Abstract
Osteosarcoma (OS) is the most frequent primary malignant tumour of bone and metastases occur in 30% of cases, the 5-year survival rate is 25–30%.
Although pre- and post-operative chemotherapy has improved prognosis in osteosarcoma (OS), high toxicity and natural and acquired drug-resistance are the first cause of treatment failure. The identification of new predictive and therapeutic biomarkers may increase drug sensitivity and better control localized and metastatic disease. By the evidence that CXCR4 receptor by binding its ligand CXCL12 activates downstream critical endpoints for tumour malignancy, we first studied human OS progression correlating CXCR4 expression in OS biopsy with patient clinical data. By Real-time PCR and immunoistochemistry we found that high levels of CXCR4 gene and protein expression significantly correlated with OS progression, emphasizing the role of CXCR4/CXCL12 axis in tumour prognosis. This was supported by univariate analyses that showed a higher probability of local and/or systemic relapse in OS patients with a high CXCR4 gene expression and a significant increase of metastasis risk associated with an increasing score of CXCR4 protein staining intensity. Secondarily, to study the role of CXCR4 as a target for new therapeutic strategies, we evaluated the response of OS cells to the fully human CXCR4 antibody, MDX1338. In the study we also included AMD3100, the most studied CXCR4 antagonist.
In CXCR4-positive OS cells cultured in CXCL12-rich BM-MCS-CM (bone marrow-derived mesenchymal stem conditioned medium), a decrease of cell proliferation up to 30%–40% of control was seen after drug exposure. However, an increase of apoptosis was seen in p53-positive U2OS and 143B after CXCR4 inhibitor incubation, while no changes were seen in treated SAOS-2 cells which also present a different labeling profile. The role of p53 in apoptotic response to CXCR4 inhibitors was confirmed by p53 silencing in U2OS cell line. Our data suggest that the response to anti-CXCR4 agents could be influenced by the genetic background and labeling profile which induces a different cross-talk between tumour cells and environment. The delay in cell cycle progression associated with increased apoptosis could sensitize p53-positive cells to conventional therapy and in vivo preclinical experiments are on going with the aim to suggest new combined target therapies in human OS.
Keywords: Sarcoma, Metastasis, CXCR4 antagonists, Prognosis, Biomarkers
1. Introduction
Osteosarcoma (OS) is a primary malignant bone tumour characterized by direct formation of immature bone or osteoid tissue.
The most common subtype is conventional high grade OS, which mainly affects paediatric and young adult patients [1], [2], [3]. Although prognosis for patients with localized OS (event-free-survival up to 70%), has improved with multimodal therapy that combines surgery with pre- and post-operative chemotherapy, outcome for patients with metastatic OS is still poor [4].
Metastatic disease is mainly localized at the lung and represents the most common cause of death [5]. New prognostic and therapeutic targets are needed to reduce OS metastatic spread and improve patient survival. Several molecules and their receptors are known to be involved in migration and invasiveness of tumour cells including CXC-Chemokine Receptor-4 (CXCR4) that seems to play an important role [6]. The binding between CXCR4 and its ligand CXCL12 (SDF-1) activates downstream cascades involving many signaling pathways, such as JAK/STAT, PI3K/Akt, MAPK, JNK considered important targets for development of new therapeutic strategies. These critical points control stemness, chemotaxis and cell survival, proliferation, migration [7]. CXCL12 is a homeostatic chemokine, produced by MSC, that binding chemokine receptor regulates hematopoietic stem cell (HSC) trafficking and secondary lymphoid tissue architecture [7], [8], [9]. CXCR4, low or absent in normal cells [7], is overexpressed in many tumours predicting malignant progression and prognosis [10], [11], [12], [13], [14].
Previous results demonstrated that in human OS a significant positive correlation between CXCR4 and VEGF and between CXCR4 and MMP9 was associated with metastatic progression and survival [15], [16].
Tumour cells cultured in bone marrow-derived mesenchymal stem conditioned medium (BM-MCS-CM) increased proliferation and migration through activation of CXCR4/CXCL12 axis that could be impaired by CXCR4 antagonists [17]. The antagonist AMD3100 combined with triptolide reduced cell proliferation and increased apoptosis in U2OS cells by controlling multiple signaling growth pathways [18].
The first part of the paper studied human OS progression correlating CXCR4 expression in surgical specimens with patient clinical data.
The second aim was to study the role of CXCR4 as a target for new therapeutic strategies. We evaluated the response of OS cells to the fully human CXCR4 antibody MDX1338, that induces apoptosis in leukemia cells [19]. We also included in the study the non-competitive inhibitor AMD3100, approved by FDA for the mobilization of HSC, that is the most studied CXCR4 antagonist [20].
2. Materials and methods
2.1. Tumour specimens
CXCR4 gene and protein expression was evaluated in 48 primary OS biopsies provided by the Rizzoli Orthopaedic Institute BioBank and diagnosed by expert pathologists (Tab 1). Thirty-five were conventional high grade OS and 13 were parosteal low grade OS. Thirty patients were male and 18 female, with a median age of 19 years. Thirty patients developed metastasis and/or local recurrence, of these 29 were high grade OS. Twenty-four high grade and 1 low grade OS patients died of disease or drug toxicity (Table 1). Follow-up was considered from the date of diagnosis to the first event (metastasis or local relapse) or to the last follow-up (minimum 4 years).
Table 1.
Clinical features of 48 OS patients.
| Variables | Number of patients (%) |
|---|---|
| Sex | |
| Man | 30 (62.5%) |
| Woman | 18 (37.5%) |
| Median age | 19 (range 1–73 years) |
| Tumor grade | |
| High Grade | 35 (73%) |
| Low grade | 13 (27%) |
| Clinical course | |
| Disease free | 18 (37.5%) |
| Relapsed | 30 (62.5%) |
| Local recurrence | 8 (16.6%) |
| Metastasis | 26 (54%) |
| Both | 4 (8.3%) |
| Outcome | |
| Alive | 23 (48%) |
| Dead | 25 (52%) |
All patients underwent wide local excision of the primary tumour and the patients with high grade OS received neoadjuvant and adjuvant chemotherapy. Paraffin-embedded and frozen material was available for each patient both and the percentage of tumour cells estimated after hematoxylin-eosin staining was equal or more than 90%. Ten healthy bone tissues from non-cancer patients were used as control. The research protocol was approved by Rizzoli Institute ethic committee where the tumour samples were collected. All patients or guardians for pediatric patients provided written informed consent to the study.
2.2. Cell lines and culture conditions
Human OS cell lines U2OS (pRB+/+.p53+/+), 143B (pRB+/+, p53+/+) and SAOS-2 (pRB-/-, p53-/-) (HTB-93, CRL-8303 and HTB-85, respectively), were obtained from the America Type Culture Collection (ATCC, Manassas, VA, USA). Cells were seeded at a density of 2 × 105 per well in 6-well plates in 2 ml of αMEM medium conditioned for 72 h by mesenchymal cells originating from bone marrow of the same patient (BM-MCS-CM) [21) and incubated at 37 °C, 5% CO2.
After 24 h cells were treated with AMD3100 or Plerixafor (-Aldrich, Milano, IT), the non-competitive inhibitor of CXCR4, and with MDX1338 or Ulucuplumab (provided by Bristol-Myers Squibb, New York, USA) the anti-CXCR4 human monoclonal antibody, diluted in BM-MSC-CM to final concentrations of 5 µg/ml, 20 µg/ml, 30 µg/ml for AMD3100 and 0.001 µg/ml, 0.005 µg/ml, 0.05 µg/ml and 0.5 µg/ml for MDX1338. Control cells were incubated with BM-MSC-CM alone.
2.3. RNA extraction
RNA extraction from OS cell lines, 48 frozen tissues and ten normal tissues was performed using TRIzol reagent (Invitrogen Carlsbad, CA, USA) according to the manufacturer's protocol. Total RNA was stored at −80 °C in RNAsecure reagent (Ambion Inc., Austin, TX, USA) and the concentration was measured with Nanodrop spectrophotometer with a 260/280 resulted ratio of 1.8. Quality and purity were identified by a denatured gel electrophoresis.
