Abstract
Resistance to 157 different herbicides and 88% of known sites of action has been observed, with many weeds resistant to two or more modes. Coupled with tighter environmental regulation, this demonstrates the need to identify new modes of action and novel herbicides. The plant sphingolipid biosynthetic enzyme, inositol phosphorylceramide synthase (IPCS), has been identified as a novel, putative herbicide target. The non-mammalian nature of this enzyme offers the potential of discovering plant specific inhibitory compounds with minimal impact on animals and humans, perhaps leading to the development of new non-toxic herbicides. The best characterised and most highly expressed isoform of the enzyme in the model-dicot Arabidopsis, AtIPCS2, was formatted into a yeast-based assay which was then utilized to screen a proprietary library of over 11,000 compounds provided by Bayer AG. Hits from this screen were validated in a secondary in vitro enzyme assay. These studies led to the identification of a potent inhibitor that showed selectivity for AtIPCS2 over the yeast orthologue, and activity against Arabidopsis seedlings. This work highlighted the use of a yeast-based screening assay to discover herbicidal compounds and the status of the plant IPCS as a novel herbicidal target.
Subject terms: Plant sciences, Biochemistry
Introduction
First discovered in Saccharomyces cerevisiae, inositol phosphorylceramide synthase (IPCS or Aur1p in yeast) catalyses the transfer of phosphorylinositol from the phosphoglycerolipid phosphatidylinositol to the C-1 hydroxyl group of (phyto)ceramide, thereby generating the complex sphingolipid inositol phosphorylceramide (IPC)1. IPC is subsequently a precursor for the generation of the other, more complex, sphingolipids: mannosylinositol phosphorylceramide (MIPC) and mannosyldiinositol phosphorylceramide [M(IP)2C]2. These have been shown to be vital for the localization and endocytosis of plasma membrane proteins in S. pombe3. In addition, aside from maintaining the structural integrity of the plasma membrane, sphingolipids have been demonstrated to play crucial roles in a number of eukaryotic cell processes including apoptosis4,5, cell differentiation6, cell cycle arrest7, cell signalling8, angiogenesis9 and senescence10.
The sphingolipid biosynthetic pathway, and the enzymes involved, show conservation in all kingdoms of the Eukaryota up to the formation of dihydrosphingosine11. Subsequently there is divergence, dihydrosphingosine is N-acylated to produce dihydroceramide which is then desaturated to give ceramide in mammals and protozoa. In contrast, in plants and fungi, phytosphingosine, generated from the hydroxylation of dihydrosphingosine, is N-acylated to give phytoceramide. Subsequently, these intermediary metabolites are transported into the Golgi apparatus where sphingomyelin synthase catalyzes the production of sphingomyelin, the major sphingolipid in mammals, and IPCS generates IPC in plants, fungi and protozoa11.
This divergence in sphingolipid biosynthesis has been exploited to investigate the protozoal IPCS as a therapeutic target for the Neglected Tropical Diseases, Chagas disease12–14 and leishmaniais15–18. In plants, the activity of IPCS was first characterized in Phaseolus vulgaris19 and its role as a negative regulator of programmed cell death in plants was validated in Arabidopsis thaliana20 and Eucalyptus grandis21. In Oryza sativa, IPCS has been shown to play a role in plant response to abiotic stress, particularly in response to drought, cold and salinity22.
Despite the fact that hundreds of herbicides are widely used, these only exhibit 25 modes of action. In fact, merely 6 modes of action, targeting 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, acetolactate synthase (ALS), photosystem (PS) II, synthetic auxins, acetyl CoA carboxylase (ACCase) and cell division, acount for 75% of the herbicide market23. It has been over 30 years since a herbicide with a new mode of action was introduced onto the market and, with the growing problem of herbicide resistance24 and the destabilizing effect of climate change on crop yield25, it is now necessary to identify new herbicidal modalities to ameliorate the challenge of feeding a rapidly increasing global population set to reach 9–10 billion in 205026.
As previously reported20–22, inhibition of the plant IPCS would lead to a buildup of the enzyme substrate, the Programmed Cell Death (PCD; apoptosis) mediator phytoceramide27. The functional divergence of IPCS from the equivalent mammalian enzyme, sphingomyelin synthase (SMS), could allow the identification of specific, non-toxic inhibitors. This possibility has, to date, lead to the identification of 5 potent inhibitors of the fungal IPCS (aureobasidin A28 (AbA), khafrefungin29, rustimicin30, pleofungin31 and haplofungin32) with low nano-molar IC50 values against Saccharomyces cerevisiae. However, currently, no inhibitor of the plant orthologue has been identified.
In this study, the well characterized Arabidopsis thaliana enzyme AtIPCS220, the most highly expressed of the 3 IPCS isoforms33, was used to complement S. cerevisiae lacking AUR1. The yeast utilized was engineered to enhance compound sensitivity through reduced expression of several efflux pumps, and thereby allow efficient hit identification in a cell-based high throughput screening (HTS) assay used for 11,440 bioactive compounds. A secondary enzyme-based assay facilitated the validation of hits as inhibitors of the enzyme, and enabled comparison of their activity against the yeast orthologue, Aur1p. This allowed the identification of hits that exhibited selectivity for IPCS2 from A. thaliana. The most potent selective compound was tested in vivo against seedlings and demonstrated herbicidal activity.
