Abstract
The inborn errors of heme biosynthesis, the Porphyrias, include eight major disorders resulting from loss-of-function (LOF) or gain-of-function (GOF) mutations in eight of the nine heme biosynthetic genes. The major sites of heme biosynthesis are the liver and erythron, and the underlying pathophysiology of each of these disorders depends on the unique biochemistry, cell biology, and genetic mechanisms in these tissues. The porphyrias are classified into three major categories: 1) the acute hepatic porphyrias (AHPs), including Acute Intermittent Porphyria (AIP), Hereditary Coproporphyria (HCP), Variegate Porphyria (VP), and 5-Aminolevlulinic Acid Dehydratase Deficient Porphyria (ADP); 2) a hepatic cutaneous porphyria, Porphyria Cutanea Tarda (PCT); and 3) the cutaneous erythropoietic porphyrias, Congenital Erythropoietic Porphyria (CEP), Erythropoietic Protoporphyria (EPP), and X-Linked Protoporphyria (XLP). Their modes of inheritance include autosomal dominant with markedly decreased penetrance (AIP, VP, and HCP), autosomal recessive (ADP, CEP, and EPP), or X-linked (XLP), as well as an acquired sporadic form (PCT). There are severe homozygous dominant forms of the three AHPs. For each porphyria, its phenotype, inheritance pattern, unique genetic principles, and molecular genetic heterogeneity are presented. To date, >1000 mutations in the heme biosynthetic genes causing their respective porphyrias have been reported, including low expression alleles and genotype/phenotype correlations that predict severity for certain porphyrias. The tissue-specific regulation of heme biosynthesis and the unique genetic mechanisms for each porphyria are highlighted.
1. Introduction
The porphyrias are a group of eight metabolic disorders, each resulting from the defective activity of a specific enzyme in the heme biosynthesis pathway (1, 2). The genetics of these inborn errors are of particular interest as they encompass many genetic mechanisms and principles (Table 1), including multiple modes of inheritance, molecular genetic heterogeneity, genotype/phenotype correlations, high prevalence and low penetrance, modifying genes, pharmacogenetics, as well as environmental factors.
Table 1.
The Porphyrias Illustrate Many Genetic Principles
| 1. Multiple Modes of Inheritance: |
| Autosomal Dominant (AIP, HCP, & VP) & Recessive (AHPs, CEP, & EPP) |
| X-Linked (XLP, Rarely CEP) |
| Rare Homozygous Dominant Types (AIP, HCP, & VP) |
| Sporadic/Acquired (PCT) |
| Prevalence & Penetrance (AHPs) |
| 2. Multiple Mutations Causing Each Disease (All) |
| Loss- or Gain-of-Function Mutations (All) |
| Common Low Expression Allele (EPP) |
| Regulation of Heme Biosynthesis & Regulatory Mutations |
| Negative Feedback Repression (AHPs) |
| Variable Expressivity (All) |
| Genotype/Phenotype Correlations (CEP) |
| Benign Variants (All) |
| 3. X-Chromosomal Inactivation (XLP) |
| 4. Modifying Genes Responsible for Expression & Severity (AHPs, EPP) |
| 5. Pharmacogenetic/Ecogenetic Disorders (AHPs) |
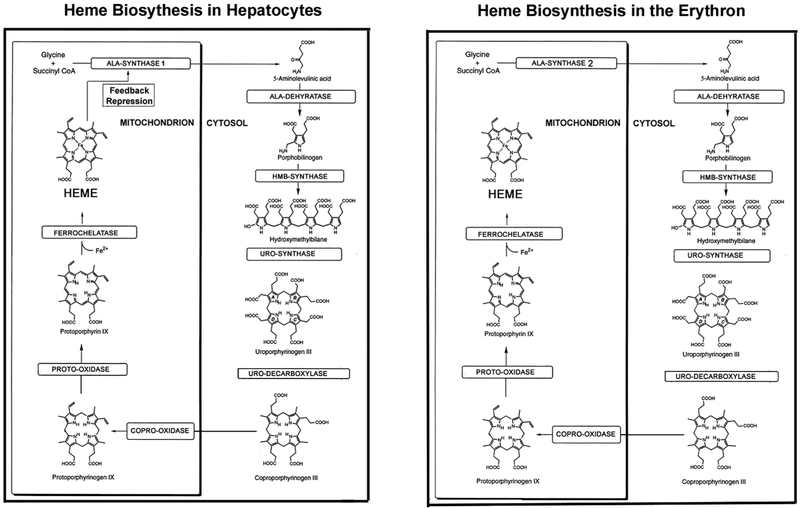
Heme biosynthesis occurs in almost all cell types, but most heme is produced in the bone marrow erythron (~85%) primarily for hemoglobin synthesis, and in hepatocytes (~15%) for critical hemoproteins, including the cytochrome P450 enzymes for drug metabolism (1). Heme biosynthesis in each of these major sites is under different regulatory mechanisms, which account for the unique pathophysiology of the hepatic and erythropoietic porphyrias. As shown in Figure 1, there are eight enzymatic steps of the heme biosynthesis pathway that are encoded by nine genes, as the first step in the pathway has two genes, 5-aminolevulinic acid synthase 1 (ALAS1), a housekeeping gene located at chromosome 3p21.1, and an erythroid-specific gene (ALAS2) on the X-chromosome at Xp11.21. The cDNAs and genes encoding all the heme biosynthetic enzymes have been cloned and sequenced (Table 2), and the 3-dimensional structures of their respective encoded enzymes have been solved for human, eukaryotic, and/or prokaryotic species (3-10).
Fig 1.
The human heme biosynthetic pathway in hepatocytes (left) and erythroid cells (right). The eight enzymatic steps, their porphyrin precursor and porphyrin substrates, and metabolic products are shown. Note that the first enzyme in the pathway in hepatocytes, ALAS1 is regulated by negative feedback, repression by the concentration of the end product heme in the free heme pool. In contrast, the erythroid-specific ALAS2 enzyme is responsible for synthesis of 5’Aminolevulinic acid (ALA) in the erythron and its expression is under control of erythroid regulatory elements. The subsequent enzymatic steps in the pathway are similar, some of the enzymes having unique housekeeping and erythroid specific promoters. Modified from Anderson et al.: Disorders of heme biosynthesis: X-Linked sideroblastic anemia and the Porphyrias. Online Metabolic and Molecular Bases of Inherited Diseases (https://ommbid.mhmedical.com).
Table 2.
Characteristics of The Heme Biosynthesis Genes
| Gene | Gene Symbol |
Chromosome Position |
Gene Length (kb) |
Exons | cDNA (bp) |
Encoded Protein Amino Acids |
|---|---|---|---|---|---|---|
| Aminolevulinic Acid Synthase 1 | ALAS1 | 3p21.1 | ~20 | 12 | ~2458 | 641 |
| Aminolevulinic Acid Synthase 2 | ALAS2 | Xp11.21 | ~26 | 12 | ~2044 | 588 |
| ALA-Dehydratase Deficiency | ALAD | 9q33.1 | ~19 | 12 | ~3200 | 331 |
| Hydroxymethylbilane Synthase | HMBS | 11q23.3 | ~13 | 15 | ~1526 | 362 |
| Uroporphyrinogen Synthase | UROS | 10q25.2-q26.3 | ~39 | 10 | ~1371 | 266 |
| Uroporphyrinogen Decarboxylase | UROD | 1p34.1 | ~8 | 10 | ~1408 | 368 |
| Coproporphyrinogen Oxidase | CPOX | 3q12 | ~18 | 7 | ~2728 | 455 |
| Protoporphyrinogen Oxidase | PPOX | 1q22 | ~9 | 13 | ~1716 | 478 |
| Ferrochelatase | FECH | 18q21.3 | ~46 | 11 | ~7277 | 424 |
Human Gene Mutation Database 2018.2: www.hgmd.org ∣ Aug 15, 2018
Genes listed in pathway sequence for ALAS to FECH
Unique to this pathway is the fact that there is a negative-feedback regulation of ALAS1 expression in the liver depending on the cellular “free heme pool” concentration (See Phillips et al. in this volume; Fig. 1). This is analogous to cholesterol biosynthesis where the first enzyme in the pathway (HMG-CoA reductase) is regulated by the hepatic cholesterol concentration (11). The hepatic negative-feedback regulation of ALAS1 is particularly relevant to the genetics, biochemistry, and pathogenesis of the acute hepatic porphyrias (AHPs). Analogously, the unique regulation of the erythroid–specific expression of ALAS2 is relevant to the erythropoietic porphyrias. ALAS2 has several regulatory elements in its promoter, including a GATA1-binding site (12), and iron response elements (IREs). The 5’ IRE binds the iron response proteins (IRPs) when cellular iron levels are low, thereby inhibiting the translation of ALAS2. Mutations in the terminal exon of the ALAS2 gene lead to truncation or elongation of the mature ALAS2 polypeptide resulting in its gain-of-function (GOF) and subsequent accumulation of erythrocyte protoporphyrin IX (PPIX), resulting in XLP [(13); see below]. Conversely, ALAS2 loss-of-function (LOF) mutations result in X-linked sideroblastic anemia.
The porphyrias can be categorized based on their major clinical features and primary sites of abnormal porphyrin or porphyrin precursor accumulation. As shown in Table 3, there are four acute hepatic porphyrias (AHPs), each due to LOF mutations that decrease the activity of a particular heme biosynthetic enzyme. These include three autosomal dominant disorders, Acute Intermittent Porphyria (AIP), Hereditary Coproporphyria (HCP), Variegate Porphyria (VP), and the ultra-rare autosomal recessive 5-Aminolevulinic Acid Dehydratase Deficient Porphyria (ADP), due to mutations in the hydroxymethylbilane synthase (HMBS), coproporphyrinogen oxidase (CPOX), protoporphyrinogen oxidase (PPOX), and 5-Aminolevulinic acid dehydratase (ALAD) gene, respectively. The most common porphyria, Porphyria Cutanea Tarda (PCT), is a hepatic disorder that has primarily cutaneous manifestations, and therefore, is classified as a hepatic cutaneous porphyria. This disorder has sporadic and familial subtypes, both postulated to be due to the induced deficiency of uropophyrinogen decarboxylase (UROD) by an enzyme inhibitor, uroporphomethene, which occurs in patients with hepatic iron overload (14-16). The third group is the erythropoietic cutaneous porphyrias, which include autosomal recessive Congenital Erythropoietic Porphyria (CEP) and Erythropoietic Protoporphyria (EPP) and the X-linked disorder, X-Linked Protoporphyria (XLP). CEP and EPP result from LOF mutations in the uroporphyrinogen synthase (UROS) and ferrochelatase (FECH) genes, respectively, while XLP results from GOF mutations in the ALAS2 gene. Below, the inheritance, genetic principles, and molecular genetics of the AHPs, PCT, and the erythropoietic porphyrias are presented.
