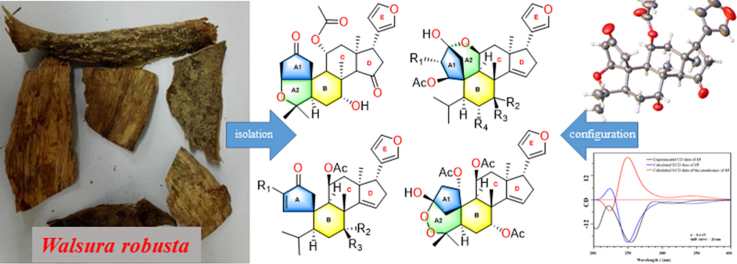
Abstract
Screening active natural products, rapid identification, and accurate isolation are of great important for modern natural lead compounds discovery1. We hereby reported the isolation of seven new neotecleanin-type limonoids (1—7), seven new limonoids with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system (8—14), and two new precursors (15—16) together with four known limonoids (17—20) from the root barks of Walsura robusta. Their structures, including their absolute configurations, were elucidated based on analyses of HR-ESI-MS, 1D/2D NMR, ECD spectrum calculations and single-crystal X-ray diffraction techniques. Compounds 2, 8, 9, 11, 13, 14, 18 showed significant anti-inflammatory activities in LPS-induced RAW 264.7 cell line, BV2 microglial cells, and Propionibacterium acnes-stimulated THP-1 human monocytic cells. Walrobsin M (11) exhibited anti-inflammatory activity with IC50 value of 7.96±0.36 μmol/L, and down-regulated phosphorylation levels of ERK and p38 in a dose-dependent manner.
KEY WORDS: Walsura robusta, limonoid, Neotecleanin-type, ECD spectrum calculation, Single-crystal X-ray diffraction, Anti-inflammatory activity, Propionibacterium acnes, THP-1 human monocytic cell
Graphical abstract
Twenty A/B spiro-type limonoids (1—20) including seven new neotecleanin-type liminoids (1—7), seven novel limonoids (8—14) with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system, and two key precursors (15—16) were isolated from the root barks of Walsura robusta. Walrobsin M (11) significantly inhibited inflammatory activity with IC50 value of 7.96 μmol/L, and down-regulated phosphorylation levels of ERK and p38 in a dose-dependent manner.
1. Introduction
Natural products (secondary metabolites) revealing interesting properties as complex molecules and/or are widely recognized as an excellent source for target drug candidate discovery1. Plants of the genus Walsura from Meliaceae2 are rich sources of bioactive limonoid derivatives with complex and diverse structures3., 4., 5., 6., 7., 8., 9.. Three Walsura species are widely distributed in south of China, such as Yunnan, Guangxi, and Hainan provinces10., 11.. In recent years, the studies of Walsura species discovered some biologically active limonoids with unprecedented carbon skeletons, such as antimalarial walsuronoid A12, 11β-HSD1-inhibited walsucochinoids D and E7, neuroprotective walsucochins A and B13 and anti-inflammatory walrobsins A and B10. In recent years, more and more new limonoids from Walsura genus have been reported. However, few A/B spiro-type limonoids with significant anti-inflammatory activities were reported. High abundances of cedrelone type limonoid and low abundances of A/B spiro-type limonoid have made research on these compounds difficult3., 4., 11.. In our previous study, two novel limonoids with unprecedented 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system were isolated and reported, and walrobsin A showed significant anti-inflammatory activity by inhibiting the expression of iNOS and IL-1β10. In our efforts to discover novel bioactive limonoids in Walsura, we decided to rapidly filter “impurious”14., 15., 16. limonoids and targeted the A/B spiro-type limonoids based on the characteristic ultraviolet absorption distinction between cedrelones, including cedrelone, 11β-acetoxycedrelone, 11β-hydroxycedrelone (Supporting Information Fig. S1) and walrobsins to guide the isolation based on HPLC—DAD method for the first time.
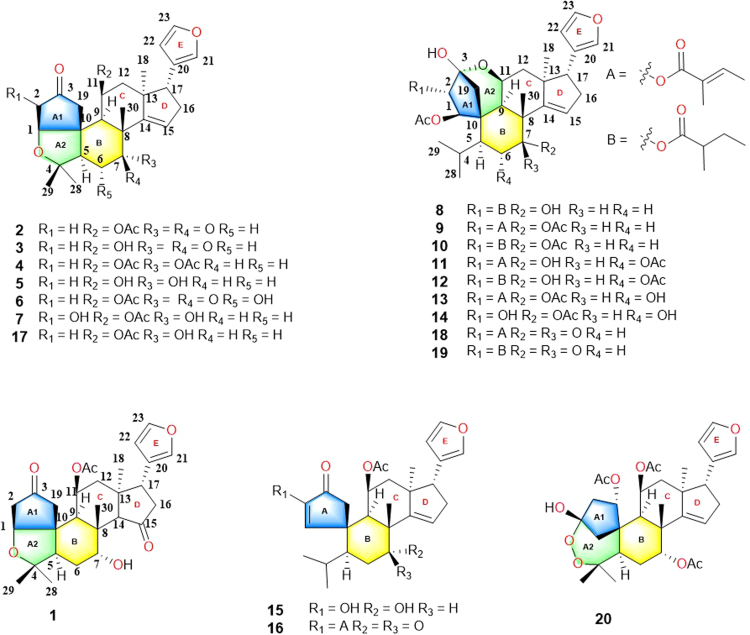
In this study, four types of A-ring rearranged limomoids were isolated and identified, including 8 neotecleanin-type limonoids (7 new compounds 1—7, and a known compound 17), 9 limonoids with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system (7 new compounds 8—14, and 2 known compounds 18 and 19), and 2 key new precursors (15—16) together with the first known limonoid peroxide (20) (Fig. 1). Their planar structures, relative configurations and absolute configurations were assigned by comprehensive comparisons and analyses of NMR, HR-ESI-MS and X-ray crystallography. Compounds 1—3, 8—14, and 18—20 were screened their inflammatory activities in three models including LPS-induced RAW 264.7, BV2 and Propionibacterium acnes-stimulated THP-1 cell lines. Walrobsin M (11) exhibited significant anti-inflammatory activity in the P. acnes-induced THP-1 cell line with IC50 value of 7.96±0.36 μmol/L (retinoic acid as the positive control), and reduced the phosphorylation levels of ERK and p38 in a dose-dependent manner in WWestern blot experience. Herein, we described the isolation, structural identification, and biological evaluation of isolated limonoids.
Figure 1.
The structures of compounds 1—20.
2. Result and discussion
Twenty A/B spiro-type limonoids (Fig. 1) including 7 new limonoids with neotecleanin-type limonoids (1—7), 7 novel limonoids with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system (8—14), and 2 key precursors (15—16) along with 4 known limonoids (17—20) were isolated from the root barks of Walsura robusta based on the application of HPLC with DAD detector and preparative HPLC instruments. In this study, the starting point is that the crude extracts of the root barks of W. robusta was detected by HPLC with the reference substance cedrelone, 11β-acetoxycedrelone and 11β-hydroxycedrelone (Supporting Information Fig. S1) after regular liquid—liquid extraction. Then, the major constituents, including cedrelone, 11β-acetoxycedrelone and 11β-hydroxycedrelone, were located and filtered by preparative HPLC instruments with UV detector. Thirdly, the trace fractions of the constituent was enriched and re-verified by HPLC. Subsequently, careful isolation by comprehensive column chromatography followed by HPLC and purified by preparative HPLC afforded analytically pure compounds 1—20. Their structures, including their absolute configurations, were elucidated based on analyses of HR-ESI-MS, 1D/2D NMR, ECD spectrum calculations and single-crystal X-ray diffraction techniques.
Compound 1 was obtained as colorless crystals. The molecular formula of 1 was determined as C28H36O7 based on the positive model HR-ESI-MS ion peak at m/z 502.2802 [M+NH4]+ (Calcd. 502.2799) and 13C NMR data, incorporating 11 indices of hydrogen deficiency. The diagnostic 1D NMR data implied the presence of a β-substituted furan moiety (δH 7.39, 7.11, 6.15, s, 3H each s; δC 143.5, 139.5, 122.6, 110.5), 2 carbonyl carbons (δC 220.8, 217.5) and an acetyl group (δH 2.09, s; δC 170.1, 21.6), which accounted for 6 out of the 11 indices of hydrogen deficiency. The aforementioned data suggested that 1 is a limonoid possessing a pentacyclic framework similar to walsuranin B17.