Results
Primary high throughput screening using a yeast-based assay
Fungi such as Saccharomyces cerevisiae possess multiple genes linked to pleiotropic drug resistance, including those encoding a range of ATP-binding cassette (ABC) transporters and the transcription factors required for their expression34. These extrusion pumps can be over-expressed in response to drug treatment, leading to decreased intracellular drug concentrations and subsequent drug resistance35, while multiple deletions of these functions render yeast cells significantly more sensitive to a range of toxic compounds including antifungal agents used in agriculture and medicine8. To increase the sensitivity of the yeast-based assay platform, an S. cerevisiae strain was utilised that lacked PDR1, PDR3, PDR16 and PDR17. This combination of pdr deletions was shown to confer significant hypersensitivity to a range of compounds (Supplementary Information 1). PDR136 and PDR337 encode paralogous Zn(II)2Cys6 zinc finger regulators, which control the transcription of ABC drug efflux pump-encoding genes including PDR538,39, SNQ240, PDR1041, PDR1541 and YOR142 through binding to cis-acting PDREs (pleiotropic drug resistance elements)40,41,43,44. PDR16 and PDR17 encode a pair of paralogous phosphatidylinositol transport proteins that also confer drug hypersensitivity when deleted45.
In the quadruple pdr1∆ pdr3∆ pdr16∆ pdr17∆ strain, AUR1 was deleted and replaced by a HIS3 selectable marker, with growth supported by expression of the essential AUR1 gene from the plasmid pRS316-AUR1 under uracil selection. In this background, galactose-inducible expression of AtIPCS2 from plasmid pESC-LEU was found to complement loss of pRS316-AUR1 when the yeast were cultured in the presence of 5-fluoroorotic acid. This made the yeast dependent upon the presence of galactose for growth, thus demonstrating dependence on the expression of the plant enzyme (Fig. 1, Supplementary Information 2)20–22. Assay of microsomal extracts from the complemented yeast demonstrated IPCS activity in vitro and confirmed that the plant activity is insensitive to the fungal Aur1p inhibitor aureobasidin A (AbA)28,33 (Fig. 2).
Figure 1.
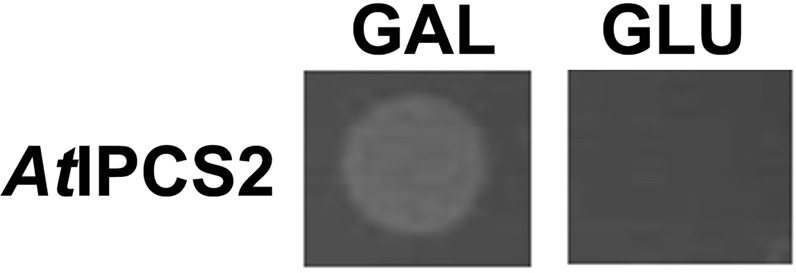
Yeast (MSYD23) dependent on expression of AtIPCS2 from a galactose inducible promotor were, as expected, viable when grown in the presence of galactose (GAL), but not glucose (GLU). The relevant section of each agar plate (GAL and GLU) is illustrated, the full plates are shown in Supplementary Information 2.
Figure 2.
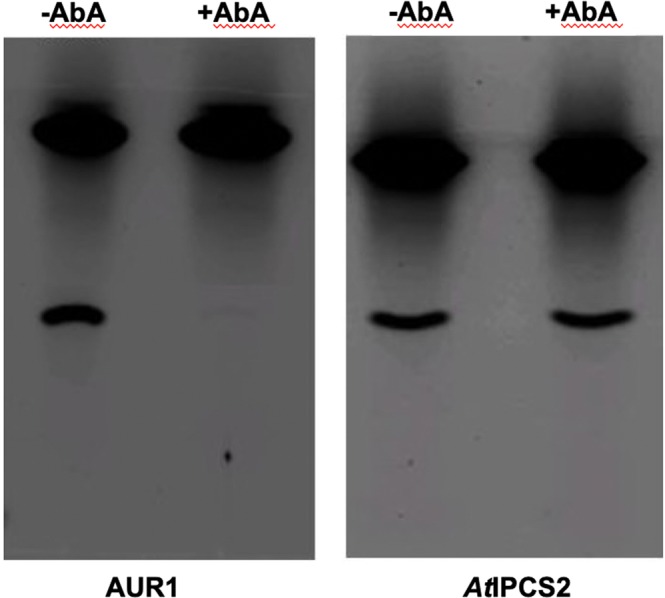
HPTLC separation of in vitro assayed Aur1p and AtIPCS2 showing the production of NBD-IPC in the presence (+) and absence (−) of AbA. Only the fungal enzyme Aur1p is sensitive to the compound. NBD-Cer is the substrate, NBD-C6-phytoceramide.
