Abstract
As d-amino acids play important roles in the physiological metabolism of bacteria, combination of d-amino acids with antibiotics may provide synergistic antibacterial activity. The aim of the study was to evaluate in vitro and in vivo activity of d-serine alone and in combination with β-lactams against methicillin-resistant Staphylococcus aureus (MRSA) strains, and to explore the possible sensitization mechanisms. The activity of d-serine, β-lactams alone and in combinations was evaluated both in vitro by standard MICs, time–kill curves and checkerboard assays, and in vivo by murine systemic infection model as well as neutropenic thigh infection model. An in vitro synergistic effect was demonstrated with the combination of d-serine and β-lactams against MRSA standard and clinical strains. Importantly, the combinations enhanced the therapeutic efficacy in the animal models as compared to β-lactam alone groups. Initial mechanism study suggested possible revision of d-alanine-d-alanine residue to d-alanine-d-serine in peptidoglycan by adding of d-alanine in the medium, which may cause decreased affinity to PBPs during transpeptidation. In conclusion, d-serine had synergistic activity in combination with β-lactams against MRSA strains both in vitro and in vivo. Considering the relatively good safety of d-serine alone or in combination with β-lactams, d-serine is worth following up as new anti-MRSA infection strategies.
KEY WORDS: MRSA, d-Serine, β-Lactams, Combination, Synergistic effect
Graphical abstract
In this study, the authors found that d-serine had synergistic activity with β-lactams (represented by oxacillin and meropenem) against MRSA strains both in vitro and in vivo, demonstrated by results in MIC determination/checkerboard assay, time–kill curve analysis, murine systemic infection model and neutropenic thigh infection model.
1. Introduction
Staphylococcus aureus (S. aureus) is one of the most important clinical organisms among Gram-positive bacteria. It is a leading cause of skin and soft tissue infections (SSTIs), bacteremia and infective endocarditis1., 2.. Methicillin-resistant S. aureus (MRSA) strain has been a heavy burden worldwide and caused a range of serious infections with poor clinical outcomes3., 4., 5.. However, grim scenario of drug discovery for new antibiotics has been presented for the last few decades as pharmaceutical companies lack interest in this field, owing to difficulty in recouping drug discovery costs from antibiotics which developed resistance within a decade or so6. Therefore, there is a critical need to develop new treatment strategies against these MRSA life-threatening infections.
d-Amino acids play important roles in bacterial physiology7., 8.. d-alanine (d-Ala) and d-glutamate (d-Glu) are components of bacterial peptidoglycan9. d-amino acids could also influence peptidoglycan composition, amount and strength, both via their incorporation into the polymer and by regulating enzymes that synthesize and modify it8., 10., 11.. Up to date, researches mainly focused on the effects of d-amino acids on biofilm, finding that d-amino acids could not only prevent biofilm formation, but also disrupt existing biofilms12., 13., 14., 15.. In addition, d-amino acids were also able to enhance the activity of rifampin against biofilm formation in S. aureus, and to increase the efficacies of colistin, ciprofloxacin and amikacin against Pseudomonas aeruginosa16., 17.. However, less studies focused on the effects of d-amino acids on planktonic bacteria, except Tong et al.18 showed that the application of d-cysteine (d-Cys), d-aspartic acid (d-Asp) and d-Glu could significantly improve the antibacterial activity of nisin against planktonic bacteria of S. mutans.
d-Serine (d-Ser) was reported to be able to replace d-Ala residue of peptidoglycan stem peptides and increase susceptibility of methicillin in MRSA19. In order to determine whether d-Ser can improve susceptibility of β-lactams against MRSA strains, we investigated the activity of d-Ser, β-lactams (e.g., oxacillin and meropenem) alone and in combination against MRSA strains, including clinical and standard isolates, both in vitro and in vivo.
2. Materials and methods
2.1. Bacterial strains
Three standard MRSA strains, 17 clinical MRSA strains and 1 standard methicillin-susceptible S. aureus (MSSA) strain, 18 clinical MSSA strains, randomly selected from our S. aureus strain collection from hospitals in China during 2005–2013 were included in the current study. MLST was performed as described by Enright et al.20 previously. The seven housekeeping gene sequences were compared with known alleles from the MLST database (https://pubmlst.org/saureus/), and the allelic profiles and ST types were determined based on the database. The polymorphic X region of spa gene was amplified as previously described21, and the spa type was determined by submitting the sequencing data to the S. aureus type database (http://spaserver.ridom.de). The genotypic features of the isolates are shown in Supporting Information Table S1.
2.2. Antibiotics, d-amino acids and culture medium
d-Amino acids were purchased from Sigma–Aldrich (St. Louis, MO, USA). The stock solutions were prepared in water (for in vitro experiments) or 0.85% NaCl (for in vivo experiments), and sterilized by filtration after adjusting pH to 7.0. Antibiotics were purchased from National Institute for Food and Drug Control (National Institutes for Food and Drug Control, Beijing, China).
2.3. Laboratory animals
CD-1 (ICR) mice (female, 18–20 g for systemic infection model, and 24–26 g for neutropenic thigh infection model) were purchased from Vital River Laboratories (Beijing, China). All animals were housed under controlled humidity (30%–70%), temperature (22 ± 3 °C) and a 12-h light-dark cycle. Animals had free access to food and water during the study. All the animal studies complied with the ARRIVE guidelines, and all experiments were approved by Animal Research Committee of the Institute of Medicinal Biotechnology (Beijing, China).
2.4. Minimal inhibitory concentration (MIC) determination
MICs were determined by broth microdilution method as recommended22. The final inoculum in each well was about 5×105 CFU/mL. The microtiter plates were incubated at 35 °C for 24 h, and the results were recorded by naked eyes.
