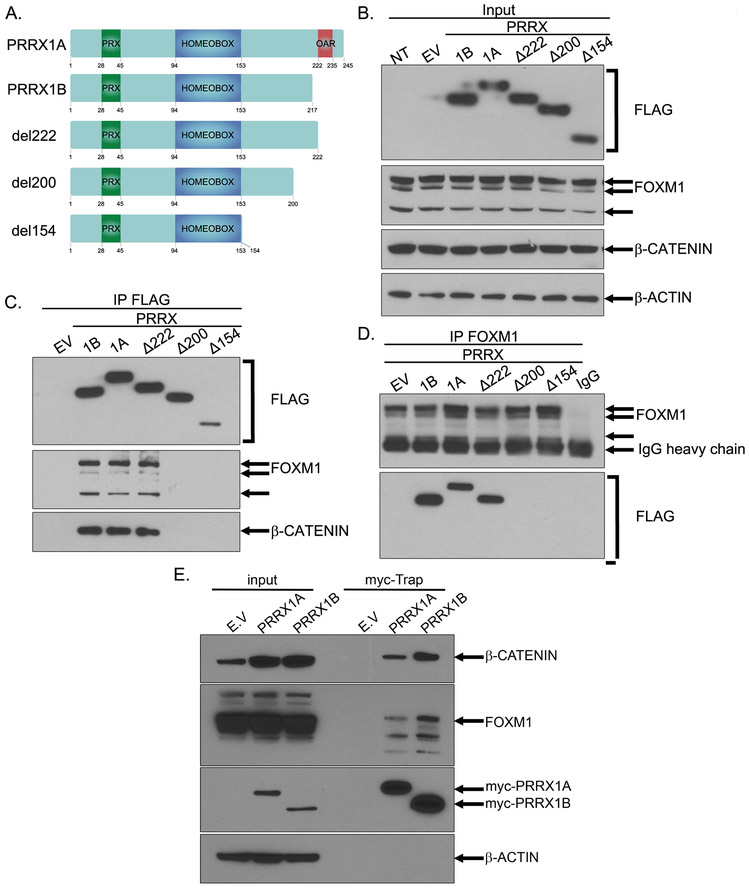
To map further the interaction between FOXM1 and PRRX1, we generated plasmids encoding FLAG-tagged PRRX1A, PRRX1B or PRRX1 deletion mutants lacking different C-terminal regions (Fig.3A). The PRRX1Δ222 mutant lacks the C-terminal region containing the “otp, aristaless, and rax” (OAR) domain, while PRRX1Δ200 and PRRX1Δ154 lack a C-terminal region extending up to the homeobox domain (Fig.3A). The expression of the FLAG-tagged PRRX1 constructs was assessed in HEK293T cells following transfection with the different plasmids (Fig.3B). We immunoprecipitated the WT or mutant PRRX1 using a FLAG antibody and observed that endogenous FOXM1 bound all forms of FLAG-tagged PRRX1 except for PRRX1Δ200 and PRRX1Δ154 (Fig.3C). β-CATENIN, a known FOXM1 binding partner (20), was also bound to all forms of PRRX1 interacting with FOXM1 i.e. PRRX1A, PRRX1B and PRRX1Δ222 (Fig.3C). To support further these results, we immunoprecipitated endogenous FOXM1 and demonstrated that it binds to FLAG-tagged PRRX1A, PRRX1B and PRRX1Δ222 (Fig.3D). The interaction between FOXM1, β-CATENIN and PRRX1 isoforms (A and B) was confirmed in PANC1 cells stably expressing myc-tagged PRRX1 constructs (Fig.3E). Collectively, these experiments indicate that the PRRX1A 200-222aa and PRRX1B 200-217aa regions are crucial for interaction with FOXM1 and β-CATENIN.
Figure 3. PRRX1 isoforms interact with FOXM1 through their 200-222/217aa region.
A. Schematic representation of the N-terminal FLAG-tagged PRRX1 wild type and deletion mutant constructs. B-D. HEK293T cells were transiently transfected with the FLAG-tagged constructs shown in (A) or the empty vector. Then, the cells were lysed at 48 hours post-transfection. B. Western Blot analysis of the indicated protein expression levels in the protein lysates (Input). C. The PRRX1 constructs were immunoprecipitated (IP) using a FLAG antibody and the expression levels of the indicated proteins was analyzed by Western Blot. D. The endogenous FOXM1 was immunoprecipitated and co-immunoprecipitation of PRRX1 constructs was analyzed by Western Blot. E. Myc-Trap® immunoprecipitation was performed on stable populations of PANC1 cells expressing empty vector (E.V.), myc-PRRX1A or myc-PRRX1B. The expression levels of the indicated proteins was analyzed by Western blot in total lysates (Input) and IP (Myc-Trap).
The ability of homeobox proteins to homodimerize or heterodimerize has been postulated as a possible mechanism of isoform specific transcriptional activity (21–23). Co-immunoprecipitation assays with myc-tagged and FLAG-tagged PRRX1 isoforms constructs were performed to evaluate PRRX1 isoform dimerization. Homodimers of both PRRX1A and PRRX1B, together with PRRX1A and PRRX1B heterodimers, were observed (Supplementary Fig.3A). These experiments were repeated with PRRX1 deletion mutants and we observed that myc-tagged PRRX1A bound to Flag-tagged PRRX1A and PRRX1Δ222 but not to PRRX1Δ200 and PRRX1Δ154 (Supplementary Fig.3B). Thus, the same region that was required for FOXM1 interaction (i.e. 200-222/217aa) is essential for PRRX1 isoform dimerization.
PRRX1 limits DNA damage in pancreatic tumor cells
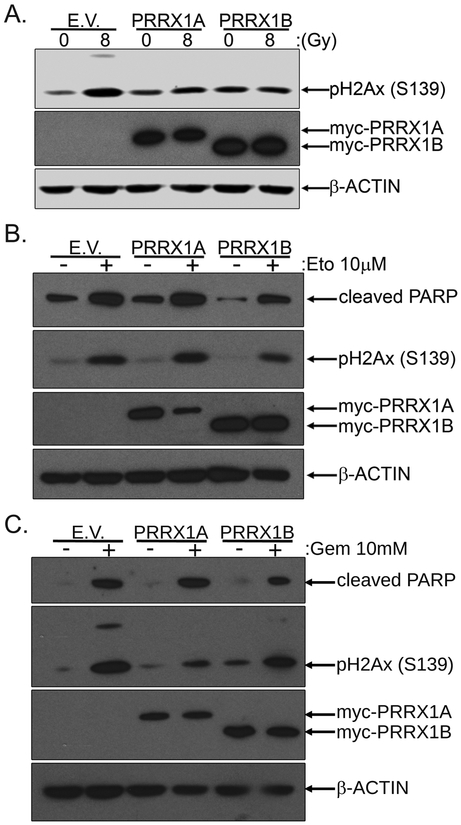
Our results thus far establish that PRRX1 and FOXM1 interact at the biochemical level. FOXM1 regulates genes involved in DNA damage repair and thus play a role critical role in the DNA damage response (DDR)(24–27). In order to determine the potential functional involvement of PRRX1 in the DDR, we irradiated PANC1 cells expressing myc-tagged PRRX1A, PRRX1B or the empty vector (E.V.). We evaluated the levels of DNA damage by analyzing histone H2Ax phosphorylation at Ser-139 (pH2Ax), which is induced following DNA damage. The irradiation (IR) of control (E.V.) PANC1 cells induced DNA damage as revealed by increased pH2Ax levels 2h post-irradiation (Fig.4A). Lower pH2Ax levels were observed in irradiated PANC1 cells expressing myc-PRRX1A and myc-PRRX1B compared to irradiated control (E.V.) PANC1 cells (Fig.4A). This result suggests that PRRX1 isoforms may potentially limit DNA damage induction in pancreatic tumor cells following irradiation.