Yeast complemented with the well characterised AtIPCS2, and an AUR1 control, were subsequently formatted into a 96-well plate. Following statistical validation by calculation of Z factor46 in the presence of positive (cycloheximide) and negative (DMSO) controls, the assay was used in HTS of a focused library of 11,440 bioactive compounds. All assay plates were required to have a calculated Z factor ≥0.5 for the data to be progressed. Following in duplicate screening at 10 µM against AtIPCS2 complemented yeast and the AUR1 control, compounds exhibiting ≥80% inhibition and ≥50% selectivity for AtIPCS2 were taken forward. After eliminating false positives (non-reproducible hits; 2.6%), 106 target directed hits were identified, a hit rate of 0.9% (Fig. 3, Supplementary Information 3). It is notable that a significant minority of compounds increased yeast proliferation and that this phenotype was more profound in the AtIPCS2 complemented yeast (negative inhibition; Fig. 3), whilst this is an interesting observation these were not analysed further. Dose response analyses (50 µM to 68 nM), using the same assay platform, demonstrated that the majority of the inhibitory compound hits (89 of 106) had an IC50 of less than 10 µM (Supplementary Information 4).
Figure 3.
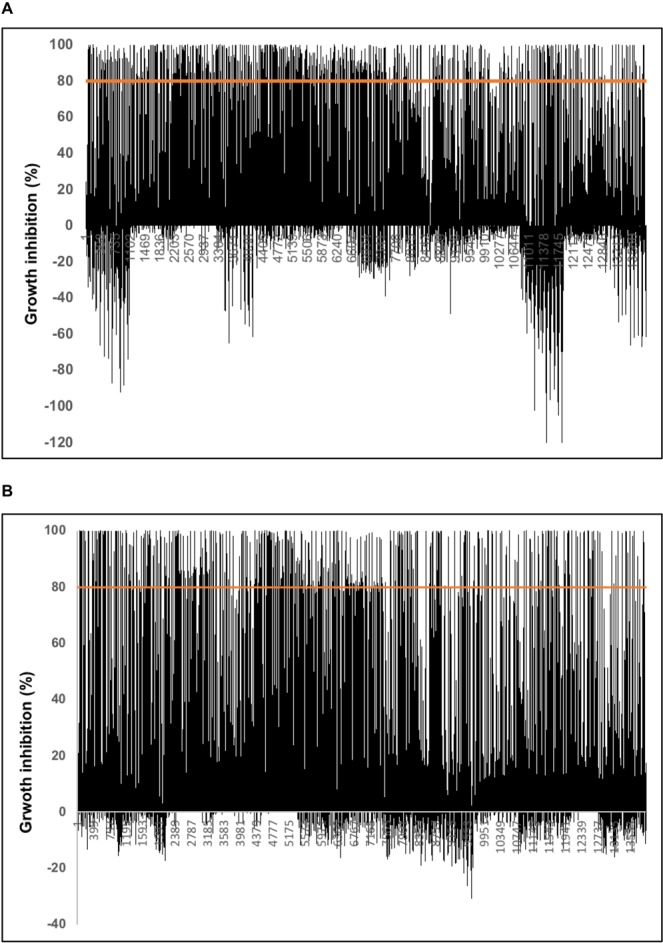
Growth inhibition for compounds (10 µM) against AtIPCS2 (A) and Aur1p (B) complemented yeast. After eliminating false positives (2.6%), 106 target directed hits were identified, a hit rate of 0.9%.
Secondary screening using an in vitro biochemical assay
In the secondary screening stage the previously described microsomal-based in vitro IPCS assay was adapted and utilised17. Initially, all 106 selective hits from the primary screen were tested, in duplicate, at 10 µM. 16 compounds which, reproducibly, showed ≥30% inhibition were carried forward for in triplicate dose response (100 µM to 46 nM) analyses and IC50 determination against AtIPCS2 and, as a control, AUR1. All were active to some degree against the Arabidopsis enzyme, whilst none showed inhibition of the fungal orthologue, demonstrating that selective AtIPCS2 inhibitors had been identified. 4 compounds demonstrated IC50 values < 10 µM (Compound 1, 4.02 µM; 2, 4.75 µM; 3, 8.41 µM; and 4, 9.84 µM; Supplementary Information 5). The structures of compounds 2–4 are withheld due to intellectual property reasons, leaving the most active (Compound 1, a phenylamidine carrying an acetonitrile functional group) to be taken forward (Fig. 4). The structural integrity of Compound 1 was confirmed using mass spectrometry and 1H and 13C spectroscopy which showed that it was a 3:2 mixture of E:Z amidine isomers (see Supplementary Information 6).
Figure 4.
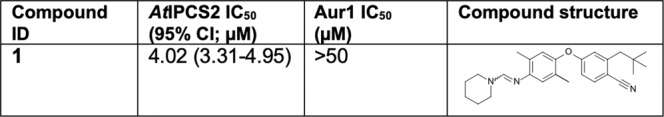
Compound 1 structure and activity in the biochemical assay against AtIPCS2 and Aur1p.
