Abstract
3-hydroxypropanoic acid (3-HP) is a valuable platform chemical with a high demand in the global market. 3-HP can be produced from various renewable resources. It is used as a precursor in industrial production of a number of chemicals, such as acrylic acid and its many derivatives. In its polymerized form, 3-HP can be used in bioplastic production. Several microbes naturally possess the biosynthetic pathways for production of 3-HP, and a number of these pathways have been introduced in some widely used cell factories, such as Escherichia coli and Saccharomyces cerevisiae. Latest advances in the field of metabolic engineering and synthetic biology have led to more efficient methods for bio-production of 3-HP. These include new approaches for introducing heterologous pathways, precise control of gene expression, rational enzyme engineering, redirecting the carbon flux based on in silico predictions using genome scale metabolic models, as well as optimizing fermentation conditions. Despite the fact that the production of 3-HP has been extensively explored in established industrially relevant cell factories, the current production processes have not yet reached the levels required for industrial exploitation. In this review, we explore the state of the art in 3-HP bio-production, comparing the yields and titers achieved in different microbial cell factories and we discuss possible methodologies that could make the final step toward industrially relevant cell factories.
Keywords: 3-hydroxypropanoic acid, glycerol, biosynthesis, cell factory, synthetic biology, metabolic engineering
Introduction
The development of microbial cell factories is fueled by aspirations to develop sustainable processes based on renewable resources. The goal is mitigation of the negative environmental consequences of production of fuels, chemicals and other materials.
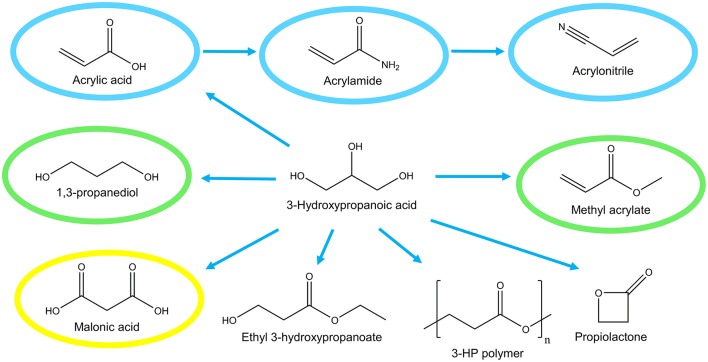
The development of microbial cell factories for production of the platform chemical 3-hydroxypropanoic acid (3-HP) has attracted much attention in the last decade. 3-HP is a non-chiral (optically inactive), small three-carbon molecule and a structural isomer of lactic acid. Specifically, 3-HP is a precursor for the production of a number of valuable chemicals including acrylic acid and bioplastics (Figure 1). Bio-based production of 3-HP also has the potential to “turn waste into a resource” since several metabolic pathways exist for converting glycerol, a by-product of biodiesel production, into 3-HP.
Figure 1.
Potential industrial uses of 3-hydroxypropanoic acid. Global market of derived compounds are indicated; >1,000 M$ (blue), 100–1,000 M$ (green), <100 M$ (yellow).
Here, we will briefly introduce the metabolic pathways that can be used to produce 3-HP from glycerol. Based on work accumulated over the last decade, we will present current knowledge pertaining to different production hosts, enzymes, and strategies for optimization of production pathway and host metabolism as well as process engineering for attaining high-level production of 3-HP. Finally, we will discuss how recent developments in synthetic biology and metabolic engineering might form the basis for further improvements of production strains and the eventual goal of industrially viable and sustainable 3-HP production.
Metabolic Pathways for Synthesis of 3-HP Starting From Glycerol
Metabolic Pathways Found in Nature
A number of microorganisms have been reported to naturally produce 3-HP using various pathways and diverse substrates such as glycerol, glucose, CO2, and uracil. Several reviews have described these in detail (Kumar et al., 2013a; de Fouchécour et al., 2018), and in this review we will focus only on the pathways for which glycerol is the substrate. Two pathways are known for conversion of glycerol into 3-HP: the CoA-dependent pathway and the CoA-independent pathway (Figure 2).
Figure 2.

Synthesis of 3-hydropropanoic acid from glycerol via the CoA-independent and -dependent pathways.
The CoA-dependent pathway has been most extensively studied in Lactobacillus reuteri and proceeds with conversion of glycerol to 3-hydroxypropanal (3-HPA) catalyzed by coenzyme B12-dependent glycerol hydratase (PduCDE). 3-HPA is subsequently converted to 3-HP via 3-hydroxypropanoyl-CoA and 3-hydroxypropanoyl-phosphate, catalyzed by the enzymes propionaldehyde dehydrogenase (PduP), phosphotransacylase (PduL), and propionate kinase (PduW), respectively (Dishisha et al., 2014).
In the CoA-independent pathway, glycerol is converted to 3-HP in 2 steps. As in the case of the CoA-dependent pathway, glycerol is first converted to 3-HPA, while in the second step, 3-HPA is converted directly to 3-HP in a reaction catalyzed by aldehyde dehydrogenase (Kumar et al., 2013a). While the CoA-independent pathway is by far the most employed for production of 3-HP using engineered bacteria, it appears to have little relevance in nature. This is possibly due to low activity of aldehyde dehydrogenase in wild type bacteria (Zhu et al., 2009).
Production Hosts
A number of microorganisms have been used for the production of 3-HP, both natural isolates as well as engineered microorganisms (Table 1). For the selection of an appropriate production host, there are several parameters to consider. The microorganism should demonstrate tolerance to organic acids, specifically 3-HP, as well as potentially toxic impurities in crude glycerol. As one of the widely used glycerol dehydratases is coenzyme B12-dependent, the production host should preferentially be capable of synthesizing coenzyme B12. Addition to the production medium instead of in situ synthesis would significantly increase the overall production cost.
Table 1.
Overview of bacterial species applied for production of 3-HP.
| Bacterial species | Natural producer | B12 synthesis |
|---|---|---|
| Lactobacillus reuteri | Yes | Yes |
| Klebsiella pneumonia | Yes | Yes |
| Escherichia coli | No | No |
| Pseudomonas denitrificans | No | Yes |
| Shimwellia blattae | Noa | Yes |
| Corynebacterium glutamicum | No | No |
| Bacillus subtilis | No | No |
| Synechococcus elongates | No | No |
Naturally produces 3-HPA.
Lactobacillus reuteri is capable of naturally producing 3-HP from glycerol via the CoA-dependent pathway (Luo et al., 2011). In one study, three L. reuteri strains were evaluated, and all were found to produce 3-HP (Burgé et al., 2015). It also was shown that 3-HPA is toxic at a concentration of 5 g/L while 2.5 g/L 3-HP is not (as long as pH is maintained above 5). Notably, L. reuteri is capable of synthesizing coenzyme B12 (Burgé et al., 2015) and has been used for the bioconversion of biodiesel-derived glycerol into 3-HP (14 g/L) and 1,3-propanediol (1,3-PDO) (Dishisha et al., 2015). To our knowledge, there have been no attempts to engineer L. reuteri strains to improve production. Among lactic acid bacteria, production of 3-HP is not restricted to L. reuteri. Of 67 lactic acid bacteria isolates tested, 22 isolates belonging to Lactobacillus diolivorans and Lactobacillus collinoides were positive for 3-HP production from glycerol (Garai-Ibabe et al., 2008). Additionally, L. diolivorans is capable of using biodiesel-derived glycerol for the production of 3-HPA (Lindlbauer et al., 2017).
Klebsiella pneumoniae is a pathogenic bacterium that has been widely used for production of 3-HP. Along with E. coli, K. pneumoniae is one of the most frequently used hosts for strain development by genetic engineering for improved 3-HP production. In fact, the highest 3-HP titer reported, 83.8 g/L, was obtained in K. pneumoniae by the combining of optimized expression of aldehyde dehydrogenase (K. pneumoniae PuuC), blocking of lactic acid synthesis (ldh1, ldh2, and pta mutant), and optimization of the fermentation conditions (Li et al., 2016). An important asset of K. pneumoniae is its capability of producing coenzyme B12 (Luo et al., 2011).
E. coli is commonly used for metabolic engineering to produce a wide variety of compounds. It has also been widely used as a chassis for the production of 3-HP. The highest 3-HP titer reported in E. coli so far is 71.9 g/L, using a strain in which, besides introduction of glycerol dehydratase and aldehyde dehydrogenase, the central metabolism was modified to reduce by-product formation (Chu et al., 2015). It has been indicated that careful choice of the strain could be of importance. A comparison of nine E. coli strains in which a heterologous pathway for 3-HP synthesis was introduced, demonstrated differences in 3-HP production, as well as in enzyme level and activity (Sankaranarayanan et al., 2014). A drawback of using E. coli is the fact that it is not naturally capable of producing coenzyme B12. However, insertion of the Pseudomonas denitrificans genes for coenzyme B12 synthesis (more than 25 genes in 6 operons) on three plasmids led to the production of coenzyme B12 under both anaerobic and aerobic conditions (Ko et al., 2014).
While K. pneumoniae and E. coli are by far the most widely used production hosts, a number of other bacteria have been engineered for 3-HP production as well. P. denitrificans synthesizes coenzyme B12 in aerobic conditions where NAD(P)+ is efficiently regenerated (Zhou et al., 2013). Introduction of glycerol dehydratase and glycerol dehydratase reactivase from K. pneumoniae allowed for the production of 3.4 g/L 3-HP. By further introducing the K. pneumoniae aldehyde dehydrogenase, the yield increased to 4.9 g/L (Zhou et al., 2013).
Another bacterium able to synthesize coenzyme B12, Shimwellia blattae, is a 1,3-PDO producer using a native coenzyme B12-dependent glycerol dehydratase. Introduction of various genes including aldehyde dehydrogenase from Pseudomonas putida KT2442 enabled the production of 0.26 g/L Poly-3HP using crude glycerol from biodiesel production as the substrate (Heinrich et al., 2013).
Corynebacterium glutamicum was engineered to produce 3-HP from glucose and xylose. By a combination of efforts, an impressive titer of 62.6 g/L 3-HP was obtained in a fed-batch fermentation (Chen et al., 2017). The steps taken to achieve this titer included enabling efficient production of glycerol from glucose, introduction of the K. pneumoniae pduCDEGH genes encoding diol dehydratase and its reactivase and Cupriavidor necator aldehyde dehydrogenase and modification in sugar uptake and glycolytic flux. In Bacillus subtilis, introduction of glycerol dehydratase along with its reactivases, and aldehyde dehydrogenase from K. pneumoniae, enabled the production of 3-HP from glycerol. Upon inactivation of glycerol kinase and optimization of growth conditions, a 3-HP titer of 10 g/L was obtained in a shake flask culture (Kalantari et al., 2017). A drawback of both of these bacteria is their inability to synthesize coenzyme B12, which thus has to be supplemented to the production medium.