2.5. Checkerboard assay
Seventeen clinical MRSA isolates and 3 standard MRSA isolates (S. aureus ATCC 33591, ATCC 43300 and N315) were used. The test concentrations were d-Ser: 0, 2.5, 5, 10, 20, 40, 80, and 100 mmol/L (The concentrations of d-Ser for MIC determination were 62.5, 125, 250, 500, 1000, and 2000 mmol/L); oxacillin (OXA): 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 mg/L and meropenem (MEM): 0.06, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 mg/L. The combination effect of d-Ser with OXA or MEM was determined by calculating the fractional inhibitory concentration index (FICI) using the concentration combinations with highest combination effects (Eq. (1)):
| (1) |
The antimicrobial combination was defined to be synergistic when the FICI was ≤0.5; indifferent when 0.5 < FICI < 4; antagonistic when FICI≥423. The experiments were performed in duplicate on different days.
2.6. Time–kill curves
Time–kill curve assays were performed with standard MRSA ATCC 43300 and N315, as well as clinical MRSA isolates (0603 and 0850), according to method described by Lu et al.24 with minor modifications. Briefly, an overnight culture of each isolate was diluted with 3 mL CAMH broth to a final concentration of ~106 CFU/mL. Then d-Ser (at 20 mmol/L), antibiotics (at the lowest concentrations that can show synergistic effects when combined with d-Ser) alone and in combinations were added. Viable cell counts were determined at 0, 2, 4, 6, 8 and 24 h after incubation at 35 °C by plating 10 μL serial diluted samples onto MH agar plates in triplicate. The results were recorded as log10 CFU/mL. Synergy was defined as ≥2 log10 CFU/mL decrease at 24 h incubation in the combination treatment in comparison to single antibiotic alone exposure23.
2.7. Murine systemic infection model
Representative MRSA strains, ATCC 43300, N315 and 0850, were used in this model. After intraperitoneal injection of 0.5 mL bacterial suspension (100% minimum lethal dose) in 5% mucin, mice were administered with d-Ser, antibiotics alone or in combinations through subcutaneous injection at 1 and 6 h after infection (8–10 animals/group). The doses of OXA were 2.5, 5, and 10 mg/kg for N315 strain and 5, 10, and 25 mg/kg for ATCC 43300; the doses of MEM were 0.5, 1, and 2.5 mg/kg for N315 and 25, 50, and 100 mg/kg for 0850. Animal deaths were recorded for 7 days and the survival rates were calculated.
2.8. Murine neutropenic thigh infection model
The experiment was carried out according to previously described methods with some modifications25., 26.. Briefly, CD-1 (ICR) female mice were rendered neutropenic by intraperitoneally dosed with cyclophosphamide on day 4 (150 mg/kg) and day 1 (100 mg/kg) prior to infection27. The right thighs were then infected intramuscularly with 100 μL of overnight cultures of MRSA N315 (4–7 × 105 CFU per thigh). Mice then received: (1) no treatment (control group); (2) d-Ser alone at 4 mmol/kg (administrated at 2, 10, 18, 26, 34 and 42 h post-infection); (3) OXA alone at 20 or 50 mg/kg (administrated at the same time-points as d-Ser); (4) OXA+d-Ser 4 mmol/kg; (5) MEM alone at 25 or 50 mg/kg (administrated at 2, 8, 14, and 20 h after infection); (6) MEM+d-Ser 4 mmol/kg. The OXA and MEM doses were chosen according to previous reports28., 29.. Mice were sacrificed at 24 h for MEM groups and 48 h for OXA groups after infection. Right thigh muscles were then aseptically excised, homogenized, serially diluted and plated on MH agar plates for CFU counts. Bacterial colony counts were expressed as mean log10 CFU/thigh (±SEM).
2.9. The docking assay
Penicillin-binding protein 2a (PBP2a), a protein exists in MRSA strains, can lead to resistance by its low affinity with β-lactams but normal affinity to natural substrate as well as existing activity on catalyzing cell wall synthesis. As we speculated that the effect of d-Ser might be related with PBP2a, we compared the binding activity of natural substrate with PBP2a as well as changed substrate after adding d-Ser with PBP2a by docking assay. Using crystal structure of PBP2a (PBD entry: 1VQQ) as receptor, we compared the binding activity of d-Ala-d-Ala and d-Ala-d-Ser by Discovery Studio 4.5. The binding modes for dipeptides towards the binding site of PBP2a were generated by MOE (Molecular Operating Environment) version 2009.10. The docking was performed through the “Dock” module and scored using LibDock scoring system. The higher the score, the better affinity of the dipeptide with the protein.
2.10. Statistical analysis
Statistical analysis was performed by SPSS v16.0. P values were calculated using one-way ANOVA to compare the differences between each pair of groups. P < 0.05 was considered statistically significant.
3. Results
3.1. In vitro activity of d-Ser in combinations against MRSA and MSSA strains
The MICs of 12 different β-lactams alone and in combination with d-Ser at 20 mmol/L against MRSA ATCC 43300 are summarized in Table 1. The MICs of β-lactams against ATCC 43300 were from 8 to > 1024 mg/L. Interestingly, the MICs of the tested β-lactams against the studied MRSA strain were significantly reduced (8 to >128-folds) with addition of d-Ser at 20 mmol/L. MEM (meropenam, usually not used alone in MRSA infection) and OXA (oxacillin, traditionally considered “inactive” against MRSA) were then chosen as the representative antibiotics in further evaluation for the purpose of reusing them.
Table 1.