Figure 4. PRRX1 isoforms limit DNA damage induced by IR, Etoposide and Gemcitabine.
PANC1 cells were transiently transfected with the pcDNA3.1(+), pcDNA3.1-myc-PRRX1A or pcDNA3.1-myc-PRRX1B plasmids. A. Then 48 hours post-transfection, the cells were irradiated with 0 or 8Gy and lysed 2 hours following irradiation. The levels of the DNA damage marker, phosphorylated H2Ax (Ser139), was analyzed by Western Blot. B. At 24 hours following transfection, the cells were treated with Etoposide 10μM or DMSO for 24 hours. The levels of cleaved PARP and phosphorylated H2Ax (Ser139), respectively a marker of apoptosis and DNA damage, were analyzed by Western Blot. C. At 24 hours following transfection, the cells were treated with Gemcitabine 10μM or DMSO for 24 hours. The cleaved PARP and phospho-H2Ax (Ser139) were analyzed by Western Blot.
Next, we evaluated PRRX1’s contribution to the DDR by using another DNA damaging agent, Etoposide, a topoisomerase II inhibitor, known to induce DNA double strand breaks (DSB). Control PANC1 cells showed increased pH2Ax levels following Etoposide treatment for 24h (Fig.4B). In agreement with irradiation-mediated DNA damage, the expression of myc-PRRX1A or myc-PRRX1B in PANC1 cells reduced the Etoposide-induced increase of pH2Ax (Fig.4B). Finally, we determined the impact of PRRX1 expression in PANC1 cells following treatment with Gemcitabine. Gemcitabine is a deoxycytidine analog that interferes with DNA replication leading to stalled replication forks and activation of the DDR (28). Gemcitabine treatment for 24h induced pH2Ax in PANC1 cells (Fig.4C). Similar to IR and Etoposide, Gemcitabine-induced increase of pH2Ax in cells expressing either myc-PRRX1A or myc-PRRX1B was lower than in cells expressing the empty vector (Fig.4C). It is noteworthy that expression of myc-PRRX1B was able to limit the apoptosis induced by Etoposide and Gemcitabine as revealed by cleaved PARP levels (Fig.4B-C). These results suggest the PRRX1 isoforms may limit the level of DNA damage under independent experimental conditions.
PRRX1 regulates genes involved in DNA damage repair
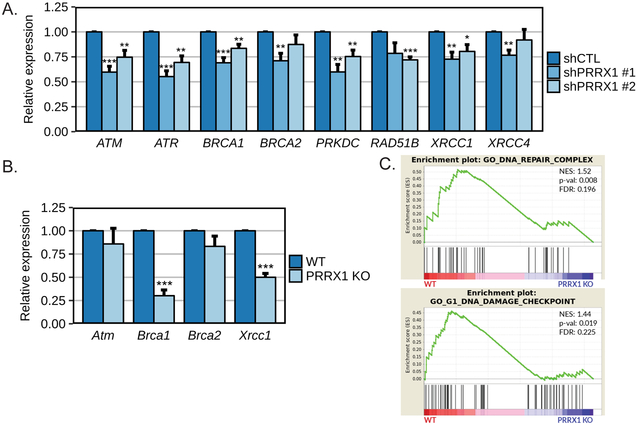
We pursued our investigation of the role of PRRX1 in the DDR by evaluating if PRRX1 regulates target genes involved in DNA repair. In that regard, we analyzed the expression of a set of key genes implicated in different DNA repair pathways, including many that were previously shown to be regulated by FOXM1 (25,29). First, we observed reduced expression of ATM, ATR, BRCA1 and PRKDC (DNA-PK) in PANC1 cells after knockdown of PRRX1 with two independent shRNAs (Fig.5A). Of note, expression of BRCA2, XRCC1 and XRCC4 were reduced only with shPRRX1 #1, which showed greater efficiency of PRRX1 knockdown (Fig.5A and 1C). We also noticed lower expression levels of RAD51B following PRRX1 knockdown, but only in PANC1-shPRRX1 #2 cells (Fig.5A). Next, we evaluated the expression of some of these genes in PRRX1 WT and KO MEFs. We observed that PRRX1 loss diminished the expression level of Brca1 and Xrcc1, but not Atm and Brca2 (Fig.5B). We further confirmed that knockdown of PRRX1 in PANC1 cells reduced BRCA1 protein levels (Supplementary Fig. 5). These collective results show that depletion of PRRX1 expression decreases expression of key DNA repair genes and suggests PRRX1 may play a broader role in the DDR. We analyzed a publically-available RNA-sequencing dataset (GSE112365) comparing BT549 human breast cancer cells with CRISPR-mediated knockout of PRRX1 relative to untreated controls. Interestingly, Gene Set Enrichment Analysis (GSEA) revealed that gene sets related to DNA repair were enriched in the WT BT549 cells expressing PRRX1 compared to PRRX1KO cells (Fig.5C).
Figure 5. Modulation of PRRX1 expression is associated with regulation of DNA damage response genes.
The relative mRNA expression levels of the indicated genes was measured by qPCR A. in PANC1 cells stably expressing either a non-targeting shRNA (shCTL) or shRNA targeting PRRX1 (n=4 independent experiments) and B. in MEFs established from wild type (WT) or PRRX1 KO mice (n=3 independent experiments). *=p<0.05, **=p<0.01 and ***=p<0.001. C. DNA repair enrichment plots from GSEA analysis of GSE112365 published dataset for BT549 human breast cancer cells wild type or KO for PRRX1.