Table 3.
Classification of the Porphyrias: Their Enzyme Defects & Inheritance
| Classification | Enzyme Deficiency | Inheritance* |
|---|---|---|
| Acute Hepatic Porphyrias (AHPs): | ||
| Acute Intermittent Porphyria | HMB-Synthase | AD |
| Hereditary Coproporphyria | COPRO-Synthase | AD |
| Variegate Porphyria | PROTO-Oxidase | AD |
| ALA-Dehydratase Deficiency | ALA-Dehydratase | AR |
| Hepatic Cutaneous Porphyria (PCT): | ||
| Porphyria Cutanea Tarda Type 1 | Hepatic UROD Deficiency | Sporadic |
| Porphyria Cutanea Tarda Type 2 | Systemic UROD Deficiency | AD |
| Erythropoietic Cutaneous Porphyrias: | ||
| Congenital Erythropoietic Porphyria | URO-Synthase | AR |
| Erythropoietic Protoporphyria | Ferrochelatase | AR |
| X-Linked Protoporphyria | ALA-Synthase 2 | X-L |
AD = Autosomal Dominant; AR = Autosomal Recessive; X-L= X-Linked
2. The Porphyrias Illustrate Many Genetic Principles
2.1. Multiple Modes of Inheritance, Prevalence and Penetrance:
As noted above, the eight major porphyrias are inherited as autosomal dominant (AIP, HCP, and VP), autosomal recessive (ADP, CEP, and EPP), and X-linked (XLP) traits. Notably, the autosomal dominant AHPs have remarkably low penetrance. For example, the estimated prevalence of AIP is remarkably high, 1 in ~1,700 Caucasians, whereas the estimated penetrance of an acute attack is about 1% (17, 18). Also, there are very rare and severe homozygous dominant forms of AIP (19-22), HCP (23-25), and VP (26-28). Notably, PCT has two major subtypes that are clinically indistinguishable: sporadic Type 1 and familial Type 2. Type 2 patients have half-normal UROD activity in all cells, whereas the Type 1 sporadic patients only have deficient hepatic UROD activity secondary to iron overload, porphyrin accumulation, and the formation of an UROD inhibitor (14).
2.2. Multiple Mutations Underlying Each Disease
In each of the eight major porphyrias, there is remarkable molecular genetic heterogeneity with all mutation types represented (see Table 4). Only a few mutations are relatively common due to CpG dinucleotide mutational hotspots in the gene (29) or due to founder effects, e.g. HMBS W198X in Sweden and Norway (30). An exception is the most common mutation in CEP, c.217T>C (p.C73R), which occurs in about 30% of unrelated patients and is not at a CpG dinucleotide, nor is it a founder mutation in a certain population (31).
Table 4.
Reported Mutations Causing The Porphyrias (HGMD v2018.2)
| HGMD Porphyria |
ALAD ADP |
HMBS AIP |
CPOX HCP |
PPOX VP |
UROS CEP |
UROD PCT |
FECH EPP |
ALAS2 XLP |
|
|---|---|---|---|---|---|---|---|---|---|
| Mutation Type: | |||||||||
| Missense | 9 | 140 | 43 | 69 | 30 | 71 | 54 | 1 | |
| Nonsense | 0 | 33 | 6 | 18 | 1 | 8 | 26 | 1 | |
| Splicing substitutions (Consensus) | 0 | 76 | 2 | 22 | 2 | 10 | 39 | 0 | |
| Splicing substitutions (Cryptic) | 2 | 15 | 2 | 7 | 2 | 3 | 7 | 0 | |
| Regulatory substitutions | 0 | 7 | 0 | 4 | 6 | 1 | 2 | 0 | |
| Small deletions | 1 | 84 | 7 | 36 | 2 | 16 | 40 | 3 | |
| Small insertions/duplications | 0 | 41 | 7 | 21 | 2 | 7 | 10 | 0 | |
| Small indels | 0 | 8 | 1 | 3 | 1 | 1 | 1 | 0 | |
| Gross deletions | 0 | 13 | 3 | 3 | 2 | 5 | 13 | 1 | |
| Gross insertions/duplications | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | |
| Complex rearrangements | 0 | 2 | 0 | 0 | 1 | 0 | 3 | 0 | |
| Repeat variations | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| TOTAL | 12 | 421 | 71 | 184 | 51 | 122 | 195 | 6 | |
2.2.1. Loss and Gain of Function Mutations
Most mutations cause LOF (i.e., decreased enzymatic activity) of its encoded heme biosynthetic enzyme. These mutations alter the enzyme’s kinetics or stability or both. The exception is the GOF mutations in the terminal exon of the ALAS2 gene, which increase the ALAS2 enzymatic activity and result in high levels of PPIX, causing XLP (13, 32, 33).
2.2.2. Common Low Expression Allele
In EPP, over 95% of patients are compound heterozygotes for a pathogenic LOF allele and the common low expression allele, c.315-48T>C [also known as IVS3-48T>C (34, 35)]. This low expression (or hypomorphic) allele causes alternative splicing and markedly reduces the amount of normal FECH mRNA it expresses (34). This allele is relatively common, occurring in ~5% of Europeans, about ~2% of Africans, and up to 33% of Asians (GnomAD).
2.2.3. Promoter Alterations
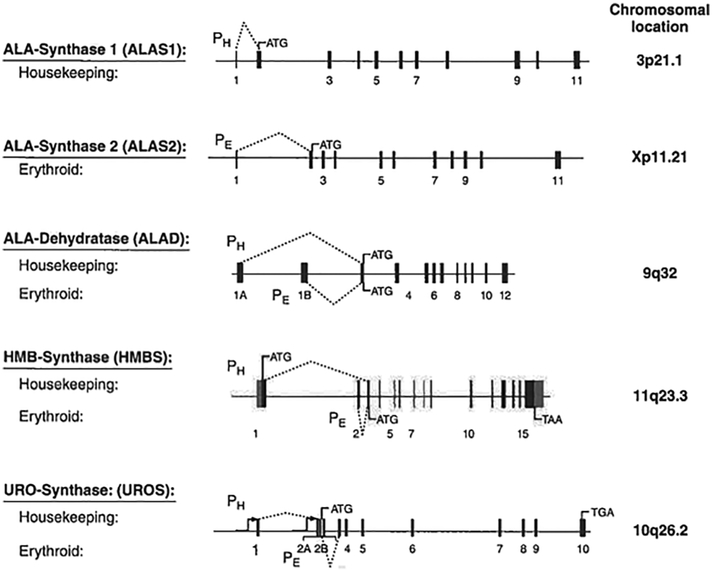
The ALAD, HMBS, and UROS enzymes are each encoded by a single gene with unique housekeeping and erythroid promoters (Fig. 2). For HMBS, the housekeeping and erythroid-specific promoters encode two isozymes. The upstream housekeeping promoter directs a transcript containing exons 1 and 3-15, while the downstream erythroid-specific promoter produces a transcript containing the exon 2-15 sequence. The two mRNAs encode the 42-kDa housekeeping and 40-kDa erythroid-specific HMBS isozymes, due to the housekeeping isozyme having 17 additional N-terminal amino acids. Therefore, mutations that alter exon 1 of the housekeeping gene or the splicing of exon 1 to exon 3 result in a variant form of AIP in which the liver enzyme has half-normal activity, but erythroid cells have normal activity (36, 37) (Fig. 3). This form of AIP is known as the “erythroid variant”, whereas mutations in exons 3-15 result in half-normal activity of both isozymes, referred to as “classic” (or “non-erythroid variant”) AIP. In contrast, the ALAD and UROS promoters encode the same enzyme (Fig. 2).
Fig 2.
The first four genes in the human heme biosynthetic pathway have unique housekeeping (PH) and erythroid-specific (PE) promoters. The dotted lines indicate the exons transcribed by each promoter. The ALAS1 and ALAS2 genes are on different chromosomes and are regulated by negative feedback repression (ALAS1) or by erythroid transcription factors, and the iron response element (ALAS2).
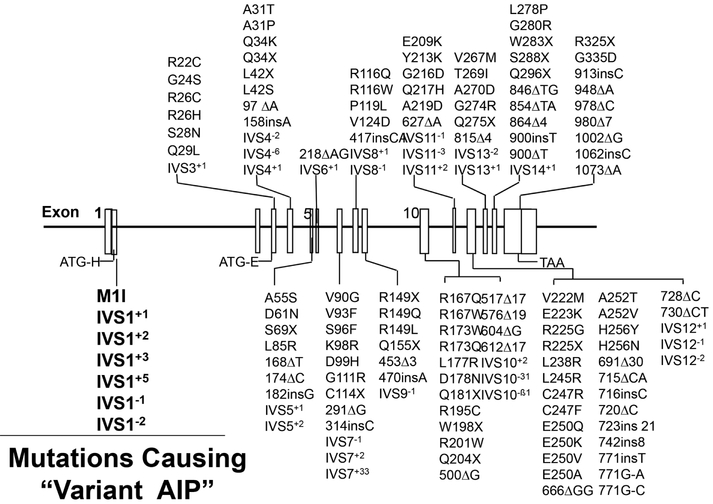
Fig 3.
Schematic of the HMBS gene indicating the numbered exons (black bars) and introns. Various known mutations are indicated by their exonic/intronic positions. Note that mutations that alter the sequence of the housekeeping gene’s initiation of transcription in exon 1 (ATG-H) or that alter the normal splicing of exon 1 to exon 3, result in a “variant form of AIP” in which the housekeeping enzyme is not synthesized, whereas, the erythroid-specific enzyme is expressed at normal levels. Thus, patients who have mutations causing “variant AIP” will have half-normal activity of the housekeeping HMBS isozyme and AIP, but normal activity of the HMBS enzyme in circulating erythrocytes.