In an engineered cyanobacterium Synechococcus elongatus CO2 was converted via glycerol to 3-HP, albeit at very low titer (31.7 mg/L). Synechococcus elongatus is unable to synthesize coenzyme B12, but the problem was alleviated by producing 3-HP under anaerobic conditions using the oxygen-sensitive B12-independent glycerol dehydratase from Clostridium butyricum (Wang et al., 2015). The use of glycerol as an additional carbon source in a strain engineered to assimilate glycerol has been suggested for increasing productivity (Kanno and Atsumi, 2017).
The Suite of Enzymes Used in Heterologous 3-HP Synthesis Pathways
As mentioned above, there are two principal pathways for production of 3-HP from glycerol. In both CoA-dependent and -independent pathways, the initial step is the conversion of glycerol to 3-HPA. In the CoA-dependent pathway, 3-HPA is converted via 3-hydroxypropanoyl-coenzyme A (3-HP-CoA) and 3-hydroxypropanoyl-phosphate (3-HP-P) to 3-HP, catalyzed by propionaldehyde dehydrogenase, phosphotransacylase, and propionate kinase, respectively (Dishisha et al., 2014). In the CoA-independent pathway, 3-HPA is converted directly to 3-HP by the action of aldehyde dehydrogenase (Kumar et al., 2013a). Both of the pathways have characteristics that are of importance for the design of cell factory and process conditions, for example, oxygen-sensitivity of enzymes and the need for co-factors such as coenzyme B12, and NAD+. In the following, we will describe the enzymes that make up these two pathways. Where available, kinetic data for the enzymes is presented in Table 2, to facilitate comparison.
Table 2.
Summary of characterized enzymes of relevance in 3-HP production.
| Source | gene / protein | KM (mM) | Vmax (U/mg) | kcat (s−1) | kcat/KM x 103 (M−1S−1) | Other characteristics | References |
|---|---|---|---|---|---|---|---|
| B12-dependent glycerol/diol dehydratase | |||||||
| K. pneumoniae | dhaB123 | n.a. | n.a. | n.a. | n.a. | ||
| K. pneumoniae | pduCDE | n.a. | n.a. | n.a. | n.a. | ||
| L. reuteri | pduCDE | 0.60 | 130.1 | n.a. | n.a. | pHopt 8.5 | Qi et al., 2006 |
| C. freundii | gldABC | 0.59 | 142.9 | n.a. | n.a. | pHopt 8.5 | Qi et al., 2006 |
| L. brevis | dhaB | n.a. | n.a. | n.a. | n.a. | pHopt 7.0 | Kwak et al., 2013 |
| B12-independent glycerol dehydratase | |||||||
| C. butyricum | n.a. | n.a. | n.a. | n.a. | B12-independent | ||
| Propionaldehyde dehydrogenase | |||||||
| L. reuteri | pduP | 1.18 | 0.35 | n.a. | n.a. | broad substrate specificity | Luo et al., 2011 |
| n.a. | n.a. | n.a. | n.a. | Inhibition by 3-HPA (from 7 mM). NAD+ preference | Sabet-Azad et al., 2013 | ||
| Aldehyde dehydrogenase | |||||||
| E. coli K-12 | aldH | n.a. | n.a. | 28.5 | 58.6 | NAD+ preference | Jo et al., 2008 |
| n.a. | 38.16 | n.a. | 57.28 | Chu et al., 2015 | |||
| 0.49 | 38.10 | n.a. | n.a. | Ko et al., 2012 | |||
| E. coli JM109 | aldA | 0.31 | 28.4 | n.a. | n.a. | NAD+ preference | Zhu et al., 2009 |
| C. necator | gabD4 | n.a. | 55.12 | n.a. | 71.48 | Chu et al., 2015 | |
| C. necator | gabD4 E209Q/E269Q | n.a. | 78.07 | n.a. | 162.34 | Chu et al., 2015 | |
| K. pneumoniae DSM 2026 | puuC | 0.48 | 22.25 | 19.81 | 41.44 | pHopt 8.0 | Raj et al., 2010 |
| 0.48 | 27.73 | n.a. | n.a. | Ko et al., 2012 | |||
| A. brasilense | KGSADH | 1.6 | n.a. | 15 | 9) | NAD+ preference | Park et al., 2017 |
| A. brasilense | KGSADH 108-QR | 0.17 | n.a. | 6 | 35 | NAD+ preference | Park et al., 2017 |
| Aldehyde oxidase | |||||||
| Pseudomonas sp. AIU 362 | aloD | 6.7 | 41.8 | n.a. | n.a. | NAD+-independent | Li et al., 2014 |
Glycerol and Diol Dehydratase
Coenzyme B12-Dependent Dehydratases
Coenzyme B12-dependent glycerol dehydratase (EC 4.2.1.30) and diol dehydratase (EC 4.2.1.28) are isofunctional enzymes that catalyse the dehydration of 1,2-diols to the corresponding aldehyde (e.g., glycerol to 3-HPA). Both glycerol dehydratase and diol dehydratase are composed of three subunits that form a dimer of a heterotrimer (α2β2γ2; Shibata et al., 1999; Yamanishi et al., 2002). In mycobacteria, the α and β subunits are fused in a single polypeptide (Liu et al., 2010). Both of the enzymes catalyze a radical process in which an adenosyl radical is formed by homolytic cleavage of the Co-C bond in coenzyme B12 (Daniel et al., 1998). The adenosyl radical abstracts a hydrogen from the substrate, glycerol, to generate a substrate radical. The substrate radical, upon rearrangement, re-abstracts a hydrogen to form the final product and regenerate coenzyme B12 (Daniel et al., 1998). If a radical side reaction takes place, coenzyme B12 is not regenerated, and instead, a catalytically inactive cobalamin species is formed, which binds tightly to the enzyme and thereby inactivates it (Toraya, 2003). Radical side reaction can be induced by both glycerol and oxygen, making the enzyme sensitive to oxygen (Wei et al., 2014). The inactive hydratase can be re-activated by the action of glycerol/diol dehydratase reactivase in the presence of ATP and intact coenzyme B12 (Mori and Toraya, 1999). Glycerol/diol dehydratase reactivase consists of two subunits that form a heterotetramer (α2β2; Liao et al., 2003). The reactivase binds ATP and hydrolyzes it to ADP. The ADP-bound reactivase forms a complex with the inactivated glycerol dehydratase. This leads to release of the damaged coenzyme B12. Subsequently, reactivase binds ATP and is released from the hydratase, which in turn binds coenzyme B12 to regenerate the active form of the enzyme (Toraya, 2003).
From the above, it can be deduced that the expression of five proteins is necessary for the efficient catalysis of glycerol to 3-HPA. In the context of engineering microorganisms for 3-HP production, the most widely used glycerol dehydratase is that of K. pneumoniae (dhaB123, and gdrAB), which has been used in E. coli (Rathnasingh et al., 2009), K. pneumoniae (Wang et al., 2013), B. subtilis (Kalantari et al., 2017), and S. elongatus (Wang et al., 2015). The use of other dehydratases has also been reported. In E. coli, the glycerol dehydratase from Lactobacillus brevis encoded by dhaB123 and its reactivase dhaR12 were used (Kwak et al., 2013). The diol dehydratase (pduCDE) and activator (pduGH) from K. penumoniae were used in C. glutamicum (Chen et al., 2017).
Considering the problem of enzyme instability, it is somewhat surprising that relatively few glycerol dehydratases have been tested. These enzymes have, by now, been discovered in many bacteria and this should provide a resource for enzyme discovery endeavors. In fact, for three selected glycerol dehydratases, the α subunit was systematically swapped, which led to identification of several combinations with improved stability and activity (Qi et al., 2006). Interestingly, fusion of the α and β subunit of K. pneumoniae glycerol dehydratase led to an increase in the kcat (albeit with concomitant increase in KM; Wang et al., 2009). As mentioned, mycobacteria contain a glycerol dehydratase variant in which the α and β subunits are fused, but to our knowledge they have not been characterized or used for 3-HP production. Enzyme engineering could also prove beneficial for generation of enzyme variants with improved properties such as increased stability and activity. As an example, by rational engineering, improved resistance to mechanism-based inactivation was conferred to a glycerol dehydratase from Klebsiella oxytoga (Yamanishi et al., 2012).
Coenzyme B12-Independent Glycerol Dehydratase
This class of glycerol dehydratases is of particular interest since it negates the need for the rather costly coenzyme B12. This enzyme also performs a radical catalysis, but instead of coenzyme B12, it uses S-adenosylmethionine as a co-factor (Raynaud et al., 2003). This glycerol dehydratase is a homodimer and it is thus structurally simpler than its coenzyme B12-dependent counterpart that is composed of three subunits (Raynaud et al., 2003). While it would seem the obvious enzyme of choice, it is important to note that it is extremely sensitive to oxygen and its use requires production under strict anaerobic conditions (Raynaud et al., 2003).
Considering the fact that coenzyme B12 is not produced in most of the microorganisms that have been used as potential 3-HP production hosts, the use of a coenzyme B12-independent dehydratase would be desirable. So far, its use has only been reported in a few studies. The C. butyricum glycerol dehydratase was used in the cyanobacterium S. elongatus for production of 3-HP under anaerobic conditions. As expected, it was not functional under aerobic conditions (Wang et al., 2015). In case of E. coli, the coenzyme B12-independent dehydratase was used in the context of 1,3-PDO production. Although the reported 1,3-PDO production was relatively low, an accumulation of 3-HPA was observed, thus demonstrating functionality of the glycerol dehydratase (Dabrowski et al., 2012). Furthermore, it was used for engineering E. coli to produce poly (3-Hydroxypropanoate) (Andreeßen et al., 2010). The scarcity of reported use of this enzyme could be due to difficulties in engineering 3-HP producing strains using this dehydratase. Alternatively, and perhaps more likely, is that reports are scarce because of the requirement of strict anaerobic conditions for the enzyme to be functional, which is not compatible with growth of most production microorganisms.
Enzymes for Converting 3-HPA to 3HP
Enzymes in the CoA-Dependent Pathway
In the CoA-dependent pathway, 3-HPA is converted via 3-HP-CoA and 3-HP-P into 3-HP. This requires three enzymes: propionaldehyde dehydrogenase, phosphotransacylase, and propionate kinase, respectively (Dishisha et al., 2014). The propionaldehyde dehydrogenase requires the cofactor NAD+, while one ATP is generated in the conversion of 3-HP-P to 3-HP. This pathway has been described in several organisms including L. reuteri and K. pneumoniae (Luo et al., 2012; Dishisha et al., 2014). The propionaldehyde dehydrogenase PduP from L. reuteri has been purified and characterized in vitro and shown to exhibit activity toward a broad spectrum of aldehydes, including 3-HPA, using either NAD+ (preferred) or NADP+ as a cofactor (Luo et al., 2011). Lactobacillus reuteri PduP was also reported to display substrate inhibition for 3-HPA (at 7 mM; Sabet-Azad et al., 2013).
