Abstract
Epidemiological population studies highlight the presence of substantial individual variability in reading skill, with approximately 5–10% of individuals characterized as having specific reading disability (SRD). Despite reported substantial heritability, typical for a complex trait, the specifics of the connections between reading and the genome are not understood. Recently, the SETBP1 gene has been implicated in several complex neurodevelopmental syndromes and disorders that impact language. Here, we examined the relationship between common polymorphisms in this gene, reading, and reading associated behaviors using data from an ongoing project on the genetic basis of SRD (n=135). In addition, an exploratory analysis was conducted to examine variation on SETBP1 and brain activation using functional magnetic resonance imaging (fMRI; n=73). Gene-based analyses revealed a significant association between SETBP1 and phonological working memory, with rs7230525 as the strongest associated single nucleotide polymorphism (SNP). fMRI analysis revealed that the rs7230525-T allele is associated with functional neural activation during reading and listening to words and pseudowords in the right inferior parietal lobule (IPL). These findings suggest that common genetic variation within SETBP1 is associated with reading behavior and reading-related brain activation patterns in the general population.
Keywords: targeted association, SETBP1, common genetic variants, working memory, fMRI, general population, single nucleotide polymorphism (SNP)
INTRODUCTION
Learning to read proficiently is an important milestone in childhood. Reading is a complex and slowly learned skill resulting from the experientially- and biologically-guided maturation and organization of the brain, and requiring the integration of multiple cognitive and sensory representations and processes (Norton et al., 2015). To be a successful reader, one must quickly and efficiently engage a broad circuit of interconnected brain regions. This “reading circuit” is made up of neural systems that support language as well as visual and orthographic processes, working memory, attention, motor movements and higher-level comprehension and cognition (Norton et al., 2015; Norton and Wolf, 2012). Following a probabilistic and multifactorial etiological model of reading acquisition, we suggest that the emergence of specific reading disability (SRD, also known as developmental dyslexia) may reflect a global failure of interacting mechanisms rooted at multiple levels, each with degrees of impairment that vary across children (Carroll et al., 2016; Gabrieli, 2009; Menghini et al., 2010; Pennington, 2006; Peterson and Pennington, 2015; Tamboer et al., 2014).
Evidence from epidemiological population studies suggest that SRD symptomatology likely reflects normally-distributed variation in behavior, consistent with the notion of varied degrees of impairment (Jorm et al., 1986; Shaywitz et al., 1992; Stevenson, 1988), and thus might be more accurately viewed as a dimensional, rather than a discrete developmental disorder (Fletcher, 2009). This evidence motivates the study of genetic correlates of reading skill across a broad spectrum of levels rather than limiting our approach to the low extreme variation in reading skill.
A crucial issue for understanding learning disabilities is the extent to which the genes that affect learning disabilities also affect normal variation in learning abilities (Plomin and Kovas, 2005). Behavioral-genetic analysis indicate overlapping genetic influences among cognitive abilities, and further suggest that learning disabilities are merely the quantitative extreme of the same genetic influences that contribute to the normal range of variation in learning abilities (Plomin and Kovas, 2005). These behavioral-genetic data have in turn a clear implication for molecular-genetic research. For example, according to the generalist genes hypothesis, as a gene is found to associate with a particular disability, the same gene is expected to associate with variation in the normal range of the ability (Plomin and Kovas, 2005).
Following earlier descriptions of high familial aggregation of SRD (Hallgren, 1950), substantial heritability typical of a complex trait was reported (Fisher and DeFries, 2002), with estimates ranging from 0.18 to 0.72 (Plomin and Kovas, 2005). As expected for a complex heritable disorder with heterogeneous genotype-phenotype association patterns, several SRD risk genes have been identified (Bishop, 2015; Kere, 2014; Mascheretti et al., 2017; Peterson and Pennington, 2015; Scerri and Schulte-Körne, 2010). Since the early 1980s, at least nine risk loci, termed DYX1–DYX9, on eight different chromosomes have been mapped and nine SRD-candidate genes have been replicated in at least one independent sample, i.e., DYX1C1, DCDC2, KIAA0319, C2ORF3, MRPL19, ROBO1, GRIN2B, FOXP2 and CNTNAP2 (Mascheretti et al., 2017). Among recent noteworthy findings is the documentation of a genome-wide significant association signal between SETBP1, a gene on chromosome 18q12.3, and a multivariate and multimodal developmental language disorder in a unique geographically isolated Russian-speaking population. Specifically, Kornilov and colleagues (2016) established a genome-wide association between the SETBP1 gene and syntactic complexity (complex structures and mean length of utterance in words) in 359 individuals belonging to a relatively geographically secluded sample with an elevated prevalence of developmental language disorder (DLD). This association was successfully replicated in an independent cohort of children at risk for DLD using teachers ratings of children’s linguistic and reading skills (n = 372) (Kornilov et al., 2016).
The SETBP1 gene encodes for SET binding protein 1 which has been proposed to play a key role in the mechanism of SET-related leukaemogenesis and tumorigenesis by regulatory function in the nucleus, and binds to another protein called SET which is involved in DNA replication, apoptosis, transcription and nucleosome assembly (Coccaro et al., 2017). Although little is known about its function, structural alterations in the SETBP1 gene have been implicated in several neurodevelopmental conditions. Mutations in the SETBP1 gene have formerly been shown to cause Schinzel-Giedion syndrome (SGS, MIM#269150) (Hoischen et al., 2010), which is a rare congenital syndrome characterized by distinctive facial features, severe mental retardation, epilepsy, multiple congenital malformations and higher-than-normal prevalence of neuroepithelial tumors (Schinzel and Giedion, 1978). To date, eight different mutations in SETBP1 have been reported in 19 patients with SGS (Carvalho et al., 2015; Herenger et al., 2015; Hoischen et al., 2010; Ko et al., 2013; López-González et al., 2015; Suphapeetiporn et al., 2011; Takeuchi et al., 2015). Proximal interstitial 18q deletions varying in size and encompassing the SETBP1 gene have been described among patients with moderate to severe intellectual disability (Coe et al., 2014; Hamdan et al., 2014; Marseglia et al., 2012). Crucially, SETBP1 haploinsufficiency has been consistently associated with expressive language difficulties (Filges et al., 2011; Marseglia et al., 2012).
Overall, these findings suggest that SETBP1 is involved in several complex neurodevelopmental syndromes and disorders that impact language. Converging evidence from high-risk and longitudinal studies indicate that children’s oral language proficiency is associated with variability in reading skill (Bishop and Adams, 1990; Lyytinen et al., 2001, 2005; Nathan et al., 2004; Nation and Snowling, 2004; Rescorla, 2005; Snowling et al., 2000; Stothard et al., 1998). Further, multivariate genetic analyses found strong genetic correlations between language and reading traits in both unselected (Hohnen and Stevenson, 1999; Thompson et al., 1991) and selected extreme (Bishop, 2001) twin samples. However, the precise elements of these shared genetic influences are not known.
