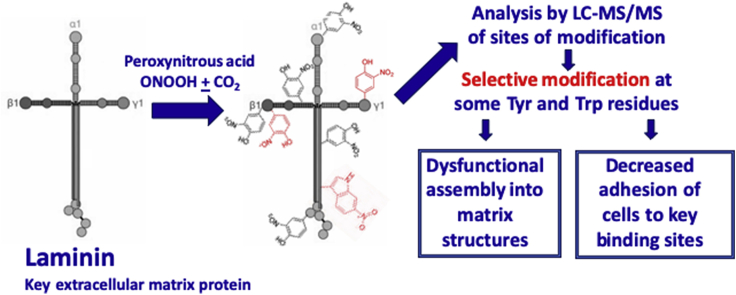
Abstract
Laminin is a major protein of the basement membrane (BM), a specialized extracellular matrix (ECM) of the artery wall. The potent oxidizing and nitrating agent peroxynitrous acid (ONOOH) is formed at sites of inflammation, and data implicate ONOOH in ECM damage and cardiovascular disease. Co-localization of 3-nitrotyrosine, a product of ONOOH-mediated tyrosine (Tyr) modification, and laminin has been reported in human atherosclerotic lesions. The sites and consequences of 3-nitrotyrosine (and related nitrated tryptophan) formation on laminin, it's self-assembly and cell interactions are poorly understood. In this study murine laminin-111 was exposed to ONOOH (1–500-fold molar excess). Nitration sites were mapped and quantified using LC-MS/MS. Mono-nitration was detected at 148 sites (126 Tyr, 22 Trp), and di-nitration at 14 sites. Label-free quantification showed enhanced nitration with increasing oxidant doses. Tyr nitration was ∼10-fold greater than at Trp. CO2 modulated damage in a site-specific manner, with most sites less extensively nitrated. 119 mono-nitration sites were identified with CO2 present, and no unique sites were detected. 23 di-nitration sites were detected, with 15 unique to the presence of CO2. Extensive modification was detected at sites involved in cell adhesion, protein-protein interactions and self-polymerization. Tyr-145 on the γ1 chain was extensively nitrated, and endothelial cells exhibited decreased adhesion to a nitrated peptide modelling this site. Modification of residues involved in self-polymerization interfered with the formation of ordered polymers as detected by scanning electron microscopy. These laminin modifications may contribute to endothelial cell dysfunction and modulate ECM structure and assembly, and thereby contribute to atherogenesis.
Keywords: Extracellular matrix, Peroxynitrous acid, Peroxynitrite, Laminin, Protein oxidation, 3-Nitrotyrosine
Abbreviations: BME, basement membrane extracts; ECM, extracellular matrix; GAG, glycosaminoglycan; LC-MS, liquid chromatography-mass spectrometry; MS, mass spectrometry; 6-nitroTrp, 6-nitrotryptophan; 3-nitroTyr, 3-nitrotyrosine; ONOOH, the physiological mixture of peroxynitrous acid and its anion peroxynitrite; ONOOCO2−, the nitrosoperoxycarbonate anion formed by addition of CO2 to peroxynitrite; PBS, phosphate-buffered saline; RSO, relative site occupancy; SDC, sodium deoxycholate; TCA, trichloroacetic acid; TFA, trifluoroacetic acid
Graphical abstract
Highlights
-
•
Laminin is a major extracellular matrix protein of the artery wall.
-
•
Peroxynitrous acid exposure gives nitration of tyrosine and tryptophan residues.
-
•
CO2 both increases and decreases damage depending of the reaction site.
-
•
LC-MS/MS used to map modifications to protein structure and functional domains.
-
•
Sites for cell adhesion, protein interactions and self-polymerization are modified.
1. Introduction
Peroxynitrous acid (ONOOH) is a powerful oxidizing and nitrating agent formed under inflammatory conditions by the diffusion-controlled reaction (with rate constant, k2, 6.7 × 109 M−1 s−1 [1]) of nitric oxide (.NO) and superoxide (O2.-) radicals (reviewed [2]). Formation of ONOOH takes place at low (pico to nano-molar) levels under normal physiological conditions, but higher levels are produced at sites of inflammation, where fluxes can reach high nanomolar to low micromolar concentrations [2,3]. A major source of ONOOH during inflammation is activated macrophages, as these cells have the capacity to produce significant levels of .NO and O2.-, due to the high activity of inducible nitric oxide synthase and NOx isoforms in the activated forms of these cells [2,3]. ONOOH is more reactive than .NO and O2.- and can induce both direct damage, via two-electron oxidation reactions (e.g. with metal-ion centres, thiols and thioethers, with rate constants, k, in the range 103 - 106 M−1 s−1), and by one-electron reactions of radicals (nitrogen dioxide, .NO2, and hydroxyl radicals, HO.) arising from limited homolysis of the -O-O- bond [2,4,5]. The peroxynitrite anion (ONOO−, pKa ∼6.8) reacts with CO2 to give the short-lived nitrosoperoxocarbonate anion (ONOOCO2−) [6,7], which may undergo homolysis to give carbonate radical anions (CO3.-) and nitrogen dioxide radical (.NO2) [8,9].
High ONOOH fluxes are likely to be present extracellularly, as macrophage-derived .NO and O2.-, and hence ONOOH, are primarily released to the extracellular environment [2,3] and ONOOH displays limited diffusion [2,3]. Due to limited nature and extent of extracellular antioxidant defenses [10], extracellular matrix (ECM) macromolecules are therefore likely to be major targets of ONOOH. Furthermore, as many ECM molecules have long half-lives (cf. an estimated half-life of 74 years for elastin [11]) and there are limited types and activities of extracellular repair mechanisms [10], modifications generated by oxidants on ECM species are likely to be both long-lived and accumulate over time, with this potentially accelerated by disease [[12], [13], [14]]. ECM materials are therefore likely to be modified by ONOOH in diseases associated with recurrent or ongoing inflammation [12,13]. This hypothesis is supported by studies that have provided evidence for the presence of such damage on ECM proteins and proteoglycans in biological samples, including human atherosclerotic lesions [[15], [16], [17], [18], [19], [20], [21]].
The ECM is composed of a meshwork of proteins that provides the three-dimensional architectural scaffold that defines tissue boundaries and biomechanical properties. Besides providing mechanical strength and elasticity for tissues, the ECM also acts as a scaffold for cell adhesion, controls cell migration and proliferation, and plays an important role in binding and modifying the activity of growth factors, cytokines and other signaling molecules [[22], [23], [24], [25]]. Thus, alterations to this highly organized network are likely to modulate both cell behavior and tissue integrity. A number of studies have demonstrated that ONOOH can induce structural and/or functional changes to ECM proteins, particularly via the detection of the long-lived product 3-nitrotyrosine (3-nitroTyr) which is formed via reaction of ONOOH with Tyr residues present on ECM proteins [[17], [18], [19], [20], [21],26,27]. 3-nitroTyr has been shown to co-localize with ECM proteins in biological samples including human atherosclerotic lesions [[18], [19], [20], [21]]. The basement membrane (BM) is a specialized ECM structure that underlies endothelial cells in the artery wall, and prevents smooth muscle cell infiltration in to the intimal space. The major protein component of BM is laminin, which plays a crucial role in the architecture of BMs [14,23]. Laminin is a heterotrimeric protein composed of one α, one β and one γ chain, each of which exists in several different isoforms (5 α, 3 or possibly 4 β, and 3 γ) that combine to form at least 16 laminin isoforms with distinct tissue distribution and/or function [22,24]. Laminins have the propensity to polymerize at physiological temperatures in the presence of Ca2+, through interactions between the N-termini of the different chains into polymeric sheets [24,28]. The C-terminus of the α-chain binds to integrins [29,30], α-dystroglycan, heparan sulfates and sulfated glycolipids on cell surfaces, and binding sites present on the laminin chains for agrin and nidogen, function as a link between type-IV collagen networks and other ECM molecules [31]. Alterations to this highly-organized network are likely to modulate both cell behavior, and tissue integrity.
In a previous study, we have shown that laminin-111 (i.e. laminin containing α1,β1, and γ1 chains) is a target for ONOOH in vitro, with the formation of both 3-nitroTyr and 6-nitroTrp detected even at low levels of ONOOH [18]. Furthermore, antibodies that recognize the laminin γ1 chain co-localize with those that recognize 3-nitroTyr in advanced human atherosclerotic lesions [18]. However, the sites of nitration on the laminin chains and the functional consequences of modifications at particular sites are unknown. In this study we have used LC-MS/MS to identify sites of nitration at Tyr and Trp residues, with label-free quantification used to assess the extent of nitration at the individual nitration sites. The effect of these modifications on binding of cells to laminin, and the self-assembly of the laminin in to 3-dimensional native structures have also been examined, and shown to be significantly perturbed.
2. Materials and methods
2.1. Materials
All chemicals were purchased from Sigma-Aldrich unless stated otherwise. MS-grade water, acetonitrile, DMSO and formic acid for LC-MS/MS was obtained from VWR (Søborg, Denmark). Murine laminin-111 (L2020) was purchased from Sigma Aldrich (Brøndby, Denmark). Protein concentrations were based on the molecular weight of laminin-111 heterotrimer (722 kDa, estimated from the amino acid composition as given in the Uniprot entries of the 3 side chains, i.e. P19137 (α1), P02469 (β1), and P02468 (γ1). Sequencing-grade trypsin (V5111) was purchased from Promega (Denmark).
2.2. ONOOH synthesis and protein nitration
ONOOH was synthesized in a two-phase system using isoamyl nitrite and H2O2. Unreacted H2O2 was eliminated using MnO2 [32]. The resulting stocks were stored at −80 °C and used immediately after defrosting; unused material was discarded. The concentration of ONOOH in stock solutions was determined spectrophotometrically prior to every experiment using λ302 nm 1705 M−1 s−1 [33]. Stock solutions of ONOOH were diluted in 0.1 M NaOH before use, and then small volumes of this stock were added to strongly buffered reaction mixtures (100 mM phosphate buffer, pH 7.4 final concentration) to minimize artifacts arising from pH changes.
2.3. Protein quantification using the bicinchoninic acid assay (BCA)
Protein concentrations were determined using the BCA assay (Pierce BCA Protein Assay Kit; ThermoFisher Scientific, Hvidovre, Denmark) in a 96-well plate format. The assay mixture contained 200 μL of the BCA reagent (solution A + B) and 20 μL of protein sample or BSA standard. After 30 min incubation at 37 °C, the absorbance was read at 562 nm using a SpectraMax i3 plate reader.
2.4. Sample preparation for LC-MS/MS
Protein samples (ONOOH-treated laminin and controls without treatment) were treated with 0.2% deoxycholic acid (DOC), 10 mM tris-(2-carboxyethyl)phosphine (TCEP, 40 mM) and 2-chloroacetamide (CAA) and incubated for 10 min at 95 °C with shaking, to simultaneously denature, reduce and alkylate proteins. Sample were then loaded onto 10 kDa cut-off filters for filter-assisted sample preparation (FASP), which was carried out as described previously [34,35]. The proteins were washed with 100 μL 0.2% DOC in 100 mM NH4HCO3 and centrifuged, then deglycosylated using PNGase F (2 U μg−1 protein, P0407S, New England Biolabs-BioNordika, Herlev, Denmark) in 100 mM NH4HCO3 + 0.2% DOC over a period of 1 h at 37 °C with mild shaking. The samples were then centrifuged, washed twice with 0.2% DOC in NH4HCO3 and re-centrifuged. Proteins were then digested with trypsin in a two-step sequential protocol. First, trypsin was added in ratio 1:100 (enzyme:protein) in 100 mM NH4HCO3 + 0.2% DOC, followed by microwaving for 40 s at 600 W in a household microwave oven. Samples were placed in a water bath to avoid excess heating. A second aliquot of trypsin was then added, in ratio 1:50, and the samples were incubated in a humid chamber for 16 h at 37 °C with mild shaking. After digestion, the resulting peptides were collected by centrifugation. Formic acid was then added to a final concentration of 1% to precipitate DOC, followed by centrifugation at 18,000 g for 15 min. The supernatant was collected, and the centrifugation step repeated twice to remove residual detergent.
2.5. LC-MS/MS analysis
Tryptic peptides were analyzed using a Dionex 3000RS LC system (Thermo Fisher Scientific) coupled on-line to a Bruker Impact II QTOF mass spectrometer equipped with an Apollo II ion source (Bruker Daltonics, Solna, Sweden). The peptides (80 μg) were loaded onto an Aeris Peptide column (2.6 μm, xb-C18, 250 × 2.1 mm, Phenomex, Værløse, Denmark), and separated by reverse-phase chromatography with a multi-step gradient of 5–45% buffer B (80% acetonitrile (ACN), 3% DMSO and 0.1% formic acid (FA)) mixed with buffer A (3% DMSO and 0.1% FA) at a flow rate of 200 μL min−1 over 120 min, followed by washing and re-equilibration steps (total run time: 150 min). LC-MS/MS data were acquired using a data-dependent method. A high resolution TOF-MS scan over a mass range of 150–2200 m/z, was followed by MS/MS scans of top 20 most intense precursor ions per cycle. Dynamic exclusion of precursor ions was set to 30 s duration after acquisition of 3 spectra, with a window of 0.05 Th. Acquisition rate was set to 4 Hz (MS) and 2–16 Hz (MS/MS, adjusted by precursor ion intensity).