2.4. Real-time PCR
CXCR4 gene expression was evaluated by RT-PCR. Reverse transcription was carried out following SuperScript™ VILO™ cDNA Synthesis Protocol (Thermo Fisher Scientific, Waltham, MA, USA), while Real-Time PCR was carried out following TaqMan Assay Protocol (Applied Biosystems, Foster City, CA, USA) (CXCR4 assay Hs00607978_sl). CXCR4 mRNA was quantified using 2−ΔΔCT comparative method (Applied Biosystems, User Bulletin no.2 P/N 4,303,859) and normalized using β-actin (ACTβ) as endogenous control (assay Hs99999903_ml), while human hMSC were used as calibrator. Human osteoblasts total RNA (Cell Application Inc. San Diego, CA, USA) was used as control for OS cell lines.
2.5. Immunohistochemistry
Hematoxylin-eosin sections from paraffin-embedded tumour samples were reviewed by pathologists and the most representative area was chosen for TMA construction using TMAMaster System (Euroclone SpA, Milano, Italy). CXCR4 protein expression was evaluated by immunohistochemistry in 35 high grade and 13 low grade OS samples. Sections were incubated overnight with rabbit monoclonal anti-CXCR4 antibody (ab2074) (Abcam, Cambridge, GB), and polyclonal anti-CXCL12 (FL-93) (Santa Cruz Biotechnology, Dallas, TX, USA) diluted 1:1000 in PBS. After, sections were washed and incubated with the streptavidin-biotin peroxidase DAB detection system (Dako, Glosturp, Denmark), according to the manufacturer's protocol.
Sample staining was scored for intensity (0=no visual staining; 1= weak/moderate; 2=strong), and percentage of positive tumour cells (0 ≤ 10%; 1 = =10%–24%; 2 = =25%–49%; 3 ≥ 50%). Cut-off levels, determined by the scores sum, were applied as 0 for negative cases, 1–3 for weak or moderate positivity in less than 50% of tumour cells, and 4–5 for moderate or strong positivity in almost 50% of tumour cells. The latter range was considered protein overexpression.
2.6. ELISA assay
CXCL12 amount was determined in BM-MSC-CM, αMEM and OS cell line supernatant by ELISA assay. Quantikine® ELISA kit (R&D Systems, McKinley Place, MN, USA) was used according to the manufacturer's protocol and the optical density was measured with Glomax Multi detection system spectrophotometry (Promega , Madison, WI, USA).
2.7. Cell growth assay
The number of adherent and viable cells was assessed microscopically using a Neubauer chamber, and viability was evaluated as the percentage of cells that excluded 0.2% trypan blue. After 48 h and 72 h from treatment, cells were washed once with 1X Dulbecco's phosphate buffered saline (PBS), harvested by trypsinization and counted.
2.8. Wound healing assay
Cell migration ability was determined using a scratch wound healing assay. OS adherent cells were incubated with 0.05 mg/ml mitomycin C (Sigma-Aldrich) for 2 h. Then, the medium was remove and the cells were incubated with AMD3100 and MDX1338 diluted in BM-MSC-CM to a final concentration of 0.05 µg/ml MDX1338 and 30 µg /ml AMD3100. A vertical line was scratched with a sterile 200 µl pipette (time 0) and the wound closure periodically monitored up to 72 h. The area was measured using IMAGEJ v.1.45r software.
2.9. Apoptosis
Apoptotic and necrotic cell death were analysed by an Annexin V-FITC apoptosis detection kit (MEBCYTO Apoptosis kit, MBL International, Woburn, MA, USA). Annexin V bound to the apoptotic cells with exposed phosphatidylserine, while propidium iodide (PI) labelled necrotic cells with membrane damage. The green (FL1) and red (FL2) fluorescence of Annexin/PI-stained cells was analysed with FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The number of viable (Annexin−/PI−), apoptotic (Annexin+/PI−), and necrotic (Annexin+/PI+) cells was determined with CellQuest Software (BD Biosciences). Briefly, after 48 h and 72 h of treatment, adherent cells were washed with PBS 1X, trypsinized, centrifuged, and washed twice with PBS. Cells were then suspended in 500 µl staining solution containing FITC-conjugated Annexin V antibody and PI. After 30 min of incubation, cells were analysed by flow cytometry. Basal apoptosis and necrosis were given by untreated cells.
2.10. Cell cycle analysis
Cells were plated at 1.5 × 105 cells per well in 6-well plates to attach overnight and cell cycle distribution analysis was performed after 48 h and 72 h of treatment.
After trypsinization and fixation with 70% ethanol, cells were stained for total DNA content with a solution containing 20 µg/ml propidium iodide. Cell cycle distribution was then analyzed with a FACScan flow cytometer (Becton Dickinson San Jose, CA, USA).
2.11. Immunofluorescence
Cells were washed and trypsinized. Then 1,000,000 cells/ml were taken and centrifuged at 1500 rpm for 5 min. Pellets were fixed with paraformaldehyde (PFA) 4% and washed with PBS 1X BSA 1% Tween20 0.5% (PAT), and then with PBS 1X TritonX 0.15%. Samples were then covered with PBS BSA 4% and incubated for 1 h with rabbit monoclonal anti-CXCR4 antibody (Abcam) diluted 1:100. After another wash with PAT, pellets were incubated with anti-rabbit antibody (GE Healthcare, Amersham, UK) diluted 1:80, washed again, and suspended with 500 µl of PBS. Proteins expression was evaluated by flow cytometry.
2.12. SiRNA duplex and transfection in U2OS cell line
A small interfering RNA (siRNA) duplex targeting p53 (TP53 Validated StealthTM, Invitrogen Paisley, UK) was used. Cells were seeded in 6-well plates (150,000 cells/well) and transfected 48 h later for 5 h with specific siRNA or control siRNA (CTRL) (Stealth siRNA Negative Control Duplex) using Lipofectamine 2000 (Invitrogen-Life Technology, Paisley, UK) according to the manufacture's protocol. After transfection, cells were incubated with MDX1338 diluted in BM-MSC-CM to a final concentration of 0.05 µg/ml for 48 h. The effect of siRNA transfection was validated by western blotting of p53 protein.
2.13. Western blotting analysis
According to standard procedures, 50 µg of protein extracts from cell lysate of U2OS, U2OS negative SiRNA control and U2OS SiRNA were prepared and analyzed by 10% SDS‑PAGE. Western blot analysis (WB) was performed by using anti‑p53 (D0-1 sc-126 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) diluted 1:1000. The signal was visualized by Immobilon Western Chemiluminiscent HRP substrate (Millipore, Billerica, MA, USA) and quantified by densitometric analysis using GS-800 imaging densitometer and Quantity One software (Bio-Rad, Hercules, CA, USA). A mouse anti-actin antibody (Sigma Chemical Co., St. Louis, MO, USA) was used as control.
2.14. Statistical analysis
Overall and disease-free survival analysis was assessed by Kaplan–Meier and Cox's regression univariate test. Mann–Whitney was used for data with non-homogeneous variance test.
Student's test was performed for in vitro experiments. All assays were performed in triplicate and for all tests p ≤ 0.05 was considered statistically significant. All statistical analyses were performed using SPSS v.19.0 (IBM Corp., Armonk, NY, USA).
3. Results
3.1. Population study
Thirty of 48 patients included in the study had metastasis and/or local relapse. In detail, 16 of the metastatic patients developed metastasis during a minimum follow-up of 4 years, while 10 presented metastasis at diagnosis or within the first 2 months. 8 patients locally relapsed (Table 1).
The average disease-free survival (DFS) was 80 months with a median of 28 months (95%CI=0–61). Average overall survival was 134.8 months with a median of 88 months (95%CI=24.5–151).
3.2. CXCR4 gene expression
CXCR4 gene expression was evaluated in 48 primary OS samples and in 10 healthy bone tissues used as control.