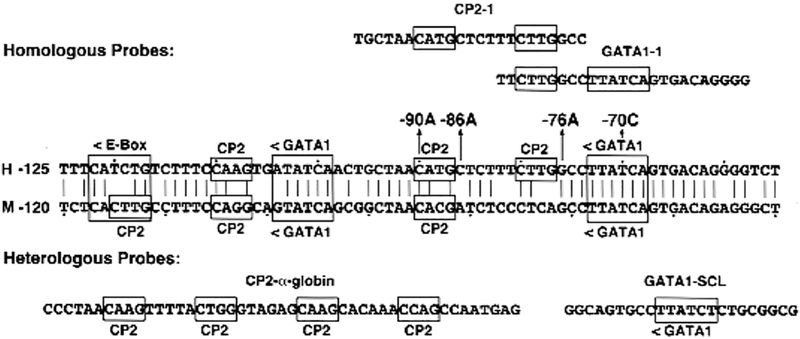
Regulatory gene defects in the 5’-promoter regions of the HMBS (38), UROS (39), and FECH genes (40) have been reported in patients with AIP, CEP, and EPP, respectively. For example, the pathogenicity of four promoter lesions in patients with CEP that involved GATA1 or CP2 erythroid transcription binding sites (Fig. 4) have been confirmed by in vitro expression, using luciferase assays (39).
Fig 4.
Partial sequence of the human and murine UROS erythroid-specific promoters showing the GATA1 and CP2 transcription factor binding sites. The location and orientation (<, >) of the GATA1, E-box, and CP2 erythroid binding elements are indicated, as are the four novel promoter mutations causing CEP. Dots are placed every tenth nucleotide. The homologous CP2 site from the α- globin gene promoter and the GATA1 site from the human stem cell leukemia (SCL) gene promoter are shown.
Additionally, hypermethylation of CpG sites within the FECH gene promoter was reported to decrease its transcriptional activity, perhaps leading to a more severe form of EPP with liver complications. Family members who had overt EPP but lacked hypermethylation of the promoter region had milder disease, suggesting that this modulated disease severity (41).
2.3. Benign Variants:
Most published mutations causing the porphyrias and other Mendelian diseases are catalogued in the Human Gene Mutation Database [HGMD, (42)]. This publically available database is updated quarterly, is available online (www.hgmd.cf.ac.uk), and provides a reference for each mutation. The database lists published mutations by disease and mutation type. However, many of these mutations have not been validated clinically and/or biochemically. In the early days of gene sequencing (1980’s and 90’s), benign variants were identified and many were thought to be pathogenic, as the entire coding region (or gene) was not sequenced to find the causative pathogenic mutation as it is today. Moreover, until recently, there were no available databases that provided the frequencies of all detected variants in various racial and ethnic populations such as GnomAD (gnomad.broadinstitute.org). Typically, benign variants are missense mutations or are located near the consensus splice-site but further into the introns (e.g., >4 nucleotides). Such mutations have been reported in HGMD as pathogenic in the past, and some have been reclassified when data indicating that they are benign variants are published [e.g., for HMBS benign variants and AIP, see (17)].
Genomic databases provide allele frequency data for variant nucleotides in all genes based on whole exome and genome sequencing of thousands of individuals in different racial and ethnic populations. They are particularly useful in identifying relatively frequent variants and are informative for classifying a variant as pathogenic or benign. For example, HMBS IVS10-31A>G was identified in a French AIP patient of Caribbean origin and originally reported as pathogenic in 1997 (43). Later, it was reclassified as a common variant that was frequent in African and Hispanic populations (44) with minor allele frequencies (MAF) of up to ~43% and ~2% in Black and Hispanic populations, respectively (based on GnomAD data).
2.4. X-Chromosomal Inactivation (XLP)
XLP, resulting from GOF mutations located in the terminal exon of ALAS2, is inherited as an X-linked trait. Affected males who inherited the X-chromosome with the GOF mutation from their heterozygous mothers (XXLP/Xnormal) have markedly increased erythrocyte PPIX levels and are clinically indistinguishable from male or female patients with EPP. However, female heterozygotes for XLP have variable clinical expressivity ranging from asymptomatic to as severely affected as their male relatives. This variability results from random X-chromosomal inactivation (45).
Since females have two X-chromosomes and males have only one, a mechanism to equalize the expression of the genes on the X-chromosomes of females occurs in early embryogenesis. One of the female’s X-chromosomes is inactivated. This “dose compensation” may vary in different cell types, but once a particular X-chromosome is inactivated, the same X will be inactivated in its daughter cells, cell division after division. Thus, if the cell inactivated the X-chromosome with the ALAS2 mutation causing XLP, only normal levels of the ALAS2 enzyme would be expressed by the active normal X-chromosome. One might expect that since the inactivation process is random, it would be 50:50. In effect, half of the cells would inactivate the X with the ALAS2 mutation and half of the cells would inactivate the normal ALAS2 on the other X-chromosome. If a 50:50 ratio for X-chromosomal inactivation occurred, the ALAS2 enzymatic activity would be increased in erythroid cells by the amount overexpressed by the GOF ALAS2 gene.
However, as one may have experienced at a casino, the 50:50 expectation may only occur about 50% of the time. The other 50% of the time, the ratio may be 60:40, 70:30, even 95:5. It is estimated that these ratios occur 40%, 30%, and 5% of the time, respectively. It has been suggested that random X-chromosomal inactivation occurs in various cell types independently, making the expression of an X-linked mutation variable by cell type, tissue, and organ. For XLP, the erythroid cells are the major cell type involved. So assume that in the earliest erythroid precursor cell in a given XLP female heterozygote, the ratio of inactivation of the chromosome with the mutant ALAS2 was 100% (a rare event!), then only the wild-type ALAS2 on the active X-chromosome would be expressed. This woman would have totally normal erythroid ALAS2 activity, normal erythrocyte PPIX levels, and no clinical manifestations, but still have a risk to pass the mutant gene to half (on average) of her male and female offspring. She would be diagnosed only by sequencing her ALAS2 gene. If the opposite occurred, and a woman had only the chromosomes with the ALAS2 mutation actively expressed, then she would have elevated ALAS2 activity, increased levels of erythrocyte ‘free’ and zinc PPIX, and would be as severely affected as her male relatives. The variation in clinical severity results from when the ratios are between the above two extreme scenarios. For example, say its 70:30 in favor of the mutation being expressed, this woman would likely have some disease manifestations including increased erythrocyte PPIX levels and photosensitivity, but likely would not be as severe as her affected male relatives. Conversely, if only 30% of erythroid cells expressed the mutant enzyme, it is likely that the erythrocyte PPIX would only be somewhat increased, but the woman may not have erythrocyte PPIX levels that caused clinical photosensitivity, or if she did, her sun exposure time would be longer until she felt the first prodromal symptoms than that of affected male relatives.
CEP resulting from GATA1 mutations also is inherited as an X-linked trait. Therefore, heterozygotes may be asymptomatic or as affected as male relatives [(46, 47); see Section 2.5 below.]
2.5. New Causative Genes
Two other genes involved in porphyria pathogenesis have been identified to date. The ATP-dependent CLP protease ATP-binding subunit (CLPX) gene located at chromosome 15q22.31 encodes a protein with mitochondrial ATPase activity, which forms a complex with the ATP-dependent CLP protease proteolytic subunit (CLPP) protein. This multimeric complex, termed CLPXP, is an ATP-dependent serine protease that degrades specific proteins, including the ALAS enzymes. A LOF mutation in CLPX reduces the proteasome activity of CLPXP and leads to increased posttranslational stability and activity of ALAS2, and consequentially, the accumulation of erythrocyte PPIX, thereby causing a form of EPP (48). The second is the GATA1 gene, which encodes a transcription factor important for erythropoiesis. Rarely, a mutation that directly alters the expression of the GATA1 gene transcription factor protein can cause an X-linked form of CEP (46, 47).
2.6. New Modifying Genes and Environmental Factors
The severity of the different porphyrias can be affected by modifying genes and the environment. To date, only a few modifying genes have been identified for the erythropoietic and hepatic porphyrias. A terminal exon mutation in the ALAS2 gene that increases ALAS2 activity was found in a family with CEP and shown to increase the severity of the patient’s photosensitivity (49). More recently, a LOF mutation in CLPX, which is involved in the degradation of the ALAS enzymes, was found to stabilize ALAS2 and increase its activity, promoting erythroid porphyrin synthesis, and consequentially, increasing erythrocyte PPIX levels and photosensitivity (48) (see Section 2.2.4). Thus, if CLPX or CLPP, which it is associated with, is mutated in any erythropoietic porphyria, it is expected to make the disease more severe. Similarly, as noted above, a very rare X-linked form of CEP resulted from a particular mutation in the X-linked GATA1 erythroid transcription factor. Hence, mutations in this gene in an erythropoietic porphyria most likely would increase disease severity, particularly in males.
In the AHPs, it was recently reported that a polymorphism in the PEPT2 gene, which is responsible for the transport of ALA from the cerebrospinal fluid (50), is also involved in the re-uptake of ALA from the urine into renal cells, which can be toxic and cause renal insufficiency (51). If these findings are confirmed, all patients with an AHP, particularly those with elevated ALA levels, should be screened for the higher affinity PEPT2 polymorphic genotypes, 1*1 or 1*2, which in this single study were associated with increased risk for renal disease.
Finally, it is clear that environmental factors play an important role in the pathogenesis of the porphyrias. For the AHPs, there is a list of porphyrinogenic drugs which patients should avoid as they can trigger an acute attack (see the Porphyrias Consortium website : https://www.rarediseasesnetwork.org/cms/porphyrias). In addition, AHP attacks can be triggered by fasting or dieting and by various hormonal changes including the use of high progesterone birth control medications. With respect to PCT, it is now appreciated that environmental agents and genetic factors that cause hepatic iron overload are precipitating factors for the cutaneous manifestations. These include significant alcohol consumption, infections such as HCV and HIV, smoking, and various cytochrome P450 variants. For the erythropoietic porphyrias, the amount of sun exposure is directly related to the degree of phototoxicity experienced by these patients.
It is likely that in the future, additional modifying genes will be identified that exacerbate AHP acute attacks or increase the severity of PCT or an erythropoietic porphyria. In addition, there may be protective variants that decrease the likelihood of an AHP attack or increase the amount of sun exposure an EPP patient can endure before the first prodromal symptom. Such advances in the identification of modifying genes will lead to better clinical management and possibly to new safe and effective treatments.