2.6. Bioinformatics
LC-MS/MS data were processed with Data Analysis (Version 4.3, Bruker Daltonics). Peak lists obtained were identified using X!Tandem (Vengeance, 2015.12.15.2) [36] and MS-GF+ (v. 10282) [37]. The search was conducted using SearchGUI (v. 3.2.20) [38]. Protein identification was performed using a concatenated target/decoy version of the Mus musculus (97.5%, 16785 sequences) SwissProt database (downloaded 21-9-2016)). Decoy sequences were created by reversing target sequences. The identification settings were as follows: Trypsin (no P rule), with a maximum of 2 missed cleavages; 0.07 Da as MS1 and 40.0 ppm as MS2 tolerances; fixed modifications: carbamidomethylation of C (+57.021464 Da), variable modifications: di-nitration of Y/W (+89.970156 Da), nitration of Y/W (+44.985078 Da), oxidation of M (+15.994915 Da), deamidation of N (+0.984016 Da). Additionally, acetylation of protein N-terminii (+42.010565 Da), pyrrolidone formation from E (−18.010565 Da), and pyrrolidone formation from Q/C (−17.026549 Da) were considered during refinement. All algorithm specific settings are listed in the Supplementary Information. Peptides and proteins were inferred from the spectrum identification results using PeptideShaker version 1.16.16 [39]. Peptide Spectrum Matches (PSMs), peptides and proteins were validated at a 1.0% False Discovery Rate (FDR) estimated using the decoy hit distribution. All validation thresholds are listed in the Supplementary Information. Post-translational modification localizations were scored using the D-score [40] and the phosphoRS score [41]. All spectra matching modified peptides were validated manually. MSMS spectra for all the nitrated species are provided in the Supplementary Data.
2.7. Feature detection and label-free quantification
Skyline software (Skyline-daily 3.6.1.10556) [42] was used for feature detection and relative MS1-based label-free quantification of peptides. Initially, a spectral library was built in Skyline with all peptide-spectrum matches assigned by PeptideShaker at a cut-off score of 0.95. Peptide filters were set to tryptic (no P rule) laminin-111 peptides with a maximum of 2 missed cleavage and 6–45 amino acids in length. MS1 ion chromatograms were extracted from raw (.d) files with high specificity within a 1 min window of a PSM and resolution set to 30.000. Extracted chromatograms were inspected and adjusted manually. For multiply-charged peptides with multiple charge states, only the most intense precursor ion was analyzed.
2.8. Scanning electron microscopy
Samples of native or modified laminin-111 were fixed in 2% glutaraldehyde in 0.05 M sodium phosphate buffer, pH 7.4. Following 3 rinses in 0.15 M sodium phosphate buffer (pH 7.4) specimens were post fixed in 1% OsO4 in 0.12 M sodium cacodylate buffer (pH 7.4) for 2 h. Following a rinse in distilled water, the specimens were dehydrated to 100% ethanol and critical point dried (Balzers CPD 030, Halmstad, Sweden) using CO2. The specimens were subsequently mounted on stubs using double adhesive carbon tape as an adhesive and sputter-coated with 6 nm gold (Leica ACE 200, Søborg, Denmark). Specimens were examined with a FEI Quanta 3D scanning electron microscope (Thermo Fisher) operated at an accelerating voltage of 2 kV.
2.9. Cell adhesion assay
Synthetic C16 peptide (KAFDITYVRLKF, synthesized by EZBiolab, Carmel, IN, USA) used for the cell adhesion assay was incubated with tetranitromethane (TNM) to give the peptide nitrated at the sole Tyr residue in high yields with minimal site products. TNM was added to the peptide solution at 10 and 100-fold molar excess over the peptide concentration, and incubated at 21 °C with shaking for 1 h, followed by purification using solid-phase extraction on a LiChrolut RP-18 cartridge (Merck, Darmstadt, Germany). Eluted peptides were dried down and stored at −80 °C until analysis or use. Tissue culture plates (96-wells) were coated with 2.5 μg peptide in 50 μL PBS (pH 7.2) overnight at 4 °C. Plates were washed twice with PBS to remove unbound peptides. In order to minimize non-specific binding of cells to the plastic surface, plates were blocked with 3% (w/v) BSA in PBS for 1 h at 37 °C. Commercial human coronary artery endothelial cells (HCAEC) obtained from Cell Applications (San Diego, CA, USA) were grown in MesoEndo Endothelial Cell Growth Media (Cell Applications). HCAEC (passage 4) were detached from plates using 0.1% (w/v) trypsin-EDTA for 1 min at 37 °C before deactivation of trypsin with growth media. Cells were pelleted by centrifugation (5 min at ∼232 g), aspirated and washed once with serum-free medium before resuspension in media containing 5 μM Calcein-AM, followed by incubation at 37 °C for 30 min. Cells were then resuspended in medium to 2.5 × 105 cells mL−1, before seeding 50 μL of the cell suspension on to native or modified peptides. After allowing cells to adhere for 90 min, non-adherent cells were removed by washing gently with PBS (containing 1 mM CaCl2 and Mg2+). Adherent cells were then lysed by incubation with 1% Triton X-100 for 30 min. Calcein-AM fluorescence in the lysate was then measured in a SpectraMax i3 plate reader with λex 494 nm and λem 517 nm.
2.10. Data processing and statistics
All data processing, visualization and statistical tests were performed with the R scripting and statistical environment R (R version 3.5.1 (2018-07-02)), using ggplot (version 3.0.0) for data visualization. Occupancy of nitration sites was calculated from peak areas of extracted MS1 chromatograms obtained from Skyline (see above). For each nitration site identified, peak areas of peptides carrying nitration at this site were summed and given as a percentage of the total (summed) peak areas of all peptides (modified and unmodified) covering the site. Occupancy of di-nitration sites was calculated separately.
Agglomertive hierarchical cluster analysis and principal component analysis was performed by using the factoextra package (version 1.0.5) with occupancies calculated as described above. The distance matrix was computed using the Manhattan method, and Ward's method was used for hierarchical clustering. Number of clusters was determined by Monte Carlo bootstrapping with 100 samples.
Normality of the distribution of occupancies was assessed using a Shapiro-Wilks test. Differences in occupancy with or without bicarbonate was tested with Student's t-test with the Benjamini-Hochberg correction for multiple hypothesis testing with a cutoff false discovery rate (FDR) value of 5%. For cell adhesion and polymerization assays, statistical significance was tested using one-way ANOVA with post hoc analysis using Dunnett's multiple comparison test. P < 0.05 was taken as significant.
3. Results
3.1. Screening of nitration sites in laminin generated by high excesses of ONOOH
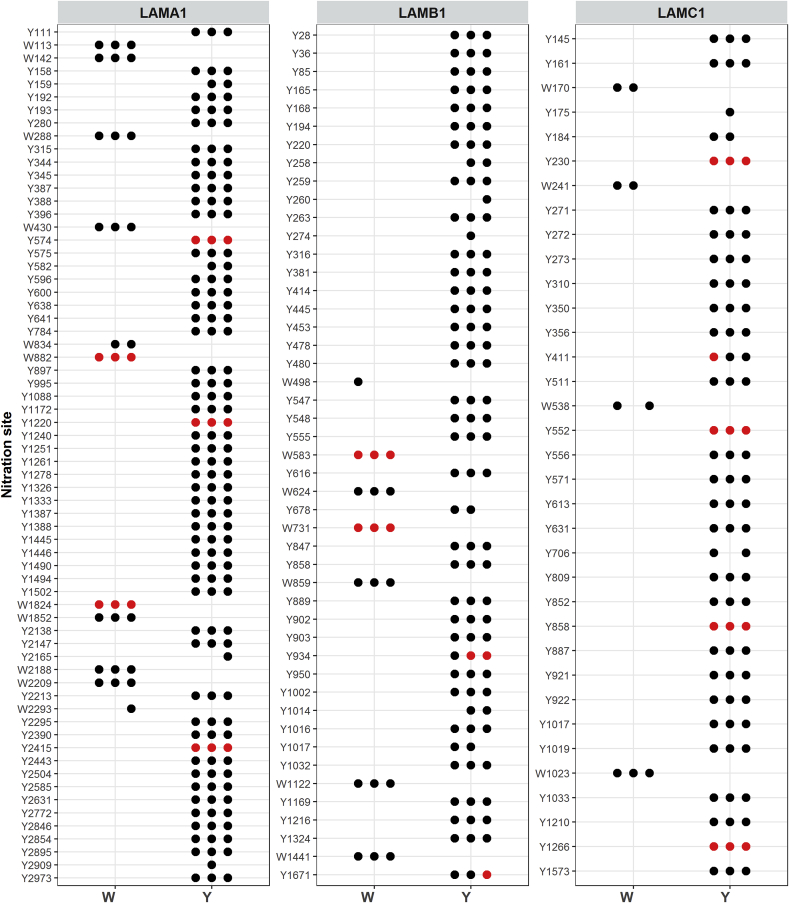
Initial studies were carried out with high concentrations of ONOOH to establish a suitable protocol to identify sites of nitration on Tyr and Trp and to establish modifications that could be used in subsequent targeted analyses (see workflow in Fig. 1). Thus, isolated murine laminin-111 (1.2 μM) was exposed to a 500-fold molar excess of ONOOH (in 0.1 M phosphate buffer, pH 7.4), purified and digested using a filter-assisted sample preparation (FASP) protocol, with the resulting peptides analyzed by LC-MS/MS. Modifications were identified using a database search, with mono- and di-nitration of Tyr and Trp residues, and oxidation of Met, included as variable modifications. The percentage coverage of the untreated and modified laminin-111 was ∼80% for all three chains (Supplementary Fig. 1). Peptide spectral matches (PSMs) of modified peptides were validated manually, and only peptides where modifications could be unambiguously localized were considered. With these constraints, 148 sites involving Tyr and Trp residues were identified (Fig. 2, black symbols). No nitration was detected in any of 3 control replicates. In addition to mono-nitration, 14 sites of di-nitration were also identified (Fig. 2; red symbols), with these including di-nitration at Trp residues. This is, to our knowledge, the first observation of Trp di-nitration on proteins, and indicates extensive damage at these sites. Oxidation was also detected at some Met residues, but this was not investigated in detail; a number of these Met modifications, as well as amidation arising from the use of deglycosylation enzymes, were also detected in the control samples (data not shown).
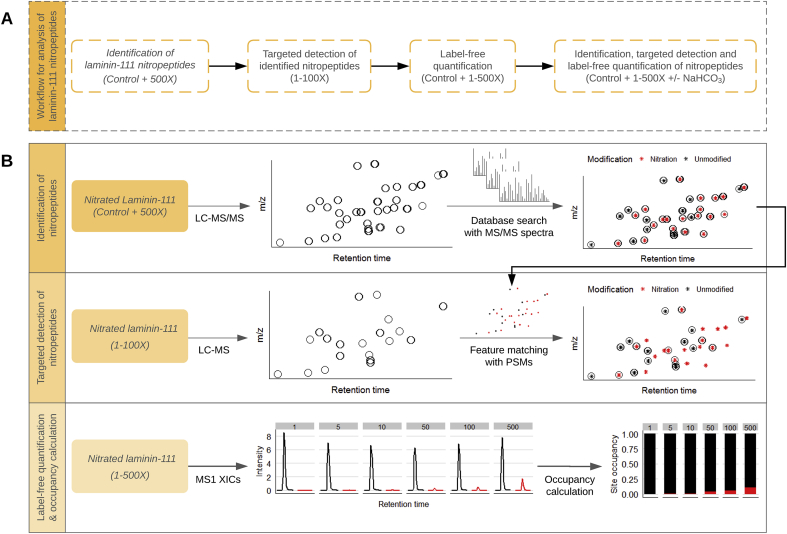
Fig. 1.
Workflow for the detection of nitration sites in laminin-111 using initial non-targeted analysis with high concentrations of ONOOH (panel A), and subsequent targeted analysis (panel B) based on the modifications detected with high levels of ONOOH.
Fig. 2.
Identification of laminin-111 nitration sites at Trp (W) and Tyr (Y) after exposure to ONOOH. Isolated laminin-111 was exposed to ONOOH in 500 times molar excess for 20 min at 37 °C, and resulting modifications were identified by LC-MS/MS. Dots indicate positive identification of the given modification at the given site in a biological replicate. Colors indicate identification of nitration (black) or di-nitration (red), respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Nitration was identified at both Tyr (126) and Trp (22) residues (Fig. 2). This corresponds to modification at 63% of the 199 Tyr residues and 44% of 50 Trp residues present in laminin-111 (using data from Uniprot entries P19137 (α1), P02469 (β1), and P02468 (γ1)). These data indicate that Tyr is the predominant target for aromatic nitration in laminin by ONOOH, though significant modification also occurs at Trp. Of the 14 sites detected with di-nitration, 10 are on Tyr and 4 on Trp, suggesting that some Tyr and Trp residues are subject to extensive modification. Limited nitration was also identified on nidogen-1, another ECM component with a high-binding affinity for laminin [43]; this was detected as a persistent, but minor, contaminant in the preparations of laminin employed. With the high fold molar excesses of ONOOH employed, 7 nitration sites, which are all at Tyr residues, were identified on this protein (Supplementary Fig. 2).
3.2. Site-specific detection of laminin nitration with low molar excesses of ONOOH
Subsequent to the above screening study to identify potential sites of modification, further studies were carried out using lower molar excesses of ONOOH (equimolar to 100-fold), with the data obtained from the experiments using the 500-fold excess used to perform a targeted search for modifications (see Fig. 1). Whilst missing or alternative fragmentation pathways for peptides is often a limiting factor in the identification of low-abundant peptides using search engines, the defined and relatively simple nature of these samples, highly consistent chromatography, and the use of a QTOF spectrometer, minimized this problem by allowing use of a targeted detection approach to detect any modified peptides (Supplementary Fig. 3) [44]. This approach allowed detection of nitrated peptides, identified in the highly modified samples (see above), at low excesses of ONOOH with high sensitivity.