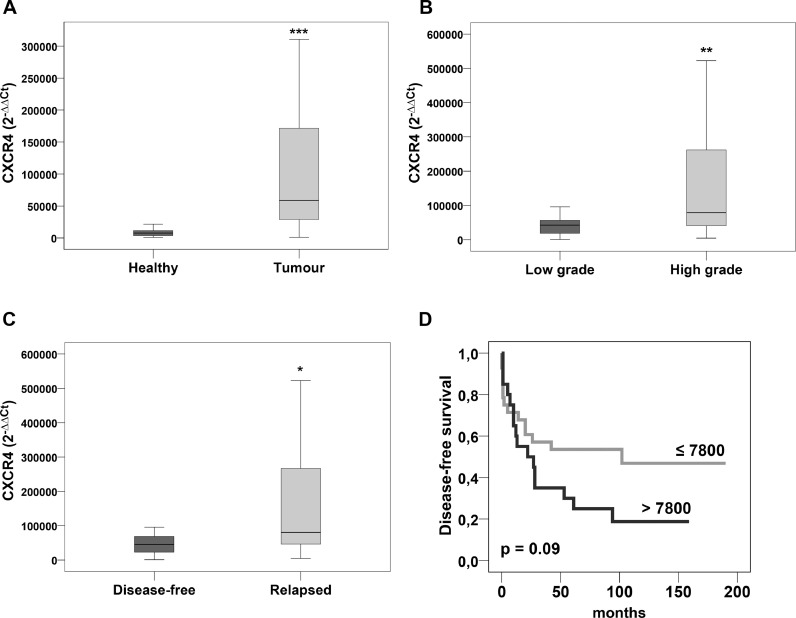
Mann–Whitney test demonstrated significantly higher mRNA levels in tumour compared to healthy tissue (p = =0.0005) (Table 2, Fig. 1A), and in 35 high grade compared 13 low grade OS (p = =0.009) (Table 2, Fig. 1B). No significant differences were seen between CXCR4 expression and other clinical parameters (gender, age, site, size, outcome).
Table 2.
CXCR4 gene expression in human OS.
| 25th P | Median 2−ΔΔCt | 75th P | P = | |
|---|---|---|---|---|
| Healthy control | 3247.75 | 7812.00 | 14,033.25 | 0.0005 |
| Tumour tissue | 27,517.7 | 58,910.5 | 175,985.0 | |
| High grade OS | 33,225.4 | 79,023.8 | 264,883.8 | |
| Low grade OS | 12,109.0 | 42,494.0 | 58,910.5 | 0.009 |
| Relapsed OS | 42,828.2 | 80,275.4 | 277,710.9 | |
| Disease-free OS | 21,892.6 | 45,569.0 | 72,444.5 | 0.02 |
| Dead | 36,230.7 | 68,082.7 | 222,589.4 | |
| Alive | 26,249 | 55,975.2 | 163,055.1 | 0.35 |
Fig. 1.
Relative levels of CXCR4. mRNA expression. Mann–Whitney analysis revealed statistical significant differences between (A) primary OS and healthy bone tissue, (B) low and high grade OS, (C) disease-free and relapsed OS. (D) Kaplan–Meier analysis based on CXCR4 expression showed a higher probability of disease-free survival in patients with low CXCR4 mRNA levels. Cut-off rounded to the 50°percentile. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
When patients were divided according to clinical follow-up in terms of both metastasis progression and local recurrence, the 30 relapsed OS presented significantly higher CXCR4 mRNA levels than the 18 disease-free patients (p = =0.02) (Table 2, Fig. 1C).
Accordingly, using a cut-off of 7800 (2−ΔΔCT) corresponding to the 50° percentile of tumour population, Kaplan–Meier analysis demonstrated that patients with high CXCR4 expression had a higher probability to relapse compared to patients with a lower expression (p = =0.09) (Fig. 1D).
3.3. CXCR4 protein expression
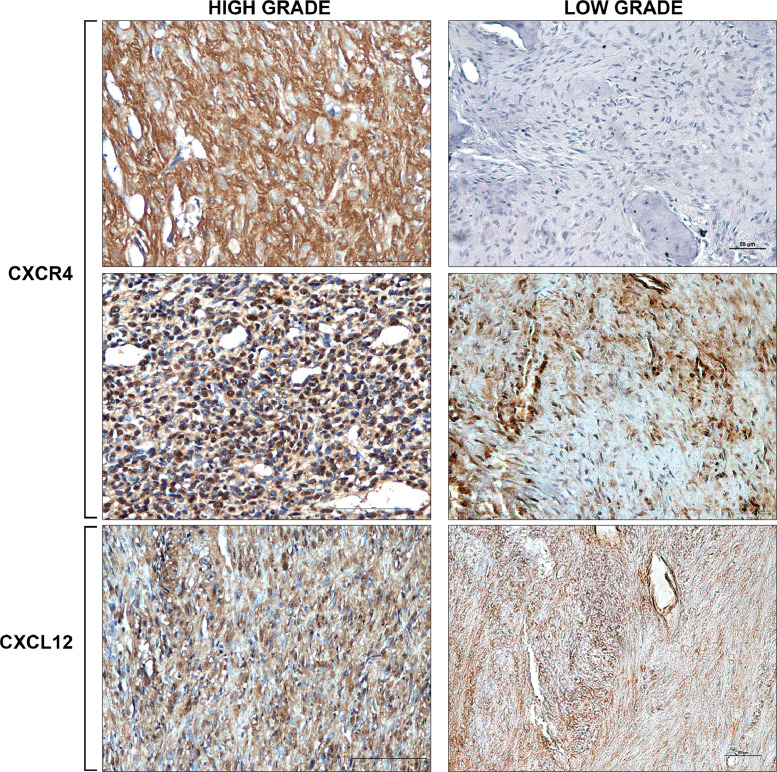
Immunohistochemistry analysis performed on 48 paraffin-embedded OS tissues showed that 19 of 35 high grade OS (54%) had a moderate to strong immunoreactivity in at least 50% of tumour cells (range 4–5). The remaining 16 samples (46%) presented a moderate CXCR4 expression in a percentage of tumour cells ranging from 25% to 49% (score 3) (Fig. 2).
Fig. 2.
Representative immunostaining of CXCR4 protein. CXCR4 was moderately to strongly expressed in cytoplasm and nucleus of high grade OS cells. In low grade OS CXCR4 was negative or week with a focal distribution. A week and diffuse distribution was seen for CXCL12 reactivity in all cases (IHC 20X).
In low grade OS CXCR4 expression was negative or weak/moderate with a focal distribution in less than 25% of tumour cells (score 0–2) (Fig. 2).
CXCR4 staining was localized both at nuclear and cytoplasmatic level, while an exclusive cytoplasmatic weak and diffuse staining distribution was seen for CXCL12 in all tumour tissues (Fig. 2).
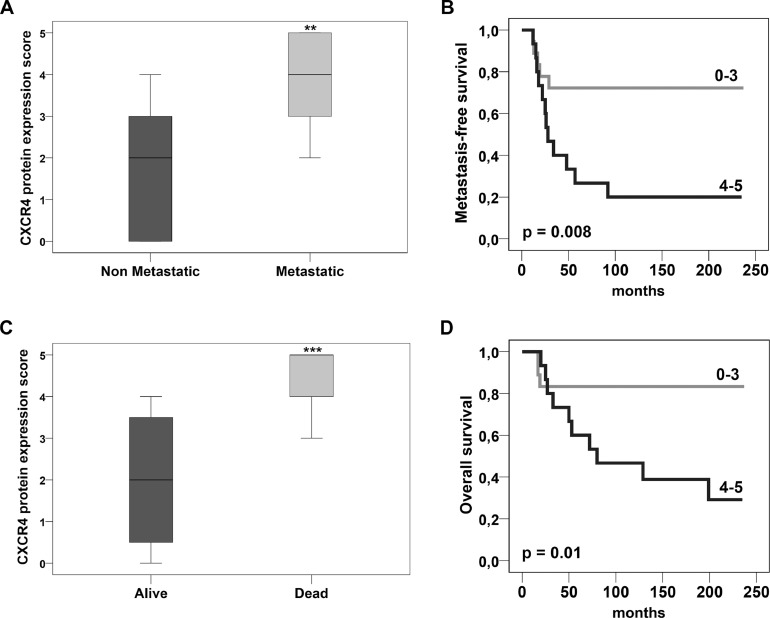
Based on staining intensity score (range 1–5), univariate Cox's analysis demonstrated a 2-fold increased metastasis risk for each increasing score (95% CI = 1. 2–3.4; p == 0.008). Accordingly, Mann–Whitney analysis revealed statistically significant higher CXCR4 staining levels in metastatic compared to non metastatic OS (p = =0.001) (Fig. 3A). Kaplan–Meier curves showed a higher probability of metastasis in the patient subset with protein overexpression (range 4–5) (log rank=6.930; p = =0.008) (Fig. 3B).