MICs of β-lactams in combination with d-Ser against MRSA ATCC43300.
| Antibiotics | MIC (mg/L) at d-Ser of |
Fold reduced | |
|---|---|---|---|
| 0 mmol/L | 20 mmol/L | ||
| Cefepime | 32 | 2 | 16 |
| Cefuroxime | 16 | 1 | 16 |
| Cephalothin | 16 | 0.25 | 64 |
| Cefixime | >1024 | 8 | >128 |
| Ceftazidime | 64 | 8 | 8 |
| Cefotaxime | 64 | 1 | 64 |
| Ceftriaxone | 128 | 2 | 64 |
| Ampicillin | 16 | 2 | 8 |
| Oxacillin | 16 | 0.125 | 128 |
| Penicillin | 16 | 2 | 8 |
| Meropenem | 8 | 0.125 | 64 |
| Ertapenem | 8 | 0.125 | 64 |
In contrast, d-Ser showed very limited sensitization effect on oxacillin and meropenem against MSSA strains. MICs of the antibiotics against the 19 MSSA strains were generally 2–4 folds reduced (Supporting Information Table S2).
3.2. Checkerboard assay
The checkerboard assay was conducted using d-Ser and the representative β-lactams (MEM and OXA) against all studied MRSA strains and the results are summarized in Table 2. d-Ser alone showed minor antibacterial activity with MICs of 500–2000 mmol/L. OXA and MEM alone had weak inhibition effects on bacterial growth, as MICs can be as high as 1024 and 128 mg/L respectively. However, when combined with d-Ser, the MICs of OXA and MEM against the MRSA strains were reduced in a concentration dependent manner of d-Ser. MICs of ≤0.25 mg/L for OXA and ≤0.06 mg/L for MEM were observed when combined with 100 mmol/L d-Ser against the studied MRSA strains (data not shown). FICIs were calculated using the concentration combinations with highest combination effects, that is 1/256–1/32 MIC of OXA or 1/256–1/2 MIC of MEM in combination with 10–100 mmol/L d-Ser. FICIs were 0.024–0.216 and 0.018–0.580 for OXA/d-Ser combination and MEM/d-Ser combination respectively. According to the results, synergistic effects existed in 20 (OXA/d-Ser combination) and 19 (MEM/d-Ser combination) MRSA strains.
Table 2.
MICs and FIC indexes of d-Ser with β-lactam antibiotics against MRSA strains.
| Strains | OXA/d-Ser |
MEM/d-Ser |
||||
|---|---|---|---|---|---|---|
| MIC in single use (mg/L)/(mmol/L) | MIC in combination (mg/L)/(mmol/L) | FIC index | MIC in single use (mg/L)/(mmol/L) | MIC in combination (mg/L)/(mmol/L) | FIC index | |
| MRSA 0501 | 512/500 | 4/40 | 0.088 | 128/500 | 4/40 | 0.111 |
| MRSA 0516 | 1024/2000 | 4/40 | 0.024 | 64/2000 | 1/40 | 0.036 |
| MRSA 0520 | 256/2000 | 2/40 | 0.028 | 16/2000 | 0.25/100 | 0.066 |
| MRSA 0533 | 512/2000 | 4/40 | 0.028 | 32/2000 | 0.25/40 | 0.028 |
| MRSA 0603 | 256/500 | 2/10 | 0.028 | 16/500 | 0.25/40 | 0.096 |
| MRSA 0616 | 64/500 | 1/100 | 0.216 | 4/500 | 2/40 | 0.580 |
| MRSA 0623 | 512/2000 | 8/40 | 0.036 | 32/2000 | 0.25/20 | 0.018 |
| MRSA 0629 | 512/1000 | 4/40 | 0.048 | 32/1000 | 0.5/20 | 0.036 |
| MRSA 0637 | 512/2000 | 8/40 | 0.036 | 32/2000 | 0.125/40 | 0.024 |
| MRSA 0826 | 1024/1000 | 8/40 | 0.048 | 32/1000 | 0.25/100 | 0.108 |
| MRSA 0832 | 512/1000 | 2/40 | 0.044 | 32/1000 | 1/40 | 0.071 |
| MRSA 0836 | 512/500 | 16/20 | 0.071 | 32/500 | 1/40 | 0.111 |
| MRSA 0844 | 1024/2000 | 4/40 | 0.024 | 32/2000 | 1/40 | 0.051 |
| MRSA 0845 | 512/2000 | 8/40 | 0.036 | 32/2000 | 0.25/100 | 0.058 |
| MRSA 0848 | 512/2000 | 4/40 | 0.028 | 32/2000 | 1/40 | 0.051 |
| MRSA 0850 | 512/500 | 2/40 | 0.084 | 32/500 | 2/20 | 0.103 |
| MRSA 0852 | 512/1000 | 8/40 | 0.056 | 32/1000 | 0.25/100 | 0.108 |
| ATCC 33591 | 256/1000 | 8/20 | 0.051 | 32/1000 | 2/20 | 0.083 |
| ATCC43300 | 64/500 | 0.25/20 | 0.044 | 4/500 | 0.06/40 | 0.095 |
| MRSA N315 | 64/500 | 0.5/40 | 0.088 | 8/500 | 0.25/40 | 0.111 |
The test concentrations were d-Ser: 0, 2.5, 5, 10, 20, 40, 80, and 100 mmol/L; oxacillin (OXA): 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 mg/L and meropenem (MEM): 0.06, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 mg/L. We found that with the increase of d-Ser concentration, the MICs of the antibiotics decreased further, MICs of oxacillin and meropenem were as low as ≤0.25 mg/L and ≤0.06 mg/L with 100 mmol/L d-Ser.