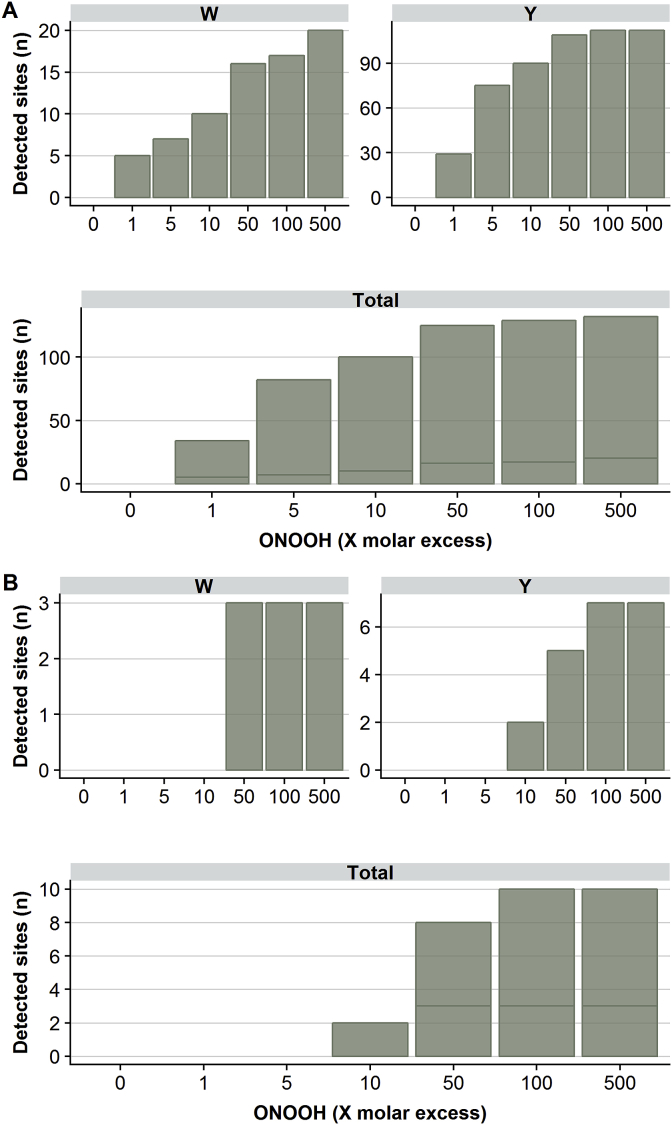
With equimolar concentrations of laminin-111 and ONOOH, 34 nitration sites were detected (Fig. 3A). At this level of oxidant, no nitration sites could be identified by a standard, non-directed database search using MS/MS spectra (i.e. without use of the database of modifications identified with the 500-fold molar excess of ONOOH). An increasing number of nitration sites were detected in response to increasing concentrations of ONOOH, with the majority of the total number of identified nitration sites detected in samples treated with ONOOH at a 50-fold molar excess or greater. In contrast to detection of Tyr sites, which reached a near plateau at this molar excess of ONOOH, the detection of nitration at Trp sites increased in a near linear manner with ONOOH concentration. Di-nitration of both Tyr and Trp was observed mainly at higher levels of ONOOH, (50–500 fold excesses; Fig. 3B), although di-nitration was detected at 2 sites with a 10-fold molar excess of ONOOH. These data indicate that high oxidant levels are usually required for di-nitration, though this is not universally true, and that both Tyr and Trp side chains are susceptible to di-nitration.
Fig. 3.
Number of sites of Trp (W) and Tyr (Y) modification (vertical axes) detected on laminin-111 after exposure to ONOOH. Isolated laminin-111 was exposed to ONOOH in concentrations ranging from equimolar to 500-fold molar excess (as indicated on the horizontal axes) for 20 min at 37 °C, with the resulting modified peptides detected by LC-MS using chromatographic and precursor mass features from the 500-fold molar excess samples used as a template. Panel A): Number of nitration sites detected by LC-MS at different concentrations of ONOOH at Trp (upper left panel), Tyr (upper right) and all residues (lower panel). Panel B): Number of di-nitration sites detected by LC-MS at different concentrations of ONOOH at Trp (upper left panel), Tyr (upper right panel) and all residues (lower panel). Only modifications detected in all three independent replicates from ONOOH treatment, and none of the control samples are included; statistical analysis is therefore not appropriate.
3.3. Quantification of Tyr and Trp nitration sites in laminin treated with ONOOH
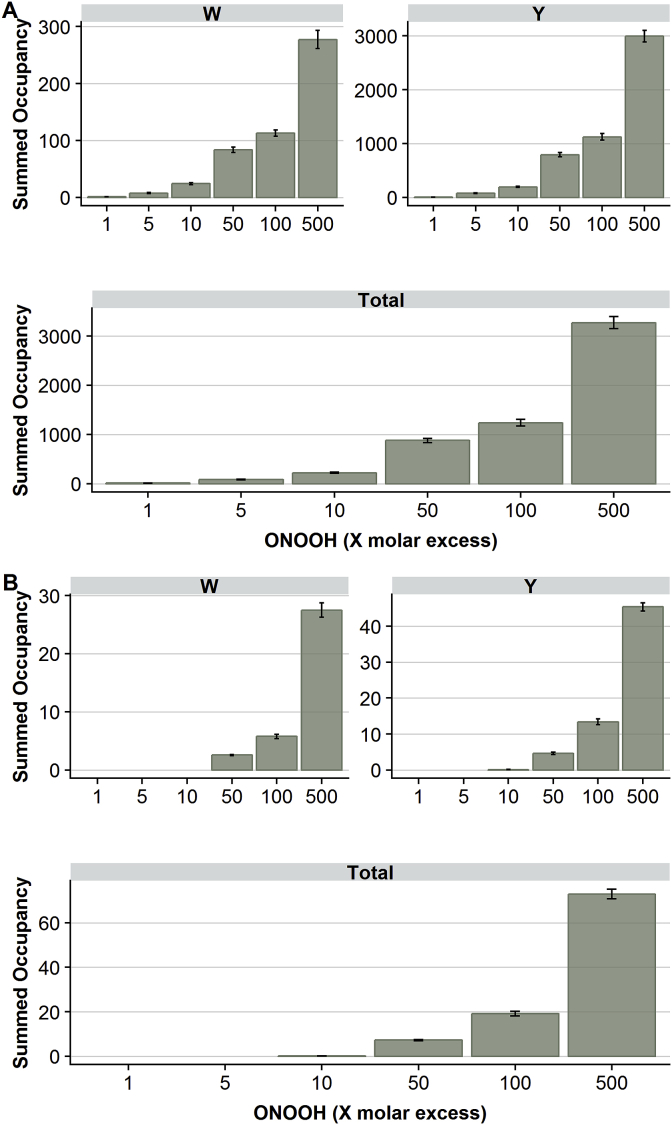
Label-free relative quantification of nitrated peptides, and corresponding unmodified species, was performed based on integrated MS1 peak areas using the Skyline software. All MS1 peaks with retention time and mass matching that of a manually validated PSM were considered for quantification. To assess the extent of nitration at an individual site, relative site occupancy (RSO) values were calculated. The overall extent of nitration was estimated by summing the occupancy of all nitration sites at each molar excess of ONOOH (Fig. 4). A dose-dependent increase in overall laminin-111 mono- and di-nitration was observed, consistent with a previous study that reported the overall (total) extent of laminin-111 nitration [18]. The overall yield of nitration at Tyr to give 3-nitroTyr was ∼11-fold higher than for nitration at Trp to give 6-nitroTrp, at all ONOOH concentrations. Thus, nitration of Trp and Tyr residues appear to respond similarly (i.e. in parallel) with increasing molar excesses of ONOOH. A similar pattern was observed for the di-nitration, although the total extent of di-nitration at Tyr residues was only ∼2-fold higher than for Trp residues.
Fig. 4.
Quantification of total extent of laminin-111 nitration. Panel A): Total laminin-111 nitration was estimated by summing the occupancies for all 3-nitroTyr sites (upper right panel) formed from Tyr residues (Y), and 6-nitroTrp sites (upper left panel) formed from Trp residues (W), or all identified nitration sites (lower panel). Panel B): Total laminin-111 di-nitration was estimated by summing the occupancies of all 3,5-dinitroTyr sites (upper right panel), 4,6-dinitroTrp sites (upper left panel) or all identified di-nitration sites (lower panel). Only modifications detected in all three independent replicates from ONOOH treatment, and none of the control samples are included; statistical analysis is therefore not appropriate.
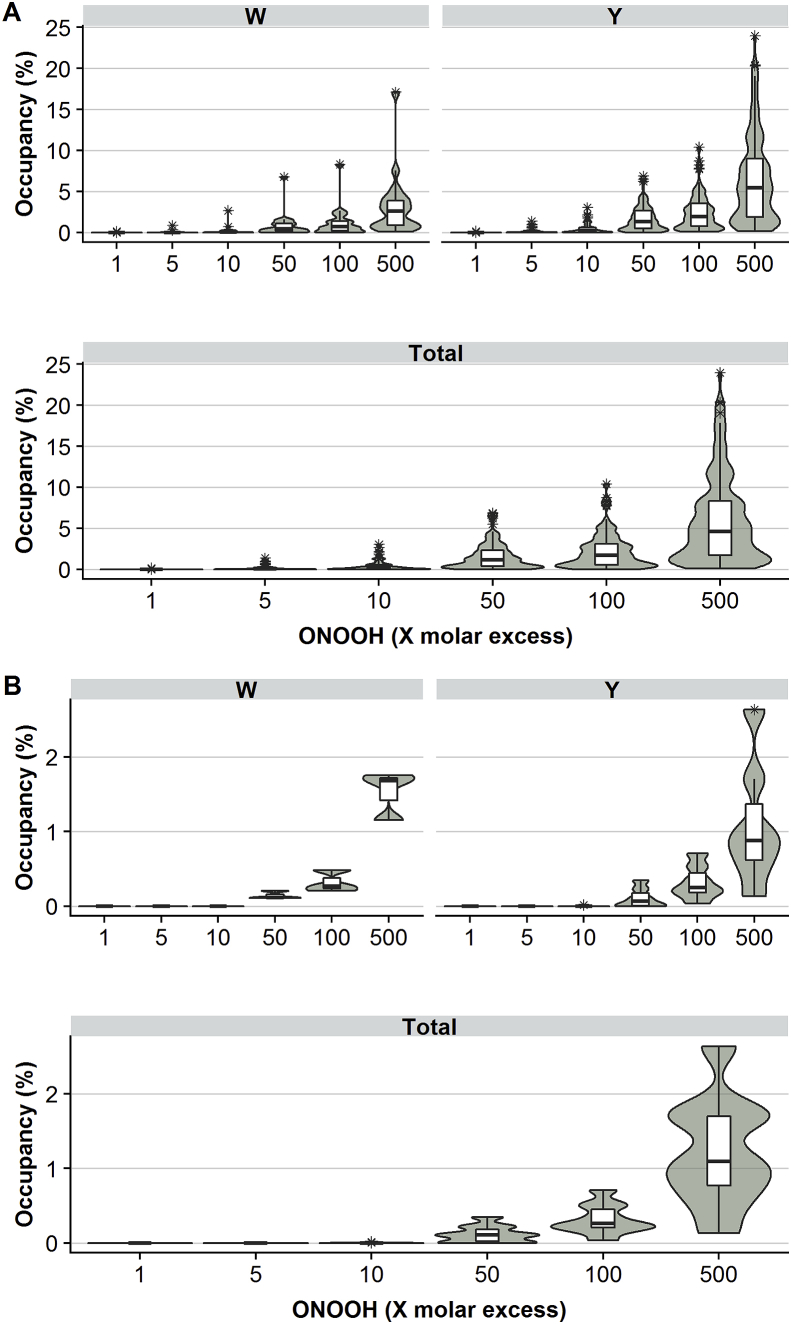
Extensive nitration was detected at multiple individual Tyr sites. At high oxidant excesses (500-fold), the mean occupancy was ∼6.5%, and 61 of the 113 quantified sites had occupancies of over 5% (Fig. 5A). At this same level of oxidant exposure, the mean percentage occupancy of Trp nitration ∼3.2%. The mean occupancy for Trp residues at lower levels of ONOOH was ∼2-fold less than that of Tyr. High occupancy was however observed for some Trp sites, with 3 of 20 Trp sites having >5% occupancy at a 500-fold molar excess of ONOOH. Trp1824 of the α1 chain was observed to be heavily nitrated at all levels of ONOOH, reaching 17.1% occupancy with a 500-fold molar excess of ONOOH. As the mean occupancy for all Trp residues at this oxidant excess was 3.2%, this indicates that selective nitration occurs at some Trp sites. In a similar manner, many Tyr nitration sites had occupancies at or below 5.5%, despite a mean value of 6.5%, as a limited number of sites had occupancy levels of >20%. The site-specific extent of di-nitration formation was, unsurprisingly, low compared to mono-nitration (Fig. 5B). However, it is interesting to note that the extent of di-nitration at a specific site was not markedly different between Tyr and Trp targets. This indicates that while Tyr side chains may be more susceptible to nitration, the two targets are equally susceptible to di-nitration.
Fig. 5.