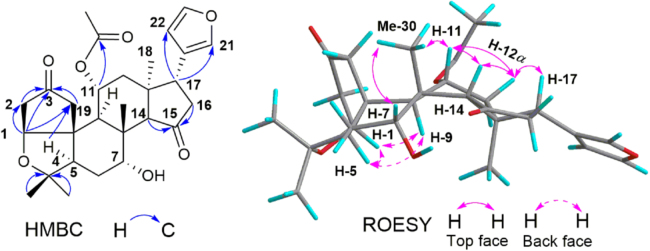
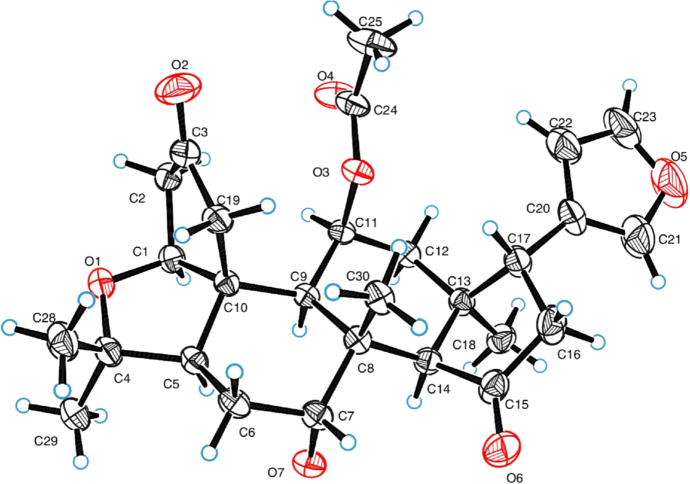
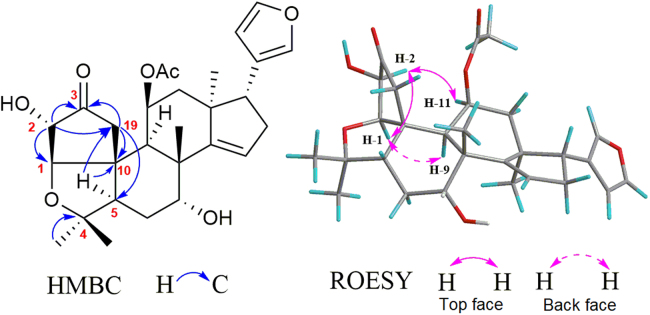
The planar structure of 1 was assigned form its 2D NMR data (Fig. 2). Two carbonyl groups in rings A1 and D were assigned by the HMBC correlations from H2-19 (δH 2.90, d, J=19.0 Hz; 2.50, d, J=19.0 Hz) and H2-2 (δH 2.42, m; 2.35, dt, J=18.0, 3.0 Hz) to C-3 (δC 217.5), and from H2-16 (δH 2.52, d, J=10.0) and H-14 (δH 2.88, s) to C-15 (δC 220.8), respectively. The acetyl group was located at C-11 (δC 72.5) based on the HMBC correlation from H-11 (δH 5.20, brd s) to acetyl carbonyl carbon. The located position of β-substituted furan moiety was determined at C-17 (δC 38.9) by the HMBC correlations from H-17 (δH 3.76, t, J=10.0 Hz) to C-20, C-21 and C-22. The ROESY correlations between H-17, H-12α (δH 2.26, dt, J=16.0, 2.5 Hz), H-14 and H-11, between Me-30 (δH 1.31, s) and H-7 (δH 3.84, m) indicated that these protons were co-facial and α-oriented. The ROESY correlations between H-1, H-5 (δH 2.15, m) and H-9 (δH 1.87, d, J=2.5 Hz) suggested that they were α-orientation. The ring A2 was speculated as a tetrahydrofuran ring built by the ether bond between C-1 and C-4. However, this speculation could not be confirmed according to the unobserved HMBC correlation from H-1 (δH 4.30, d, J=3.0 Hz) to C-4 (δC 81.1). The spectroscopically elucidated structure of 1 was ultimately confirmed though a single-crystal X-ray diffraction study using Cu Kα radiation [Flack parameter of 0.01], and the absolute configuration of 1 was assigned as 1S, 5R, 7R, 8 R, 9S, 10R, 11R, 13S, 14S and 17R (Fig. 3).
Figure 2.
Selective HMBC and ROESY correlations of compound 1.
Figure 3.
ORTEP drawing of the compound 1.
Compound 2, a white amorphous powder, exhibited the molecular formula of C28H34O6 as deduced from the (+)-HR-ESI-MS ion at m/z 484.2692 [M+NH4]+ (Calcd. 484.2694, C28H38NO6) and 13C NMR data. Comparison of the 1D and 2D NMR data (Supporting Information Fig. S2) of 1 and 2 showed similarity except for the presence of an additional olefinic hydrogen in 1H NMR data and two additional olefinic carbons in 13C NMR. Thus, the afore-mentioned information suggested that they were closely related analogues featuring identical carbon frameworks. The olefinic bond was located as C-14 (δC 150.7) and C-15 (δC 128.3) as reported walrobsin A10, which was confirmed by the HMBC correlation from H-15 (δH 6.22, brd s) to C-16 (δC 34.7) and C-17 (δC 52.2). The corresponding carbonyl groups were located at C-3 (δC 216.4) and C-7 (δC 207.4) instead of C-3 and C-14. The HMBC correlations from H-5 (δH 2.17, dd, J=18.5, 3.5 Hz), H2–6 (δH 2.78, t, J=15.0 Hz; 2.44 m) and H-9 (δH 2.55 overlapped) to C-7 confirmed the conclusion, and the structure of 2 were thus determined as depicted. The molecular weight of compound 3 showed 42 Da less than 2 as deduced from the (+)-HR-ESI-MS ion at m/z 425.2324 [M+H]+(Calcd. 425.2323, C26H33O5) and 13C NMR data of 3. Comparison of the 1H NMR data (Supporting Information Fig. S3) of 2 and 3 exhibited similarity except for the absence of an acetyl group. Thus, the structure of 3 was determined as 11-deacetyl derivative of 2.
Compounds 4 and 5, white amorphous powders, showed that their molecular formulas were C30H38O7 and C26H34O5 deduced from the (+)-HR-ESI-MS ions at m/z 528.2957 [M+NH4]+ and 444.2742 [M+NH4]+, respectively, and also from their 13C NMR data. Comparison of the 1D NMR data of 4 and 5 with those of walsuranin B (17)17 suggested that 4 was a 7-acetyl derivative of 17 and 5 was an 11-deacetyle derivative of 17, respectively. Analysis of the 1D NMR data and ESI-MS data of 4 and 5 confirmed this deduction by the presence of diagnostic resonances of 2 acetyloxy groups (δH 1.98, 1.97 each 3Hs; δC 170.5, 21.6; 169.8, 21.3) in 4 and the absence of acetyloxy group in 5. The key HMBC correlation of 4 from H-7 (δH 5.24, t, J=2.5 Hz) to an acetyl group (δC 169.8) determined its structure as 7-acetyl derivative of 17 as shown in Fig. 1. Compared to the 1H NMR data of 4, the absence of two acetyl groups in that of 5 indicated that 5 was an 11-deacetyle derivative of 17, which was consistent with the HMBC correlations from H-7 (δH 3.96, t, J=3.0 Hz) to C-8 (δC 45.9), C-6 (δC 26.4) and from H-11 to C-9 (δC 43.2) and C-12 (δC 45.7).
Compound 6, a white amorphous powder, displayed a molecular ion at m/z 502.2803 [M+NH4]+ (Calcd. 502.2799) in the (+)-HR-ESI-MS data and determined its molecular formula as C28H36O7 together with 13C NMR data. Comparison of the HR-ESI-MS data of 6 and 2 suggested that 6 was 16 Da greater than that of 2 and was identified as the oxidative product of 2. The 1H NMR and 13C NMR spectra of 6 were quite similar to those of 2 except for the chemical shift of H-6 down shifted from δH 2.47 to 4.07 and the chemical shift of C-6 down shifted from δC 38.2 to 68.4. Thus, compound 6 was deduced as 5-hydroxy derivative of 2, which was determined by HMBC correlations from H-7 (δH 3.92, brd s), H-5 (δH 2.51, m) to C-6 (δC 68.4). In ROESY spectra, the correlation between H-6 (δH 4.07, m) and H-7, Me-30 (δH 1.39, s), H2-19 (δH 2.96, d, J=19.0 Hz; 2.56, J=19.0 Hz) suggested the 6-OH and 7-OH were β-orientation. The structure of 6 was determined as shown and named as walrobsin H.
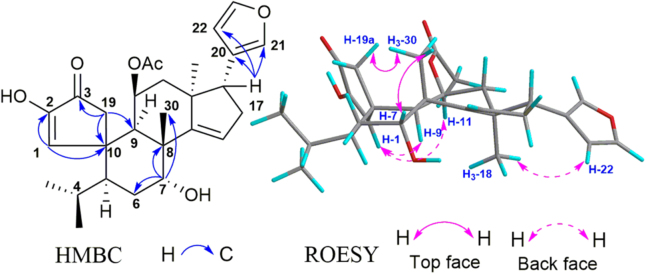
The 1H NMR and 13C NMR data of 7 were similar to those of walsuranin B (17), with the major difference being the chemical shift of H-2 down shifted from δH 2.13 to 3.51 and the chemical shift of C-2 down shifted from δC 45.2 to 58.6. The molecular weight of 7 exhibiting 16 Da greater than that of 17 in ESI-MS data indicated that compound 7 is a 2-oxidation product of 17. Its molecular formula was determined as C28H36O7 based on a molecular ion at m/z 502.2801 [M+NH4]+ (Calcd. C28H40NO7 502.2799) in positive HR-ESI-MS spectra. The special A1-A2 ring structure could be determined by the key HMBC correlations of H-1/C-2, C-3, C-19, H-2/C-10, H-19/C-9, C-10, Me-28,29/C-4, C-5, and H-5/C-19. The location of OH was assigned at C-2 based on the key HMBC correlations from H-1 (δH 3.79, d, J=2.0 Hz) and H2-19 (δH 2.82, d, J=19.0 Hz; δH 2.73, d, J=19.0 Hz) to C-2, and from H-2 to C-1 (δC 64.4), C-3 (δC 205.7) and C-19 (δC 37.6). The vicinal coupling constant (2.0 Hz) implied cis configuration between H-1 and H-2, which was also determined by the correlations of H-1/H-2, H-1/H-9 and H-2/H-11. Thus, the planar structure and relative configuration of 7 were determined as shown in Fig. 4.
Figure 4.
Selective HMBC and ROESY correlations of 7.