In vivo screening
In vivo testing of the phenylamidine Compound 1 was undertaken against Arabidopsis seedlings grown on agar. Dose response analyses (Fig. 5) showed that treatment restricted growth and led to purple leaf patches at 11 µM and above. Examination of treated 7 day old seedlings grown on agar containing 10 and 40 µM of Compound 1 showed plants with clear purple patches associated with anthocyanin biosynthesis in response to stress47, and an absence of lateral root development compared to the DMSO control (Fig. 6).
Figure 5.
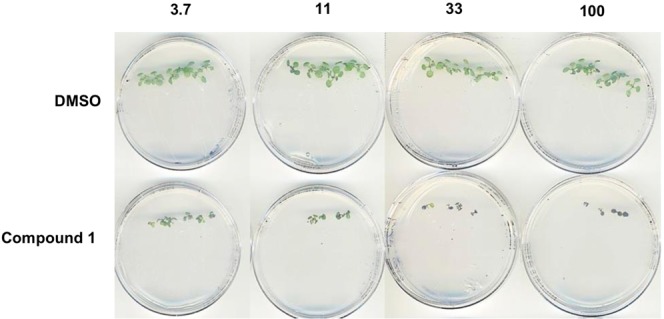
Compound 1 tested against Arabidopsis seedlings at 3.7 µM, 11 µM, 33 µM and 100 µM. Treatment restricted growth and led to purple leaf patches at 11 µM and above compared to the DMSO control.
Figure 6.
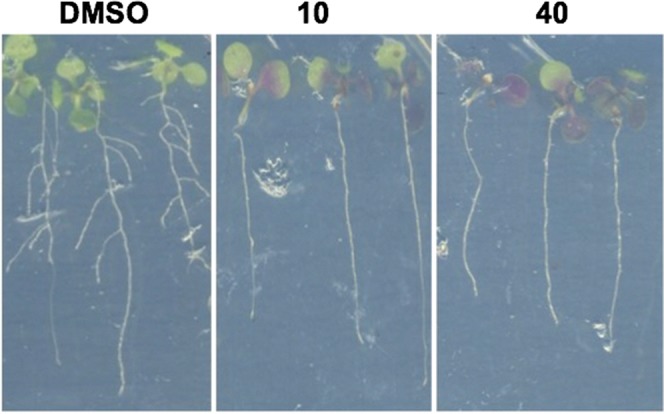
From left to right, 7 day old Arabidopsis seedlings grown on agar containing DMSO (vehicle), 10 µM and 40 µM of Compound 1. Treated plants had clear purple patches and an absence of lateral root development compared to the DMSO control.
Discussion
With herbicide resistance increasing24 and climate change effecting on crop yield25, the need to identify new herbicide targets and lead molecules to address these challenges is pressing. One major hurdle to overcome in this search for a new herbicide is to ensure identified chemicals have acceptable toxicity profiles which are safe to the user and the environment48. The divergence in the sphingolipid biosynthetic pathway between mammals and plants, where the former produce SM and the latter IPC20,33, may present an opportunity to identify molecules with such a profile.
Following the recent publication of our successful HTS campaign against a protozoan IPCS49, this study is the first report of HTS for inhibitors of an enzyme in the plant sphingolipid synthetic pathway, the non-mammalian AtIPCS2 – the most highly expressed and best characterised isoform in the model dicot Arabidopsis, which catalyses the synthesis of IPC20,33. The role of this enzyme in phytoceramide homeostasis20,33 and therefore PCD50, coupled with the product, IPC, functioning as the precursor for the synthesis of glycosylinositol phosphorylceramide (GIPC; 25% of plasma membrane lipid51), makes IPCS an attractive target for the discovery of new, non-toxic, herbicidal agents. However, given the multi-transmembrane nature of the enzyme20,33 assay development is challenging. Therefore, to facilitate HTS, we developed a novel cell-based assay utilising an AtIPCS2 complemented S. cerevisiae strain lacking 4 extrusion pumps linked to pleiotropic drug resistance (PDR1, PDR3, PDR16 and PDR17) to increase sensitivity, and utilised this system to screen a library of 11,440 bioactive compounds. Counter screening against Aur1p (the yeast orthologue) formatted in the same assay yielded 106 selective hits, of these 4 demonstrated IC50 values < 10 µM in a secondary in vitro enzyme assay and minimal activity against yeast Aur1p (>50 µM). The most active was a phenylamidine, Compound 1 (IC50 < 5 µM), which has been patented by Bayer as a fungicide52. Previous phylogenic analyses33 have shown that the three IPCS isoforms in Arabidopsis are closely related. Further, focused, sequence analyses demonstrated that whilst AtIPCS2 orthologues are highly conserved within the monocots and eudicots, there is distance between the two clades (see Supplementary Information 7). This indicated that selective inhibition of the enzyme (for example in a weed species) maybe feasible. Future studies should examine the selectivity of Compound 1 for AtIPCS2 over the other two isoforms and other plant orthologues to establish selectively in Planta.