3.3. Time–kill curve analysis of OXA/d-Ser and MEM/d-Ser
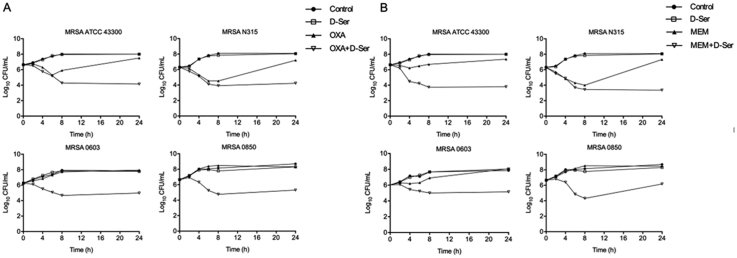
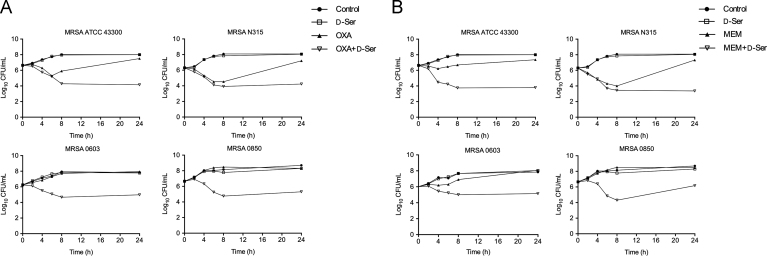
As shown in Fig. 1, OXA and MEM alone at sub-MIC levels showed modest bactericidal activity. However, the OXA/d-Ser and MEM/d-Ser combinations demonstrated enhanced bactericidal activities against all tested MRSA strains, with viable cell counts significantly reduced by 2.97–3.36 and 2.31–3.96 log10 CFU/mL respectively at 24 h compared with the corresponding OXA and MEM alone groups. These data suggest synergistic bactericidal activities of the combinations.
Figure 1.
Time–kill curves against MRSA strains ATCC43300, N315, 0603 and 0850 for combination of d-Ser (20 mmol/L, equivalent to 1/25 MIC) with OXA (Panel A) and MEM (Panel B). For Panel A, the OXA doses were: 1/32 MIC for MRSA ATCC 43300, 1/4 MIC for MRSA N315, 1/32 MIC for MRSA 0603, 1/8 MIC for MRSA 0850. For Panel B, the MEM doses were: 1/4 MIC for MRSA ATCC43300, 1/2 MIC for MRSA N315, 1/2 MIC for MRSA 0603, 1/2 MIC for MRSA 0850.
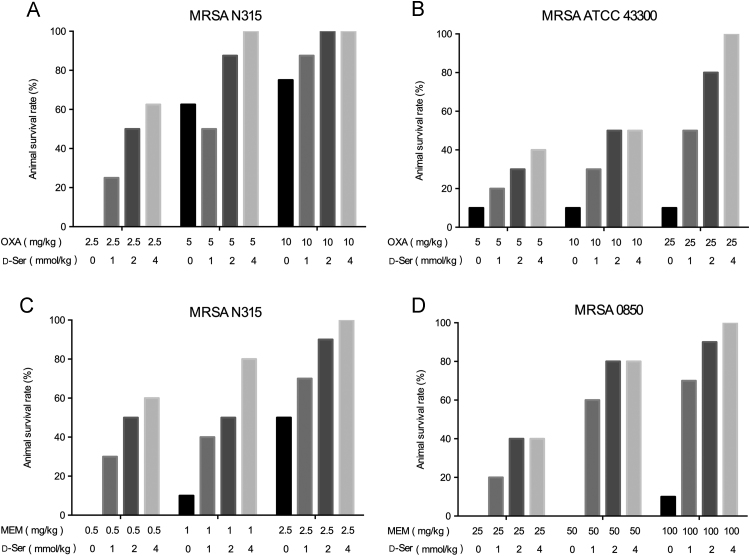
3.4. OXA/d-Ser and MEM/d-Ser combinations enhanced animal survival rates in murine systemic infection model
Generally, the combination of OXA or MEM with d-Ser increased animal survival rates as compared to OXA and MEM alone groups in a concentration dependent manner of d-Ser (Fig. 2). Combination of d-Ser (4 mmol/kg) with OXA significantly increased the animal survival rates, from 0% to 62.5% (OXA at 2.5 mg/kg), 62.5% to 100% (OXA at 5 mg/kg), 75% to 100% (OXA at 10 mg/kg) for MRSA N315 strain infection (Fig. 2A), and from 10% to 40% (OXA at 5 mg/kg), 10% to 50% (OXA at 10 mg/kg), 10% to 100% (OXA at 25 mg/kg) for MRSA ATCC43300 strain infection (Fig. 2B). The combination of d-Ser (4 mmol/kg) with MEM also significantly increased animal survival rates, from 0% to 60% (MEM at 0.5 mg/kg), 10% to 80% (MEM at 1 mg/kg), 50% to 100% (MEM at 2.5 mg/kg) for MRSA N315 strain infection (Fig. 2C), and from 0% to 40% (MEM at 25 mg/kg), 0% to 80% (MEM at 50 mg/kg), 10% to 100% (MEM at 100 mg/kg) for MRSA 0850 strain infection (Fig. 2D). d-Ser alone at 1, 2 and 4 mmol/kg had no protection on animals with 100% mortality due to tested MRSA strain infections (data not shown).
Figure 2.
Animal survival rates of OXA, MEM alone and in combination with d-Ser in murine systemic infection model. Panel A: OXA±d-Ser against MRSA N315; Panel B: OXA±d-Ser against MRSA ATCC 43300; Panel C: MEM±d-Ser against MRSA N315; Panel D: MEM±d-Ser MRSA 0850. The infection doses were 7.91×103 CFU per mouse for panel A, 6.5×105 CFU per mouse for panel B, 1.87×104 CFU per mouse for panel C and 1.2×105 CFU per mouse for panel D.