Laminin-111 nitration site occupancies (peak areas of peptides carrying nitration at Tyr (Y)/Trp (W)/Tyr + Trp sites, summed and given as a percentage of the total, summed, peak areas of all modified and unmodified peptides containing these residues) after exposure of the parent protein to different molar excesses of ONOOH. The distribution of site occupancies is presented in the form of boxplots, indicating the median and quartiles (25th and 75th). Values more than 1.5 times the interquartile range away from the quartile edges (indicated by whiskers) are plotted and regarded as outliers. The violin plots surrounding boxplots represent kernel probability densities, i.e. the width of the shaded area represents the proportion of the data located there. Panel A): Distribution of nitration site occupancies for Trp residues (upper left panel), Tyr residues (upper right panel) and all nitration sites (lower panel). Panel B): Distribution of di-nitration site occupancies for Trp sites (upper left panel), Tyr sites (upper right panel) and all nitration sites (lower panel).
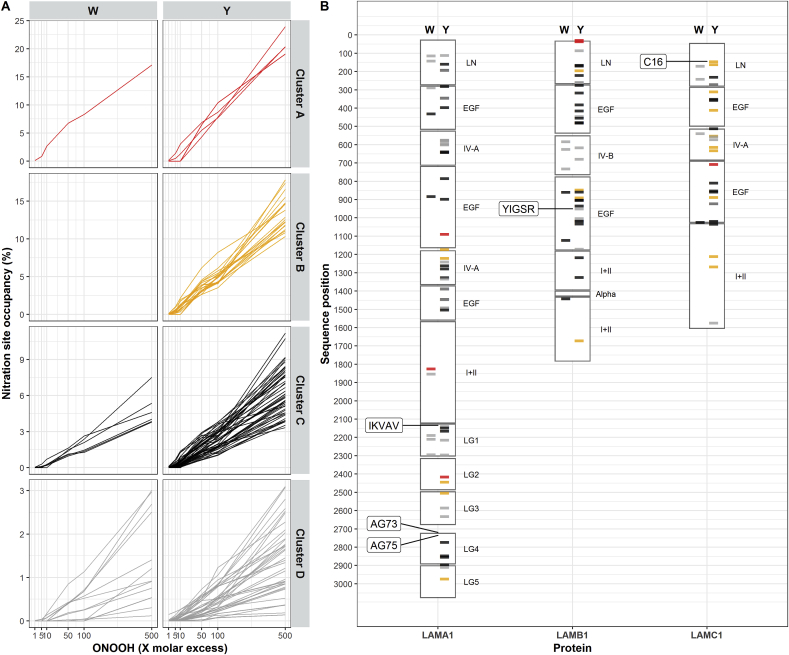
A Shapiro-Wilks test showed a clear lack of normality in the distribution of occupancies at all molar excesses of ONOOH, and for both Trp and Tyr residues (Fig. 5, Supplementary Table 1). The occupancies displayed a positive skew distribution (mode < median < mean) across variables, consistent with the conclusion that nitration occurs more readily at some Tyr and Trp sites than others. To examine sites that are selectively nitrated, these were clustered based on their occupancy at all molar excesses of ONOOH using hierarchical clustering. This analysis separated the detected nitration sites into 4 groups, with two small (group A: 6 sites; group B: 19 sites) and two large groups (group C: 56 sites; group D: 52 sites) (Supplementary Figs. 4 and 5). Groups A and B contain nitration sites that are extensively damaged (when compared to the other two groups) throughout the ONOOH concentration range assayed (Fig. 6A) whereas groups C and D comprise low occupancy nitration sites (c.f. the differences in the vertical axis scales, which indicates the % nitration of the particular amino acid site, in Fig. 6, panel A). Despite differences in the extent of modifications, all sites displayed a dose-dependent increase in nitration occupancy with increasing molar excesses of oxidant. However, a principal component analysis indicates that variance in occupancy at lower levels of ONOOH (1–10-fold) does not correlate fully with the variance in occupancy at high levels of ONOOH (50–500-fold, Supplementary Fig. 4). This suggests that some sites, even within a cluster, are more susceptible to low levels of ONOOH. The identified sites of nitration are relatively evenly distributed throughout the laminin-111 sequence, though clusters of high occupancy sites appear, in some cases, in close proximity (Fig. 6B). Interestingly, several high occupancy sites occur within regions with well-described functionality, including the cell binding domains, and the N-terminal domains involved in laminin polymerization.
Fig. 6.
Clustering and positions of laminin-111 nitration sites based on their extent of modification. Panel A): Overview of 6-nitroTrp (left panels) and 3-nitroTyr (right panels) formation, given as site occupancies, in response to increasing molar excesses of ONOOH (as indicated on the horizontal axis) within their cluster of site occupancy level. Panel B): Position of laminin-111 nitration sites in the sequences of the 3 laminin chains (horizontal axis: LAMA1 = α1, LAMB1 = β1, LAMC1 = γ1). Bars indicate the sites of nitration with the color indicating the assigned cluster and thus extent of modification (red, cluster A; yellow, cluster B; black, cluster C; grey, cluster D, in order of decreasing extent of modification, see text for further details). Left hand columns, modifications at Trp residues (W), right hand columns, modifications at Tyr residues (Y). Annotations to the right of the chain sequences in panel B indicate sites with known biological function. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Effect of CO2 on the extent of laminin nitration
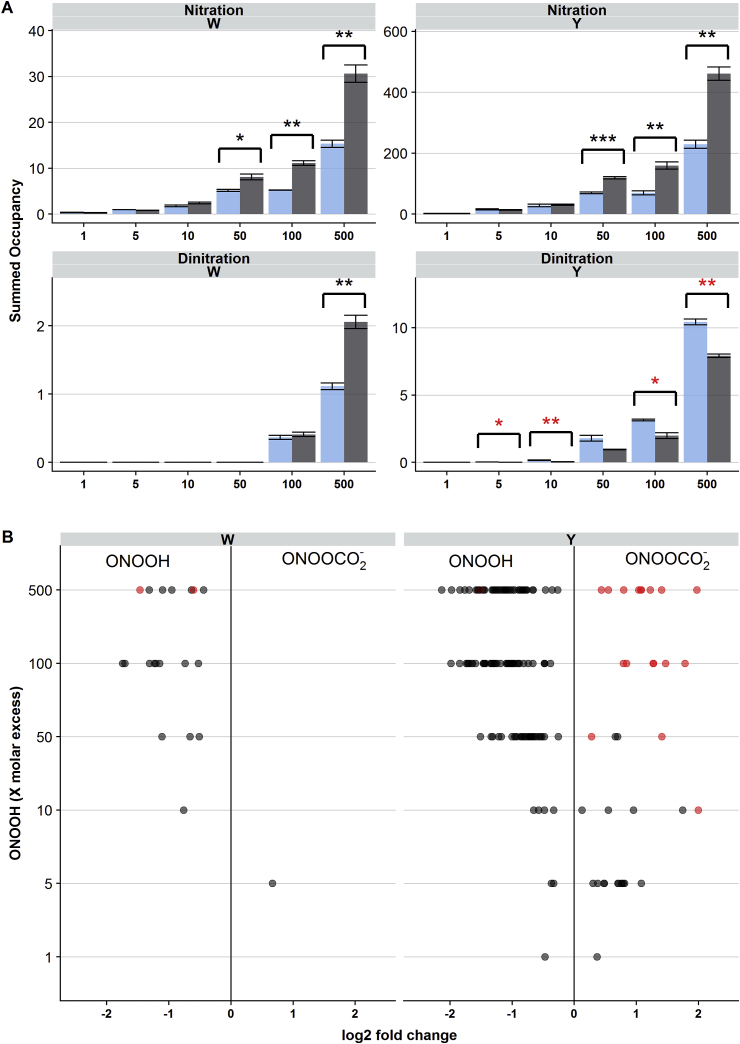
We have previously reported that CO2 can have a protective effect with regard to the overall extent of 3-nitroTyr and 6-nitroTrp formation in laminin-111 exposed to ONOOH in vitro [18]. To investigate whether this protective effect of CO2 is universal (i.e. occurs equally at all sites), or is specific to particular locations, laminin-111 was exposed to a 500-fold molar excesses ONOOH (as above) in both the absence or presence of 25 mM sodium bicarbonate (NaHCO3, which gives a concentration of CO2 of ∼1 mM; i.e. physiological levels). Samples were then analyzed in a non-targeted manner by LC-MS/MS followed by a database search, as described above, to identify potential nitration sites. A total of 114 sites were identified in samples nitrated in the presence of NaHCO3, with no unique nitration sites detected when compared to samples nitrated in the absence of added NaHCO3 (Supplementary Fig. 6). Di-nitration was identified at 23 sites in samples exposed to ONOOH in the presence of NaHCO3, compared to 14 sites identified in its absence, with 15 sites being uniquely di-nitrated in the presence of NaHCO3 (Fig. 7A, Supplementary Fig. 6).
Fig. 7.
Effect of CO2 on laminin-111 nitration. Panel A): the extent of laminin-111 nitration in the presence (blue bars) or absence (black bars) of CO2 was estimated by summing occupancies for all 6-nitroTrp sites (upper left panel), 3-nitroTyr sites (upper right panel), 4,6-dinitroTrp sites (lower left panel) and 3,5-dinitroTyr sites (lower right panel), respectively. Values are expressed as means ± SEM (n = 3). Asterisks indicate significant differences between samples (Students t-test, *p < 0.05, **p < 0.01, ***p < 0.001). Panel B) Differences in site-specific formation of nitration (black symbols) or di-nitration (red symbols) given as log2 fold-change between the presence and absence of CO2. Only sites where the differences were statistically significant (t-test with false discovery rate correction, p < 0.05) are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To investigate this further, laminin-111 was exposed to equimolar to 100-fold molar excesses of ONOOH in the presence of NaHCO3. Sites of nitration were detected using a targeted detection approach as described above, and the extent of modification was assessed by label-free quantification and calculation of relative site occupancies. These data were then compared to the data obtained in the absence of NaHCO3 (see above). Fig. 7 shows a volcano plot of log2 fold change in abundancies of modified peptides from samples generated in the absence and presence of NaHCO3. In agreement with previous data [18], the majority of the nitration sites (80), were less extensively modified by ONOOH in the presence of NaHCO3, indicating a protective effect of CO2. In contrast, the 7 sites that were more extensively modified in the presence of CO2 were all Tyr residues, and 6 of these were also detected as sites of di-nitration. In comparison, 3 di-nitration sites (2 Trp, 1 Tyr) were less modified in the presence of NaHCO3, and 3 displayed no difference at a 500-fold molar excess of ONOOH (data not shown). These data indicate that the protective effect of CO2 against laminin-111 nitration is relatively uniform at high molar excesses of oxidant. However, at lower oxidant excesses, both increased and decreased nitration was observed in the presence of NaHCO3, indicating that at low oxidant excesses CO2 sometimes promotes, and in other cases inhibits, nitration depending on the site in question. At equimolar concentrations of laminin-111 and ONOOH, few significant differences were observed, likely reflecting the imprecision of the quantification data for low abundances of nitrated species. At all the Tyr di-nitration sites, the extent of di-nitration was enhanced in the presence of NaHCO3, and this effect was observed at all excesses of oxidant where Tyr di-nitration could be detected. In contrast, the Trp sites displayed no significant change in di-nitration occupancy.
3.5. Effects of ONOOH on laminin polymerization
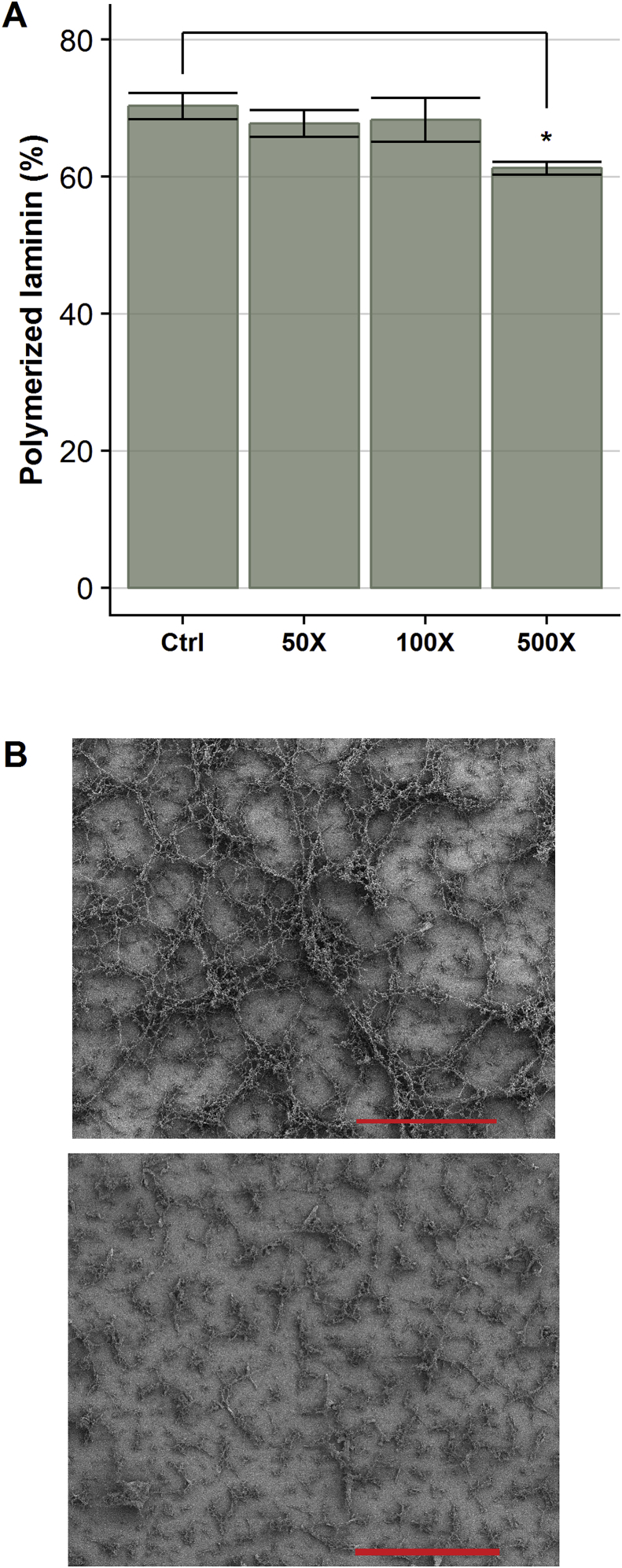
The data presented above indicate extensive nitration at the N-termini of the 3 laminin subunits where the LN domains are located. These domains have been implicated in laminin polymerization [45,46], a process critical to basement membrane assembly and cell function [47,48]. We therefore hypothesized that laminin-111 nitration in these domains would interfere with self-polymerization. In order to test this, laminin-111 was pre-treated with 50–500-fold molar excesses of ONOOH, then diluted in pre-heated acidic buffer containing CaCl2 (or EDTA as negative control), to initiate protein polymerization. Monomeric and polymerized laminin were then separated by centrifugation, solubilized and denatured in SDS and the protein quantified. Laminin-111 treated with a 500-fold molar excess ONOOH displayed a small, but significant decrease in polymerization compared to native laminin-111 (Fig. 8A). When treated with 50 or 100-fold molar excesses of ONOOH, laminin-111 sedimented to the same degree as native laminin-111, indicating no marked changes in the quantity of material that could be sedimented. Scanning electron microscopy (SEM) was used to investigate the structure of the polymers formed from native and nitrated laminin. Laminin-111 was either left untreated or nitrated with a 500-fold molar excess of ONOOH, followed by polymerization on to glass coverslips for SEM imaging. Native laminin-111 formed a well-ordered and homogenous mesh (Fig. 8B, top panel), whereas nitrated laminin-111 formed a heterogenous mixture of unconnected aggregates (Fig. 8B, bottom panel). The structures detected for the native laminin-111 polymers are consistent with previous findings using similar conditions and techniques [31]. Although nitrated laminin-111 did form aggregates, consistent with the sedimentation assay, the observed structures are in stark contrast to those generated by native laminin-111. This indicates that while nitrated laminin can form polymers, the native structure is significantly perturbed.