FOXM1 inhibition reduces growth and induces apoptosis of pancreatic tumor cells
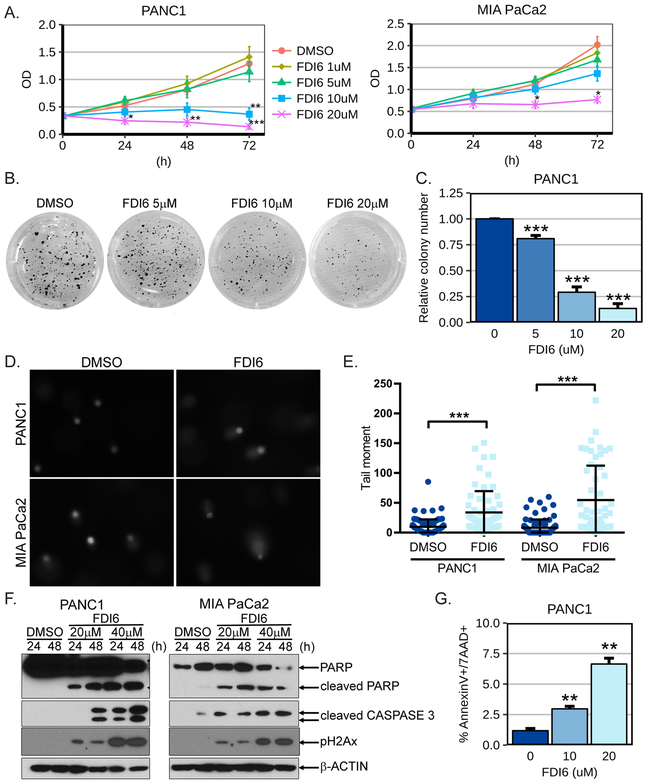
Knockdown of PRRX1 in PANC1 cells significantly decreased their anchorage-independent growth (Supplementary Fig.4). Since there is no current PRRX1-directed therapy, we therefore explored the therapeutic potential of the small molecule FOXM1 inhibitor FDI6 (16) in human pancreatic cancer cells as a translational proof-of-principle approach. We performed first WST1 viability assays in PANC1 and MIA PaCa2 cells following treatment with FDI6 at different doses. We observed significant reduction of PANC1 and MIA PaCa2 cell viability following treatment with FDI6 at 10μM and 20μM at accepted doses (16) (Fig.6A). Next, we analyzed the anchorage-independent growth of PANC1 cells following treatment with FDI6. We noticed a dose-dependent decrease of colony formation in soft-agarose by PANC1 cells treated with FDI6 compared to vehicle treated cells (Figs.6B and 6C). We then determined if the growth reduction observed following FDI6 treatment is in part due to induction of apoptosis from DNA damage. We first confirmed that FDI6 increased levels of DNA damage through comet assay in PANC1 and MIA PaCa2 cells (Fig. 6D-E). We further observed that treatment of PANC1 and MIA PaCa2 cells with FDI6 for 24 hrs. and 48 hrs. increased cleavage of PARP and of CASPASE 3, suggesting induction of apoptosis (Fig.6D). Interestingly, FDI6 treatment also increased the levels of pH2Ax in a dose and time dependent manner (Fig. 6D). This suggests that as with PRRX1 knockdown, FOXM1 inhibition induces DNA damage. FDI6-induced apoptosis was also observed in other human pancreatic cancer cell lines i.e. AsPC1, BxPC3, Capan1 and Capan2 (Supplementary Fig.6). The induction of apoptosis in PANC1 cells following FDI6 treatment was substantiated further by increased number of AnnexinV/7-AAD positive cells in FDI6 vs. vehicle treated cells (Fig.6E). Taken together, our results show that specific inhibition of FOXM1 by FDI6 decreases pancreatic cancer cell proliferation and induces their apoptotic cell death.
Figure 6. FOXM1 inhibition reduces PDAC cell growth and induces apoptosis.
A. The cell viability of PANC1 (left panel) and MIA PaCa2 (right panel) human pancreatic cancer cells was analyzed by WST1 assay following the indicated treatments with the specific FOXM1 inhibitor FDI6. PANC1 (n=5-7) and MIA PaCa2 (n=3-4). B-C. Soft agarose assay of PANC1 cells treated with the indicated doses of FDI6. B. Images from a representative experiment are shown. C. Plot of the relative colony number formed by PANC1 cells (n=3-4). D-E. PANC1 and MIA PaCa2 cells were treated for 48 hours with 20 μM FDI6 and comet assay was performed. Representative images are shown in D and quantification of individual comet tail moments in E. F. The PANC1 and MIA PaCa2 cells were treated 24 and 48 hours. with the indicated doses of the FOXM1 inhibitor FDI6. Western Blot analysis of cleaved PARP and cleaved CASPASE 3. G. PANC1 cells were treated 48 hours with the indicated doses of FDI6 and analyzed by flow cytometry for apoptotic cell death using Annexin-V/7-AAD staining. The % of cells positive for both AnnexinV and 7-AAD is shown (n=3 independent experiments). *=p<0.05, **=p<0.01 and ***=p<0.001.
Treatment with the FOXM1 inhibitor FDI6 sensitizes pancreatic cancer cells to select chemotherapeutic drugs
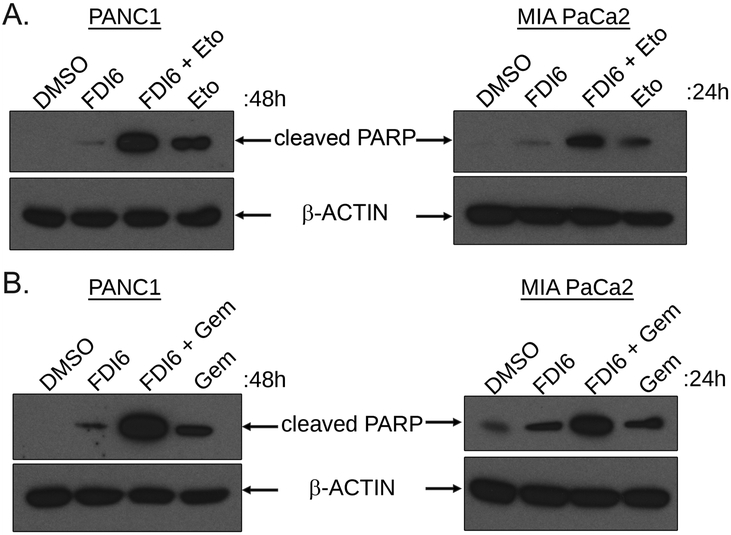
We next explored the therapeutic potential for FOXM1 inhibition in pancreatic cancer cells by evaluating the efficiency of FDI6 in combination with select chemotherapeutic agents. Consistent with our previous results (Fig.6F and supplementary Fig.6), the treatment of PANC1 and MIA PaCa2 cells with FDI6 alone increased the levels of cleaved PARP (Fig.7A-B). Treatment with single agent Etoposide (Fig.7A) or Gemcitabine (Fig.7B) induced apoptosis in PANC1 and MIA-PaCa2 cells as revealed by the increased cleavage of PARP. Interestingly, the levels of cleaved PARP were increased further when FDI6 treatment was combined with Etoposide (Fig.7A) or Gemcitabine (Fig.7B) compared to either single agent treatment. Thus, our results suggest that FOXM1 inhibition by FDI6 sensitizes pancreatic cancer cells to Etoposide and Gemcitabine induced apoptosis.
Figure 7. FOXM1 inhibition sensitizes PDAC cells to Etoposide and Gemcitabine.