3. The AHPs
3.1. Clinical Manifestations of the AHPs
The AHPs include the three autosomal dominant disorders, AIP, HCP, VP, and the ultra-rare autosomal recessive ADP (Table 3). All four acute porphyrias present with acute neurovisceral attacks, which typically start with a prodome of ‘brain fog’, insomnia, and fatigue, and then crippling abdominal pain that progresses to include tachycardia, hypertension, motor weakness, and seizures. The attacks are triggered by various porphyrinogenic factors, including fasting, cytochrome P450-inducing drugs, and hormonal fluctuations, all of which induce the hepatic expression of ALAS1, the first and rate-limiting enzyme of the heme biosynthetic pathway. When hepatic ALAS1 is induced, the respective enzyme deficiency becomes rate-limiting, leading to decreased heme production and depletion of the hepatic ‘free’ heme pool. This leads to derepression and further induction of hepatic ALAS1, resulting in the marked elevation of the putative neurotoxic porphyrin precursors, ALA and PBG. Patients with HCP and VP may also present with cutaneous photosensitivity.
3.2. Genetics of the AHPs
As noted above, three of the AHPs, AIP, HCP, and VP, are autosomal dominant disorders with incomplete penetrance, while ADP is inherited as an autosomal recessive disease with variable clinical severity depending on the amount of residual ALAD activity (Table 3). Of the three autosomal dominant AHPs, AIP is by far the most common, with an estimated prevalence of 1 in ~1600-1700 (17, 18). Each disorder develops clinical manifestations due to LOF mutations in the respective heme biosynthetic gene, HMBS, CPOX, PPOX, and ALAD (Tables 2 and 3). For the three autosomal dominant AHPs, LOF mutations lead to half-normal activity of the respective enzyme, whereas in ADP, patients typically have LOF mutations that result in markedly decreased, but not absent ALAD activity. Prokaryotic expression studies have shown that patients with clinically severe ADP had two ALAD mutations with <8% of expressed wild-type activity, whereas those with less severe disease had one mutation with significant residual activity (ranging from 19-70% of expressed wild-type activity) and a severe ALAD mutation (<8% of expressed wild-type activity) on the other allele (52).
To date, >685 mutations for the four AHPs have been reported in their respective genes [HGMD version 2018.2; (42)], (see Table 4 for types of AHP mutations)]. There are 415 HMBS mutations causing AIP, 69 CPOX mutations causing HCP, and 182 PPOX mutations causing VP. While most AHP mutations are “private,” identified in a single or few families, certain mutations are more common as they occur at CpG dinucleotides, hot spots for mutations (29). For the HMBS gene, these include the mutations encoding p.R173W and p.R167Q. Table 5 indicates the most common mutations causing AIP, HCP, and VP, based on the Mount Sinai Porphyria Diagnostic Laboratory’s experience. Founder mutations occur in certain populations and are inherited for generations. For example, in Sweden and Norway, HMBS c.593G>A (p.W198X) is a common founder mutation dating back multiple generations (53). There are also founder mutations for AIP in the Netherlands [c.346C>T, p.R116W; (54)], Switzerland and France [c.848G>A, p.W283X; (55)], Argentina [c.331G>A, p.G111R; (56)], and the Murcia region of Spain [c.669_698del; (57)]. Although there are no specific founder or common mutations for HCP, for VP, there are an estimated 30,000 to 40,000 patients in South Africa with the founder mutation c.175C>T (p.R59W), which was brought to the Cape by a Dutch couple who arrived in 1680 (58). It should be noted that AIP patients who have HMBS exon 1 mutations have normal erythrocytic HMBS activity, as this exon is not included in the alternatively-spliced erythroid HMBS transcript (Figs. 2 and 3; also see Principles Section above).
Table 5.
Most Common Mutations: Acute Porphyrias – Mount Sinai Experience Since 1/1/07
| AIP / HMBS | HCP / CPOX | VP / PPOX | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | % of 315 | Mutation | % of 29 | Mutation | % of 54 | ||||||
| cDNA | Amino Acid |
Unrelated Probands |
cDNA | Amino Acid |
Unrelated Probands |
cDNA | Amino Acid |
Unrelated Probands |
|||
| c.517C>T | p.R173W | 13.0 | c.601G>A | p.E201K | 13.8 | c.1303C>T | p.Q435X | 5.6 | |||
| c.500G>A | p.R167Q | 7.6 | c.565G>A | p.G189S | 6.9 | c.503G>A | p.R168H | 5.6 | |||
| c.848G>A | p.W283X | 6.3 | c.626T>A | p.V209D | 6.9 | c.503G>T | p.R168L | 5.6 | |||
| c.331G>A | p.G111R | 3.2 | c.395C>T | p.A132V | 3.4 | c.565C>T | p.Q189X | 5.6 | |||
| c.673C>T | p.R225X | 2.9 | c.520G>A | p.A174T | 3.4 | c.217C>G | p.L73V | 3.7 | |||
| c.499C>T | p.R167W | 2.5 | c.980A>G | p.H327R | 3.4 | c.454C>T | p.R152C | 3.7 | |||
| c.973C>T | p.R325X | 2.2 | c.40G>C | p.G14R | 1.9 | ||||||
| c.76C>T | p.R26C | 1.9 | |||||||||
| c.992C>T | p.A331V | 1.9 | |||||||||
| c.1084delT | c.1084delT | 1.9 | |||||||||
| TOTAL: | 45.5% | 52% | 34% | ||||||||
Several previously reported pathogenic HMBS mutations causing AIP have been shown to be benign variants (17). To verify the pathogenicity of the genes causing the AHPs, an International Collaborative has been established between the Porphyrias Consortium of the NIH-supported Rare Disease Clinical Research Network (https://www.rarediseasesnetwork.org/cms/porphyrias) and the European Porphyrias Network (EPNET: http://porphyria.eu). All previously reported and new mutations causing the AHPs are being validated both clinically and biochemically (i.e. in patients with acute attacks and markedly elevated ALA and/or PBG values). Each mutation will be classified as pathogenic, likely pathogenic, or benign, based on clinical and biochemical evidence (see Chen et al., ‘Collaborative’ in this issue). This information will be available in a public database.
Of particular importance, clinical manifestations for the three autosomal dominant AHPs do not occur in most mutation carriers. These asymptomatic individuals are termed ‘latent heterozygotes,’ and although they have never had an acute attack, they should be counseled to avoid the precipitating factors that can trigger an attack, including the use of porphyrinogenic drugs, dieting or fasting, and certain hormonal changes resulting from the use of high progesterone birth control medications. Previous studies have estimated that the penetrance of symptomatic AIP acute attacks ranged from <10% to ~30% (59-61). More recently, using samples from the blood bank or large population-based exome/genome databases, the prevalence of HMBS mutations was estimated to be 1 in ~1700 Caucasians (17, 18). In contrast, the prevalence of symptomatic AIP patients, based on data from porphyria specialist centers in Europe, was estimated to be 1 in ~200,000 Caucasians (17, 18). Together, these studies indicate that the penetrance of AIP is estimated to be only ~1% of HMBS heterozygotes (17, 18). The fact that the penetrance of clinical manifestations is so low among unrelated AHP heterozygotes, but much higher (~23%) within AHP families who have at least one member with recurrent attacks, strongly suggests that there may be important modifying genes and/or other environmental factors that trigger the severe acute attacks (18). To date, little serious effort has been undertaken to systematically search for such modifying genes and other factors.
3.3. Homozygous Dominant (HD)-AHPs
Rare cases of patients with biallelic mutations in their HMBS, PPOX, and CPOX genes, causing HD-AIP, HD-VP, and HD-HCP, respectively, have been reported (19-25, 27, 62). These patients have profound deficiency of their respective AHP enzyme, resulting in severe disease with marked neurological and/or cutaneous manifestations that begin in infancy or early childhood with growth retardation and short stature. Interestingly, with the exception of one HD-HCP patient (24), acute neurovisceral attacks have not been reported in HD-AHP patients.
3.3.1. HD-AIP
To date, five HD-AIP patients have been reported (19-22, 63). These patients had low levels of residual HMBS activity, typically <4% of normal (64), and constitutively elevated urinary ALA and PBG. In addition to gross psychomotor retardation, these patients have early-onset ataxia, nystagmus, and dystonia. The neurological impairment is progressive and patients typically die in childhood. Interestingly, of the five HD-AIP patients reported to date, four had mutations encoding p.R167Q, p.R167W, or p.R173Q. One was homozygous for p.R167W (19), two siblings were heterozygous for p.R167Q and p.R167W (21), while one was a compound heterozygote for p.R167W and p.R173Q (20).
3.3.2. HD-VP
HD-VP patients who have biallelic PPOX mutations have been reported to have between 5 and 20% of normal PPOX activity (27). They present in infancy with psychomotor delay, nystagmus, severe cutaneous photosensitivity and photomutilation, and skeletal deformities of the hand. Of the 11 HD-VP patients reported to date, eight have had both of their PPOX mutations identified (28, 65). Most were compound heterozygotes, while two unrelated HD-VP patients, both with consanguineous parents, were homozygous for the PPOX mutations encoding p.D349A and p.A433P (65). Of interest, although the p.R59W mutation is frequent in South Africa, no homozygotes have been identified to date, presumably because this mutation is severe and nearly abolishes PPOX activity, and therefore, is not compatible with life (58).
3.3.3. HD-HCP
Only two biochemically-confirmed HD-HCP patients have been reported to date, both young females with <10% of normal CPOX activity and marked elevation of coproporphyrins, with predominance of the III isomer (23, 24). One presented with CEP-like symptoms, including skin fragility and erythrodontia (23), while the other had skin pigmentation, hypertrichosis and episodic acute neurovisceral attacks with markedly elevated urinary ALA and PBG during the attacks (24). A clinically distinct subtype of HD-HCP, Harderoporphyria, has been described in four families to date (25, 62, 66, 67). Patients with Harderoporphyria present with neonatal jaundice and hemolytic anemia and accumulate harderoporphyrin in their feces, an intermediate metabolite in the conversion of coproporphyrin to PPIX. Of interest, HD-HCP patients identified to date were either homozygous for the CPOX mutation encoding p.K404E or carried this mutation in trans- with another pathogenic CPOX mutation (62, 66, 67). Only one patient was homozygous for the CPOX mutation encoding p.H327R (25). Mutagenesis studies have shown that CPOX amino acids residues Y399 to K405 are directly involved in the enzyme’s oxidative decarboxylation of harderoporphyrinogen (62), while molecular modeling studies based on the CPOX crystal structure support that the p.H327R change also interferes with this enzymatic step (68).