Given tight links between language and reading, we hypothesized that variants in the SETBP1 gene may contribute to individual differences in reading-related skills. Therefore, the current study examined the relationship between common variation in the SETBP1 gene, performance on reading and language assessments, and brain activation in a sample of young children. Our behavioral battery assessed children’s abilities across multiple reading (word reading, pseudoword reading, passage comprehension) and language tasks (phonological awareness, spelling, and oral language skills). In addition, we used functional magnetic resonance imaging (fMRI) to examine whether variability in this gene is associated with patterns of neural activation as children read or listened to words and pseudowords. This task has previously been shown to recruit language and reading circuitry and to discriminate good from poor readers (Frost et al., 2009; Preston et al., 2016, 2012, 2010; Pugh et al., 2014; Jasińska et al., 2016, 2017; Landi et al., 2013). We present both words and pseudowords as these stimuli have been shown to similarly engage neural circuitry for reading, but differ in terms of the demands they put on sematic and memory systems (Jobard et al., 2003; Taylor et al., 2013). We also examine activation patterns for both reading and listening, as both brain regions that have been linked primarily to reading (e.g., the “visual word form area” ) and regions more broadly implicated in language processing (e.g., inferior frontal gyrus, inferior parietal cortex, perisylvian regions, insula and cerebellum) are implicated in specific reading disability (Frost et al., 2009; Landi et al., 2010; Preston et al., 2016). Moreover, extant work suggests that the degree to which individuals co-activate neural regions for processing of printed and spoken words is related to reading proficiency (Frost et al., 2009; Preston et al., 2016).
Given that the SETBP1 gene has been associated with expressive language abilities (Filges et al., 2011; Marseglia et al., 2012) and syntactic complexity (Kornilov et al., 2016), we hypothesize that variants in this gene may also be associated with reading-related patterns of neural activation. Such a finding would contribute to building a causal model of the mechanisms by which SETBP1 impacts language and/or reading abilities. To the best of our knowledge, no study has yet performed gene-based associations to examine whether variants in SETBP1 have an impact on individual differences in reading-related skills and on the patterns of brain activation required for reading. This combined “gene-brain-behavior” approach can provide new insights into the biological underpinnings of a complex neurocognitive phenotype, such as reading ability and its underlying componential cognitive skills.
MATERIALS AND METHODS
Participants
One hundred and thirty-five children ages 5–12 (79 males, 56 females, mean age = 8.16 ± 1.27 years, 116 right-handed, 15 left-handed, 4 missing handedness data) representing a broad range of reading and language abilities participated in large-scale study on the relations between brain function and reading skill. Inclusion criteria for this study required native English language, standardized performance IQ greater than or equal to 80, no history of severe developmental or neuropsychological disorders, normal or corrected to normal vision, and normal hearing. With respect to race and ethnicity, the vast majority of the participants (n=116) were Caucasian; of the remaining 19 participants, two participants were of African-American ethnicity, three participants were of Hispanic ethnicity, four participants were of Asian ethnicity, eight participants were of mixed ethnicity, and there were two participants for which information was not available. All 135 children were included in the gene – behavior targeted association analyses.
A subset of 73 participants (29 males, 44 females, mean age = 8.82 ± 1.27 years, 61 right-handed, 9 left-handed, 3 missing handedness data) with complete behavioral, genetic and imaging data were available for gene – brain targeted association analyses. As in the full sample, the majority of the fMRI participants were Caucasian (n=64); of the remaining participants, 2 were of African-American ethnicity, 1 was of Hispanic ethnicity, 2 were of Asian ethnicity, and 4 were of mixed ethnicity.
This study was approved by the Yale University Institutional Review Board. Written informed consent and verbal assent were obtained from parents and their participating children, respectively.
Behavioral Measures
The behavioral battery administered to the participants included assessments of cognitive, language, and reading skills, and evaluations of educational and neuropsychological history. Assessments included subtests from the Woodcock Johnson III (Woodcock et al., 2001) assessing reading and spelling, lexical knowledge, language development and comprehension knowledge; the Comprehensive Test of Phonological Processing (CTOPP) (Wagner et al., 1999) for an assessment of reading-related phonological processing skills; Test of Word Reading Efficiency (Rashotte et al., 1999) which provides a measure of an individual’s ability to pronounce printed words and phonemically regular pseudowords accurately and fluently; Peabody Picture Vocabulary Test (Dunn, 1997) measuring the receptive vocabulary; Gray Oral Reading Test (Wiederholt, 2001) assessing oral reading fluency and comprehension; and Wechsler Abbreviated Scale of Intelligence (Wechsler, 1999).
DNA Collection, Genotyping, and Quality Control
Sterile Oragene™ saliva collection kits (DNA Genotek, Inc.) were used to obtain saliva samples during behavioral testing sessions and DNA was extracted from the samples according to manufacturer’s protocol.
DNA libraries were prepared for microarray genotyping on Illumina’s HumanCoreExome v1 panel according to the manufacturer’s protocol. Microarray genotyping was carried out by Illumina, Inc. (San Diego, CA, U.S.A.) using the company’s FastTrak service. Allele calling was performed using GenomeStudio for Windows software, and clustering parameters were manually reviewed and adjusted when necessary to improve genotype calling.
Following conventional quality assurance procedures, samples and markers were evaluated for their call rates. All samples had a call rate above 95%. SNP markers with call rate below 95% were excluded from the dataset, and the respective genotypes were set to missing.
Gene – Behavior Targeted Association Analyses
As mentioned above, the current paper reports on the results of analyses that targeted a novel candidate gene SETBP1. Correspondingly, these analyses were performed in a targeted-association fashion. However, to utilize the richness of genome-wide data available, our analyses relied on pairwise identity-by-state (IBS) relatedness matrix, estimated for our sample using a larger set of 360,000 SNPs, to directly control for relatedness and population stratification as a random effect. All analyses also controlled for children’s age and sex. Thirty-four SNPs within SETBP1 were available for analysis; however, rs617459, rs663651 and rs3085861 were found to be in linkage disequilibrium (r2 values > .90; see Supplementary Table 1), as such, we removed rs617459 and rs3085861 from our analyses to reduce multiple-testing burden.
