Abstract
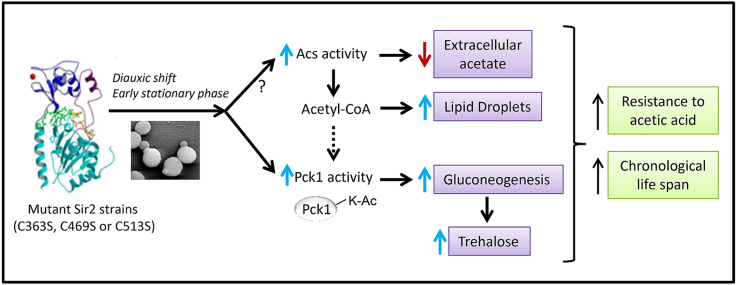
Yeast Sir2 is an NAD-dependent histone deacetylase related to oxidative stress and aging. In a previous study, we showed that Sir2 is regulated by S-glutathionylation of key cysteine residues located at the catalytic domain. Mutation of these residues results in strains with increased resistance to disulfide stress. In the present study, these mutant cells were highly resistant to acetic acid and had an increased chronological life span. Mutant cells had increased acetyl-CoA synthetase activity, which converts acetic acid generated by yeast metabolism to acetyl.CoA. This could explain the acetic acid resistance and lower levels of this toxic acid in the extracellular media during aging. Increased acetyl-CoA levels would raise lipid droplets, a source of energy during aging, and fuel glyoxylate-dependent gluconeogenesis. The key enzyme of this pathway, phosphoenolpyruvate carboxykinase (Pck1), showed increased activity in these Sir2 mutant cells during aging. Sir2 activity decreased when cells shifted to the diauxic phase in the mutant strains, compared to the WT strain. Since Pck1 is inactivated through Sir2-dependent deacetylation, the decline in Sir2 activity explained the rise in Pck1 activity. As a consequence, storage of sugars such as trehalose would increase. We conclude that extended longevity observed in the mutants was a combination of increased lipid droplets and trehalose, and decreased acetic acid in the extracellular media. These results offer a deeper understanding of the redox regulation of Sir2 in acetic acid resistance, which is relevant in some food and industrial biotechnology and also in the metabolism associated to calorie restriction, aging and pathologies such as diabetes.
Keywords: Sirtuin, Redox regulation, Aging, Acetic acid, Metabolism, Yeast
Abbreviations: Acs, acetyl-CoA synthetase; LD, Lipid droplets; Pck1, phosphoenolpyruvate carboxykinase; ROS, reactive oxygen species; Sod, superoxide dismutase
Graphical abstract
Highlights
-
•
Mutation of key redox-regulated Cys from yeast Sir2 increases chronological life span.
-
•
Sir2 mutants are highly resistant to acetic acid stress compared to the WT strain.
-
•
Increased Acs activity in Sir2 mutants could justify acetic acid resistance.
-
•
Sir2 mutants show higher Pck1 activity and trehalose stores.
-
•
Low extracellular acetic acid and high trehalose extends longevity in Sir2 mutants.
1. Introduction
Under conditions of rich media and high glucose concentration, S. cerevisiae ferments glucose to ethanol, also producing acetic acid. The ethanol and the acetic acid excreted to the medium inhibit the growth of other microorganisms. When glucose becomes limiting, yeasts enter the diauxic shift and metabolism changes from glucose fermentation to ethanol respiration, which has been accumulating during the fermentative phase. Finally, when all carbon sources have been exhausted, cells enter the stationary phase (G0). In the course of the diauxic shift, and also at the stationary phase, reactive oxygen species (ROS) generated by aerobic metabolism cause oxidative damage to cell components such as proteins, DNA, and lipids. However, ROS may also act in signaling events and culminate in adaptive responses. In proteins, one of the most reactive groups of amino acid side chains is thiols (-SH) from cysteine residues. Different oxidants can react with thiols to yield a variety of products [1] that can be detected by proteomic approaches [2]. They include i) sulfenic acid (-SOH), which can react to other thiols to generate mixed disulfides with glutathione (Prot-S-SG), also known as S-glutathionylation, ii) intramolecular disulfides (-S-S-), and iii) intermolecular disulfides between proteins. Sulfenic acid may become further oxidized to sulfinic (-SO2H) or sulfonic (-SO3H) acids in the presence of stronger oxidants or prolonged exposure to mild oxidants. When reversible, post-translational modifications of reactive cysteine residues of proteins may act as a regulatory mechanism [3,4]. The thioredoxin and the glutaredoxin systems reduce disulfide bonds, and both systems require NADPH as the final electron donor.
Sirtuins are NAD+-dependent histone deacetylases present from bacteria to mammals, and yeast Sir2 was the first sirtuin described [5]. In humans, there are seven mammalian Sir2 orthologues (Sirt1–Sirt7); the biochemical activities of Sirt1 are the most similar to Sir2. Sirt1 participates in a great variety of processes. Directly or indirectly, Sirt1 regulates insulin secretion, gluconeogenesis, circadian rhythms, adiponectin production, and oxidative stress [6]. Sirt1 has also been involved in different pathologies, including cancer and neurodegenerative diseases [7]. Both selective Sirt1 activators and inhibitors are available, and initial clinical trials have been carried out (reviewed in Ref. [8]). Yeast Sir2 is involved in important functions such as silencing of telomeres, rDNA and the mating type loci, maintaining genome integrity and aging [9,10]. In mammals, the role of Sirt1 in antioxidant defense has been described as dependent on FoxO3a deacetylation [11,12]. Yeast cells have four forkhead transcription factors, three (Hcm1, Fkh1, Fkh2) of which have been related to Sir2 upon oxidative stress [[13], [14], [15]]. Up-regulation of yeast Sir2 has been observed upon exogenous oxidative stress [16,17] or endogenously generated stress [18]. Thus, the relationship of sirtuins with stress resistance is well known. In this context, it has been described that the cellular redox status modulates Sirt1 activity [17,19]. In mammalian [20,21] and zebrafish [22] Sirt1, several cysteines have been identified as S-glutathionylation targets, mostly located in the catalytic domain and near the binding site of the cofactor NAD+. Although key cysteine residues involved in these processes have been identified, none of them is conserved in yeast Sir2.
Recently, we showed yeast Sir2 is S-glutathionylated upon strong disulfide stress [23]. Monothiol glutaredoxins Grx3 and Grx4 are able to S-deglutathionylate it, acting as the physiological regulators of Sir2. Three key cysteines (C363, C469 and C513) located at the catalytic domain have been identified as S-glutathionylation targets. Such post-translational modification results in enzyme inactivation, as is described in zebrafish [22], and mammalian Sirt1 [20,21]. Nevertheless, redox modification of key cysteine residues has been described activating many proteins. Among them are transcription factors like oxyR from E. coli [24], Yap1 from S. cerevisiae [25], Pap1 from S. pombe [26], or enzymes such as bacterial Hsp33 [27] and typical 2-Cys peroxiredoxins [28].
Like mammalian Sirt1, yeast Sir2 is described as deacetylating not only histones, but proteins involved in metabolism and related to aging. Sir2 deacetylates, both in vitro and in vivo, phosphoenolpyruvate carboxykinase (Pck1), a key enzyme in the gluconeogenesis pathway, resulting in the loss of its catalytic activity [29,30]. Acetylation of Lys514 is essential for enzyme activity and the ability of cells to grow in nonfermentable carbon sources [29]. Pck1 inactivation by Sir2 decreases intracellular glucose stores, like trehalose and glycogen, which are necessary for chronological longevity. In humans, activation of Pck1 through acetylation has also been described during fasting and is related to increased gluconeogenesis and longevity [29].
Previously, we showed that Sir2 mutant strains lacking one of three cysteine residues identified as targets of redox regulation are more resistant to strong diamide stress [23]. In the present study, these strains were also more resistant to acetic acid stress and showed increased chronological life span. Our new results may help to elucidate the role of redox-regulated cysteines of Sir2 in the metabolic changes that could explain such phenotypes.
2. Materials and methods
2.1. Yeast strains, genetic methods and culture conditions
Yeast strains used in this study are listed in Table 1. All the mutants are derived from the strain CML128 (MATa ura3-52 his4 leu2-3,112 trp1) considered the wild-type (WT) strain in this work [31]. Standard protocols were used for DNA manipulations and transformation of yeast cells. Strains Sir2-C363S, Sir2-C469S and Sir2-C513S, generated by site-directed mutagenesis of Sir2 cysteine residues [23] were also used. To analyze intracellular pH, WT, C363S, C469S and C513S strains were transformed with pYES-PACT1-pHluorin (URA3) plasmid, which contains the pHluorin (a pH-sensitive variant of the green fluorescent protein) open reading frame, under the control of the constitutive S. cerevisiae actin (ACT1) gene promoter and the URA3 gene as a selectable marker [5,6]. The plasmid pYES-PACT1-pHluorin (URA3) was kindly provided by Dr. Ariño (Universitat Autònoma de Barcelona, Spain). A selection of transformants was performed on solid synthetic medium, and expression of the pHluorin gene in the selected transformants was checked by fluorescence microscopy. Plasmid pDM312 (RDN1–NTS2::mURA3–LEU2) was a gift from Dr. Moazed at Harvard Medical School [32]. mURA3 silencing strains were generated by transformation with pDM312 cut with SmaI to integrate at NTS2. All transformations were performed with the lithium acetate method [33], and correct integration was confirmed by PCR. Cells were grown at 30 °C by incubation in a rotary shaker using SC medium (0.67% yeast nitrogen base, 2% glucose plus dropout mixture, and auxotrophic requirements). Specific supplements were omitted for selection of the corresponding plasmid-carrying cells. When indicated, YPD medium containing 2% glucose (1% yeast extract, 2% peptone, 2% glucose), YPD medium containing 0.5% glucose (1% yeast extract, 2% peptone, 0.5% glucose) or YPG medium (1% yeast extract, 2% peptone and 3% glycerol) were also used. Diamide, acetic acid, sorbic acid, benzoic acid and sodium chloride were purchased from Sigma. Exponentially growing cells were obtained at OD600 = 0.5–1 (day 0); day 1 cells were obtained 24 h after.
Table 1.
Yeast strains used in this work.
| Strain | Relevant genotype | Source |
|---|---|---|
| CML128 | MATa ura3-52 his4 leu2-3,112 trp1 | [31] |
| NVLL4 | CML128 sir2::natMX4 | [23] |
| NVLL5 | NVLL4 [pNVLL2(SIR2 C363S)]::LEU2 | [23] |
| NVLL6 | NVLL4 [pNVLL3(SIR2 C469S)]::LEU2 | [23] |
| NVLL7 | NVLL4 [pNVLL4(SIR2 C513S)]::LEU2 | [23] |
| BQS281 | CML128 pYES-PACT1 pHluorin::URA3 | This work |
| BQS283 | NVLL5 pYES-PACT1 pHluorin::URA3 | This work |
| BQS284 | NVLL6 pYES-PACT1 pHluorin::URA3 | This work |
| BQS285 | NVLL7 pYES-PACT1 pHluorin::URA3 | This work |
| BQS304 | CML128 RDN1-NTS2::mURA3 | This work |
| BQS305 | NVLL5 RDN1-NTS2::mURA3 | This work |
| BQS306 | NVLL6 RDN1-NTS2::mURA3 | This work |
| BQS307 | NVLL7 RDN1-NTS2::mURA3 | This work |
| BQS308 | NVLL4 RDN1-NTS2::mURA3 | This work |
2.2. pHluorin calibration and pHluorin measurements
Strains transformed with the plasmid pYES-PACT1-pHluorin were grown to an OD600 of 0.5–0.75 in SC medium, centrifuged and resuspended in PBS containing 0.1 M digitonin (Sigma), which permeabilizes cell membranes. After 10 min, cells were washed with PBS at 4 °C and resuspended in citric acid/Na2HPO4 buffer of pH values ranging from 5.0 to 8.0. Cell fluorescence was measured using a fluorimeter (Shimadzu RF-5000). pHluorin fluorescence emission was measured at 508 nm providing excitation bands of 395 nm and 475 nm [34,35]. A calibration curve was generated plotting the ratio of emission intensity resulting from excitation at 395 and 475 nm (395/415 ratio) against the corresponding buffer pH [35]. Background fluorescence for a WT culture not expressing pHluorin was subtracted from the measurements. All pH determination experiments were repeated, at least, three times (biological replicates). To analyze changes in intracellular pH upon acetic acid stress, strains were grown to an OD600 of 0.5 in SC medium, centrifuged and resuspended in SC medium plus the indicated amount of acetic acid (pH adjusted to 4.5). After 5 and 10 min, fluorescence emission (395/415 ratio) was measured and converted to intracellular pH according to the calibration curve previously obtained.