The CoA-dependent pathway has been used in several engineered bacteria. The K. pneumoniae pduPLW genes were introduced in an E. coli strain harboring the K. pneumoniae glycerol dehydratase. Here they were shown to be functional (although the obtained titer was lower than that obtained using an aldehyde dehydrogenase from Azospirillum brasilense; Honjo et al., 2015). In K. pneumoniae, overexpression of the first gene in the pathway, pduP, led to a 4-fold (0.72 g/L) increase in the 3-HP titer (Luo et al., 2011). It could be speculated that the native K. pneumoniae CoA-dependent pathway enzymes were responsible for the conversion of 3-HP-CoA to 3-HP, and that over-expression of all three genes in the pathway would further increase the titer.
It remains to be seen whether the CoA-dependent pathway could be an alternative to the more widely used aldehyde dehydrogenases for the conversion of 3-HPA to 3-HP. Although both pathways use an NAD(P)+, ATP is generated in the CoA-dependent pathway. However, this should be balanced against the metabolic burden of over-expressing three enzymes instead of one. Further, it is unclear if the substrate inhibition exhibited by L. reuteri PduP at a relatively low concentration of 3-HPA is a general trait of propionaldehyde dehydrogenase, or whether a more robust enzyme can be discovered or engineered.
Aldehyde Dehydrogenase
Aldehyde dehydrogenase (EC 1.2.1.3) has decidedly been the enzyme of choice for conversion of 3-HPA to 3-HP in engineered cells. It catalyses the conversion of an aldehyde to its corresponding carboxylic acid using NAD(P)+ as a co-factor. In the context of the 3-HP production pathway, it is of utmost importance that enzyme activity is sufficient to ensure that no accumulation of the toxic intermediate 3-HPA takes place. Aldehyde dehydrogenases are present in most organisms, but generally their activity is not high enough to sustain a high production of 3-HP (Raj et al., 2008; Zhu et al., 2009). Consequently, many studies have focused on discovering suitable aldehyde dehydrogenases. Earlier studies mainly employed the aldehyde dehydrogenase AldH from E. coli K-12 in both, E. coli and K. pneumoniae (Raj et al., 2008; Zhu et al., 2009). In a study by Chu and co-workers, an indirect comparison (based on 3-HP production in E. coli) was made amongst 17 aldehyde dehydrogenases from various organisms, benchmarked against E. coli AldH. One aldehyde dehydrogenase performing better than AldH, GabD4 from C. necator, was identified (Chu et al., 2015). The enzyme was further improved by enzyme engineering, substantially increasing its Vmax and kcat/KM, and used for high-titer production of 3-HP in both E. coli (71.9 g/L) and C. glutamicum (62.6 g/L) (Chu et al., 2015; Chen et al., 2017).
The α-ketoglutaric semialdehyde dehydrogenase (KGSADH) from A. brasilense has also been used in several studies. KGSADH was evaluated against E. coli AldH and PuuC from K. pneumoniae (Ko et al., 2012). This study indicated higher activity (Vmax) of KGSADH in extracts of the K. pneumoniae strain, while in contrast it had the lowest Vmax and affinity for 3-HPA when enzymes purified from E. coli were evaluated (Ko et al., 2012). Whether this discrepancy is due to e.g., stability in vivo is unclear. The structure of KGSADH has been solved and has provided a basis for rational enzyme engineering. This has yielded enzyme variants with improved activity toward 3-HPA (Park et al., 2017; Son et al., 2017; Seok et al., 2018).
PuuC from K. pneumoniae has also been used in several production hosts, including K. pneumoniae and B. subtilis (Li et al., 2016; Kalantari et al., 2017). Substrate specificity was evaluated against 3-HPA and three other aldehydes indicating broad substrate specificity (and lowest activity when 3-HPA was used; Raj et al., 2010). Comparative studies indicated that E. coli AldH and C. necator GabD4 E209Q/E269Q were better alternatives for converting 3-HPA to 3-HP (Huang et al., 2012; Chen et al., 2017). Nevertheless, PuuC was successfully used in an engineered K. pneumoniae strain that produced 3-HP with a titer of 83.8 g/L (highest reported) in a bioreactor (Li et al., 2016).
Several other aldehyde dehydrogenases have been tested in the context of 3-HP production. E. coli YneI was shown to selectively target 3-HPA over other aldehydes (Luo et al., 2013), although it was not tested against succinic semialdehyde which was previously reported as its primary substrate (Kurihara et al., 2010). The aldehyde dehydrogenase DhaS from B. subtilis has also been suggested as an aldehyde dehydrogenase that is specific for 3-HPA (Su et al., 2015).
Due to the oxygen-sensitivity of glycerol dehydratase, 3-HP production is often performed under anaerobic or microaerophilic conditions, where the regeneration of NAD+ is diminished. Thus, efficient 3-HP production becomes a compromise between assuring optimal glycerol dehydratase activity (low oxygen level) and NAD+-dependent aldehyde dehydrogenase activity (high oxygen level). An interesting strategy was suggested to overcome the limitation imposed by NAD+, namely, the use of an aldehyde oxidase in place of dehydrogenase. Li and co-workers characterized the NAD+-independent aldehyde oxidase from Pseudomonas sp. AIU 362 and found it to exhibit a broad substrate specificity but relatively low affinity for 3-HP. When expressed in K. pneumoniae, a relatively low 3-HP titer was obtained (Li et al., 2014). As can be seen in Table 2, the affinity of aldehyde oxidase to 3-HPA is several-fold lower than the described aldehyde dehydrogenases, with a KM in the range where 3-HPA becomes toxic to the cell. Considering the importance of maintaining a low concentration of 3-HPA, better enzymes would likely be needed to successfully exploit aldehyde oxidase for 3-HP production. To the best of our knowledge, no other attempts have been made at identifying a more suitable aldehyde oxidase for the conversion of 3-HPA to 3-HP.
As outlined in this section, many enzyme variants have been used in the design of 3-HP-producing strains. While some of the enzymes have been characterized biochemically, it is obvious from Table 2 that this is not a norm in the field. Nevertheless, a better understanding of the catalytic properties of these enzymes could provide a platform for more optimal selection of enzymes for constructing 3-HP pathways.
Strain Engineering
There are several parameters to consider when engineering bacteria to produce 3-HP. As covered in sections Production Hosts and The Suite of Enzymes Used in Heterologous 3-HP Synthesis Pathways, a suitable host should be established and in most cases some or all of the enzymes needed for converting glycerol to 3-HP should be introduced. This provides a basis for 3-HP production, but is not enough to ensure the desired high titers. There are several hurdles that need to be overcome, and these include, but are not limited to, proper balancing of enzyme activities to prevent buildup of the toxic intermediate 3-HPA, efficient channeling of substrate into product, co-factor regeneration, prevention of by-product formation and countering of stress. In the following, we will explore challenges and strategies that have been applied in the engineering of strains for 3-HP production.
Improving 3-HP Production by Optimizing Expression of the Production Pathway
As illustrated above, a number of different enzymes have been used for the construction of synthetic operons to confer the ability to produce 3-HP to various bacteria. In a number of these studies, it was shown that further optimization of the expression of the pathway genes could enhance 3-HP production.
A critical parameter is to ensure that the intermediate product 3-HPA is kept at a low concentration. 3-HPA (also known as reuterin) is a broad range antimicrobial compound. Its minimal inhibitory concentrations for various bacteria are in the range of <1.9–50 mM, and it exerts its effect via modification of thiol groups of proteins and small molecules (Cleusix et al., 2007; Schaefer et al., 2010). Considering the fact that 3-HPA is toxic to bacteria even at minute concentrations, it is evident that balancing enzyme activities to prevent 3-HPA accumulation is critical. To this end, different approaches have been attempted to fine-tune the expression levels of glycerol dehydratase and aldehyde dehydrogenase. The order in which the genes encoding aldehyde dehydrogenase and glycerol dehydratase are arranged was shown to impact 3-HP production. The rationale here is that the gene adjacent to the promoter in an operon is normally more expressed. Specifically, the arrangement favoring aldehyde dehydrogenase expression led to a higher 3-HP production due to the diminished build-up of 3-HPA (Li et al., 2013a). Different promoters for driving the expression of aldehyde dehydrogenase gene has also been successfully evaluated (Li et al., 2016).
While these approaches are simple, they do not offer a high degree of tunability, and a number of more advanced approaches have been applied for optimization of expression levels. Optimization of expression level can be done by applying various 5′ untranslated regions (5′UTRs) in the genetic constructs, which leads to differences in the translation rate. Using this approach, a better balance of the enzyme activities was obtained by fine-tuning expression of dhaB1 encoding a glycerol dehydratase subunit. Testing only four different UTRs led to construction of a strain that enabled a 2.4-fold improvement of 3-HP titer in shake flask experiments (Lim et al., 2016). Several tools are available for the prediction of translation rate of 5′UTRs, including the UTR Designer (Seo et al., 2013), and the RBS Calculator v2.0 (Espah Borujeni et al., 2014). To the same end, a study showed potential in tuning translation by modulation of the Shine-Dalgarno sequence for which an online tool, EMOPEC, is available (Bonde et al., 2016). For a selection of 106 Shine-Dalgarno sequences, the measured protein level was within 2-fold of the predicted level in 91 % of cases (Bonde et al., 2016). While in silico prediction of expression level is a very attractive venue, it should be noted that even for the good predictors, the number of false predictions appear to be significant (Bonde et al., 2016). Nevertheless, these computational approaches should allow for design and evaluation of smaller, more focused libraries, compared to random mutagenesis libraries.
In most studies, researchers have taken advantage of plasmids for the expression of relevant genes in the production host. While these studies provide proof of principle, they might not be optimal for industrial production due to the well-known problem of plasmid instability in bioreactors (Gao et al., 2014). Consequently, the generation of genetically stable strains where expression cassettes are integrated into the genome will be a necessity. Additionally, the elimination of plasmids might improve the production via a reduced metabolic burden associated with plasmid maintenance and replication (Silva et al., 2012). To construct a plasmid-free strain, the E. coli aldH was inserted in the genome of K. pneumoniae. While 3-HP production was improved (over parent strain) the 3-HP titer upon production in shake flasks was low (Wang and Tian, 2017). In B. subtilis, a plasmid-free strain allowed the production of 3-HP in shake flasks with a titer of 10 g/L (Kalantari et al., 2017).