Fig. 3.
CXCR4 protein expression. (A) Mann–Whitney analysis showed higher CXCR4 protein levels in metastatic than in non metastatic OS. (B) Based on staining intensity score (range 1–5) metastasis-free survival was significantly higher in patients with CXCR4 low expression. (C) Higher CXCR4 staining levels were present in alive compared to deceased patients. (D) Overall survival probability was significantly higher in patients with no or low CXCR4 expression. Range 0–3 indicates a low or absent immunoreactivity; range 4–5 indicates a moderate to strong immunoreactivity. ** p ≤ 0.01; *** p ≤ 0.001.
Significantly higher CXCR4 staining levels were also found in alive compared to deceased patients (p = =0.0001) (Fig. 3C), concomitant with a significantly higher probability of overall survival in patients with no or low/moderate expression of CXCR4 (range 0–3) (log rank=6.53; p = =0.01) (Fig. 3D).
3.4. OS cell sensitivity to CXCR4 antagonists
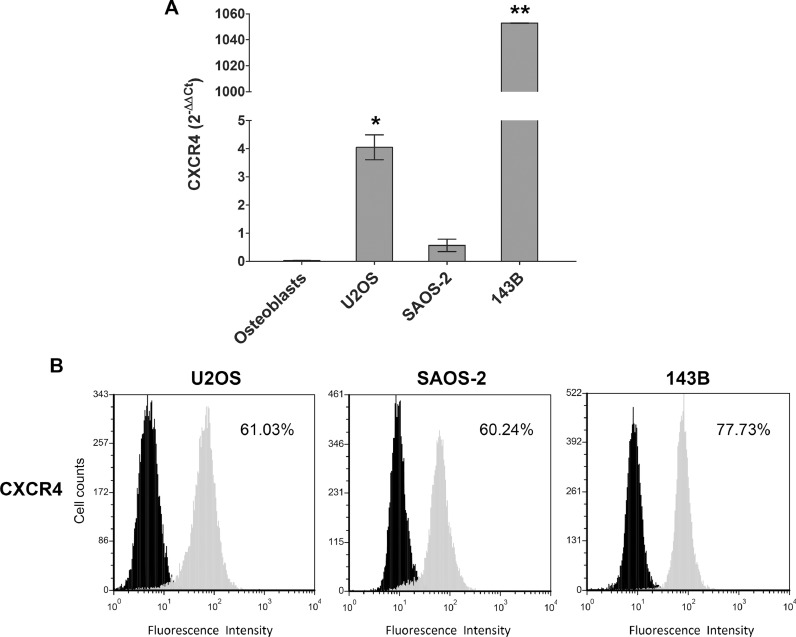
CXCR4 mRNA expression (2−ΔΔCT) was significantly higher in U2OS (4.05, p = =0.04) and 143B (1052.7, p = =0.01) than in osteoblasts (0.03), while in SAOS-2 the difference of expression did not reach statistical significance (0.57, p == 0.6). The fold change was respectively of 129.69, 33,922. 73 and 19 (Fig. 4A). By FACS analysis, a high percentage of CXCR4-positive cells was present in all cell lines (Fig. 4B).
Fig. 4.
CXCR4 expression in OS cell lines. (A) CXCR4 mRNA levels in OS cell lines and osteoblasts by RT-PCR. (B) CXCR4 protein expression by FACS analysis in OS cells. Black= negative control; Grey=Ab anti-CXCR4; Each value indicates the average of three independent experiments, * p ≤ 0.05, ** p ≤ 0.01.
Although ELISA analysis showed that CXCL12 was measurable in cell supernatant (197 pg/ml in U2OS, 100 pg/ml in 143B, and 190 pg/ml in SAOS-2), the activity of CXCR4 antagonists was evaluated in cells cultured in BM-MSC-CM that produced a higher quantity of CXCL12 ligand than non conditioned αMEM medium (1071.37 pg/ml and 307.56 pg/ml respectively).
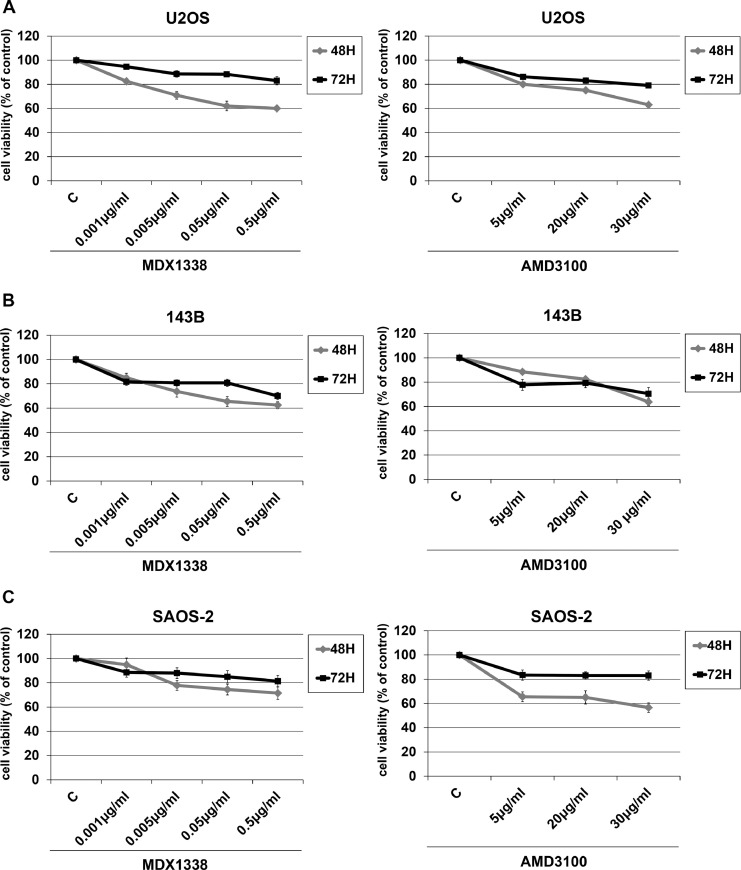
The OS cells responded to 0.5 µg/ml MDX1338 with a cell growth decrease up to 30% (SAOS-2) and 40% (U2OS and 143B) of control after 48 h of treatment. Similarly, the response to 30 µg/ml AMD3100 caused a 40% maximum decrease at 48 h for all OS cell lines (Fig. 5).
Fig. 5.
Sensitivity of OS cells to CXCR4 antagonists. Cells were exposed to increasing doses of MDX1338 and AMD3100 for 48 h and 72 h. A proliferation decrease of 30–40% compared to non treated cells occurred at 48 h at the doses of 0.5 µg/ml and 30 µg/ml respectively by counting with trypan blue. Each point indicates the average of three independent experiments. C= non treated cells.
3.5. Apoptosis and cell cycle distribution
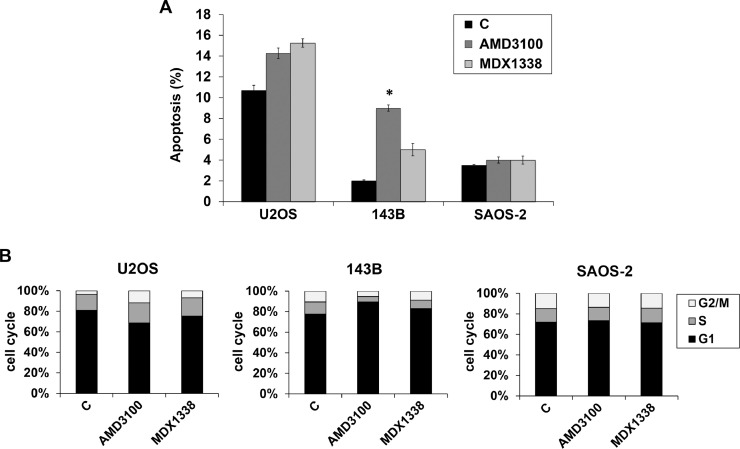
By using Annexin V-FITC assay an increase of apoptosis was seen in U2OS and 143B after 48 h of 0.05 µg/ml MDX1338 and 30 µg/ml AMD3100, slightly more evident in 143B after AMD3100 exposure (p = =0.05) (Fig. 6A). No changes in apoptotic fraction were seen in treated SAOS-2 cells.