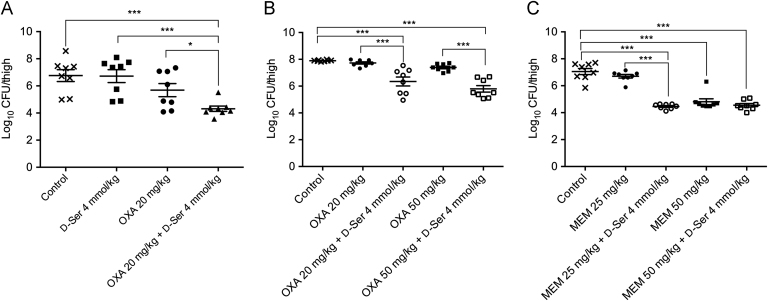
3.5. In vivo antibacterial activity of OXA/d-Ser and MEM/d-Ser combinations in murine neutropenic thigh infection model
As shown in Fig. 3A, d-Ser alone at 4 mmol/kg didn׳t reduce colony counts in thigh vs. the control group, OXA alone at 20 mg/kg had weak activity in curing thigh infection as CFU counts had no significant difference vs. the untreated control group (P > 0.05). However, combination of d-Ser at 4 mmol/kg significantly enhanced the antibacterial activity of OXA (P < 0.001 vs. untreated control and d-Ser alone groups, and P < 0.05 vs. OXA alone group). The antibacterial activity of OXA/d-Ser and MEM/d-Ser combinations was then evaluated in detail. As shown in Fig. 3B, OXA alone at 20 mg/kg or 50 mg/kg were ineffective in curing thigh infections, with CFU counts similar to the control group (P > 0.05). Addition of d-Ser at 4 mmol/kg significantly enhanced the antibacterial activity of OXA, with CFU counts significantly reduced compared to OXA alone groups (P<0.001). With addition of 4 mmol/kg d-Ser, 50 mg/kg OXA group showed further enhanced antibacterial activity than 20 mg/kg OXA group, with CFU counts of 1.85×106 CFU per thigh vs. 8.79×106 CFU per thigh respectively (P > 0.05). As shown in Fig. 3C, MEM alone at 25 mg/kg was ineffective in this animal model, while the addition of d-Ser (4 mmol/kg) significantly enhanced the antibacterial activity (P < 0.001 vs. control or MEM alone). MEM alone at 50 mg/kg was effective in reducing MRSA counts, while addition of d-Ser (4 mmol/kg) showed a little further effect, with CFU counts of 2.86 ×105 CFU per thigh vs. 4.79×104 CFU per thigh respectively (P > 0.05). Notably, with addition of 4 mmol/kg d-Ser, 50 mg/kg MEM group showed no further enhanced antibacterial activity than 25 mg/kg MEM group (P > 0.05).
Figure 3.
Efficacy of OXA, MEM alone and in combination with d-Ser in murine neutropenic thigh infection model caused by MRSA N315. Infection doses: 4.0 × 105 CFU per thigh for panel A, 7.0 × 105 CFU per thigh for panel B and 6.3 × 105 CFU per thigh for panel C. Oneway-ANOVA test was used for statistical analysis. *P < 0.05, ***P < 0.001.
3.6. Effects of d-Ala and d-Glu on the sensitization effect of d-Ser
As shown in Supporting Information Table S3, in the absence of d-Ala or d-Glu, 20 mmol/L d-Ser significantly sensitized the antibacterial activity of OXA or MEM, with MICs decreased from 16- to >32-fold. However, with the adding of 20 mmol/L d-Ala in the medium, the sensitization effect of d-Ser almost disappeared, while adding of d-Glu at concentration as high as 80 mmol/L didn׳t show any obvious effect.
3.7. The docking results of dipeptides with PBP2a
As shown in Supporting Information Table S4, the LibDock_Score of d-Ala-d-Ala and d-Ala-d-Ser were 70.9502 and no poses respectively, hence we predicted that d-Ala-d-Ala had a good affinity with PBP2a, while d-Ala-d-Ser had no (or very weak) affinity with the same protein.
3.8. The sensitization activity of d-Ser with β-lactam antibiotics with relatively selective affinities for different PBPs
Cefaclor and cephradine are representative compounds which have relatively selective affinities for PBP3, while cefoxitin for PBP4. As shown in Supporting Information Table S5, the level of sensitization activity of d-Ser varied with β-lactam antibiotics. Addition of 40 mmol/L d-Ser could enhance the antibacterial activity of all three cephalosporins, with MICs reduced by 2–128 times. Notably, MICs of cefoxitin in combination with 40 mmol/L d-Ser were reduced to larger degrees than cefaclor and cephradine. The reduced folds of cefoxitin with addition of 40 mmol/L d-Ser were generally 8–16 times larger than cefaclor, and 4–8 times larger than cephradine.