The molecular formula of walrobsin J (8), C33H46O8 with 11 degrees of unsaturation, was deduced from its HR-ESI-MS data with a molecular ion at m/z 593.3085 [M+H]+ (Calcd. for C33H46O8Na, 593.3085). Similar NMR data indicated that 8 was a limonoid the same as 19, except for a 2 amu greater than that of 19 in positive model HR-ESI-MS data, as well as the loss of a carbonyl carbon and the appearance of an oxygen carbon (δC 71.4) in 13C NMR spectrum. Thus, the structure of 8 was determined as shown in Fig. 5, which was elucidated by the key HMBC correlations from H-5 (δH 2.10, m), H-6 (δH 1.68, m) and H3-30 (δH 1.36, s) to C-7 (δC 71.4), and the key ROESY correlation between H3-30 (δH 1.36, s) and H-7 (δH 3.95, s).
Figure 5.
Selective HMBC and ROESY correlations of 8.
The molecular formula of walrobsin K (9) and walrobsin L (10) as C35H46O9 and C35H48O9, respectively, according to their HR-ESI-MS data m/z 611.3212 [M+H]+ (Calcd. for C35H47O9, 611.3215) and m/z 613.3370 [M+H]+ (Calcd. for C35H49O9, 613.3372). Compound 10 was determined as the 7β-acetylate-walrobsin C based on the HMBC correlation from 7-OAc (δH 2.18, s) to C-7 (δC 77.4) and an additional 42 mass unit in HR-ESI-MS data. Walrobsin K (9) was subsequently determined as shown in Fig. 2, whose substitutional group at C-1 (δC 86.6) was “A” instead of “B” in 10.
Walrobsin M (11) and walrobsin N (12) showed same molecular formulas as 9 and 10, respectively, according to their HR-ESI-MS data m/z 649.2980 [M+Na]+ (Calcd. for C35H46O10Na, 649.2983) and m/z 651.3138 [M+Na]+ (Calcd. for C35H48O10Na, 651.3140). Compound 11 differed form 9 only in the location of OAc, which was confirmed by the key HMBC correlation from OAc to C-6 (δC 72.4) instead of C-7. The configuration of OAc was in α position, which was assigned by the ROESY data between H-6 (δH 5.33, dd, J=12.0, 3.0 Hz) and H-4 (δH 2.39, overlapped). The similar condition occurred between 12 and 10, and its structure was determined as shown in Fig. 1.
The HR-ESI-MS data (m/z 627.3166 [M+Na]+; Calcd. for C35H47O10Na, 627.3164) showed an additional oxygen atom in the molecular formula of walrobsin O (13) compared to 9. Compound 13 was assigned as 6α-O-walrobsin K, which was confirmed by the additional oxygen carbon in 13C NMR data, the further key HMBC correlations from H-4 (δH 1.61, d, J=12.0 Hz), H-7 (δH 5.29, d, J=3.0 Hz) to C-6 (δC 68.2), and the key ROESY correlation between H-4 and H-6 (δH 4.25, dd, J=11.5, 3.0 Hz). Walrobsin P (14) differed from 13 in the substituted group at C-1 (δC 91.4), which was confirmed by the disappearance of “A” group in 1H NMR and 13C NMR data, and the loss of 82 amu in its HR-ESI-MS data. Herein, the structures of walrobsins O and P (13 and 14) were established as shown in Fig. 1.
Comprehensive analysis of the experimental ECD of type I compounds 1—7 and 17 (Supporting Information Fig. S17), allowed the establishment of same absolute configuration of chiral centers as 1S, 5R, 8R, 9S, 10R, 11R, 13S, and 17R. The assignment of same absolute configuration of chiral centers in type II compounds 8—14, 18 and 19 as 1R, 2S, 3R, 5S, 8R, 9R, 10S, 11S, 13S, and 17R using the same method.
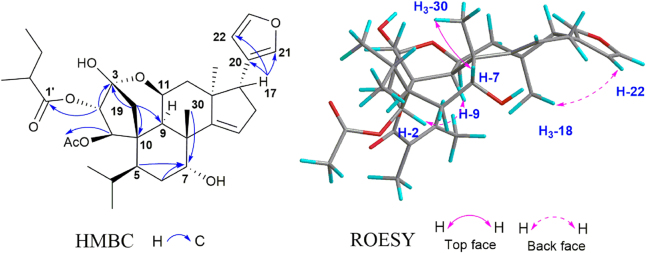
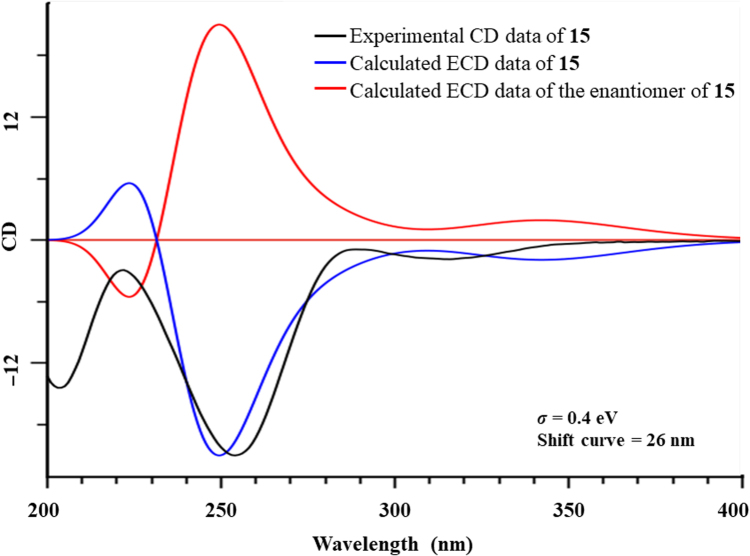
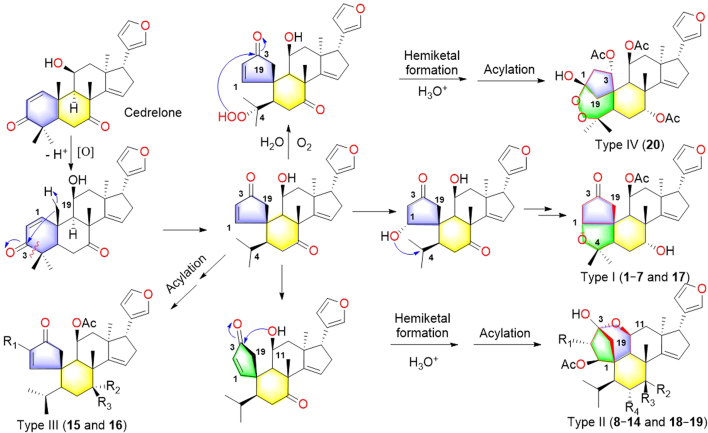
Walrobsin Q (15), a white amorphous powder, was determined to have a molecular formula of C28H36O6 by the ion at m/z 469.2586 [M+H]+ (Calcd. for C28H37O6, 469.2585) in positive model of HR-ESI-MS and 13C NMR data. Comparison of the 1H and 13C NMR data of 15 with walsuranin C17, an characteristic isopropyl motif and a furan ring group, suggested that they were structural analogs. Furthermore, one down field shifted proton resonance at δH 3.95 (t, J=3.0 Hz, H-7) and one corresponding carbon signal at δC 73.1 were appeared in 1H and 13C NMR spectra, respectively. The above information suggested that the carbonyl group located at C-7 in walsuranin C was reduced as hydroxyl group to form walrobsin Q, which was determined by the HMBC correlations from H-7 to C-6 (δC 37.7), C-8 (δC 52.1) and C-30 (δC 29.0) (Fig. 6). The penta-spirocyclic system of A-ring at C-10, and the presence of α,β-unsaturated carbonyl group in A-ring was similar to walsuranin C validated by the obvious HMBC correlations form H-1 to C-2, C-3, C-10, and from H-19 to C-3, C-9 and C-10. The planar structure of 15 was arbitrarily assigned as in Fig. 6. The similar 1D NMR data indicated that they shared the same relative configuration in skeleton core. The key ROESY correlations of H-1/H-9, H-11, and H-18/H-22 showed that these protons were co-facial and determined as α-orientation. The correlations of H-30/H-19a, H-7 were also observed in ROESY spectra and assigned these protons as β-orientation. The hydroxyl group were assigned as α-oriented by the ROESY correlations H-7/H-30. Compound 16 have a molecular formula of C31H38O6 by the HR-ESI-MS data and 13C NMR spectra. Comparison of the 1H and 13C NMR data between 16 and walsuranin C17, the absence of an acetyl group at C-11 and the appearance of an additional tigloyl group (δH 7.16, m; 1.91, s; 1.88, d, J=7.0, 1.5 Hz; δC 161.1, 149.8, 127.0, 14.8 12.0) observed in 1H NMR and 13C NMR data, which were also consistent with the HR-ESI-MS data (16, Calcd. for C31H38O6Na, 529.2561; walsuranin C, C28H34O6Na, 489.2253). Thus, the planar and relative structures of 15 and 16 were determined as shown. Unfortunately, compounds 15 and 16 were not crystallized in aqueous methanol solvent. The absolute configuration of 15 were determined as 1E, 5S, 7R, 8R, 9R, 10S, 11S, 13S, 14E and 17R by the well-matched experimental ECD data and the calculated ECD data under time-dependent density functional theory18 (Fig. 7). The ECD data of 15 and 16 detected in acetonitrile solvent were well matched indicating that they shared the same absolute configuration and that of 16 was assigned as 2′E, 1E, 5S, 8R, 9R, 10S, 11 S, 13S, 14E and 17R (Supporting Information Fig. S132). The hypothetical biosynthesis pathway of all isolated compounds was illustrated in Scheme 1. Four types A-ring rearranged limonoids started from cedrelone type limonoids based on comprehensive free radical reaction, oxidation, acylation and hemiketal formation reactions under the help of some enzymes in plant cells (Scheme 1).
Figure 6.
Selective HMBC and ROESY correlations of 15.
Figure 7.