4. Hepatic Cutaneous Porphyria: Porphyria Cutanea Tarda (PCT)
4.1. Clinical Manifestations of PCT
PCT, the most common human porphyria, is classified as a hepatic cutaneous porphyria since it presents with cutaneous bullous lesions, but the primary site of porphyrin accumulation is the liver. Clinically, PCT is characterized by the development of blisters on sun-exposed skin, similar to those of CEP. The disease becomes active when patients are exposed to predisposing factors that cause hepatic iron overload, including excess alcohol consumption, estrogen use, infections (HCV, HIV, etc.), and smoking (1, 2, 69). It has been postulated that the hepatic activity of UROD is markedly reduced during active disease due to the formation of uroporphomethene, an iron-oxidized product of uroporphyrinogen, which acts as a reversible inhibitor of UROD activity (14-16). This leads to marked accumulation of porphyrins, predominantly consisting of uroporphyrin and 7-carboxylate porphyrin in the liver and urine.
4.2. Genetics of PCT
PCT occurs both in a Type 1 sporadic subtype, in the absence of a UROD mutation, and a Type 2 familial subtype, in which a pathogenic UROD mutation is inherited in an autosomal dominant pattern (Table 3). To date, over 120 UROD mutations have been reported to cause PCT in HGMD version 2018.2 (see Table 4 for mutation types). Of clinically and biochemically documented cases of PCT, about 75-80% have the Type I sporadic subtype with no UROD mutation, while the remaining 20-25% have the Type 2 familial form with a pathogenic UROD mutation (see Weiss et al., this volume). Type 1 sporadic PCT patients have normal hepatic UROD activity except during active disease when the UROD inhibitor is formed, while Type 2 familial PCT patients had half-normal UROD activity systemically (70). Clinically, the two forms are indistinguishable. While the half-normal activity of UROD predisposes to PCT, additional susceptibility factors are required to activate disease in Type 2 familia l patients, like the Type 1 sporadic patients.
As iron overload is a major predisposing factor, patients who have hemochromatosis (HFE) gene mutations are at higher risk for developing PCT. In fact, >50% of PCT patients with active disease are reported to carry HFE mutations, most commonly the mutation encoding p.C282Y or p.H63D (69).
4.3. Hepatoerythropoietic Porphyria
Biallelic UROD LOF mutations cause Hepatoerythropoietic Porphyria (HEP), a rare condition that presents with CEP-like manifestations, including hemolytic anemia, severe skin blistering that begins in infancy, discolored teeth (erythrodontia), and reddish-colored urine (1). Patients typically have 15-20% of normal UROD activity.
4.4. Dual Porphyrias
To date, >15 patients have been identified to have clinical and biochemical features of two major porphyrias (71-73). The majority of these patients have combined deficiency of UROD and HMBS, CPOX, or PPOX, consistent with the fact that PCT is the most common porphyria. Other combinations, including dual deficiencies of CPOX and HMBS; UROS and CPOX; CPOX and ALAD; UROS and UROD; CPOX and PPOX, have been reported. However, it should be noted that only a few cases have been confirmed by genetic testing.
5. Erythropoietic Cutaneous Porphyrias
The erythropoietic porphyrias include three disorders, CEP, EPP, and XLP (Table 3). These disorders are characterized by cutaneous photosensitivity that results from the massive accumulation of photoreactive porphyrins in the bone marrow erythroid cells and circulating erythrocytes, which when released during erythrocyte senescence gain access to blood vessel endothelial cells and other organs, particularly the liver in EPP and XLP, as PPIX is excreted via the hepatobiliary system.
5.1. CEP
5.1.1. Clinical Manifestations of CEP
CEP is a rare disorder, with less than 250 cases reported to date. Clinically, CEP is characterized by marked cutaneous photosensitivity, with blistering and formation of vesicles on sun-exposed skin. Recurrent vesicles and secondary infection can lead to cutaneous scarring as well as skin and bone loss that is disfiguring. Other prominent symptoms include red urine since birth, reddish-brownish discoloration of the teeth (erythrodontia) and hemolytic anemia. Clinical severity can vary, from non-immune hydrops fetalis in utero to transfusion-dependent severe disease, or later-onset disease with only moderate or mild cutaneous photosensitivity in adulthood. Biochemically, CEP patients display markedly elevated erythrocyte and urinary uroporphyrin I and coproporphyrin I isomers, which are nonphysiologic and phototoxic porphyrins that accumulate, as they are not further metabolized to heme.
5.1.2. Genetics of CEP:
CEP is an autosomal recessive disorder due to LOF mutations in the UROS gene (Table 3). To date, > 50 UROS mutations have been reported in HGMD, of which ~60% are missense (Table 4). The most common missense mutation encoding p.C73R occurs in ~35% of genotyped CEP patients, which can result in non-immune hydrops fetalis or newborns with severe anemia, particularly when homozygous (31). Among the mutations causing CEP, there are four verified regulatory mutation in the promoter (39). These mutations, located within a 20 bp region of the promoter (−90 to −70 from the initiation ATG), involve transcription factor binding sites for GATA1 and CP2 (Fig. 4). They have been expressed in luciferase constructs to demonstrate their pathogenicity (39). Genotype/phenotype correlations predict disease severity, as the activity of the encoded UROS enzyme deficiency correlates with clinical severity. For example, the mutation encoding p.C73R has <0.1% of wild-type UROS activity when prokaryotically expressed, while L4F, which causes transfusion-independent later-onset CEP, has >3% of wild-type UROS activity [Table 6; (74)]. These genotype/phenotype correlations are particularly useful to predict transfusion dependency and the need for early hematopoietic stem cell transplantation, when feasible.
Table 6.
Genotype/Phenotype Correlations in CEP
| Phenotype | Genotype | Residual Activity Expressed in E.coli Alleles/Total |
|
|---|---|---|---|
| Hydrops Fetalis/ Newborn Demise: | |||
| C73R/C73R | <0.1 / <0.1 | = <0.1 | |
| Transfusion Dependent: | |||
| Severe | C73R/T228M | <0.1 / <0.1 | = <0.1 |
| Transfusion Independent: | |||
| Moderate | T62A/E249X | <0.1 / 1.8 | = 1.8 |
| G225S/T228L | 2.1 / <0.1 | = 2.1 | |
| L4F/C73R | 2.9 / <0.1 | = 2.9 | |
| L4F/Deletion | 2.9 / 0 | = 2.9 | |
| L4F/IVS2+1 | 2.9 / ? | = 2.9 | |
| Mild | Y19C/G225S | 2.7 / 2.1 | = 4.8 |
| V99A/Ins211A | 3.7 / 1.7 | = 5.4 | |
| C73R/A104V | <1.0 / 5.6 | = 5.6 | |
| A66V/C73R | 14.5 / <0.1 | = 14.5 | |
| L4F/V82F | 2.9 / 24.2 | = 27.1 | |
Desnick, RJ & Astrin, KH: Br. J. Haematol. 117:779–795, 2002
To date, three cases of the CEP phenotype have been reported to be caused by a mutation encoding p.R216W in the X-linked GATA1 gene (46, 47). GATA1, a transcriptional factor that is critical for normal erythropoiesis and megakaryocyte development, regulates the expression of various erythroid-specific genes, including UROS, ALAS2, and the α- and β- globins. All three unrelated CEP male patients were hemizygous for the p.R216W GATA1 mutation that is located in the N-terminal zinc finger domain, which mediates the interaction with cofactors to stabilize the binding of the GATA1 transcription factor to promoter binding sites of its target genes. Two of the patients did not have UROS mutations, while one was heterozygous for the UROS mutation encoding p.D113V (46). Clinically, these patients had a photosensitive bullous dermatosis, scarring of the skin, microcytic anemia, and markedly increased levels of uroporphyrin I, similar to CEP patients with UROS mutations. However, they had additional hematological abnormalities, including increased fetal hemoglobin and thrombocytopenia, which were unique to the GATA1 mutation-positive patients (46, 47).
Additionally, a non-inherited mild form of CEP secondary to myeloid malignancy, most commonly myelodysplastic syndrome (MDS), has been reported in a small number of patients (75, 76). Sarkany et al. demonstrated in four of these patients that germline UROS and GATA1 mutations were not detectable, erythrocyte UROS activities were normal, and that only a small fraction of their circulating erythrocytes were uroporphyric (75). As MDS causes genomic instability, it is highly probable that a minor clone of erythropoietic cells contained an acquired somatic UROS mutation that was not detected due to the small number of mutation-positive cells relative to normal erythroid cells.
5.2. Erythropoietic Portoporphyria
5.2.1. Clinical Manifestations of EPP
EPP, the most common childhood porphyria, is primarily characterized by extremely painful photosensitivity that is accompanied by marked elevation of free PPIX in plasma and erythrocytes. A mild microcytic anemia occurs in ~20 to 30% of patients. ~20% of patients experience cholelithiasis, while ~5% of patients develop liver failure due to hepatobiliary involvement, requiring liver transplantation. Since the PPIX released from erythroid cells is lipophilic, it accumulates in the vascular endothelial cells and liver and is excreted via the hepatobiliary system. The accumulation of PPIX in the liver and biliary tract leads to cholestasis and hepatobiliary fibrosis, and eventually, liver failure.