Fig. 8.
Oxidative modification interferes with laminin-111 self-polymerization. Oxidized and native laminin-111 was dissolved in 50 mM sodium acetate, pH 4 supplemented with 10 mM CaCl2, and incubated for 60 min to promote polymerization. Panel A) Polymerized laminin-111 was seperated from non-polymerized proteins by centrifugation, and protein content in the 2 fractions was quantified by BCA assay. The percent polymerization was calculated as: . Values are expressed as means ± SEM (n = 3). Panel B) Scanning electron micrographs at 1,000X magnification were recorded for polymerized laminin-111, either left untreated (top image) or exposed to a 500-fold molar excess of ONOOH (bottom image). Scale bar = 100 μm.
3.6. Effect of nitration on cell adhesion to a cell-binding laminin peptide
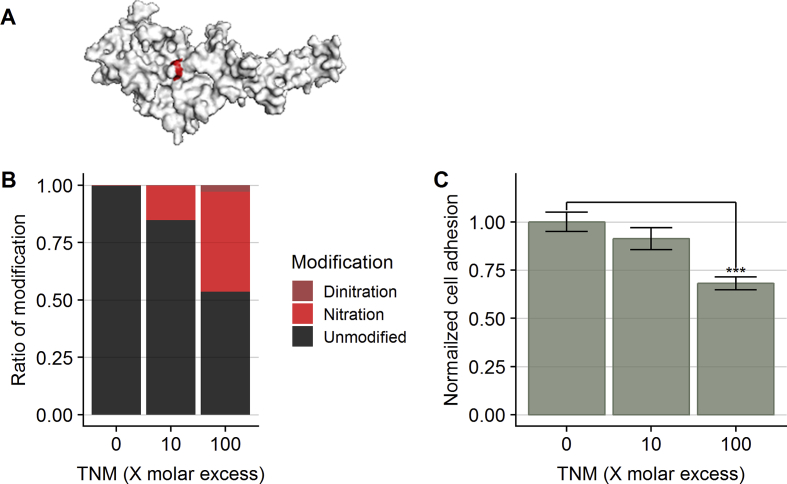
The data reported above demonstrate that nitration induced by ONOOH occurs within functional domains on laminin-111, including at Tyr145 on the γ1 chain (LAMC1-Y145). This Tyr is located in a sequence that corresponds to the C16 peptide, a laminin-derived peptide with cell-binding properties [49]. This suggests that Tyr145 on the C16 peptide is an important site for endothelial cell adhesion on the γ1 chain. Together with Arg147 and Phe141, the Tyr145 residue constitutes the solvent exposed part of the C16 peptide in the laminin γ1 structure (Fig. 9A). Nitration of Tyr145 may therefore modulate cell adhesion, and this might explain (at least in part) the decreased endothelial cell adhesion to nitrated laminin-111, reported previously [18]. To test this hypothesis, we nitrated synthetic C16 peptide with tetranitromethane (TNM) at low (10-) or high (100-) molar excesses. The extent of peptide modification was determined by LC-MS/MS, with the 100-fold molar excess of TNM resulting in 46% modification of the peptide, with this being primarily (43%) mono-nitration at the single Tyr.
Fig. 9.
Nitration reduces cell adhesion to laminin-derived C16 peptide. A) Crystal structure of the N-terminal domain of the laminin- chain shows solvent exposure of Tyr-145 in the C16 sequence (structure from Ref. [62]). The C16 peptide was left untreated (0X) or nitrated with tetranitromethane (TNM), either in 10-fold (10X) or 100-fold (100X) molar excess to the peptide, and used to coat 96-well cell culture plates. HCAECs stained with Calcein-AM were allowed to adhere to the pre-coated 96-well plates for 90 min. After removal of nonadherent cells, the adherent cells were quantified using Calcein-AM fluorescence. Values are expressed as means ± SEM (n = 3). Asterisks indicate significant difference from control samples (ANOVA with Dunnett's correction for multiple testing, ***p < 0.001).
Human coronary artery endothelial cell (HCAEC), preloaded with the fluorescent dye calcein-AM, were seeded on to tissue culture plates pre-coated with native or TNM-treated C16 peptide. After 90 min to allow adherence, non-bound cells were washed off, and the number of adherent cells quantified by determination of the calcein-AM fluorescence after cell lysis. HCAECs plated onto C16 peptide subjected to extensive nitration (100-fold excess of TNM) displayed a significant decrease in adherence (32%) compared to the untreated peptide (Fig. 9B). Mild nitration (10-fold excess of TNM) showed a trend towards a decrease, but this was not statistically significant.
4. Discussion
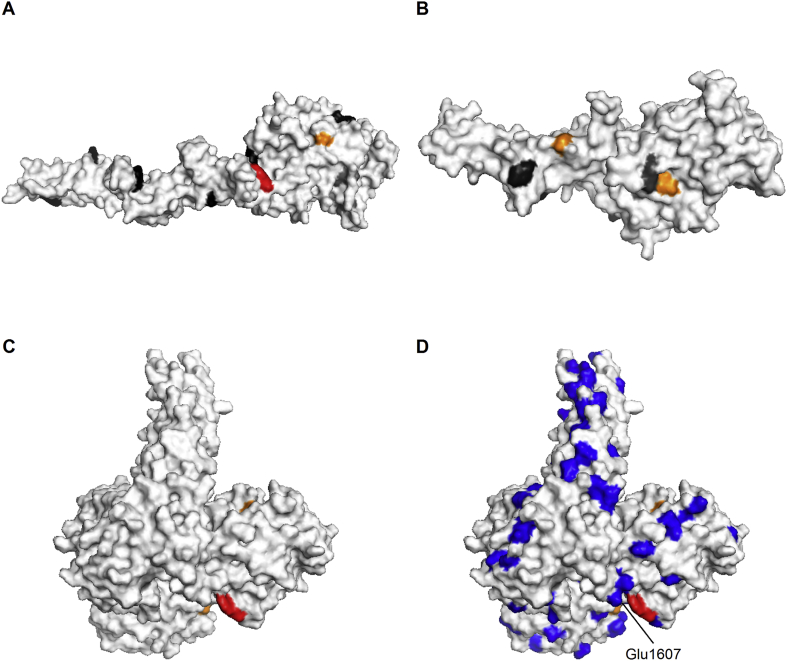
Nitration, particularly at Tyr and Trp, is a post-translational protein modification that can cause dramatic changes in protein structure and function (reviewed [50]). Here we have used LC-MS/MS to map laminin-111 nitration sites formed in response to treatment with ONOOH in vitro in both the absence and presence of CO2. The data presented here show that a large proportion of the Tyr and Trp residues on laminin-111 sites can be nitrated by ONOOH at concentrations ranging from equimolar (i.e. single hit) to 500-fold molar excesses of the oxidant. With increasing concentrations of oxidant, both the number of nitration sites detected, and the extent of modification at each site, increased. Cluster analysis has revealed the existence of 4 groups of nitration sites, with 2 subgroups of extensively nitrated aromatic sites. The relative site occupancy of the residues in each of these clusters, at the various molar excesses of ONOOH examined, are presented in Supplementary Table 2. The presence of 25 highly modified sites, 6 of which have particularly high occupancies (i.e. extents of modification), suggests selective reaction of ONOOH at these sites. Solvent exposure, positioning in a loop structure and proximity to acidic residues have been suggested as factors that increase the likelihood of nitration at an aromatic residue [51]. Due to its large, complex structure and heterotrimeric nature, the complete structure of laminin-111 has not been resolved. However, several partial structures exist. From these structures we conclude that 2 of the most extensively nitrated sites, Tyr2415 in the α1 chain and Tyr36 of the β1 chain, meet all 3 of these criteria (Fig. 10), suggesting that the requirements for selective nitration proposed by Ischiropoulos and Beckman hold true, at least in part, for laminin-111 [51,52]. Several other surface-exposed Tyr residues also appear to be heavily nitrated (i.e. those in groups A and B), but these do not appear to be present in loop structures or in the immediate proximity to acidic residues. However, some nitration sites that only display low or moderate nitration in response to treatment with ONOOH (i.e. groups C and D) appear to be highly surface exposed, and some also meet one or two of the other parameters, which are inconsistent with their low extents of modification. This may be explained by the partial nature of the structures that are available for the N-terminal regions where these sites are located, with these sites appearing to be surface-exposed in the partial structures (due to the absence of the remaining residues), whereas in reality they may not be surface exposed. Another plausible explanation is that these N-terminal regions may be involved in interactions with other laminin molecules, and hence protected by protein-protein interactions.
Fig. 10.
Mapping of nitration sites to laminin-111 crystal structures. Nitration sites identified are mapped to resolved crystal structures of laminin-111 domains. Colors indicate the cluster assigned to the nitration sites based on their extent of modification: Red: Cluster A, very extensively modified sites, Orange: Cluster B, extensively modified sites, Black: Cluster C, moderately modified sites, Grey: Cluster C, mildly modified sites. The crystal structures shown are resolved for: A) the N-terminal domain of the laminin- chain [62]; B) the N-terminal domain of the laminin- chain [62]; C) the C-terminal part of the laminin-111 heterotrimer [65]; D) as C), but with all the acidic residues indicated in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Although the majority of nitration sites identified on ONOOH-treated laminin-111 are at Tyr rather than Trp residues, this difference can be partially explained by the lower overall abundance of Trp in the primary sequence of laminin-111 compared to Tyr (199 Tyr, 50 Trp; see above). However, the mean occupancy of Trp nitration sites was still determined to be 2-fold lower that of Tyr, despite the rate constants for reaction of (free) Trp and Tyr residues with ONOOH being similar (1.3 × 102 M−1 s−1 [53] and 3.6 × 102 M−1 s−1 [54], respectively). This is in agreement with previous data [18], where the ratio of 6-nitroTrp to Trp was ∼3 fold less than that of 3-nitroTyr to Tyr. As Trp residues are hydrophobic in nature, and hence typically display limited solvent exposure, this may hinder reaction with ONOOH, consistent with the lower extent of modification at most Trp sites. However, as buried Trp residues contribute greatly to protein structure and stability [55], even low levels of nitration at such sites may be structurally and functionally disruptive. In this light, the observation of Trp nitration at low oxidant levels and the observation of moderate (>5% occupancy) modification of several Trp sites at higher ONOOH molar excesses, suggests that Trp residues in laminin-111 may be important targets, possibly with effects on structure and function. Additionally, a single Trp site, Trp1824 located in the coiled-coil domain of the α1 chain, appears to be extensively damaged by ONOOH. No data on the accessibility of this site is available, but these data indicate that it is likely to be surface exposed. As surface-exposed Trp residues are often involved in intermolecular interactions (e.g. with nucleic acids, carbohydrates or other proteins [55]), Yamakura and Ikeda [56] have suggested that even though Trp nitration may be less common than Tyr nitration, it may be disruptive to protein function and thus of particular biological importance. Since the coiled-coil region has been reported to have cell binding properties [57,58], it is possible that α1-Trp1824 is involved in cell adhesion, and accordingly that nitration at this site may interfere with cell adhesion to laminin-111.
