Abstract
Drug-metabolizing enzymes, transporters, and nuclear receptors are essential for the absorption, distribution, metabolism, and excretion (ADME) of drugs and xenobiotics. MicroRNAs participate in the regulation of ADME gene expression via imperfect complementary Watson—Crick base pairings with target transcripts. We have previously reported that Cytochrome P450 3A4 (CYP3A4) and ATP-binding cassette sub-family G member 2 (ABCG2) are regulated by miR-27b-3p and miR-328-3p, respectively. Here we employed our newly established RNA bioengineering technology to produce bioengineered RNA agents (BERA), namely BERA/miR-27b-3p and BERA/miR-328-3p, via fermentation. When introduced into human cells, BERA/miR-27b-3p and BERA/miR-328-3p were selectively processed to target miRNAs and thus knock down CYP3A4 and ABCG2 mRNA and their protein levels, respectively, as compared to cells treated with vehicle or control RNA. Consequently, BERA/miR-27b-3p led to a lower midazolam 1′-hydroxylase activity, indicating the reduction of CYP3A4 activity. Likewise, BERA/miR-328-3p treatment elevated the intracellular accumulation of anticancer drug mitoxantrone, a classic substrate of ABCG2, hence sensitized the cells to chemotherapy. The results indicate that biologic miRNA agents made by RNA biotechnology may be applied to research on miRNA functions in the regulation of drug metabolism and disposition that could provide insights into the development of more effective therapies.
Abbreviations: ABCG2, ATP-binding cassette sub-family G member 2;, ADME, absorption, distribution, metabolism, and excretion, BERA, bioengineered RNA agent;, CYP, cytochrome P450, E. coli, Escherichia coli;, FPLC, fast protein liquid chromatography, LC--MS/MS, liquid chromatographytandem mass spectroscopy;, microRNA, miR or miRNA, ncRNA, noncoding RNA;, PAGE, polyacrylamide gel electrophoresis, RNAi, RNA interference;, RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction, RXRα, retinoid X receptor α;, tRNA, transfer RNA, 3′-UTR, 3′-untranslated region;, VDR, vitamin D receptor
KEY WORDS: Bioengineered RNA, miR-27b, miR-328, CYP3A4, ABCG2, Drug disposition
Graphical abstract
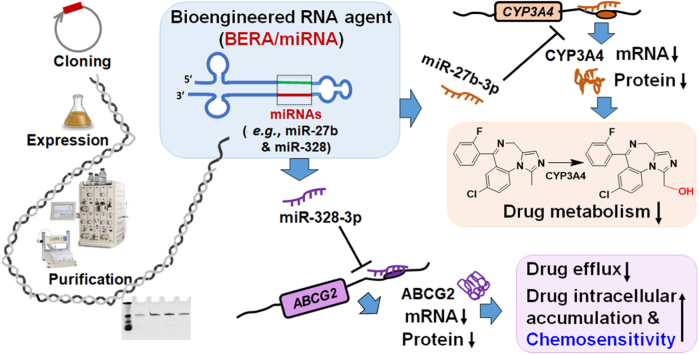
A novel RNA biotechnology was used to produce bioengineered miRNA agents (BERA), namely BERA/miR-27b-3p and BERA/miR-328-3p, which were selectively processed to target miR-27-3p and -328-3p to modulate the expression of CYP3A4 enzyme and ABCG2 transporter, respectively, in human cells, and subsequently alter cellular drug metabolism and disposition capacity.
1. Introduction
Drug-metabolizing enzymes, transporters, nuclear receptors, and transcription factors are essential for the absorption, distribution, metabolism, and excretion (ADME) of drugs and xenobiotics. Knowledge of the mechanisms involved in expression and function of ADME genes is a prerequisite for predicting drug efficacy and safety profiles, thus it is critical for the practice of personalized or precision medicine1., 2., 3., 4., 5., 6.. Bioinformatics and experimental studies suggest that miRNAs, a large class of genome derived, short noncoding RNA (ncRNA) molecules participate in the regulation of ADME gene expression7. MiRNAs are capable of inducing translation suppression and/or mRNA cleavage of target genes via imperfect complementary Watson—Crick base pairings with targeted elements that are usually located within the 3′-untranslated regions (3′-UTRs) of target transcripts8., 9., 10..
Many miRNAs have been shown to modulate the expression of specific ADME genes (see recent reviews7., 11., 12.). For instance, miR-27b-3p has been shown to negatively regulate the expression of cytochrome P450s (CYPs) 1B1 in human carcinoma cells13., 14.. Previous studies have also demonstrated that miR-27b-3p regulates the expression of CYP3A415., 16., which is the most abundant hepatic and intestinal drug metabolizing enzyme in humans17. Changes in CYP3A4 expression and function will undoubtedly lead to a broader variability in drug metabolism as CYP3A4 contributes to the biotransformation of more than 50% of the drugs used in clinic18. MiR-27b-3p could also influence other drug metabolizing enzymes and transporters via its regulation to certain nuclear receptors, such as vitamin D receptor (VDR/NR1I1)15., 19. and retinoid X receptor α (RXRα/NR2B1)20.
MiR-328-3p and some other miRNAs (e.g., miR-520h, -519c, -328, -181a, and -487a)21., 22., 23., 24., 25. have been shown to regulate the expression of ABCG2, also known as breast cancer resistance protein (BCRP). ABCG2 is an important transporter in ADME and multidrug resistance (MDR), which plays a critical role in cellular transport of chemotherapeutic drugs such as mitoxantrone, doxorubicin, and topotecan. We have revealed previously that miR-328-3p could regulate ABCG2 expression level and consequently improve the chemosensitivity of ABCG2-overexpressing drug-resistant MCF7/MX100 cells21. Our further comparative studies have demonstrated that miR-328-3p and -519c are more effective than miR-520h in the regulation of ABCG2 expression in human breast cancer cells. As a result, both miR-328-3p and -519c are able to significantly elevate intracellular accumulation of mitoxantrone, an ABCG2 substrate22. These findings suggest that understanding the potential role of miRNAs in epigenetic regulation of ADME genes may offer clues to develop more effective therapeutic strategy, e.g., using miRNAs to combat resistance in chemotherapy.