Experimental ECD spectra of compound 15 overlaid with the calculated ECD spectra of 15 and its enantiomers.
Scheme 1.
Hypothetical biosynthetic pathways of four types A-ring rearranged limonoids 1—20.
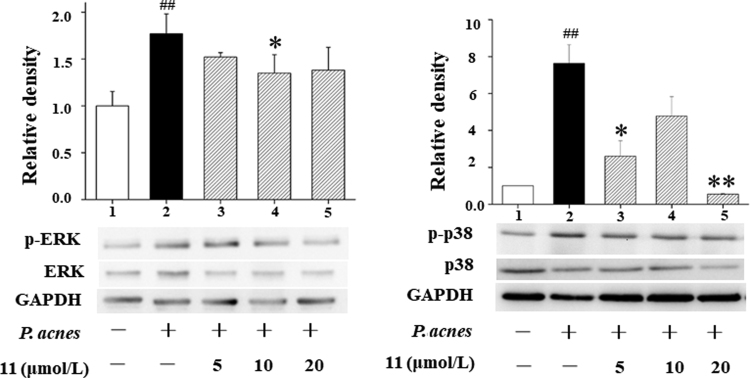
In the anti-inflammatory activity evaluation, compounds 1—3, 8—14, and 18—20 were tested in three inflammatory models including LPS-induced RAW 264.7, BV2 and P. acnes-stimulated THP-1 cell line. Limonoids with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system showed better significant anti-inflammatory activity than other A ring rearranged limonoids in this study as shown in Table 5 implying that the hexatomic oxygen heterocycle is essential. Furthermore, compound 11 exhibited significant anti-inflammatory activity in the P. acnes induced THP-1 cell line with IC50 value 7.96±0.36 μmol/L (retinoic acid as the positive control). In Western blot experience, walrobsin M (11) could reduce the phosphorylation levels of ERK and p38 in a dose-dependent manner (Fig. 8). These results reinforced the significance in the discovery of new anti-inflammatory leading-drugs and/or cosmetic ingredients.
Table 5.
Anti-inflammatory activities of selective compounds on LPS-induced RAW 264.7, BV2 and P. acnes stimulated YHB-1. macrophages.
| No. | IC50a |
||
|---|---|---|---|
| RAW 264.7 | BV2 | THB-1 | |
| 1 | >50 | >50 | >50 |
| 2 | 41.15±2.35 | 21.19±1.43 | >50 |
| 3 | >50 | >50 | >50 |
| 8 | 28.29±0.95 | 15.09±2.10 | 28.5±0.35 |
| 9 | 25.69±1.38 | 22.25±1.36 | 18.01±0.24 |
| 10 | >50 | >50 | >50 |
| 11 | 16.58±1.46 | 20.36±1.37 | 7.96±0.36 |
| 12 | >50 | >50 | >50 |
| 13 | 30.72±2.56 | NTa | 20.05±0.78 |
| 14 | 52.46±3.25 | >50 | 15.97±0.29 |
| 18 | 9.2±0.64 | NT | NT |
| 19 | 52.76±3.23 | >50 | >50 |
| 20 | >50 | >50 | >50 |
| l-NMMAb | 48.15±1.56 | >50 | NA |
| Retinoic acidc | NA | NA | 15.31±1.13 |
NT, not tested. NA, not applicable.
(μmol/L, mean±SD, n = 3).
Positive control substance in RAW 264.7 and BV2 cell line.
Positive control substance in THB-1 cell line.
Figure 8.
Effects of 11 on the MAPK signaling pathway in Propionibacterium ances-stimulated THP-1 cells. The changes of ERK and p38 proteins were examined by Western blot. The data were in three independent experiments. ##P<0.01 compared to control cells; **P<0.01 and *P<0.05 compared to only P. ances-stimulated cells.
3. Conclusions
As an illustrative case study, 20 mg level A/B spiro-type limonoids including 7 new neotecleanin-type limonoids (1—7), 7 novel limonoids with 5-oxatricyclo[5.4.0.11., 4.]hendecane ring system (8—14), and 2 key precursors (15—16) along with four known limonoids (17—20) were isolated from the root barks of W. robusta. In the anti-inflammatory evaluation, compounds 2, 8, 9, 11, 13, 14 and 18 showed significant anti-inflammatory activities in LPS-induced RAW 264.7 cell line, BV2 microglial cells, and P. acnes-stimulated THP-1 human monocytic cells. Walrobsin M (11) significantly inhibited inflammatory activity with IC50 value of 7.96±0.36 μmol/L, and down-regulated phosphorylation levels of ERK and p38 in a dose-dependent manner. Our results further proved a valuable strategy for discovery of trace and/or bioactive compounds.
4. Experimental
4.1. General experimental procedures
Optical rotations were measured with a JASCO P-1020 polarimeter in solvent MeOH at the sodium D line (589 nm) at 25°C. The UV spectra were obtained on a UV-2450 UV—Vis spectrophotometer. Nuclear magnetic resonance (NMR) spectra were on a Bruker AVIII-500 NMR spectrometer (1H: 500 MHz, 13C: 125 MHz) (Bruker, Karlsruhe, Germany), with tetramethylsilane (TMS) as an internal standard. Chemical shift values (δ) are given in parts per million (ppm) and coupling constants in Hertz (Hz). Electrospray ionization (ESI) and high-resolution electrospray ionization (HR-ESI-MS) were carried out an Agilent 1100 series LC/MSD ion trap mass spectrometer and an Agilent 6529B Q-TOF instrument (Agilent Technologies, Santa Clara, CA, USA), respectively. HPLC analyses were performed using an Agilent 1260 system equipped with a RP-C18 column (250 mm × 4.6 mm, 5 μm, Agilent, Santa Clara, CA, USA). Preparative high-performance liquid chromatography (Pre-HPLC) was performed on a Shimadzu LC-8A system equipped with a Shim-pack RP-C18 column (200 mm × 20 mm i.d., 10 μm, Shimadzu, Tokyo, Japan) detected by a binary channel UV detector at 210 and 230 nm. The parameters of flow rate and column temperature were set as 10.0 mL/min and 25 °C , respectively. All solvents used were of analytical grade. Silica gel (200—300 mesh; Qingdao Haiyang Chemical Co., Ltd., Qingdao, China), MCI (Mitsubishi, Tokyo, Japan) and YMC RP-C18 silica (40—63 μm; Milford, MA, USA) were used for column chromatography. Fractions obtained from repetitive column chromatography (CC) were monitored by thin-layer chromatography (TLC) with precoated silica gel GF254 plates (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China). Spots were observed under UV lamp at 254 and/or 365 nm, and then visualized by heating silica gel plates sprayed with vanillin-sulfuric acid.
4.2. Plant material
Air-dried root barks of W. robusta, which were deposited in the Department of Natural Medicinal Chemistry, China Pharmaceutical University (China, accession number 2015-GSS), were collected from Xishuangbanna, China, in September 2015, and were authenticated by Professor Shuncheng Zhang, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, China.
4.3. Extraction and isolation
The dried powder of fruits of W. robusta (5.0 kg) was extracted three times (3 × 5 L) with 95% EtOH—H2O under reflux, and the crude (500 g) was suspended in H2O and extracted with petroleum ether (PE) (3 times with 1 L each) and EtOAc (3 times with 1 L each), successively. The EtOAc extract (100.0 g) was chromatographed on a silica gel column (100 mush, 300 g), eluted with a gradient of CH2Cl2—MeOH (100:1, 50:1, 25:1, 10:1 and 5:1, v/v) to give six fractions (A—F), which were combined based on TLC (vanillin-sulfuric acid as color-developing agent). Medium polarity fraction E (≈10.0 g) was chromatographed over a middle chromatogram isolated (MCI) column eluted with a gradient system of MeOH—H2O (50:5, 75:25 and 95:5, v/v) to afford three subfractions (EA—EC), respectively. EB fraction (≈4.5 g) was sequentially purified by columns of RP-C18 silica gel (MeOH—H2O, 50%—75%, v/v) and then further separated over semi-preparative HPLC to “filter” the major and “impurity” ingredients including cedrelone, 11β-acetoxycedrelone and 11β-hydroxycedrelone, with walrobsins A (18) and B (19) as “light” target as shown in Fig. 1. Finally, 20 new compounds (1—16) and 4 known compounds (17—20) were obtained from the “filtered” extracts. Walrobsins C-R (1—16) and known compounds (17—20) were yielded as 1 (5 mg), 2 (20 mg), 3 (7 mg), 4 (4 mg), 5 (5 mg), 6 (4 mg), 7 (10 mg), 8 (3 mg), 9 (10 mg), 10 (14 mg), 11 (20 mg), 12 (30 mg), 13 (5 mg), 14 (5 mg), 15 (4 mg), 16 (2 mg), 17 (30 mg), 18 (40 mg), 19 (35 mg) and 20 (14 mg), respectively (detailed procedure for extraction and isolation, see Supporting Information).
4.3.1. Walrobsin C (1)
Colorless crystals; [α]23D —18.6 (c 0.139, MeOH); UV (MeOH) λmax (logε) 210 (4.07) nm; IR vmax 3435, 2965, 1727, 1378, 1233, 1171, 1042, 873, 790, 602 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 519.18 [M+Cl]-; positive ESI-MS m/z 502.24 [M+NH4]+; HR-ESI-MS m/z 502.2802 [M+NH4]+ (Calcd. for C28H40NO7, 502.2799).
Table 1.
1H NMR spectroscopic data (δ) for compounds 1—7a (δ in ppm, J in Hz).