Fig. 6.
Apoptosis and cell cycle. (A) By FACS analysis an increase of apoptosis was seen in U2OS and 143B after 48 h of 0.05 µg/ml MDX1338 and 30 µg/ml AMD3100 exposure. No changes in apoptotic fraction were seen in treated SAOS-2 cells. (B) Cell cycle analysis distribution of G1, S and G2/M phase at 48 h of 0.05 µg/ml MDX1338 and 30 µg/ml AMD3100 exposure shows a lengthening of G2/M and G1 in U2OS and 143B respectively. No differences were seen in cell cycle progression of treated SAOS-2 compared to control; Each value indicates the average of three independent experiments, *p ≤ 0.05.
Concomitantly, BrDU incorporation showed a different cell cycle distribution (Fig. 6B). U2OS responded to 0.05 µg/ml MDX1338 and 30 µg/ml AMD3100 with cell accumulation in S phase (from 14% in non treated cells to 17% and 18% in treated cells respectively) and in G2/M phase (from 3% to 7% and 11% respectively), associated with a decrease in G1 phase (from 77% to 71% and 64% respectively). In contrast, 143B incubated with MDX13338 and AMD310 presented increase of cells in G1 (from 73% to 78% and 84% respectively) accompanied by decrease in S (from 11% to 7% and 5% respectively) and G2/M (10% to 8.3% and 5% respectively). No evident cell cycle perturbation occurred in treated SAOS-2 cells.
3.6. Cell migration
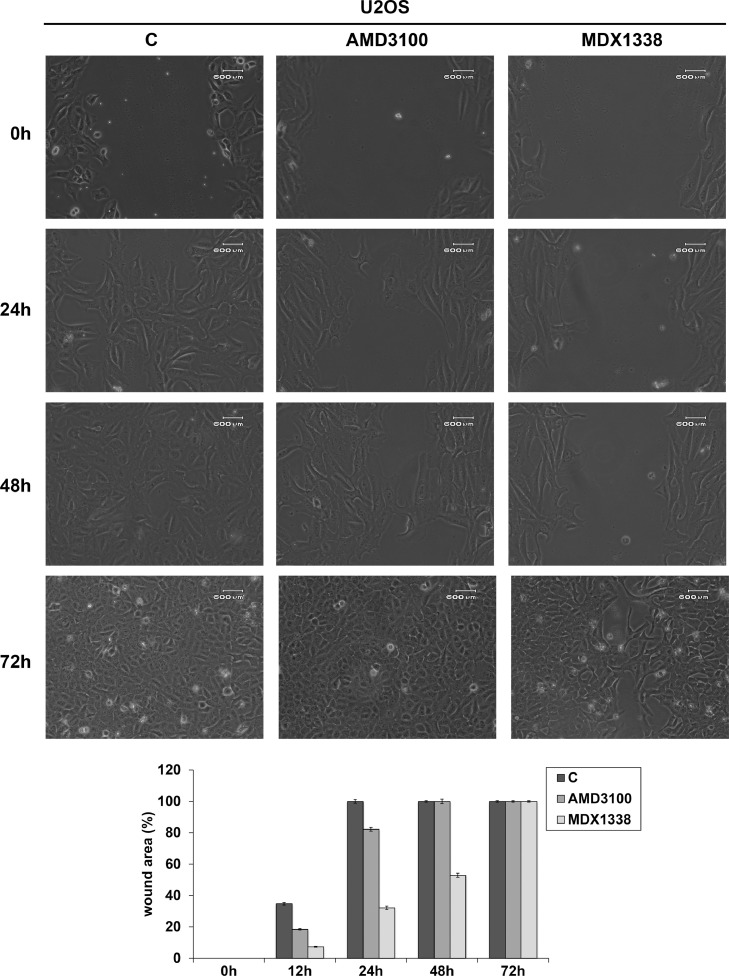
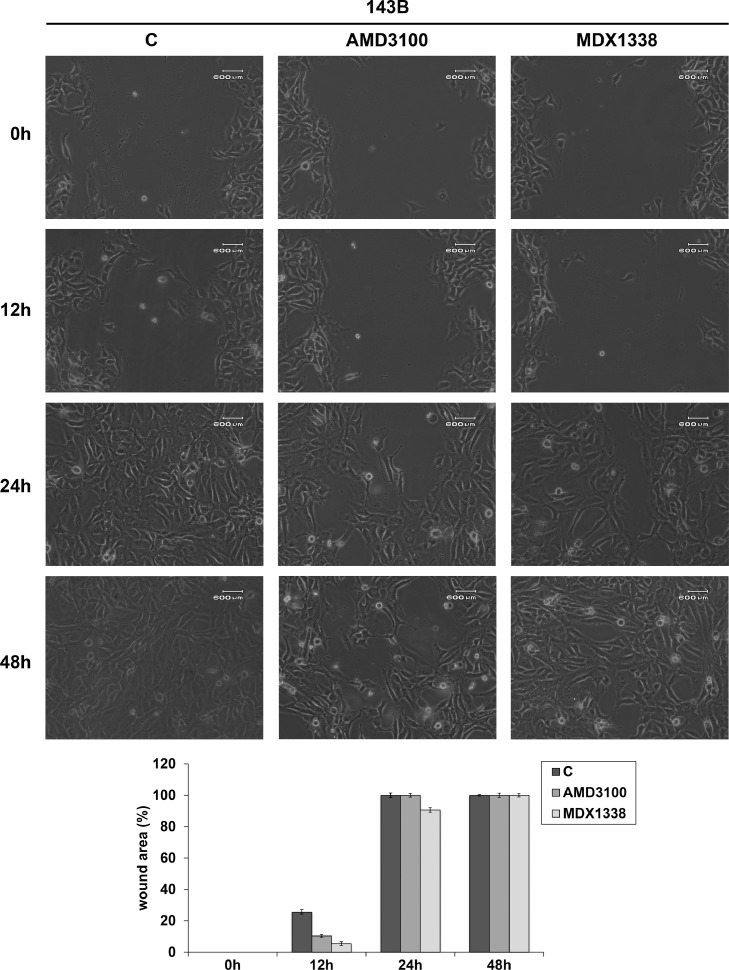
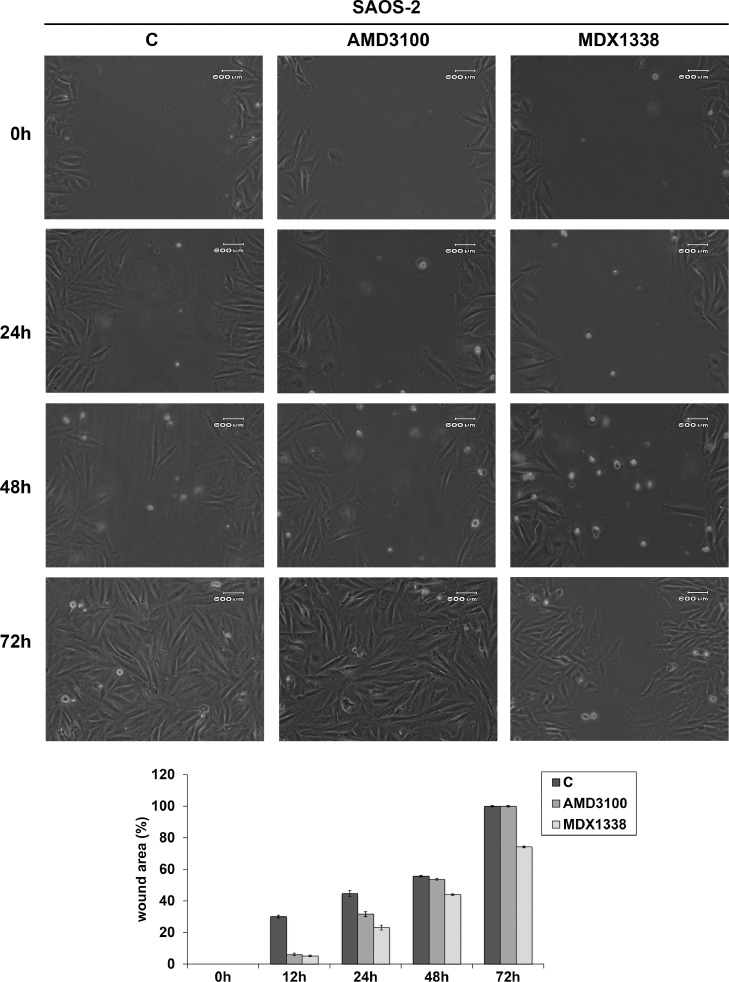
U2OS responded to AMD3100 and MDX1338 and with an evident decrease of cell motility with respect to non treated cells respectively up to 24 h and 48 h (82.2% and 52.8% of wound closure), reaching complete healing at 48 h and 72 h of treatment (Fig. 7). 143B showed a wound closure of 10.4% and 5.4% at 12 h and 100% and 90.6% at 24 h of AMD3100 and MDX1338 treatment respectively (Fig. 8). SAOS-2 responded to the exposure of two CXCR4 inhibitors with a similar migration rate up to 24 h. However, MDX1338 treatment delayed the wound closure more than AMD3100 (43.9% versus 55.6% at 48 h and 74% versus 100% at 72 h respectively) (Fig. 9), reaching a complete healing only at 96 h.