Nevertheless, currently used RNA interfering (RNAi) miRNA/siRNA materials are limited to the use of viral or non-viral vector-based short hairpin RNA (shRNA) or miRNA expressing systems and chemically engineered miRNA mimics26. The shRNA/miRNA expressing vectors, plasmids or other systems are literally DNA based agents, and the efficiency in targeting relies not only on the effectiveness of gene delivery but also the capability of host cells or organisms to process the DNA into target miRNA/siRNA molecules. On the other hand, chemically engineered RNA mimics carry a wide range of artificial modifications, which is in contrast to natural RNAs produced in live cells that have no or minimal posttranscriptional modifications. Those synthetic RNA agents with extensive artificial modification are undoubtedly to exhibit distinct structures, physicochemical properties, biologic/pharmacological activities, and safety profiles. To produce natural RNA molecules that may better capture the properties of cellular RNAs, we have recently established and further refined a novel tRNA/pre-miRNA-based ncRNA bioengineering technology27., 28., which are proven to be useful for the production of many ready-to-use RNAi agents (e.g., miRNAs and siRNAs)19., 27., 29., 30., 31., 32.. These bioengineered RNAi agents (BERA) should represent a new class of RNAi materials for basic and translational research26.
In the present study, using BERA/miR-27b-3p and -328-3p as examples, we demonstrated that both BERA/miRNAs were selectively processed to mature miRNAs in human carcinoma cells and thus suppress CYP3A4 and ABCG2 mRNA and their protein levels, respectively, as compared to cells treated with vehicle or control RNA that has been verified as a valid control for assessing the actions of BERAs through various RNA sequencing and functional studies27., 29., 30., 31., 32.. As a result, BERA/miR-27b-3p led to a lower midazolam 1′-hydroxylase activity suggesting the reduction of CYP3A4-mediated metabolism. Likewise, BERA/miR-328-3p readily facilitated the intracellular accumulation of anticancer drug mitoxantrone, a classic substrate of ABCG2, hence sensitizing the cancer cells to chemotherapy. These findings illustrated the utilities of BERA/miRNA molecules for studying the functions of miRNAs in the regulation of ADME, which may offer clue to the development of improved therapies.
2. Materials and methods
2.1. Chemicals and materials
Midazolam (MDZ) and its metabolite 1′-hydroxymidazolam (1′-OH-MDZ) were purchased from Cambridge Isotope Laboratories (Tewksbury, MA, USA) and Cayman Chemical Company (Ann Arbor, MI, USA), respectively. 1-α-Hydroxycholecalciferol (1α-VD3) was bought from EMD Millipore (Billerica, MA, USA). Mitoxantrone was bought from Sigma—Aldrich (St. Louis, MO, USA). Fetal bovine serum, RPMI 1640, F-12K medium, trypsin, phosphate-buffered saline (PBS), and antibiotics were bought from Invitrogen (Carlsbad, AL, USA). Thiazolyl blue tetrazolium bromide (MTT), dimethyl sulfoxide (DMSO), and bovine serum albumin (BSA) were bought from VWR (Radnor, PA, USA). Anti-CYP3A4 antibody and rabbit anti-mouse IgG were purchased from BD Biosciences-Discovery Labware. Anti-GAPDH and anti-VDR antibody were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Peroxidase-conjugated goat anti-rabbit IgG was bought from Jackson ImmunoResearch (West Grove, PA, USA). Primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). Chlorzoxazone, 6-hydroxychlorzoxazone, phenacetin and warfarin sodium purchased from Sigma—Aldrich (St. Louis, MO, USA). Acetaminophen was bought from MP Biomedicals, LLC (Solon, OH, USA). Dextromethorphan hydrobromide and dextrophan were purchased from ICN Biomedicals, Inc. (Aurora, OH, USA). Diclofenac sodium was purchased from TCI America (Portland, OR, USA), and 4′-hydroxydiclofenac was from Toronto Research Chemicals, Inc. (Toronto, ON, CA). All other chemicals and organic solvent were purchased from Sigma—Aldrich or Themo Fisher Scientific Inc. (Waltham, MA, USA).
2.2. Expression and FPLC purification of BERA/miR-27b-3p and BERA/miR-328-3p
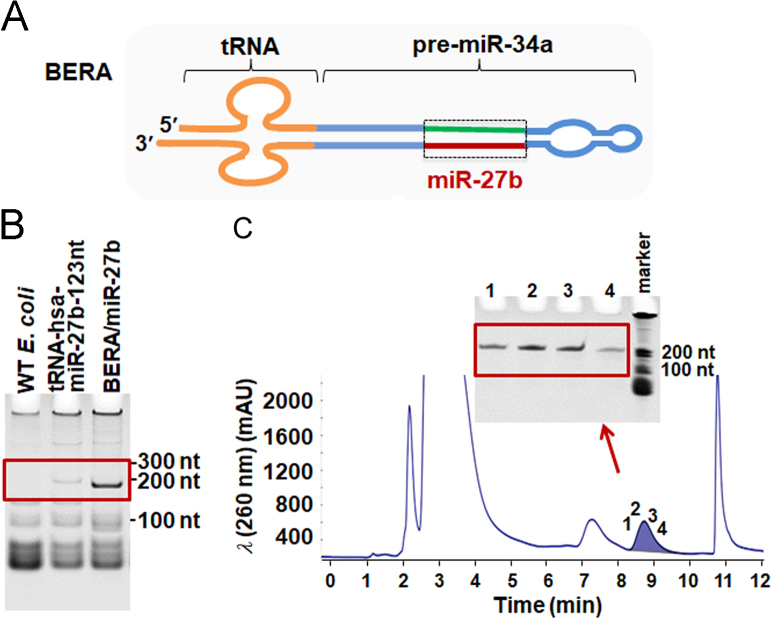
The production of BERA/miR-27b-3p, BERA/miR-328-3p, and control RNAs was carried out using the ncRNA bioengineering technology as we described recently27., 29., 31., 32.. The DNA fragment encoding tRNA/pre-miR-27b-123nt was cloned into the vector pBSMrnaSeph33 linearized by SalI and AatII (New England Biolabs, Ipswich, MA, USA), as described19. Likewise, plasmids were constructed to express tRNA/pre-miR-34a (Fig. 1A) based BERA/miR-27b27 and BERA/miR-32828 in this study. Plasmids were amplified in DH5α strain and confirmed by sequencing analyses (Genscript, Piscataway, NJ, USA). Recombinant ncRNAs were expressed in HST08 E. coli strain (Clontech, Mountain View, CA, USA). Total bacterial RNAs were isolated by phenol extraction and verified by denaturing urea (8 mol/L) polyacrylamide (8%) gel electrophoresis (PAGE) analysis.
Figure 1.
