ABSTRACT
Background
Infancy is a crucial period for establishing the intestinal microbiome. This process may be influenced by vitamin A (VA) status because VA affects intestinal immunity and epithelial integrity, factors that can, in turn, modulate microbiome development.
Objectives
The aim of this study was to determine if neonatal VA supplementation (VAS) affected the abundance of Bifidobacterium, a beneficial commensal, or of Proteobacteria, a phylum containing enteric pathogens, in early (6–15 wk) or late (2 y) infancy. Secondary objectives were to determine if VAS affected the abundance of other bacterial taxa, and to determine if VA status assessed by measuring plasma retinol was associated with bacterial abundance.
Methods
Three hundred and six Bangladeshi infants were randomized by sex and birthweight status (above/below median) to receive 1 VA dose (50,000 IU) or placebo within 48 h of birth. Relative abundance at the genus level and above was assessed by 16S rRNA gene sequencing. A terminal restriction fragment-length polymorphism assay was used to identify Bifidobacterium species and subspecies at 6 wk.
Results
Linear regression showed that Bifidobacterium abundance in early infancy was lower in boys (median, 1st/3rd quartiles; 0.67, 0.52/0.78) than girls (0.73, 0.60/0.80; P = 0.003) but that boys receiving VAS (0.69, 0.55/0.78) had higher abundance than boys receiving placebo (0.65, 0.44/0.77; P = 0.039). However this difference was not seen in girls (VAS 0.71, 0.54/0.80; placebo 0.75, 0.63/0.81; P = 0.25). VAS did not affect Proteobacteria abundance. Sex-specific associations were also seen for VA status, including positive associations of plasma retinol with Actinobacteria (the phylum containing Bifidobacterium) and Akkermansia, another commensal with possible health benefits, for girls in late infancy.
Conclusions
Better VA status in infancy may influence health both in infancy and later in life by promoting the establishment of a healthy microbiota. This postulated effect of VA may differ between boys and girls. This trial was registered at clinicaltrials.gov as NCT02027610.
Keywords: vitamin A, retinol, retinol-binding protein, Bifidobacterium, microbiota, microbiome, infant, Akkermansia
Introduction
Early infancy is an important period for development of the intestinal microbiome (1, 2). Facultative anaerobic bacteria are the initial colonizers of the colon but are soon followed by strict anaerobes such as Bifidobacterium longum subsp. infantis, a dominant commensal of breastfed infants that preferentially utilizes human milk oligosaccharides (3, 4). Successful colonization by strict anaerobes depends on an oxygen barrier between the blood and the gut lumen that is provided by the physical integrity and metabolic activity of the colonic epithelium, as recently reviewed (5). Impaired development of this barrier in early infancy could affect this colonization. Early infancy is a high-risk period for development of vitamin A deficiency (VAD) (6) because, in part, infants are typically born with low VA stores (7, 8). VAD can impair the development of this intestinal barrier because VA is required for normal proliferation of intestinal epithelial cells and for goblet cell differentiation (9–12). Mucus from goblet cells protects the epithelium from damage by luminal bacteria (13). VA is also required for regeneration of damaged mucosal epithelium in the gut (14–16). Thus, VAD during the neonatal period can potentially impair epithelial integrity and decrease colonization by strict anaerobes such as B. longum subsp. infantis, or by other taxa.
In addition to being required for epithelial integrity and repair, VA is also required for the normal development of the mucosal immune system. In particular, many aspects of adaptive mucosal immunity are affected by VA, including the development of regulatory T cells (17), memory B cells, and antibody-secreting plasma cells (particularly involving IgA production) (18), and targeting of these cells to the intestinal mucosa via expression of chemokine receptor type 9 and α-4/β-7 integrin on these cells (19). This targeting is particularly important for IgA-producing plasma cells which develop, beginning in the neonatal period, in response to antigen stimulation in the intestinal mucosa and then return to this site to produce secretory IgA that is transported across the epithelium into the gut lumen to coat a subset of intestinal bacteria, including both commensals and potential pathogens (20). It is thus not surprising that VAD increases the risk of invasive bacterial infections (21), and that VA-dependent effects on both T cells (required for some IgA responses) and B cells have been shown to directly affect gut microbial composition (18, 22). Determining how the immune system regulates the commensal microbiome is an active area of investigation, but recent evidence indicates that coating of commensals with IgA can help secure niche space near the mucosal surface and that such coating can also promote a tolerogenic response to these bacteria by intestinal dendritic cells (23, 24). VAD may thus impair both tolerogenic responses to commensals and protective responses to pathogens.