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 4.30 d (3.0) | 4.33 d (3.5) | 4.36 brd s | 4.32 d (3.0) | 4.34 brd s | 4.31 d (3.0) | 3.79 d (2.0) |
| 2a | 2.42 m | 2.44 (overlapped) | 2.78 d (18.0) | 2.33 (overlapped) | 2.71 dd (17.5, 3.0) | 2.37 d (18.5) | 3.51 d (2.0) |
| 2b | 2.35 dt (18.0,3.0) | 2.23 dd (18.5, 3.5) | 2.45 (overlapped) | 2.16 dd (18.0, 3.0) | 2.35 (overlapped) | 2.10 dt (18.5, 3.0) | |
| 5 | 2.51 m | 2.17 dd (16.0, 3.0) | 2.16 dd (15.5 3.0) | 2.33 dd (13.5, 3.0) | 2.52 (overlapped) | 2.51 m | 2.39 dd (13.0, 2.0) |
| 6a | 1.91 dt (13.0, 2.0) | 2.78 t (15.0) | 2.80 dd (15.5, 14.0) | 1.89 td (14.0, 2.5) | 1.85 (overlapped) | 4.07 m | 2.05 m |
| 6b | 1.60 m | 2.44 (overlapped) | 2.47 (overlapped) | 1.78 dd (14.0, 3.0) | 1.65 m | ||
| 7 | 3.84 m | 5.24 t (2.5) | 3.96 t (3.0) | 3.92 brd s | 3.89 m | ||
| 9 | 1.87 d (2.5) | 2.55 (overlapped) | 2.31 d (5.5) | 2.53 d (5.5) | 2.38 d (5.5) | 2.46 d (5.5) | 2.58 d (6.5) |
| 11 | 5.20 brd s | 5.14 dt (9.0, 5.5) | 4.34 dt (9.0, 5.5) | 5.16 dt (9.0, 5.5) | 4.34 (overlapped) | 5.13 td (9.0, 6.0) | 5.42 td (10.0, 5.0) |
| 12a | 2.26 dt (16.0, 2.5) | 2.73 dd (14.0, 10.0) | 2.50 (overlapped) | 2.80 dd (14.0, 9.5) | 2.48 (overlapped) | 2.80 dt (14.0, 9.5) | 2.79 dd (14.0, 9.0) |
| 12b | 1.45 dt (16.0, 4.0) | 1.37 dd (14.0, 5.5) | 1.58 dd (13.0, 7.0) | 1.35 dd (14.0, 5.5) | 1.53 dd (13.0, 7.0) | 1.32 dt (16.0, 6.0) | 1.28 (overlapped) |
| 15 | 2.88 s (H-14) | 6.22 brd s | 6.24 t (2.5) | 5.53 t (2.5) | 5.71 t (2.5) | 5.70 brd d (3.0) | 5.69 brd d (3.0) |
| 16a | 2.52 d (10.0) | 2.55 (overlapped) | 2.57 dd (15.5, 2.0) | 2.42 (overlapped) | 2.57 ddd (16.0,11.0, 2.0) | 2.58 (overlapped) | 2.55 ddd (11.0, 9.5, 2.0) |
| 16b | 2.44 (overlapped) | 2.47 (overlapped) | 2.50 (overlapped) | 2.51 (overlapped) | 2.47 ddd (15.5, 7.5, 3.5) | ||
| 17 | 3.76 t (10.0) | 2.84 dd (11.0, 7.5) | 2.89 dd (11.0, 7.0) | 2.83 dd (11.0, 7.0) | 2.92 dd (11.0, 7.0) | 2.89 dd (11.0, 7.0) | 2.87 dd (11.0,7.0) |
| 18 | 0.76 s | 0.80 s | 0.80 s | 0.82 s | 0.82 s | 0.84 s | 0.83 s |
| 19a | 2.90 d (19.0) | 3.15 d (19.0) | 3.36 d (19.0) | 3.03 d (19.0) | 3.13 d (19.0) | 2.96 d (19.0) | 2.82 d (19.0) |
| 19b | 2.50 d (19.0) | 2.64 d (19.0) | 2.55 d (19.0) | 2.46 d (19.0) | 2.33 d (19.0) | 2.56 d (19.0) | 2.73 d (19.0) |
| 21 | 7.11 s | 7.24 s | 7.27 s | 7.24 s | 7.28 s | 7.25 s | 7.25 s |
| 22 | 6.15 s | 6.23 s | 6.28 s | 6.26 s | 6.28 s | 6.26 s | 6.25 s |
| 23 | 7.39 s | 7.36 s | 7.39 s | 7.37 s | 7.40 s | 7.39 s | 7.38 s |
| 28 | 1.27 s | 1.18 s | 1.26 s | 1.23 s | 1.28 s | 1.37 s | 1.36 s |
| 29 | 1.12 s | 1.26 s | 1.18 s | 1.13 s | 1.14 s | 1.37 s | 1.28 s |
| 30 | 1.31 s | 1.55 s | 1.60 s | 1.40 s | 1.35 s | 1.39 s | 1.43 s |
| 11-OAc | 2.09 s | 1.99 s | 1.98 s | 1.96 s | 2.00 s | ||
| 7-OAc | 1.97 s |
NMR data (δ) were measured at 500 MHz in CDCl3 for 1–7.
Table 4.
13C NMR spectroscopic data (δ) for compounds 1—16a (δ in ppm).
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 84.4 | 84.1 | 84.4 | 84.6 | 84.8 | 84.5 | 64.4 | 86.4 | 86.6 | 86.4 | 86.0 | 86.2 | 86.1 | 91.4 | 137.0 | 153.0 |
| 2 | 45.5 | 45.1 | 45.8 | 45.4 | 46.3 | 44.7 | 58.6 | 86.1 | 86.3 | 86.3 | 86.5 | 86.0 | 86.6 | 83.3 | 151.9 | 150.5 |
| 3 | 217.5 | 216.4 | 217.3 | 217.5 | 219.1 | 217.5 | 205.7 | 102.1 | 102.0 | 102.1 | 102.0 | 102.1 | 102.1 | 102.4 | 204.6 | 201.3 |
| 4 | 81.1 | 80.7 | 80.6 | 80.8 | 80.9 | 81.3 | 73.7 | 27.3 | 27.3 | 27.3 | 28.2 | 28.1 | 28.1 | 28.2 | 25.4 | 26.6 |
| 5 | 52.6 | 59.5 | 59.6 | 54.6 | 52.9 | 58.0 | 48.3 | 37.1 | 38.2 | 39.1 | 41.1 | 41.1 | 45.0 | 41.2 | 44.2 | 51.7 |
| 6 | 27.7 | 38.1 | 38.2 | 26.1 | 26.4 | 68.4 | 29.3 | 24.3 | 23.6 | 23.6 | 72.4 | 72.6 | 68.2 | 72.7 | 27.0 | 37.7 |
| 7 | 71.3 | 207.4 | 208.1 | 75.9 | 73.3 | 77.0 | 72.7 | 71.4 | 77.4 | 73.8 | 72.6 | 72.4 | 77.7 | 72.5 | 73.1 | 209.4 |
| 8 | 42.4 | 51.5 | 51.8 | 43.6 | 45.9 | 46.6 | 44.4 | 43.5 | 41.6 | 41.6 | 43.4 | 43.5 | 42.2 | 43.4 | 43.7 | 52.1 |
| 9 | 47.0 | 46.5 | 47.4 | 43.3 | 43.2 | 41.3 | 42.0 | 37.9 | 39.2 | 37.3 | 37.4 | 37.3 | 37.1 | 37.8 | 40.3 | 48.4 |
| 10 | 53.9 | 53.5 | 53.8 | 53.7 | 54.4 | 54.4 | 47.6 | 48.7 | 48.6 | 48.6 | 49.0 | 49.1 | 49.0 | 48.1 | 47.2 | 47.8 |
| 11 | 72.5 | 71.4 | 68.6 | 71.2 | 67.9 | 70.7 | 70.5 | 68.7 | 68.6 | 68.6 | 68.2 | 68.3 | 68.3 | 68.4 | 71.5 | 68.6 |
| 12 | 36.9 | 43.2 | 46.8 | 43.5 | 45.7 | 42.6 | 43.3 | 39.7 | 40.1 | 40.1 | 39.9 | 39.9 | 40.1 | 39.5 | 42.4 | 46.4 |
| 13 | 40.9 | 46.9 | 47.7 | 46.3 | 47.0 | 46.3 | 45.9 | 46.6 | 46.6 | 46.6 | 46.5 | 46.5 | 46.5 | 46.7 | 46.3 | 46.6 |
| 14 | 60.6 | 150.7 | 151.7 | 156.3 | 160.0 | 158.9 | 159.3 | 159.6 | 157.6 | 157.5 | 158.5 | 158.5 | 157.2 | 158.7 | 158.9 | 150.4 |
| 15 | 220.8 | 128.3 | 127.8 | 121.8 | 122.2 | 122.7 | 122.2 | 120.4 | 119.6 | 119.7 | 120.8 | 120.9 | 120.6 | 120.8 | 121.9 | 127.2 |