2.3. In vivo Sir2 silencing activity
In vivo Sir2 telomeric silencing activity was performed by quantifying transcriptional silencing at the naturally occurring telomere linked gene, YFR057W [36]. Sir2-dependent YFR057W gene expression was measured by quantitative real-time PCR using the TaqMan System (Applied Biosystems). Total RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer's recommendations. First-strand cDNA synthesis was performed on 1 μg DNase-treated RNA using the SuperScript IV First-Strand Synthesis System (Invitrogen), and 50 ng of cDNA was used for each individual real-time PCR assay in a 48-cycle, three-step PCR reaction using the iCycler (Bio-Rad). Amplification was performed using the TaqMan Universal PCR Master Mix kit (Applied Biosystems). Quantification was completed using iCycler IQ Real-Time detection system software. Actin (ACT1) was used as an internal control. Relative expression ratios were calculated based on ΔCq values with efficiency correction based on multiple samples [37]. Data represent three technical repeats of each analysis from, at least, three different experiments.
In vivo Sir2 ribosomal DNA (rDNA) silencing was analyzed using strains transformed with plasmid pDM312 (RDN1–NTS2::mURA3–LEU2). Silencing of NTS2 reporter within rDNA was assessed by monitoring the growth of 5-fold serial dilutions of cells plated on Ura-depleted SC medium (- Ura). Ura-containing SC medium (+Ura) was used as plating control.
2.4. Enzyme activities
Cells were grown to the desired OD600 and centrifuged, and cell extracts were obtained with a Mini-Bead beater cell disrupter (Biospec) using glass beads. All activity assays were measured at 25 °C. Catalase activity was measured by H2O2 consumption at 240 nm [38]. Alcohol dehydrogenase was measured spectrophotometrically by NADH formation [39]. Citrate synthase was measured in a coupled assay to reduce 5,5-dithiobis-(2-nitrobenzoic acid) [40]. Glutathione reductase was performed by measuring NADPH consumption at 340 nm [41]. Acetyl CoA synthetase activity was measured using an enzymatic method coupled with malate dehydrogenase and citrate synthase [42]. Phosphoenolpyruvate carboxykinase activity was measured in the OAA formation direction coupled with malate dehydrogenase as described [43]. Superoxide dismutase (SOD) activities were assayed in a native gel by chromogenic staining (zymogram) as described [44], with modifications [13]. In brief, cells were homogenized in 50 mM Tris-HCl buffer, pH 8.5 and after centrifugation, 5% (w/v) saccharose and 0.05% (w/v) Bromophenol blue were added. 25 μg of cell extract were applied directly to a nondenaturing 10% polyacrylamide gel. Following electrophoresis, the gel was stained in a solution containing 50 mM Tris-HCl buffer, pH 7.8, 125 μg/ml thiazolyl blue tetrazolium bromide, 75 μg/ml phenazine methosulfate, and 188 μg/ml MgCl2. The blue tetrazolium stain for O2− was developed by exposure to light. Staining was absent at sites of O2− scavenging.
2.5. Microscopic analysis of lipid droplets
Lipid droplets were visualized by fluorescence microscopy using BODIPY™ 493/503 (Invitrogen, Inc., USA) prepared as a 10 mM stock solution in DMSO and kept at −80 °C [45]. BODIPY 493/503 is a green emitting fluorophore for high-contrast labeling of yeast lipid droplets and shows very high quantum yield and photostability [46]. Aliquots of 5 μl of cellular suspension and 5 μl of 10 mM BODIPY 493/503 were mounted onto a polylysine-treated glass slide. Microscopy was performed on Olympus DP30BW using a 60x oil immersion objective.
2.6. Other analyses
To measure resistance to stress, both cell growth and viability were determined under control conditions or with the stressing agent added to the media at the indicated concentrations. Cell growth was monitored in 0.5 ml cultures in 24-well plates incubated at the indicated temperature and constant agitation in a PowerWave XS microplate spectrophotometer (Biotek). Plates were sealed with Breathe Easy membranes (Diversified Biotech, USA). Optical density (OD600 nm) was recorded every 30 min for 20_25 h and the values were analyzed with Gen 5 1.06 software. Total iron quantitation from yeast cells was performed following the method previously described [47]. Intracellular glutathione (GSH) levels were measured with the ThiolTracker™ Violet fluorescent thiol probe (Invitrogen) following the manufacturer's recommendations. Cell extracts were obtained as previously described [48], separated in SDS-polyacrylamide gels, and transferred to polyvinylidene difluoride membranes. Western blot analysis was performed by standard methods and the anti-Sir2 antibody (Santa Cruz Biotechnology) was used. Images were acquired in a ChemiDoc XRS System (Bio-Rad) and analyzed with Quantity One software (Bio-Rad). Ethanol from the extracellular medium was measured spectrophotometrically at 340 nm using the Enzytec™ fluid ethanol kit (Thermo Fisher Scientific). Acetic acid from the extracellular medium was quantified spectrophotometrically at 340 nm using the Acetic Acid kit from Megazyme. Intracellular trehalose was measured according to the Trehalose assay kit from Megazyme.
2.7. Statistical analysis
Data are presented as means ± SD from, at least, three independent experiments. Statistical analysis was performed using two-tailed Student t-test. The P-values lower than 0.05(*), 0.01(**) or 0.005 (***) were considered significant.
3. Results
3.1. Lack of redox-regulated Sir2 cysteines increases longevity and acetic acid resistance
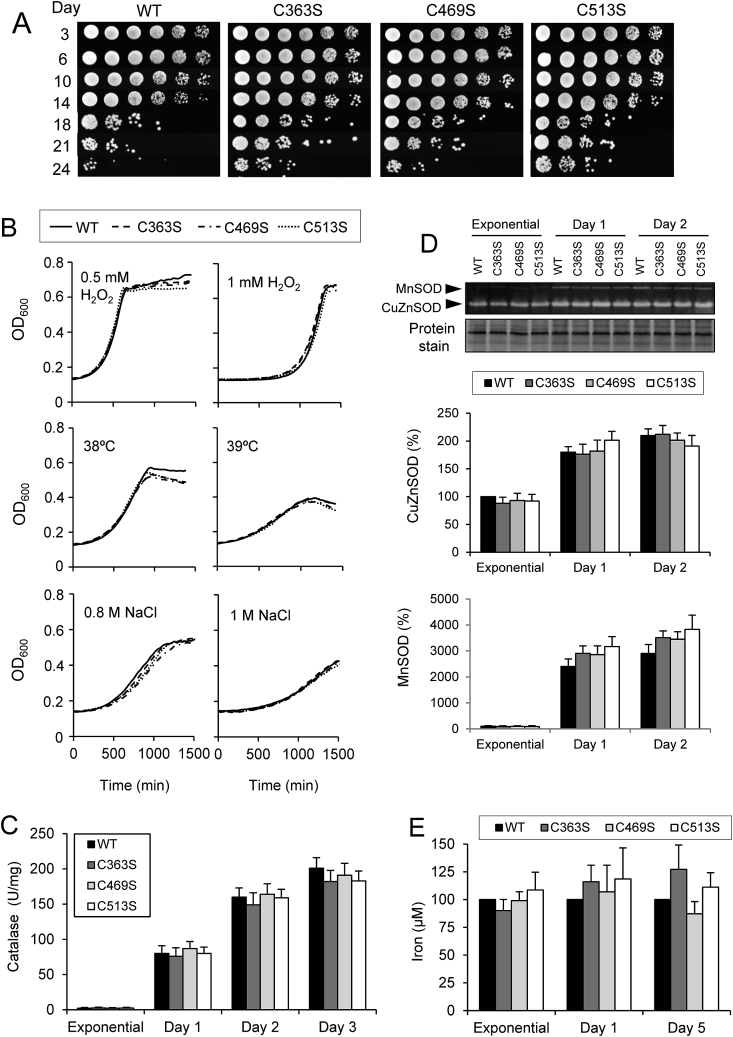
In a previous study [23], we have reported that yeast Sir2 activity is post-translationally regulated by reversible S-glutathionylation, with monothiol Grx3 and Grx4 acting as the physiological thiol-reductases. Our study also provides evidence that the redox state of three key cysteine residues of Sir2 (C363, C469 and C513), located at the catalytic domain (Supplementary Figs. 1A–C), influences yeast resistance to disulfide stresses (diamide and diethyl maleate). To more deeply explore the physiological role of such Sir2 cysteine residues, mutant strains in which one of these cysteine residues was mutated to serine (C363S, C469S and C513S) were grown in SC medium and survival along stationary phase (chronological life span) was analyzed and quantified (Fig. 1A). We observed a significantly increased survival in the mutant strains compared with the WT strain. After 24 days, only ∼0.01% of cells survived in the WT strain in comparison with ∼0.1% in C469S and ∼0.5–1% in both C363S and C513S strains. Such increased longevity may be due to a variety of factors, including increased resistance to several stresses. Although mutant cells, expressing physiological levels of Sir2, were more resistant to high disulfide stress [23], no growth differences were detected when H2O2 was used as stressing agent, heat shock or osmotic stress (Fig. 1B). In this context, several activities related to resistance to oxidative stress were also measured. As shown, no significant differences in catalase (Fig. 1C), superoxide dismutases (Sod 1 and Sod 2) (Fig. 1D), or intracellular iron were detected (Fig. 1E). Similarly, glutathione reductase activity (Supplementary Fig. 2A) and levels of glutathione (GSH) (Supplementary Fig. 2B), two important parameters under oxidative stress, showed similar levels between mutant and WT cells.
Fig. 1.
Increased chronological life span of mutant Sir2 strains is not due to higher antioxidant capacity. Cultures of WT, C363S, C469S and C513S cells were grown in SC medium. (A) Viability was measured during chronological aging by plating serial dilutions (1:5) in YPD plates. (B) Growth curves were measured upon oxidative stress (0.5 mM or 1 mM H2O2), heat stress (38 °C or 39 °C) and osmotic stress (0.8 M or 1 M NaCl). The data are the averages of two independent experiments with two replicas each. (C) Catalase activity was analyzed in whole extracts from the exponential phase, days 1, 2 and 3 of culture. (D) Cell lysates from cultures at exponential phase, days 1 and 2 were analyzed by native electrophoresis and SOD staining. Total Coomassie blue protein staining was used as a loading control. Bands corresponding to MnSOD and Cu/ZnSOD activities were measured with a densitometer, and the relative intensity was calculated. (E) Cultures grown at exponential phase, days 1 and 5 were digested with nitric acid and total iron indicated with respect to the exponential WT strain. From C to E, statistical analysis was performed comparing mutant with WT cells. The data are represented as the means ± SD from at least three independent experiments.