5.2.2. Genetics of EPP
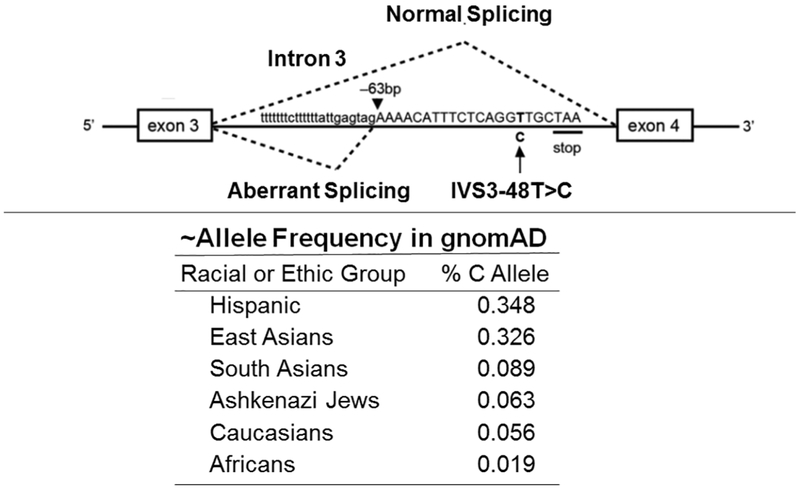
EPP is an autosomal recessive disorder due to mutations in the ferrochelatase (FECH) gene (Table 3). Its prevalence is estimated at 1 in ~50,000 Caucasians (61). To date, >190 LOF FECH mutations have been reported, including ~7% that are large deletions, which are sequencing cryptic and require gene dosage analysis [Table 4; (77)]. Notably, ~95% of EPP patients have a pathogenic LOF FECH mutation on one chromosome and the common ‘low expression allele’, IVS3-48A>G, on the other (32-34). As shown in Figure 5, the IVS3-48T>C change leads to the increased use of an alternative 3’-splice site that is 63 nucleotides upstream of the normal site, leading to an aberrant transcript with 63 additional nucleotides. Two in-frame stop codons in this additional sequence target this aberrant transcript for nonsense-mediated decay. As a result, the ‘low expression allele’ has ~30% of normal FECH activity (34). The allele frequency of the ‘low expression allele’ ranges from ~2% in Africans to 6% in Caucasians and 30-35% in East Asians and Hispanics, based on GenomAD data. Only a small number of patients with EPP have two LOF FECH mutations, which leads to a more severe form of the disease and seasonal palmer keratoderma. The hyperkeratosis ranged in severity from a waxy keratoderma over the whole palm to mild hyperkeratosis of the first interdigital web (78). To date, over 20 patients have been reported in 16 unrelated EPP families. In these patients, ~85% of the mutations were missense, and most patients were compound heterozygotes (78).
Fig 5.
Alternative splicing of the FECH IVS-48T/C intronic sequence. The presence of the IVS-48T/C variant modulates the splicing efficiency of a constitutive cryptic acceptor splice-site. Note that the IVS-48C variant alters splicing such that 63 intronic base pairs are included in the mutant allele resulting in an abnormal transcript and decreased expression (35% of normal) for this allele. The minor allele frequency for the IVS-48C mutation in various racial and ethic groups from gnomAD is shown. Gouya et al., Nature Genet. 30:27-28, 2002.
Recently, a dominant mutation in the CLPX gene was reported that caused an EPP-like phenotype (48). CLPX is a mitochondria AAA+ (or ATPase associated with a variety of cellular activities) unfoldase that promotes heme biosynthesis by activation of ALAS 1 and 2. Importantly, CLPX associates with CLPP to form CLPXP, an ATP-dependent serine protease that mediates the heme-induced turnover of ALAS 1 and 2 (79, 80). Within the CLPXP complex, CLPX binds substrate, unfolds stable tertiary structure in the substrate, and then traslocates the unfolded polypeptide chain into the proteolytic compartment of CLPP. A dominant CLPX mutation encoding p.G98D in the active site was shown to reduce its ATPase activity and to weaken its interaction with CLPP, resulting in reduced proteasome activity. This leads to increases the post-translational stability of ALAS2, leading to the abnormal accumulation of PPIX and clinical photosensitivity (48).
A non-congenital late-onset form of EPP also may develop as a rare complication of MDS or a myeloproliferative disorder (MPD) (81-83). One patient with MPD had clonal expansion of hematopoietic cells with an acquired somatic mutation that resulted in the complete deletion of one FECH allele. This patient had the IVS-48T>C ‘low expression allele’ on the remaining FECH allele, resulting in the EPP phenotype (82).
5.3. XLP
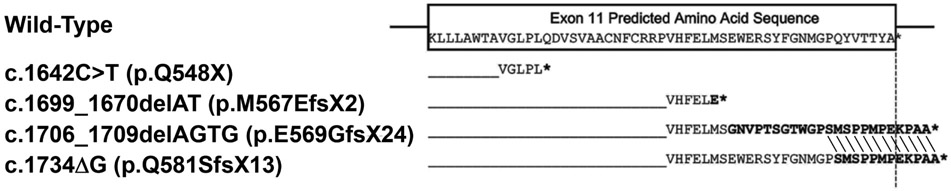
XLP is clinically indistinguishable from EPP, although biochemically, XLP patients accumulate both Zn-PPIX and free protoporphyrin, whereas EPP patients predominantly accumulate free-PPIX. As XLP is an X-linked disorder, most symptomatic patients are males. Females can be asymptomatic or as severely affected as their male relatives, due to random X-chromosomal inactivation (see Genetic Principles Section). Unlike the other porphyrias, XLP is due to GOF mutations in the erythroid-specific ALAS2 gene [Table 3; (13)]. To date, six XLP mutations have been reported, all of which reside in the terminal exon 11 of ALAS2 (13, 32, 84). These mutations are nonsense or frameshift lesions that prematurely truncate or extend the carboxy-terminal residues (Fig. 6). The truncations and extensions lead to structural alterations of the carboxyl-terminus, which normally shutters the active site, to abnormally remain open (4, 84). This results in increased ALAS2 activity [2 to 3–fold in in vitro; (84)], and consequentially, increased production and accumulation of PPIX.
Fig 6.
Variations in the carboxy-terminal mutant sequences of XLP mutations. The partial wild-type exon 11 ALAS2 sequence is boxed. The hybrid sequences of wild-type ALAS2 and the sequence of several GOD mutations are aligned below the wild-type sequence. The termination codons are denoted by asterisks. Note that for the pE569GfsX24 and pQ581SfsX13 mutations, the last 12 mutated residues bolded are identical and different from the wild-type sequence. Balwani et al., Molec. Med. 19:26-35, 2013.
As noted above, s GOF mutation in the ALAS2 gene has been shown to increase the severity of other porphyrias, including CEP. In four unrelated CEP patients with UROS mutations encoding p.C73R/p.P248Q, one had moderately severe disease and a novel ALAS2 exon 11 gain-of-function mutation (c.1757A>T), while the other three lacking ALAS2 mutations had milder disease (49).
6. Summary
The porphyrias, as a group of eight metabolic disorders, illustrate many genetic principles of Mendelian disorders. There are multiple modes of inheritance in the eight disorders: three AHPs are inherited as autosomal dominant traits with markedly reduced penetrance, while most erythropoietic cutaneous porphyrias are inherited as fully penetrant autosomal recessive or X-linked traits, the latter in XLP males. Although about 20% of patients with PCT have a LOF mutation in their UROD gene, most patients have the sporadic or acquired form secondary to factors that induce hepatic iron overload and the formation of the UROD inhibitor, uroporphomethene. Together, the eight porphyrias result from a total of >1000 mutations in the eight heme biosynthetic genes. Multiple mutations of various types occur in each disease, giving rise to the molecular genetic heterogeneity and variable severity in a given porphyria due to the level of the mutation’s residual activity in vivo. While most of these mutations are LOF mutations that result in decreased enzymatic activity, XLP is caused by GOF mutations that alter the carboxyl-terminal structure of the ALAS2 protein resulting in a more catalytically active enzyme. The three autosomal dominant AHPs have very low penetrance, suggesting that modifying genes (predisposing and/or protective) and/or environmental factors are responsible for the penetrance as well as the onset and severity of these disorders. While genetic sequencing of the heme biosynthetic genes has identified the disease-causing mutations in almost all porphyria patients, the nature of the primary genetic defects for a few patients with AHPs and erythopoietic porphyrias remain to be determined, perhaps by whole genome sequencing. Such studies identified the roles of the ALAS2 and CLPX genes. Future studies of the porphyrias may identify other heme transport and/or degradation genes that may provide additional understanding of the pathophysiology of the porphyrias and identify new targets for their therapy.
Acknowledgements
This work was supported in part by the Department of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, and by the NIH-supported Porphyrias Consortium (U54 DK0839), which is a part of the NCATS Rare Diseases Clinical Research Network (RDCRN) of the National Institutes of Health. RDCRN is an initiative of the Office of Rare Diseases Research (ORDR), NCATS, funded through collaboration between NCATS and the NIDDK.
Footnotes
Conflicts of Interest
MY and RJD are past recipients of research grants from Alnylam Pharmaceuticals and Recordati Rare Diseases. They are co-inventors of a patent licensed to Alnylam Pharmaceuticals for RNAi therapy of the AHPs. RJD is a consultant for Alnylam Pharmaceuticals, Mitsubishi Tanabe Pharma Development America and Recordati Rare Disease. BC has no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Makiko Yasuda, Email: makiko.yasuda@mssm.edu.
Brenden Chen, Email: Brenden.chen@mssm.edu.
Robert J. Desnick, Email: Robert.Desnick@mssm.edu.