Considering that both multicopy vectors and strong promoters have been used for the expression of relevant genes, one foreseeable difficulty in shifting to a single (or a few) genome-integrated expression cassette will be to ensure sufficient protein synthesis in the cell. A recent study described an approach based on translational coupling between the gene of interest and an antibiotic resistance cassette, thus making a high rate of translation selectable via an increased resistance to an antibiotic (Rennig et al., 2018). With limited screening, the production of two selected proteins (a nanobody and an affibody) in E. coli was improved 2- and 10-fold, respectively (Rennig et al., 2018). The methodology was shown to also work in Gram-positive bacteria B. subtilis and Lactococcus lactis where production of a sialidase and a tyrosine ammonia lyase was improved 2- and 8-fold, respectively (Ferro et al., 2018).
Engineering Host Metabolism for Improving 3-HP Production
Besides integrating an efficient pathway for the conversion of glycerol to 3-HP, it is often necessary to further modulate the host metabolism, in order to direct the substrate into the production pathway more efficiently, eliminate unwanted by-products and/or reduce stress.
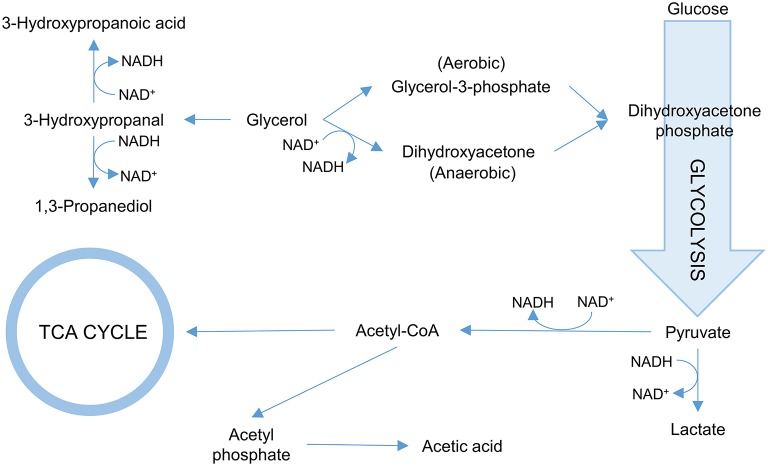
One of the more common approaches is to ensure that glycerol is converted more efficiently into 3-HP. To this end, targeting the first step in the glycerol utilization pathways is common. In E. coli, two pathways for glycerol utilization exist. Glycerol is converted to the glycolytic metabolite dihydroxyacetone phosphate either via glycerol-3-phosphate catalyzed by glycerol kinase under aerobic conditions or via dihydroxyacetone catalyzed by glycerol dehydrogenase under anaerobic conditions (Durnin et al., 2009; Figure 3). Inactivating the gene glpK (which encodes glycerol kinase in E. coli) increased the titer of 3-HP 1.6-fold in an aerobic fed batch fermentation when co-fed with glucose and glycerol. Under the conditions used (aerobic), further inactivating the gene encoding glycerol dehydrogenase had no effect (Kim et al., 2014). Similarly, the 3-HP titer increased in B. subtilis co-fed on glycerol and glucose upon inactivation of glpK (Kalantari et al., 2017). Inactivation of glpK is possible in process setups where glycerol is not the sole carbon source. When cells grow only on glycerol, targeting glpK becomes less trivial as the fluxes toward biomass accumulation and 3-HP production need to be balanced. In E. coli, the glpK was placed under control of an inducible promoter and fine-tuning of its expression led to an increase in both 3-HP titer and yield on glycerol (Jung et al., 2014). In that study, the gene glpF encoding glycerol facilitator was also overexpressed to increase the influx of glycerol in the cell, which led to a modest increase in 3-HP production (Jung et al., 2014). Interestingly, the opposite approach was evaluated by Su and co-workers. They overexpressed glycerol dehydrogenase in K. pneumoniae to stimulate growth, and found that this strain not only exhibited faster initial growth and similar final biomass yield while using less glycerol, but also exhibited increased 3-HP production (Su et al., 2014).
Figure 3.
Overview of the glycerol metabolism in bacteria. Reactions consuming NADH/NAD+ are indicated.
In natural 3-HP producers such as K. pneumoniae and L. reuteri, 3-HPA can be oxidized to 3-HP and reduced to 1,3-PDO (Zhu et al., 2009; Dishisha et al., 2014). The conversion of 3-HPA to 1,3-PDO is catalyzed by an oxidoreductase. In K. pneumoniae, the most important oxidoreductase appears to be the 1,3-propanediol reductase encoded by dhaT. However, significant amounts of 1,3-PDO are still produced in the dhaT mutant, indicating that other oxidoreductases in the cell can also catalyse this reaction (Ko et al., 2012). In a subsequent attempt to eliminate 1,3-PDO formation, four oxidoreductases were inactivated, but the quadruple mutant still retained the ability to produce 1,3-PDO (Ko et al., 2015). With respect to cofactor balance, 3-HP synthesis requires NAD+ while 1,3-PDO formation requires NADH and thus regenerates NAD+. Thus, the elimination of 1,3-PDO formation adversely affects the NAD+/NADH balance, and in fact, it was reported that the dhaT mutation under 3-HP production conditions impeded cell growth (Ko et al., 2012). In E. coli, 3-HPA is also converted to 1,3-PDO by the action of a broad-range aldehyde oxidoreductase encoded by yqhD. While the deletion of yqhD dramatically reduces 1,3-PDO formation, it is also here not fully abolished, likely due to the presence of alternate oxidoreductases (Tokuyama et al., 2014).
Other important by-products in 3-HP production are lactate and acetate. The deletion of K. pneumoniae ldhA encoding lactate dehydrogenase was reported to eliminate lactate formation (Kumar et al., 2013b). Another study reported a reduction in lactate formation upon inactivation of ldh1 and ldh2 in K. pneumoniae (Li et al., 2016). Likewise, to reduce the acetate formation, the synthesis gene pta was inactivated, leading to a reduction but not elimination of acetate formation in K. pneuomoniae (Li et al., 2016). To further optimize 3-HP producing cell factories, it is essential to assure that by-product formation is reduced to a minimum. Besides the obvious loss of carbon that could have otherwise been used for biomass and product formation, it also leads to a more complex fermentation broth thus potentially increasing costs associated with downstream purification.
The NAD+/NADH balance is of importance for the 3-HP production due to the NAD+-dependence of the employed aldehyde dehydrogenases. The synthesis of by-products 1,3-PDO and lactate also generates NAD+. Elimination of respective pathways thus further skews the NAD+/NADH balance (Figure 3). Under aerobic conditions, NAD+ is also regenerated by the electron transport chain. However, due to the oxygen-sensitivity of glycerol dehydratase, 3-HP production is often performed under microaerophilic or anaerobic conditions where NAD+ regeneration is reduced. To counter these effects, various enzymes can be used to modulate the NAD+/NADH balance. In the context of 3-HP production, over-expression of the genes encoding either NADH oxidase, NADH dehydrogenase, or glycerol-3-phosphate dehydrogenase in K. pneumoniae showed potential in stimulating 3-HP production via regenerating the NAD+ pool in all cases (Li et al., 2013b).
From an economical point of view, it would be beneficial to use a production strain capable of synthesizing coenzyme B12. However, even in K. pneumoniae which is capable thereof, it has been reported that addition of coenzyme B12 increases the glycerol dehydratase activity (Ashok et al., 2013). This could indicate that upregulation of coenzyme B12 synthesis is another parameter for increasing the glycerol dehydratase activity. Studies on Bacillus megaterium and more recently on P. denitrificans show good potential for increasing coenzyme B12 synthesis, e.g., by removing the riboswitch-based feedback inhibition system (Biedendieck et al., 2010; Nguyen-Vo et al., 2018).
The application of genome scale metabolic models is a common approach in the field of metabolic engineering and has been used successfully in multiple studies. Using a genome-scale metabolic model for K. pneumoniae, no single mutants that would improve 3-HP production were predicted. Instead, a double knockout of tpi (triose phosphate isomerase) and zwf (glucose-6-phosphate-1-dehydrogenase) involved in central metabolism was suggested. The introduction of these two mutations led to a 4.4-fold increase in the 3-HP yield compared to the parent strain (Tokuyama et al., 2014). Predictions were made of beneficial mutations in B. subtilis, but here only the more obvious candidate, glycerol kinase, was suggested as a beneficial mutation (Kalantari et al., 2017).
Another important aspect of optimizing the production strain is to improve the robustness of the strain, specifically, alleviating stress. Probably the most significant stressor is that imposed by the intermediate product 3-HPA and, to a lesser extent, the product 3-HP. Warnecke et al. developed a method to screen for regions and genes that could be involved in the tolerance to 3-HP. Starting with a genomic library of E. coli clones, each carrying a plasmid with a genomic DNA insert, they selected for increased tolerance to 3-HP and subsequently quantified the enriched plasmid DNA by microarray analysis (Warnecke et al., 2010). Using this method, they concluded that 3-HP inhibition is due to the limitations in the chorismate and threonine pathways. Surprisingly, they found that over-expression of almost any of the genes in the pathways dramatically alleviated the stress imposed by 3-HP (Warnecke et al., 2010). In a follow up study, the authors identified a 21-amino acid peptide that, when expressed, increased the 3-HP tolerance about 2.3-fold (Warnecke et al., 2012).
The potential of using omics data to identify the targets for improving 3-HP tolerance has been demonstrated. Using 2D gel-based proteomics, Liu and co-workers identified 46 up- and 23 down-regulated proteins upon challenging E. coli with 5 g/L of 3-HP. Over-expression of several of these proteins alleviated the stress imposed by 3-HP (Liu et al., 2016).
In recent years, there have been key improvements in the technology for generating targeted libraries of mutants, which is exemplified in a study by Liu and co-workers where a method termed iCREATE for iterative genome editing was used to generate 162,000 mutations in 115 genes (Liu et al., 2018). Using this approach, the production of 3-HP via the Malonyl-CoA pathway was increased 60-fold (Liu et al., 2018). However, for its successful employment, appropriate screening and/or selection techniques need to be in place. Small molecule biosensors based on transcription factors or riboswitches can couple metabolite concentration to a measurable output such as fluorescence for selection or selectable output such as antibiotic resistance (Liu et al., 2017). It is, thus, of particular importance that a 3-HP-inducible system was identified in P. putida recently and demonstrated to be functional when expressed in E. coli (Hanko et al., 2017). This biosensor has already found application for screening in the directed evolution of the aldehyde dehydrogenase KGSADH which improved the catalytic efficiency of the enzyme 2.8-fold (Seok et al., 2018).