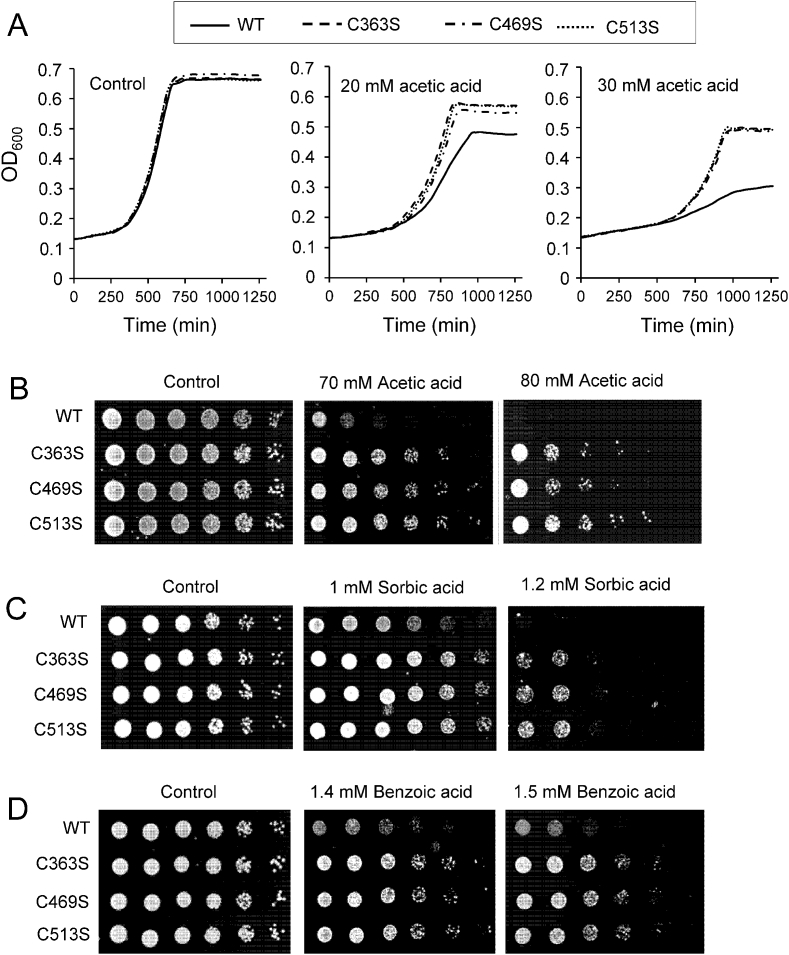
It has also been described that resistance to acetic acid correlates with longevity [[49], [50], [51]] although this is not always fulfilled [52]. Thus, to analyze acetic acid tolerance, WT and mutant strains were grown in SC medium without (control) or with acetic acid addition at the indicated concentration. In all conditions, pH was adjusted to 4.5 (the normal pH of SC medium), and cell growth measured for 20 h. As shown in Fig. 2A, all three mutant (C363S, C469S and C513S) strains showed an improved response upon acetic acid treatment compared with WT, but no significant differences were observed under control conditions. Increased survival of mutant cells, compared with WT, was also observed when cells were plated in media containing acetic acid (Fig. 2B).
Fig. 2.
Mutant Sir2 strains show increased acetic acid tolerance. Cultures of WT, C363S, C469S and C513S cells were grown in SC medium. (A) Growth curves of WT, C363S, C469S, and C513S strains were measured when grown in SC medium without or with 20 or 30 mM acetic acid. The data are the averages of two independent experiments with two replicas each. (C) The viability of WT, C363S, C469S and C513S cells grown in SC medium was measured by plating serial dilutions (1:5) in SC plates containing 70 or 80 mM acetic acid.
We determined viability to other weak organic acids, such as benzoic and sorbic acid, which are used as food preservatives to inhibit the growth of spoilage yeasts, including S. cerevisiae [53]. Again, our three mutant strains were more resistant to such acids (Fig. 2C and D).
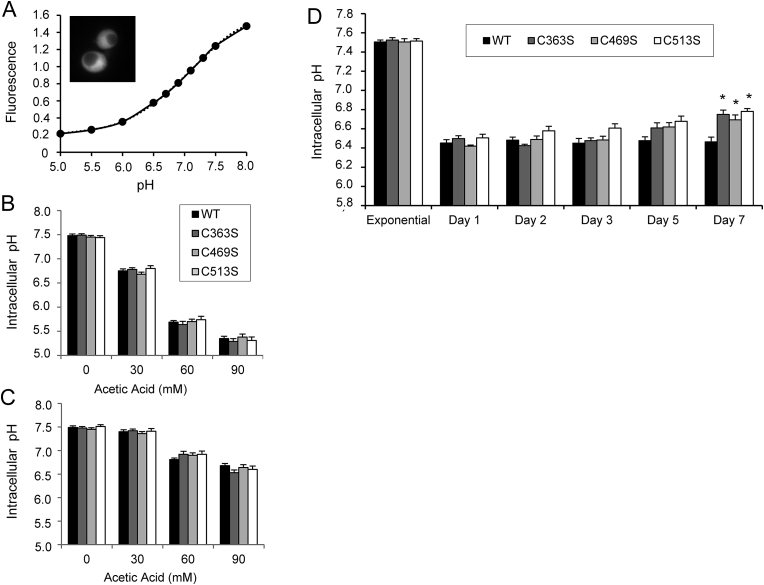
3.2. Acetic acid tolerance in Sir2 mutant strains is not due to changes in intracellular pH
Although the adaptive response and resistance to these weak organic acids are complex [54,55], it has been published that the capacity of yeast cells to maintain cytosolic pH (pHc) plays a key role in acetic acid tolerance [56]. To investigate whether an alteration occurs in the pHc of mutant strains before and after exposure to the acid, WT and all three mutant strains were transformed with pYES-PACT1-pHluorin plasmid, carrying a pH-sensitive variant of the green fluorescent protein [34,35]. A calibration curve was performed (Fig. 3A) to analyze the effects of increasing acetic acid concentrations on pHc. Fig. 3B shows the pHc values of cells 5 min after exposure to medium containing 0, 30, 60 or 90 mM acetic acid. Non-stressed cells showed no differences among strains (pHc ∼ 7.5). After 30, 60 or 90 mM acetic acid, pHc decreased to ∼6.8, 5.7 and 5.3 respectively, but no significant differences were observed among strains (Fig. 3B). Cells were able to recover rapidly, and after 10 min, cells treated with 30 mM acetic acid showed the same pHc as untreated cells. Cells treated with 60 and 90 mM showed a pHc of 6.9 and 6.6, respectively (Fig. 3C). Again, no significant differences between strains were observed during recovery. These results indicated that in exponentially growing cells, acetic acid tolerance is not due to the capacity to maintain and/or recover pHc upon acute acid stress.
Fig. 3.
Intracellular pH shift upon exposure to different concentrations of acetic acid and in chronologically aged cells. WT, C363S, C469S and C513S cells expressing pHluorin were cultivated in SC medium. (A) Calibration curve performed with WT cells expressing pHluorin grown in SC medium to exponential phase. The results are the average of two independent experiments with almost identical results. (B) pHc drop upon exposure to different concentrations of acetic acid for 5 min. (C) Recovery of pHc after 10 min upon exposure to different concentrations of acetic acid. (D) Shift in the pHc in chronologically aged cells at different days of culture. In all cases, statistical analysis was performed comparing mutant with WT cells. From B to D, the data are represented as the means ± SD from, at least, three independent experiments.
3.3. Sir2 mutants show increased acetyl-CoA synthetase activity and lipid droplets during/along aging
Using cells transformed with pHluorin, we analyzed pHc throughout chronological aging. As shown in Fig. 3D, pHc dropped abruptly from 7.5 at the exponential phase to 6.5 at the diauxic shift (day 1). This slightly acidic intracellular pH was maintained for a few days; however, at day 5 and statistically significant at day 7, mutant cells showed an increased pHc compared with WT cells. We were unable to follow this experiment beyond day 7 because cells started to accumulate lipofuscin, a well-known marker of aged cells with a broad spectrum of autofluorescence [57,58].
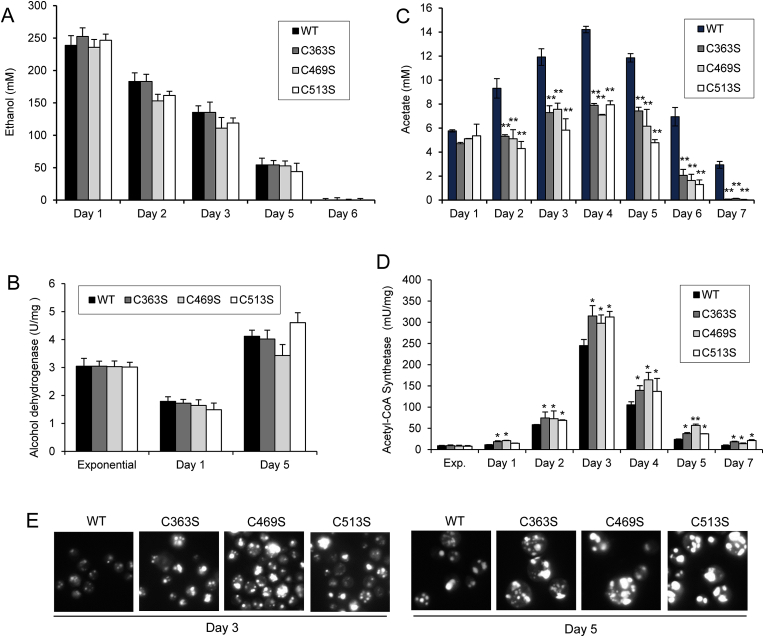
To understand such increased pHc in the mutant cells along aging, we analyzed the metabolic pathways that could be involved. Ethanol and acetate are two well-known by-products of yeast glucose fermentation that are transiently accumulated in the culture medium. In fact, after the diauxic shift, these C2 compounds are used as substrates of a respiration-based metabolism that, in addition to functional mitochondria, requires the involvement of the glyoxylate/gluconeogenesis pathways. As expected, levels of ethanol in the culture medium were very high on day 1 (diauxic shift) and decreased to undetectable levels after 6 days of culture (Fig. 4A). No significant differences between WT and Sir2 mutant strains were detected. Ethanol levels are the result of the activities of alcohol dehydrogenase (Adh) 1 and 2. At exponential phase, Adh activity is mainly due to the fermentative Adh 1 enzyme, which converts acetaldehyde to ethanol. When glucose decreases, ADH1 is repressed and ADH2 induced. Adh2 activity catalyzes the opposite reaction, which is necessary for ethanol respiratory metabolism. Alcohol dehydrogenase activity showed no differences between strains along aging (Fig. 4B), which agress with the similar levels of ethanol detected.
Fig. 4.
Ethanol and acetate metabolism in chronologically aged cells. Cultures of WT, C363S, C469S and C513S cells were grown in SC medium and different parameters measured at the indicated days of growth. (A) Ethanol levels in the extracellular media. (B) Alcohol dehydrogenase activity from cell extracts. (C) Acetic acid levels in the extracellular media. (D) Acetyl-CoA synthetase activity from cell extracts. From A to D, statistical analyses were performed comparing Sir2 mutants with WT cells. The data are represented as the means ± SD from at least, three independent experiments. (E) Representative images of lipid droplets by fluorescence microscopy are shown.