References
- 1.Puy H, Gouya L, Deybach JC. Porphyrias. Lancet 2010;375:924–937. [DOI] [PubMed] [Google Scholar]
- 2.Anderson KE, Sassa S, Bishop DF, Desnick RJ: Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias In: Scriver C, Beaudet A, Sly W, Valle D, eds. The Metabolib and Molecular Bases of Inherited Disease. New York: McGraw-Hill, 2001; 2961–3062. [Google Scholar]
- 3.Bung N, Roy A, Chen B, Das D, Pradhan M, Yasuda M, New MI, et al. Human hydroxymethylbilane synthase: Molecular dynamics of the pyrrole chain elongation identifies step-specific residues that cause AIP. Proc Natl Acad Sci U S A 2018;115:E4071–E4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown BL, Kardon JR, Sauer RT, Baker TA. Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme. Structure 2018;26:580–589 e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitby FG, Phillips JD, Kushner JP, Hill CP. Crystal structure of human uroporphyrinogen decarboxylase. EMBO J 1998;17:2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills-Davies N, Butler D, Norton E, Thompson D, Sarwar M, Guo J, Gill R, et al. Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis. Acta Crystallogr D Struct Biol 2017;73:9–21. [DOI] [PubMed] [Google Scholar]
- 7.Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, et al. Crystal structure of human uroporphyrinogen III synthase. EMBO J 2001;20:5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J 2003;22:6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J Biol Chem 2006;281:38625–38633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure 1997;5:1501–1510. [DOI] [PubMed] [Google Scholar]
- 11.Smith JR, Osborne TF, Brown MS, Goldstein JL, Gil G. Multiple sterol regulatory elements in promoter for hamster 3-hydroxy-3-methylglutaryl-coenzyme A synthase. J Biol Chem 1988;263:18480–18487. [PubMed] [Google Scholar]
- 12.Chiabrando D, Mercurio S, Tolosano E. Heme and erythropoieis: more than a structural role. Haematologica 2014;99:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, Badminton MN, Elder GH, et al. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet 2008;83:408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips JD, Bergonia HA, Reilly CA, Franklin MR, Kushner JP. A porphomethene inhibitor of uroporphyrinogen decarboxylase causes porphyria cutanea tarda. Proc Natl Acad Sci U S A 2007;104:5079–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benton CM, Lim CK, Moniz C, Jones DJ. Liquid chromatography-tandem mass spectrometry of porphyrins and porphyrinogens in biological materials: separation and identification of interfering poly(ethylene) glycol by travelling wave ion mobility spectrometry/tandem mass spectrometry. Biomed Chromatogr 2013;27:1782–1787. [DOI] [PubMed] [Google Scholar]
- 16.Danton M, Lim CK. Porphomethene inhibitor of uroporphyrinogen decarboxylase: analysis by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Biomed Chromatogr 2007;21:661–663. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Solis-Villa C, Hakenberg J, Qiao W, Srinivasan RR, Yasuda M, Balwani M, et al. Acute Intermittent Porphyria: Predicted Pathogenicity of HMBS Variants Indicates Extremely Low Penetrance of the Autosomal Dominant Disease. Hum Mutat 2016;37:1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenglet H, Schmitt C, Grange T, Manceau H, Karboul N, Bouchet-Crivat F, Robreau AM, et al. From a dominant to an oligogenic model of inheritance with environmental modifiers in acute intermittent porphyria. Hum Mol Genet 2018;27:1164–1173. [DOI] [PubMed] [Google Scholar]
- 19.Solis C, Martinez-Bermejo A, Naidich TP, Kaufmann WE, Astrin KH, Bishop DF, Desnick RJ. Acute intermittent porphyria: studies of the severe homozygous dominant disease provides insights into the neurologic attacks in acute porphyrias. Arch Neurol 2004;61:1764–1770. [DOI] [PubMed] [Google Scholar]
- 20.Beukeveld GJ, Wolthers BG, Nordmann Y, Deybach JC, Grandchamp B, Wadman SK. A retrospective study of a patient with homozygous form of acute intermittent porphyria. J Inherit Metab Dis 1990;13:673–683. [DOI] [PubMed] [Google Scholar]
- 21.Llewellyn DH, Smyth SJ, Elder GH, Hutchesson AC, Rattenbury JM, Smith MF. Homozygous acute intermittent porphyria: compound heterozygosity for adjacent base transitions in the same codon of the porphobilinogen deaminase gene. Hum Genet 1992;89:97–98 [DOI] [PubMed] [Google Scholar]
- 22.Picat C, Delfau MH, de Rooij FW, Beukeveld GJ, Wolthers BG, Wadman SK, Nordmann Y, et al. Identification of the mutations in the parents of a patient with a putative compound heterozygosity for acute intermittent porphyria. J Inherit Metab Dis 1990;13:684–686. [DOI] [PubMed] [Google Scholar]
- 23.Doss MO, Gross U, Lamoril J, Kranl C, Jacob K, Doss M, da Silva V, et al. Compound heterozygous hereditary coproporphyria with fluorescing teeth. Ann Clin Biochem 1999;36 ( Pt 5):680–682. [DOI] [PubMed] [Google Scholar]
- 24.Grandchamp B, Phung N, Nordmann Y. Homozygous case of hereditary coproporphyria. Lancet 1977;2:1348–1349. [DOI] [PubMed] [Google Scholar]
- 25.Hasanoglu A, Balwani M, Kasapkara CS, Ezgu FS, Okur I, Tumer L, Cakmak A, et al. Harderoporphyria due to homozygosity for coproporphyrinogen oxidase missense mutation H327R. J Inherit Metab Dis 2011;34:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank J, McGrath J, Lam H, Graham RM, Hawk JL, Christiano AM. Homozygous variegate porphyria: identification of mutations on both alleles of the protoporphyrinogen oxidase gene in a severely affected proband. J Invest Dermatol 1998;110:452–455. [DOI] [PubMed] [Google Scholar]
- 27.Hift RJ, Meissner PN, Todd G, Kirby P, Bilsland D, Collins P, Ferguson J, et al. Homozygous variegate porphyria: an evolving clinical syndrome. Postgrad Med J 1993;69:781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauppinen R, Timonen K, von und zu Fraunberg M, Laitinen E, Ahola H, Tenhunen R, Taketani S, et al. Homozygous variegate porphyria: 20 y follow-up and characterization of molecular defect. J Invest Dermatol 2001;116:610–613. [DOI] [PubMed] [Google Scholar]
- 29.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet 1988;78:151–155. [DOI] [PubMed] [Google Scholar]
- 30.Floderus Y, Shoolingin-Jordan PM, Harper P. Acute intermittent porphyria in Sweden. Molecular, functional and clinical consequences of some new mutations found in the porphobilinogen deaminase gene. Clin Genet 2002;62:288–297. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, Wang X, Lam HM, Aita VM, Jugert FK, Goerz G, Merk HF, et al. C73R is a hotspot mutation in the uroporphyrinogen III synthase gene in congenital erythropoietic porphyria. Ann Hum Genet 1998;62:225–230. [DOI] [PubMed] [Google Scholar]
- 32.Balwani M, Doheny D, Bishop DF, Nazarenko I, Yasuda M, Dailey HA, Anderson KE, et al. Loss-of-function ferrochelatase and gain-of-function erythroid-specific 5-aminolevulinate synthase mutations causing erythropoietic protoporphyria and x-linked protoporphyria in North American patients reveal novel mutations and a high prevalence of X-linked protoporphyria. Mol Med 2013;19:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balwani M, Naik H, Anderson KE, Bissell DM, Bloomer J, Bonkovsky HL, Phillips JD, et al. Clinical, Biochemical, and Genetic Characterization of North American Patients With Erythropoietic Protoporphyria and X-linked Protoporphyria. JAMA Dermatol 2017;153:789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouya L, Deybach JC, Lamoril J, Da Silva V, Beaumont C, Grandchamp B, Nordmann Y. Modulation of the phenotype in dominant erythropoietic protoporphyria by a low expression of the normal ferrochelatase allele. Am J Hum Genet 1996;58:292–299. [PMC free article] [PubMed] [Google Scholar]
- 35.Gouya L, Puy H, Robreau AM, Lyoumi S, Lamoril J, Da Silva V, Grandchamp B, et al. Modulation of penetrance by the wild-type allele in dominantly inherited erythropoietic protoporphyria and acute hepatic porphyrias. Hum Genet 2004;114:256–262. [DOI] [PubMed] [Google Scholar]
- 36.Chen CH, Astrin KH, Lee G, Anderson KE, Desnick RJ. Acute intermittent porphyria: identification and expression of exonic mutations in the hydroxymethylbilane synthase gene. An initiation codon missense mutation in the housekeeping transcript causes "variant acute intermittent porphyria" with normal expression of the erythroid-specific enzyme. J Clin Invest 1994;94:1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grandchamp B, Picat C, Kauppinen R, Mignotte V, Peltonen L, Mustajoki P, Romeo PH, et al. Molecular analysis of acute intermittent porphyria in a Finnish family with normal erythrocyte porphobilinogen deaminase. Eur J Clin Invest 1989;19:415–418. [DOI] [PubMed] [Google Scholar]
- 38.Brancaleoni V, Granata F, Colancecco A, Tavazzi D, Cappellini MD, Di Pierro E. Seven novel genetic mutations within the 5'UTR and the housekeeping promoter of HMBS gene responsible for the non-erythroid form of acute intermittent porphyria. Blood Cells Mol Dis 2012;49:147–151. [DOI] [PubMed] [Google Scholar]
- 39.Solis C, Aizencang GI, Astrin KH, Bishop DF, Desnick RJ. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J Clin Invest 2001;107:753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiorentino V, Brancaleoni V, Granata F, Graziadei G, Di Pierro E. The assessment of noncoding variant of PPOX gene in variegate porphyria reveals post-transcriptional role of the 5' untranslated exon 1. Blood Cells Mol Dis 2016;61:48–53. [DOI] [PubMed] [Google Scholar]
- 41.Onaga Y, Ido A, Uto H, Hasuike S, Kusumoto K, Moriuchi A, Numata M, et al. Hypermethylation of the wild-type ferrochelatase allele is closely associated with severe liver complication in a family with erythropoietic protoporphyria. Biochem Biophys Res Commun 2004;321:851–858. [DOI] [PubMed] [Google Scholar]
- 42.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 2017;136:665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puy H, Deybach JC, Lamoril J, Robreau AM, Da Silva V, Gouya L, Grandchamp B, et al. Molecular epidemiology and diagnosis of PBG deaminase gene defects in acute intermittent porphyria. Am J Hum Genet 1997;60:1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robreau-Fraolini AM, Puy H, Aquaron C, Bogard C, Traore M, Nordmann Y, Aquaron R, et al. Porphobilinogen deaminase gene in African and Afro-Caribbean ethnic groups: mutations causing acute intermittent porphyria and specific intragenic polymorphisms. Hum Genet 2000;107:150–159. [DOI] [PubMed] [Google Scholar]