The data presented here indicate that the presence of HCO3−/CO2 is generally protective in terms of nitration, and that this is true for both Tyr and Trp residues. This conclusion is in agreement with previous studies that have reported on the total level of modification, but not the sites at which these occur, under similar reaction conditions [18,19,21]. The site-specific data presented here indicate, for the first time, that CO2 is uniformly protective across the large number of sites modified with high molar excesses of ONOOH. Thus, the majority of sites displayed significantly less nitration in the presence of CO2, some were equally nitrated in both conditions, and only a single nitration site displayed a higher degree of modification in the presence of CO2. However, with low concentrations of oxidant, the presence of CO2 can result in a mixture of both protection from, and aggravation of, damage depending on the specific target. In contrast, di-nitration appears to be mostly aggravated in the presence of CO2, with both more being identified, and an increased extent of modification detected at several sites, despite CO2 being protective overall in terms of total modification of Tyr and Trp residues. The decreased extent of nitration observed in the presence of CO2 may be due to electrostatic repulsion between the negatively-charged oxidant ONOOCO2− (or the charged radical CO3.- derived therefrom) and negatively-charged amino acids in the protein. Negatively-charged, protein-bound, sugar polymers appear to have similar effects [21]. It has also been proposed that presence of CO2 reduces the yield of reactive radicals formed by homolysis, due to an accelerated isomerization to nitrite [59]. The lower rate constants for reaction of CO3.- compared to HO. with target residues may also play a role (cf. data in Refs. [60,61]). However, Santos et al. have reported that with pure L-Tyr, the presence of CO2 results in higher extents of formation of 3-nitroTyr [8], with this ascribed to the higher specificity of CO3.- compared to HO.. Thus, there is no universal consensus as to whether CO2 is protective or not. Thus, we speculate that if CO3.- is not affected by electrostatic repulsion arising from charged groups on the protein surface, it may result in a higher degree of damage, exemplified by the observed increased formation of 3,5-dinitroTyr at some sites.
Interestingly, and of potential importance, several of the highly nitrated Tyr sites are located in regions of known biological function, including well-established cell binding sites. Percentage occupancy data for these sites, at various ONOOH concentrations, are presented in Supplementary Table 2. The Tyr nitration sites γ1-Y145 (i.e. Tyr145 on the γ1 chain) and β1-Y950, are located within the sequences of two biologically-active laminin peptides, C16 and YIGSR respectively, both of which support endothelial cell adhesion [49,62,63]. We found that β1-Y950, in the YIGSR binding site, had a maximum occupancy of ∼0.9%, whereas γ1-Y145, in the C16 binding site, reached 11.1% occupancy with a 500-fold molar excess of ONOOH. Modification at γ1-Y145 was detected with equimolar or higher concentrations of ONOOH, indicating that it is a sensitive target for modification (cluster B, see Fig. 6). Although other Tyr residues near the YIGSR cell binding site display higher extents of nitration, and hence may affect the affinity of cells for the YIGSR sites, it is more likely that nitration of γ1-Y145, in the C16 binding site, is the cause of the decreased endothelial cell adhesion to ONOOH-treated laminin reported previously [18]. The current data indicate that HCAECs have a decreased adhesion to nitrated C16, compared to the native peptide, and that the extent of cell adhesion decreases with an increased extent of C16 nitration. This suggests that nitration of γ1-Y145, together with the extensively-nitrated Trp residue (Trp1824), which is also present in a cell adhesion region of laminin-111, are the cause of the decreased cell adhesion to nitrated laminin-111 [18]. Additionally, Tyr2415, a major target of ONOOH, is placed immediately next to an acidic residue (Glu1605) critical to integrin binding and cell adhesion [64]. The recently published crystal structure of the laminin-111 C-terminal region shows that Glu1605 is surface-exposed and can therefore bind integrins directly [65]. We hypothesize that due to its close proximity to this site, nitration of Tyr2415 may interfere with integrin binding and thereby cell adhesion.
The LC-MS/MS data reported here also show that extensive nitration occurs within the N-terminal regions of the 3 laminin subunits (the LN domains). These domains are implicated in laminin polymerization [66], a process critical to basement membrane assembly and cell function [47,48]. The current data suggest that although the overall extent of laminin polymerization was only moderately affected by ONOOH, the structure of these polymers is markedly perturbed, compared to those from native laminin-111. Under the conditions used here (acidic buffer in the presence of CaCl2) laminin-111 forms polymeric structures resembling those of natural basement membranes [45,67]. It is therefore likely that the formation of nitrated laminin will affect both basement membrane integrity, and cell behavior/attachment. These altered structures and diminished cell adhesion, observed here and previously [18], may result in alterations to the structure and strength of the fibrous cap of cardiovascular atherosclerotic lesions, with this damage resulting in an increased propensity of lesions to rupture. Furthermore, these data indicate that relatively modest levels of modification at a significant number of sites (each of which may be modified to only a few %) may together be responsible for the diminished cell adhesion (cf. the 30% reduction), and the effects on laminin structure (as evidenced by markedly altered polymerization), rather than very high extents of modification at a small number of particularly susceptible individual sites.
In a recent study by Nybo et al, modifications to laminin-111 by HOCl and myeloperoxidase-derived oxidants were investigated [68]. These treatments resulted in aggregation and backbone fragmentation of the polymer analogous to those seen previously with ONOOH [18]. Tandem mass spectrometry (LC-MS/MS) was employed to map the resulting oxidations and chlorination of the laminin chains [68]. The authors showed that Tyr residues could be chlorinated, and that both Met and Trp residues were subject to extensive oxidation. Interestingly, this study also found extensive modification of Tyr2415 on the α1 chain. This suggests that this residue is highly susceptible to both nitration and chlorination, and it may be speculated that both types of modification can interfere with integrin binding to the α1 laminin chain.
5. Conclusions
This study has shown that laminin-111 can be nitrated by ONOOH at numerous Tyr and Trp sites, and that nitration occurs in a site-specific manner. Consistent with previous findings, the effects of ONOOH have been shown to be modulated by the presence of CO2, with this resulting in an overall diminished extent of nitrative modifications to the protein [18,19,21]. However, increased di-nitration was detected with ONOOCO2−, suggesting an intricate modulation of ONOOH reactivity by CO2. Several high-occupancy nitration sites have been detected in functionally important sites/domains, including cell binding sites and regions involved in laminin self-polymerization, with these changes resulting in altered polymer formation and diminished endothelial cell binding with the latter arising, at least in part via nitration at Tyr145 on the γ1 chain.
Acknowledgements
The authors are grateful to the Novo Nordisk Foundation (grant: NNF13OC0004294) and the Danish Council for Independent Research (Det Frie Forskningsråd, grant: DFF-7014-00047) for financial support. ARW was supported by the Independent Research Fund Denmark – Natural Sciences (FNU) and a Villum Foundation grant to the VILLUM Center for Bioanalytical Sciences at SDU.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101226.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nauser T., Koppenol W.H. The rate constant of the reaction of superoxide with nitrogen monoxide: approaching the diffusion limit. J. Phys. Chem. A. 2002;106(16):4084–4086. [Google Scholar]; T. Nauser, W.H. Koppenol, The rate constant of the reaction of superoxide with nitrogen monoxide: Approaching the diffusion limit, Journal of Physical Chemistry A 106(16) (2002) 4084-4086.
- 2.Ferrer-Sueta G., Campolo N., Trujillo M., Bartesaghi S., Carballal S., Romero N., Alvarez B., Radi R. Biochemistry of peroxynitrite and protein tyrosine nitration. Chem. Rev. 2018;118(3):1338–1408. doi: 10.1021/acs.chemrev.7b00568. [DOI] [PubMed] [Google Scholar]; G. Ferrer-Sueta, N. Campolo, M. Trujillo, S. Bartesaghi, S. Carballal, N. Romero, B. Alvarez, R. Radi, Biochemistry of peroxynitrite and protein tyrosine nitration, Chem Rev 118(3) (2018) 1338-1408. [DOI] [PubMed]
- 3.Radi R., Peluffo G., Alvarez M.N., Naviliat M., Cayota A. Unraveling peroxynitrite formation in biological systems. Free Radic. Biol. Med. 2001;30(5):463–488. doi: 10.1016/s0891-5849(00)00373-7. [DOI] [PubMed] [Google Scholar]; R. Radi, G. Peluffo, M.N. Alvarez, M. Naviliat, A. Cayota, Unraveling peroxynitrite formation in biological systems, Free Radic Biol Med 30(5) (2001) 463-488. [DOI] [PubMed]
- 4.Alvarez B., Ferrer-Sueta G., Freeman B.A., Radi R. Kinetics of peroxynitrite reaction with amino acids and human serum albumin. J. Biol. Chem. 1999;274(2):842–848. doi: 10.1074/jbc.274.2.842. [DOI] [PubMed] [Google Scholar]; B. Alvarez, G. Ferrer-Sueta, B.A. Freeman, R. Radi, Kinetics of peroxynitrite reaction with amino acids and human serum albumin, J Biol Chem 274(2) (1999) 842-848. [DOI] [PubMed]
- 5.Koppenol W.H., Bounds P.L., Nauser T., Kissner R., Ruegger H. Peroxynitrous acid: controversy and consensus surrounding an enigmatic oxidant. Dalton Trans. 2012;41(45):13779–13787. doi: 10.1039/c2dt31526b. [DOI] [PubMed] [Google Scholar]; W.H. Koppenol, P.L. Bounds, T. Nauser, R. Kissner, H. Ruegger, Peroxynitrous acid: controversy and consensus surrounding an enigmatic oxidant, Dalton Trans 41(45) (2012) 13779-13787. [DOI] [PubMed]
- 6.Denicola A., Freeman B.A., Trujillo M., Radi R. Peroxynitrite reaction with carbon dioxide/bicarbonate: kinetics and influence on peroxynitrite-mediated oxidations. Arch. Biochem. Biophys. 1996;333(1):49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]; A. Denicola, B.A. Freeman, M. Trujillo, R. Radi, Peroxynitrite reaction with carbon dioxide/bicarbonate: kinetics and influence on peroxynitrite-mediated oxidations, Arch Biochem Biophys 333(1) (1996) 49-58. [DOI] [PubMed]
- 7.Lymar S.V., Hurst J.K. Rapid reaction between peroxonitrite ion and carbon dioxide: implications for biological activity. J. Am. Chem. Soc. 1995;117(34):8867–8868. [Google Scholar]; S.V. Lymar, J.K. Hurst, Rapid reaction between peroxonitrite ion and carbon dioxide: Implications for biological activity, J Am Chem Soc 117(34) (1995) 8867-8868.