Infants born in several developing countries are at risk of VAD, and VA supplementation (VAS) between 6 and 59 mo of age reduces the risk of death from common infections, including diarrheal disease (25). Neonatal VAS has also been evaluated for decreasing infant mortality, and we recently conducted a randomized controlled trial of high-dose (50,000 IU) VAS in newborn Bangladeshi infants to determine if this intervention affected thymic function and T cell–mediated immune responses, as described in our study design manuscript (26). As an add-on to that study we also assessed the intestinal microbiome in early infancy (at 6, 11, and 15 wk of age) and at 2 y of age (clinicaltrials.gov: NCT02027610). Our primary hypothesis in that study, the results of which are reported here, was that VAS would increase the relative abundance of the commensal Bifidobacterium, the dominant genus in this cohort (27), and decrease the abundance of Proteobacteria, a phylum containing common enteric pathogens, based on the rationales outlined above. Although an effect seemed most likely in early infancy, we hypothesized that an imprinting effect on the immune system could also affect the microbiome composition at 2 y of age. Our initial study was designed to examine the interaction of VAS with sex of the infant and birthweight status (as a proxy for nutritional status at birth) (26) and we thus examined the effects of VAS on Bifidobacterium and Proteobacteria based on these interactions. In addition, we conducted an exploratory analysis to examine the association of VAS at birth on all microbial taxa at these ages. Finally, we also determined VA status at 15 wk and 2 y of age by measuring plasma retinol and retinol-binding protein (RBP) and separately evaluated, in an exploratory analysis, the association of VA status with the microbiome at these ages.
Methods
Study design
Many details of the study can be found in our study design manuscript (26). In brief, the initial study (NCT015839720) was a randomized controlled trial among healthy newborns in Dhaka, Bangladesh to examine the effect of VA [50,000 IU VA or identical placebo (PL) capsule within 48 h of birth] on immune function through 15 wk of age. A second study funded microbiome work and a follow-up visit for these study participants at ∼2 y of age (NCT02027610). Stool samples were collected at 6, 11, and 15 wk, and 2 y of age to measure the gut microbiota. At the 2-y visit children were categorized into older (n = 126, age 27.2–38.4 mo) or younger (n = 123, age 22.7–26.7 mo) age groups for certain analyses. For study visits in early infancy body length was measured to the nearest 0.1 cm and weight to the nearest 10 g. The same standard was used for height at 2 y but weight was recorded only to the nearest 500 g.
Plasma retinol and RBP analysis
Plasma retinol was measured in a subset of samples, plasma RBP in all samples, and plasma retinol estimated in all samples based on the use of linear regression analysis to predict retinol concentration from RBP based on this subset (28). Analytic details are provided in the Supplemental Materials.
Gut microbiota assay
Whole stool samples were collected from all infants at home by a caregiver and stored in an insulated container with a cold pack provided by study staff. Chilled stool samples were delivered to the clinic site the same or the next day in conjunction with the indicated study visits. Samples were frozen at −70°C until DNA was prepared as previously described (27). Stool DNA samples were randomized, and a mixed-template 16S V4 amplicon library was prepared and sequenced. Raw paired-end sequences were analyzed with the open-source software Quantitative Insights Into Microbial Ecology (QIIME) version 1.8 (29) as has been reported earlier (27). To estimate Bifidobacterium species and subspecies in 6-wk stool samples, we used Bifidobacterium-specific terminal restriction fragment length polymorphism assay and Bifidobacterium longum subsp. longum-infantis ratio assay, respectively, as has been reported earlier (27). Detailed amplicon library preparation, QIIME analysis pipeline, and Bifidobacterium species and subspecies assays are described in the Supplemental Material.
Statistical analysis
Statistical analyses to identify treatment effects for the preplanned outcome variables, genus Bifidobacterium and phylum Proteobacteria relative abundance, were performed with SAS 9.4 statistical software (SAS Institute Inc.). A mixed linear model procedure (MIXED) was used to identify differences between the VA and PL groups with all study visits in early infancy (6, 11, and 15 wk) included in the same analysis. A generalized linear model (GLM) was used for the Bifidobacterium species–level data, which were available at 6 wk only, and for the genus Bifidobacterium and phylum Proteobacteria data at 2 y. In addition to treatment group and age category, sex and birth weight category [above or below median birth weight (BWM) for each sex] were included in all statistical analyses per the original study design. The statistical model included treatment group, sex, BWM, age, as well as all higher-order interactions. Key maternal and infant characteristics at baseline were evaluated to identify differences between treatment groups for possible inclusion in the statistical model. The prevalence of delivery type (cesarian compared with vaginal) differed between groups, and thus it was included as a covariate without interaction terms (delivery type was examined for interactions with age but none were found significant). Statistical significance for group effects was set at P < 0.05. Interactions with treatment group at P < 0.10 were examined for significant group effects (P < 0.05) within age, sex, or BWM categories (e.g., an overall group effect at a specific age, or group effect for 1 sex at a specific age). Some group comparisons were made with Student's ttest or a chi-squared test.