Fig. 7.
Wound healing assay of U20S. 30 µg/ml AMD3100 and 0.05 µg/ml MDX1338 exposure caused a relevant decrease in cell migration respectively up to 24 h and 48 h of treatment compared to control. At 48 h and 72 h migration rate shifted towards control values (100% of closure). Histograms show the percentage of wound closure. C= non treated cells.
Fig. 8.
Wound healing assay of 143B Cells responded to the treatment with a relevant cell migration slowdown up to 12 h. At 24 h the percentage of wound closure approached the control value. Histograms show the percentage of wound closure. C= non treated cells.
Fig. 9.
Wound healing assay of SAOS-2. MDX1338 reduced cell motility in a more marked and long-term way than AMD3100, delaying the wound closure. C= non treated cells.
3.7. siRNAp53 in U2OS cells
To confirm the involvement of wt-p53 in the apoptotic response of OS cells to MDX1338 treatment, we used siRNA approach in the U2OS cell line.
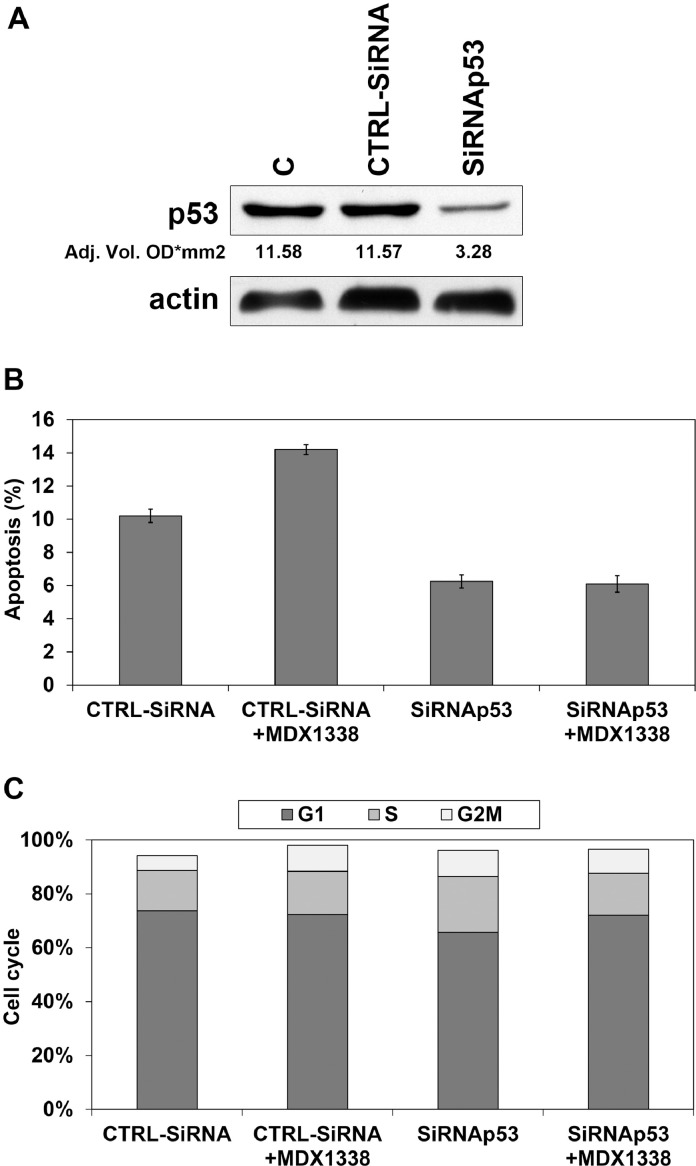
Transfection of siRNA duplexes targeting p53 markedly reduced the protein level after 48 h from the transfection, while p53 levels were not affected by CTRL siRNA (Fig. 10A).
Fig. 10.
Response of U2OS to MDX1338 after p53 silencing. A) Western Blot analysis of p53 showed a marked reduction of protein expression after MDX1338 incubation. p53 levels were not affected by CTRL siRNA. B) CTRL siRNA transfected cells exposed to MDX1338 for 48 h responded by increasing apoptotic fraction, while no differences occurred in p53siRNA transfected cells. C) A slight delay in cell cycle progression was seen in both CTRL siRNA and p53 siRNA transfected cells after MDX1338 treatment.
Actin = reference protein; C= control; CTRLsiRNA= Negative Control siRNA,.
After 48 h of MDX1338 exposure an increase of apoptotic fraction associated to the increase of G2/M phase (Fig. 10B, C) was seen only in U2OS transfected with CTRLsiRNA. Although p53siRNA transfected cells responded with a slight increase of G1 cells, no differences in apoptosis were seen (Fig. 10B, C).
4. Discussion
In osteosarcoma adjuvant and neo-adjuvant chemotherapy includes doxorubicin, methotrexate and cisplatin as first choice drugs and ifosfamide in poor responder patients (second-line therapy). The first cause of treatment failure is high toxicity and natural and acquired drug-resistance that occurs in 30–40% of OS patients and is associated to metastatic progression [2], [4], [5]. Thus, it is necessary to identify new prognostic and predictive biomarkers that may represent targets for new anti-neoplastic agents able to increase drug sensitivity and better control localized and metastatic disease.
CXCR4 overexpression has been found in many tumours [10], [11], [22], and there is evidence that CXCR4/CXCL12 axis activates downstream multiple pathways that play an important role in tumour progression [7], [8], [23].
In this study we demonstrated that in OS the expression of CXCR4 significantly increased as histological grade and aggressiveness increased (high grade versus low grade; relapsed versus disease-free). The possible prognostic role was supported by univariate analyses that showed a higher probability of local and/or systemic relapse in OS patients with levels of CXCR4 gene expression above the cut-off and a significant increase of metastatic risk associated with an increasing score of CXCR4 protein staining intensity. These results agree with previous studies that found a correlation between chemokine expression and tumour development [22], [24], [25], [26]. Furthermore, the gene expression profile study (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE32981) showed a significant CXCR4 expression in metastatic compared to non metastatic OS patients [27].
In a series of synovial sarcoma the loss of CXCR4 nuclear expression was related with improvement of overall survival [28].
The prognostic role of CXCR4 and its involvement in activation of the most important growth signalling pathways makes it suitable as target for more personalize therapies. This is emphasized by the results on the activity of CXCR4 inhibitors to suppress metastatic spread of OS cells playing a complementary role to current chemotherapy [29], [30].
The antagonist AMD3100, originally approved as a mobilizing agent of hemopoietic precursors [31], [32], is considered an effective chemosensitizing agent [17], [18], [33], [34], [35] that reduces cell survival, migration and angiogenesis through inhibition of CXCR4 downstream targets [17], [33], [36].