Combining approaches such as iCREATE for generating diversity, and biosensors for detection or selection of improved variants should hold high promises for moving bacterial 3-HP production strains to industrially relevant levels of production.
Process Engineering for Optimizing 3-HP Production
In a majority of the studies reviewed for this paper, the ability of the recombinant strains to produce 3-HP was tested in 1.5–5 L bioreactors operated in fed-batch mode (Table 3; Supplementary Table 1). A fed-batch operation typically entails growing the cells until the end of the exponential phase. This is followed by the addition of feed medium containing the substrates, thus keeping the growth rate of the microorganism at a desired level (Liu, 2017). A fed-batch mode of operation has certain advantages over batch fermentation. For instance, a fed-batch operation has the ability to prevent overflow metabolism, that is, the phenomenon where fast-growing cells, in the presence of oxygen, utilize fermentation instead of respiration to produce energy (Basan et al., 2015). Another advantage of a fed-batch operation is its ability to maintain the substrate concentration at a low level (de Fouchécour et al., 2018). This is particularly important when the substrate is glycerol. It was reported that no growth of K. pneumoniae cells could be seen above 110 and 133 g/L glycerol (extrapolated values) in aerobic and anaerobic conditions, respectively, with glycerol concentrations higher than 40 g/L showing significant growth inhibition (Cheng et al., 2005). Consequently, high titers of 3-HP cannot be obtained from glycerol in a batch fermentation. In L. reuteri, the conversion of glycerol to 3-HPA was reported to be about 10 times faster than the subsequent conversion of 3-HPA to 3-HP and 1,3-PDO, resulting in the accumulation of the toxic intermediate 3-HPA. To circumvent this problem, a fed-batch fermentation process was established. It produced 3-HP and 1,3-PDO without any accumulation of 3-HPA. This was achieved by identifying a maximum value of specific glycerol consumption rate at which only 1,3-PDO and 3-HP are produced, and no accumulation of 3-HPA takes place. By maintaining the glycerol amount in the bioreactor such that its specific consumption rate stayed under this maximum value, a 3-HP titer of 14 g/L with a productivity of 0.25 g/L.h was obtained (Dishisha et al., 2014, 2015). This underlines how the knowledge of cell metabolism can be used to solve production problems by means of process engineering.
Table 3.
Summary of studies reporting the highest 3-HP titer, yield, and productivity in various production hosts.
| Strain | Carbon source | Fermentation conditions | Titera | Yielda | Produc-tivitya | Reference |
|---|---|---|---|---|---|---|
| K. pneumoniae Δldh1Δldh2Δpta_pTAC-puuC | Glycerol | 5-L bioreactor, fed-batch fermentation, pH 7.0, 37°C, 1.5 vvm air, 400 rpm | 83.8 | n.a. | 1.16 | Li et al., 2016 |
|
K. pneumoniae Kp4 ΔldhAΔdhaT_ lac-aldH |
Glycerol | 5-L bioreactor, fed-batch fermentation, pH 7.0, 37°C, 2.2 L/min air, 450 rpm | 61.9 | 0.58 | 1.62b | Jiang et al., 2018 |
| E. coli W3110 ΔackA-ptaΔyqhD_ T7-dhaB-gdrAB-gabD4 | Glycerol | 5-L bioreactor, fed-batch fermentation, pH 7.0, 35°C, 1 vvm air, 500 rpm, vitamin B12 added externally | 71.9 | n.a. | 1.8 | Chu et al., 2015 |
| E. coli W ΔackA-ptaΔyqhD_tac-dhaB-gdrAB-KGSADH | Glycerol and glucose | 5-L bioreactor, fed-batch fermentation, pH 7.0, 37°C, 1 vvm air, 500 rpm, coenzyme B12 added externally | 40.5 | 0.97 g/g | 1.35 | Lim et al., 2016 |
| L. reuteri RPRB3007c | Glycerol | 3-L bioreactor, anaerobic fed-batch fermentation, pH 7.0, 37°C, 500 rpm | 10.6 | n.a. | 1.08 | Dishisha et al., 2014 |
|
B. subtilis 168 trp+ ΔglpK_pHyperspank-dhaB-gdrAB-puuC |
Glucose | Shake flask, 37°C, 200 rpm, coenzyme B12 added externally | 10 | 0.79 g/g | n.a. | Kalantari et al., 2017 |
| L. reuteri DSM 20016 and G. oxydans DSM 50049d | Glycerol | 3-L bioreactor, Step 1: anaerobic fed-batch fermentation, pH 5.5, 37°C, 200 rpm. Step 2: aerobic batch fermentation, pH 5.5, 28°C, 1 L/min air, 800 rpm. | 23.6 | 0.98 | n.a. | Dishisha et al., 2015 |
| K. pneumoniae and G. oxydanse | Glycerol | 7-L bioreactor, fed-batch fermentation, Step 1: pH 7.0, 37°C, 0.2 vvm air, 150 rpm. Step 2: pH 5.5, 28°C, 0.5 vvm air, 600 rpm. | 60.5 | 0.51 | 1.12 | Zhao et al., 2015 |
Unless otherwise mentioned, the units of titer, yield, and productivity are g/L, mol3−HP/molGlycerol and g/L.h, respectively
Calculated based on the published data
Mutated catabolite repression element (CRE) in the upstream region of the pdu operon
L. reuteri converted glycerol to 3-HP and 1,3-propanediol followed by G. oxydans converting 1,3-propanediol to 3-HP
K. pneumoniae converted glycerol to 1,3-propanediol followed by G. oxydans converting 1,3-propanediol to 3-HP
adhE, alcohol dehydrogenase; aldA, aldehyde dehydrogenase A (E. coli); aldH, γ-glutamyl-γ-aminobutyraldehyde dehydrogenase (E. coli); aldHk, NAD+-dependent homolog of E. coli aldH; CRE, catabolite repression element; dhaB, glycerol dehydratase; dhaR, dhaB reactivating factor in L. brevis; dhaS, putative aldehyde dehydrogenase from B. subtilis; dhaT, 1,3-propanediol oxidoreductase; DO, dissolved oxygen; frdA, succinate dehydrogenase; gabD4, aldehyde dehydrogenase (Cupriavidus necator); gdrA, gdrB, glycerol dehydratase reactivase; glpF, glycerol uptake facilitator protein; glpK, glycerol kinase; KGSADH, α-ketoglutaric semialdehyde dehydrogenase (A. brasilense); ldhA, lactate dehydrogenase; n.a. , not available; pduP, propionaldehyde dehydrogenase; puuC, γ-glutamyl-γ-aminobutyraldehyde dehydrogenase (K. pneumoniae); yqhD, NADPH-dependent aldehyde reductase/alcohol dehydrogenase.
The challenge concerning the requirement of anaerobic conditions for coenzyme B12 generation and aerobic conditions for efficient NAD+ regeneration for production of 3-HP from glycerol was tackled by electro-fermentation in a recently published study (Kim et al., 2017). Certain microorganisms possess the ability to deliver electrons to solid electrodes when grown under anaerobic conditions. These exoelectrogens, as these microorganisms are called, can be used to control the redox state in the cells independent of the electron transport chain or fermentation. Electro-fermentation utilizes this property of exoelectrogens to drive the unbalanced fermentations. Anodic electro-fermentation has been used for bioconversion of glycerol to 3-HP using recombinant K. pneumoniae over-expressing KGSADH from A. brasilense. An electrical potential of + 0.5 V vs. Ag/AgCl was applied to the anode, with 2-hydroxy-1,4-naphthoquinone used for shuttling the electrons between the bacteria and the anode. The transfer of electrons from the bacteria to the anode during anaerobic fermentation led to a decrease in NADH/NAD+ ratio in the cells, and hence to an enhanced 3-HP production as compared to the fermentative control (Kim et al., 2017). Although the 3-HP titer reached was low, the study nevertheless demonstrated that it is possible to improve NAD+ regeneration under anaerobic conditions. With further optimization, this novel strategy might pave the way for providing new opportunities to enhance 3-HP production from glycerol. It might similarly enhance other bioconversions where there is a need to control the intracellular redox state of a cell factory (Kim et al., 2017; de Fouchécour et al., 2018).
In a fermentation operation, the process parameters such as aeration rate, medium composition, etc. play an important role in determining the final outcome. While there are ample reports on strain development, generally less focus has been put on the optimization of the production conditions. Conversion of glycerol to 3-HP being an oxidative reaction, the aeration rate does affect the final yield of the product (de Fouchécour et al., 2018). In one study, aerobic, micro-aerobic and anaerobic conditions were tested in batch fermentations performed to convert glycerol to 3-HP using recombinant K. pneumoniae. They reported that amongst the three conditions tested, the micro-aerobic condition yielded the highest 3-HP titer (2.2 g/L; Zhu et al., 2009). In a more recent study, the transcript levels of the genes encoding the enzymes of the CoA-independent pathway of glycerol conversion to 3-HP and the genes coding for the enzymes of the formate hydrogen lyase pathway were analyzed to understand the intracellular response of K. pneumoniae under aerobic, micro-aerobic and anaerobic growth conditions (Huang et al., 2016). The transcription of the glycerol dehydratase operon was downregulated in the presence of oxygen, while aldehyde dehydrogenase-, hydrogenase- and formate dehydrogenase-coding genes were found to be upregulated. The authors suggested that in the presence of oxygen, the formate hydrogen lyase pathway consumed the excess NADH generated due to the over-expression of aldH (Huang et al., 2016). In another study aimed at enhancing the coproduction of 3-HP and 1,3-PDO in K. pneumoniae, the production of by-products such as lactate, ethanol, succinate, and acetate was reduced by disrupting their synthesis (Ko et al., 2017). However, the disruption of pta-ackA gene of the acetate pathway led to a reduction in cell growth, glycerol uptake, and 3-HP and 1,3-PDO production. The authors bypassed the disruption of this gene for reducing acetate production by testing various agitation speeds between 200 and 600 rpm. They reported an increase in 3-HP yield from 0.18 to 0.38 mol3−HP/molglycerol, respectively, which could be attributed to an enhanced cell growth rate because of better oxygen transfer at 600 rpm. Although acetate production was not completely abolished, they did manage to reduce it from 163 mM (at 400 rpm) to only 80 mM (at 600 rpm; Ko et al., 2017). Recently, a production of 61.9 g/L 3-HP with a yield of 0.58 mol3−HP/molglycerol in 38 h in a 5 L bioreactor operated in fed-batch mode by using an engineered K. pneumoniae strain was reported (Jiang et al., 2018). The aeration rate was reduced to half of the initial rate once the cell biomass OD reached close to the maximum value. Furthermore, using the same strategy, they were able to scale-up the process in a 300 L bioreactor and reported a titer of 54.5 g/L for 3-HP with a yield of 0.58 mol3−HP/molglycerol in 51 h.