Bioengineering of new BERA/miR-27b-3p molecule. (A) Schematic illustration of the tRNA/pre-miR-34a carrier used for the production of BERA/miR-27b-3p. (B) Urea-PAGE analysis of total bacterial RNAs showed that chimeric miR-27b-3p was heterogeneously expressed in E. coli using the tRNA/pre-miR-34a scaffold. Total RNAs isolated from untransformed HST08 (WT) E. coli were used as control, and our previously constructed tRNA-hsa-miR-27b-123nt plasmid19 transformed HST08 E. coli was utilized for comparison. MiR-27b-3p was expressed at much higher levels when using the tRNA/pre-miR-34a carrier. (C) Representative FPLC traces during the purification of BERA/miR-27b-3p. Inserts were corresponding urea-PAGE analyses of collected fractions (1, 2, 3 and 4) eluted at 8.7 min, which confirmed the purity of isolated recombinant ncRNAs.
Anion exchange FPLC purification of recombinant ncRNAs was conducted on a NGC QUEST 10PLUS FPLC system (Bio-Rad) equipped with a fraction collector19., 27., 31., 32.. Following urea-PAGE analysis, FPLC fractions containing pure target RNAs were pooled and precipitated with ethanol. The pure RNA precipitates were reconstituted with nuclease-free water, desalted and then concentrated with Amicon ultra-0.5 mL centrifugal filters (30 KD; EMD Millipore, Billerica, MA, USA). RNA concentrations were determined with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific), and RNA purities were further determined by a high performance liquid chromatography (HPLC) assay19., 27., 31., 32..
2.3. Cell culture and transfection
Human colon carcinoma cell line LS-180 and human choriocarcinoma cell line BeWo were purchased from American Tissue Culture Collection (Manassas, VA, USA) with authentication, and mitoxantrone-selected human breast cancer cell line MCF-7/MX100 was obtained from Dr. Susan E. Bates at National Cancer Institute (Bethesda, MD, USA). LS-180, BeWo, and MCF-7/MX100 cells were maintained in EMEM, F-12K, and RPMI 1640 media, respectively, containing 10% FBS, at 37 °C and in a humidified atmosphere with 5% CO2 and 95% air. The recombinant ncRNAs and control RNAs were transfected into the cells within the logarithmic growth phases by using Lipofectamine 3000 reagent, according to the manufacturer׳s instructions.
2.4. Reverse transcription quantitative real-time PCR (RT-qPCR)
Cells were treated with recombinant ncRNAs or control RNAs for 48 h. Total RNAs were isolated with Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA), from which the cDNAs were synthesized using NxGen M-MuLV reverse transcriptase (Lucigen, Middleton, WI, USA), with random hexamers or respective stem-loop primers (Supporting Information Table S1). qPCR analyses were carried out using iTaq™ universal SYBR® Green supermix (Bio-Rad, Hercules, CA, USA) and gene-specific primers (Supporting Information Table S1) on a CFX96 Touch real-time PCR system (Bio-Rad, Hercules, CA, USA), as described previously19., 21., 22.. 18S and GAPDH were used as internal control for the assessment of mRNA levels, and miRNAs were normalized to U74 and U6. Cells were treated in triplicate and assayed separately. The comparative threshold cycle (Ct) method with the formula 2−ΔΔCt was used to calculate the relative gene expression.
2.5. Western blot analysis
LS-180 cells were treated with 2 μmol/L 1α-VD3 or vehicle for 72 h to induce CYP3A4 expression34, and then transfected with BERA/miR-27b-3p or control RNA for 48 h. Cell lysates were prepared for immunoblot analysis using RIPA lysis buffer (Sigma—Aldrich, St. Louis, MO, USA) containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein concentrations were determined by BCA Protein Assay Kit (Themo Fisher Scientific Inc., Waltham, MA, USA). 30 μg/lane whole cell proteins were resolved on a 10% SDS-PAGE and then transferred onto polyvinylidene fluoride membranes (Bio-Rad). The membranes were blocked in 5% non-fat milk in TBST buffer for 2 h, and immunoblot was thus performed by incubating the membranes with the corresponding primary antibodies in TBST at 4 °C overnight. After washing with TBST three times, the resulting membranes were incubated with secondary antibodies conjugated with horseradish peroxidase for 2 h at room temperature. The resultant membranes were washed extensively before analysis. The protein bands were detected by an enhanced chemiluminescence detection system (Bio-Rad), and visualized by a ChemiDoc MP Imaging System (Bio-Rad). ABCG2 expression levels were detected in a similar manner.
2.6. CYPs activity assay
A liquid chromatography—tandem mass spectrometry (LC—MS/MS) cocktail assay was used to determine the cellular activity of five major CYPs in LS-180 cells, as we described previously19., 35.. Briefly, LS-180 cells were first treated with 2 μmol/L of 1α-VD3 for 72 h, and then transfected with 40 nmol/L BERA/miR-27b-3p or control RNA. At 48 h post-transfection, cocktail substrates (3 μmol/L phenacetin, 2 μmol/L diclofenac, 2 μmol/L dextromethorphan, 3 μmol/L chlorzoxazone and 1 μmol/L midazolam) were added to the media, and cells were maintained at 37 °C for 90 min. Then 100 μL of media were collected and extracted with 1 mL ethyl acetate containing 10 nmol/L harmaline and 5 nmol/L warfarin as internal standard for positive and negative modes respectively. The extracts were evaporated and reconstituted with 100 μL methanol-water (20:80, v/v). The samples were analyzed by the LC—MS/MS method35.
2.7. Flow cytometry analysis
Intracellular drug accumulation was investigated using a FacsCanto II Flow Cytometer (BD Biosciences, Franklin Lakes, NJ, USA), as reported22. Briefly, cells (1 × 106 cells per well) were incubated with phenol red-free RPMI 1640 medium containing 10% FBS and 800 nmol/L of mitoxantrone at 37 °C for 1 h. Cells incubated in the absence of drug were used as negative controls. Cells were then washed twice with ice-cold PBS buffer, re-suspended, and subjected to flow cytometry analyses with a 635 nm red diode laser and a 670 nm band pass filter. Drug efflux ability was reflected by the fluorescence of drug accumulated within cells. Flow cytometry data were analyzed by FlowJo software (BD Biosciences). Relative intracellular drug accumulation was calculated by normalized geometric mean of BERA/miR-328-3p group to the control group after subtraction of the basal fluorescence (negative control).