4. Discussion
MRSA has the ability to cause a range of serious problems since its first emergence and it now still maintains a high infection rate worldwide. β-Lactam antibiotics, widely used in the treatment of infections caused by S. aureus, have a limited effect in treating infections caused by MRSA strains. This highlights the need of development of novel treatments. Research institutes, including our laboratory, have been searching for novel targets30, potential alternatives to antibiotics31, as well as effective combinations of antibiotics with agents such as manuka honey, isoliquiritigenin, plant essential oils and plant extracts24., 30., 31., 32., 33., 34..
d-Ser was reported to be able to increase the susceptibility of MRSA strain to methicillin by replacing d-Ala residue of the peptidoglycan stem peptides19. In order to determine the usability of d-Ser as sensitizer for β-lactams against MRSA strains, we evaluated the in vitro and in vivo antibacterial activity of d-Ser with different β-lactams (especially OXA and MEM) against different MRSA strains, including standard and clinical isolates. The MIC results demonstrated that d-Ser had sensitization effects with all tested β-lactams. Checkerboard assay with d-Ser/OXA or d-Ser/MEM combinations further confirmed the synergistic effects of the combinations in clinical MRSA isolates. The synergistic bactericidal activity of the combinations was then demonstrated by time–kill curve analysis. More importantly, the in vivo antibacterial activity of the combinations was manifested in murine systemic infection and neutropenic thigh infection models. Notably, in murine neutropenic thigh infection model, with addition of 4 mmol/kg d-Ser, the antimicrobial activity of OXA at 20/50 mg/kg and MEM at 25/50 mg/kg showed no significant difference. Considering the potential resistance development in future, the lowest effective concentration is recommended, and more precise data are needed for future preclinical as well as clinical studies.
Besides, d-Ser also had sensitization activity in combination with ceftaroline. Ceftaroline, currently used as treatment of critical S. aureus infections, received FDA approval several years ago, now facing challenges that non-susceptible S. aureus is on the rise worldwide35., 36.. We studied and found that addition of 40 mmol/L d-Ser could enhance the antibacterial activity of ceftaroline against MRSA strains, with the MICs reduced by 2–16 times (data not shown). The results suggested that d-Ser might have a promising application in future.
β-Lactams inhibit bacterial growth by competing with d-Ala-d-Ala termini to bind with active sites of PBPs and interfering with cell wall assembly37. Methicillin resistance of S. aureus is associated with PBP2a38, which can mediate cell wall assembly with low β-lactam affinity39. Our results demonstrated that the sensitization effect of d-Ser in MSSA (Supporting Information Table S2) was much weaker than in MRSA strains (Table 1), suggesting the possible involvement of PBP2a in d-Ser sensitization in MRSA.
It was reported that addition of d-Ser could replace d-Ala in d-Ala-d-Ala termini of the peptidoglycans, and result in sensitization of the bacteria to methicillin19. In consistent with this report, our study showed that the sensitization effect of d-Ser on OXA and MEM against MRSA could be abolished with addition of the same concentration of d-Ala, while addition of d-Glu with even higher concentration had no effect (Supporting Information Table S3). Herein, the sensitization mechanism of d-Ser on β-lactams against MRSA may be related to the replacement of d-Ala residue in the peptidoglycans and the formation of d-Ala-d-Ser. d-Ala-d-Ser may not be recognized by PBPs, especially PBP2a, and result in unnormal transpeptidation process as well as cell wall assembly, leading to increased sensitivity of MRSA to β-lactams. Results by Thorsing et al.40 also demonstrated that the exposure to thioridazine can lead to sensitization of MRSA strain to β-lactams through change of intracellular amino acids, which lead to unnormal peptidoglycan precursor formation. To verify the hypothesis that we made on PBP2a, docking study between protein and dipeptides (d-Ala-d-Ala or d-Ala-d-Ser) was conducted. The binding activity of d-Ala-d-Ala and d-Ala-d-Ser to PBP2a predicted by the dock module (Supporting Information Table S4) showed that compared with d-Ala-d-Ala, d-Ala-d-Ser had no detectable affinity to PBP2a.
PBP2a cannot accounts for the full mechanism of resistance present in MRSA strains, though it dose play an important role. Researches had been done that different β-lactam antibiotics had relatively selective affinities for PBPs41. Sieradzki et al.42 found that cephradine, a relatively selective inhibitor of PBP3, was effective in reducing methicillin resistance, indicating that PBP3 might participate in methicillin resistance through some cooperative functioning with PBP2a. However, recent researches through genome sequencing and molecular genetic studies revealed that PBP4 could mediate S. aureus resistance of β-lactam antibiotics43., 44.. Our results (Supporting Information Table S5) demonstrated that although d-Ser had sensitization activity with all three antibiotics, d-Ser in combination with cefoxitin, a relatively selective inhibitor of PBP4, had a more profound sensitization activity than relatively selective inhibitors of PBP3, i.e., cefaclor and cephradine. We hypothesize that several PBPs might participate in β-lactam resistance, while PBP4 might act primarily as an efficient transpeptidase in addition to PBP2a.
d-Ser is well known for its nephrotoxicity due to oxidative stress caused by hydrogen peroxide, a byproduct of d-amino acid oxidase (DAAO)-mediated metabolism of d-Ser45., 46.. However, in our initial safety experiment in mice, d-Ser alone and in combination with MEM or OXA didn׳t demonstrate obvious toxicity. Administration of d-Ser 10, 20, and 40 mmol/kg, MEM 100 mg/kg+d-Ser 4 mmol/kg or OXA 100 mg/kg+d-Ser 4 mmol/kg didn׳t cause any death in mice over 7 days. No significant body weight change was observed between treatment and control groups during the 7 days experiment period. And there was also no significant difference of urea nitrogen and creatinine in serum between the treatment and control groups at 24 and 48 h after dosing (Supporting Information Fig. S1). Co-administration of DAAO inhibitors with d-Ser was believed to be able to minimize its metabolism by DAAO, and hence to improve d-Ser bioavailability and reduce nephrotoxic effects. Indeed, oral administration of DAAO inhibitor CBIO in conjunction with d-Ser enhanced the plasma and brain levels of d-Ser in rats compared to the oral administration of d-Ser alone47. Hence, we expected that co-administration of DAAO inhibitors with d-Ser+antibiotic combinations can reduce the effective dose of d-Ser and the possible toxicity caused by oxidation of d-Ser.