The growth medium used for cultivating a microorganism plays an important role in influencing the production process. The current record holder for the highest 3-HP titer, addressed this issue by testing different media for the production of 3-HP in K. pneumoniae in shake flasks (Li et al., 2016). A gradient concentration of each component of the media (except CaCl2) was individually analyzed. The ameliorated medium enabled an increase of 80.5% in the production of 3-HP. They also tested the effect of pH on 3-HP production and found pH 7.0 to be the most appropriate for 3-HP production (Li et al., 2016). To optimize the growth medium for 3-HP production using L. reuteri, 30 different media with variable amounts of sugar beet and wheat processing coproducts (used as the carbon source), yeast extract, tween 80 and vitamin B12 were tested. The authors reported an increase in 3-HP production yield by 70 %, accompanied with a decreased 3-HPA titer (Couvreur et al., 2017).
Another way to solve the earlier-mentioned challenge of conflicting requirements for coenzyme B12 generation and NAD+ regeneration, is to divide the conversion of glycerol to 3-HP into 2 steps. First, glycerol is converted into either 3-HPA or a mixture of 3-HP and 1,3-PDO. The second step involves the conversion of 3-HPA and 1,3-PDO to 3-HP. This two-step strategy was employed in a study where wild-type L. reuteri resting cells first converted glycerol to 1,3-PDO and 3-HP in equimolar amounts. In the subsequent step, the supernatant was fed to the resting cells of Gluconobacter oxydans which oxidized 1,3-PDO to 3-HP, yielding a 3-HP titer of 23.6 g/L (Dishisha et al., 2015). In a similar study, Zhao et al. employed K. pneumoniae in the first step to convert glycerol into 1,3-PDO. In the second step, the resting cells of G. oxydans were introduced in the same bioreactor after heat inactivating K. pneumoniae. This led to the conversion of 1,3-PDO to 3-HP with a yield of 0.52 mol/mol on glycerol and 0.94 mol/mol on 1,3-PDO, and a 3-HP titer of 60.5 g/L (Zhao et al., 2015). The advantage of using two bioconversion steps is that each uncoupled step can be optimized separately. As attractive as it may appear because of high yields, the industrial implementation of this two-step strategy may not be as cost-effective and economical as a single step fermentation because of higher associated costs. Nevertheless, this strategy could well be a first step for achieving an integrated continuous process for the production of 3-HP, which in addition would be highly desirable because of its low operating costs (de Fouchécour et al., 2018).
Conclusions and Perspectives
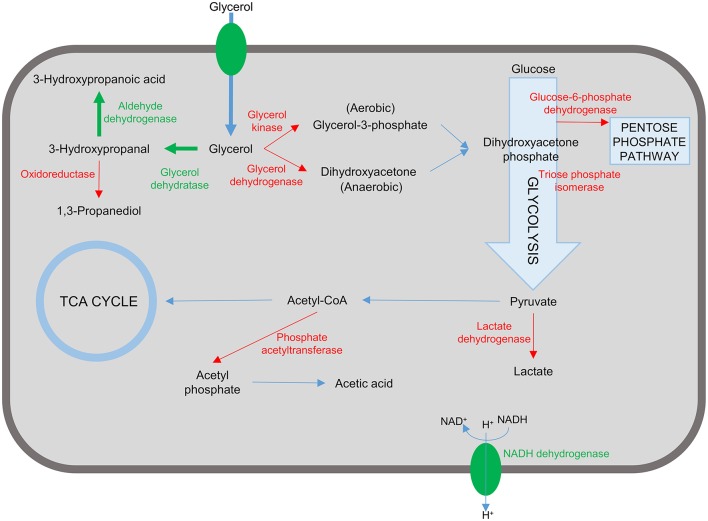
As outlined in this review considering the case of 3-HP production in bacterial cell factories, there are many elements to the successful design of a suitable production strain, ranging from the choice of organism, pathway, and enzymes to engineering of the cell for improved flux through the production pathway and the ability to tolerate the stress imposed by intermediate, by-, and final products (Figure 4). Most of the studies described in this review have targeted some of these elements. The highest titer recorded so far is 83.8 g/L which was achieved in K. pneumoniae by optimizing the expression of aldehyde dehydrogenase in the production pathway and increasing flux through the production pathway by blocking synthesis of the by-products lactic acid and acetic acid (Li et al., 2016). This impressive feat was based on rational design and accomplished by combining some of the successful approaches outlined above in very few engineering steps.
Figure 4.
Representation of targets in the bacterial cell for engineering a 3-hydroxypropanoic acid producing strain. Reactions and proteins in green represents targets that are introduced or over-expressed to confer/improve 3-HP production. Unwanted reactions and the corresponding enzymes are shown in red. Inactivation of these targets have been shown to improve 3-HP production and/or reduce/eliminate unwanted by-product formation.
For bacterial 3-HP production to reach a level of economic viability, further improvements in titer, and productivity will be necessary. By now, many different enzyme variants have been used for constructing 3-HP production pathways but in many cases the enzymes have not been characterized biochemically. A better understanding of the properties of the enzymes with respect to catalytic properties, pH range, and stability could prove instrumental in identifying the optimal enzymes and would also help to identify enzyme qualities to improve by enzyme engineering.
The impressive body of research on strain engineering has identified numerous challenges and solutions such as balancing enzyme activities, preventing by-product formation and alleviating stresses. With this accumulating knowledge base pertaining to 3-HP cell factory design, it seems likely that it will be possible to further tweak production by combining all of the successful approaches described herein. To facilitate this work, powerful engineering techniques are continually emerging that allow the design of large combinatorial libraries of targeted mutations. Notably, the recent development of a 3-HP biosensor can be used in combination with adaptive laboratory evolution for identifying as-of-yet unknown mechanisms for further fine-tuning of the production strains. While it seems reasonable that further improvements in titer will be achieved in the coming years, there should be an increased focus to also develop industrially relevant process designs and subsequent upscaling in order to provide proof of concept.
In conclusion, the massive developments in synthetic biology and metabolic engineering could be expected to soon deliver the long-sought goal of converting the abundant by-product glycerol into the much desired value-added chemical 3-HP.
Author Contributions
CJ, AG, and AK performed literature search and drafted the manuscript. All authors revised the manuscript and approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Kirsten Leistner for proofreading and revising the manuscript.
Footnotes
Funding. This work was supported by grants from the Novo Nordisk Foundation (Grant no. NNF16OC0021474 and NNF10CC1016517) to IM.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00124/full#supplementary-material
References
- Andreeßen B., Lange A. B., Robenek H., Steinbüchel A. (2010). Conversion of glycerol to poly(3-hydroxypropionate) in recombinant Escherichia coli. Appl. Environ. Microbiol. 76, 622–626. 10.1128/AEM.02097-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok S., Sankaranarayanan M., Ko Y., Jae K. E., Ainala S. K., Kumar V., et al. (2013). Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol. Bioeng. 110, 511–524. 10.1002/bit.24726 [DOI] [PubMed] [Google Scholar]
- Basan M., Hui S., Okano H., Zhang Z., Shen Y., Williamson J. R., et al. (2015). Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 528, 99–104. 10.1038/nature15765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedendieck R., Malten M., Barg H., Bunk B., Martens J. H., Deery E., et al. (2010). Metabolic engineering of cobalamin (vitamin B12) production in Bacillus megaterium. Microb. Biotechnol. 3, 24–37. 10.1111/j.1751-7915.2009.00125.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M. T., Pedersen M., Klausen M. S., Jensen S. I., Wulff T., Harrison S., et al. (2016). Predictable tuning of protein expression in bacteria. Nat. Methods. 13, 233–236. 10.1038/nmeth.3727 [DOI] [PubMed] [Google Scholar]
- Burgé G, Saulou-Bérion C., Moussa M., Pollet B., Flourat A., Allais F., et al. (2015). Diversity of Lactobacillus reuteri strains in converting glycerol into 3-Hydroxypropionic acid. Appl. Biochem. Biotechnol. 177, 923–939. 10.1007/s12010-015-1787-8 [DOI] [PubMed] [Google Scholar]
- Chen Z., Huang J., Wu Y., Wu W., Zhang Y., Liu D. (2017). Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab. Eng. 39, 151–158. 10.1016/j.ymben.2016.11.009 [DOI] [PubMed] [Google Scholar]
- Cheng K. K., Liu H. J., Liu D. H. (2005). Multiple growth inhibition of Klebsiella pneumoniae in 1,3-propanediol fermentation. Biotechnol. Lett. 27, 19–22 10.1007/s10529-004-6308-8 [DOI] [PubMed] [Google Scholar]
- Chu H. S., Kim Y. S., Lee C. M., Lee J. H., Jung W. S., Ahn J. H., et al. (2015). Metabolic engineering of 3-hydroxypropionic acid biosynthesis in Escherichia coli. Biotechnol. Bioeng. 112, 356–364. 10.1002/bit.25444 [DOI] [PubMed] [Google Scholar]
- Cleusix V., Lacroix C., Vollenweider S., Duboux M., Le Blay G. (2007). Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 7:101. 10.1186/1471-2180-7-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur J., Teixeira A., Allais F., Spinnler H. E., Saulou-Bérion C., Clément T. (2017). Wheat and sugar beet coproducts for the bioproduction of 3-Hydroxypropionic Acid by Lactobacillus reuteri DSM17938. Fermentation 3:32 10.3390/fermentation3030032 [DOI] [Google Scholar]
- Dabrowski S., Pietrewicz-Kubicz D., Zabłotna E., Długołecka A. (2012). 1,3-propanediol production by Escherichia coli expressing genes of dha operon from Clostridium butyricum 2CR371.5. Acta Biochim. Pol. 59, 357–361 10.18388/abp.2012_2121 [DOI] [PubMed] [Google Scholar]
- Daniel R., Bobik T. A., Gottschalk G. (1998). Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 22, 553–566 10.1111/j.1574-6976.1998.tb00387.x [DOI] [PubMed] [Google Scholar]
- de Fouchécour F., Sánchez-Castañeda A. K., Saulou-Bérion C., Spinnler H. E. (2018). Process engineering for microbial production of 3-hydroxypropionic acid. Biotechnol. Adv. 36, 1207–1222 10.1016/j.biotechadv.2018.03.020 [DOI] [PubMed] [Google Scholar]