| 16 | 43.0 | 34.7 | 34.8 | 34.3 | 34.4 | 34.3 | 34.3 | 34.2 | 34.3 | 34.3 | 34.3 | 34.3 | 34.3 | 34.2 | 34.4 | 34.7 |
| 17 | 38.9 | 52.2 | 52.0 | 52.1 | 51.7 | 52.1 | 52.2 | 51.3 | 51.3 | 51.2 | 51.3 | 51.3 | 51.3 | 51.2 | 51.8 | 51.7 |
| 18 | 27.8 | 21.7 | 21.2 | 21.0 | 20.4 | 21.1 | 21.1 | 18.9 | 18.7 | 18.9 | 19.4 | 19.5 | 19.5 | 19.4 | 20.6 | 21.3 |
| 19 | 41.9 | 41.6 | 42.0 | 42.4 | 43.0 | 43.0 | 37.6 | 37.1 | 37.2 | 37.3 | 37.0 | 37.2 | 39.0 | 35.9 | 39.6 | 39.8 |
| 20 | 122.6 | 124.0 | 124.3 | 124.0 | 124.0 | 123.7 | 123.7 | 124.0 | 124.4 | 124.3 | 124.0 | 124.0 | 124.1 | 124.0 | 123.9 | 124.3 |
| 21 | 139.9 | 140.1 | 140.1 | 140.0 | 140.0 | 140.1 | 140.0 | 140.0 | 140.1 | 140.3 | 140.1 | 140.1 | 140.1 | 140.0 | 140.0 | 139.8 |
| 22 | 110.5 | 111.1 | 111.1 | 111.1 | 111.0 | 111.0 | 111.0 | 111.1 | 111.3 | 111.2 | 111.2 | 111.1 | 111.2 | 111.0 | 111.1 | 111.0 |
| 23 | 143.5 | 143.0 | 142.9 | 143.0 | 143.0 | 143.2 | 143.1 | 142.9 | 142.8 | 142.8 | 142.8 | 142.9 | 142.8 | 142.9 | 143.3 | 142.7 |
| 28 | 31.2 | 23.9 | 31.4 | 31.3 | 31.4 | 33.7 | 33.0 | 19.6 | 19.4 | 23.1 | 25.1 | 25.1 | 25.6 | 24.9 | 25.8 | 24.8 |
| 29 | 24.1 | 31.3 | 23.9 | 24.0 | 24.2 | 23.8 | 27.5 | 23.2 | 23.1 | 19.4 | 18.3 | 18.3 | 18.5 | 18.2 | 19.7 | 19.1 |
| 30 | 20.6 | 30.4 | 30.8 | 29.0 | 29.7 | 28.6 | 28.8 | 29.0 | 29.2 | 29.1 | 27.8 | 27.8 | 29.0 | 28.0 | 28.0 | 29.0 |
| 1′ | 179.1 | 170.4 | 179.0 | 170.2 | 179.0 | 170.4 | 166.1 | |||||||||
| 2′ | 41.1 | 127.8 | 41.1 | 127.7 | 41.1 | 127.7 | 127.0 | |||||||||
| 3′ | 26.7 | 140.6 | 26.8 | 140.6 | 26.7 | 140.7 | 149.8 | |||||||||
| 4′ | 11.6 | 14.9 | 16.7 | 14.8 | 11.6 | 14.9 | 14.8 | |||||||||
| 5′ | 16.7 | 12.2 | 11.6 | 12.2 | 16.6 | 12.2 | 12.0 | |||||||||
| 11-OAc | 21.6 | 21.5 | 21.6 | 21.6 | 22.4 | 21.3 | 21.3 | 21.3 | 21.7 | 21.8 | 21.6 | 21.8 | 21.5 | |||
| 11-OAc | 170.1 | 170.4 | 170.5 | 170.5 | 170.8 | 169.9 | 170.4 | 170.4 | 170.0 | 170.2 | 170.1 | 172.8 | 169.8 | |||
| 7-OAc | 21.3 | 21.3 | 170.2 | 21.4 (6-OAc) | 21.4 (6-OAc) | 21.3 | 21.3 (6-OAc) | |||||||||
| 7-OAc | 169.8 | 170.0 | 21.3 | 170.0 (6-OAc) | 170.0 (6-OAc) | 171.7 | 170.3 (6-OAc) |
NMR data (δ) were measured at 125 MHz in CDCl3 for 1–16.
4.3.2. Walrobsin D (2)
White powder; [α]23D —54.2 (c 0.197, MeOH); UV (MeOH) λmax (logε) 209 (4.72) nm; IR vmax 3234, 2970, 2933, 1742, 1638, 1453, 1381, 1236, 1012, 820, 609 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 501.28 [M+Cl]—; positive ESI-MS m/z 484.28 [M+NH4]+; HR-ESI-MS m/z 484.2692 [M+NH4]+ (Calcd. for C28H38NO6, 484.2694).
4.3.3. Walrobsin E (3)
White powder; [α]23D —81.3 (c 0.174, MeOH); UV (MeOH) λmax (logε) 208 (4.63) nm; IR vmax 3429, 2933, 1741, 1454, 1383, 1196, 1157, 1011, 823, 602 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 459.10 [M+Cl]—; positive ESI-MS m/z 425.21 [M+H]+; HR-ESI-MS m/z 425.2324 [M+H]+ (Calcd. for C26H33O5, 425.2323).
4.3.4. Walrobsin F (4)
White powder; [α]23D —22.7 (c 0.165, MeOH); UV (MeOH) λmax (logε) 208 (4.69) nm; IR vmax 3436, 2930, 1741, 1375, 1236, 1030, 943, 873, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 545.24 [M+Cl]—; positive ESI-MS m/z 528.25 [M+NH4]+; HR-ESI-MS m/z 528.2957 [M+NH4]+ (Calcd. for C30H42NO7, 528.2956).
4.3.5. Walrobsin G (5)
White powder; [α]23D —68.4 (c 0.371, MeOH); UV (MeOH) λmax (logε) 215 (4.43) nm; IR vmax 3434, 2926, 1736, 1456, 1381, 1113, 1038, 1000, 874 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 461.21 [M+Cl]—; positive ESI-MS m/z 444.23 [M+NH4]+; HR-ESI-MS m/z 444.2742 [M+NH4]+ (Calcd. for C26H38NO5, 444.2744).
4.3.6. Walrobsin H (6)
White powder; [α]23D —9.0 (c 0.126, MeOH); UV (MeOH) λmax (logε) 207 (4.70) nm; IR vmax 3436, 2931, 1740, 1382, 1238, 1079, 1028, 1002, 873, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 519.21 [M+Cl]—; positive ESI-MS m/z 502.25 [M+NH4]+; HR-ESI-MS m/z 502.2803 [M+NH4]+ (Calcd. for C28H40NO7, 502.2799).
4.3.7. Walrobsin I (7)
White powder; [α]23D —22.7 (c 1.10, MeOH); UV (MeOH) λmax (logε) 221 (4.13) nm; IR vmax 3467, 2936, 1740, 1456, 1381, 1237, 1159, 1035, 1001, 873, 819, 773, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 1, Table 4; negative ESI-MS m/z 519.23 [M+Cl] —; positive ESI-MS m/z 507.26 [M+Na]+; HR-ESI-MS m/z 502.2801 [M+NH4]+ (Calcd. for C28H40NO7, 502.2799).
4.3.8. Walrobsin J (8)
White powder; [α]23D —0.6 (c 0.204, MeOH); UV (MeOH) λmax (logε) 209 (4.68) nm; IR vmax 3436, 2968, 1744, 1461, 1384, 1304, 1229, 1063, 1031, 874, 602 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 605.37 [M+Cl]—; positive ESI-MS m/z 588.31 [M+NH4]+; HR-ESI-MS m/z 593.3086 [M+Na]+ (Calcd. for C33H46NaO8, 593.30859).
Table 2.
1H NMR spectroscopic data (δ) for compounds 8—14a (δ in ppm, J in Hz).