The phenylamidines were first identified in the 1960s as pesticides for the control of plant fungal pathogens52 and specific variants have subsequently been patented for use as herbicides53. In vivo screening of the identified phenylamidine, Compound 1, against wild type Col-0 Arabidopsis seedlings demonstrated dose dependent effects with decreased lateral root development, and distinctive purple leaf patches associated with anthocyanin biosynthesis in response to stress47. The mode of action of herbicidal phenylamidines have not been published, but the phenotypic effects reported here for Compound 1 are consistent with those expected for an IPCS inhibitor.
In conclusion, using a novel HTS approach the first inhibitor (Compound 1 - a phenylamidine) of plant IPCS was identified and shown, in vivo, to induce the plant stress response. This low molecular weight compound is ideal for further development towards use in agriculture, and further studies are planned to investigate this possibility.
Methods
Yeast strains
The diploid Saccharomyces cerevisiae strain MSYD20 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 pdr1∆::KanMX4/pdr1∆::KanMX4 pdr3∆::KanMX4/pdr3∆::KanMX4 pdr16∆::KanMX4/pdr16∆::KanMX4 pdr17∆::KanMX4/pdr17∆::KanMX4) is homozygous for knockouts of four genes conferring drug hypersensitivity and was made by sequential crosses and tetrad dissection starting with the single MATa pdr1∆::KanMX4 (Y04381), MATα pdr3∆::KanMX4 (Y13029), MATa pdr16∆::KanMX4 (Y01981) and pdr17∆::KanMX4 (Y11180) strains obtained from the Euroscarf collection (http://www.euroscarf.de). MSYD20 was made heterozygous for aur1::HIS3 (knocking out the AUR1 gene encoding yeast IPCS) by transformation with a PCR product amplified from pFA6a-HIS3 template using the following two primers (plasmid-specific sequences, upper case; AUR1 flanking sequences, lower case), generating MSYD23.
AUR1-F1
atatcctacaggttgcggttttcatattttaaaaagcttttaatcattcctttgcgtCGGATCCCCGGGTTAATTAA
AUR1-R1
atttatatgtatctacataagaccaaccgtatccgtaattgcagataaaatactcaGAATTCGAGCTCGTTTAAAC
pFA6a-HIS3 was generated by replacing the AscI-MfeI interval of pFA6a-TRP154 that carries the TRP1 gene with the BssHII-EcoRI fragment of YDpH55 encoding HIS3, enabling amplification of a HIS3-containing gene knockout fragment that cannot recombine with existing KanMX4 gene knockouts such as those present in MSYD20. Sporulation and tetrad analysis of MSYD23 confirmed that AUR1 is an essential gene since no viable aur1::HIS3 segregants could be obtained. MSYD23 was next transformed with pRS316-AUR1, made by inserting a copy of AUR1 into the URA3 plasmid pRS31656. Strain MSY23-3C (MATa pdr1Δ::KanMX, pdr3Δ::KanMX4 pdr16Δ::KanMX4 pdr17Δ::KanMX4 aur1Δ::HIS3 [pRS316-AUR1]), lacking the four genes conferring drug hypersensitivity and in which growth was supported by AUR1 expression from pRS31615, was identified following sporulation and tetrad analysis of MSYD23 transformed with pRS316-AUR1. MSY23-3C was verified by appropriate diagnostic PCR and could not grow in the presence of 5FOA as expected.
Primary screening, yeast cell-based assay
The complete open reading frames of AtIPCS2 and AUR1p were amplified from cDNA or GenEZ ORF Clones (GenScript®) using Phusion Flash® PCR master mix (ThermoFisher) according to manufacturer’s guidelines. Primers for In-Fusion® cloning (Clontech) were:
AtIPCS2F ctcactatagggcccATGACACTTTATATTCGTCGT
AtIPCS2R tccatgtcgacgcccTCACGCGCCATTCATTGTGTT
AUR1F ctcactatagggcccATGGCAAACC
AUR1R tccatgtcgacgcccTTAAGCCCTC
These open reading frames were cloned into the pESC-LEU vector (Agilent) and verified by sequence analyses, creating pESC-LEU_AtIPCS2 and pESC-LEU_AUR1. In this vector, expression of the open reading frame was under the control of a galactose-inducible promoter. All plasmids were subsequently transformed into the MSY23-3C S. cerevisiae strain as previously described15 and selected on SD -TRP -URA -LEU agar (0·17% Bacto yeast nitrogen base, 0·5% ammonium sulphate, 2% glucose, containing the appropriate nutritional supplements) at 30 °C. Yeast were then ‘cured’ of the pRS316-AUR1 plasmid by selection on SGR -TRP –LEU +FOA agar (0·17% Bacto yeast nitrogen base, 0·5% ammonium sulphate, 0.1% galactose, 1% raffinose, 0.1% 5-Fluoroorotic Acid Monohydrate (FOA) containing the appropriate nutritional supplements) at 30 °C, creating MSY23-3C pESC-LEU_AtIPCS2; and MSY23-3C pESC-LEU_AUR1. Following PCR validation and propagation in SGR -TRP -LEU, frozen stocks of both yeast lines were created (OD600 = 10).