- Dishisha T., Pereyra L. P., Pyo S. H., Britton R. A., Hatti-Kaul R. (2014). Flux analysis of the Lactobacillus reuteri propanediol-utilization pathway for production of 3-hydroxypropionaldehyde, 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Microb. Cell Fact. 13:76. 10.1186/1475-2859-13-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishisha T., Pyo S. H., Hatti-Kaul R. (2015). Bio-based 3-hydroxypropionic- and acrylic acid production from biodiesel glycerol via integrated microbial and chemical catalysis. Microb. Cell Fact. 14:200. 10.1186/s12934-015-0388-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnin G., Clomburg J., Yeates Z., Alvarez P. J., Zygourakis K., Campbell P., et al. (2009). Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol. Bioeng. 103, 148–161. 10.1002/bit.22246 [DOI] [PubMed] [Google Scholar]
- Espah Borujeni A., Channarasappa A. S., Salis H. M. (2014). Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42, 2646–2659. 10.1093/nar/gkt1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro R., Rennig M., Hernández-Rollán C., Daley D. O., Nørholm M. H. H. (2018). A synbio approach for selection of highly expressed gene variants in Gram-positive bacteria. Microb. Cell Fact. 17:37. 10.1186/s12934-018-0886-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu C., Ding Y., Sun C., Zhang R., Xian M., et al. (2014). Development of genetically stable Escherichia coli strains for poly(3-hydroxypropionate) production. PLoS ONE. 9:e97845. 10.1371/journal.pone.0097845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai-Ibabe G., Ibarburu I., Berregi I., Claisse O., Lonvaud-Funel A., Irastorza A., et al. (2008). Glycerol metabolism and bitterness producing lactic acid bacteria in cidermaking. Int. J. Food Microbiol. 121, 253–261. 10.1016/j.ijfoodmicro.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Hanko E. K. R., Minton N. P., Malys N. (2017). Characterisation of a 3-hydroxypropionic acid-inducible system from Pseudomonas putida for orthogonal gene expression control in Escherichia coli and Cupriavidus necator. Sci. Rep. 7:1724. 10.1038/s41598-017-01850-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich D., Andreeßen B., Madkour M. H., Al-Ghamdi M. A., Shabbaj I. I., Steinbüchel A. (2013). From waste to plastic: synthesis of poly(3-hydroxypropionate) in Shimwellia blattae. Appl. Environ. Microbiol. 79, 3582–3589. 10.1128/AEM.00161-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo H., Tsuruno K., Tatsuke T., Sato M., Hanai T. (2015). Dual synthetic pathway for 3-hydroxypropionic acid production in engineered Escherichia coli. J. Biosci. Bioeng. 120, 199–204. 10.1016/j.jbiosc.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li Z., Shimizu K., Ye Q. (2012). Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol by a recombinant strain of Klebsiella pneumoniae. Bioresour. Technol. 103, 351–359. 10.1016/j.biortech.2011.10.022 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li Z., Ye Q. (2016). Transcriptional regulation of genes involved in 3-hydroxypropionic acid production in response to aeration of recombinant Klebsiella pneumoniae. Appl. Biochem. Biotechnol. 178, 1129–1140 10.1007/s12010-015-1933-3 [DOI] [PubMed] [Google Scholar]
- Jiang J., Huang B., Wu H., Li Z., Ye Q. (2018). Efficient 3-hydroxypropionic acid production from glycerol by metabolically engineered Klebsiella pneumoniae. Bioresour. Bioprocess. 5:34 10.1186/s40643-018-0218-4 [DOI] [Google Scholar]
- Jo J. E., Mohan Raj S., Rathnasingh C., Selvakumar E., Jung W. C., Park S. (2008). Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-Hydroxypropionaldehyde as a substrate. Appl. Microbiol. Biotechnol. 81, 51–60. 10.1007/s00253-008-1608-x [DOI] [PubMed] [Google Scholar]
- Jung W. S., Kang J. H., Chu H. S., Choi I. S., Cho K. M. (2014). Elevated production of 3-hydroxypropionic acid by metabolic engineering of the glycerol metabolism in Escherichia coli. Metab. Eng. 23, 116–122. 10.1016/j.ymben.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Kalantari A., Chen T., Ji B., Stancik I. A., Ravikumar V., Franjevic D., et al. (2017). Conversion of glycerol to 3-Hydroxypropanoic acid by genetically engineered Bacillus subtilis. Front. Microbiol. 8:638. 10.3389/fmicb.2017.00638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno M., Atsumi S. (2017). Engineering an obligate photoautotrophic cyanobacterium to utilize glycerol for growth and chemical production. ACS Synth. Biol. 6, 69–75. 10.1021/acssynbio.6b00239 [DOI] [PubMed] [Google Scholar]
- Kim C., Kim M. Y., Michie I., Jeon B. H., Premier G. C., Park S., et al. (2017). Anodic electro-fermentation of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae L17 in a bioelectrochemical system. Biotechnol. Biofuels. 10:199. 10.1186/s13068-017-0886-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim S. K., Park Y. C., Seo J. H. (2014). Enhanced production of 3-hydroxypropionic acid from glycerol by modulation of glycerol metabolism in recombinant Escherichia coli. Bioresour. Technol. 156, 170–175. 10.1016/j.biortech.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Ko Y., Ashok S., Ainala S. K., Sankaranarayanan M., Chun A. Y., Jung G. Y., et al. (2014). Coenzyme B12 can be produced by engineered Escherichia coli under both anaerobic and aerobic conditions. Biotechnol. J. 9, 1526–1535. 10.1002/biot.201400221 [DOI] [PubMed] [Google Scholar]
- Ko Y., Ashok S., Seol E., Ainala S. K., Park S. (2015). Deletion of putative oxidoreductases from Klebsiella pneumoniae J2B could reduce 1,3-propanediol during the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioproc E. 20, 834–843. 10.1007/s12257-015-0166-1 [DOI] [Google Scholar]
- Ko Y., Ashok S., Zhou S., Kumar V., Park S. (2012). Aldehyde dehydrogenase activity is important to the production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae. Process Biochem. 47, 1135–1143 10.1016/j.procbio.2012.04.007 [DOI] [Google Scholar]
- Ko Y., Seol E., Sundara Sekar B., Kwon S., Lee J., Park S. (2017). Metabolic engineering of Klebsiella pneumoniae J2B for co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol: Reduction of acetate and other by-products. Bioresour. Technol. 244, 1096–1103. 10.1016/j.biortech.2017.08.099 [DOI] [PubMed] [Google Scholar]
- Kumar V., Ashok S., Park S. (2013a). Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol. Adv. 31, 945–961. 10.1016/j.biotechadv.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Kumar V., Sankaranarayanan M., Durgapal M., Zhou S., Ko Y., Ashok S., et al. (2013b). Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol using resting cells of the lactate dehydrogenase-deficient recombinant Klebsiella pneumoniae overexpressing an aldehyde dehydrogenase. Bioresour. Technol. 135, 555–563. 10.1016/j.biortech.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Kurihara S., Kato K., Asada K., Kumagai H., Suzuki H. (2010). A putrescine-inducible pathway comprising PuuE-YneI in which γ-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192, 4582–4591. 10.1128/JB.00308-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S., Park Y. C., Seo J. H. (2013). Biosynthesis of 3-hydroxypropionic acid from glycerol in recombinant Escherichia coli expressing Lactobacillus brevis dhaB and dhaR gene clusters and E. coli K-12 aldH. Bioresour Technol. 135, 432–439. 10.1016/j.biortech.2012.11.063 [DOI] [PubMed] [Google Scholar]
- Li Y., Ge X., Tian P. (2013a). Gene arrangements in expression vector affect 3-hydroxypropionic acid production in Klebsiella pneumoniae. Indian J. Microbiol. 53, 418–424. 10.1007/s12088-013-0390-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu L., Tian P. (2014). NAD+-independent aldehyde oxidase catalyzes cofactor balanced 3-hydroxypropionic acid production in Klebsiella pneumoniae. Biotechnol. Lett. 36, 2215–2221. 10.1007/s10529-014-1590-6 [DOI] [PubMed] [Google Scholar]
- Li Y., Su M., Ge X., Tian P. (2013b). Enhanced aldehyde dehydrogenase activity by regenerating NAD+ in Klebsiella pneumoniae and implications for the glycerol dissimilation pathways. Biotechnol Lett. 35, 1609–1615. 10.1007/s10529-013-1243-1 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang X., Ge X., Tian P. (2016). High production of 3-hydroxypropionic acid in Klebsiella pneumoniae by systematic optimization of glycerol metabolism. Sci. Rep. 6:26932. 10.1038/srep26932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D. I., Reiss L., Turner I., Dotson G. (2003). Structure of glycerol dehydratase reactivase: a new type of molecular chaperone. Structure. 11, 109–119 10.1016/S0969-2126(02)00935-8 [DOI] [PubMed] [Google Scholar]
- Lim H. G., Noh M. H., Jeong J. H., Park S., Jung G. Y. (2016). Optimum rebalancing of the 3-Hydroxypropionic acid production pathway from glycerol in Escherichia coli. ACS Synth. Biol. 5, 1247–1255 10.1021/acssynbio.5b00303 [DOI] [PubMed] [Google Scholar]
- Lindlbauer K. A., Marx H., Sauer M. (2017). 3-Hydroxypropionaldehyde production from crude glycerol by Lactobacillus diolivorans with enhanced glycerol uptake. Biotechnol. Biofuels. 10:295. 10.1186/s13068-017-0982-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Han X., Xian M., Ding Y., Liu H., Zhao G. (2016). Development of a 3-hydroxypropionate resistant Escherichia coli strain. Bioengineered. 7, 21–27. 10.1080/21655979.2015.1122143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Liang L., Choudhury A., Bassalo M. C., Garst A. D., Tarasava K., et al. (2018). Iterative genome editing of Escherichia coli for 3-hydroxypropionic acid production. Metab. Eng. 47, 303–313. 10.1016/j.ymben.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Liu S. (2017). Cell cultivation in Bioprocess Engineering, 2nd Edn, ed Liu S. (Amsterdam: Elsevier; ), 699–782. [Google Scholar]