A. PANC1 (left panel) and MIA PaCa2 (right panel) cells were treated with FDI6 20μM with or without Etoposide (Eto) 10μM for 48 hours (PANC1) or 24 hours (MIA PaCa2). The cells were lysed and cleaved PARP was analyzed by Western Blot. B. PANC1 (left panel) and MIA PaCa2 (right panel) cells were treated with FDI6 20μM with or without Gemcitabine (Gem) 10μM (PANC1) or 5μM (MIA PaCa2) for 48 hours or 24 hours respectively. The cells were lysed and the levels of cleaved PARP was analyzed by Western blot.
DISCUSSION
The importance of homeodomain transcription factors in development is well established. Indeed, Prrx1−/− mice die soon after birth due to defects in skeletogenesis and display craniofacial, limb, mandible and vascular abnormalities (30–35). Studies suggest unequal compensation between Prrx1 and Prrx2 that may potentially be explained by the difference of transcriptional activity observed between Prrx1 isoforms (36,37). Herein, we demonstrate for the first time that PRRX1A and PRRX1B form homodimers and heterodimers through their 200-222/217 aa region which might impact their transcriptional activity. For example, homodimerization of the homeodomain protein HOXA13 has been shown to be required for its transcriptional activity (21). Furthermore, the homodimerization of the paired homeodomain protein AXL4 or its heterodimerization with two members of the same family of transcription factors, CART1 (22) and Goosecoid (23), modulate their transcriptional activity and DNA binding specificity. Therefore, our observation of PRRX1 dimerization provides new insights for a possible explanation of PRRX1 isoform-specific functions.
The distinct roles of each PRRX1 isoforms could also be dependent on the involvement of different cofactors. Herein, we have identified the forkhead transcription factor FOXM1 as a novel PRRX1 interaction partner. The PRRX1(A/B) 200-222/217aa region required for PRRX1 dimerization is also necessary for PRRX1 binding to FOXM1. It is noteworthy that PRRX1 binding to FOXM1 is dependent upon the FOXM1 forkhead domain and since this domain is well conserved throughout the FOX family of TFs, PRRX1 might bind conceivably to other FOX proteins. Interestingly, other Forkhead and homeodomain transcription factors can also bind to each other. FOXA2 has been shown to interact with the homeodomain TFs PDX1 (38) and Engrailed (39). Additionally, PITX2 and OCT4, a paired and a POU homeodomain transcription factor, respectively, were shown to interact with FOXC1 and FOXD3, respectively (40,41). In this study, we demonstrated that both PRRX1A and PRRX1B synergistically cooperate with FOXM1 in the regulation of FOXM1-dependent transcriptional activity and the regulation of Tenascin-C, a known PRRX1 target gene (34). We identified also that FoxM1 and FoxA2 may be regulated by PRRX1. Finally, we showed that both PRRX1 and FOXM1 are highly expressed and co-localize in PDAC tissues. We speculate that the PRRX1-FOXM1 complex may increase FOXM1 stability, thus leading to increased transcriptional activity of FOXM1, as has been noted for other FOXM1 binding partners (42). This is highlighted by our observation that PANC1 cells transfected with shPRRX1 have decreased expression of canonical FOXM1 targets CDC25B and CCNB1.
Functionally, we demonstrate that PRRX1 expression limits DNA damage induced by irradiation, the DNA damaging agent Etoposide or the chemotherapeutic agent Gemcitabine in pancreatic cancer cells. Moreover, we demonstrate that several genes (i.e. Atm, Atr, Brca1, Brca2, DNA-pk, Xrcc1 Xrcc4) involved in the DNA damage response and DNA repair pathways are associated with modulation of PRRX1 expression. Interestingly, FOXM1 has been shown to regulate DNA repair genes such as Brca2, Xrcc1, and Brip1 and is involved in the homologous recombination DNA repair pathway (25,26,43,44). Furthermore, FOXM1 has been associated with cancer cells mediated resistance to irradiation, Doxorubicin and cisplatin (24,44–46). Taken together, this leads to the premise that PRRX1 expression, partially with its FOXM1 interaction, may contribute to pancreatic cancer cell resistance to DNA damage-inducing treatments. Indeed, genomic analysis of pancreatic cancer has revealed oncogenic mutations in genes that aggregate into the DNA damage repair pathways (47,48). Study of the mutational landscape of pancreatic cancer has identified four molecular subtypes of pancreatic cancers (squamous, pancreatic progenitor, immunogenic and ADEX) and reaffirmed the importance of DNA repair pathways (49).
The thiazole antibiotics such as Thiostrepton have been used to inhibit FOXM1 transcriptional activity, although their use raises concerns of off-target effects (50,51). Thiostrepton and other proteasome inhibitors have been shown to inhibit FOXM1 that highlights the need for more specific FOXM1 inhibitors (52). The recent discovery of FDI6, a small molecule inhibitor of FOXM1 DNA binding, with high specificity (16) provides a new opportunity to investigate the potential of FOXM1 targeting therapy. Hence, we evaluated the effect of FDI6 on human pancreatic cancer cells and demonstrate that FDI6 reduces the growth of PDAC cells and induces their apoptotic cell death. Moreover, treatment with the FOXM1 inhibitor FDI6 sensitizes pancreatic cancer cells to Etoposide and Gemcitabine chemotherapies. Our results suggest that targeting FOXM1, and through its interplay with PRRX1, could be investigated in vivo for PDAC treatment.
In summary, our results provide new insights into PRRX1 regulation and functions by demonstrating that PRRX1 isoforms dimerize and by identifying FOXM1 as a novel PRRX1 interaction partner. We describe a potential PRRX1-FOXM1 axis in which PRRX1 not only regulates FOXM1 expression but also cooperates with FOXM1 in the potential regulation of target genes. Finally, we describe for the first time the involvement of PRRX1 in the DNA-damage response and our work supports the therapeutic potential of targeting the PRRX1-FOXM1 axis in PDAC via the use of a specific inhibitor of FOXM1.
MATERIALS AND METHODS
Cell culture and drug treatments
HEK293T, HEK293FT and the human pancreatic cancer cell lines AsPC1, BxPC3, Capan1, Capan2, MIA PaCa2 and PANC1 were obtained from ATCC (American Type Culture Collection) and maintained in a humidified 5% CO2 atmosphere at 37°C. The mouse embryonic fibroblast (MEF) cells established from PRRX1 WT or KO mice was kindly provided by Kaori Ihida-Stansbury (UPenn, Philadelphia, PA). The Capan1 cells were grown in DMEM (Corning) supplemented with 20% fetal bovine serum (FBS; SH3007103, GE) and 100U/mL penicillin-streptomycin (15140122, Gibco, ThermoFisher) while AsPC1 and BxPC3 cells were cultured in RPMI1640 (Corning) supplemented with 10% FBS and 100U/mL penicillin-streptomycin. All other cell lines were grown in DMEM supplemented with 10% FBS and 100U/mL penicillin-streptomycin. During the course of this study all cell lines were tested for mycoplasma contamination. Etoposide and the FOXM1 inhibitor, FDI6, were purchased from Selleck Chem (S1225) and Axon MedChem (2384) respectively. Gemcitabine HCl solution was purchased from Hospira, inc (Lake Forest, IL, USA). A Gammacell 40 Cesium 137 Irradiation unit was used to deliver 8Gy.