2.8. Mitoxantrone cytotoxicity
Cytotoxicity studies were performed as described previously31. In brief, cells were seeded at 5000 cells/well in 96-well plate and cultured overnight. Attached cells were treated with 5 nmol/L of BERA/miR-328-3p or control RNA plus various concentrations of mitoxantrone (0–1000 nmol/L). After 72 h, cell viability was determined using MTT assay. Mitoxantrone chemosensitivity data were fit to a normalized, inhibitory dose-response model with variable slope, Y = 100/(1+10 ^[(LogIC50−X)×Hill slope]) (GraphPad Prism, San Diego, CA, USA). All experiments were carried out in quadruplicate and repeated once with separate cultures.
2.9. Statistical analysis
All values were expressed as mean±SD. Unpaired Student׳s t-test, one-way or two-way analysis of variance was used to perform statistics analysis. Difference was considered as significant if P value was less than 0.05.
3. Results
3.1. Expression of BERA/miR-27b-3p and BERA/miR-328-3p in E. coli and purification to a high homogeneity by anion exchange FPLC
The expression of pre-miR-27b-123nt using tRNA scaffold was very low19, and thus we employed the tRNA/pre-miR-34a based carrier27 for the production of miR-27b-3p (Fig. 1A). In particular, we replaced the 22-nt miR-34a-5p with 21-nt miR-27b-3p, as well as their complementary sequences accordingly (Supporting Information Table S1). To assess to what degree target BERA/miR-27b-3p was heterogeneously expressed in bacteria, total RNAs were extracted from HST08 E. coli at 16 h post-transformation with BERA/miR-27b-3p expression plasmid, and then subjected to RNA electrophoresis. As shown in Fig. 1B, a 200 nt RNA band appeared, indicating a successful expression of target BERA/miR-27b-3p. As expected, the level of BERA/miR-27b-3p was much higher than that of tRNA-hsa-miR-27b-3p-123nt19 (Fig. 1B), supporting the advantages of tRNA/pre-miRNA-based platform in producing target miRNA/siRNA agents.
An anion-exchange FPLC method27., 30. was utilized to purify the target BERA/miR-27b-3p. Gradient elution with low to high concentration of sodium chloride buffer successfully separated the recombinant BERA/miR-27b-3p, which was eluted at 8.7 min (Fig. 1C), from other small RNAs, tRNAs, and 5S ribosomal RNA using a UNO Q6 column. The purity of isolated recombinant ncRNAs in individual fractions were confirmed by the urea-PAGE analyses (Fig. 1C), and the final BERA/miR-27b-3p showed a high degree of homogeneity (>95%) by HPLC analysis.
BERA/miR-328-3p was expressed by using an optimized pre-miR-34a derivative and purified by a streamlined FPLC method, as we reported very recently32. As a result, BERA/miR-328-3p accounted >50% of total bacterial RNAs, and we readily obtained >10 mg of final BERA/miR-328-3p molecule (>98% pure by HPLC) from 1 L bacterial fermentation.
3.2. BERA/miR-27b-3p is processed to mature miRNA in human LS-180 cells and effectively reduces VDR and CYP3A4 expression levels
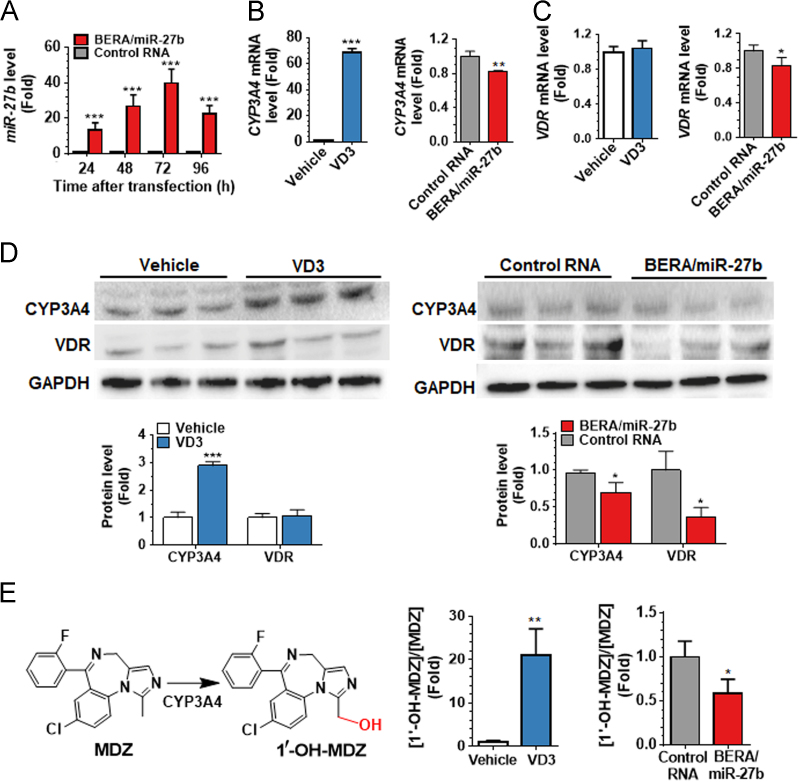
To assess whether mature miR-27b-3p can be produced from BERA/miR-27b-3p in LS-180 cells, a selective stem-loop RT-qPCR assay was used to quantitate mature miR-27b-3p. The data showed that miR-27b-3p level was 10- to 40-fold higher in LS-180 cells transfected with BERA/miR-27b-3p than control RNA, and the high levels of mature miR-27b-3p (>20 fold) persisted 96 h post-transfection (Fig. 2A). These results demonstrated that BERA/miR-27b-3p was successfully transfected into LS-180 cells and processed to mature miR-27b-3p.
Figure 2.
BERA/miR-27b-3p is processed to miR-27b-3p in human LS-180 cells, effectively reduces VDR and CYP3A4 protein expression levels, and thus alters cellular CYP3A4 drug metabolism capacity. LS-180 cells was transfected with 50 nmol/L BERA/miR-27b-3p or control RNA for 24, 48, 72 and 96 h. Levels of miR-27b-3p in BERA/miR-27b-3p treated cells were significantly higher than control RNA and the high level of miR-27b-3p persisted 96 h (A). Treatment of 1α-VD3 greatly induced CYP3A4 mRNA (B) and protein (C) expression, whereas had no effect on VDR mRNA (B) or protein (C) levels. Following the induction by 1α-VD3, CYP3A4 (B) and VDR (C) mRNA levels, as well as their protein levels (D) were significantly reduced by BERA/miR-27b-3p, compared to control RNA (C). CYP3A4 enzymatic activity, as measured by [1′-OH-MDZ]/[MDZ] metabolic ratio after exposure to MDZ for 1.5 h (at 48 h post-transfection), was dramatically increased by 1α-VD3 treatment. 1′-OH-MDZ formation was significantly lower in LS-180 cells treated with BERA/miR-27b-3p than control RNA (E). Values are mean ± SD of triplicate treatments. *P<0.05; **P<0.01; ***P<0.001, compared to vehicle or control RNA group (Student׳s t-test).