| No. | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|
| 1 | 5.16 t (3.0) | 5.31 t (3.0) | 5.11 t (3.0) | 5.23 brd s | 4.80 d (3.5) | 5.21 t (3.0) | 4.47 brd s |
| 2 | 4.80 d (3.5) | 4.90 d (3.5) | 4.83 d (3.5) | 4.80 d (3.5) | 5.17 t (3.0) | 4.82 d (3.0) | 3.83 d (3.5) |
| 4 | 2.44 (overlapped) | 2.50 m | 2.44 m | 2.39 (overlapped) | 2.42 m | 2.29 m | |
| 5 | 2.10 m | 2.51 m | 2.10 m | 2.10 (overlapped) | 2.10 (overlapped) | 1.61 d (12.0) | 2.17 d (12.0) |
| 6a | 1.78 m | 2.51 m | 2.50 m | 5.30 dd (12.0, 3.0) | 4.08 d (3.0) | 4.25 dd (11.5, 3.0) | 5.29 dd (12.0, 2.0) |
| 6b | 2.37 m | 1.60 m | |||||
| 7 | 3.95 s | 5.27 d (3.5) | 5.20 d (3.5) | 4.07 d (3.0) | 5.31 dd (12.0, 3.0) | 5.29 d (3.0) | 4.09 d (3.0) |
| 9 | 2.12 (overlapped) | 2.18 m | 2.10 m | 2.23 d (5.5) | 2.10 d (6.5) | 2.10 d (6.5) | |
| 11 | 5.14 td (8.5, 6.0) | 4.52 dt (7.5, 7.0) | 4.46 td (7.5, 7.0) | 4.47 td ( 8.5. 6.0) | 2.23 d ( 6.5) | 4.46 td (8.5, 6.0) | 4.57 td (8.5, 6.0) |
| 12a | 2.86 dd (12.5, 6.5) | 2.33 m | 2.28 dd (12.5, 8.5) | 2.29 dd (13.0, 9.0) | 2.28 dd (13.0, 9.0) | 2.31 (overlapped) | 2.41 m |
| 12b | 1.37 dd (14.0, 5.5) | 1.72 m | 1.65 m | 1.64 dd (13.0, 9.0) | 1.69 (overlapped) | 1.62 (overlapped) | 1.65 dd (13.0,9.0) |
| 15 | 5.63 brd (3.5) | 5.50 brd s | 5.45 brd s | 5.59 t (2.5) | 5.60 t (3.0) | 5.49 brd s | 5.59 brd d (3.0) |
| 16a | 2.60 dd (15.5 11.0) | 2.22 dd (12.5, 2.5) | 2.35 ddd (15.0, 7.0, 3.5) | 2.58 ddd (16.0,11.0, 2.0) | 2.59 ddd (16.0,11.0, 2.0) | 2.36 dd (16.0, 12.0) | 2.57 ddd (15.0,11.0, 2.0) |
| 16b | 2.38 (overlapped) | 1.73 m | 1.63 m | 2.40 (overlapped) | 2.45 (overlapped) | 2.31 (overlapped) | 2.44 (overlapped) |
| 17 | 2.92 dd (11.0, 7.0) | 2.93 dd (11.0, 7.5) | 2.88 dd (11.0, 7.5) | 2.91 dd (11.0, 7.0) | 2.92 dd (11.0, 7.0) | 2.85 dd (11.0, 7.0) | 2.91 dd (11.0 7.0) |
| 18 | 0.76 s | 0.79 s | 0.74 s | 0.76 s | 0.76 s | 0.74 s | 0.72 s |
| 19a | 2.01 (overlapped) | 2.22 m | 2.10 m | 2.17 d (12.5) | 2.17 d (12.5) | 2.18 d (12.5) | 2.01 d (12.5) |
| 19b | 1.81 (overlapped) | 1.73 m | 1.64 m | 2.12 (overlapped) | 2.12 d (12.5) | 2.05 d (12.5) | 1.94 d (12.5) |
| 21 | 7.26, s | 7.29 s | 7.25 s | 7.25 s | 7.26 s | 7.25 s | 7.25 s |
| 22 | 6.27, s | 6.33 s | 6.27 s | 6.28 s | 6.27 s | 6.38 s | 6.28 s |
| 23 | 7.39, s | 7.43 s | 7.38 s | 7.38 s | 7.39 s | 7.38 s | 7.37 s |
| 28 | 0.83 d (7.0) | 0.94 d (7.0) | 0.93 d (7.0) | 1.08 d (7.0) | 1.09 d (7.0) | 1.12 d (7.0) | 1.13 d (7.0) |
| 29 | 0.95 d (7.0) | 0.88 d (7.0) | 0.82 d (7.0) | 0.82 d (7.0) | 0.83 d (7.0) | 1.00 d (7.0) | 0.82 d (7.0) |
| 30 | 1.36 s | 1.44 s | 1.39 s | 1.42 s | 1.42 s | 1.40 s | 1.44 s |
| 2′ | 2.52 (overlapped) | 1.60 m | 2.50 m | ||||
| 3′ | 1.70 (m) | 7.01 m | 2.50 m | 6.96 m | 1.67 m | 6.96 m | |
| 1.50 (m) | 1.60 m | 1.51 m | |||||
| 4′ | 0.92 t (7.0) | 1.89 s | 0.93 t (7.0) | 1.83 s | 0.92 t (7.0) | 1.83 s | |
| 5′ | 1.17 d (7.0) | 1.88 d (6.0) | 1.19 d (7.0) | 1.80 d (6.0) | 1.18 d (7.0) | 1.82 d (6.0) | |
| 7-OAc | 2.10 s | 2.18 s | 2.12 s (6-OAc) | 2.13 s (6-OAc) | 2.16 s | 2.15 s | |
| 11-OAc | 2.02 s | 2,12 s | 2,13 s | 2.04 s | 2.13 s | ||
| OH | 5.94 s | 5.54 s | 5.91 s |
NMR data (δ) were measured at 500 MHz in CDCl3 for 8–14.
4.3.9. Walrobsin K (9)
White powder; [α]23D —7.9 (c 0.211, MeOH); UV (MeOH) λmax (logε) 216 (4.86) nm; IR vmax 3434, 2966, 1743, 1370, 1253, 1161, 1029, 733, 605 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 645.17 [M+Cl] —; positive ESI-MS m/z 628.21 [M+NH4]+; HR-ESI-MS m/z 611.3212 [M+H]+ (Calcd. for C35H47O9, 611.3215).
4.3.10. Walrobsin L (10)
White powder; [α]23D —1.9 (c 0.208, MeOH); UV (MeOH) λmax (logε) 208 (4.68) nm; IR vmax 3412, 2965, 1750, 1711, 1458, 1260, 1225, 1035, 873, 605 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 611.27 [M — H]—; positive ESI-MS m/z 635.36 [M+Na]+; HR-ESI-MS m/z 635.3190 [M+Na]+ (Calcd. for C35H48O9Na, 635.3191).
4.3.11. Walrobsin M (11)
White powder; [α]23D +39.4 (c 0.208, MeOH); UV (MeOH) λmax (logε) 215 (4.87), 277 (3.79) nm; IR vmax 3436, 2934, 1722, 1370, 1252, 1163, 1030, 874, 736, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 625.31 [M–H] —, 661.30 [M+Cl]—; positive ESI-MS m/z 644.26 [M+NH4]+; HR-ESI-MS m/z 649.2980 [M+Na]+ (Calcd. for C35H46O10Na, 649.2983).
4.3.12. Walrobsin N (12)
White powder; [α]23D +61.4 (c 0.158, MeOH); UV (MeOH) λmax (logε) 207 (4.75) nm; IR vmax 3444, 2935, 1742, 1461, 1370, 1229, 1163, 1030, 874, 799, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 663.11 [M+Cl]—; positive ESI-MS m/z 629.16 [M+H]+; HR-ESI-MS m/z 651.3138 [M+Na]+ (Calcd. for C35H48O10Na, 651.3140).
4.3.13. Walrobsin O (13)
White powder; [α]23D +48.1 (c 0.201, MeOH); UV (MeOH) λmax (logε) 212 (4.85) nm; IR vmax 3446, 2934, 2360, 1741, 1373, 1254, 1163, 1030, 874, 733, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 661.25 [M+Cl]—; positive ESI-MS m/z 644.29 [M+NH4]+; HR-ESI-MS m/z 627.3166 [M+H]+ (Calcd. for C35H47O10, 627.3164).
4.3.14. Walrobsin P (14)
White powder; [α]23D +55.3 (c 0.210, MeOH); UV (MeOH) λmax (logε) 208 (4.64) 270 (3.70) nm; IR vmax 3435, 2935, 1722, 1371, 1248, 1161, 1026, 874, 798, 736, 601 cm—1; 1H and 13C NMR (CDCl3), see Table 2, Table 4; negative ESI-MS m/z 579.21 [M+Cl]—; positive ESI-MS m/z 562.28 [M+NH4]+; HR-ESI-MS m/z 545.2744 [M+H]+ (Calcd. for C30H41O9, 545.2745).
4.3.15. Walrobsin Q (15)
White powder; [α]23D —55.8 (c 0.575, MeOH); UV (MeOH) λmax (logε) 215 (4.43), 257 (4.44) nm; IR vmax 3435, 2957, 1705, 1386, 1242, 1028, 874, 786, 600 cm—1; 1H and 13C NMR (CDCl3), see Table 3, Table 4; negative ESI-MS m/z 503.16 [M+Cl]—; positive ESI-MS m/z 486.26 [M+NH4]+; HR-ESI-MS m/z 469.2586 [M+H]+ (Calcd. for C28H37O6, 469.2585).
Table 3.
1H NMR spectroscopic data (δ) for compounds 15 and 16a (δ in ppm, J in Hz).
| No. | 15 | 16 | No. | 15 | 16 |
|---|---|---|---|---|---|
| 1 | 6.24 s | 6.27 s | 19 | 3.06 d (19.0), 2.35 d (19.0) | 3.64 d (19.0), 2.60 d (19.0) |
| 4 | 1.48 m | 1.72 m | 21 | 7.24 s | 7.26 s |
| 5 | 1.90 (overlapped) | 2.48 m | 22 | 6.26 s | 7.28 s |
| 6a | 1.91 m | 2.73 t (14.5) | 23 | 7.37 s | 7.38 s |
| 6b | 1.67 m | 2.44 m | |||
| 7 | 3.95 t (3.0) | 28 | 0.87 d (7.0) | 0.90 d (7.0) | |
| 9 | 2.68 d (6.5) | 2.20 d (4.5) | 29 | 0.80 d (7.0) | 0.86 d (7.0) |
| 11 | 4.57 td (9.0, 6.0) | 4.34 m | 30 | 1.32 s | 1.63 s |
| 12a | 2.67 dd (13.5, 9.0) | 2.80 dd (14.0, 8.5) | 3′ | 7.16 m | |
| 12b | 1.33 (overlapped) | 1.69 dd (13.5, 6.5) | |||
| 15 | 5.69 brd d (3.0) | 6.28 brd s | 4′ | 1.91 s | |
| 16a | 2.55 ddd (15.5, 11.0, 2.0) | 2.58 (overlapped) | 5′ | 1.88 dd (7.0,1.5) | |
| 16b | 2.48 ddd (15.5, 7.0, 3.0) | 2.42 (overlapped) | |||
| 17 | 2.87 dd (11.0, 7.5) | 2.92 dd (11.0,7.0) | OAc | 1.90 s | |
| 18 | 0.81 s | 0.86 s |
NMR data (δ) were measured at 500 MHz in CDCl3 for 15 and 16.
4.3.16. Walrobsin R (16)
White powder; [α]23D —35.8 (c 0.053, MeOH); UV (MeOH) λmax (logε) 212 (4.27), 257 (4.34) nm; IR vmax 3419, 2931, 2310, 1715, 1646, 1454, 1384, 1253, 1114, 1022, 951, 723, 615 cm—1; 1H and 13C NMR (CDCl3), see Table 3, Table 4; negative ESI-MS m/z 547.31 [M+Cl]—; positive ESI-MS m/z 524.29 [M+NH4]+; HR-ESI-MS m/z 529.2559 [M+Na]+ (Calcd. for C31H38O6Na, 529.2561).