Data from 32 SNPs within SETBP1 were analyzed separately for each phenotype, using the Efficient Mixed-Model Association eXpedited algorithm, EMMAX (Kang et al., 2010) method, as implemented in GoldenHelix SVS under the additive model. Briefly, mixed linear modeling as applied to genetic association involves 1) estimating a genetic relationship matrix that models genome-wide structure of the sample (i.e., evaluating empirically the extent to which each pair of individuals is similar genetically), 2) evaluating the contribution of this structure to phenotypic variance in its random effects part, and 3) testing for association between individual markers and phenotypes in its fixed effects part (with effect sizes interpreted identically to unstandardized regression coefficients (B) in a linear regression framework, i.e., B corresponds to the change in the phenotype for each copy of the minor allele, while controlling for relatedness). Recent studies show that mixed linear modeling techniques provide the best correction for population stratification and cryptic relatedness from all of the currently available methods (Kang et al., 2010; Zhou and Stephens, 2012). Gene-based empirical association p-values were estimated with the Versatile Gene-based Association Study-2 Version 2 (VEGAS2v02) web platform, https://vegas2.qimrberghofer.edu.au/ (Mishra and Macgregor, 2015), considering the 100% most significant SNPs from the gene. Based on SNP association p-values the software calculates empirical gene-based p-values using a simulation procedure. According to the ethnic make-up of our sample (cf. ‘Participants’ paragraph), estimation of inter-marker linkage disequilibrium was based on the Utah residents with Northern and Western European Ancestry (CEU) sub-population from the 1000 Genomes Project phase III. Gene boundaries were set to ±0 kb of the gene.
fMRI Paradigm
The fMRI task used in this study was a cue-target identity task with an event-related protocol. The display presented to participants included a picture of an animal or common object that was paired with a visual or auditory linguistic stimulus. The picture was initially presented with an empty box below it and remained on the screen during presentation of a series of trials containing a word (e.g. DRESS), pseudoword (e.g. DREAK), or consonant string (e.g. DRLST); on reading trials, the printed stimulus appeared in the box below the picture for 2000 ms, and on listening trials the auditory stimulus was presented via MR-compatible headphones. All real words were high frequency and all pseudowords were pronounceable in English; all were monosyllabic and 4–5 letters in length. The picture remained constant for approximately one quarter of the run before changing to a different picture and set of trials. Participants were asked to respond to each trial with a match/mismatch judgment via button press, with one button to indicate that the picture and word matched (match condition) and another button to indicate that the picture and word did not match (mismatch condition). A fixation cross was displayed during rest periods. The task included 32 trials for each condition and each condition occurred in all runs. (For additional details of this task, see Frost et al., 2009; Pugh et al., 2013; Jasińska et al., 2016).
fMRI Acquisition
Acquisition of brain images was conducted using a Siemens Sonata 1.5-Tesla MRI Scanner. Twenty axial-oblique anatomic images (TE 11 ms; TR 420 ms; FOV 20 × 20 cm; 6mm slice thickness, no gap; 256 × 256 × 1 NEX) parallel to the intercommissural line were acquired prior to functional imaging. A single-shot gradient echo, echo-planar pulse sequence (FA 80˚; TE 50 ms; TR 2000 ms; FOV 20 × 20 cm; 6mm slice thickness, no gap; 64 × 64 × 1 NEX) was used for acquisition of activation images at the twenty slice locations used for the anatomic images. Jittered interstimulus intervals of 4, 5, 6, and 7s durations were used for trial presentation, with occasional longer intervals (i.e., null trials). High-resolution anatomical images were acquired for 3D co-registration (sagittal MPRAGE acquisition, FA 8˚; TE 3.65 ms; TR 2000 ms; FOV 256 × 256 mm; 1 mm slice thickness, no gap; 256 × 256 × 1 NEX; 160 slices total). A maximum of 10 imaging runs were acquired for each participant.
fMRI Data Analysis
Processing and statistical analysis of brain images was performed via the Analysis of Functional Neuroimages software package, AFNI (Cox, 1996). The preprocessing pipeline included correction for slice acquisition time (3dTshift), motion correction (3dvolreg), and affine transformation (3dWarp) to a standardized reference space defined by the Montreal Neurological Institute (MNI) by mapping the participant’s high-resolution anatomical scan to the ‘Colin27’ brain (Ashburner, 2007; Holmes et al., 1998). A 6 mm FWHM Gaussian filter was then applied for spatial smoothing (3dmerge).
A multiple regression analysis was conducted to estimate the hemodynamic response at the single subject level with six movement parameters treated as nuisance regressors. A generalized least squares time series fit with a restricted maximum likelihood estimation of the temporal auto-correlation structure (3dREMLfit) was used in the regression.
Group by condition analysis was performed to test effects of genotype, lexicality, and modality via AFNI’s 3dMVM pipeline (Chen et al., 2014). Blood oxygen level dependent (BOLD) responses were compared between ancestral ‘T’ allele homozygotes and derived ‘C’ allele carriers or each lexicality condition (words and pseudowords) within each modality (auditory and visual). Age, gender, and IQ were included as covariates. AFNI’s 3dClustSim program was employed for cluster-extent correction. The empirical spatial autocorrelation function (ACF) was estimated from the data using AFNI’s 3dFWHMx program. To determine the minimum cluster size corresponding to a corrected p-threshold of .05, 3dClustSim was run using the empirical ACF with a conservative cluster-forming threshold of p < .001 and 10,000 iterations, yielding a minimum cluster size of 24.5 voxels.
RESULTS
Gene – Behavior Targeted Association Analyses
EMMAX analyses performed separately for each of the behavioral traits and interrogated SNPs revealed nominally significant (p’s < .05) associations of 14 SNPs within SETBP1 with behavioral reading-related traits (Supplementary Table 2). After implementing VEGAS2v02, the overall gene-based test for SETBP1 was statistically significant (gene-based test statistic=121.708, empirical gene-based p-value=1.599−5, number of simulations=1,000,000) and the association between rs7230525 and CTOPP memory for digits (CTOPP-MD) subtest generated the strongest signal of association (β=1.074, top SNP p-value=0.007). For this SNP, the ancestral ‘T’ allele1 was associated with poorer performance on the memory for digits subtest relative to the derived ‘C’ allele.2
Gene – Brain Targeted Association Analyses
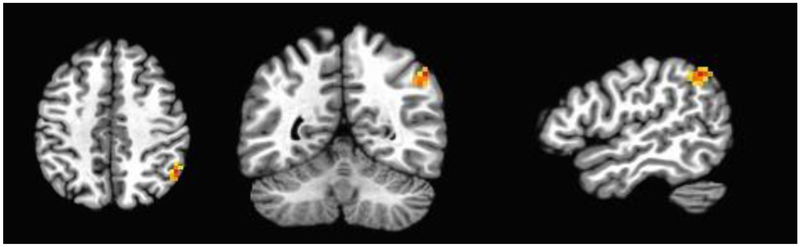
To assess brain activation patterns associated with rs7230525 we performed whole brain analyses to test for effects of genotype (ancestral ‘T’ allele homozygotes vs. derived ‘C’ allele carriers), modality (reading vs. listening) and lexicality (word vs. pseudoword) with age, gender, and IQ as covariates. This analysis revealed a significant 3-way gene by lexicality by modality interaction (peak voxel: F=20.546, p < .001; cluster size = 36, corresponding to cluster-extent-corrected p-value < .05) in the right inferior parietal lobule (R. IPL; Figure 1)3.
Figure 1: Statistical parametric map showing the cluster of voxels in the R. IPL in which a significant gene by lexicality by modality interaction was identified.
Voxel-wise threshold p= .001, cluster-corrected threshold = 24.5 voxels. Peak voxel coordinates (Talairach space) −49.5, 52.5, 41.5, cluster size = 36 voxels.