Our previous studies revealed that the well-conserved miR-27b-3p can regulate CYP3A4 expression in human carcinoma cells via targeting of the 3′-UTR of CYP3A4 and VDR/NR1I115. Therefore, we assessed the effectiveness of BERA/miR-27b-3p in the modulation of CYP3A4 expression in LS-180 cells. Firstly, we evaluated whether CYP3A4 was induced in LS-180 cells by 1α-VD3. RT-qPCR analyses (Fig. 2B) using gene-specific primers confirmed that CYP3A4 mRNA levels were increased sharply in cells following the exposure to VD3, whereas there was no difference in VDR mRNA levels between cells treated with VD3 and vehicle control (Fig. 2C). The treatment of BERA/miR-27b-3p led to a modest reduction (20%) of both VDR and CYP3A4 mRNA level, as compared to the control RNA treated LS-180 cells (Fig. 2B & C). Further immunoblot analyses revealed a 3-fold increase in CYP3A4 protein level in LS-180 cells after VD3 induction (Fig. 2D). Following the transfection of VD3-treated LS-180 cells with BERA/miR-27b-3p, CYP3A4 protein levels were reduced by 30% and VDR levels was suppressed by over 60%, as compared to control RNA treatment (Fig. 2D), indicating the modulation of target ADME gene expression by miR-27b.
3.3. BERA/miR-27b-3p consequently alters the cellular drug metabolism capacity of CYP3A4
Furthermore, we investigated the consequent influence on cellular drug metabolism capacity. MDZ, a widely used CYP3A4 probe substrate36, was utilized to evaluate cellular CYP3A4 enzymatic activity. MDZ and 1′-OH-MDZ, the major metabolite generated by CYP3A437, were quantified by a sensitive and specific LC—MS/MS method35. Our data revealed that the [1′-OH-MDZ]/[MDZ] metabolic ratio was increased more than 20 folds in VD3-treated LS-180 cells (Fig. 2E), suggesting an induction of CYP3A4 enzymatic activity. [1′-OH-MDZ]/[MDZ] metabolic ratio was 40% lower in the BERA/miR-27b-3p treated LS-180 cells than control RNA treated cells (Fig. 2E), indicating a decrease of drug metabolizing activity with the reduction of CYP3A4 protein levels (Fig. 2D).
3.4. BERA/miR-328-3p is processed to target miRNA in human MCF7/MX100 cells and effectively suppresses ABCG2 expression
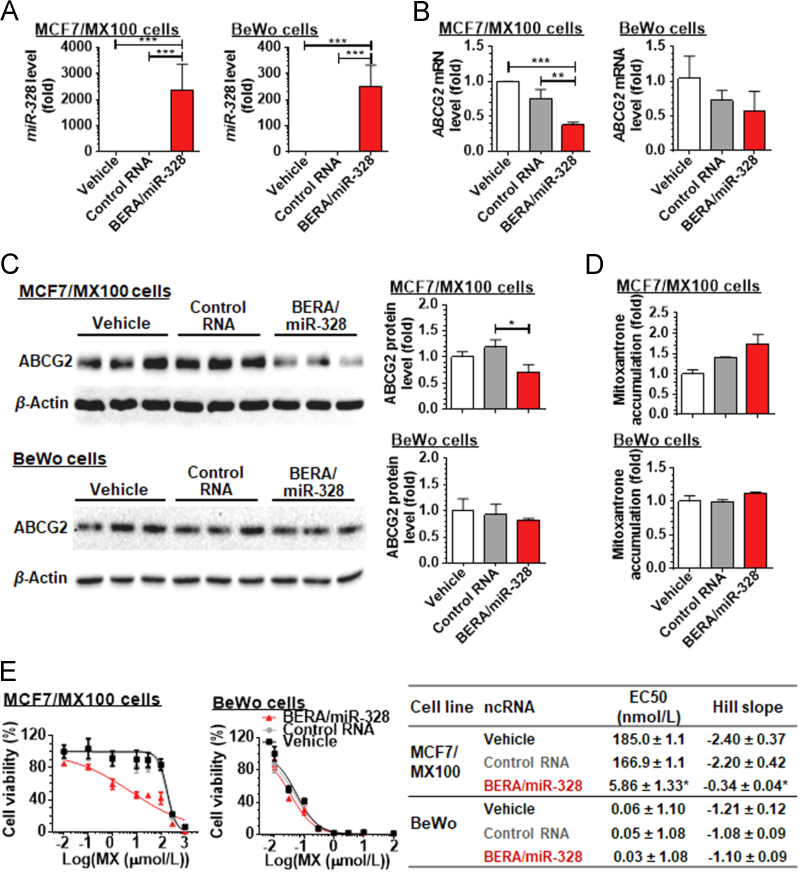
To examine if BERA/miR-328-3p could be processed to target miR-328-3p in human MCF7/MX100 and BeWo cells, we quantitated mature miR-328-3p using selective stem-loop RT-qPCR assay 48 h post transfection. Our data revealed a 2400-fold higher level of mature miR-328-3p in MCF7/MX100 cells treated with BERA/miR-328-3p than the control RNA. Similarly, BERA/miR328-3p-treated BeWo cells showed 250-fold higher levels of miR-328-3p than those cells treated with RNA control (Fig. 3A). These results demonstrated that BERA/miR-328-3p was successfully transfected into human MCF7/MX100 and BeWo cells and processed to target miR-328-3p.
Figure 3.