When required, MSY23-3C pESC-LEU_AtIPCS2 and MSY23-3C pESC-LEU_AUR1 were thawed on ice and diluted 1:20 with SGR -TRP -LEU. Using a Biomek FXp automated workstation (Beckman Coulter) 198 µl was aliquoted into 96-well plates (Thermo Scientific) before the addition of 2 ul of compounds (to the desired concentration) and controls – DMSO (negative; Sigma Aldrich) and cycloheximide (to 10 µM; positive; Sigma Aldrich). Following incubation at 30 °C for 24 hours optical density (OD600) was measured (Biotek Synergy H4 with Gen5™). All assays were carried out in duplicate and inhibition (%) calculated.
Secondary screening, biochemical assay
Microsomal material was prepared from MSY23-3C pESC-LEU_AtIPCS2 and MSY23-3C pESC-LEU_AUR1 as previously described33. IPCS turnover was assayed using HPTLC (Merck) and imaged using a Fuji FLA−3000 reader and AIDA Image Analyser® software (version 3.52) as previously described17. Subsequently, a 96-well plate assay was formatted based on the protocol described by Mina et al.17. Following optimisation of substrate concentration and incubation time, each compound at the desired concentration (100 µM to 46 nM; in triplicate), was incubated in 96-well plates (Corning® Costar®) in phosphate buffer (71.4 mM, pH 7.0) with soybean PI (100 µM, final concentration, Avanti), NBD-C6-phytoceramide (15 µM; ThermoFisher) and microsomal membranes (0.3–0.4 Units17). Following incubation for 60 minutes (or 40 minutes for MSY23-3C pESC-LEU_AUR1 membranes) at 30 °C the reaction was quenched by the addition of 200 µl methanol per well, the reaction product separated using exchange chromatography in 96-well filter plates (Millipore)17 and the fluorescence measured at Ex460/Em540 using a fluorescence Microplate Reader (Biotek Synergy H4 with Gen5™). Analyses were carried out using GraphPad Prism 7.
In vivo screening, Arabidopsis seedlings
A. thaliana (Col0) seedlings were grown for 10 days on 0.8% Murashige and Skoog (MS) agar and then transferred to 1.2% MS agar containing compounds at the desired concentrations or DMSO as a control. Plants were grown at 20 °C under 16 hour day/8 hour night photoperiod.
Supplementary information
Acknowledgements
E.C.P. was supported by a Biotechnology and Biological Research Council CASE studentship with Bayer Crop Sciences (BB/K012703/1). J.G.M. and P.W.D. were also supported by the Biotechnology and Biological Research Council (BB/M024156/1); P.W.D. and P.G.S. are also supported by the Medical Research Council (MR/P027989/1). We thank Durham University’s Wolfson Research Institute for Health and Wellbeing, and Biophysical Sciences Institute for support.
Author Contributions
E.C.P. constructed, validated and utilised the yeast assay, and performed the in vivo experiments; E.C.P. and J.G.M. analysed the data; M.J.R.S. constructed the yeast line utilised; S.D.L. and P.L. managed all aspects of the screening process; M.R.K. supervised the in vivo experiments; S.D.L., P.G.S and P.W.D. conceived, directly managed the project and analysed data; E.C.P. and P.W.D. wrote the manuscript and constructed figures.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Patrick G. Steel, Email: p.g.steel@durham.ac.uk
Paul W. Denny, Email: p.w.denny@durham.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44544-1.