5. Conclusions
In summary, bacterial resistance is an increasingly serious problem. While new antibiotic development is time-consuming, combination therapy of bioactive molecules with existing antibiotics is a good way to solve this conflict. Considering the great in vitro and in vivo activity as well as relatively good safety of OXA/d-Ser and MEM/d-Ser combinations against MRSA, d-Ser/β-lactam combinations may have the potential to be new treatment strategies against MRSA infections.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers 81621064 and 81361138020), CAMS Initiative for Innovative Medicine (Grant No. 2016-I2M-3-014, China), PUMC Youth Fund (grant number 3332013145, China) and National Mega-project for Innovative Drugs (Grant No. 2018ZX09721001, China). We are very grateful to Dr. Chung-Dar Lu from University of Massachusetts Lowell for valuable suggestions and helpful discussions during the study. We also thank Meng Wu from Chinese Academy of Medical Sciences & Peking Union Medical College (Beijing, China) for his technical assistance in dipeptides binding activity comparison.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.apsb.2019.01.017.
Contributor Information
Congran Li, Email: cong5885@aliyun.com.
Xuefu You, Email: xuefuyou@imb.pumc.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Talan D.A., Krishnadasan A., Gorwitz R.J., Fosheim G.E., Limbago B., Albrecht V. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis. 2011;53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 2.Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens R.M., Morrison M.A., Nadle J., Petit S., Gershman K., Ray S. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Dantes R., Mu Y., Belflower R., Aragon D., Dumyati G., Harrison L.H. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med. 2013;173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen D.B., Lessa F.C., Belflower R., Mu Y., Wise M., Nadle J. Invasive methicillin-resistant Staphylococcus aureus infections among patients on chronic dialysis in the United States, 2005–2011. Clin Infect Dis. 2013;57:1393–1400. doi: 10.1093/cid/cit546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhary A.S. A review of global initiatives to fight antibiotic resistance and recent antibiotics׳ discovery. Acta Pharm Sin B. 2016;6:552–556. doi: 10.1016/j.apsb.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cava F., de Pedro M.A., Lam H., Davis B.M., Waldor M.K. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cava F., Lam H., de Pedro M.A., Waldor M.K. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell Mol Life Sci. 2011;68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radkov A.D., Hsu Y.P., Booher G., VanNieuwenhze M.S. Imaging Bacterial Cell Wall Biosynthesis. Annu Rev Biochem. 2018;87:991–1014. doi: 10.1146/annurev-biochem-062917-012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caparrós M., Pisabarro A.G., de Pedro M.A. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam H., Oh D.C., Cava F., Takacs C.N., Clardy J., de Pedro M.A. d-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodkin-Gal I., Romero D., Cao S., Clardy J., Kolter R., Losick R. d-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochbaum A.I., Kolodkin-Gal I., Foulston L., Kolter R., Aizenberg J., Losick R. Inhibitory effects of d-amino acids on Staphylococcus aureus biofilm development. J Bacteriol. 2011;193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramón-Peréz M.L., Diaz-Cedillo F., Ibarra J.A., Torales-Cardeña A., Rodríguez-Martínez S., Jan-Roblero J. d-amino acids inhibit biofilm formation in Staphylococcus epidermidis strains from ocular infections. J Med Microbiol. 2014;63:1369–1376. doi: 10.1099/jmm.0.075796-0. [DOI] [PubMed] [Google Scholar]
- 15.Yang H., Wang M., Yu J., Wei H. Aspartate inhibits Staphylococcus aureus biofilm formation. FEMS Microbiol Lett. 2015;362:1–7. doi: 10.1093/femsle/fnv025. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez C.J., Jr, Akers K.S., Romano D.R., Woodbury R.L., Hardy S.K., Murray C.K. d-amino acids enhance the activity of antimicrobials against biofilms of clinical wound isolates of Staphylococcus aureus and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58:4353–4361. doi: 10.1128/AAC.02468-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.She P., Chen L., Liu H., Zou Y., Luo Z., Koronfel A. The effects of d-tyrosine combined with amikacin on the biofilms of Pseudomonas aeruginosa. Microb Pathog. 2015;86:38–44. doi: 10.1016/j.micpath.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Tong Z., Zhang L., Ling J., Jian Y., Huang L., Deng D. An in vitro study on the effect of free amino acids alone or in combination with nisin on biofilms as well as on planktonic bacteria of Streptococcus mutans. PLoS One. 2014;9:e99513. doi: 10.1371/journal.pone.0099513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Jonge B.L., Gage D., Xu N. The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:3151–3155. doi: 10.1128/AAC.46.10.3151-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koreen L., Ramaswamy S.V., Graviss E.A., Naidich S., Musser J.M., Kreiswirth B.N. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.〈https://clsi.org/〉. Clinical and Laboratory Standards Institute. Performance Standard for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA, 2015.