BERA/miR-328-3p is processed to miR-328-3p in human cancer cells and consequently knocks down ABCG2 levels, leading to higher intracellular accumulation of mitoxantrone and greater sensitivity to mitoxantrone. Human MCF7/MX100 and BeWo cells were transfected with 10 nmol/L BERA/miR-328-3p, control RNA or vehicle for 48 or 72 h. Level of miR-328-3p in BERA/miR-328-3p treated cells was significantly higher than tRNA control (A). (n = 3/group; ***P<0.001, **P<0.01, compared to the vehicle or tRNA control; one-way ANOVA). As a result, ABCG2 mRNA (B) and protein (C) levels were significantly reduced in BERA/miR-328-3p treated MCF/MX100 cells but not in BeWo cells. (***P<0.001, **P<0.01, compared to the vehicle or control RNA; n=3 per group; one-way ANOVA). Flow cytometry analyses revealed that compared to the vehicle control, MCF-7/MX100 cells transfected with BERA/miR-328-3p had significantly higher mitoxantrone fluorescence intensity (D). (*P<0.05, compared to the vehicle control; n=3/group; one-way ANOVA). BERA/miR-328-3p significantly sensitized MCF7/MX100 cells but not BeWo cells to mitoxantrone. The estimated EC50 and Hill slope for mitoxantrone (MX) cytotoxicity against BERA/miR-328-3p- and control tRNA-treated MCF7/MX100 and BeWo cells (***P<0.001, compared to vehicle and control RNA; n=4/group; one-way ANOVA).
To evaluate the impact of BERA/miR-328-3p on ABCG2 expression, a validated target for miR-328-3p21., 22., qPCR and immunoblot analyses were carried out to examine ABCG2 mRNA and its protein levels using specific primers and antibody, respectively. While low concentrations of control RNA (10 nmol/L) showed no or minimal effects on ABCG2 mRNA or its protein expression, as compared to vehicle control (Fig. 3B and C), BERA/miR-328-3p significantly reduced the levels of ABCG2 mRNA and its protein by approximately 50% and 40%, respectively, in MCF7/MX100 cells as compared to control RNA treatment. Surprisingly, no significant changes in ABCG2 mRNA and its protein expression levels by BERA/miR-328-3p treatment were shown in BeWo cells.
3.5. BERA/miR-328-3p is able to increase the intracellular accumulation of mitoxantrone and enhance the sensitivity of human MCF7/MX100 cells to mitoxantrone
To investigate to what degree drug disposition may be altered in cells with the change of ABCG2 protein expression by BERA/miR-328-3p, we conducted flow cytometry analyses to directly determine intracellular accumulation of mitoxantrone, an ABCG2 substrate anticancer drug22. The data showed that, MCF-7/MX100 cells transfected with BERA/miR-328-3p had higher mitoxantrone fluorescence intensity as compared to control RNA and vehicle treated cells (Fig. 3D), despite that it does not reach to statistical significance. By contrast, there was no difference in intracellular mitoxantrone fluorescence between BERA/miR-328-3p-treated BeWo cells and controls, consistent with the finding on an unchanged ABCG2 protein expression (Fig. 3C).
Lastly, we examined if recombinant BERA/miR-328-3p could alter the sensitivity of human carcinoma cells to mitoxantrone. MTT assays were employed to determine the mitoxantrone cytotoxicity against cancer cells after transfection with BERA/miR-328-3p, control RNA, and vehicle. Our data showed that BERA/miR-328-3p transfected MCF7/MX100 cells were more sensitive to mitoxantrone than control RNA and vehicle-treated cells (Fig. 3E). The greater chemosensitivity was also indicated by a significantly lower EC50 value in cells transfected with BERA/miR-328-3p (5.86 nmol/L) than control RNA (166.9 nmol/L) and vehicle control (185.0 nmol/L) (Fig. 3E). In contrast, BERA/miR-328-3p was unable to sensitize BeWo cells to mitoxantrone treatment, which is agreement with the finding on the unchanged ABCG2 protein expression and intracellular drug accumulation (Fig. 2C and D). Together, these findings indicated that modulation of ABCG2 transporter expression with biologic miR-328-3p molecule could facilitate intracellular drug accumulation and thus sensitize the drug-resistant MCF7/MX100 cells to chemotherapy.
4. Discussion
Evidence is emerging that noncoding miRNAs play important roles in the control of drug disposition and response whose research necessitates the development and utilization of proper genetic38., 39. and pharmacological40 tools. Chemically engineered miRNA mimics have been predominately utilized for a large number of studies due to the easy accessibility, even though their exact chemical modifications are not disclosed to users and readers, and it remains unknown how excessive artificial modifications will alter the higher-order structures, activities and toxicities of miRNAs. Inspired by protein bioengineering technology that offers large-scale biologic proteins for research and therapy, we have recently established a novel tRNA/pre-miRNA based RNA bioengineering technology27., 28. for the production of highly-structured, stable natural miRNA reagents with minimal posttranscriptional modifications29., 30. which was utilized for the production of miR-27b-3p and miR-328-3p in the present study.
Previous studies have demonstrated that miR-27b-3p regulated CYP3A4 expression via direct and indirect targeting, using either plasmid-based miRNA expression materials15 or early version recombinant miRNA just using tRNA as scaffold19. It is also noteworthy, compared to the miR-27-3p agent produced with tRNA carrier19, BERA/miR-27-3p made in current study using the novel tRNA/pre-miR-34a based RNA bioengineering platform27., 28. (Fig. 1) is characterized by a much greater level of expression in E. coli and thus higher overall yield of target BERA/miRNA. As the tRNA/pre-miR-34a carrier is refined further28, the BERA/miR-328-3p was produced at surprisingly high levels, i.e., >50% of total bacterial RNA and >10 mg of pure BERA/miR-328-3p from 1 L fermentation. These results support the consistent high-yield, large-scale and cost-effective production of biologic RNAi molecules through fermentation using our novel tRNA/pre-miRNA based technology that should be addition to existing tools for RNA research and drug development.
To assess the biological activity of BERA/miR-27b-3p in the regulation of CYP3A4-mediated drug metabolism, CYP3A4 mRNA and its protein levels as well as CYP3A-dependent midazolam 1′-hydroxylation activity were firstly induced by VD3 treatment in human colon carcinoma LS-180 to mimic the physiological transcriptional activation in human enterocyte34. BERA/miR-27b-3p was subsequently transfected into LS-180 cells which was indeed processed into high levels of mature miR-27b-3p (Fig. 2). This led to the interference of VDR-CYP3A4 signaling pathway, as manifested by a significant suppression of the mRNA and protein levels of both VDR and CYP3A4. Consequently, BERA/miR-27b-3p treatment caused a reduction of CYP3A4 enzymatic activity, as manifested by midazolam 1′-hydroxylation. By contrast, enzymatic activities of CYP1A2, 2C9, 2D6, and 2E1 measured simultaneously using the cocktail LC—MS/MS method35 were below the quantification limits, supporting the low or lack of expression of CYP1A2, 2C9, 2D6, and 2E1 in LS-180 cells.