References
- 1.Nagiec MM, et al. Sphingolipid synthesis as a target for antifungal drugs complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. Journal of Biological Chemistry. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 2.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Progress in lipid research. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Nakase M, et al. Mannosylinositol phosphorylceramide is a major sphingolipid component and is required for proper localization of plasma-membrane proteins in Schizosaccharomyces pombe. J Cell Sci. 2010;123:1578–1587. doi: 10.1242/jcs.059139. [DOI] [PubMed] [Google Scholar]
- 4.Sweeney EA, Inokuchi J-i, Igarashi Y. Inhibition of sphingolipid induced apoptosis by caspase inhibitors indicates that sphingosine acts in an earlier part of the apoptotic pathway than ceramide. FEBS letters. 1998;425:61–65. doi: 10.1016/S0014-5793(98)00198-7. [DOI] [PubMed] [Google Scholar]
- 5.Kolesnick R, Fuks Z. Radiation and ceramide-induced apoptosis. Oncogene. 2003;22:5897. doi: 10.1038/sj.onc.1206702. [DOI] [PubMed] [Google Scholar]
- 6.Favale NO, Santacreu BJ, Pescio LG, Marquez MG, Sterin-Speziale NB. Sphingomyelin metabolism is involved in the differentiation of MDCK cells induced by environmental hypertonicity. Journal of lipid research. 2015;56:786–800. doi: 10.1194/jlr.M050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesley UV, Hatcher JF, Dempsey RJ. Sphingomyelin synthase 1 regulates Neuro-2a cell proliferation and cell cycle progression through modulation of p27 expression and Akt signaling. Molecular neurobiology. 2015;51:1530–1541. doi: 10.1007/s12035-014-8829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki, M., Aoki, H., Ramanathan, R., Hait, N. C. & Takabe, K. Sphingosine-1-phosphate signaling in immune cells and inflammation: roles and therapeutic potential. Mediators of inflammation2016 (2016). [DOI] [PMC free article] [PubMed]
- 9.Camaré Caroline, Vanucci-Bacqué Corinne, Augé Nathalie, Pucelle Mélanie, Bernis Corinne, Swiader Audrey, Baltas Michel, Bedos-Belval Florence, Salvayre Robert, Nègre-Salvayre Anne. 4-Hydroxynonenal Contributes to Angiogenesis through a Redox-Dependent Sphingolipid Pathway: Prevention by Hydralazine Derivatives. Oxidative Medicine and Cellular Longevity. 2017;2017:1–11. doi: 10.1155/2017/9172741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astarita G, et al. Methamphetamine accelerates cellular senescence through stimulation of de novo ceramide biosynthesis. PloS one. 2015;10:e0116961. doi: 10.1371/journal.pone.0116961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young SA, Mina JG, Denny PW, Smith TK. Sphingolipid and ceramide homeostasis: potential therapeutic targets. Biochem Res Int. 2012;2012:248135. doi: 10.1155/2012/248135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueiredo JM, Mendonca-Previato L, Previato JO, Heise N. Characterization of the inositol phosphorylceramide synthase activity from Trypanosoma cruzi. Biochemical Journal. 2005;387:519–529. doi: 10.1042/BJ20041842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueiredo JM, et al. Molecular and functional characterization of the ceramide synthase from Trypanosoma cruzi. Mol Biochem Parasitol. 2012;182:62–74. doi: 10.1016/j.molbiopara.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koeller CM, Heise N. The Sphingolipid Biosynthetic Pathway Is a Potential Target for Chemotherapy against Chagas Disease. Enzyme Res. 2011;2011:648159. doi: 10.4061/2011/648159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denny PW, Shams-Eldin H, Price HP, Smith DF, Schwarz RT. The protozoan inositol phosphorylceramide synthase a novel drug target that defines a new class of sphingolipid synthase. Journal of Biological Chemistry. 2006;281:28200–28209. doi: 10.1074/jbc.M600796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mina JG, Mosely JA, Ali HZ, Denny PW, Steel PG. Exploring Leishmania major inositol phosphorylceramide synthase (LmjIPCS): insights into the ceramide binding domain. Org Biomol Chem. 2011;9:1823–1830. doi: 10.1039/c0ob00871k. [DOI] [PubMed] [Google Scholar]
- 17.Mina JG, et al. A plate-based assay system for analyses and screening of the Leishmania major inositol phosphorylceramide synthase. International Journal of Biochemistry & Cell Biology. 2010;42:1553–1561. doi: 10.1016/j.biocel.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 18.MINA JOHN G. M., DENNY P. W. Everybody needs sphingolipids, right! Mining for new drug targets in protozoan sphingolipid biosynthesis. Parasitology. 2017;145(2):134–147. doi: 10.1017/S0031182017001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bromley PE, Li YO, Murphy SM, Sumner CM, Lynch DV. Complex sphingolipid synthesis in plants: characterization of inositolphosphorylceramide synthase activity in bean microsomes. Archives of Biochemistry and Biophysics. 2003;417:219–226. doi: 10.1016/S0003-9861(03)00339-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, et al. An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. The Plant Cell. 2008;20:3163–3179. doi: 10.1105/tpc.108.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangwanda R, Myburg AA, Naidoo S. Transcriptome and hormone profiling reveals Eucalyptus grandis defence responses against Chrysoporthe austroafricana. BMC genomics. 2015;16:319. doi: 10.1186/s12864-015-1529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao P, et al. Characterization and expression analysis of inositolphosphorylceramide synthase family genes in rice (Oryza sativa L.) Genes & Genomics. 2017;39:485–492. doi: 10.1007/s13258-016-0489-0. [DOI] [Google Scholar]
- 23.Lindell, S. et al. HPPD herbicide-safener combinations as resistance breaking solutions for 21st century agriculture. Discovery and Synthesis of Crop Protection Products, Maienfisch, P. & Stevenson, T. editors, American Chemical Society Symposium Series1024, Chapter 16, 219–231 (2015).
- 24.Heap I. Global perspective of herbicide‐resistant weeds. Pest management science. 2014;70:1306–1315. doi: 10.1002/ps.3696. [DOI] [PubMed] [Google Scholar]
- 25.Arnell NW, et al. The consequences of CO2 stabilisation for the impacts of climate change. Climatic Change. 2002;53:413–446. doi: 10.1023/A:1015277014327. [DOI] [Google Scholar]
- 26.UN. http://reliefweb.int/sites/reliefweb.int/files/resources/FAO_2013_stats_yrbook.pdf (2013).