- 23.Verma P. Methods for determining bactericidal activity and antimicrobial interactions synergy testing, time–kill curves, and population analysis. In: Schwalbe R., Steele-Moore L., Goodwin A.C., editors. Antimicrobial Susceptibility Testing Protoccols. CRC Press; Boca Raton: 2007. pp. 275–298. [Google Scholar]
- 24.Lu X., Yang X., Li X., Lu Y., Ren Z., Zhao L. In vitro activity of sodium new houttuyfonate alone and in combination with oxacillin or netilmicin against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8:e68053. doi: 10.1371/journal.pone.0068053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuluaga A.F., Salazar B.E., Rodriguez C.A., Zapata A.X., Agudelo M., Vesga O. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis. 2006;6:55. doi: 10.1186/1471-2334-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labrou M., Michail G., Ntokou E., Pittaras T.E., Pournaras S., Tsakris A. Activity of oxacillin versus that of vancomycin against oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates evaluated by population analyses, time–kill assays, and a murine thigh infection model. Antimicrob Agents Chemother. 2012;56:3388–3391. doi: 10.1128/AAC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dandekar P.K., Tessier P.R., Williams P., Nightingale C.H., Nicolau D.P. Pharmacodynamic profile of daptomycin against Enterococcus species and methicillin-resistant Staphylococcus aureus in a murine thigh infection model. J Antimicrob Chemother. 2003;52:405–411. doi: 10.1093/jac/dkg337. [DOI] [PubMed] [Google Scholar]
- 28.Sugihara K., Sugihara C., Matsushita Y., Yamamura N., Uemori M., Tokumitsu A. In vivo pharmacodynamic activity of tomopenem (formerly CS-023) against Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus in a murine thigh infection model. Antimicrob Agents Chemother. 2010;54:5298–5302. doi: 10.1128/AAC.00267-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janardhanan J., Meisel J.E., Ding D., Schroeder V.A., Wolter W.R., Mobashery S. In vitro and in vivo synergy of the oxadiazole class of antibacterials with β-lactams. Antimicrob Agents Chemother. 2016;60:5581–5588. doi: 10.1128/AAC.00787-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W., Liu C., Zhu N., Lin Y., Jiang J., Wang Y. Identification of anti-Gram-negative bacteria agents targeting the interaction between ribosomal proteins L12 and L10. Acta Pharm Sin B. 2018;8:772–783. doi: 10.1016/j.apsb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2015;5:310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins R.E., Cooper R. Synergy between oxacillin and manuka honey sensitizes methicillin-resistant Staphylococcus aureus to oxacillin. J Antimicrob Chemother. 2012;67:1405–1407. doi: 10.1093/jac/dks071. [DOI] [PubMed] [Google Scholar]
- 33.Gaur R., Gupta V.K., Singh P., Pal A., Darokar M.P., Bhakuni R.S. Drug resistance reversal potential of isoliquiritigenin and liquiritigenin isolated from Glycyrrhiza glabra against methicillin-resistant Staphylococcus aureus (MRSA) Phytother Res. 2016;30:1708–1715. doi: 10.1002/ptr.5677. [DOI] [PubMed] [Google Scholar]
- 34.Lahmar A., Bedoui A., Mokdad-Bzeouich I., Dhaouifi Z., Kalboussi Z., Cheraif I. Reversal of resistance in bacteria underlies synergistic effect of essential oils with conventional antibiotics. Microb Pathog. 2017;106:50–59. doi: 10.1016/j.micpath.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Biedenbach D.J., Alm R.A., Lahiri S.D., Reiszner E., Hoban D.J., Sahm D.F. In vitro activity of ceftaroline against Staphylococcus aureus isolated in 2012 from Asia-Pacific countries as part of the AWARE surveillance program. Antimicrob Agents Chemother. 2015;60:343–347. doi: 10.1128/AAC.01867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biedenbach D.J., Hoban D.J., Reiszner E., Lahiri S.D., Alm R.A., Sahm D.F. In vitro activity of ceftaroline against Staphylococcus aureus isolates collected in 2012 from Latin American countries as part of the AWARE surveillance program. Antimicrob Agents Chemother. 2015;59:7873–7877. doi: 10.1128/AAC.01833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guignard B., Entenza J.M., Moreillon P. β-Lactams against methicillin-resistant Staphylococcus aureus. Curr Opin Pharmacol. 2005;5:479–489. doi: 10.1016/j.coph.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Katayama Y., Ito T., Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers H.F., Hartman B.J., Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985;76:325–331. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorsing M., Klitgaard J.K., Atilano M.L., Skov M.N., Kolmos H.J., Filipe S.R. Thioridazine induces major changes in global gene expression and cell wall composition in methicillin-resistant Staphylococcus aureus USA300. PLoS One. 2013;8:e64518. doi: 10.1371/journal.pone.0064518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers H.F., Sachdeva M. Binding of β-lactam antibiotics to penicillin-binding proteins in methicillin-resistant Staphylococcus aureus. J Infect Dis. 1990;161:1170–1176. doi: 10.1093/infdis/161.6.1170. [DOI] [PubMed] [Google Scholar]
- 42.Sieradzki K., Tomasz A. Suppression of β-lactam antibiotic resistance in a methicillin-resistant Staphylococcus aureus through synergic action of early cell wall inhibitors and some other antibiotics. J Antimicrob Chemother. 1997;39 Suppl A:47–51. doi: 10.1093/jac/39.suppl_1.47. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton S.M., Alexander J.A.N., Choo E.J., Basuino L., da Costa T.M., Severin A. High-level resistance of Staphylococcus aureus to β-lactam antibiotics mediated by penicillin-binding protein 4 (PBP4) Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02727-16. :e02727-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee S.S., Chen L., Gilbert A., da Costa T.M., Nair V., Datta S.K. PBP4 mediates β-lactam resistance by altered function. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00932-17. :e00932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams R.E., Lock E.A. Sodium benzoate attenuates d-serine induced nephrotoxicity in the rat. Toxicology. 2005;207:35–48. doi: 10.1016/j.tox.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Chung S.P., Sogabe K., Park H.K., Song Y., Ono K., Abou El-Magd R.M. Potential cytotoxic effect of hydroxypyruvate produced from d-serine by astroglial d-amino acid oxidase. J Biochem. 2010;148:743–753. doi: 10.1093/jb/mvq112. [DOI] [PubMed] [Google Scholar]
- 47.Ferraris D., Duvall B., Ko Y.S., Thomas A.G., Rojas C., Majer P. Synthesis and biological evaluation of d-amino acid oxidase inhibitors. J Med Chem. 2008;51:3357–3359. doi: 10.1021/jm800200u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material