The function of BERA/miR-328-3p in the regulation of ABCG2 and subsequent effects on drug disposition and chemosensitivity were examined. Upon introduction of target miR-328-3p into the cells, ABCG2 mRNA and its protein levels were knocked down in MCF7/MX100 cells (Fig. 3), leading to a higher level of intracellular accumulation of mitoxantrone. In contrast, ABCG2 expression was not modulated by BERA/miR-328-3p in BeWo cell line which is another well-recognized model for the study of ABCG241. The distinct biological effects of miRNA on its target in different type of cells are likely related to cell-specific regulatory elements including target mRNA abundance42., 43.. ABCG2 protein level was proven to be dramatically higher in MCF7/MX100 cells than in BeWo cells (Supporting Information Fig. S1). Meanwhile, the inability of miR-328-3p to modulate ABCG2 expression in BeWo cells might be attributable to other factors that are more specific and/or important for placental ABCG2 gene regulation, e.g., the hypoxia signaling transcription factors44 that has been shown to play a critical role in controlling overall placental ABCG2 expression levels. Further investigations are warranted to understand particular cell properties and cellular conditions for microRNA-mediated regulation and the context-dependent post-transcriptional regulatory network45. Nevertheless, the successful suppression of ABCG2 by biologic miR-328-3p molecule in ABCG2-mediated drug resistant MCF7/MX100 cells was able to sensitize the cells to chemotherapy (Fig. 3), supporting the notion that intervention of efflux transporters with miRNAs11., 46. or chemical inhibitors46 may help to improve the efficacy of chemotherapeutics.
In summary, this study demonstrated a successful production of biologically active miR-27b-3p and miR-328-3p agents using the novel tRNA/pre-miRNA-based RNA bioengineering technology. Our results showed that the BERA/miR-27b-3p was processed to mature miR-27b-3p in human carcinoma LS-180 cells in a time-dependent manner. Consequently, biologic miR-27b-3p was capable to suppress VDR and CYP3A4 protein expression, leading to a lower cellular drug metabolism capacity. In addition, BERA/miR-328-3p was effective to modulate ABCG2 expression in drug resistant MCF7/MX100 cells, which subsequently sensitized the cells to chemotherapeutic drug. These findings support that bioengineered miRNA agents can be utilized for the study of miRNA functions in regulation of drug metabolism and disposition, and interference of efflux transporter may improve the outcome of chemotherapy.
Acknowledgments
This work was supported in part by the National Institutes of Health [Grant No. R01GM113888 (Aiming Yu), USA]. Xin Li was supported by Visiting Scholar Programs from China Scholarship Council (201608440507, USA) and Guangzhou Medical University, National Natural Science Foundation of China (81603191, China) and Natural Science Foundation of Guangdong Province (2015A030310153, China). Ye Tian was supported by the 3102018zy053 from Fundamental Research Funds for the Central Universities (China). The authors appreciate the access to the Flow Cytometry and Molecular Pharmacology Shared Resources funded by the UC Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute (Grant No. P30CA093373, USA).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.apsb.2018.12.002.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Lu A.Y. Drug-metabolism research challenges in the new millennium: individual variability in drug therapy and drug safety. Drug Metab Dispos. 1998;26:1217–1222. [PubMed] [Google Scholar]
- 2.Yu A.M., Pan Y.Z. Noncoding microRNAs: small RNAs play a big role in regulation of ADME? Acta Pharm Sin B. 2012;2:93–101. doi: 10.1016/j.apsb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Tracy T.S., Chaudhry A.S., Prasad B., Thummel K.E., Schuetz E.G., Zhong X.B. Interindividual variability in cytochrome P450-mediated drug metabolism. Drug Metab Dispos. 2016;44:343–351. doi: 10.1124/dmd.115.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manikandan P., Nagini S. Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets. 2018;19:38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 6.Gu X., Xiao Q., Ruan Q., Shu Y., Dongre A., Iyer R. Comparative untargeted proteomic analysis of ADME proteins and tumor antigens for tumor cell lines. Acta Pharm Sin B. 2018;8:252–260. doi: 10.1016/j.apsb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu A.M., Ingelman-Sundberg M., Cherrington N.J., Aleksunes L.M., Zanger U.M., Xie W. Regulation of drug metabolism and toxicity by multiple factors of genetics, epigenetics, lncRNAs, gut microbiota, and diseases: a meeting report of the 21st International Symposium on Microsomes and Drug Oxidations (MDO) Acta Pharm Sin B. 2017;7:241–248. doi: 10.1016/j.apsb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J. Combinatorial microRNA target predictions. Nat Gen. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 9.Yokoi T., Nakajima M. microRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 10.Ingelman-Sundberg M., Zhong X.B., Hankinson O., Beedanagari S., Yu A.M., Peng L. Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos. 2013;41:1725–1731. doi: 10.1124/dmd.113.053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu A.M., Tian Y., Tu M.J., Ho P.Y., Jilek J.L. MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos. 2016;44:308–319. doi: 10.1124/dmd.115.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano M., Nakajima M. Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Expert Opin Drug Metab Toxicol. 2018;14:493–504. doi: 10.1080/17425255.2018.1472237. [DOI] [PubMed] [Google Scholar]
- 13.Tsuchiya Y., Nakajima M., Takagi S., Taniya T., Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 14.Chuturgoon A.A., Phulukdaree A., Moodley D. Fumonisin B1 modulates expression of human cytochrome P450 1b1 in human hepatoma (Hepg2) cells by repressing Mir-27b. Toxicol Lett. 2014;227:50–55. doi: 10.1016/j.toxlet.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Pan Y.Z., Gao W., Yu A.M. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekström L., Skilving I., Ovesjö M.L., Aklillu E., Nylén H., Rane A. miRNA-27b levels are associated with CYP3A activity in vitro and in vivo. Pharmacol Res Perspect. 2015;3:e00192. doi: 10.1002/prp2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apellániz-Ruiz M., Inglada-Pérez L., Naranjo M.E., Sánchez L., Mancikova V., Currás-Freixes M. High frequency and founder effect of the CYP3A4*20 loss-of-function allele in the Spanish population classifies CYP3A4 as a polymorphic enzyme. Pharmacogenomics J. 2015;15:288–292. doi: 10.1038/tpj.2014.67. [DOI] [PubMed] [Google Scholar]