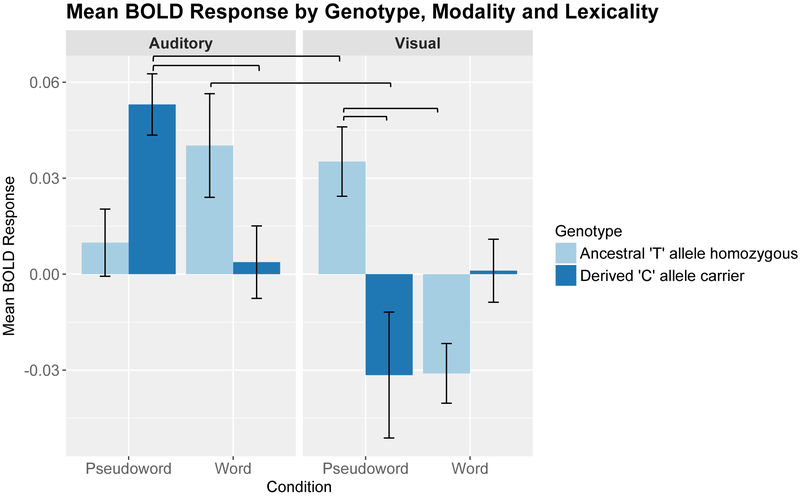
Post-hoc analyses within this cluster revealed a significant genotype effect for pseudowords in the reading modality only, such that activation was increased for ancestral ‘T’ allele homozygotes relative to the derived ‘C’ allele carriers. We also observed a significant lexicality effect within the listening condition for the derived ‘C’ allele carriers only, such that activation was increased for pseudowords relative to words, and a lexicality effect during reading for the ancestral allele ‘T’ homozygotes with greater activation for pseudowords relative to words. Finally, modality effects were present in the word condition for ancestral ‘T’ allele homozygotes only, with increased activation to words during listening relative to reading, and in the pseudoword condition for derived ‘C’ allele carriers, with increased activation to pseudowords during listening relative to reading. Bar plots summarizing these effects are shown in Figure 2.
Figure 2: Bar plots representing the mean blood oxygen level dependent (BOLD) response as a function of genotype, lexicality, and modality.
Error bars represent ± 1 SD from the mean. Brackets indicate significant contrasts.
DISCUSSION
Our study investigated whether genetic variants in the SETBP1 gene are associated with individual differences in reading and reading-related skills and patterns of neural activation in ways that are relevant for children’s reading ability in a sample of developing readers. To address these research questions we used a combined gene-brain-behavior approach with the aim to unravel new information about the biological underpinnings of the development of reading and reading-related skills. Overall, we found that multiple variants spanning the SETBP1 gene are associated with individual differences in reading-related skills and patterns of reading-related activation in a developing brain.
Targeted association analyses established an association between rs7230525 and children’s performance on phonological working memory (pWM) for non-alphabetic items (CTOPP-MD), such that ancestral allele ‘T’ homozygotes had poorer pWM skills compared to the derived ‘C’ allele carriers. Previous studies showed that non-alphabetic tasks predict later reading performance (Lervåg and Hulme, 2009; Parrila et al., 2004) without being biased by reading experience (Rakhlin et al., 2013) or early differences in reading ability as for alphabetic items (Bowey and Muller, 2005). Deficits in pWM have been consistently documented in language-based learning disabilities and usually persist throughout life, suggesting a deficit rather than a developmental delay model (Perrachione et al., 2017; Swanson et al., 2009). Phonological WM is thought to support a wide range of linguistic behaviors, including novel word learning and vocabulary development, maintenance of information during sentence and discourse processing, and the acquisition of reading skill. At the cognitive level, pWM is implicated in the transient storing of all relevant representations, thus allowing grapheme-to-phoneme conversion and phoneme blending, necessary for adequate reading development (Perrachione et al., 2017). According to the sluggish attentional shifting hypothesis (Hari and Renvall, 2001), subjects with SRD seem to have a prolonged ‘cognitive window’ (or ‘time or input chunk’) within which the temporal order of successive items is easily confused (Franceschini et al., 2013; Gori and Facoetti, 2014; Marseglia et al., 2012; Vidyasagar and Pammer, 2010). This deficit may subsequently distort proper development of cortical representations that are essential for reading acquisition (Hari and Renvall, 2001). Moreover, genetic studies have shown that variation in reading performance is explained by specific genes and by a set of genes in common with pWM (Christopher et al., 2016; van Leeuwen et al., 2009), suggesting a common underlying genetic factor. It is therefore plausible to hypothesize a specific detrimental effect of SRD-candidate genes upon the phonological loop (Baddeley, 2003), which has been shown to be distinctly impaired in subjects with SRD compared to normal readers (Swanson et al., 2009), rather than upon the sequence processing in general.
Interestingly, we also observed significant interactions among genotype at rs7230525, lexicality, and modality, on patterns of brain activation in the right IPL. Within this region, ancestral allele ‘T’ homozygotes (who performed more poorly on the pWM task), showed greater activation during pseudoword reading compared to word reading, and greater activation during pseudoword reading relative to the derived ‘C’ allele carriers (who had better performance on the pWM task). These findings are partially consistent with findings from previous neuroimaging studies of good and poor readers (who generally differ on phonological skills tapped by the pWM task as well). For example, several studies have found increased engagement of right temporo-parietal regions in dyslexic readers relative to typically developing readers during reading tasks (Sarkari et al., 2002; Shaywitz et al., 1998), and these right parietal brain-behavior relations hold in larger population-based samples with a broader distribution of reading ability (Pugh et al., 2013). Such compensatory right hemisphere activation may be particularly evident for more difficult to process stimuli (here pseudowords), and for younger and/or impaired readers who may require additional support from right hemisphere reading network homologues. Further, research following sluggish attentional shift based models of dyslexia (Hari and Renvall, 2001), has found atypical right parietal function for poor readers, particularly for difficult to process stimuli including mixed case strings and pseudowords (Wimmer et al., 2002).
Within this same right parietal region, the ‘C’ allele carriers (who had better performance on the pWM task) showed greater activation when listening to spoken pseudowords relative to reading pseudowords, and during listening conditions only, showed greater activation for pseudowords relative to words. Although lexicality effects during listening, and modality effects have been observed during similar tasks (Jobard et al., 2003; Rumsey et al., 1997; Taylor et al., 2013), we are more cautious in our interpretation of these findings with respect to genotype group. First, because our fMRI task was optimized for examining reading in school age children (ages 7–11) and thus contained high-frequency words, the listening condition in particular may be too simple to reveal skill related effects (Preston et al., 2010). Second, although the direction of the lexicality effect in the listening condition in minor allele carriers is consistent with what has been observed more broadly (i.e., higher activation for pseudowords), these findings are more commonly observed in different regions (e.g., inferior frontal gyrus) and most of this work has been done with adult participants (Newman and Twieg, 2001; Perrachione et al., 2017; Xiao et al., 2005). A deviation from previous findings with adults may be expected in children because they may not yet have developed a fully automatized lexical decision response and may therefore recruit more attentional resources and rely upon more suprasegmental features for processing stimulus sets that contain novel pseudowords (Nora et al., 2017; Weiss-Croft and Baldeweg, 2015). However, this interpretation does not explain why this pattern was not observed in major allele carriers, or the broader modality by lexicality by genotype interactions we observed.