Generation of plasmids
Prrx1 C-terminal deletion mutants were generated by PCR amplification of pIRES2-EGFP-FLAG-Prrx1 (4) using primers listed in Table 1. A C-terminal V5-tagged FOXM1 expression vector (pIRES2-EGFP-Foxm1-V5) was generated by PCR amplification of Foxm1 from mus musculus ORF (6417437, Open Biosystem) using primers listed in Table 1. Foxm1 N-terminal deletion mutants were generated from pIRES2-EGFP-Foxm1-V5 using primers in Table 1. The truncated Prrx1 and Foxm1 sequences were cloned into pIRES2-EGFP vector using XhoI and EcoRI restriction enzymes. Expression vectors for Myc-tagged Prrx1a, Prrx1b and Foxm1 were generated by PCR amplification of the corresponding pIRES2-EGFP plasmids using primers listed in table 1 and subcloned into pcDNA3.1(+) (V790-20, Invitrogen) using NheI and EcoRI.
Table 1:
Primers for Subcloning
| Primer | 5’-3’ sequence | notes |
|---|---|---|
| FW Prrx1 | ACTGACTCGAGGCCACCATGGATTACAAGGATGACGACGATAAGACCTCCAGCTACGGGCAC | XhoI, Kosak, FLAG-tag |
| RV Prrx1d222 | AATCGGAATTCTCAGGCCATGTTGATACCCTG | EcoRI |
| RV Prrx1d200 | AATCGGAATTCTCAGTACGGAGAGGCTGTCCC | EcoRI |
| RV Prrx1d154 | AATCGGAATTCTCACTCATTCCTGCGGAACTT | EcoRI |
| FW Foxm1 WT | TGATCCTCGAGGCCACCATGAGAACCAGCCCCCGC | XhoI, Kosak |
| FW Foxm1d232 | TGATCCTCGAGGCCACCATGGAGAGGCCACCCTACT | XhoI, Kosak |
| FW Foxm1d325 | TGATCCTCGAGGCCACCATGGAACCAGGGTCTCCACA | XhoI, Kosak |
| RV Foxm1 V5 | GCTCAGAATTCCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTCTAAGGGATGAACTGAGACCAGTTGAT | V5-tag (GKPIPNPLLGLDST), EcoRI |
| FW Myc-Prrx1 (A and B) | ACTGAGCTAGCGCCACCATGGAACAAAAACTCATCTCAGAAGAGGATCTGACCTCCAGCTACGGGCAC | NheI, Kosak, Myc-tag (EQKLISEEDL), |
| RV Myc-Prrx1A | AATCGGAATTCTCAGTTGACTGTTGGCACCTGG | EcoRI |
| RV Myc-Prrx1B | AATCGGAATTCTTAGAATCCGTTATGAAGCCCCTCG | EcoRI |
| FW Foxm1-3xMyc | TGATCGCTAGCGCCACCATGAGAACCAGCCCCCGCCGGCCA | NheI, Kosak |
| RV Foxm1-3xMyc | GCTCAGAATTCCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCCAGATCCTCTTCTGAGAT GAGTTTTTGTTCCAGATCCTCTTCTGAGATGAGTTTTTGTTCAGGGATGAACTGAGACCAGTTGATGTT |
3xMyc, EcoRI |
Transfection
HEK293T or PANC1 cells were transiently transfected with Lipofectamine 2000 (11668019, ThermoFisher) according to manufacturer’s instructions. Stable populations of PANC1-pcDNA3.1(+), PANC1-pcDNA3.1-myc-Prrx1a and PANC1-pcDNA3.1-myc-Prrx1b cells were selected (10 days) with 1mg/mL of G418 (goldbio)
Luciferase assay
Experiments were performed as described previously (53). Briefly, PANC1 cells were transfected with Lipofectamine 2000 according to the manufacturer’s instructions. Experiments were done in 24 well plates using 0.1μg/well of 6xFOXM1 luciferase (0.1μg/well) (19) or Tenascin-C (TN7; −247/+121) luciferase (54,55) with 0.004μg/well of pRL-SV40 (Promega). The expression vectors pcDNA3.1(+), pcDNA3.1-myc-Prrx1a, pcDNA3.1-myc-Prrx1b and pcDNA3.1-Foxm1-3xmyc plasmids were used at 0.1μg/well. Luciferase activities were measured using the Dual-Luciferase Reporter Assay system (E1910, Promega).
Lentiviral production and transduction
Lentiviruses were produced in HEK293FT cells using the following shRNAs purchased from Sigma-Aldrich: shNonTarget (SHC016), shPRRX1 #1 (TRCN0000020648) and shPRRX1 #2 (TRCN0000020646). Briefly, HEK293FT cells in a 10cm dish were transfected using Lipofectamine 2000 with 10µg of shRNA plasmid and 5µg of the lentiviral packaging plasmids psPAX2 and pMD2.G. Then, 48 hours following transfection the media containing lentiviruses was collected and filtered through a 0.45µm filter. PANC1 cells were infected with the viral suspension containing 8μg/mL of polybrene (TR-1003-G, Millipore) for 1 hour at 5% CO2 and 37°C. Complete media containing 8μg/mL of polybrene was added and the cells were grown for 48 hours. Infected cells were selected (10 days) with 2μg/mL puromycin (Goldbio) for generation of stable PANC1-shCTL, PANC1-shPRRX1 #1 and PANC1-shPRRX1 #2 populations.
Quantitative PCR
Total RNA was extracted using the GeneJET purification kit (K0732, Thermo Scientific) and 2μg of RNA was used to synthesize cDNA with the High-Capacity cDNA Reverse Transcription kit (4368814, Applied Biosystems™, Thermo Scientific). The PCR reactions were performed in triplicate on the StepOnePlus system (Applied Biosystem) using Power SYBR Green (4367659, ThermoFisher). Primer efficiency was determined and relative gene expression was measured using the primers listed in Table 2.