- 27.Berkey R, Bendigeri D, Xiao S. Sphingolipids and plant defense/disease: the “death” connection and beyond. Front Plant Sci. 2012;3:68. doi: 10.3389/fpls.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidler SA, Radding JA. The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A (LY295337) Antimicrobial agents and chemotherapy. 1995;39:2765–2769. doi: 10.1128/AAC.39.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandala SM, et al. Khafrefungin, a novel inhibitor of sphingolipid synthesis. Journal of Biological Chemistry. 1997;272:32709–32714. doi: 10.1074/jbc.272.51.32709. [DOI] [PubMed] [Google Scholar]
- 30.Mandala SM, et al. Rustmicin, a potent antifungal agent, inhibits sphingolipid synthesis at inositol phosphoceramide synthase. Journal of Biological Chemistry. 1998;273:14942–14949. doi: 10.1074/jbc.273.24.14942. [DOI] [PubMed] [Google Scholar]
- 31.Yano T, et al. Pleofungins, novel inositol phosphorylceramide synthase inhibitors, from Phoma sp. SANK 13899. Journal of Antibiotics. 2007;60:136. doi: 10.1038/ja.2007.13. [DOI] [PubMed] [Google Scholar]
- 32.Ohnuki T, Yano T, Takatsu T. Haplofungins, new inositol phosphorylceramide synthase inhibitors, from Lauriomyces bellulus SANK 26899 II. Structure elucidation. Journal of Antibiotics. 2009;62:551–557. doi: 10.1038/ja.2009.73. [DOI] [PubMed] [Google Scholar]
- 33.Mina J, et al. Functional analyses of differentially expressed isoforms of the Arabidopsis inositol phosphorylceramide synthase. Plant Mol Biol. 2010;73:399–407. doi: 10.1007/s11103-010-9626-3. [DOI] [PubMed] [Google Scholar]
- 34.Prasad R, Goffeau A. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu Rev Microbiol. 2012;66:39–63. doi: 10.1146/annurev-micro-092611-150111. [DOI] [PubMed] [Google Scholar]
- 35.St Georgiev V. Membrane transporters and antifungal drug resistance. Curr Drug Targets. 2000;1:261–284. doi: 10.2174/1389450003349209. [DOI] [PubMed] [Google Scholar]
- 36.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. Journal of Biological Chemistry. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 37.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 38.Decottignies A, Kolaczkowski M, Balzi E, Goffeau A. Solubilization and characterization of the overexpressed PDR5 multidrug resistance nucleotide triphosphatase of yeast. J Biol Chem. 1994;269:12797–12803. [PubMed] [Google Scholar]
- 39.Katzmann DJ, Burnett PE, Golin J, Mahe Y, Moye-Rowley WS. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/MCB.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahé Y, et al. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol. 1996;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 41.Wolfger H, Mahe Y, Parle-McDermott A, Delahodde A, Kuchler K. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 1997;418:269–274. doi: 10.1016/S0014-5793(97)01382-3. [DOI] [PubMed] [Google Scholar]
- 42.Katzmann DJ, et al. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/MCB.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeRisi J, et al. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000;470:156–160. doi: 10.1016/S0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 44.Katzmann DJ, Hallstrom TC, Mahe Y, Moye-Rowley WS. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 45.van den Hazel HB, et al. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J-H, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of biomolecular screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 47.Chalker-Scott L. Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol. 1999;70:1–9. doi: 10.1562/0031-8655(1999)070<0001:Esoaip>2.3.Co;2. [DOI] [Google Scholar]
- 48.Corsi, C. & Lamberth, C. New paradigms in crop protection research: Registrability and cost of goods. Discovery and Synthesis of Crop Protection Products, Maienfisch, P. & Stevenson, T. editors, American Chemical Society Symposium Series1024, Chapter 16, 25–37 (2015).
- 49.Norcliffe JL, et al. Identifying inhibitors of the Leishmania inositol phosphorylceramide synthase with antiprotozoal activity using a yeast-based assay and ultra-high throughput screening platform. Sci Rep. 2018;8:3938. doi: 10.1038/s41598-018-22063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang H, et al. Ceramides modulate programmed cell death in plants. Genes & development. 2003;17:2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rennie EA, et al. Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. The Plant Cell. 2014;26:3314–3325. doi: 10.1105/tpc.114.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duerr, D., Aebi, H. & Ebner, L. Inventors; Ciba Limited, assignee. Method for protecting plants from fungi. United States patent US 3,284,286 (1966 Nov 8); and Luemmen, P. et al. Pesticidal phenoxy substituted phenylamidine derivatives, PCT International Application WO 2007/031508 (2007 March 22).
- 53.Kuhn, B. et al. Inventors; Bayer AG, assignee. Use of n2-phenylamidines as herbicides and herbicidal agents comprising the same. International patent WO 2008/110278 A3 (2008 Sept 18).
- 54.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast (Chichester, England) 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 56.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