- Liu Y., Gallo A. A., Bajpai R. K., Chistoserdov A., Nelson A. T., Segura L. N., et al. (2010). The diversity and molecular modelling analysis of B12-dependent and B12-independent glycerol dehydratases. Int. J. Bioinform. Res. Appl. 6, 484–507 10.1504/IJBRA.2010.037988 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu Y., Wang M. (2017). Design, optimization and application of small molecule biosensor in metabolic engineering. Front. Microbiol. 8:2012. 10.3389/fmicb.2017.02012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. H., Kim C. H., Heo S. Y., Oh B. R., Hong W. K., Kim S., et al. (2012). Production of 3-hydroxypropionic acid through propionaldehyde dehydrogenase PduP mediated biosynthetic pathway in Klebsiella pneumoniae. Bioresour. Technol. 103, 1–6. 10.1016/j.biortech.2011.09.099 [DOI] [PubMed] [Google Scholar]
- Luo L. H., Seo J. W., Baek J. O., Oh B. R., Heo S. Y., Hong W. K., et al. (2011). Identification and characterization of the propanediol utilization protein PduP of Lactobacillus reuteri for 3-hydroxypropionic acid production from glycerol. Appl. Microbiol. Biotechnol. 89, 697–703. 10.1007/s00253-010-2887-6 [DOI] [PubMed] [Google Scholar]
- Luo L. H., Seo J. W., Heo S. Y., Oh B. R., Kim D. H., Kim C. H. (2013). Identification and characterization of Klebsiella pneumoniae aldehyde dehydrogenases increasing production of 3-hydroxypropionic acid from glycerol. Bioprocess Biosyst. Eng. 36, 1319–1326. 10.1007/s00449-012-0880-4 [DOI] [PubMed] [Google Scholar]
- Mori K., Toraya T. (1999). Mechanism of reactivation of coenzyme B12-dependent diol dehydratase by a molecular chaperone-like reactivating factor. Biochemistry. 38, 13170–13178 10.1021/bi9911738 [DOI] [PubMed] [Google Scholar]
- Nguyen-Vo T. P., Ainala S. K., Kim J. R., Park S. (2018). Analysis, characterization of coenzyme B12 biosynthetic gene clusters and improvement of B12 biosynthesis in Pseudomonas denitrificans ATCC 13867. FEMS Microbiol Lett. 1:365 10.1093/femsle/fny211 [DOI] [PubMed] [Google Scholar]
- Park Y. S., Choi U. J., Nam N. H., Choi S. J., Nasir A., Lee S. G., et al. (2017). Engineering an aldehyde dehydrogenase toward its substrates, 3-hydroxypropanal and NAD+, for enhancing the production of 3-hydroxypropionic acid. Sci. Rep. 7:17155. 10.1038/s41598-017-15400-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Sun L., Luo Z., Wu J., Meng X., Tang Y., et al. (2006). Rational design of glycerol dehydratase: Swapping the genes encoding the subunits of glycerol dehydratase to improve enzymatic properties. Chinese Sci. Bull. 51:2977 10.1007/s11434-006-2219-5 [DOI] [Google Scholar]
- Raj S. M., Rathnasingh C., Jo J. E., Park S. (2008). Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem. 43, 1440–1446 10.1016/j.procbio.2008.04.027 [DOI] [Google Scholar]
- Raj S. M., Rathnasingh C., Jung W. C., Selvakumar E., Park S. (2010). A Novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC Gene of Klebsiella pneumoniae DSM 2026 that Utilizes 3-Hydroxypropionaldehyde as a Substrate. Biotechnol. Bioproc. E. 15, 131–138 10.1007/s12257-010-0030-2 [DOI] [Google Scholar]
- Rathnasingh C., Raj S. M., Jo J. E., Park S. (2009). Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol. Bioeng. 104, 729–739. 10.1002/bit.22429. [DOI] [PubMed] [Google Scholar]
- Raynaud C., Sarçabal P., Meynial-Salles I., Croux C., Soucaille P. (2003). Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. U.S.A. 100, 5010–5015 10.1073/pnas.0734105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennig M., Martinez V., Mirzadeh K., Dunas F., Röjsäter B., Daley D. O., et al. (2018). TARSyn: Tunable antibiotic resistance devices enabling bacterial synthetic evolution and protein production. ACS Synth. Biol. 7, 432–442. 10.1021/acssynbio.7b00200 [DOI] [PubMed] [Google Scholar]
- Sabet-Azad R., Linares-Pastén J. A., Torkelson L., Sardari R. R., Hatti-Kaul R. (2013). Coenzyme A-acylating propionaldehyde dehydrogenase (PduP) from Lactobacillus reuteri: kinetic characterization and molecular modeling. Enzyme Microb. Technol. 53, 235–242. 10.1016/j.enzmictec.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan M., Ashok S., Park S. (2014). Production of 3-hydroxypropionic acid from glycerol by acid tolerant Escherichia coli. J. Ind. Microbiol. Biotechnol. 41, 1039–1050. 10.1007/s10295-014-1451-2 [DOI] [PubMed] [Google Scholar]
- Schaefer L., Auchtung T. A., Hermans K. E., Whitehead D., Borhan B., Britton R. A. (2010). The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156, 1589–1599. 10.1099/mic.0.035642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S. W., Yang J. S., Kim I., Yang J., Min B. E., Kim S., et al. (2013). Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 15, 67–74. 10.1016/j.ymben.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Seok J. Y., Yang J., Choi S. J., Lim H. G., Choi U. J., Kim K. J., et al. (2018). Directed evolution of the 3-hydroxypropionic acid production pathway by engineering aldehyde dehydrogenase using a synthetic selection device. Metab. Eng. 47, 113–120. 10.1016/j.ymben.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Shibata N., Masuda J., Tobimatsu T., Toraya T., Suto K., Morimoto Y., et al. (1999). A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Structure 7, 997–1008 10.1016/S0969-2126(99)80126-9 [DOI] [PubMed] [Google Scholar]
- Silva F., Queiroz J. A., Domingues F. C. (2012). Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 30, 691–708. 10.1016/j.biotechadv.2011.12.005 [DOI] [PubMed] [Google Scholar]
- Son H. F., Park S., Yoo T. H., Jung G. Y., Kim K. J. (2017). Structural insights into the production of 3-hydroxypropionic acid by aldehyde dehydrogenase from Azospirillum brasilense. Sci. Rep. 7:46005. 10.1038/srep46005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Li Y., Ge X., Tian P. (2015). 3-Hydroxypropionaldehyde-specific aldehyde dehydrogenase from Bacillus subtilis catalyzes 3-hydroxypropionic acid production in Klebsiella pneumoniae. Biotechnol. Lett. 37, 717–724. 10.1007/s10529-014-1730-z [DOI] [PubMed] [Google Scholar]
- Su M. Y., Li Y., Ge X. Z., Tian P. F. (2014). Insights into 3-hydroxypropionic acid biosynthesis revealed by overexpressing native glycerol dehydrogenase in Klebsiella pneumoniae. Biotechnol Biotechnol Equip. 28, 762–768 10.1080/13102818.2014.944419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama K., Ohno S., Yoshikawa K., Hirasawa T., Tanaka S., Furusawa C., et al. (2014). Increased 3-hydroxypropionic acid production from glycerol, by modification of central metabolism in Escherichia coli. Microb. Cell Fact. 13:64. 10.1186/1475-2859-13-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toraya T. (2003). Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem. Rev. 103, 2095–2127 10.1021/cr020428b [DOI] [PubMed] [Google Scholar]
- Wang K., Tian P. (2017). Engineering Plasmid-Free Klebsiella Pneumoniae for Production of 3-Hydroxypropionic Acid. Curr. Microbiol. 74, 55–58. 10.1007/s00284-016-1153-2 [DOI] [PubMed] [Google Scholar]
- Wang X., Sa N., Wang F. H., Tian P. F. (2013). Engineered constitutive pathway in Klebsiella pneumoniae for 3-hydroxypropionic acid production and implications for decoupling glycerol dissimilation pathways. Curr. Microbiol. 66, 293–299. 10.1007/s00284-012-0271-8 [DOI] [PubMed] [Google Scholar]
- Wang X. D., Zhang X. E., Guo Y. C., Zhang Z. P., Cao Z. A., Zhou Y. F. (2009). Characterization of glycerol dehydratase expressed by fusing its α- and β-subunits. Biotechnol. Lett. 31, 711–717. 10.1007/s10529-009-9911-x [DOI] [PubMed] [Google Scholar]
- Wang Y., Tao F., Ni J., Lia C., Xu P. (2015). Production of C3 platform chemicals from CO2 by genetically engineered cyanobacteria. Green Chem. 17, 3100–3110 10.1039/C5GC00129C [DOI] [Google Scholar]
- Warnecke T. E., Lynch M. D., Karimpour-Fard A., Lipscomb M. L., Handke P., Mills T., et al. (2010). Rapid dissection of a complex phenotype through genomic-scale mapping of fitness altering genes. Metab. Eng. 12, 241–250. 10.1016/j.ymben.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Warnecke T. E., Lynch M. D., Lipscomb M. L., Gill R. T. (2012). Identification of a 21 amino acid peptide conferring 3-hydroxypropionic acid stress-tolerance to Escherichia coli. Biotechnol. Bioeng. 109, 1347–1352. 10.1002/bit.24398 [DOI] [PubMed] [Google Scholar]
- Wei X., Meng X., Chen Y., Wei Y., Du L., Huang R. (2014). Cloning, expression, and characterization of coenzyme-B12-dependent diol dehydratase from Lactobacillus diolivorans. Biotechnol. Lett. 36, 159–165 10.1007/s10529-013-1346-8 [DOI] [PubMed] [Google Scholar]
- Yamanishi M., Kinoshita K., Fukuoka M., Saito T., Tanokuchi A., Ikeda Y., et al. (2012). Redesign of coenzyme B12 dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols. FEBS J. 279, 793–804. 10.1111/j.1742-4658.2012.08470.x [DOI] [PubMed] [Google Scholar]
- Yamanishi M., Yunoki M., Tobimatsu T., Sato H., Matsui J., Dokiya A., et al. (2002). The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 269, 4484–4494 10.1046/j.1432-1033.2002.03151.x [DOI] [PubMed] [Google Scholar]
- Zhao L., Lin J., Wang H., Xie J., Wei D. (2015). Development of a two-step process for production of 3-hydroxypropionic acid from glycerol using Klebsiella pneumoniae and Gluconobacter oxydans. Bioprocess Biosyst. Eng. 38, 2487–2495 10.1007/s00449-015-1486-4 [DOI] [PubMed] [Google Scholar]
- Zhou S., Catherine C., Rathnasingh C., Somasundar A., Park S. (2013). Production of 3-hydroxypropionic acid from glycerol by recombinant Pseudomonas denitrificans. Biotechnol. Bioeng. 110, 3177–3187. 10.1002/bit.24980 [DOI] [PubMed] [Google Scholar]
- Zhu J. G., Ji X. J., Huang H., Du J., Li S., Ding Y. Y. (2009). Production of 3-hydroxypropionic acid by recombinant Klebsiella pneumoniae based on aeration and ORP controlled strategy. Korean J. Chem. Eng. 26:1679 10.1007/s11814-009-0240-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.