Table 2:
Primers for qPCR
| Primer | Species | FW (5’-3’) | RV (5’-3’) |
|---|---|---|---|
| Atm | mouse | GTCTCCGATATGCCAGTCTTTTCAG | CTTCTGCCACTTCCAGTATGCT |
| Brca1 | mouse | GCCTCACTTTAACTGACGCA | TGACTTCAAATTCATGCACATTCAG |
| Brca2 | mouse | GAGGAGGAGTGACTGTTGACG | CTTCTGTCTTTTACTGTGCCATCTG |
| Cyclophilin | mouse | ATGGTCAACCCCACCGTGT | TTCTGCTGTCTTTGGAACTTTGTC |
| Foxa2 | mouse | CACTCGGCTTCCAGTATGCT | CGTTCATGTTGCTCACGGAAGA |
| Foxb2 | mouse | TAGAGAACATCATCGGGCGG | CTACCGAATCCATCACGCCA |
| Foxg1 | mouse | TCCCTCTACTGGCCCATGTC | CGTGGTCCCGTTGTAACTCA |
| Foxm1 | mouse | AGCACTGAGAGAAAGCGCATG | TGGCAGATGTCTCTCGAACAAA |
| Foxo1 | mouse | CGCTTGGACTGTGACATGGA | AATGTAGCCTGCTCACTAACTCT |
| Foxp1 | mouse | CCTCCTAACAATTCAGCCGG | GTTGCTGCTTGTGGTTTCCT |
| Foxp2 | mouse | GCCTCGCACACTCTCTATGG | ATTTGCACTCGACATTGGGC |
| Prrx1a | mouse | ACAGCCTCTCCGTACAGCGC | AGTCTCAGGTTGGCAATGCT |
| Prrx1b | Mouse/human | CATCGTACCTCGTCCTGCTC | GCCCCTCGTGTAAACAACAT |
| Xrcc1 | mouse | AGCCAGGACTCGACCCATTG | GCCGAGCCATCATTGCCAAT |
| β-ACTIN | human | TTGCCGACAGGATGCAGAA | GCCGATCCACACGGAGTACT |
| ATM | human | TTGACCTTCCGAGTGCAGTG | ACTTCTTTCTTTCGTTCTGTAGCTC |
| ATR | human | ACATTTGTGACTGGAGTAGAAGATT | TCCACAATTGGTGACCTGGG |
| BRCA1 | human | GCTCTTCGCGTTGAAGAAGT | TCAACTCCAGACAGATGGGAC |
| BRCA2 | human | CTGTGTACCCTTTCGCACAAC | ATTCTTGACCAGGTGCGGTA |
| FOXA2 | human | AGCCCGAGGGCTACTCC | CGTGTTCATGCCGTTCATCC |
| CDC25B | human | CCTCCCTGTCGTCTGAATCC | GGAGTTGGTGATGTTCCGAAG |
| CCNB1 | human | GGAGAGGTTGATGTCGAGCAA | CAGGTAATGTTGTAGAGTTGGTGT |
| FOXB2 | human | CTGGCAAGGGTAGCTTCTGG | CGCGTGCAAGTGAGTATGGT |
| FOXM1 | human | ACTTTAAGCACATTGCCAAGC | CGTGCAGGGAAAGGTTGT |
| FOXO1 | human | AGTGGATGGTCAAGAGCGTG | GCACACGAATGAACTTGCTGT |
| FOXP1 | human | GCTTACTTCCGACGCAACG | TCATCCACTGTCCATACTGCC |
| FOXP2 | human | CCGGGAAGTTTGCTCTAACATTTC | TGCTTACTTCAGAGCTGGTGTC |
| PRKDC | human | TGAACACCATGTCCCAAGAGG | CAGTACGATTAGCGCCCTTATACA |
| PRRX1A | human | ACAGCGTCTCCGTACAGCGC | AGTCTCAGGTTGGCAATGCT |
| RAD51B | human | GAAGACAGATTCTTATTGCCAAGTC | GCAAGATGAACAGGTTTGCAC |
| XRCC1 | human | GTGGTCCTACAGTTGGAGAAGG | CTCCACGAAAGCTGAGCCAT |
| XRCC4 | human | ATGTCTCATTCAGACTTGGTTCC | CTGCAATGGTGTCCAAGCAA |
Western blot
Cells lysis was performed as described previously (56) in Triton, High salt or RIPA (1% Nonidet-P40, 50mM Tris, pH 7.5, 150mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 5mM EDTA, 40mM β-glycerophosphate, 10mM NaF, 5% glycerol, 0.2mM orthovanadate, 1X cOmplete™-EDTA-free protease inhibitor cocktail) lysis buffer. Western blot was performed using the NuPAGE Bis-Tris precast gel system (ThermoFisher) with MOPS SDS running buffer. Proteins were detected immunologically following electrotransfer onto PVDF membranes (IPVH00010 or IPFL00005, Millipore). The cleaved CASPASE 3 (9661, 1:200), cleaved PARP D64E10 XP (5625, 1:750), PARP (9542, 1:1,500) and phospho H2Ax (Ser139) 20E3 (9718, 1:2,000) antibodies were purchased from Cell Signaling. The following antibodies were obtained from Santa Cruz Biotechnology FOXM1 C-20 (Sc-502, 1:1,000), FOXM1 G-5 (Sc-376471, 1:1,000), and BRCA1 (Sc-642, 1:1000). The antibodies against β-CATENIN (610153, 1:1,000), Myc-tag 9E1 (9e1-100, 1:2,000) and V5-tag (46-0705, 1:2,000) were purchased respectively from BD Biosciences, Chromotek (Planegg-Martinsried, Germany) and ThermoFisher. The FLAG M2 (F1804, 1:10,000) and β-ACTIN (A5316, 1:15,000) was obtained from Sigma-Aldrich while GAPDH (Mab374, 1:10,000) was obtained from Millipore. HRP (NA931V, NA934V, NA935V, GE) or IRDye (925-32219, 925-68072, LI-COR) conjugated secondary antibodies were used and blots were visualized with chemiluminescence reagent (RPN2232, GE) or Odyssey® system (LI-COR).
Immunoprecipitation
Immunoprecipitation was performed as described previously (56). Briefly, 1mg of protein lysate was pre-cleared 30 minutes at 4°C with beads (161-4023, Bio-Rad or bmab-20, Chromotek). Samples were incubated 3 hours with 1.5μg of antibody (FLAG M2 or FOXM1 C-20) or 1 hour with 25μL MycTrap® beads (ytma-20, Chromotek) at 4°C. Samples containing antibodies were incubated for an additional hour with 55μL of beads (161-4023, Bio-Rad). Beads were washed three times with lysis buffer and resuspended in Laemmli 2X prior to Western Blot.
Animal studies
All animal work was approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Immunohistochemistry and immunofluorescence
Paraffin embedded zinc-formalin fixed tissue was sectioned and antigen retrieval was performed with citrate pH6.0 buffer in a pressure cooker. Slides were blocked with serum-free protein block (X0909, Dako) for 1 hour at RT and incubated overnight at 4°C with the following antibodies FOXM1 (AF3975, Novus, 1:100), PRRX1 (NBP2-13816, Novus, 1:50) or GFP (ab13970, abcam, 1:250). Immunohistochemistry was performed using Biotinylated secondary antibodies, ABC kit (PK-6100) and DAB (SK-4100) from Vector Labs. For immunofluorescence, slides were incubated with Cy-conjugated secondary antibodies (Jackson ImmunoResearch, 1:300) for 1 hour at RT and nuclei were stained with DAPI. Image were acquired on a Nikon E600 microscope and processed with ImageJ (1.51w) software. Antibody fidelity was verified by staining murine PDAC tissue in the absence of primary antibody (Supplementary Figure 1A). Human PDAC tissue was obtained from US Biomax, Inc. (PA483).