Several limitations of the current study should be noted. First, our sample size, while considerable for combined gene-brain-behavior analyses, is still quite modest in terms of its statistical power to detect moderate and small effect sizes. Further, our behavioral battery and fMRI task were optimized for examining reading and reading related-sub-skills (e.g., phonological processing), and thus do not directly index language processes and abilities that have previously been associated with SETBP1. Thus, while our findings extend previous work on SETBP1 into the domain of reading and related skills, they are exploratory and additional studies with larger samples are needed to verify our findings.
Despite these limitations, the current study contributes to a growing literature that stresses the importance of considering common genetic variants in understanding the etiology of cognitive differences, especially in samples drawn from the general population. Although such variants might not target a particular cognitive skill or process because of their critical role in brain function, they appear to be pleiotropic in their impact, affecting multiple components of reading skills. Specifically, the present study extends existing work on the SETBP1 gene by examining relations between multiple variants in this gene and reading- related skills at the level of both behavior and neural function. Here, the ancestral 'T' allele of the SETBP1-rs7230525 SNP was associated with lower scores on phonological working memory and functional neural activation in a region that supports reading (i.e., right IPL). These findings suggest that variants within SETBP1 may represent a genetic predisposition for poorer phonological memory performance as well as increased activation in brain regions that support compensatory phonological processing and attentional engagement. According to previous studies suggesting the involvement of the SETBP1 gene in several complex neurodevelopmental syndromes and disorders, we suggest that variants within this gene may be more relevant in the general population and associated distribution of reading skills and brain activations than any single rare mutation, which can be a powerful causal factor in a single family or a few families, but is unlikely to be generalizable to the general population (Landi et al., 2013; Mozzi et al., 2017).
Supplementary Material
Highlights:
rs7230525-T allele is associated with lower scores on phonological working memory
rs7230525-T allele is associated with functional neural activation in the right IPL
Common SETBP1 variants affect multiple components of reading
Acknowledgements
The authors thank all children who took part in this study. Sara Mascheretti was supported by 2016/2017 Progetto Professionalità "Ivano Becchi" - Fondazione Banca Del Monte Di Lombardia. Meaghan Perdue was supported by NSF IGERT grant DGE-1144399. fMRI data collection and analysis, and collection of saliva were supported by NIH R01 HD 48830 and NIH R03 HD 053409. Genotyping and genetic data processing were supported by NIH P50 HD 052120.
Footnotes
Conflicts of interest: None
We will use the ancestral/derived labels to refer to respective alleles. In the present study, the ancestral ‘T’ allele was also the major allele (with 0.70 frequency), and the derived ‘C’ allele was also the minor allele (with the complementary 0.30 frequency).
To confirm that our gene–behavior targeted association results remain consistent when accounting for participants’ ethnicity, we ran a follow-up analyses including only Caucasian participants (n=116). The results were similar (Supplementary Table 3); after implementing VEGAS2v02, the overall gene-based test for SETBP1 was statistically significant (gene-based test statistic=113.297, empirical gene-based p-value=4.999−5, number of simulations=1,000,000) and the association between rs7230525 and CTOPP-MD subtest generated the strongest signal of association (β=1.245, top SNP p-value=0.006). For this SNP, the ancestral ‘T’ allele was associated with poorer performance on the memory for digits subtest relative to the derived ‘C’ allele.
To confirm that our fMRI results remain consistent when accounting for participants’ ethnicity we ran two follow-up MRI analyses. In the first, we included only Caucasian participants (N=64); the results were similar, highlighting the significant 3-way gene by lexicality—by modality interaction (peak voxel: F=19.119, p< .001; cluster size = 11) in the R. IPL. In the second, we used the top five PCA components (estimated for the sample using the full set of genetic data under the additive model after LD-pruning) as covariates in the fMRI analysis. Results of this analysis were consistent with our original results, showing the same 3-way interaction (peak voxel: F=24.971, p< .001; cluster size = 24) in the R. IPL.
REFERENCES
- Ashburner J, 2007. A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113. 10.1016/J.NEUROIMAGE.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Baddeley A, 2003. Working memory and language: an overview. J. Commun. Disord. 36, 189–208. [DOI] [PubMed] [Google Scholar]
- Bishop DVM, 2015. The interface between genetics and psychology: lessons from developmental dyslexia. Proceedings. Biol. Sci. 282, 20143139 10.1098/rspb.2014.3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DV, 2001. Genetic influences on language impairment and literacy problems in children: same or different? J. Child Psychol. Psychiatry. 42, 189–98. [PubMed] [Google Scholar]
- Bishop DV, Adams C, 1990. A prospective study of the relationship between specific language impairment, phonological disorders and reading retardation. J. Child Psychol. Psychiatry. 31, 1027–50. [DOI] [PubMed] [Google Scholar]
- Bowey JA, Muller D, 2005. Phonological recoding and rapid orthographic learning in third-graders’ silent reading: a critical test of the self-teaching hypothesis. J. Exp. Child Psychol. 92, 203–19. 10.1016/j.jecp.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Carroll JM, Solity J, Shapiro LR, 2016. Predicting dyslexia using prereading skills: the role of sensorimotor and cognitive abilities. J. Child Psychol. Psychiatry. 57, 750–8. 10.1111/jcpp.12488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho E, Honjo R, Magalhães M, Yamamoto G, Rocha K, Naslavsky M, Zatz M, Passos-Bueno MR, Kim C, Bertola D, 2015. Schinzel-Giedion syndrome in two Brazilian patients: Report of a novel mutation in SETBP1 and literature review of the clinical features. Am. J. Med. Genet. Part A 167, 1039–1046. 10.1002/ajmg.a.36789 [DOI] [PubMed] [Google Scholar]