On the contrary, acetate levels in the culture medium were lower in the Sir2 mutant strains, compared with the WT strain, being statistically significant from day 2 until day 7, when levels were almost undetectable in the mutant cells (Fig. 4C). To understand why Sir2 mutants displayed this phenotype, we measured acetyl-CoA synthetase (Acs) activity along aging. This enzyme is responsible for generating acetyl-CoA from the acetate previously excreted to the culture medium. As shown in Fig. 4D, Acs activity was higher in Sir2 mutant cells compared with WT cells. In the exponential phase, Acs activity was very low, and no significant differences were observed among strains (not shown).
Lipids droplets (LD), also referred to as lipid bodies, function primarily as organelles for lipid storage, although they have been described as highly dynamic organelles [46]. The neutral lipids (triacylglycerol and cholesteryl esters) stored in LD are used as energy reserve and require acetyl-CoA for their production. We visualized LD using BODIPY 493/503 at days 3 and 5 of cell culture (Fig. 4E). The results showed increased lipid accumulation in the mutant cells, which agrees with their higher Acs activity.
3.4. Chronologically-aged mutant cells have increased phosphoenolpyruvate carboxykinase activity and trehalose stores
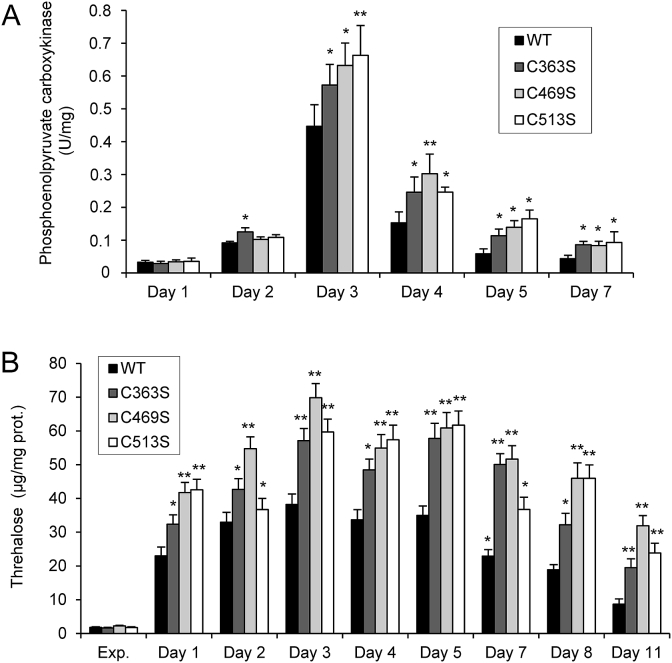
At the stationary phase, the storage of sugar as glycogen and trehalose is important for survival. After the diauxic shift, acetyl-CoA obtained from acetate and ethanol is used to fuel the glyoxylate and TCA cycles. During growth on C2 compounds, the glyoxylate cycle plays a key role for anaplerosis of oxaloacetate, which is the substrate of phosphoenolpyruvate carboxykinase (Pck1), the most important gluconeogenic enzyme. Fig. 5 shows significantly increased Pck1 activity in the mutant strains during chronological aging, compared with the WT strain. In line with an upregulation of gluconeogenesis, increased intracellular trehalose stores were consistently detected in the mutant cells (Fig. 5B).
Fig. 5.
Gluconeogenesis is increased in chronologically aged Sir2 mutant cells. Cultures of WT, C363S, C469S and C513S cells were grown in SC medium and parameters measured at the indicated days of growth. (A) Phosphoenolpyruvate decarboxylase activity from cell extracts. (B) Trehalose levels from cell extracts. Statistical analyses were performed comparing Sir2 mutants with WT cells. The data are represented as the means ± SD from, at least, three independent experiments.
3.5. Mild oxidative stress increases Sir2 activity through redox regulation
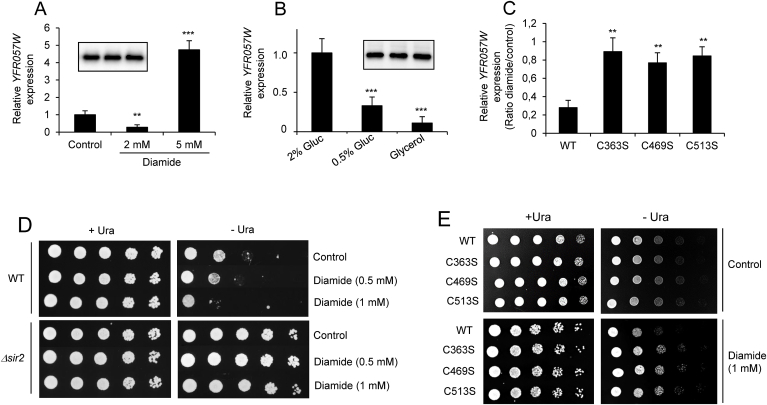
Increased Pck1 activity in Sir2 mutant cells suggested decreased Sir2 activity during chronological aging compared with WT Sir2 [29,30]. In vivo, Sir2 activity may be measured by its silencing activity at the telomeric locus. Transcription of a naturally occurring telomere-linked gene, YFR057W [59], was assayed by quantitative real-time PCR (Supplementary Fig. 3A). An increase in YFR057W mRNA levels in Δsir2 cells or in WT cells upon the addition of 5 mM nicotinamide (a Sir2 inhibitor) indicated that the silencing of this gene was strictly Sir2-dependent (Supplementary Fig. 3B) in accordance with previous results [36]. We published that, upon a high disulfide stress (treatment with 5 mM diamide for 1 h), Sir2 is inactivated by glutathionylation of one or more of the key cysteine residues studied in the present work (C363, C469 or C513) [23]. Here, we analyzed the effect of milder stress on WT Sir2 activity. As shown in Fig. 6A, when WT cultures were treated with 2 mM diamide for 1 h, a clear decrease in YFR057W expression was detected, resulting in a 5-fold increase in telomeric silencing, compared with untreated cells. As described above, a 5-fold reduction in telomeric silencing occurred upon treatment with 5 mM diamide (Fig. 6A). Such variations in YFR057W mRNA levels were not due to changes in the amount of Sir2 protein, which remained unchanged after treatments (Fig. 6A). To investigate whether other kinds of moderate stresses may also increase Sir2 activity, WT cells were grown on a medium containing 0.5% glucose or 3% glycerol as a carbon source and compared with the conventional medium containing 2% glucose. It is known that lower levels of glucose (calorie restriction) increases yeast oxidative metabolism. Glycerol is fully metabolized through mitochondrial oxidation. In both cases, cells are exposed to more oxidative metabolism, which generates mild oxidative stress compared with standard conditions (2% glucose) [58]. As shown in Fig. 6B, when cells were grown in the presence of 0.5% glucose or 3% glycerol, we found a 3- and 10-fold increment in telomeric silencing, respectively. Again, Sir2 levels did not change significantly. Such increase in telomeric silencing after mild stress was not observed when Sir2 mutant cells were treated with 2 mM diamide for 1 h (Fig. 6C).
Fig. 6.
Wild-type Sir2 but not mutant Sir2 cells increase silencing activities upon mild oxidative stress. (A) Cultures of WT cells grown exponentially in SC medium were treated with 2 or 5 mM diamide for 60 min. Expression of YFR057W gene was determined by quantitative real-time PCR. Inset, levels of Sir2 were assessed by Western Blot anti-Sir2. (B) WT cells were grown exponentially in media containing 2% glucose, 0.5% glucose or 3% glycerol as carbon source. Expression of YFR057W gene was determined by quantitative real-time PCR. Inset, levels of Sir2 were assessed by Western Blot anti-Sir2. (C) Cultures of WT, C363S, C469S and C513S exponentially grown in SC were treated with 2 mM diamide for 60 min. Expression of YFR057W gene was determined by quantitative real-time PCR. Results are expressed as the ratio of values from diamide-treated vs. untreated cells. In all experiments from A to C, ACT1 gene expression was used as an internal control to normalize expression levels. The data are represented as the means ± SD from, at least, three independent experiments. (D) Cultures of WT cells grown exponentially in SC medium were treated with 0.5 or 1 mM diamide for 60 min, and silencing of NTS2 reporter within rDNA was assessed by monitoring the growth of 5-fold serial dilutions of cells plated on Ura-depleted SC medium (- Ura). Ura-containing SC medium (+Ura) was used as a control. (E) Cultures of WT, C363S, C469S and C513S exponentially grown in SC were treated with 1 mM diamide for 60 min. Silencing of NTS2 reporter within rDNA was assessed by monitoring the growth of 5-fold serial dilutions of cells plated on Ura-depleted SC medium (- Ura). Ura-containing SC medium (+Ura) was used as a control.
To analyze whether Sir2 activation after moderate stress can be detected at a target locus other than telomeres, silencing was assessed at rDNA (Supplementary Fig. 4). In budding yeast, 100_200 copies of a 9.1-kb rDNA repeat exist as a tandem array. Each rDNA unit yields a 35S precursor rRNA and a 5S rRNA, separated by two non-transcribed spacers, NTS1 and NTS2. NTS1 silencing requires Fob1 protein, while NTS2 silencing is Fob1-independent [60]. WT cells transformed with pDM312 plasmid were untreated or treated with 0.5 or 1 mM diamide for 1 h, serially diluted and spotted on medium containing uracil as a plating control, and on a medium lacking uracil to monitor the expression of the reporter gene. As shown in Fig. 6D, low levels of diamide decrease viability in uracil-depleted plates, indicating increased Sir2 rDNA silencing activity. As a control, Δsir2 cells transformed with pDM312 showed no silencing at this location. To analyze the role of Sir2 cysteines 363, 469 and 513 in silencing, all three mutant strains transformed with pDM312 were submitted to 1 mM diamide stress and rDNA silencing was assayed (Fig. 6E). As shown, compared with WT cells, no increased silencing was detected in the mutant cells.
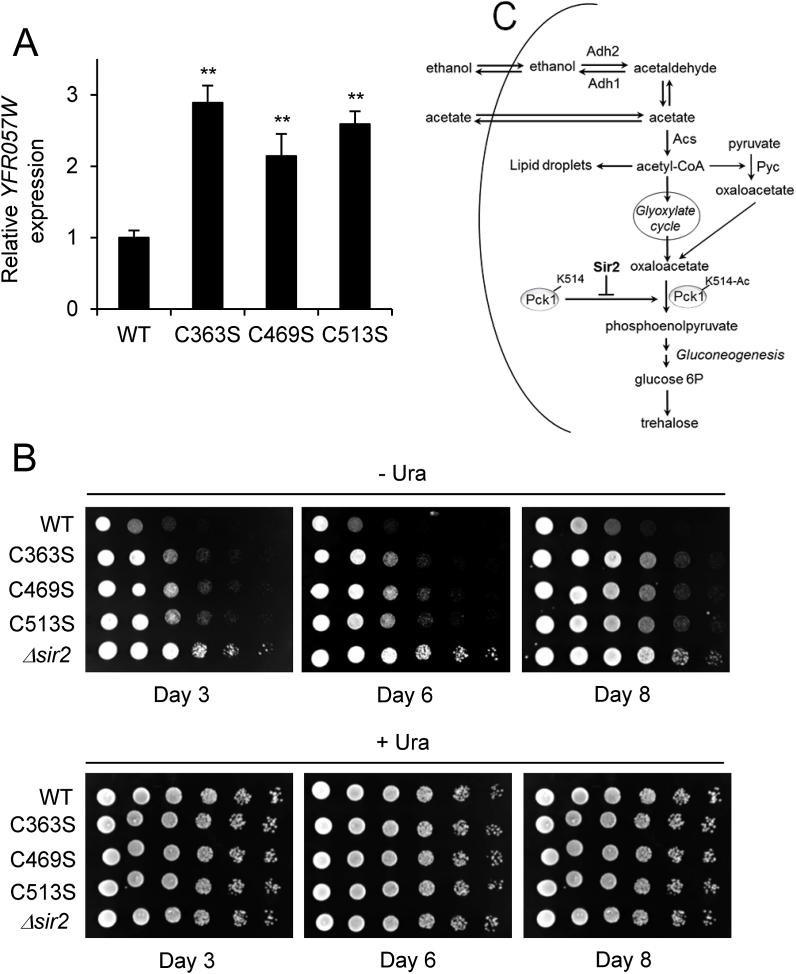
3.6. Chronologically-aged Sir2 mutant cells have decreased Sir2-dependent silencing activity
As described above, WT Sir2 was activated either by external mild disulfide stress or a physiologically generated stress. This activation did not occur in the Sir2 mutant cells (Fig. 6C and E). To explore if this effect may be physiologically relevant in the context of our research, we measured Sir2 silencing activity along chronological aging. As shown in Fig. 7A, at diauxic shift (day 1), mutant cells showed from 2- to 3-fold decreased telomeric silencing activity compared with WT cells. ACT1 gene expression was used as an internal control to normalize expression levels. Telomeric silencing could not be measured with confidence along the stationary phase by quantitative real-time PCR due to expression variability of reference genes. To bypass such a problem, we measured rDNA silencing activity. As shown in Fig. 7B, mutant strains showed decreased rDNA silencing activity (increased growth in plates lacking uracil) along chronological aging, compared with WT strain. Δsir2 strain was used as a control.