- 18.Wrighton S.A., Stevens J.C. The human hepatic cytochromes P450 involved in drug metabolism. Crit Rev Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 19.Li M.M., Wang W.P., Wu W.J., Huang M., Yu A.M. Rapid production of novel pre-microRNA agent hsa-mir-27b in Escherichia coli using recombinant RNA technology for functional studies in mammalian cells. Drug Metab Dispos. 2014;42:1791–1795. doi: 10.1124/dmd.114.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji J., Zhang J., Huang G., Qian J., Wang X., Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Pan Y.Z., Morris M.E., Yu A.M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Pan Y.Z., Seigel G.M., Hu Z.H., Huang M., Yu A.M. Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, -519c and -520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem Pharmacol. 2011;81:783–792. doi: 10.1016/j.bcp.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To K.K., Leung W.W., Ng S.S. Exploiting a novel miR-519c-HuR-ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp Cell Res. 2015;338:222–231. doi: 10.1016/j.yexcr.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Jiao X., Zhao L., Ma M., Bai X., He M., Yan Y. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2) Breast Cancer Res Treat. 2013;139:717–730. doi: 10.1007/s10549-013-2607-x. [DOI] [PubMed] [Google Scholar]
- 25.Ma M.T., He M., Wang Y., Jiao X.Y., Zhao L., Bai X.F. MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2) Cancer Lett. 2013;339:107–115. doi: 10.1016/j.canlet.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Ho P.Y., Yu A.M. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA. 2016;7:186–197. doi: 10.1002/wrna.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q.X., Wang W.P., Zeng S., Urayama S., Yu A.M. A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucl Acids Res. 2015;43:3857–3869. doi: 10.1093/nar/gkv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho P.Y., Duan Z., Batra N., Jilek J.L., Tu M.J., Qiu J.X. Bioengineered noncoding RNAs selectively change cellular mirnome profiles for cancer therapy. J Pharmacol Exp Ther. 2018;365:494–506. doi: 10.1124/jpet.118.247775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M.M., Addepalli B., Tu M.J., Chen Q.X., Wang W.P., Limbach P.A. Chimeric microRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos. 2015;43:1129–1136. doi: 10.1124/dmd.115.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W.P., Ho P.Y., Chen Q.X., Addepalli B., Limbach P.A., Li M.M. Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther. 2015;354:131–141. doi: 10.1124/jpet.115.225631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P.C., Tu M.J., Ho P.Y., Jilek J.L., Duan Z., Zhang Q.Y. Bioengineered NRF2-siRNA is effective to interfere with NRF2 pathways and improve chemosensitivity of human cancer cells. Drug Metab Dispos. 2018;46:2–10. doi: 10.1124/dmd.117.078741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho P.Y., Duan Z., Batra N., Jilek J.L., Tu M.J., Qiu J.X. Bioengineered ncRNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther. 2018;365:494–506. doi: 10.1124/jpet.118.247775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponchon L., Beauvais G., Nonin-Lecomte S., Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc. 2009;4:947–959. doi: 10.1038/nprot.2009.67. [DOI] [PubMed] [Google Scholar]
- 34.Thummel K.E., Brimer C., Yasuda K., Thottassery J., Senn T., Lin Y. Transcriptional control of intestinal cytochrome P-4503A by 1α, 25-dihydroxy vitamin D3. Mol Pharmacol. 2001;60:1399–1406. doi: 10.1124/mol.60.6.1399. [DOI] [PubMed] [Google Scholar]
- 35.Jilek J.L., Tian Y., Yu A.M. Effects of MicroRNA-34a on the pharmacokinetics of cytochrome P450 probe drugs in mice. Drug Metab Dispos. 2017;45:512–522. doi: 10.1124/dmd.116.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen L.R., Gabel-Jensen C., Wubshet S.G., Kongstad K.T., Janfelt C., Badolo L. Characterization of midazolam metabolism in locusts: the role of a CYP3A4-like enzyme in the formation of 1′-OH and 4-OH midazolam. Xenobiotica. 2016;46:99–107. doi: 10.3109/00498254.2015.1051604. [DOI] [PubMed] [Google Scholar]
- 37.Pillai V.C., Strom S.C., Caritis S.N., Venkataramanan R. A sensitive and specific CYP cocktail assay for the simultaneous assessment of human cytochrome P450 activities in primary cultures of human hepatocytes using LC—MS/MS. J Pharm Biomed Anal. 2013;74:126–132. doi: 10.1016/j.jpba.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson S., Antonsson M., Elebring M., Jansson-Löfmark R., Weidolf L. Drug metabolism and pharmacokinetic strategies for oligonucleotide- and mRNA-based drug development. Drug Discov Today. 2018;23:1733–1745. doi: 10.1016/j.drudis.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 39.Lauschke V.M., Barragan I., Ingelman-Sundberg M. Pharmacoepigenetics and toxicoepigenetics: novel mechanistic insights and therapeutic opportunities. Annu Rev Pharmacol Toxicol. 2018;58:161–185. doi: 10.1146/annurev-pharmtox-010617-053021. [DOI] [PubMed] [Google Scholar]
- 40.Jackson S.M., Manolaridis I., Kowal J., Zechner M., Taylor N.M.I., Bause M. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018;25:333–340. doi: 10.1038/s41594-018-0049-1. [DOI] [PubMed] [Google Scholar]
- 41.Evseenko D.A., Paxton J.W., Keelan J.A. ABC drug transporter expression and functional activity in trophoblast-like cell lines and differentiating primary trophoblast. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1357–R1365. doi: 10.1152/ajpregu.00630.2005. [DOI] [PubMed] [Google Scholar]
- 42.Arvey A., Larsson E., Sander C., Leslie C.S., Marks D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahadi A., Sablok G., Hutvagner G. miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA-target interaction data. Nucl Acids Res. 2017;45:e42. doi: 10.1093/nar/gkw1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francois L.N., Gorczyca L., Du J., Bircsak K.M., Yen E., Wen X. Down-regulation of the placental BCRP/ABCG2 transporter in response to hypoxia signaling. Placenta. 2017;51:57–63. doi: 10.1016/j.placenta.2017.01.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erhard F., Haas J., Lieber D., Malterer G., Jaskiewicz L., Zavolan M. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24:906–919. doi: 10.1101/gr.166702.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi Y.H., Yu A.M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20:793–807. doi: 10.2174/138161282005140214165212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.