- 8.Santos C.X., Bonini M.G., Augusto O. Role of the carbonate radical anion in tyrosine nitration and hydroxylation by peroxynitrite. Arch. Biochem. Biophys. 2000;377(1):146–152. doi: 10.1006/abbi.2000.1751. [DOI] [PubMed] [Google Scholar]; C.X. Santos, M.G. Bonini, O. Augusto, Role of the carbonate radical anion in tyrosine nitration and hydroxylation by peroxynitrite, Arch. Biochem. Biophys. 377(1) (2000) 146-152. [DOI] [PubMed]
- 9.Goldstein S., Czapski G., Lind J., Merenyi G. Carbonate radical ion is the only observable intermediate in the reaction of peroxynitrite with CO(2) Chem. Res. Toxicol. 2001;14(9):1273–1276. doi: 10.1021/tx0100845. [DOI] [PubMed] [Google Scholar]; S. Goldstein, G. Czapski, J. Lind, G. Merenyi, Carbonate radical ion is the only observable intermediate in the reaction of peroxynitrite with CO(2), Chem Res Toxicol 14(9) (2001) 1273-1276. [DOI] [PubMed]
- 10.Halliwell B., Gutteridge J.M. The antioxidants of human extracellular fluids. Arch. Biochem. Biophys. 1990;280(1):1–8. doi: 10.1016/0003-9861(90)90510-6. [DOI] [PubMed] [Google Scholar]; B. Halliwell, J.M. Gutteridge, The antioxidants of human extracellular fluids, Arch Biochem Biophys 280(1) (1990) 1-8. [DOI] [PubMed]
- 11.Shapiro S.D., Endicott S.K., Province M.A., Pierce J.A., Campbell E.J. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon. J. Clin. Investig. 1991;87(5):1828–1834. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]; S.D. Shapiro, S.K. Endicott, M.A. Province, J.A. Pierce, E.J. Campbell, Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon, J Clin Invest 87(5) (1991) 1828-1834. [DOI] [PMC free article] [PubMed]
- 12.Chuang C.Y., Degendorfer G., Davies M.J. Oxidation and modification of extracellular matrix and its role in disease. Free Radic. Res. 2014;48(9):970–989. doi: 10.3109/10715762.2014.920087. [DOI] [PubMed] [Google Scholar]; C.Y. Chuang, G. Degendorfer, M.J. Davies, Oxidation and modification of extracellular matrix and its role in disease, Free Radic Res 48(9) (2014) 970-989. [DOI] [PubMed]
- 13.Rees M.D., Kennett E.C., Whitelock J.M., Davies M.J. Oxidative damage to extracellular matrix and its role in human pathologies. Free Radic. Biol. Med. 2008;44(12):1973–2001. doi: 10.1016/j.freeradbiomed.2008.03.016. [DOI] [PubMed] [Google Scholar]; M.D. Rees, E.C. Kennett, J.M. Whitelock, M.J. Davies, Oxidative damage to extracellular matrix and its role in human pathologies, Free Radic Biol Med 44(12) (2008) 1973-2001. [DOI] [PubMed]
- 14.Hohenester E., Yurchenco P.D. Laminins in basement membrane assembly. Cell Adhes. Migrat. 2013;7(1):56–63. doi: 10.4161/cam.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]; E. Hohenester, P.D. Yurchenco, Laminins in basement membrane assembly, Cell Adh Migr 7(1) (2013) 56-63. [DOI] [PMC free article] [PubMed]
- 15.Sucu N., Unlu A., Tamer L., Aytacoglu B., Ercan B., Dikmengil M., Atik U. 3-Nitrotyrosine in atherosclerotic blood vessels. Clin. Chem. Lab. Med. 2003;41(1):23–25. doi: 10.1515/CCLM.2003.005. [DOI] [PubMed] [Google Scholar]; N. Sucu, A. Unlu, L. Tamer, B. Aytacoglu, B. Ercan, M. Dikmengil, U. Atik, 3-Nitrotyrosine in atherosclerotic blood vessels, Clin Chem Lab Med 41(1) (2003) 23-25. [DOI] [PubMed]
- 16.Beckman J.S., Ye Y.Z., Anderson P.G., Chen J., Accavitti M.A., Tarpey M.M., White C.R. Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry. Biol. Chem. Hoppe-Seyler. 1994;375(2):81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]; J.S. Beckman, Y.Z. Ye, P.G. Anderson, J. Chen, M.A. Accavitti, M.M. Tarpey, C.R. White, Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry, Biol. Chem. Hoppe-Seyler 375(2) (1994) 81-88. [DOI] [PubMed]
- 17.Chuang C.Y., Degendorfer G., Hammer A., Whitelock J.M., Malle E., Davies M.J. Oxidation modifies the structure and function of the extracellular matrix generated by human coronary artery endothelial cells. Biochem. J. 2014;459:313–322. doi: 10.1042/BJ20131471. [DOI] [PubMed] [Google Scholar]; C.Y. Chuang, G. Degendorfer, A. Hammer, J.M. Whitelock, E. Malle, M.J. Davies, Oxidation modifies the structure and function of the extracellular matrix generated by human coronary artery endothelial cells, Biochem J 459 (2014) 313-322. [DOI] [PubMed]
- 18.Degendorfer G., Chuang C.Y., Hammer A., Malle E., Davies M.J. Peroxynitrous acid induces structural and functional modifications to basement membranes and its key component, laminin. Free Radical Biol. Med. 2015;89:721–733. doi: 10.1016/j.freeradbiomed.2015.09.018. [DOI] [PubMed] [Google Scholar]; G. Degendorfer, C.Y. Chuang, A. Hammer, E. Malle, M.J. Davies, Peroxynitrous acid induces structural and functional modifications to basement membranes and its key component, laminin, Free radical biology & medicine 89 (2015) 721-733. [DOI] [PubMed]
- 19.Degendorfer G., Chuang C.Y., Kawasaki H., Hammer A., Malle E., Yamakura F., Davies M.J. Peroxynitrite-mediated oxidation of plasma fibronectin. Free Radical Biol. Med. 2016;97:602–615. doi: 10.1016/j.freeradbiomed.2016.06.013. [DOI] [PubMed] [Google Scholar]; G. Degendorfer, C.Y. Chuang, H. Kawasaki, A. Hammer, E. Malle, F. Yamakura, M.J. Davies, Peroxynitrite-mediated oxidation of plasma fibronectin, Free radical biology & medicine 97 (2016) 602-615. [DOI] [PubMed]
- 20.Degendorfer G., Chuang C.Y., Mariotti M., Hammer A., Hoefler G., Hagglund P., Malle E., Wise S.G., Davies M.J. Exposure of tropoelastin to peroxynitrous acid gives high yields of nitrated tyrosine residues, di-tyrosine cross-links and altered protein structure and function. Free Radic. Biol. Med. 2018;115:219–231. doi: 10.1016/j.freeradbiomed.2017.11.019. [DOI] [PubMed] [Google Scholar]; G. Degendorfer, C.Y. Chuang, M. Mariotti, A. Hammer, G. Hoefler, P. Hagglund, E. Malle, S.G. Wise, M.J. Davies, Exposure of tropoelastin to peroxynitrous acid gives high yields of nitrated tyrosine residues, di-tyrosine cross-links and altered protein structure and function, Free Radic Biol Med 115 (2018) 219-231. [DOI] [PubMed]
- 21.Kennett E.C., Rees M.D., Malle E., Hammer A., Whitelock J.M., Davies M.J. Peroxynitrite modifies the structure and function of the extracellular matrix proteoglycan perlecan by reaction with both the protein core and the heparan sulfate chains. Free Radic. Biol. Med. 2010;49(2):282–293. doi: 10.1016/j.freeradbiomed.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; E.C. Kennett, M.D. Rees, E. Malle, A. Hammer, J.M. Whitelock, M.J. Davies, Peroxynitrite modifies the structure and function of the extracellular matrix proteoglycan perlecan by reaction with both the protein core and the heparan sulfate chains, Free Radic Biol Med 49(2) (2010) 282-293. [DOI] [PMC free article] [PubMed]
- 22.Miner J.H., Yurchenco P.D. Laminin functions in tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]; J.H. Miner, P.D. Yurchenco, Laminin functions in tissue morphogenesis, Annu Rev Cell Dev Biol 20 (2004) 255-284. [DOI] [PubMed]
- 23.Pozzi A., Yurchenco P.D., Iozzo R.V. The nature and biology of basement membranes. Matrix Biol. 2017;57–58:1–11. doi: 10.1016/j.matbio.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Pozzi, P.D. Yurchenco, R.V. Iozzo, The nature and biology of basement membranes, Matrix Biology 57-58 (2017) 1-11. [DOI] [PMC free article] [PubMed]
- 24.Rasmussen D.G.K., Karsdal M.A. Laminins. In: Karsdal M.A., Leeming D.J., Henriksen K., Bay-Jensen A.-C., editors. Biochemistry of Collagens, Laminins and Elastin: Structure, Function and Biomarkers. 2016. pp. 163–196. [Google Scholar]; D.G.K. Rasmussen, M.A. Karsdal, Laminins, in: M.A. Karsdal, D.J. Leeming, K. Henriksen, A.-C. Bay-Jensen (Eds.), Biochemistry of Collagens, Laminins and Elastin: Structure, Function and Biomarkers 2016, pp. 163-196.
- 25.Yurchenco P.D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect Biol. 2011;3(2) doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]; P.D. Yurchenco, Basement membranes: cell scaffoldings and signaling platforms, Cold Spring Harb Perspect Biol 3(2) (2011). [DOI] [PMC free article] [PubMed]
- 26.Kennett E.C., Davies M.J. Degradation of extracellular matrix by peroxynitrite/peroxynitrous acid. Free Radic. Biol. Med. 2008;45(5):716–725. doi: 10.1016/j.freeradbiomed.2008.05.027. [DOI] [PubMed] [Google Scholar]; E.C. Kennett, M.J. Davies, Degradation of extracellular matrix by peroxynitrite/peroxynitrous acid, Free Radic. Biol. Med. 45(5) (2008) 716-725. [DOI] [PubMed]
- 27.Robert L. Aging of the vascular wall and atherogenesis: role of the elastin-laminin receptor. Atherosclerosis. 1996;123(1–2):169–179. doi: 10.1016/0021-9150(96)05804-2. [DOI] [PubMed] [Google Scholar]; L. Robert, Aging of the vascular wall and atherogenesis: role of the elastin-laminin receptor, Atherosclerosis 123(1-2) (1996) 169-179. [DOI] [PubMed]
- 28.Yurchenco P.D., Tsilibary E.C., Charonis A.S., Furthmayr H. Laminin polymerization in vitro. Evidence for a two-step assembly with domain specificity. J. Biol. Chem. 1985;260(12):7636–7644. [PubMed] [Google Scholar]; P.D. Yurchenco, E.C. Tsilibary, A.S. Charonis, H. Furthmayr, Laminin polymerization in vitro. Evidence for a two-step assembly with domain specificity, J Biol Chem 260(12) (1985) 7636-7644. [PubMed]
- 29.Malinda K.M., Nomizu M., Chung M., Delgado M., Kuratomi Y., Yamada Y., Kleinman H.K., Ponce M.L. Identification of laminin alpha1 and beta1 chain peptides active for endothelial cell adhesion, tube formation, and aortic sprouting. Faseb J. 1999;13(1):53–62. [PubMed] [Google Scholar]; K.M. Malinda, M. Nomizu, M. Chung, M. Delgado, Y. Kuratomi, Y. Yamada, H.K. Kleinman, M.L. Ponce, Identification of laminin alpha1 and beta1 chain peptides active for endothelial cell adhesion, tube formation, and aortic sprouting, Faseb J 13(1) (1999) 53-62. [PubMed]
- 30.Yamada M., Sekiguchi K. Molecular basis of laminin-integrin interactions. Curr. Top. Membr. 2015;76:197–229. doi: 10.1016/bs.ctm.2015.07.002. [DOI] [PubMed] [Google Scholar]; M. Yamada, K. Sekiguchi, Molecular Basis of Laminin-Integrin Interactions, Curr Top Membr 76 (2015) 197-229. [DOI] [PubMed]
- 31.Hochman-Mendez C., Cantini M., Moratal D., Salmeron-Sanchez M., Coelho-Sampaio T. A fractal nature for polymerized laminin. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0109388. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Hochman-Mendez, M. Cantini, D. Moratal, M. Salmeron-Sanchez, T. Coelho-Sampaio, A fractal nature for polymerized laminin, PLoS One 9(10) (2014) e109388. [DOI] [PMC free article] [PubMed]
- 32.Uppu R.M., Pryor W.A. Synthesis of peroxynitrite in a two-phase system using isoamyl nitrite and hydrogen peroxide. Anal. Biochem. 1996;236:242–249. [PubMed] [Google Scholar]; R.M. Uppu, W.A. Pryor, Synthesis of peroxynitrite in a two-phase system using isoamyl nitrite and hydrogen peroxide, Anal Biochem 236 (1996) 242-249. [PubMed]
- 33.Bohle D.S., Hansert B., Paulson S.C., Smith B.D. Biomimetic synthesis of the putative cytotoxin peroxynitrite, ONOO–, and its characterization as a tetramethylammonium salt. J. Am. Chem. Soc. 1994;116:7423–7424. [Google Scholar]; D.S. Bohle, B. Hansert, S.C. Paulson, B.D. Smith, Biomimetic Synthesis of the Putative Cytotoxin Peroxynitrite, ONOO-, and its Characterization as a Tetramethylammonium Salt, J. Am. Chem. Soc. 116 (1994) 7423-7424.
- 34.Wisniowski P., Carmichael I., Fessenden R.W., Hug G.L. Evidence for b scission in the oxidation of amino acids. J. Phys. Chem. A. 2002;106:4573–4580. [Google Scholar]; P. Wisniowski, I. Carmichael, R.W. Fessenden, G.L. Hug, Evidence for b scission in the oxidation of amino acids, J. Phys. Chem. A 106 (2002) 4573-4580.