- 45.Brancaleoni V, Balwani M, Granata F, Graziadei G, Missineo P, Fiorentino V, Fustinoni S, et al. X-chromosomal inactivation directly influences the phenotypic manifestation of X-linked protoporphyria. Clin Genet 2016;89:20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Pierro E, Russo R, Karakas Z, Brancaleoni V, Gambale A, Kurt I, Winter SS, et al. Congenital erythropoietic porphyria linked to GATA1-R216W mutation: challenges for diagnosis. Eur J Haematol 2015;94:491–497. [DOI] [PubMed] [Google Scholar]
- 47.Phillips JD, Steensma DP, Pulsipher MA, Spangrude GJ, Kushner JP. Congenital erythropoietic porphyria due to a mutation in GATA1: the first trans-acting mutation causative for a human porphyria. Blood 2007;109:2618–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yien YY, Ducamp S, van der Vorm LN, Kardon JR, Manceau H, Kannengiesser C, Bergonia HA, et al. Mutation in human CLPX elevates levels of delta-aminolevulinate synthase and protoporphyrin IX to promote erythropoietic protoporphyria. Proc Natl Acad Sci U S A 2017;114:E8045–E8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.To-Figueras J, Ducamp S, Clayton J, Badenas C, Delaby C, Ged C, Lyoumi S, et al. ALAS2 acts as a modifier gene in patients with congenital erythropoietic porphyria. Blood 2011;118:1443–1451. [DOI] [PubMed] [Google Scholar]
- 50.Hu Y, Shen H, Keep RF, Smith DE. Peptide transporter 2 (PEPT2) expression in brain protects against 5-aminolevulinic acid neurotoxicity. J Neurochem 2007;103:2058–2065. [DOI] [PubMed] [Google Scholar]
- 51.Tchernitchko D, Tavernier Q, Lamoril J, Schmitt C, Talbi N, Lyoumi S, Robreau AM, et al. A Variant of Peptide Transporter 2 Predicts the Severity of Porphyria-Associated Kidney Disease. J Am Soc Nephrol 2017;28:1924–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruno M, Furuyama K, Akagi R, Horie Y, Meguro K, Garbaczewski L, Chiorazzi N, et al. Highly heterogeneous nature of delta-aminolevulinate dehydratase (ALAD) deficiencies in ALAD porphyria. Blood 2001;97:2972–2978. [DOI] [PubMed] [Google Scholar]
- 53.Tjensvoll K, Bruland O, Floderus Y, Skadberg O, Sandberg S, Apold J. Haplotype analysis of Norwegian and Swedish patients with acute intermittent porphyria (AIP): Extreme haplotype heterogeneity for the mutation R116W. Dis Markers 2003;19:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Rooij FW, Kavelaars FG, Koole-Lesuis H, Wilson JH. Evidence for an ancestral founder of the common R116W mutation in the hydroxymethylbilane synthase gene in acute intermittent porphyria in The Netherlands. Cell Mol Biol (Noisy-le-grand) 2009;55:64–69. [PubMed] [Google Scholar]
- 55.Schneider-Yin X, Hergersberg M, Goldgar DE, Rufenacht UB, Schuurmans MM, Puy H, Deybach JC, et al. Ancestral founder of mutation W283X in the porphobilinogen deaminase gene among acute intermittent porphyria patients. Hum Hered 2002;54:69–81. [DOI] [PubMed] [Google Scholar]
- 56.De Siervi A, Rossetti MV, Parera VE, Astrin KH, Aizencang GI, Glass IA, Batlle AM, et al. Identification and characterization of hydroxymethylbilane synthase mutations causing acute intermittent porphyria: evidence for an ancestral founder of the common G111R mutation. Am J Med Genet 1999;86:366–375. [PubMed] [Google Scholar]
- 57.Guillen-Navarro E, Carbonell P, Glover G, Sanchez-Solis M, Fernandez-Barreiro A. Novel HMBS founder mutation and significant intronic polymorphism in Spanish patients with acute intermittent porphyria. Ann Hum Genet 2004;68:509–514. [DOI] [PubMed] [Google Scholar]
- 58.Meissner PN, Dailey TA, Hift RJ, Ziman M, Corrigall AV, Roberts AG, Meissner DM, et al. A R59W mutation in human protoporphyrinogen oxidase results in decreased enzyme activity and is prevalent in South Africans with variegate porphyria. Nat Genet 1996;13:95–97. [DOI] [PubMed] [Google Scholar]
- 59.Kauppinen R, Mustajoki P. Prognosis of acute porphyria: occurrence of acute attacks, precipitating factors, and associated diseases. Medicine (Baltimore) 1992;71:1–13. [PubMed] [Google Scholar]
- 60.Mykletun M, Aarsand AK, Stole E, Villanger JH, Tollanes MC, Baravelli C, Sandberg S. Porphyrias in Norway. Tidsskr Nor Laegeforen 2014;134:831–836. [DOI] [PubMed] [Google Scholar]
- 61.Elder G, Harper P, Badminton M, Sandberg S, Deybach JC. The incidence of inherited porphyrias in Europe. J Inherit Metab Dis 2013;36:849–857. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt C, Gouya L, Malonova E, Lamoril J, Camadro JM, Flamme M, Rose C, et al. Mutations in human CPO gene predict clinical expression of either hepatic hereditary coproporphyria or erythropoietic harderoporphyria. Hum Mol Genet 2005;14:3089–3098. [DOI] [PubMed] [Google Scholar]
- 63.Hessels J, Voortman G, van der Wagen A, van der Elzen C, Scheffer H, Zuijderhoudt FM. Homozygous acute intermittent porphyria in a 7-year-old boy with massive excretions of porphyrins and porphyrin precursors. J Inherit Metab Dis 2004;27:19–27. [DOI] [PubMed] [Google Scholar]
- 64.Edixhoven-Bosdijk A, de Rooij FW, de Baar-Heesakkers E, Wilson JH. Residual activity of human porphobilinogen deaminase with R167Q or R167W mutations: an explanation for survival of homozygous and compound heterozygous acute intermittent porphyrics. Cell Mol Biol (Noisy-le-grand) 2002;48:861–866. [PubMed] [Google Scholar]
- 65.Roberts AG, Puy H, Dailey TA, Morgan RR, Whatley SD, Dailey HA, Martasek P, et al. Molecular characterization of homozygous variegate porphyria. Hum Mol Genet 1998;7:1921–1925. [DOI] [PubMed] [Google Scholar]
- 66.Lamoril J, Puy H, Whatley SD, Martin C, Woolf JR, Da Silva V, Deybach JC, et al. Characterization of mutations in the CPO gene in British patients demonstrates absence of genotype-phenotype correlation and identifies relationship between hereditary coproporphyria and harderoporphyria. Am J Hum Genet 2001;68:1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nordmann Y, Grandchamp B, de Verneuil H, Phung L, Cartigny B, Fontaine G. Harderoporphyria: a variant hereditary coproporphyria. J Clin Invest 1983;72:1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee DS, Flachsova E, Bodnarova M, Demeler B, Martasek P, Raman CS. Structural basis of hereditary coproporphyria. Proc Natl Acad Sci U S A 2005;102:14232–14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bissell DM, Anderson KE, Bonkovsky HL. Porphyria. N Engl J Med 2017;377:862–872. [DOI] [PubMed] [Google Scholar]
- 70.Kushner JP, Barbuto AJ, Lee GR. An inherited enzymatic defect in porphyria cutanea tarda: decreased uroporphyrinogen decarboxylase activity. J Clin Invest 1976;58:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akagi R, Inoue R, Muranaka S, Tahara T, Taketani S, Anderson KE, Phillips JD, et al. Dual gene defects involving delta-aminolaevulinate dehydratase and coproporphyrinogen oxidase in a porphyria patient. Br J Haematol 2006;132:237–243. [DOI] [PubMed] [Google Scholar]
- 72.Poblete-Gutierrez P, Badeloe S, Wiederholt T, Merk HF, Frank J. Dual porphyrias revisited. Exp Dermatol 2006;15:685–691. [DOI] [PubMed] [Google Scholar]
- 73.van Tuyll van Serooskerken AM, de Rooij FW, Edixhoven A, Bladergroen RS, Baron JM, Joussen S, Merk HF, et al. Digenic inheritance of mutations in the coproporphyrinogen oxidase and protoporphyrinogen oxidase genes in a unique type of porphyria. J Invest Dermatol 2011;131:2249–2254. [DOI] [PubMed] [Google Scholar]
- 74.Desnick RJ, Astrin KH. Congenital erythropoietic porphyria: advances in pathogenesis and treatment. Br J Haematol 2002;117:779–795. [DOI] [PubMed] [Google Scholar]
- 75.Sarkany RP, Ibbotson SH, Whatley SD, Lawrence CM, Gover P, Mufti GJ, Murphy GM, et al. Erythropoietic uroporphyria associated with myeloid malignancy is likely distinct from autosomal recessive congenital erythropoietic porphyria. J Invest Dermatol 2011;131:1172–1175. [DOI] [PubMed] [Google Scholar]
- 76.Podlipnik S, Guijarro F, Combalia A, To-Figueras J, Badenas C, Costa D, Rozman M, et al. Acquired erythropoietic uroporphyria secondary to myelodysplastic syndrome with chromosome 3 alterations: a case report. Br J Dermatol 2017. [DOI] [PubMed] [Google Scholar]
- 77.Whatley SD, Mason NG, Holme SA, Anstey AV, Elder GH, Badminton MN. Gene dosage analysis identifies large deletions of the FECH gene in 10% of families with erythropoietic protoporphyria. J Invest Dermatol 2007;127:2790–2794. [DOI] [PubMed] [Google Scholar]
- 78.Holme SA, Whatley SD, Roberts AG, Anstey AV, Elder GH, Ead RD, Stewart MF, et al. Seasonal palmar keratoderma in erythropoietic protoporphyria indicates autosomal recessive inheritance. J Invest Dermatol 2009;129:599–605. [DOI] [PubMed] [Google Scholar]
- 79.Kardon JR, Yien YY, Huston NC, Branco DS, Hildick-Smith GJ, Rhee KY, Paw BH, et al. Mitochondrial ClpX Activates a Key Enzyme for Heme Biosynthesis and Erythropoiesis. Cell 2015;161:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kubota Y, Nomura K, Katoh Y, Yamashita R, Kaneko K, Furuyama K. Novel Mechanisms for Heme-dependent Degradation of ALAS1 Protein as a Component of Negative Feedback Regulation of Heme Biosynthesis. J Biol Chem 2016;291:20516–20529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aplin C, Whatley SD, Thompson P, Hoy T, Fisher P, Singer C, Lovell CR, et al. Late-onset erythropoietic porphyria caused by a chromosome 18q deletion in erythroid cells. J Invest Dermatol 2001;117:1647–1649. [DOI] [PubMed] [Google Scholar]
- 82.Goodwin RG, Kell WJ, Laidler P, Long CC, Whatley SD, McKinley M, Badminton MN, et al. Photosensitivity and acute liver injury in myeloproliferative disorder secondary to late-onset protoporphyria caused by deletion of a ferrochelatase gene in hematopoietic cells. Blood 2006;107:60–62. [DOI] [PubMed] [Google Scholar]
- 83.Lim HW, Cooper D, Sassa S, Dosik H, Buchness MR, Soter NA. Photosensitivity, abnormal porphyrin profile, and sideroblastic anemia. J Am Acad Dermatol 1992;27:287–292. [DOI] [PubMed] [Google Scholar]
- 84.Bishop DF, Tchaikovskii V, Nazarenko I, Desnick RJ. Molecular expression and characterization of erythroid-specific 5-aminolevulinate synthase gain-of-function mutations causing X-linked protoporphyria. Mol Med 2013;19:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]