Soft agarose assay
The Soft Agarose Assay was performed as described previously (53,56). Briefly, 15,000 PANC1, PANC1-shCTL, PANC1-shPRRX1 #1 or PANC1-shPRRX1 #2 cells were suspended in a mixture of DMEM and 0.7% agarose and seeded on a layer of DMEM-agarose mix in 6 well plates. For PANC1 treatments, DMEM media containing the FOXM1 inhibitor (FDI6) or vehicle (DMSO) was added on the surface of the agarose and changed every day. Cells were grown 3 weeks before colonies were stained with MTT (475989, Millipore) and photographed (Nikon D3100). Colonies were quantified using ImageJ (1.51w) software for three independent experiments done in duplicate.
Cell viability and apoptosis assays
For cell viability analysis, MIA PaCa2 and PANC1 cells were seeded in 96 well plates at 5,000cells/well and treated with FDI6 or vehicle (DMSO). Cell viability was assessed by incubating the cells in WST1 (11644807001, Sigma-Aldrich) for 2 hours at 37°C and measuring the OD at 450 and 605nm. For the detection of apoptosis, PANC1 cells were seeded in 12 well plates and treated with FDI6 or vehicle (DMSO). Apoptosis was measured using the APC-AnnexinV and 7-AAD kit from BioLegend (640930) according to the manufacturer’s instructions. Data were acquired on a BD accuri C6 flow cytometer and analyzed using FlowJo software.
Comet assay
Comet assay was performed as previously described (57–59). Briefly, cells were treated with FDI6 and trypsinized, resulting in a suspension of single epithelial cells. Cells were added to 1% low-melting agarose at 37°C at a cell density of 1 × 105/mL. The low gelling 2-Hydroxyethylagarose (Sigma-Aldrich) and cell mixture was applied to Comet slides (Trevigen, Gaithersburg, MD) and incubated in pre-chilled alkaline lysis buffer overnight at 4°C as described (57). After lysis, horizontal electrophoresis was performed for 25 minutes at 12 volts. DNA content was stained with 10 μg/mL propidium iodide for 20 minutes in the dark at room temperature. Comet slides were visualized under a Nikon E600 fluorescence microscope. Digital images were taken with iVision software. Comets were analyzed with OpenComet software version 1.3.0. (57).
Gene set enrichment analysis (GSEA)
GSEA was performed on publicly available RNA-Sequencing data (GSE112365) using Broad Institute guidelines as established previously (60). Significantly associated gene sets had nominal Pvalues lower than 0.05 and false discovery rates lower than 0.25 with 1,000 permutations and weighted enrichment scoring.
Statistical Analysis
Experiments were performed at least 3 independent times with 2 or more technical replicates. R studio (1.1.383; R version 3.4.2) software was used to perform statistical analysis and visualize the data. The Shapiro-Wilk test was used to assess normality of the data. Data from two groups were analyzed with the two-tailed unpaired Student’s T-Test if normally distributed or otherwise compared using the Man-Whitney U test. Significant differences were defined as P<0.05. Values are expressed as mean ± SD.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Zheng Fu (Virginia Commonwealth University) for providing the 6xFOXM1 luciferase reporter construct. We thank Dr. Kaori Ihida-Stansbury (University of Pennsylvania) for the PRRX1 wild type and Knockout mouse embryonic fibroblasts (MEFs). The Tenascin-C (TN7) luciferase construct was kindly provided by Dr. Edward E. Morrisey (University of Pennsylvania). We are thankful to the Molecular Pathology and Imaging Core, the Cell Culture and iPS Core and the Human-Microbial Analytic and Repository Core Facilities. This work was supported by the NIH/NIDDK R01 DK060694 (BM, MR, JRP, AKR), the Center for Molecular Studies in Digestive and Liver Diseases (NIH P30 DK050306), the American Cancer Society, Fonds de recherche en santé du Québec P-Marchand-35978 (BM), National Pancreas Foundation (MR), German Cancer Aid Foundation (Max Eder Program, Deutsche Krebshilfe 111273 to MR), AGA-Actavis Research Award in Pancreatic Disorders (MR), NCI F32 CA221094 (JRP) and the NIH loan repayment program (JRP).
Footnotes
CONFLICTS OF INTEREST
None
REFERENCES
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014. June 1;74(11):2913–21. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2018. [Internet]. American Cancer Society; 2018. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf [Google Scholar]
- 3.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell. 2012. January;148(1–2):349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reichert M, Takano S, von Burstin J, Kim S-B, Lee J-S, Ihida-Stansbury K, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013. February 1;27(3):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern MJ, Witte DP, Valerius MT, Aronow BJ, Potter SS. A novel murine homeobox gene isolated by a tissue specific PCR cloning strategy. Nucleic Acids Res. 1992. October 11;20(19):5189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kern MJ, Argao EA, Birkenmeier EH, Rowe LB, Potter SS. Genomic organization and chromosome localization of the murine homeobox gene Pmx. Genomics. 1994. January 15;19(2):334–40. [DOI] [PubMed] [Google Scholar]
- 7.Takano S, Reichert M, Bakir B, Das KK, Nishida T, Miyazaki M, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 2016. January 15;30(2):233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ocaña OH, Córcoles R, Fabra Á, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell. 2012. December;22(6):709–24. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer. 2013. July 23;109(2):307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L, Zhang Y, Lin S, Sun A, Chen R, Ding Y, et al. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):10534–44. [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J, Fu Z, Wei J, Lu W, Feng J, Zhang S. PRRX1 promotes epithelial–mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer. Med Oncol [Internet]. 2015. January [cited 2016 Oct 9];32(1). Available from: http://link.springer.com/10.1007/s12032-014-0393-x [DOI] [PubMed] [Google Scholar]
- 12.Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Dev Camb Engl 2016. December 15;143(24):4558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, et al. A Novel FoxM1-Caveolin Signaling Pathway Promotes Pancreatic Cancer Invasion and Metastasis. Cancer Res. 2012. February 1;72(3):655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Xie D, Cui J, Li Q, Gao Y, Xie K. FOXM1c Promotes Pancreatic Cancer Epithelial-to-Mesenchymal Transition and Metastasis via Upregulation of Expression of the Urokinase Plasminogen Activator System. Clin Cancer Res. 2014. March 15;20(6):1477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong X, Li L, Li Z, Le X, Huang C, Jia Z, et al. Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer Res. 2013. July 1;73(13):3987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gormally MV, Dexheimer TS, Marsico G, Sanders DA, Lowe C, Matak-Vinković D, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014. November 12;5:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003. December;4(6):437–50. [DOI] [PubMed] [Google Scholar]
- 18.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005. May;7(5):469–83. [DOI] [PubMed] [Google Scholar]