- 35.Erde J., Loo R.R., Loo J.A. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J. Proteome Res. 2014;13(4):1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Erde, R.R. Loo, J.A. Loo, Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments, J Proteome Res 13(4) (2014) 1885-1895. [DOI] [PMC free article] [PubMed]
- 36.Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]; R. Craig, R.C. Beavis, TANDEM: matching proteins with tandem mass spectra, Bioinformatics 20(9) (2004) 1466-1467. [DOI] [PubMed]
- 37.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Kim, P.A. Pevzner, MS-GF+ makes progress towards a universal database search tool for proteomics, Nat Commun 5 (2014) 5277. [DOI] [PMC free article] [PubMed]
- 38.Vaudel M., Barsnes H., Berven F.S., Sickmann A., Martens L. SearchGUI: an open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011;11(5):996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]; M. Vaudel, H. Barsnes, F.S. Berven, A. Sickmann, L. Martens, SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches, Proteomics 11(5) (2011) 996-999. [DOI] [PubMed]
- 39.Vaudel M., Burkhart J.M., Zahedi R.P., Oveland E., Berven F.S., Sickmann A., Martens L., Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015;33(1):22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]; M. Vaudel, J.M. Burkhart, R.P. Zahedi, E. Oveland, F.S. Berven, A. Sickmann, L. Martens, H. Barsnes, PeptideShaker enables reanalysis of MS-derived proteomics data sets, Nat Biotechnol 33(1) (2015) 22-24. [DOI] [PubMed]
- 40.Vaudel M., Breiter D., Beck F., Rahnenfuhrer J., Martens L., Zahedi R.P. D-score: a search engine independent MD-score. Proteomics. 2013;13(6):1036–1041. doi: 10.1002/pmic.201200408. [DOI] [PubMed] [Google Scholar]; M. Vaudel, D. Breiter, F. Beck, J. Rahnenfuhrer, L. Martens, R.P. Zahedi, D-score: a search engine independent MD-score, Proteomics 13(6) (2013) 1036-1041. [DOI] [PubMed]
- 41.Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 2011;10(12):5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]; T. Taus, T. Kocher, P. Pichler, C. Paschke, A. Schmidt, C. Henrich, K. Mechtler, Universal and confident phosphorylation site localization using phosphoRS, J Proteome Res 10(12) (2011) 5354-5362. [DOI] [PubMed]
- 42.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]; B. MacLean, D.M. Tomazela, N. Shulman, M. Chambers, G.L. Finney, B. Frewen, R. Kern, D.L. Tabb, D.C. Liebler, M.J. MacCoss, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments, Bioinformatics 26(7) (2010) 966-968. [DOI] [PMC free article] [PubMed]
- 43.Mayer U., Nischt R., Poschl E., Mann K., Fukuda K., Gerl M., Yamada Y., Timpl R. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO J. 1993;12(5):1879–1885. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; U. Mayer, R. Nischt, E. Poschl, K. Mann, K. Fukuda, M. Gerl, Y. Yamada, R. Timpl, A single EGF-like motif of laminin is responsible for high affinity nidogen binding, Embo J 12(5) (1993) 1879-1885. [DOI] [PMC free article] [PubMed]
- 44.Weisser H., Choudhary J.S. Targeted feature detection for data-dependent shotgun proteomics. J. Proteome Res. 2017;16(8):2964–2974. doi: 10.1021/acs.jproteome.7b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Weisser, J.S. Choudhary, Targeted Feature Detection for Data-Dependent Shotgun Proteomics, J Proteome Res 16(8) (2017) 2964-2974. [DOI] [PMC free article] [PubMed]
- 45.Yurchenco P.D., Cheng Y.S., Colognato H. Laminin forms an independent network in basement membranes. J. Cell Biol. 1992;117(5):1119–1133. doi: 10.1083/jcb.117.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]; P.D. Yurchenco, Y.S. Cheng, H. Colognato, Laminin forms an independent network in basement membranes, J Cell Biol 117(5) (1992) 1119-1133. [DOI] [PMC free article] [PubMed]
- 46.Odenthal U., Haehn S., Tunggal P., Merkl B., Schomburg D., Frie C., Paulsson M., Smyth N. Molecular analysis of laminin N-terminal domains mediating self-interactions. J. Biol. Chem. 2004;279(43):44504–44512. doi: 10.1074/jbc.M402455200. [DOI] [PubMed] [Google Scholar]; U. Odenthal, S. Haehn, P. Tunggal, B. Merkl, D. Schomburg, C. Frie, M. Paulsson, N. Smyth, Molecular analysis of laminin N-terminal domains mediating self-interactions, J Biol Chem 279(43) (2004) 44504-44512. [DOI] [PubMed]
- 47.Schuger L., Yurchenco P., Relan N.K., Yang Y. Laminin fragment E4 inhibition studies: basement membrane assembly and embryonic lung epithelial cell polarization requires laminin polymerization. Int. J. Dev. Biol. 1998;42(2):217–220. [PubMed] [Google Scholar]; L. Schuger, P. Yurchenco, N.K. Relan, Y. Yang, Laminin fragment E4 inhibition studies: basement membrane assembly and embryonic lung epithelial cell polarization requires laminin polymerization, Int J Dev Biol 42(2) (1998) 217-220. [PubMed]
- 48.Yang Y., Palmer K.C., Relan N., Diglio C., Schuger L. Role of laminin polymerization at the epithelial mesenchymal interface in bronchial myogenesis. Development. 1998;125(14):2621–2629. doi: 10.1242/dev.125.14.2621. [DOI] [PubMed] [Google Scholar]; Y. Yang, K.C. Palmer, N. Relan, C. Diglio, L. Schuger, Role of laminin polymerization at the epithelial mesenchymal interface in bronchial myogenesis, Development 125(14) (1998) 2621-2629. [DOI] [PubMed]
- 49.Ponce M.L., Nomizu M., Delgado M.C., Kuratomi Y., Hoffman M.P., Powell S., Yamada Y., Kleinman H.K., Malinda K.M. Identification of endothelial cell binding sites on the laminin gamma 1 chain. Circ. Res. 1999;84(6):688–694. doi: 10.1161/01.res.84.6.688. [DOI] [PubMed] [Google Scholar]; M.L. Ponce, M. Nomizu, M.C. Delgado, Y. Kuratomi, M.P. Hoffman, S. Powell, Y. Yamada, H.K. Kleinman, K.M. Malinda, Identification of endothelial cell binding sites on the laminin gamma 1 chain, Circ Res 84(6) (1999) 688-694. [DOI] [PubMed]
- 50.Bartesaghi S., Radi R. Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biol. 2018;14:618–625. doi: 10.1016/j.redox.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Bartesaghi, R. Radi, Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration, Redox Biol 14 (2018) 618-625. [DOI] [PMC free article] [PubMed]
- 51.Ischiropoulos H., Beckman J.S. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J. Clin. Investig. 2003;111(2):163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Ischiropoulos, J.S. Beckman, Oxidative stress and nitration in neurodegeneration: cause, effect, or association?, J Clin Invest 111(2) (2003) 163-169. [DOI] [PMC free article] [PubMed]
- 52.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]; H. Ischiropoulos, Biological selectivity and functional aspects of protein tyrosine nitration, Biochem. Biophys. Res. Commun. 305 (2003) 776-783. [DOI] [PubMed]
- 53.Padmaja S., Ramazenian M.S., Bounds P.L., Koppenol W.H. Reaction of peroxynitrite with L-tryptophan. Redox Rep. 1996;2(3):173–177. doi: 10.1080/13510002.1996.11747045. [DOI] [PubMed] [Google Scholar]; S. Padmaja, M.S. Ramazenian, P.L. Bounds, W.H. Koppenol, Reaction of peroxynitrite with L-tryptophan, Redox Rep 2(3) (1996) 173-177. [DOI] [PubMed]
- 54.Nakagawa H., Takusagawa M., Arima H., Furukawa K., Kinoshita T., Ozawa T., Ikota N. Selective scavenging property of the indole moiety for the nitrating species of peroxynitrite. Chem. Pharm. Bull. (Tokyo) 2004;52(1):146–149. doi: 10.1248/cpb.52.146. [DOI] [PubMed] [Google Scholar]; H. Nakagawa, M. Takusagawa, H. Arima, K. Furukawa, T. Kinoshita, T. Ozawa, N. Ikota, Selective scavenging property of the indole moiety for the nitrating species of peroxynitrite, Chem Pharm Bull (Tokyo) 52(1) (2004) 146-149. [DOI] [PubMed]
- 55.Santiveri C.M., Jimenez M.A. Tryptophan residues: scarce in proteins but strong stabilizers of beta-hairpin peptides. Biopolymers. 2010;94(6):779–790. doi: 10.1002/bip.21436. [DOI] [PubMed] [Google Scholar]; C.M. Santiveri, M.A. Jimenez, Tryptophan residues: scarce in proteins but strong stabilizers of beta-hairpin peptides, Biopolymers 94(6) (2010) 779-790. [DOI] [PubMed]
- 56.Yamakura F., Ikeda K. Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species. Nitric Oxide. 2006;14(2):152–161. doi: 10.1016/j.niox.2005.07.009. [DOI] [PubMed] [Google Scholar]; F. Yamakura, K. Ikeda, Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species, Nitric Oxide 14(2) (2006) 152-161. [DOI] [PubMed]
- 57.Sanz L., Garcia-Bermejo L., Blanco F.J., Kristensen P., Feijoo M., Suarez E., Blanco B., Alvarez-Vallina L. A novel cell binding site in the coiled-coil domain of laminin involved in capillary morphogenesis. EMBO J. 2003;22(7):1508–1517. doi: 10.1093/emboj/cdg150. [DOI] [PMC free article] [PubMed] [Google Scholar]; L. Sanz, L. Garcia-Bermejo, F.J. Blanco, P. Kristensen, M. Feijoo, E. Suarez, B. Blanco, L. Alvarez-Vallina, A novel cell binding site in the coiled-coil domain of laminin involved in capillary morphogenesis, Embo J 22(7) (2003) 1508-1517. [DOI] [PMC free article] [PubMed]
- 58.Santos-Valle P., Guijarro-Munoz I., Cuesta A.M., Alonso-Camino V., Villate M., Alvarez-Cienfuegos A., Blanco F.J., Sanz L., Alvarez-Vallina L. The heterotrimeric laminin coiled-coil domain exerts anti-adhesive effects and induces a pro-invasive phenotype. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039097. [DOI] [PMC free article] [PubMed] [Google Scholar]; P. Santos-Valle, I. Guijarro-Munoz, A.M. Cuesta, V. Alonso-Camino, M. Villate, A. Alvarez-Cienfuegos, F.J. Blanco, L. Sanz, L. Alvarez-Vallina, The heterotrimeric laminin coiled-coil domain exerts anti-adhesive effects and induces a pro-invasive phenotype, PLoS One 7(6) (2012) e39097. [DOI] [PMC free article] [PubMed]
- 59.Serrano-Luginbuehl S., Kissner R., Koppenol W.H. Reaction of CO2 with ONOO(-): one molecule of CO2 is not enough. Chem. Res. Toxicol. 2018;31(8):721–730. doi: 10.1021/acs.chemrestox.8b00068. [DOI] [PubMed] [Google Scholar]; S. Serrano-Luginbuehl, R. Kissner, W.H. Koppenol, Reaction of CO2 with ONOO(-): One Molecule of CO2 Is Not Enough, Chem Res Toxicol 31(8) (2018) 721-730. [DOI] [PubMed]
- 60.Buxton G.V., Greenstock C.L., Helman W.P., R.A.B. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms, and hydroxyl radicals (.OH/.O-) in aqueous solution. J. Phys. Chem. Ref. Data. 1988;17:513–886. [Google Scholar]; G.V. Buxton, C.L. Greenstock, W.P. Helman, R. A.B., Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms, and hydroxyl radicals (.OH/.O-) in aqueous solution, J. Phys. Chem. Ref. Dafa 17 (1988) 513-886.
- 61.Chen S.N., Hoffman M.Z. Rate constants for the reaction of the carbonate radical with compounds of biochemical interest in neutral aqueous solution. Radiat. Res. 1973;56(1):40–47. [PubMed] [Google Scholar]; S.N. Chen, M.Z. Hoffman, Rate constants for the reaction of the carbonate radical with compounds of biochemical interest in neutral aqueous solution, Radiat Res 56(1) (1973) 40-47. [PubMed]
- 62.Carafoli F., Hussain S.A., Hohenester E. Crystal structures of the network-forming short-arm tips of the laminin beta1 and gamma1 chains. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0042473. [DOI] [PMC free article] [PubMed] [Google Scholar]; F. Carafoli, S.A. Hussain, E. Hohenester, Crystal structures of the network-forming short-arm tips of the laminin beta1 and gamma1 chains, PLoS One 7(7) (2012) e42473. [DOI] [PMC free article] [PubMed]
- 63.Boateng S.Y., Lateef S.S., Mosley W., Hartman T.J., Hanley L., Russell B. RGD and YIGSR synthetic peptides facilitate cellular adhesion identical to that of laminin and fibronectin but alter the physiology of neonatal cardiac myocytes. Am. J. Physiol. Cell Physiol. 2005;288(1):C30–C38. doi: 10.1152/ajpcell.00199.2004. [DOI] [PubMed] [Google Scholar]; S.Y. Boateng, S.S. Lateef, W. Mosley, T.J. Hartman, L. Hanley, B. Russell, RGD and YIGSR synthetic peptides facilitate cellular adhesion identical to that of laminin and fibronectin but alter the physiology of neonatal cardiac myocytes, Am J Physiol Cell Physiol 288(1) (2005) C30-C38. [DOI] [PubMed]
- 64.Ido H., Nakamura A., Kobayashi R., Ito S., Li S., Futaki S., Sekiguchi K. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin gamma chains in integrin binding by laminins. J. Biol. Chem. 2007;282(15):11144–11154. doi: 10.1074/jbc.M609402200. [DOI] [PubMed] [Google Scholar]; H. Ido, A. Nakamura, R. Kobayashi, S. Ito, S. Li, S. Futaki, K. Sekiguchi, The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin gamma chains in integrin binding by laminins, J Biol Chem 282(15) (2007) 11144-11154. [DOI] [PubMed]
- 65.Pulido D., Hussain S.A., Hohenester E. Crystal structure of the heterotrimeric integrin-binding region of laminin-111. Structure. 2017;25(3):530–535. doi: 10.1016/j.str.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; D. Pulido, S.A. Hussain, E. Hohenester, Crystal Structure of the Heterotrimeric Integrin-Binding Region of Laminin-111, Structure 25(3) (2017) 530-535. [DOI] [PMC free article] [PubMed]
- 66.Yurchenco P.D., Cheng Y.S. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J. Biol. Chem. 1993;268(23):17286–17299. [PubMed] [Google Scholar]; P.D. Yurchenco, Y.S. Cheng, Self-assembly and calcium-binding sites in laminin. A three-arm interaction model, J Biol Chem 268(23) (1993) 17286-17299. [PubMed]
- 67.Barroso M.M., Freire E., Limaverde G.S., Rocha G.M., Batista E.J., Weissmuller G., Andrade L.R., Coelho-Sampaio T. Artificial laminin polymers assembled in acidic pH mimic basement membrane organization. J. Biol. Chem. 2008;283(17):11714–11720. doi: 10.1074/jbc.M709301200. [DOI] [PubMed] [Google Scholar]; M.M. Barroso, E. Freire, G.S. Limaverde, G.M. Rocha, E.J. Batista, G. Weissmuller, L.R. Andrade, T. Coelho-Sampaio, Artificial laminin polymers assembled in acidic pH mimic basement membrane organization, J Biol Chem 283(17) (2008) 11714-11720. [DOI] [PubMed]
- 68.Nybo T., Dieterich S., Gamon L.F., Chuang C.Y., Hammer A., Hoefler G., Malle E., Rogowska-Wrzesinska A., Davies M.J. Chlorination and oxidation of the extracellular matrix protein laminin and basement membrane extracts by hypochlorous acid and myeloperoxidase. Redox Biol. 2019;20:496–513. doi: 10.1016/j.redox.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; T. Nybo, S. Dieterich, L.F. Gamon, C.Y. Chuang, A. Hammer, G. Hoefler, E. Malle, A. Rogowska-Wrzesinska, M.J. Davies, Chlorination and oxidation of the extracellular matrix protein laminin and basement membrane extracts by hypochlorous acid and myeloperoxidase, Redox Biol. 20 (2019) 496-513. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.