Fig. 7.
Mutant Sir2 strains show decreased silencing during chronological aging. (A) Cultures of WT cells were grown in SC medium and at day 1, expression of YFR057W gene was determined by quantitative real-time PCR. ACT1 gene expression was used as an internal control to normalize expression levels. The data are represented as the means ± SD from, at least, three independent experiments. (B) Cultures of WT, C363S, C469S and C513S were grown in SC and at days 3, 6 and 8, silencing of NTS2 reporter within rDNA was assessed by monitoring the growth of 5-fold serial dilutions of cells plated on SC -Ura medium. SC medium (complete, +Ura) was used as a control. (C) A simplified scheme of S. cerevisiae metabolism showing main pathways affected in the Sir2 mutant strains in chronologically aged cells.
4. Discussion
Yeast Sir2 has been related to aging and oxidative stress. However, despite the huge amount of information available, its regulatory mechanisms are just starting to be elucidated. Recent results from our group have shown that yeast Sir2 is S-glutathionylated upon strong disulfide stress (5 mM diamide), reducing its catalytic activity [23]. We identified three key cysteines (C363, C469 and C513) located at the catalytic domain as the major targets of redox regulation. Cysteine 363 is adjacent to the catalytic histidine 364. Cysteine 469 is located in the catalytic pocket and could be part of a putative nuclear export sequence. Cysteine 513 is one of the 20 amino acids involved in ligand_protein interaction and located in the pocket that contains the active site, but not excessively buried (Supplementary Fig. 1). In the present study we demonstrated that Sir2 mutant strains lacking one of these three cysteine residues were more resistant to acetic acid and showed increased chronological life span. Both phenotypes are complex and different pathways may be involved.
Survival to stationary phase has been extensively associated with resistance to oxidative stress [58]. However, although the role of Sir2 in this context is well_known, no differences in catalase, MnSod, CuZnSod, glutathione reductase, GSH or iron were detected between WT and mutant cells. In agreement, no growth differences were detected when H2O2 was used as a stressing agent. At the stationary phase, cells become extremely resistant not only to oxidative stress but other stresses, including heat shock and osmotic stress. Again, no different response in growth upon such treatments was detected among strains. Thus, we hypothesize that the redox regulation of the three cysteines studied in the present work is not involved in the direct regulation of enzymes and parameters analyzed. It is possible that interaction with the yeast forkhead transcription factors (Hcm1, Fkh1, Fkh2) involved in oxidative stress resistance [[13], [14], [15]] remains unchanged in these mutant strains.
Tolerance to acetic and other weak acids involves genomic and transcriptome responses [61,62]. Understanding acetic acid tolerance of S. cerevisiae is important in the field of food and industrial biotechnology. Acetic acid, like other weak acids, such as sorbic acid and benzoic acid, has been traditionally used as a preservative agent in food and beverages by arresting the growth of yeasts and other fungi. In addition, hydrolysates of lignocellulosic biomass are considered renewable feedstock for microbial fermentations [63]. In those hydrolysates, the acetic acid concentrations can be extremely high and become a strong inhibitor of microbial growth and fermentation.
At pH 4.5, the ratio between acetate and acetic acid is close to one [64]. Acetate can be transported through a transporter-mediated mechanism, whereas the undissociated form can diffuse across the plasma membrane by simple diffusion and may trigger regulated cell death [65]. Inside the cytoplasm, acetic acid (pKa = 4.76) also dissociates into a proton and its counterion, decreasing the intracellular pH and accumulating acetate anions. A variety of mechanisms have evolved to adapt to acetic acid toxicity. The capacity to maintain and recover intracellular pH is a key factor [56,66]. A direct correlation between the initial pHc and the pHc drop after exposure to the acid has been described [56]. We demonstrated that, under exponentially growing conditions, all strains have the same intracellular pH (pHc∼7.5). Upon acetic acid stress, pHc drop was indistinguishable among strains as well as pHc recovery, which was very fast. This indicates that redox regulation of Sir2 is involved in maintaining neither the pHc under the control of exponentially growing conditions nor the vacuolar H + -ATPases, which rapidly pump protons out of the cytosol [61,66]. When pHc was measured along chronological aging, decay in one pH unit was observed after day 1. This was not unexpected since the metabolic change occurring at the diauxic shift generates acetic acid that can be detected extracellularly. In any case, pHc drop was unchanged between strains, and only after day 5, mutant strains started to recover pHc better than WT cells.
Why did Sir2 mutant strains show these phenotypes? We demonstrated that chronologically-aged mutant cells presented lower levels of acetate in the extracellular medium because they had increased acetate consumption. Acs activity was higher in Sir2 mutant cells, compared with WT cells, after 1 day of growth with maximum activity on day 3. In yeast, two Acs isoenzymes, Acs1 and Acs2, exist. Although they differ concerning kinetic properties and cellular localization, to date, there is no experimental demonstration of reversible acetylation involved in their enzymatic activity [67]. It has been published that overexpression of ACS2 results in higher resistance to acetic acid as measured by increased growth rate and shorter lag phase relative to the WT strain [68]. Increased Acs activity detected in the Sir2 mutant strains would generate higher levels of acetyl-CoA required for the production of neutral lipids, such as triacylglycerol and cholesteryl esters, which are stored in LD in the cytosol [46]. LD, also known as lipid bodies, lipid particles or oil bodies, are found in most eukaryotic and some prokaryotic cells and serve as an energy reserve, which may contribute to increase longevity in Sir2 mutant cells.
It has been published that a lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors [51]. In Δsir2 mutant cells, gluconeogenic pathway is enhanced due to the increase in Pck1 activity, the main flux-controlling enzyme of this route. Lack of Sir2 increases the acetylated active form of Pck1 due to the lack of Sir2-targeted deacetylation [50]. In addition, resveratrol supplementation to WT cells activates Sir2-mediated deacetylation of Pck1, reducing its activity and as a consequence decreasing gluconeogenesis and longevity [69]. Sir2 deacetylates Pck1 both in vitro and in vivo, and Pck1 acetylation of Lys514, determined by mass spectrometry, was proved to be crucial for enzymatic activity [29]. In agreement, loss of Pck1 activity blocked the extension of yeast chronological life span caused by water starvation. Despite sharing minimal primary structural identity with yeast Pck1, human Pck1 is also activated by acetylation. Although the latter might be deacetylated by multiple deacetylases, yeast Pck1 is exclusively deacetylated by Sir2 [29].
In the present work, we demonstrated that cells lacking one of key redox-regulated cysteines of Sir2 resulted in increased Pck1 activity and, as a consequence, increased trehalose levels. As we proved, WT-Sir2 silencing activity (at both the telomere and the rDNA) was activated under mild disulfide stress. However, this activation did not occur in the Sir2 mutant strains (Fig. 6). Interestingly, these results were obtained after exogenously added diamide and were similar after in vivo stress generated during the diauxic and the postdiauxic shifts, pointing out the importance of these cysteine residues on Sir2 regulation. Mutant cells lacking one of the key cysteine residues have decreased Sir2 silencing activity, both telomeric and at rDNA (Fig. 7A and B). Therefore, it is proposed that mutant strains would have increased Pck1 activity due to decreased Sir2-dependent Pck1 deacetylation, compared with WT cells. Thus, in the mutant cells, gluconeogenesis was fueled by i) increased acetyl-CoA produced from acetate metabolism and ii) enhanced Pck1 activity.
As described in Fig. 7C, acetyl-CoA and Pck1 play a crucial role. Acetyl-CoA generated by Acs supplies glyoxylate pathway, which generates oxaloacetate, and is necessary to increase lipid stores. Moreover, acetyl-CoA is an allosteric activator of pyruvate carboxylase (another key enzyme of the gluconeogenesis pathway), which catalyzes the conversion of pyruvate to oxaloacetate [70]. Oxaloacetate is channeled through the gluconeogenic flux by Pck1, increasing the storage of sugar such as trehalose, needed for cell survival during chronological aging.
In the context of mammalian metabolism, fasting activates the expression of Sirt1, promoting the production of acetyl-CoA by deacetylating Acs1 [71]. The increased acetyl-CoA favors the transcription of PCK1 [72] and activation of Pck1 through acetylation by acetyltransferases like Nut 4 and TIP60 [29]. Like in yeast, increased Pck1 activity ensures a rapid adaptation of the gluconeogenic flux to energy status during fasting and may have a role in longevity. Pck1 activity is also important in the context of pathological status like diabetes [73,74]. To better understand the basis, unraveling the redox regulatory mechanism of key cysteines in human Sirt1 may be important for clarifying how glycemic levels are maintained within a narrow range.
5. Conclusions
Sir2 redox regulation modulates acetic acid metabolism and chronological life span. We have provided evidence that a lack of key redox_regulated cysteines of Sir2 decreased its activity when cells enter to diauxic phase and during the early stationary phase. We propose that the stress generated by the metabolic change occurring at diauxic shift increases Sir2 silencing activity, and such activation occurs to a lesser extent in the mutant Sir2 proteins because they lack one of the redox regulated cysteines. Decreased Sir2 activity, compared with WT cells, inevitably would lead to higher Pck1 acetylation which resulted in increased Pck1 activity. In addition, increased acetyl-CoA synthetase activity observed in the Sir2 mutant cells would generate acetyl-CoA pushing the glyoxylate-requiring gluconeogenesis. In agreement, we reported increased trehalose levels, and decreased acetate in the extracellular medium, both factors known to participate in cell survival during aging. The results presented in this work also open the possibility of exploring the potential effect of sirtuin redox regulation in i) fasting and its role in diabetes and aging, and ii) acetic acid resistance and its role in biotechnology and food processing.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Roser Pané for technical assistance. We also thank Dr. Ariño for providing plasmid containing pHluorin, and Dr. Moazed for plasmid pDM312. This work was supported by grants TR625 from the University of Lleida and SGR2009-00196 from the Generalitat de Catalunya). Núria Vall-llaura received a Ph.D. fellowship from the Generalitat de Catalunya.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101229.
Contributor Information
Núria Vall-llaura, Email: nuria.vall-llaura@cmb.udl.cat.
Noèlia Mir, Email: noeliamirrebull@gmail.com.
Lourdes Garrido, Email: lourdes7garrido@gmail.com.
Celia Vived, Email: celiavived@hotmail.com.
