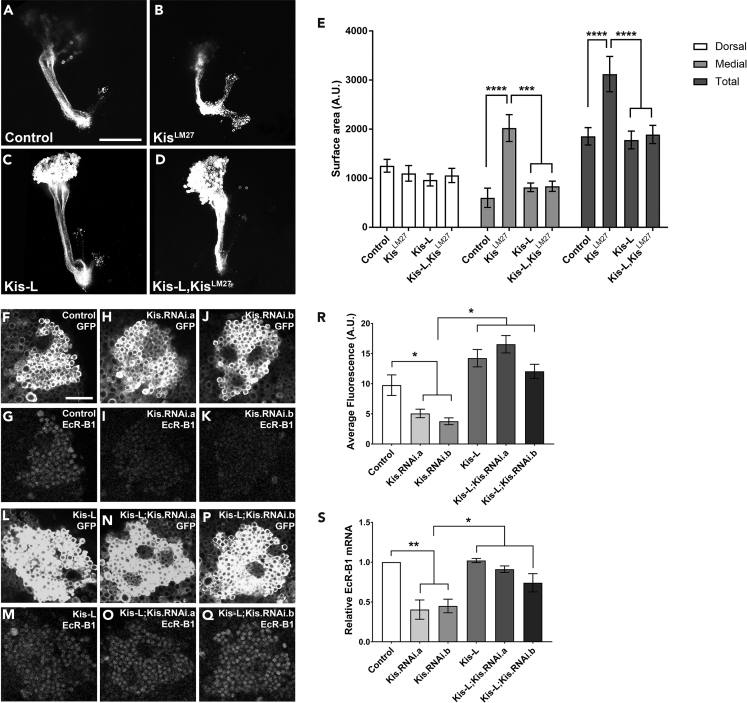
Figure 1.
Kis Is Required for Developmental Axon Pruning and EcR Expression
(A–D) Representative images of MARCM-generated MB clones expressing membrane-bound GFP using the 201y-Gal4 driver 18–22 h APF. (A) control (w1118; FRT40A), (B) kis null mutant (KisLM27,FRT 40A), (C) Kis overexpression (UAS:Kis-L,FRT40A), and (D) Kis rescue (UAS:Kis-L,KisLM27,FRT40A).
(E) Quantification of dorsal, medial, and total MB lobe surface areas in MARCM animals (from left to right, n = 12, 11, 10, 12 MBs).
(F–Q) Representative images of pupal Kenyon cells using the elav-Gal4,UAS:mCD8-GFP driver 18–22 h APF. (F) control (w1118) membrane bound GFP, (G) control (w1118) EcR-B1 protein staining, Kis knockdowns: (H) GFP and (I) EcR-B1 in UAS:Kis.RNAi.a and (J) GFP and (K) EcR-B1 in UAS:Kis.RNAi.b. (L) Kis overexpression (UAS:Kis-L) GFP, and (M) EcR-B1 protein staining and Kis rescues: (N) GFP and (O) EcR-B1 in UAS:Kis-L; UAS:Kis.RNAi.a, and (P) GFP and (Q) EcR-B1 in UAS:Kis-L,UAS:Kis.RNAi.b.
(R) Quantification of α-EcR-B1 fluorescence intensity within pupal Kenyon cells (from left to right, n = 10, 10, 10, 10, 7, 10 MBs).
(S) Abundance of ecr-b1 mRNA isolated from pupal heads analyzed by RT-qPCR using the elav-Gal4 driver (number of biological replicates from left to right, 3, 3, 4, 3, 3, 3; 10 heads/biological replicate).
Scale bars: 10 μm in (A) and 20 μm in (F). Statistical significance is represented by * = p < 0.05, ** = p < 0.01, *** = p < 0.001, and **** = p < 0.0001. Error bars represent the SEM.