- 19.Major ML, Lepe R, Costa RH. Forkhead Box M1B Transcriptional Activity Requires Binding of Cdk-Cyclin Complexes for Phosphorylation-Dependent Recruitment of p300/CBP Coactivators. Mol Cell Biol. 2004. April 1;24(7):2649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N, Wei P, Gong A, Chiu W-T, Lee H-T, Colman H, et al. FoxM1 Promotes β-Catenin Nuclear Localization and Controls Wnt Target-Gene Expression and Glioma Tumorigenesis. Cancer Cell. 2011. October;20(4):427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Larsen CA, Stadler HS, Ames JB. Structural basis for sequence specific DNA binding and protein dimerization of HOXA13. PloS One. 2011;6(8):e23069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu S, Tucker SC, Zhao Q, deCrombrugghe B, Wisdom R. Physical and genetic interactions between Alx4 and Cart1. Dev Camb Engl. 1999. January;126(2):359–69. [DOI] [PubMed] [Google Scholar]
- 23.Tucker SC, Wisdom R. Site-specific heterodimerization by paired class homeodomain proteins mediates selective transcriptional responses. J Biol Chem. 1999. November 5;274(45):32325–32. [DOI] [PubMed] [Google Scholar]
- 24.Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, Weekes D, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2013. September 26;32(39):4634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007. February;27(3):1007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranski OA, Kalinichenko VV, Adami GR. Increased FOXM1 expression can stimulate DNA repair in normal hepatocytes in vivo but also increases nuclear foci associated with senescence. Cell Prolif. 2015. February;48(1):105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW-F. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014. November;1839(11):1316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007. April 3;6(4):1239–48. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Wu X, Yang L, Xiao F, Zhang H, Zhou A, et al. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clin Cancer Res Off J Am Assoc Cancer Res. 2012. November 1;18(21):5961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Dev Camb Engl. 1998. October;125(19):3831–42. [DOI] [PubMed] [Google Scholar]
- 31.Lu MF, Cheng HT, Kern MJ, Potter SS, Tran B, Diekwisch TG, et al. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Dev Camb Engl 1999. February;126(3):495–504. [DOI] [PubMed] [Google Scholar]
- 32.Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995. May 15;9(10):1237–49. [DOI] [PubMed] [Google Scholar]
- 33.Lu M-F, Cheng H-T, Lacy AR, Kern MJ, Argao EA, Potter SS, et al. Paired-Related Homeobox Genes Cooperate in Handplate and Hindlimb Zeugopod Morphogenesis. Dev Biol. 1999. January;205(1):145–57. [DOI] [PubMed] [Google Scholar]
- 34.Ihida-Stansbury K, McKean DM, Gebb SA, Martin JF, Stevens T, Nemenoff R, et al. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ Res. 2004. June 11;94(11):1507–14. [DOI] [PubMed] [Google Scholar]
- 35.Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, et al. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch Int J Pathol. 2000. January;436(1):12–9. [DOI] [PubMed] [Google Scholar]
- 36.Norris RA, Kern MJ. The Identification of Prx1 Transcription Regulatory Domains Provides a Mechanism for Unequal Compensation by thePrx1 and Prx2 Loci. J Biol Chem. 2001. July 20;276(29):26829–37. [DOI] [PubMed] [Google Scholar]
- 37.Norris RA, Kern MJ. Identification of domains mediating transcription activation, repression, and inhibition in the paired-related homeobox protein, Prx2 (S8). DNA Cell Biol. 2001. February;20(2):89–99. [DOI] [PubMed] [Google Scholar]
- 38.Marshak S, Benshushan E, Shoshkes M, Havin L, Cerasi E, Melloul D. Functional Conservation of Regulatory Elements in the pdx-1 Gene: PDX-1 and Hepatocyte Nuclear Factor 3beta Transcription Factors Mediate beta -Cell-Specific Expression. Mol Cell Biol. 2000. October 15;20(20):7583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foucher I. Joint regulation of the MAP1B promoter by HNF3beta/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and Fox transcription factors. Development. 2003. May 1;130(9):1867–76. [DOI] [PubMed] [Google Scholar]
- 40.Berry FB, Lines MA, Oas JM, Footz T, Underhill DA, Gage PJ, et al. Functional interactions between FOXC1 and PITX2 underlie the sensitivity to FOXC1 gene dose in Axenfeld-Rieger syndrome and anterior segment dysgenesis. Hum Mol Genet. 2006. March 15;15(6):905–19. [DOI] [PubMed] [Google Scholar]
- 41.Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, et al. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci. 2002. March 19;99(6):3663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res. 2017. 15;77(12):3135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park Y-Y, Jung SY, Jennings NB, Rodriguez-Aguayo C, Peng G, Lee S-R, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012. October;33(10):1843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khongkow P, Karunarathna U, Khongkow M, Gong C, Gomes AR, Yagüe E, et al. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene. 2014. August 7;33(32):4144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA-damage. PloS One. 2012;7(2):e31761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwok JM-M, Peck B, Monteiro LJ, Schwenen HDC, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res MCR. 2010. January;8(1):24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015. February 26;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008. September 26;321(5897):1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016. February 24;531(7592):47–52. [DOI] [PubMed] [Google Scholar]
- 50.Bhat UG, Halasi M, Gartel AL. Thiazole Antibiotics Target FoxM1 and Induce Apoptosis in Human Cancer Cells. Blagosklonny MV, editor. PLoS ONE. 2009. May 18;4(5):e5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radhakrishnan SK, Bhat UG, Hughes DE, Wang I-C, Costa RH, Gartel AL. Identification of a Chemical Inhibitor of the Oncogenic Transcription Factor Forkhead Box M1. Cancer Res. 2006. October 1;66(19):9731–5. [DOI] [PubMed] [Google Scholar]
- 52.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PloS One. 2009. August 12;4(8):e6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchand B, Tremblay I, Cagnol S, Boucher M-J. Inhibition of glycogen synthase kinase-3 activity triggers an apoptotic response in pancreatic cancer cells through JNK-dependent mechanisms. Carcinogenesis. 2012. March 1;33(3):529–37. [DOI] [PubMed] [Google Scholar]
- 54.Copertino DW, Edelman GM, Jones FS. Multiple promoter elements differentially regulate the expression of the mouse tenascin gene. Proc Natl Acad Sci U S A. 1997. March 4;94(5):1846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones FS, Meech R, Edelman DB, Oakey RJ, Jones PL. Prx1 controls vascular smooth muscle cell proliferation and tenascin-C expression and is upregulated with Prx2 in pulmonary vascular disease. Circ Res. 2001. July 20;89(2):131–8. [DOI] [PubMed] [Google Scholar]
- 56.Marchand B, Arsenault D, Raymond-Fleury A, Boisvert F-M, Boucher M-J. Glycogen Synthase Kinase-3 (GSK3) Inhibition Induces Prosurvival Autophagic Signals in Human Pancreatic Cancer Cells. J Biol Chem. 2015. February 27;290(9):5592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement M-V. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2014;2:457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1(1):23–9. [DOI] [PubMed] [Google Scholar]
- 59.Laczkó D, Wang F, Johnson FB, Jhala N, Rosztóczy A, Ginsberg GG, et al. Modeling Esophagitis Using Human Three-Dimensional Organotypic Culture System. Am J Pathol. 2017. August;187(8):1787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005. October 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.