- Chen G, Adleman NE, Saad ZS, Leibenluft E, Cox RW, 2014. Applications of Multivariate Modeling to Neuroimaging Group Analysis: A Comprehensive Alternative to Univariate General Linear Model. Neuroimage 9906 10.1016/j.neuroimage.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher ME, Keenan JM, Hulslander J, DeFries JC, Miyake A, Wadsworth SJ, Willcutt E, Pennington B, Olson RK, 2016. The genetic and environmental etiologies of the relations between cognitive skills and components of reading ability. J. Exp. Psychol. Gen. 145, 451–466. 10.1037/xge0000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro N, Tota G, Zagaria A, Anelli L, Specchia G, Albano F, 2017. SETBP1 dysregulation in congenital disorders and myeloid neoplasms. Oncotarget 8, 51920–51935. 10.18632/oncotarget.17231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BWM, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LELM, Schuurs-Hoeijmakers JH, Hoischen A, Pfundt R, Krumm N, Carvill GL, Li D, Amaral D, Brown N, Lockhart PJ, Scheffer IE, Alberti A, Shaw M, Pettinato R, Tervo R, de Leeuw N, Reijnders MRF, Torchia BS, Peeters H, Thompson E, O’Roak BJ, Fichera M, Hehir-Kwa JY, Shendure J, Mefford HC, Haan E, Gécz J, de Vries BBA, Romano C, Eichler EE, 2014. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46, 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–73. [DOI] [PubMed] [Google Scholar]
- Dunn LM, 1997. PPVT-III: Peabody Picture Vocabulary Test. American Guidance Service, Circle Pines, MN. [Google Scholar]
- Filges I, Shimojima K, Okamoto N, Röthlisberger B, Weber P, Huber AR, Nishizawa T, Datta AN, Miny P, Yamamoto T, 2011. Reduced expression by SETBP1 haploinsufficiency causes developmental and expressive language delay indicating a phenotype distinct from Schinzel-Giedion syndrome. J. Med. Genet. 48, 117–22. 10.1136/jmg.2010.084582 [DOI] [PubMed] [Google Scholar]
- Fisher SE, DeFries JC, 2002. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat. Rev. Neurosci. 3, 767–80. 10.1038/nrn936 [DOI] [PubMed] [Google Scholar]
- Fletcher JM, 2009. Dyslexia: The evolution of a scientific concept. J. Int. Neuropsychol. Soc. 15, 501–8. 10.1017/S1355617709090900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini S, Gori S, Ruffino M, Viola S, Molteni M, Facoetti A, 2013. Action Video Games Make Dyslexic Children Read Better. Curr. Biol. 23, 462–466. 10.1016/j.cub.2013.01.044 [DOI] [PubMed] [Google Scholar]
- Frost SJ, Landi N, Mencl WE, Sandak R, Fulbright RK, Tejada ET, Jacobsen L, Grigorenko EL, Todd Constable R, Pugh KR, 2009. Phonological awareness predicts activation patterns for print and speech. Ann. Dyslexia 59, 78–97. 10.1007/s11881-009-0024-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli JDE, 2009. Dyslexia: A New Synergy Between Education and Cognitive Neuroscience. Science (80-. ). 325, 280–283. 10.1126/science.1171999 [DOI] [PubMed] [Google Scholar]
- Gori S, Facoetti A, 2014. Perceptual learning as a possible new approach for remediation and prevention of developmental dyslexia. Vision Res. 99, 78–87. 10.1016/j.visres.2013.11.011 [DOI] [PubMed] [Google Scholar]
- HALLGREN B, 1950. Specific dyslexia (congenital word-blindness); a clinical and genetic study. Acta Psychiatr. Neurol. Suppl. 65, 1–287. [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi J-M, Daoud H, Nassif C, Patry L, Massicotte C, Ambalavanan A, Spiegelman D, Diallo O, Henrion E, Dionne-Laporte A, Fougerat A, Pshezhetsky AV, Venkateswaran S, Rouleau GA, Michaud JL, 2014. De Novo Mutations in Moderate or Severe Intellectual Disability. PLoS Genet. 10, e1004772 10.1371/journal.pgen.1004772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari R, Renvall H, 2001. Impaired processing of rapid stimulus sequences in dyslexia. Trends Cogn. Sci. 5, 525–532. [DOI] [PubMed] [Google Scholar]
- Herenger Y, Stoetzel C, Schaefer E, Scheidecker S, Manière M-C, Pelletier V, Alembik Y, Christmann D, Clavert J-M, Terzic J, Fischbach M, De Saint Martin A, Dollfus H, 2015. Long term follow up of two independent patients with Schinzel–Giedion carrying SETBP1 mutations. Eur. J. Med. Genet 58, 479–487. 10.1016/j.ejmg.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Hohnen B, Stevenson J, 1999. The structure of genetic influences on general cognitive, language, phonological, and reading abilities. Dev. Psychol 35, 590–603. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, Devriendt K, Amorim MZ, Revencu N, Kidd A, Barbosa M, Turner A, Smith J, Oley C, Henderson A, Hayes IM, Thompson EM, Brunner HG, de Vries BBA, Veltman JA, 2010. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 42, 483–5. 10.1038/ng.581 [DOI] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC, 1998. Enhancement of MR images using registration for signal averaging. J. Comput. Assist. Tomogr. 22, 324–33. [DOI] [PubMed] [Google Scholar]
- Jasińska KK, Molfese PJ, Kornilov SA, Mencl WE, Frost SJ, Lee M, Pugh KR, Grigorenko EL, Landi N, 2017. The BDNF Val 66 Met polymorphism is associated with structural neuroanatomical differences in young children. Behav. Brain Res. 328, 48–56. 10.1016/j.bbr.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasińska KK, Molfese PJ, Kornilov SA, Mencl WE, Frost SJ, Lee M, Pugh KR, Grigorenko EL, Landi N, 2016. The BDNF Val66Met Polymorphism Influences Reading Ability and Patterns of Neural Activation in Children. PLoS One 11, e0157449 10.1371/journal.pone.0157449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobard G, Crivello F, Tzourio-Mazoyer N, 2003. Evaluation of the dual route theory of reading: a metanalysis of 35 neuroimaging studies. Neuroimage 20, 693–712. 10.1016/S1053-8119(03)00343-4 [DOI] [PubMed] [Google Scholar]
- Jorm AF, Share DL, Maclean R, Matthews R, 1986. Cognitive factors at school entry predictive of specific reading retardation and general reading backwardness: a research note. J. Child Psychol. Psychiatry. 27, 45–54. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-Y, Freimer NB, Sabatti C, Eskin E, 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kere J, 2014. The molecular genetics and neurobiology of developmental dyslexia as model of a complex phenotype. Biochem. Biophys. Res. Commun. 452, 236–43. 10.1016/j.bbrc.2014.07.102 [DOI] [PubMed] [Google Scholar]
- Ko JM, Lim BC, Kim KJ, Hwang YS, Ryu HW, Lee JH, Kim JS, Chae J-H, 2013. Distinct neurological features in a patient with Schinzel-Giedion syndrome caused by a recurrent SETBP1 mutation. Childs. Nerv. Syst. 29, 525–9. 10.1007/s00381-013-2047-2 [DOI] [PubMed] [Google Scholar]