Based on the multiple activities of the CXCR4/CXCL12 axis other antagonists have been studied [29] including the fully human CXCR4 antibody, MDX1338, tested in vitro and in vivo for hematologic malignancy [37] and in phase I clinical trials in patients with leukemia and multiple myeloma (NCT01120457, NCT01359657). In xenograft models, MDX1338 binding CXCR4-expressing cells blocks CXCL12, induces apoptosis and inhibits tumour growth [37]. In vitro and in vivo data obtained by non small cell lung cancer demonstrated that response to human anti-CXCR4 antibody correlated with CXCR4 expression [38]. To date no data are reported about MDX1338 activity in OS cells. Although CXCR4 protein expression was present in a high percentage of tumour cells in the OS cell lines included in this study, SAOS-2 presented lower CXCR4 gene expression levels than U2OS and 143B The concomitant presence of low CXCR4 gene levels and high CXCR4 protein expression may be caused by mRNA degradation after translation accompanied by a high-half life of the protein that remains in the cellular pool.
According to the role of CXCR4/CXCL12 axis in mediating cancer cell migration [9], [17], we found that OS cells exposed to CXCR4 inhibitors delayed the wound healing compared to non-treated cells. Interestingly, MDX1338 caused a more marked and long-term slowdown than AMD3100.
In terms of cell proliferation, the response of CXCR4-positive OS cells cultured in a CXCL12-rich medium to MDX1338 was similar to that of AMD3100, with a decrease up to 30%–40% of the percentage of control.
Moreover, a modulation of cell cycle also occurred in stimulated p53-positive U2OS and 143B cell lines. MDX1338 and AMD3100 caused a slowing-down of cell cycle progression with G2/M and G1 lengthening respectively, associated with an increase of apoptotic fraction.
These results agree with recent data that demonstrated that CXCR4 down-regulation induced apoptosis in OS cell lines, and indicated CXCR4 as a potential target for OS therapy [39]. Our study revealed that there is a delay in cell cycle progression suggesting checkpoint activation that strengthen the cell response to anticancer agents. This could sensitize p53-positive OS cells to radiation or chemotherapy [40].
No effects were seen for apoptosis in p53-negative SAOS-2 cells that in basal conditions differ also for a slower proliferation rate with presence of a more mature osteoblastic labeling profile compared to U2OS and 143B [41]. These data suggest that the different response of SAOS-2 cells to anti-CXCR4 agents could be influenced both by the lack of p53 and , as some Authors stated, by a predominant osteoblastic differentiation that may promote a different cross-talk between tumour cells and environment [41], [17].
To support the role of p53 in OS cells response to MDX1338 we transfected U2OS with p53 siRNA that markedly decreased the p53 protein level. Although p53siRNA transfected cells responded with a slight increase of G1 phase, the percentage of apoptotic cells did not change suggesting a cytostatic role of MDX3100 in cells with a reduced expression of p53.
5. Conclusions
Our data support the prognostic role of CXCR4 in human OS malignant progression and demonstrate that similarly to AMD3100, MDX1338 reduced OS cell proliferation and decreased cell motility in a more marked and long-term way.
Moreover, the delay in cell cycle progression associated with increased apoptosis in U2OS and 143B could sensitize p53-positive cells to conventional therapy. In vivo preclinical experiments are on going on xenograft models using CXCR4 antagonists alone and combined with the aim of suggesting new target therapies in human osteosarcoma.
Acknowledgments
Acknowledgments
The authors wish to thank Dr Alba Balladelli for editing the manuscript and Ms. Cristina Ghinelli for graphic work.
Funding
This study was supported by 5‰ citizen income tax contribution to the Rizzoli Orthopaedic Institute and from the Italian Ministry of Health (No. 2016-02361373).
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2019.100239.
Appendix. Supplementary materials
References
- 1.Picci P. Osteosarcoma (Osteogenic Sarcoma) Orphanet J. Rare Dis. 2007;2:6. doi: 10.1186/1750-1172-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picci P. Classic osteosarcoma. In: Manfrini M., Fabbri N., Gambarotti M., Vanel D., editors. Atlas of Muscoloskeletal Tumors and Tumorlike Lesions. Springer International Publishing; 2014. pp. 147–151. [Google Scholar]
- 3.Klein M.J., Siegal G.P. Osteosarcoma: anatomic and histologic variants. Am. J. Clin. Pathol. 2006;4:555–581. doi: 10.1309/UC6K-QHLD-9LV2-KENN. [DOI] [PubMed] [Google Scholar]
- 4.Harrison D.J., Schwartz C.I. Osteogenic sarcoma: systemic chemotherapy options for localized disease. Curr. Treat. Options Oncol. 2017;18:24. doi: 10.1007/s11864-017-0464-2. [DOI] [PubMed] [Google Scholar]
- 5.Osanan S., Zang M., Shen F., Paul P.J., Persad S., Sergi C. Osteogenic Sarcoma: a 21st century review. Anticancer Res. 2016;36:4391–4398. doi: 10.21873/anticanres.10982. [DOI] [PubMed] [Google Scholar]
- 6.Cortini M., Avnet S., Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. doi: 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Cojoc M., Peitzsch C., Trautmann F., Polishchuk L., Telegeev G.D., Dubrovska A. Emerging targets in cancer managment: role of the CXCL12/CXCR4 axis. Onco Targets Ther. 2013;6:1347–1361. doi: 10.2147/OTT.S36109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teicher B.A., Fricker S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 9.Domanska U.M., Kruizinga R.C., Nagengast W.B., Timmer-Bosscha H., Huls G., de Vries E.G., Walenkamp A.M. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur. J Cancer. 2013;49:219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Liang J.X., Gao W., Liang Y., Zhou X.M. Chemokine receptor CXCR4 expression and lung cancer prognosis: a meta-analysis. Int. J. Clin. Exp. Med. 2015;8:5163–5174. [PMC free article] [PubMed] [Google Scholar]
- 11.Du Y., Long Q., Guan B., Mu L. Prognostic value of high CXCR4 expression in renal cell carcinoma: a system review and meta-analysis, 2015. Dis. Mark. 2015 doi: 10.1155/2015/568980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y., Mao X., Fan C., Liu C., Guo A., Guan S., Jin Q., Li B., Yao F., Jin F. CXCL12-CXCR4 axis promotes the natural selection of breast cancer cell metastasis. Tumour Biol. 2014;35:7765–7773. doi: 10.1007/s13277-014-1816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanelli M.F., Chinen L.T., Begnami M.D., Costa W.L., Jr, Fregnami J.H., Soares F.A., Montagnini A.L. The influence of transforming growth factor-á, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial-mesenchymal transition on overall survival of patients with gastric cancer. Histopathology. 2012;61:153–161. doi: 10.1111/j.1365-2559.2011.04139.x. [DOI] [PubMed] [Google Scholar]
- 14.Bachet J.B., Maréchal R., Demetter P., Bonnetain F., Couvelard A., Svrcek M., Bardier-Dupas A., Hammel P., Sauvanet A., Louvet C., Paye F., Rougier P., Penna C., Vaillant J.C., André T., Closset J., Salmon I., Emile J.F., Van Laethem J.L. Contribution of CXCR4 and SMAD4 in predicting disease progression pattern and benefit from adjuvant chemotherapy in resected pancreatic adenocarcinoma. Ann. Oncol. 2012;23:2327–2335. doi: 10.1093/annonc/mdr617. [DOI] [PubMed] [Google Scholar]
- 15.Oda Y., Yamamoto H., Tamiya S., Matsuda S., Tanaka K., Yokoyama R., Iwamoto Y., Tsuneyoshi M. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod Pathol. 2006 May;19(5):738–745. doi: 10.1038/modpathol.3800587. [DOI] [PubMed] [Google Scholar]
- 16.Ren Z., Liang S., Yang J., Han X., Shan L., Wang B., Mu T., Zhang Y., Yang X., Xiong S., Wang G. Coexpression of CXCR4 and MMP9 predicts lung metastasis and poor prognosis in resected osteosarcoma. Tumour Biol. 2016;37:5089–5096. doi: 10.1007/s13277-015-4352-8. [DOI] [PubMed] [Google Scholar]