Elisa Cabiscol, Email: elisa.cabiscol@cmb.udl.cat.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]; C.C. Winterbourn, M.B. Hampton, Thiol chemistry and specificity in redox signaling, Free Radic. Biol. Med. 45 (2008) 549-561. doi:10.1016/J.FREERADBIOMED.2008.05.004. [DOI] [PubMed]
- 2.Boronat S., Domènech A., Hidalgo E. Proteomic characterization of reversible thiol oxidations in proteomes and proteins. Antioxidants Redox Signal. 2016:2016–6720. doi: 10.1089/ars.2016.6720. ars. [DOI] [PubMed] [Google Scholar]; S. Boronat, A. Domenech, E. Hidalgo, Proteomic Characterization of Reversible Thiol Oxidations in Proteomes and Proteins., Antioxid. Redox Signal. (2016) ars.2016.6720. doi:10.1089/ars.2016.6720. [DOI] [PubMed]
- 3.Brigelius-Flohé R., Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxidants Redox Signal. 2011;15:2335–2381. doi: 10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]; R. Brigelius-Flohe, L. Flohe, Basic principles and emerging concepts in the redox control of transcription factors., Antioxid. Redox Signal. 15 (2011) 2335-2381. doi:10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed]
- 4.García-Santamarina S., Boronat S., Hidalgo E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry. 2014;53:2560–2580. doi: 10.1021/bi401700f. [DOI] [PubMed] [Google Scholar]; S. Garcia-Santamarina, S. Boronat, E. Hidalgo, Reversible Cysteine Oxidation in Hydrogen Peroxide Sensing and Signal Transduction, Biochemistry. 53 (2014) 2560-2580. doi:10.1021/bi401700f. [DOI] [PubMed]
- 5.Ivy J.M., Klar A.J., Hicks J.B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=367560&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]; J.M. Ivy, A.J. Klar, J.B. Hicks, Cloning and characterization of four SIR genes of Saccharomyces cerevisiae., Mol. Cell. Biol. 6 (1986) 688-702. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=367560&tool=pmcentrez&rendertype=abstract (accessed June 30, 2014). [DOI] [PMC free article] [PubMed]
- 6.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological Insights and diseas relevance. Annu. Rev. Pathol. 2010:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. (Mammalian) [DOI] [PMC free article] [PubMed] [Google Scholar]; M.C. Haigis, D.A. Sinclair, Mammalian sirtuins: biological Insights and diseas relevance, Annu. Rev. Pathol. (2010) 253-295. doi:10.1146/annurev.pathol.4.110807.092250.Mammalian. [DOI] [PMC free article] [PubMed]
- 7.Imai S., Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Imai, L. Guarente, Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases., Trends Pharmacol. Sci. 31 (2010) 212-220. doi:10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed]
- 8.Dai H., Sinclair D.A., Ellis J.L., Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Dai, D.A. Sinclair, J.L. Ellis, C. Steegborn, Sirtuin activators and inhibitors: Promises, achievements, and challenges, Pharmacol. Ther. 188 (2018) 140-154. doi:10.1016/J.PHARMTHERA.2018.03.004. [DOI] [PMC free article] [PubMed]
- 9.Bitterman K.J., Medvedik O., Sinclair D.A. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 2003;67 doi: 10.1128/MMBR.67.3.376-399.2003. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=193872&tool=pmcentrez&rendertype=abstract 376–99, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]; K.J. Bitterman, O. Medvedik, D.A. Sinclair, Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin., Microbiol. Mol. Biol. Rev. 67 (2003) 376-99, table of contents. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=193872&tool=pmcentrez&rendertype=abstract (accessed July 23, 2015). [DOI] [PMC free article] [PubMed]
- 10.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=317077&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Kaeberlein, M. McVey, L. Guarente, The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms., Genes Dev. 13 (1999) 2570-2580. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=317077&tool=pmcentrez&rendertype=abstract (accessed June 30, 2014). [DOI] [PMC free article] [PubMed]
- 11.Wang F., Nguyen M., Qin F.X.-F., Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]; F. Wang, M. Nguyen, F.X.-F. Qin, Q. Tong, SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction., Aging Cell. 6 (2007) 505-514. doi:10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed]
- 12.Olmos Y., Sánchez-Gómez F.J., Wild B., García-Quintans N., Cabezudo S., Lamas S. SirT1 regulation of antioxidant genes is dependent on the formation of a FoxO3a/PGC-1α complex. Antioxidants Redox Signal. 2013;19:1507–1521. doi: 10.1089/ars.2012.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y. Olmos, F.J. Sanchez-Gomez, B. Wild, N. Garcia-Quintans, S. Cabezudo, S. Lamas, et al., SirT1 regulation of antioxidant genes is dependent on the formation of a FoxO3a/PGC-1α complex., Antioxid. Redox Signal. 19 (2013) 1507-1521. doi:10.1089/ars.2012.4713. [DOI] [PMC free article] [PubMed]
- 13.Rodriguez-Colman M.J., Reverter-Branchat G., Sorolla M.A., Tamarit J., Ros J., Cabiscol E. The forkhead transcription factor Hcm1 promotes mitochondrial biogenesis and stress resistance in yeast. J. Biol. Chem. 2010;285:37092–37101. doi: 10.1074/jbc.M110.174763. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.J. Rodriguez-Colman, G. Reverter-Branchat, M.A. Sorolla, J. Tamarit, J. Ros, E. Cabiscol, The forkhead transcription factor Hcm1 promotes mitochondrial biogenesis and stress resistance in yeast., J. Biol. Chem. 285 (2010) 37092-37101. doi:10.1074/jbc.M110.174763. [DOI] [PMC free article] [PubMed]
- 14.Linke C., Klipp E., Lehrach H., Barberis M., Krobitsch S. Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions. Front. Physiol. 2013;4:173. doi: 10.3389/fphys.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Linke, E. Klipp, H. Lehrach, M. Barberis, S. Krobitsch, Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions., Front. Physiol. 4 (2013) 173. doi:10.3389/fphys.2013.00173. [DOI] [PMC free article] [PubMed]
- 15.Rodríguez-Colman M.J., Sorolla M.A., Vall-Llaura N., Tamarit J., Ros J., Cabiscol E. The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor 1/Sch9 kinases. Biochim. Biophys. Acta. 2013;1833 doi: 10.1016/j.bbamcr.2013.02.015. 2004–15. [DOI] [PubMed] [Google Scholar]; M.J. Rodriguez-Colman, M.A. Sorolla, N. Vall-Llaura, J. Tamarit, J. Ros, E. Cabiscol, The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor1/Sch9 kinases., Biochim. Biophys. Acta. 1833 (2013) 2004-15. doi:10.1016/j.bbamcr.2013.02.015. [DOI] [PubMed]
- 16.Reverter-Branchat G., Cabiscol E., Tamarit J., Sorolla M.A., Angeles de la Torre M., Ros J. Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1. Microbiology. 2007;153:3667–3676. doi: 10.1099/mic.0.2007/009340-0. [DOI] [PubMed] [Google Scholar]; G. Reverter-Branchat, E. Cabiscol, J. Tamarit, M.A. Sorolla, M. Angeles de la Torre, J. Ros, Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1., Microbiology. 153 (2007) 3667-3676. doi:10.1099/mic.0.2007/009340-0. [DOI] [PubMed]
- 17.Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]; T. Prozorovski, U. Schulze-Topphoff, R. Glumm, J. Baumgart, F. Schroter, O. Ninnemann, et al., Sirt1 contributes critically to the redox-dependent fate of neural progenitors., Nat. Cell Biol. 10 (2008) 385-394. doi:10.1038/ncb1700. [DOI] [PubMed]
- 18.Sorolla M.A., Nierga C., Rodríguez-Colman M.J., Reverter-Branchat G., Arenas A., Tamarit J. Sir2 is induced by oxidative stress in a yeast model of Huntington disease and its activation reduces protein aggregation. Arch. Biochem. Biophys. 2011;510:27–34. doi: 10.1016/j.abb.2011.04.002. [DOI] [PubMed] [Google Scholar]; M.A. Sorolla, C. Nierga, M.J. Rodriguez-Colman, G. Reverter-Branchat, A. Arenas, J. Tamarit, et al., Sir2 is induced by oxidative stress in a yeast model of Huntington disease and its activation reduces protein aggregation., Arch. Biochem. Biophys. 510 (2011) 27-34. doi:10.1016/j.abb.2011.04.002. [DOI] [PubMed]
- 19.Fulco M., Schiltz R.L., Iezzi S., King M.T., Zhao P., Kashiwaya Y. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. http://www.ncbi.nlm.nih.gov/pubmed/12887892 [DOI] [PubMed] [Google Scholar]; M. Fulco, R.L. Schiltz, S. Iezzi, M.T. King, P. Zhao, Y. Kashiwaya, et al., Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state., Mol. Cell. 12 (2003) 51-62. http://www.ncbi.nlm.nih.gov/pubmed/12887892 (accessed July 17, 2014). [DOI] [PubMed]
- 20.Zee R.S., Yoo C.B., Pimentel D.R., Perlman D.H., Burgoyne J.R., Hou X. Redox regulation of sirtuin-1 by S-glutathiolation. Antioxidants Redox Signal. 2010;13:1023–1032. doi: 10.1089/ars.2010.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]; R.S. Zee, C.B. Yoo, D.R. Pimentel, D.H. Perlman, J.R. Burgoyne, X. Hou, et al., Redox regulation of sirtuin-1 by S-glutathiolation., Antioxid. Redox Signal. 13 (2010) 1023-1032. doi:10.1089/ars.2010.3251. [DOI] [PMC free article] [PubMed]
- 21.Shao D., Fry J.L., Han J., Hou X., Pimentel D.R., Matsui R. A redox-resistant sirtuin-1 mutant protects against hepatic metabolic and oxidant stress. J. Biol. Chem. 2014;289:7293–7306. doi: 10.1074/jbc.M113.520403. [DOI] [PMC free article] [PubMed] [Google Scholar]; D. Shao, J.L. Fry, J. Han, X. Hou, D.R. Pimentel, R. Matsui, et al., A redox-resistant sirtuin-1 mutant protects against hepatic metabolic and oxidant stress., J. Biol. Chem. 289 (2014) 7293-7306. doi:10.1074/jbc.M113.520403. [DOI] [PMC free article] [PubMed]
- 22.Bräutigam L., Jensen L.D.E., Poschmann G., Nyström S., Bannenberg S., Dreij K. Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 2013;110 doi: 10.1073/pnas.1313753110. 20057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]; L. Brautigam, L.D.E. Jensen, G. Poschmann, S. Nystrom, S. Bannenberg, K. Dreij, et al., Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1., Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 20057-62. doi:10.1073/pnas.1313753110. [DOI] [PMC free article] [PubMed]
- 23.Vall-Llaura N., Reverter-Branchat G., Vived C., Weertman N., Rodríguez-Colman M.J., Cabiscol E. Reversible glutathionylation of Sir2 by monothiol glutaredoxins Grx3/4 regulates stress resistance. Free Radic. Biol. Med. 2016;96:45–56. doi: 10.1016/j.freeradbiomed.2016.04.008. [DOI] [PubMed] [Google Scholar]; N. Vall-Llaura, G. Reverter-Branchat, C. Vived, N. Weertman, M.J. Rodriguez-Colman, E. Cabiscol, Reversible glutathionylation of Sir2 by monothiol glutaredoxins Grx3/4 regulates stress resistance., Free Radic. Biol. Med. 96 (2016) 45-56. doi:10.1016/j.freeradbiomed.2016.04.008. [DOI] [PubMed]