4.4. NO production bioassay
The RAW264.7 cell line and BV-2 cell line were purchased from the Chinese Academic of Sciences. The cells were cultured in DMEM containing 10% FBS with penicillin (100 U/mL) and streptomycin (100 U/mL) at 37 °C in a humidified atmosphere with 5% CO2. The cells were allowed to grow in 96-well plates with 1×105 cells/well to treat test compounds. After being incubated for 2 h, the cells were treated with 100 ng/mL of LPS for 18 h. Nitrite in culture media was measured to assess NO production using Griess reagent. The absorbance at 540 nm was measured on a microplate reader. N-monomethyl-l-arginine was used as the positive control. Cytotoxicity was determined by the MTT method after 48 h incubation with test compounds. All the experiments were performed in three independent replicates19.
4.5. Anti-bacterial test
The anti-bacterial activity of selected walrobsins 1—3, 8—14, and 19—20 were carried out using a broth microdilution method and the minimum inhibitory concentration (MIC) was determined. P. acnes at the logarithmic phase were added to the chopped meat medium containing various concentrations of walrobsins in a 96-well plate. The final inoculum concentration of P. acnes was 1.25 × 106 CFU/mL. After incubation under anaerobic conditions for 24 h, the level of microbial growth was tested using a microplate reader at 600 nm. The MIC was defined as the lowest dilution of walrobsins at which growth was inhibited completely20., 21..
4.6. MTT assay
Effects of walrobsins 1—3, 8—14, and 19—20 on the viability of THP-1 cells were determined by the MTT method. Approximately 2 × 105 cells/well were seeded in 96-well plates and treated with different concentrations of walrobsins for 36 h (5% CO2, 37 °C). Then, 20 μL MTT reagent (5 mg/mL) was added into the culture medium in each well accordingly. The formazan crystals were solubilized in 150 μL dimethyl sulfoxide after 4 h incubation. Absorbance was measured by a spectrophotometer at 570 nm excitation and 630 nm emission20., 21..
4.7. Enzyme-linked immunosorbent assay
THP-1 cells (2 × 105 cells/well) were cultured in 96-well plates in medium without FBS and incubated with different concentrations of walrobsins 1—3, 8—14, and 19—20 for 4 h. Then, THP-1 cells were stimulated by live P. acnes for 24 h and centrifuged to collect cell-free supernatants. The levels of IL-1β in culture supernatants were measured with ELISA assays according to the instructions of manufacturer. Retinoic acid was used as the positive control20., 21..
4.8. Theoretical calculated ECD of 15
Theoretical calculations of ECD spectra for that of 15 were performed with the Gaussian 09 program package. The structure of 15 was optimized with MM2, and its geometry was re-optimized at the b3lyp/6–31g(d,p) level of theory. The ECD calculations of compound 15 were performed with DFT calculations at the b3lyp/6–311+g(d,2p) level of theory with 26 nm UV correction (σ=0.40). Detailed calculated parameters were proved in Supporting Information.
4.9. X-ray crystallography of 1
Single crystals of C28H35O7 (1) were recrystallized from mixture solvent (CH2Cl2:MeOH=1:1, v:v). A suitable crystal was selected and recorded on a diffractometer using Cu Kα radiation. The crystal was kept at 291(2) K during data collection. The structure was solved with the ShelXT structure solution program using Direct Methods and refined with the ShelXL refinement package using Least Squares minimisation based on Olex2 software22. The crystallographic data of compound 1 have been deposited at the Cambridge Crystallographic Data Center with the deposition number CCDC 1880306.
Acknowledgments
Financial support for this study by the National Natural Science Foundation of China (31470416, China), the Outstanding Youth Fund of the Basic Research Program of Jiangsu Province (BK20160077, China), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R63, China), and the "Double First-Class" University project (CPU2018GY08, China).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.apsb.2019.02.009.
Contributor Information
Jun Luo, Email: luojun@cpu.edu.cn.
Lingyi Kong, Email: cpu_lykong@126.com.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Guo Z. The modification of natural products for medical use. Acta Pharm Sin B. 2017;7:119–136. doi: 10.1016/j.apsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan Q.G., Luo X.D. Meliaceous limonoids:chemistry and biological activities. Chem Rev. 2011;111:7437–7522. doi: 10.1021/cr9004023. [DOI] [PubMed] [Google Scholar]
- 3.Han M.L., Shen Y., Wang G.C., Leng Y., Zhang H., Yue J.M. 11β-HSD1 inhibitors from Walsura cochinchinensis. J Nat Prod. 2013;76:1319–1327. doi: 10.1021/np400260g. [DOI] [PubMed] [Google Scholar]
- 4.Ji K.L., Zhang P., Hu H.B., Hua S., Liao S.G., Xu Y.K. Limonoids from the leaves and twigs of Walsura yunnanensis. J Nat Prod. 2014;77:1764–1769. doi: 10.1021/np400976p. [DOI] [PubMed] [Google Scholar]
- 5.Luo X.D., Wu S.H., Ma Y.B., Wu D.G. Tetranortriterpenoids from Walsura yunnanensis. J Nat Prod. 2000;63:947–951. doi: 10.1021/np990607x. [DOI] [PubMed] [Google Scholar]
- 6.Wang G.C., Yu J.H., Shen Y., Leng Y., Zhang H., Yue J.M. Limonoids and triterpenoids as 11β-HSD1 inhibitors from Walsura robusta. J Nat Prod. 2016;79:899–906. doi: 10.1021/acs.jnatprod.5b00952. [DOI] [PubMed] [Google Scholar]
- 7.Han M.L., Shen Y., Leng Y., Zhang H., Yue J.M. New rearranged limonoids from Walsura cochinchinensis. RSC Adv. 2014;4:19150–19158. [Google Scholar]
- 8.Nugroho A.E., Okuda M., Yamamoto Y., Hirasawa Y., Wong C.P., Kaneda T. Walsogynes B–G, limonoids from Walsura chrysogyne. Tetrahedron. 2013;69:4139–4145. [Google Scholar]
- 9.Rao M.S., Suresh G., Yadav P.A., Prasad K.R., Rani P.U., Rao C.V. Piscidinols H–L, apotirucallane triterpenes from the leaves of Walsura trifoliata and their insecticidal activity. Tetrahedron. 2015;71:1431–1437. [Google Scholar]
- 10.An F.L., Sun D.M., Li R.J., Zhou M.M., Yang M.H., Yin Y. Walrobsins A and B, two anti-inflammatory limonoids from root barks of Walsura robusta. Org Lett. 2017;19:4568–4571. doi: 10.1021/acs.orglett.7b02173. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., An F.L., Huang S.S., Yang L., Gu Y.C., Luo J. Diverse tritepenoids from the fruits of Walsura robusta and their reversal of multidrug resistance phenotype in human breast cancer cells. Phytochemistry. 2017;136:108–118. doi: 10.1016/j.phytochem.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Yin S., Wang X.N., Fan C.Q., Liao S.G., Yue J.M. The first limonoid peroxide in the meliaceae family: walsuronoid A from Walsura robusta. Org Lett. 2007;9:2353–2356. doi: 10.1021/ol070735+. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Z.W., Yin S., Zhang H.Y., Fu Y., Yang S.P., Wang X.N. Walsucochins A and B with an unprecedented skeleton isolated from Walsura cochinchinensis. Org Lett. 2006;10:465–468. doi: 10.1021/ol702831e. [DOI] [PubMed] [Google Scholar]
- 14.Grkovic T., Pouwer R.H., Vial M.L., Gambini L., Noël A., Hooper J.N. NMR fingerprints of the drug-like natural-product space identify iotrochotazine A: a chemical probe to study parkinson׳s disease. Angew Chem Int Ed. 2014;53:6070–6074. doi: 10.1002/anie.201402239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Zhou J.L., Liu P., Sun S., Li P. Chemical markers׳ fishing and knockout for holistic activity and interaction evaluation of the components in herbal medicines. J Chromatogr A. 2010;1217:5239–5245. doi: 10.1016/j.chroma.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Song H.P., Wu S.Q., Qi L.W., Long F., Jiang L.F., Liu K. A strategy for screening active lead compounds and functional compound combinations from herbal medicines based on pharmacophore filtering and knockout/knockin chromatography. J Chromatogr A. 2016;1456:176–186. doi: 10.1016/j.chroma.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L. Three tetranortriterpenoids from Walsura yunnanensis. Chem Nat Compd. 2013;48:1013–1016. [Google Scholar]
- 18.O׳Boyle N.M., Vandermeersch T., Flynn C.J., Maguire A.R., Hutchison G.R. Confab-systematic generation of diverse low-energy conformers. J Cheminform. 2011;3:8. doi: 10.1186/1758-2946-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R.J., Gao C.Y., Guo C., Zhou M.M., Luo J., Kong L.Y. The anti-inflammatory activities of two major withanolides from Physalis minimavia acting on NF-κB, STAT3, and HO-1 in LPS-stimulated RAW264.7 cells. Inflammation. 2017;40:401–413. doi: 10.1007/s10753-016-0485-1. [DOI] [PubMed] [Google Scholar]
- 20.Guo M.M., An F.L., Yu H.Y., Wei X., Hong M.H., Lu Y.H. Comparative effects of schisandrin A, B, and C on Propionibacterium acnes-induced, NLRP3 inflammasome activation-mediated IL-1β secretion and pyroptosis. Biomed Pharmacother. 2017;96:129–136. doi: 10.1016/j.biopha.2017.09.097. [DOI] [PubMed] [Google Scholar]
- 21.Guo M.M., An F.L., Wei X., Hong M.H., Lu Y.H. Comparative effects of schisandrin A, B, and C on acne-related inflammation. Inflammation. 2017;40:2163–2172. doi: 10.1007/s10753-017-0656-8. [DOI] [PubMed] [Google Scholar]
- 22.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. J Appl Cryst. 2009;42:339–341. doi: 10.1107/S0021889811041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material