- 17.Fontanella R., Pelagalli A., Nardelli A., D'Alterio C., Ieranò C., Cerchia L., Lucarelli E., Scala S., Zanetti A. A novel antagonist of CXCR4 prevents bone marrow-derived mesenchymal stem cell-mediated osteosarcoma and hepatocellular carcinoma cell migration and invasion. Cancer Lett. 2016;370:100–107. doi: 10.1016/j.canlet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Jiang C., Fang X., Zhang H., Wang X., Li M., Jiang W., Tian F., Zhu L., Bian Z. AMD3100 combined with triptolide inhibit proliferation, invasion and metastasis and induce apoptosis of human U2OS osteosarcoma cells. Biomed. Pharmacother. 2017;86:677. doi: 10.1016/j.biopha.2016.12.055. 685. [DOI] [PubMed] [Google Scholar]
- 19.Kashyap M.K., Kumar D., Jones H., Amaya-Chanaga C.I., Choi M.Y., Melo-Cardenas J., Ale-Ali A., Kuhne M.R., Sabbatini P., Cohen L.J., Shelat S.G., Rassenti L.Z., Kipps T.J., Cardarelli P.M., Castro J.E. Ulocuplumab (BMS-936564/MDX1338): a fully human anti-CXCR4 antibody induces cell death in chronic lymphocytic leukemia mediated through a reactive oxygen species-dependent pathway. Oncotarget. 2016;3:2809–2822. doi: 10.18632/oncotarget.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dar A., Schajnovitz A., Lapid K., Kalinkovich A., Itkin T., Ludin A., Kao W.M., Battista M., Tesio M., Kollet O., Cohen N.N., Margalit R., Buss E.C., Baleux F., Oishi S., Fujii N., Larochelle A., Dunbar C.E., Broxmeyer H.E., Frenette P.S., Lapidot T. Rapid mobilization of hematopoietic progenitors by AMD3100 and catecholamines is mediated by CXCR4-dependent SDF-1 release from bone marrow stromal cells. Leukemia. 2011;25:1286–1296. doi: 10.1038/leu.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierini M., Dozza B., Lucarelli E., Tazzari P.L., Ricci F., Remondini D., di Bella C., Giannini S., Donati D. Efficient isolation and enrichment of mesenchymal stem cells from bone marrow. Cytotherapy. 2012;14:686–693. doi: 10.3109/14653249.2012.677821. [DOI] [PubMed] [Google Scholar]
- 22.Scala S., Ottaiano A., Ascierto P.A., Cavalli M., Simeone E., Giuliano P., Napolitano M., Franco R., Botti G., Castello G. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin. Cancer Res. 2005;11:1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- 23.Kim R.H., Li B.D.L., Chu Q.D. The role of chemokine receptor CXCR4 in the biologic behavior of human soft tissue sarcoma. Sarcoma. 2011;2011 doi: 10.1155/2011/593708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yopp A.C., Shia J., Butte J.M., Allen P.J., Fong Y., Jarnagin W.R., DeMatteo R.P., D'Angelica M.I. CXCR4 expression predicts patient outcome and recurrence patterns after hepatic resection for colorectal liver metastasis. Ann. Surg. Oncol. 2012;19(Suppl 3):S339–S346. doi: 10.1245/s10434-011-1774-4. [DOI] [PubMed] [Google Scholar]
- 25.Werner T.A., Forster C.M., Dizdar L., Verde P.E., Raba K., Schott M., Knoefel W.T., Krieg A. CXCR4/CXCR7/CXCL12 axis promotes an invasive phenotype in medullary thyroid carcinoma. Br. J. Cancer. 2017;117:1837–1845. doi: 10.1038/bjc.2017.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laverdiere C., Hoang B.H., Yang R., Sowers R., Qin J., Meyers P.A., Huvos A.G., Healey J.H., Gorlick R. Messenger RNA expression levels of CXCR4 correlate with metastatic behaviour and outcome in patients with osteosarcoma. Clin. Cancer Res. 2005;11:2561–2567. doi: 10.1158/1078-0432.CCR-04-1089. [DOI] [PubMed] [Google Scholar]
- 27.Namløs H.M., Kresse S.H., Müller C.R., Henriksen J., Holdhus R., Sæter G., Bruland O.S., Bjerkehagen B., Steen V.M., Myklebost O. Global gene expression profiling of human osteosarcomas reveals metastasis-associated chemokine pattern. Sarcoma. 2012;2012 doi: 10.1155/2012/639038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmerini E., Benassi M.S., Quattrini I., Pazzaglia L., Donati D., Benini S., Gamberi G., Gambarotti M., Picci P., Ferrari S. Prognostic and predictive role of CXCR4, IGF-1R and Ezrin expression in localized synovial sarcoma: is chemotaxis important to tumor response? Orphanet J. Rare Dis. 2015;10:6. doi: 10.1186/s13023-014-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scala S. Molecular pathways: targeting the CXCR4–CXCL12 axis-untapped potential in the tumor microenvironment. Clin. Cancer Res. 2015;21:4278–4285. doi: 10.1158/1078-0432.CCR-14-0914. [DOI] [PubMed] [Google Scholar]
- 30.Brennecke P., Arlt M.J., Campanile C., Husmann K., Gvozdenovic A., Apuzzo T., Thelen M., Born W., Fuchs B. CXCR4 antibody treatment suppresses metastatic spread to the lung of intratibial human osteosarcoma xenografts in mice. Clin. Exp. Met. 2014;31:339–349. doi: 10.1007/s10585-013-9632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Clercq E. The AMD3100 story: the path to the discovery of a stem cell mobilizer (Mozobil) Biochem. Pharmacol. 2009;77:1655–1664. doi: 10.1016/j.bcp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Rettig M.P., Ansstas G., DiPersio J.F. Mobilization of hematopoietic stem and progenitor cells using inhibitors of CXCR4 and VLA-4. Leukemia. 2012;26:34–53. doi: 10.1038/leu.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y.X., Fu Z.Z., Zhou C.H., Shan L.C., Wang Z.W., Yin F., Zheng L.P., Hua Y.Q., Cai Z.D. AMD3100 reduces CXCR4-mediated survival and metastasis of osteosarcoma by inhibiting JNK and Akt, but not p38 or Erk1/2, pathways in in vitro and mouse experiments. Oncol. Rep. 2015;34:33–42. doi: 10.3892/or.2015.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domanska U.M., Timmer-Bosscha H., Nagengast W.B., Oude Munnink T.H., Kruizinga R.C., Ananias H.J., Kliphuis N.M., Huls G., De Vries E.G., de Jong I.J., Walenkamp A.M. CXCR4 inhibition with AMD3100 sensitizes prostate cancer to docetaxel chemotherapy. Neoplasia. 2012;14:709–718. doi: 10.1593/neo.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakamatsu T., Naka N., Sasagawa S., Tanaka T., Takenaka S., Araki N., Ueda T., Nishizawa Y., Yoshikawa H., Itoh K. Deflection of vascular endothelial growth factor action by SS18–SSX and composite vascular endothelial growth factor- and chemokine (C-X-Cmotif) receptor 4-targeted therapy in synovial sarcoma. Cancer Sci. 2014;105:1124–1134. doi: 10.1111/cas.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X., Charbonneau C., Wei L., Yang W., Chen Q., Terek R.M. CXCR4-targeted therapy inhibits VEGF expression and chondrosarcoma angiogenesis and metastasis. Mol. Cancer Ther. 2013;12:1163–1170. doi: 10.1158/1535-7163.MCT-12-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhne M.R., Mulvey T., Belanger B., Chen S., Pan C., Chong C., Cao F., Niekro W., Kempe T., Henning K.A., Cohen L.J., Korman A.J., Cardarelli P.M. BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin. Cancer Res. 2013;19:357–366. doi: 10.1158/1078-0432.CCR-12-2333. [DOI] [PubMed] [Google Scholar]
- 38.Azad B.B., Chatterjee S., Lesniak W.G., Lisok A., Pullambhatla M., Bhujwalla Z.M., Pomper M.G., Nimmagadda S. A fully human CXCR4 antibody demonstrates diagnostic utility and therapeutic efficacy in solid tumor xenografts. Oncotarget. 2016;7:12344–12358. doi: 10.18632/oncotarget.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C., Ma S., Hu R., Wang X., Li M., Tian F., Jiang W., Zhu L., Bian Z. Effect of CXCR4 on apoptosis in osteosarcoma cells via the PI3K/Akt/NF-êâ signaling pathway. Cell. Physiol. Biochem. 2018;46:2250–2260. doi: 10.1159/000489593. [DOI] [PubMed] [Google Scholar]
- 40.Taylor W.R., Stark G.R. Regulation of G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 41.Pautke C., Schieker M., Tischer T., Kolk A., Neth P., Mutschler W., Milz S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 2004;24:3743–3748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.