- 24.Zheng M., Aslund F., Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. http://www.ncbi.nlm.nih.gov/pubmed/9497290 [DOI] [PubMed] [Google Scholar]; M. Zheng, F. Aslund, G. Storz, Activation of the OxyR transcription factor by reversible disulfide bond formation., Science. 279 (1998) 1718-1721. http://www.ncbi.nlm.nih.gov/pubmed/9497290 (accessed August 25, 2014). [DOI] [PubMed]
- 25.Delaunay A., Isnard A.D., Toledano M.B. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Delaunay, A.D. Isnard, M.B. Toledano, H2O2 sensing through oxidation of the Yap1 transcription factor., EMBO J. 19 (2000) 5157-5166. doi:10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed]
- 26.Calvo I.A., Ayté J., Hidalgo E. Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1. J. Cell Sci. 2013;126:2279–2284. doi: 10.1242/jcs.124370. [DOI] [PubMed] [Google Scholar]; I.A. Calvo, J. Ayte, E. Hidalgo, Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1., J. Cell Sci. 126 (2013) 2279-2284. doi:10.1242/jcs.124370. [DOI] [PubMed]
- 27.Ilbert M., Horst J., Ahrens S., Winter J., Graf P.C.F., Lilie H. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Ilbert, J. Horst, S. Ahrens, J. Winter, P.C.F. Graf, H. Lilie, et al., The redox-switch domain of Hsp33 functions as dual stress sensor., Nat. Struct. Mol. Biol. 14 (2007) 556-563. doi:10.1038/nsmb1244. [DOI] [PMC free article] [PubMed]
- 28.Kumsta C., Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Kumsta, U. Jakob, Redox-regulated chaperones., Biochemistry. 48 (2009) 4666-4676. doi:10.1021/bi9003556. [DOI] [PMC free article] [PubMed]
- 29.Lin Y., Lu J., Zhang J., Walter W., Dang W., Wan J. Protein acetylation microarray reveals that NuA 4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Y. Lin, J. Lu, J. Zhang, W. Walter, W. Dang, J. Wan, et al., Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis., Cell. 136 (2009) 1073-1084. doi:10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed]
- 30.Orlandi I., Stamerra G., Strippoli M., Vai M. During yeast chronological aging resveratrol supplementation results in a short-lived phenotype Sir2-dependent. Redox Biol. 2017;12:745–754. doi: 10.1016/j.redox.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; I. Orlandi, G. Stamerra, M. Strippoli, M. Vai, During yeast chronological aging resveratrol supplementation results in a short-lived phenotype Sir2-dependent, Redox Biol. 12 (2017) 745-754. doi:10.1016/j.redox.2017.04.015. [DOI] [PMC free article] [PubMed]
- 31.Gallego C., Garí E., Colomina N., Herrero E., Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Gallego, E. Gari, N. Colomina, E. Herrero, M. Aldea, The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast., EMBO J. 16 (1997) 7196-7206. doi:10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed]
- 32.Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Huang, D. Moazed, Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing., Genes Dev. 17 (2003) 2162-2176. doi:10.1101/gad.1108403. [DOI] [PMC free article] [PubMed]
- 33.Guthrie C G.R., Fink Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. http://www.ncbi.nlm.nih.gov/pubmed/2005781 [PubMed] [Google Scholar]; G.R. Guthrie C. and Fink, Guide to yeast genetics and molecular biology., Methods Enzymol. 194 (1991) 1-863. http://www.ncbi.nlm.nih.gov/pubmed/2005781 (accessed July 1, 2014). [PubMed]
- 34.Miesenböck G., De Angelis D.A., Rothman J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]; G. Miesenbock, D.A. De Angelis, J.E. Rothman, Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins., Nature. 394 (1998) 192-195. doi:10.1038/28190. [DOI] [PubMed]
- 35.Orij R., Postmus J., Ter Beek A., Brul S., Smits G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology. 2009;155:268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]; R. Orij, J. Postmus, A. Ter Beek, S. Brul, G.J. Smits, In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth, Microbiology. 155 (2009) 268-278. doi:10.1099/mic.0.022038-0. [DOI] [PubMed]
- 36.Ringel A.E., Ryznar R., Picariello H., Huang K., Lazarus A.G., Holmes S.G. Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet. 2013;9:e1003871. doi: 10.1371/journal.pgen.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.E. Ringel, R. Ryznar, H. Picariello, K. Huang, A.G. Lazarus, S.G. Holmes, Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination., PLoS Genet. 9 (2013) e1003871. doi:10.1371/journal.pgen.1003871. [DOI] [PMC free article] [PubMed]
- 37.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.W. Pfaffl, A new mathematical model for relative quantification in real-time RT-PCR., Nucleic Acids Res. 29 (2001) e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed]
- 38.Jakubowski W., Biliński T., Bartosz G. Oxidative stress during aging of stationary cultures of the yeast Saccharomyces cerevisiae. Free Radic. Biol. Med. 2000;28:659–664. doi: 10.1016/s0891-5849(99)00266-x. http://www.ncbi.nlm.nih.gov/pubmed/10754260 [DOI] [PubMed] [Google Scholar]; W. Jakubowski, T. Biliński, G. Bartosz, Oxidative stress during aging of stationary cultures of the yeast Saccharomyces cerevisiae., Free Radic. Biol. Med. 28 (2000) 659-664. http://www.ncbi.nlm.nih.gov/pubmed/10754260 (accessed January 9, 2019). [DOI] [PubMed]
- 39.Maitra P.K., Lobo Z. A kinetic study of glycolytic enzyme synthesis in yeast. J. Biol. Chem. 1971;246:475–488. http://www.ncbi.nlm.nih.gov/pubmed/5542016 [PubMed] [Google Scholar]; P.K. Maitra, Z. Lobo, A kinetic study of glycolytic enzyme synthesis in yeast., J. Biol. Chem. 246 (1971) 475-488. http://www.ncbi.nlm.nih.gov/pubmed/5542016 (accessed January 9, 2019). [PubMed]
- 40.Shepherd D., Garland P.B. The kinetic properties of citrate synthase from rat liver mitochondria. Biochem. J. 1969;114:597–610. doi: 10.1042/bj1140597. http://www.ncbi.nlm.nih.gov/pubmed/5820645 [DOI] [PMC free article] [PubMed] [Google Scholar]; D. Shepherd, P.B. Garland, The kinetic properties of citrate synthase from rat liver mitochondria., Biochem. J. 114 (1969) 597-610. http://www.ncbi.nlm.nih.gov/pubmed/5820645 (accessed January 9, 2019). [DOI] [PMC free article] [PubMed]
- 41.Carlberg I., Mannervik B. Glutathione reductase. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. http://www.ncbi.nlm.nih.gov/pubmed/3003504 [DOI] [PubMed] [Google Scholar]; I. Carlberg, B. Mannervik, Glutathione reductase., Methods Enzymol. 113 (1985) 484-490. http://www.ncbi.nlm.nih.gov/pubmed/3003504 (accessed January 9, 2019). [DOI] [PubMed]
- 42.Frenkel E.P., Kitchens R.L. Purification and properties of acetyl coenzyme A synthetase from bakers' yeast. J. Biol. Chem. 1977;252:504–507. http://www.ncbi.nlm.nih.gov/pubmed/13070 [PubMed] [Google Scholar]; E.P. Frenkel, R.L. Kitchens, Purification and properties of acetyl coenzyme A synthetase from bakers’ yeast., J. Biol. Chem. 252 (1977) 504-507. http://www.ncbi.nlm.nih.gov/pubmed/13070 (accessed November 14, 2016). [PubMed]
- 43.Andrade C., Sepulveda C., Cardemil E., Jabalquinto A.M. The role of tyrosine 207 in the reaction catalyzed by Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase. Biol. Res. 2010;43:191–195. doi:/S0716-97602010000200007. [PubMed] [Google Scholar]; C. Andrade, C. Sepulveda, E. Cardemil, A.M. Jabalquinto, The role of tyrosine 207 in the reaction catalyzed by Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase., Biol. Res. 43 (2010) 191-195. doi:/S0716-97602010000200007. [PubMed]
- 44.Culotta V.C., Klomp L.W., Strain J., Casareno R.L., Krems B., Gitlin J.D. The copper chaperone for superoxide dismutase. J. Biol. Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. http://www.ncbi.nlm.nih.gov/pubmed/9295278 [DOI] [PubMed] [Google Scholar]; V.C. Culotta, L.W. Klomp, J. Strain, R.L. Casareno, B. Krems, J.D. Gitlin, The copper chaperone for superoxide dismutase., J. Biol. Chem. 272 (1997) 23469-23472. http://www.ncbi.nlm.nih.gov/pubmed/9295278 (accessed January 9, 2019). [DOI] [PubMed]
- 45.Bozaquel-Morais B.L., Madeira J.B., Maya-Monteiro C.M., Masuda C.A., Montero-Lomeli M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013692. [DOI] [PMC free article] [PubMed] [Google Scholar]; B.L. Bozaquel-Morais, J.B. Madeira, C.M. Maya-Monteiro, C.A. Masuda, M. Montero-Lomeli, A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism, PLoS One. 5 (2010). doi:10.1371/journal.pone.0013692. [DOI] [PMC free article] [PubMed]
- 46.Radulovic M., Knittelfelder O., Cristobal-Sarramian A., Kolb D., Wolinski H., Kohlwein S.D. The emergence of lipid droplets in yeast: current status and experimental approaches. Curr. Genet. 2013;59:231–242. doi: 10.1007/s00294-013-0407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Radulovic, O. Knittelfelder, A. Cristobal-Sarramian, D. Kolb, H. Wolinski, S.D. Kohlwein, The emergence of lipid droplets in yeast: Current status and experimental approaches, Curr. Genet. 59 (2013) 231-242. doi:10.1007/s00294-013-0407-9. [DOI] [PMC free article] [PubMed]
- 47.Tamarit J., Irazusta V., Moreno-Cermeño A., Ros J. Colorimetric assay for the quantitation of iron in yeast. Anal. Biochem. 2006;351:149–151. doi: 10.1016/j.ab.2005.12.001. [DOI] [PubMed] [Google Scholar]; J. Tamarit, V. Irazusta, A. Moreno-Cermeño, J. Ros, Colorimetric assay for the quantitation of iron in yeast., Anal. Biochem. 351 (2006) 149-151. doi:10.1016/j.ab.2005.12.001. [DOI] [PubMed]
- 48.Rodríguez-Manzaneque M.T., Ros J., Cabiscol E., Sorribas A., Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19 doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]; M.T. Rodriguez-Manzaneque, J. Ros, E. Cabiscol, A. Sorribas, E. Herrero, Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae, Mol. Cell. Biol. 19 (1999). doi:10.1128/MCB.19.12.8180. [DOI] [PMC free article] [PubMed]
- 49.Burtner C.R., Murakami C.J., Kennedy B.K., Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]; C.R. Burtner, C.J. Murakami, B.K. Kennedy, M. Kaeberlein, A molecular mechanism of chronological aging in yeast., Cell Cycle. 8 (2009) 1256-1270. doi:10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed]
- 50.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Kaeberlein, Lessons on longevity from budding yeast., Nature. 464 (2010) 513-519. doi:10.1038/nature08981. [DOI] [PMC free article] [PubMed]
- 51.Casatta N., Porro A., Orlandi I., Brambilla L., Vai M. Lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:593–601. doi: 10.1016/j.bbamcr.2012.11.008. [DOI] [PubMed] [Google Scholar]; N. Casatta, A. Porro, I. Orlandi, L. Brambilla, M. Vai, Lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors, Biochim. Biophys. Acta - Mol. Cell Res. 1833 (2013) 593-601. doi:10.1016/j.bbamcr.2012.11.008. [DOI] [PubMed]