- Kornilov SA, Rakhlin N, Koposov R, Lee M, Yrigollen C, Caglayan AO, Magnuson JS, Mane S, Chang JT, Grigorenko EL, 2016. Genome-Wide Association and Exome Sequencing Study of Language Disorder in an Isolated Population. Pediatrics 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi N, Frost SJ, Mencl WE, Preston JL, Jacobsen LK, Lee M, Yrigollen C, Pugh KR, Grigorenko EL, 2013. The COMT Val/Met polymorphism is associated with reading-related skills and consistent patterns of functional neural activation. Dev. Sci 16, 13–23. 10.1111/j.1467-7687.2012.01180.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi N, Mencl WE, Frost SJ, Sandak R, Pugh KR, 2010. An fMRI study of multimodal semantic and phonological processing in reading disabled adolescents. Ann. Dyslexia 60, 102–121. 10.1007/s11881-009-0029-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lervåg A, Hulme C, 2009. Rapid automatized naming (RAN) taps a mechanism that places constraints on the development of early reading fluency. Psychol. Sci. 20, 1040–8. 10.1111/j.1467-9280.2009.02405.x [DOI] [PubMed] [Google Scholar]
- López-González V, Domingo-Jiménez MR, Burglen L, Ballesta-Martínez MJ, Whalen S, Piñero-Fernández JA, Guillén-Navarro E, 2015. Síndrome Schinzel-Giedion: nueva mutación en SETBP1. An. Pediatría 82, e12–e16. 10.1016/j.anpedi.2014.06.017 [DOI] [PubMed] [Google Scholar]
- Lyytinen H, Ahonen T, Eklund K, Guttorm TK, Laakso ML, Leinonen S, Leppänen PH, Lyytinen P, Poikkeus AM, Puolakanaho A, Richardson U, Viholainen H, 2001. Developmental pathways of children with and without familial risk for dyslexia during the first years of life. Dev. Neuropsychol. 20, 535–54. 10.1207/S15326942DN2002_5 [DOI] [PubMed] [Google Scholar]
- Lyytinen P, Eklund K, Lyytinen H, 2005. Language development and literacy skills in late-talking toddlers with and without familial risk for dyslexia. Ann. Dyslexia 55, 166–92. 10.1007/s11881-005-0010-y [DOI] [PubMed] [Google Scholar]
- Marseglia G, Scordo MR, Pescucci C, Nannetti G, Biagini E, Scandurra V, Gerundino F, Magi A, Benelli M, Torricelli F, 2012. 372 kb microdeletion in 18q12.3 causing SETBP1 haploinsufficiency associated with mild mental retardation and expressive speech impairment. Eur. J. Med. Genet 55, 216–221. 10.1016/j.ejmg.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Mascheretti S, De Luca A, Trezzi V, Peruzzo D, Nordio A, Marino C, Arrigoni F, 2017. Neurogenetics of developmental dyslexia: from genes to behavior through brain neuroimaging and cognitive and sensorial mechanisms. Transl. Psychiatry 7, e987 10.1038/tp.2016.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menghini D, Finzi A, Benassi M, Bolzani R, Facoetti A, Giovagnoli S, Ruffino M, Vicari S, 2010. Different underlying neurocognitive deficits in developmental dyslexia: a comparative study. Neuropsychologia 48, 863–72. 10.1016/j.neuropsychologia.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Mishra A, Macgregor S, 2015. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res. Hum. Genet 18, 86–91. 10.1017/thg.2014.79 [DOI] [PubMed] [Google Scholar]
- Mozzi A, Riva V, Forni D, Sironi M, Marino C, Molteni M, Riva S, Guerini FR, Clerici M, Cagliani R, Mascheretti S, 2017. A common genetic variant in FOXP2 is associated with language-based learning (dis)abilities: Evidence from two Italian independent samples. Am. J. Med. Genet. B. Neuropsychiatr. Genet 10.1002/ajmg.b.32546 [DOI] [PubMed] [Google Scholar]
- Nathan L, Stackhouse J, Goulandris N, Snowling MJ, 2004. The development of early literacy skills among children with speech difficulties: a test of the “critical age hypothesis”. J. Speech. Lang. Hear. Res. 47, 377–91. 10.1044/1092-4388(2004/031) [DOI] [PubMed] [Google Scholar]
- Nation K, Snowling MJ, 2004. Beyond phonological skills: broader language skills contribute to the development of reading. J. Res. Read. 27, 342–356. 10.1111/j.1467-9817.2004.00238.x [DOI] [Google Scholar]
- Newman SD, Twieg D, 2001. Differences in Auditory Processing of Words and Pseudowords: An fMRI Study. Hum. Brain Mapp. 14, 39–47. 10.1002/hbm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora A, Karvonen L, Renvall H, Parviainen T, Kim J-Y, Service E, Salmelin R, 2017. Children show right-lateralized effects of spoken word-form learning. PLoS One 12, e0171034 10.1371/journal.pone.0171034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton ES, Beach SD, Gabrieli JDE, 2015. Neurobiology of dyslexia. Curr. Opin. Neurobiol. 30, 73–8. 10.1016/j.conb.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton ES, Wolf M, 2012. Rapid automatized naming (RAN) and reading fluency: implications for understanding and treatment of reading disabilities. Annu. Rev. Psychol. 63, 427–52. 10.1146/annurev-psych-120710-100431 [DOI] [PubMed] [Google Scholar]
- Parrila R, Kirby JR, McQuarrie L, 2004. Articulation Rate, Naming Speed, Verbal Short-Term Memory, and Phonological Awareness: Longitudinal Predictors of Early Reading Development? Sci. Stud. Read 8, 3–26. 10.1207/s1532799xssr0801_2 [DOI] [Google Scholar]
- Pennington BF, 2006. From single to multiple deficit models of developmental disorders. Cognition 101, 385–413. 10.1016/j.cognition.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Perrachione TK, Ghosh SS, Ostrovskaya I, Gabrieli JDE, Kovelman I, 2017. Phonological Working Memory for Words and Nonwords in Cerebral Cortex. J. Speech. Lang. Hear. Res. 60, 1959–1979. 10.1044/2017_JSLHR-L-15-0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, Pennington BF, 2015. Developmental Dyslexia. Annu. Rev. Clin. Psychol. 11, 283–307. 10.1146/annurev-clinpsy-032814-112842 [DOI] [PubMed] [Google Scholar]
- Plomin R, Kovas Y, 2005. Generalist genes and learning disabilities. Psychol. Bull. 131, 592–617. 10.1037/0033-2909.131.4.592 [DOI] [PubMed] [Google Scholar]