- 52.Orozco H., Matallana E., Aranda A. Genetic manipulation of longevity-related genes as a tool to regulate yeast life span and metabolite production during winemaking. Microb. Cell Factories. 2013;12:1. doi: 10.1186/1475-2859-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Orozco, E. Matallana, A. Aranda, Genetic manipulation of longevity-related genes as a tool to regulate yeast life span and metabolite production during winemaking., Microb. Cell Fact. 12 (2013) 1. doi:10.1186/1475-2859-12-1. [DOI] [PMC free article] [PubMed]
- 53.Guerreiro J.F., Mira N.P., Sá-Correia I. Adaptive response to acetic acid in the highly resistant yeast species Zygosaccharomyces bailii revealed by quantitative proteomics. Proteomics. 2012;12:2303–2318. doi: 10.1002/pmic.201100457. [DOI] [PubMed] [Google Scholar]; J.F. Guerreiro, N.P. Mira, I. Sa-Correia, Adaptive response to acetic acid in the highly resistant yeast species Zygosaccharomyces bailii revealed by quantitative proteomics., Proteomics. 12 (2012) 2303-2318. doi:10.1002/pmic.201100457. [DOI] [PubMed]
- 54.Mira N.P., Palma M., Guerreiro J.F., Sá-Correia I. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb. Cell Factories. 2010;9:79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]; N.P. Mira, M. Palma, J.F. Guerreiro, I. Sa-Correia, Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid., Microb. Cell Fact. 9 (2010) 79. doi:10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed]
- 55.Longo V., Ždralević M., Guaragnella N., Giannattasio S., Zolla L., Timperio A.M. Proteome and metabolome profiling of wild-type and YCA1-knock-out yeast cells during acetic acid-induced programmed cell death. J. Proteomics. 2015;128:173–188. doi: 10.1016/j.jprot.2015.08.003. [DOI] [PubMed] [Google Scholar]; V. Longo, M. Ždralević, N. Guaragnella, S. Giannattasio, L. Zolla, A.M. Timperio, Proteome and metabolome profiling of wild-type and YCA1-knock-out yeast cells during acetic acid-induced programmed cell death., J. Proteomics. 128 (2015) 173-188. doi:10.1016/j.jprot.2015.08.003. [DOI] [PubMed]
- 56.Fernández-Niño M., Marquina M., Swinnen S., Rodríguez-Porrata B., Nevoigt E., Ariño J. The cytosolic pH of individual Saccharomyces cerevisiae cells is a key factor in acetic acid tolerance. Appl. Environ. Microbiol. 2015;81:7813–7821. doi: 10.1128/AEM.02313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; M. Fernandez-Niño, M. Marquina, S. Swinnen, B. Rodriguez-Porrata, E. Nevoigt, J. Ariño, The Cytosolic pH of Individual Saccharomyces cerevisiae Cells Is a Key Factor in Acetic Acid Tolerance., Appl. Environ. Microbiol. 81 (2015) 7813-7821. doi:10.1128/AEM.02313-15. [DOI] [PMC free article] [PubMed]
- 57.Brunk U.T., Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. http://www.ncbi.nlm.nih.gov/pubmed/11985575 [DOI] [PubMed] [Google Scholar]; U.T. Brunk, A. Terman, The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis., Eur. J. Biochem. 269 (2002) 1996-2002. http://www.ncbi.nlm.nih.gov/pubmed/11985575 (accessed January 16, 2019). [DOI] [PubMed]
- 58.Reverter-Branchat G., Cabiscol E., Tamarit J., Ros J. Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:31983–31989. doi: 10.1074/jbc.M404849200. [DOI] [PubMed] [Google Scholar]; G. Reverter-Branchat, E. Cabiscol, J. Tamarit, J. Ros, Oxidative Damage to Specific Proteins in Replicative and Chronological-aged Saccharomyces cerevisiae, J. Biol. Chem. 279 (2004) 31983-31989. doi:10.1074/jbc.M404849200. [DOI] [PubMed]
- 59.Vega-Palas M.A., Martín-Figueroa E., Florencio F.J. Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 2000;263:287–291. doi: 10.1007/s004380051170. http://www.ncbi.nlm.nih.gov/pubmed/10778747 [DOI] [PubMed] [Google Scholar]; M.A. Vega-Palas, E. Martin-Figueroa, F.J. Florencio, Telomeric silencing of a natural subtelomeric gene., Mol. Gen. Genet. 263 (2000) 287-291. http://www.ncbi.nlm.nih.gov/pubmed/10778747 (accessed June 29, 2014). [DOI] [PubMed]
- 60.Huang J., Moazed D. 2003. Association of the RENT Complex with Nontranscribed and Coding Regions of rDNA and a Regional Requirement for the Replication Fork Block Protein Fob1 in rDNA Silencing; pp. 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Huang, D. Moazed, Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing, (2003) 2162-2176. doi:10.1101/gad.1108403.large. [DOI] [PMC free article] [PubMed]
- 61.Mira N.P., Teixeira M.C., Sá-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae : a genome-wide view. OMICS A J. Integr. Biol. 2010;14:525–540. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]; N.P. Mira, M.C. Teixeira, I. Sa-Correia, Adaptive Response and Tolerance to Weak Acids in Saccharomyces cerevisiae : A Genome-Wide View, Omi. A J. Integr. Biol. 14 (2010) 525-540. doi:10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed]
- 62.Schüller C., Mamnun Y.M., Mollapour M., Krapf G., Schuster M., Bauer B.E. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]; C. Schuller, Y.M. Mamnun, M. Mollapour, G. Krapf, M. Schuster, B.E. Bauer, et al., Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae., Mol. Biol. Cell. 15 (2004) 706-720. doi:10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed]
- 63.Almario M.P., Reyes L.H., Kao K.C. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol. Bioeng. 2013;110:2616–2623. doi: 10.1002/bit.24938. [DOI] [PubMed] [Google Scholar]; M.P. Almario, L.H. Reyes, K.C. Kao, Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass, Biotechnol. Bioeng. 110 (2013) 2616-2623. doi:10.1002/bit.24938. [DOI] [PubMed]
- 64.Giannattasio S., Guaragnella N., Zdralević M., Marra E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front. Microbiol. 2013;4:33. doi: 10.3389/fmicb.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]; S. Giannattasio, N. Guaragnella, M. Zdralević, E. Marra, Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid., Front. Microbiol. 4 (2013) 33. doi:10.3389/fmicb.2013.00033. [DOI] [PMC free article] [PubMed]
- 65.Guaragnella N., Antonacci L., Passarella S., Marra E., Giannattasio S. Achievements and perspectives in yeast acetic acid-induced programmed cell death pathways. Biochem. Soc. Trans. 2011;39:1538–1543. doi: 10.1042/BST0391538. [DOI] [PubMed] [Google Scholar]; N. Guaragnella, L. Antonacci, S. Passarella, E. Marra, S. Giannattasio, Achievements and perspectives in yeast acetic acid-induced programmed cell death pathways., Biochem. Soc. Trans. 39 (2011) 1538-1543. doi:10.1042/BST0391538. [DOI] [PubMed]
- 66.Mollapour M., Calderon C.O., Hatzixanthis K., Piper P. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology. 2001;147:2635–2642. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]; M. Mollapour, C.O. Calderon, K. Hatzixanthis, P. Piper, Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives, Microbiology. 147 (2001) 2635-2642. doi:10.1099/00221287-147-10-2635. [DOI] [PubMed]
- 67.Falcón A.A., Chen S., Wood M.S., Aris J.P. Acetyl-coenzyme A synthetase 2 is a nuclear protein required for replicative longevity in Saccharomyces cerevisiae. Mol. Cell. Biochem. 2010;333:99–108. doi: 10.1007/s11010-009-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; A.A. Falcon, S. Chen, M.S. Wood, J.P. Aris, Acetyl-coenzyme A synthetase 2 is a nuclear protein required for replicative longevity in Saccharomyces cerevisiae., Mol. Cell. Biochem. 333 (2010) 99-108. doi:10.1007/s11010-009-0209-z. [DOI] [PMC free article] [PubMed]
- 68.Ding J., Holzwarth G., Penner M.H., Patton-Vogt J., Bakalinsky A.T. Overexpression of acetyl-CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance. FEMS Microbiol. Lett. 2015;362:1–7. doi: 10.1093/femsle/fnu042. [DOI] [PMC free article] [PubMed] [Google Scholar]; J. Ding, G. Holzwarth, M.H. Penner, J. Patton-Vogt, A.T. Bakalinsky, Overexpression of acetyl-CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance., FEMS Microbiol. Lett. 362 (2015) 1-7. doi:10.1093/femsle/fnu042. [DOI] [PMC free article] [PubMed]
- 69.Orlandi I., Ronzulli R., Casatta N., Vai M. Ethanol and acetate acting as carbon/energy sources negatively affect yeast chronological aging. Oxid. Med. Cell. Longev. 2013;2013:802870. doi: 10.1155/2013/802870. [DOI] [PMC free article] [PubMed] [Google Scholar]; I. Orlandi, R. Ronzulli, N. Casatta, M. Vai, Ethanol and acetate acting as carbon/energy sources negatively affect yeast chronological aging., Oxid. Med. Cell. Longev. 2013 (2013) 802870. doi:10.1155/2013/802870. [DOI] [PMC free article] [PubMed]
- 70.PRONK J.T., YDE STEENSMA H., VAN DIJKEN J.P. Pyruvate metabolism inSaccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]; J.T. PRONK, H. YDE STEENSMA, J.P. VAN DIJKEN, Pyruvate Metabolism inSaccharomyces cerevisiae, Yeast. 12 (1996) 1607-1633. doi:10.1002/(SICI)1097-0061(199612)12:16<1607::AID-YEA70>3.0.CO;2-4. [DOI] [PubMed]
- 71.Schwer B., Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metabol. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]; B. Schwer, E. Verdin, Conserved metabolic regulatory functions of sirtuins., Cell Metab. 7 (2008) 104-112. doi:10.1016/j.cmet.2007.11.006. [DOI] [PubMed]
- 72.Kim H., Zheng Z., Walker P.D., Kapatos G., Zhang K. CREBH maintains circadian glucose homeostasis by regulating hepatic glycogenolysis and gluconeogenesis. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; H. Kim, Z. Zheng, P.D. Walker, G. Kapatos, K. Zhang, CREBH Maintains Circadian Glucose Homeostasis by Regulating Hepatic Glycogenolysis and Gluconeogenesis., Mol. Cell. Biol. 37 (2017). doi:10.1128/MCB.00048-17. [DOI] [PMC free article] [PubMed]
- 73.Giralt A., Denechaud P.-D., Lopez-Mejia I.C., Delacuisine B., Blanchet E., Bonner C. E2F1 promotes hepatic gluconeogenesis and contributes to hyperglycemia during diabetes. Mol. Metab. 2018;11:104. doi: 10.1016/j.molmet.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Giralt, P.-D. Denechaud, I.C. Lopez-Mejia, B. Delacuisine, E. Blanchet, C. Bonner, et al., E2F1 promotes hepatic gluconeogenesis and contributes to hyperglycemia during diabetes, Mol. Metab. 11 (2018) 104. doi:10.1016/J.MOLMET.2018.02.011. [DOI] [PMC free article] [PubMed]
- 74.Yuan X., Dong D., Li Z., Wu B. Rev-erbα activation down-regulates hepatic Pck1 enzyme to lower plasma glucose in mice. Pharmacol. Res. 2019;141:310–318. doi: 10.1016/j.phrs.2019.01.010. [DOI] [PubMed] [Google Scholar]; X. Yuan, D. Dong, Z. Li, B. Wu, Rev-erbα activation down-regulates hepatic Pck1 enzyme to lower plasma glucose in mice, Pharmacol. Res. 141 (2019) 310-318. doi:10.1016/J.PHRS.2019.01.010. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.