- Preston JL, Felsenfeld S, Frost SJ, Mencl WE, Fulbright RK, Grigorenko EL, Landi N, Seki A, Pugh KR, 2012. Functional Brain Activation Differences in School-Age Children with Speech Sound Errors: Speech and Print Processing. J. Speech Lang. Hear. Res 55, 1068–1082. 10.1044/1092-4388(2011/11-0056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JL, Frost SJ, Mencl WE, Fulbright RK, Landi N, Grigorenko E, Jacobsen L, Pugh KR, 2010. Early and late talkers: school-age language, literacy and neurolinguistic differences. Brain 133, 2185–95. 10.1093/brain/awq163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JL, Molfese PJ, Frost SJ, Mencl WE, Fulbright RK, Hoeft F, Landi N, Shankweiler D, Pugh KR, 2016. Print-Speech Convergence Predicts Future Reading Outcomes in Early Readers. Psychol. Sci. 27, 75–84. 10.1177/0956797615611921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh KR, Frost SJ, Rothman DL, Hoeft F, Del Tufo SN, Mason GF, Molfese PJ, Mencl WE, Grigorenko EL, Landi N, Preston JL, Jacobsen L, Seidenberg MS, Fulbright RK, 2014. Glutamate and choline levels predict individual differences in reading ability in emergent readers. J. Neurosci. 34, 4082–9. 10.1523/JNEUROSCI.3907-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh KR, Landi N, Preston JL, Mencl WE, Austin AC, Sibley D, Fulbright RK, Seidenberg MS, Grigorenko EL, Constable RT, Molfese P, Frost SJ, 2013. The relationship between phonological and auditory processing and brain organization in beginning readers. Brain Lang. 125, 173–183. 10.1016/j.bandl.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhlin N, Cardoso-Martins C, Kornilov SA, Grigorenko EL, 2013. Spelling well despite developmental language disorder: what makes it possible? Ann. Dyslexia 63, 253–273. 10.1007/s11881-013-0084-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte C, Torgesen JK, Wagner R, 1999. Test of Word Reading Efficiency. PRO-ED, Austin, TX. [Google Scholar]
- Rescorla L, 2005. Age 13 language and reading outcomes in late-talking toddlers. J. Speech. Lang. Hear. Res. 48, 459–72. 10.1044/1092-4388(2005/031) [DOI] [PubMed] [Google Scholar]
- Rumsey JM, Horwitz B, Donohue BC, Nace K, Maisog JM, Andreason P, 1997. Phonological and orthographic components of word recognition. A PET-rCBF study. Brain 120 ( Pt 5, 739–59. [DOI] [PubMed] [Google Scholar]
- Sarkari S, Simos PG, Fletcher JM, Castillo EM, Breier JI, Papanicolaou AC, 2002. Contributions of magnetic source imaging to the understanding of dyslexia. Semin. Pediatr. Neurol. 9, 229–38. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Schulte-Körne G, 2010. Genetics of developmental dyslexia. Eur. Child Adolesc. Psychiatry 19, 179–97. 10.1007/s00787-009-0081-0 [DOI] [PubMed] [Google Scholar]
- Schinzel A, Giedion A, 1978. A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. Am. J. Med. Genet. 1, 361–375. 10.1002/ajmg.1320010402 [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Escobar MD, Shaywitz BA, Fletcher JM, Makuch R, 1992. Evidence that dyslexia may represent the lower tail of a normal distribution of reading ability. N. Engl. J. Med. 326, 145–50. 10.1056/NEJM199201163260301 [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA, Pugh KR, Fulbright RK, Constable RT, Mencl WE, Shankweiler DP, Liberman AM, Skudlarski P, Fletcher JM, Katz L, Marchione KE, Lacadie C, Gatenby C, Gore JC, 1998. Functional disruption in the organization of the brain for reading in dyslexia. Proc. Natl. Acad. Sci. U. S. A. 95, 2636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowling M, Bishop DV, Stothard SE, 2000. Is preschool language impairment a risk factor for dyslexia in adolescence? J. Child Psychol. Psychiatry. 41, 587–600. [DOI] [PubMed] [Google Scholar]
- Stevenson J, 1988. Which aspects of reading ability show a “hump” in their distribution? Appl. Cogn. Psychol. 2, 77–85. 10.1002/acp.2350020107 [DOI] [Google Scholar]
- Stothard SE, Snowling MJ, Bishop DV, Chipchase BB, Kaplan CA, 1998. Language-impaired preschoolers: a follow-up into adolescence. J. Speech. Lang. Hear. Res. 41, 407–18. [DOI] [PubMed] [Google Scholar]
- Suphapeetiporn K, Srichomthong C, Shotelersuk V, 2011. SETBP1 mutations in two Thai patients with Schinzel-Giedion syndrome. Clin. Genet. 79, 391–3. 10.1111/j.1399-0004.2010.01552.x [DOI] [PubMed] [Google Scholar]
- Swanson HL, Zheng Xinhua, Jerman O, 2009. Working memory, short-term memory, and reading disabilities: a selective meta-analysis of the literature. J. Learn. Disabil. 42, 260–87. 10.1177/0022219409331958 [DOI] [PubMed] [Google Scholar]
- Takeuchi A, Okamoto N, Fujinaga S, Morita H, Shimizu J, Akiyama T, Ninomiya S, Takanashi J, Kubo T, 2015. Progressive brain atrophy in Schinzel–Giedion syndrome with a SETBP1 mutation. Eur. J. Med. Genet 58, 369–371. 10.1016/j.ejmg.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Tamboer P, Vorst HCM, Oort FJ, 2014. Identifying dyslexia in adults: an iterative method using the predictive value of item scores and self-report questions. Ann. Dyslexia 64, 34–56. 10.1007/s11881-013-0085-9 [DOI] [PubMed] [Google Scholar]
- Taylor JSH, Rastle K, Davis MH, 2013. Can cognitive models explain brain activation during word and pseudoword reading? A meta-analysis of 36 neuroimaging studies. Psychol. Bull 139, 766–91. 10.1037/a0030266 [DOI] [PubMed] [Google Scholar]
- Thompson LA, Detterman DK, P.R., 1991. Associations between cognitive abilities and scholastic achievement: genetic overlap but environmental differences. Psychol Sci. 2, 158–165. [Google Scholar]
- van Leeuwen M, van den Berg SM, Peper JS, Hulshoff Pol HE, Boomsma DI, 2009. Genetic covariance structure of reading, intelligence and memory in children. Behav. Genet. 39, 245–54. 10.1007/s10519-009-9264-1 [DOI] [PubMed] [Google Scholar]
- Vidyasagar TR, Pammer K, 2010. Dyslexia: a deficit in visuo-spatial attention, not in phonological processing. Trends Cogn. Sci. 14, 57–63. 10.1016/j.tics.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Wagner RK, Torgesen JK, Rashotte CA, 1999. Comprehensive Test of Phonological Processing. PRO-ED, Austin, TX. [Google Scholar]
- Wechsler D, 1999. Wechsler Abbreviated Scale of Intelligence. The Psychological Corporation: Harcourt Brace & Company, New York. [Google Scholar]
- Weiss-Croft LJ, Baldeweg T, 2015. Maturation of language networks in children: A systematic review of 22years of functional MRI. Neuroimage 123, 269–81. 10.1016/j.neuroimage.2015.07.046 [DOI] [PubMed] [Google Scholar]
- Wiederholt JL, 2001. Gray Oral Reading Test. PRO-ED, Austin, TX. [Google Scholar]
- Wimmer H, Hutzler F, Wiener C, 2002. Children with dyslexia and right parietal lobe dysfunction: event-related potentials in response to words and pseudowords. Neurosci. Lett. 331, 211–3. [DOI] [PubMed] [Google Scholar]
- Woodcock RW, McGrew KS, Mather N, 2001. Woodcock Johnson III. Riverside Publishing, Itasca, IL. [Google Scholar]
- Xiao Z, Zhang JX, Wang X, Wu R, Hu X, Weng X, Tan LH, 2005. Differential activity in left inferior frontal gyrus for pseudowords and real words: An event-related fMRI study on auditory lexical decision. Hum. Brain Mapp. 25, 212–221. 10.1002/hbm.20105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M, 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–4. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.