Other statistical analyses were performed with R version 3.4.2 for Windows (30). Because the microbiota in early life was very similar, we combined 6, 11, and 15 bacterial sequence counts (sum of count data for subjects who had all 3 time points for microbiota data) and analyzed these time points together in these analyses. Metadata continuous variables were analyzed for normality with the Shapiro-Wilk normality test and the QQ-normal plot. Variables with the Shapiro-Wilk “W” value ≥0.95 were considered as normal. Nonnormal metadata variables were transformed by natural log, square root, square, or Box-Cox power transformation. If no appropriate transformation was found, the variables were normalized rank-transformed. Differences of homogeneity of microbial composition dispersions between VA and PL groups were determined by permutes (PERMDISP2) with the use of 999 permutations by the R package Vegan. Differences in microbial community β diversity were tested by ADONIS (perMANOVA) in the R package Vegan (31) upon controlling for the mode of delivery (vaginal or elective cesarian delivery) and breastfeeding status (exclusive or nonexclusive). Principal coordinate analysis was carried out by PhyloSeq (32). Graphs were prepared by GGplot2 (33) and GraphPad Prism (GraphPad Software, Inc.). The microbiota operational taxonomic unit table was normalized with the R package metagenomeSeq (34) by cumulative sum scaling in which nonrarified raw counts are divided by the cumulative sum of counts up to a particular quantile. Sequences belonging to an operational taxonomic unit with a shared taxonomy were combined into taxa: genus, family, and phylum levels (i.e., collapsed taxonomies) (35). The effect of VAS or VA status (plasma retinol) on microbial composition was determined by zero-inflated Gaussian mixture modeling (fitZIG) upon controlling for the confounding factors type of delivery, and breastfeeding status. The gut microbiota was found to be differentially abundant between older (n = 126; age 27.2–38.4 mo) and younger infants (n = 123; age 22.7–26.7 mo). Therefore, 2-y fitZIG mixture models were additionally adjusted for age groups. If a taxon does not have power for fitZIG mixture model analysis (meaning it was not present in the effective number of samples or it did not have at least an effective quantile of the count), the taxon was filtered out from the final result with the use of the “eff” function of the metagenomeSeq package. The SE of the differential abundant taxa was determined by multiplying the “fit$fit$stdev.unscaled” and “fit$fit$sigma” of the fitZIG model as was suggested by the metagenomeSeq developers. A false discovery rate (FDR)-adjusted P value was calculated by the Benjamin-Hochberg method at each taxonomic level. An FDR-adjusted P value (Q value) of <0.20 was considered as significant, a value similar to that used by others (36–40).
Results
Characteristics of study population
Of the 306 infants who received VA or PL at birth, 262 had 16S data available from all 3 time points in early infancy (6, 11, and 15 wk of age) (Table 1). Bifidobacterium species was determined at 6 wk of age for 280 infants, who also had 16S data available. In the overall study population, 97% of infants were born at the study hospital, 7% of births were preterm, and 60% were by cesarian delivery (as expected in hospital settings in Bangladesh) (26) (Table 1). Household characteristics (including parental education, number of siblings, and indicators of sanitation) did not differ by treatment group (Table 1). Breastfeeding was initiated for all infants and continued through 15 wk of age with an increasing rate of supplemental feeding that did not differ by treatment group (Table 1). The prevalence of moderate wasting malnutrition was <10% and of moderate stunting malnutrition was <20% (Table 1). Of the 262 infants who completed the 15-wk visit, 249 were re-enrolled at ∼2 y of age and 16S microbiota analysis was again performed. At 2 y, the study infants had somewhat higher rates of malnutrition than in early infancy (Table 1). High-dose VA supplements are provided to infants >6 mo of age as part of public health programs in Dhaka, and 83.1% of infants received ≥1, and as many as 3, such doses, but the frequency did not differ between the VA and PL groups (Supplemental Table 1).
TABLE 1.
Cohort characteristics of the study population1
| 6 wk | Early infancy2 | 2 y | ||||
|---|---|---|---|---|---|---|
| Characteristics | PL (n = 141) | VA (n = 139) | PL (n = 134) | VA (n = 128) | PL (n = 128) | VA (n = 121) |
| Sex, (male), % | 48.2 | 49.6 | 48.5 | 50.0 | 47.7 | 50.4 |
| Mode of delivery (cesarian), % | 66.7 | 54.7 | 65.7 | 54.7 | 68.8 | 56.2 |
| Place of birth (hospital), % | 96.4 | 96.4 | 96.3 | 96.9 | 96.1 | 97.5 |
| Gestational age, wk | 39.2 ± 1.58 | 39.1 ± 1.67 | 39.2 ± 1.59 | 39.1 ± 1.66 | 39.2 ± 1.58 | 39.1 ± 1.64 |
| Preterm birth (<37 wk), % | 7.19 | 7.19 | 6.82 | 7.81 | 7.14 | 6.67 |
| Birth weight, g | 2754 ± 388 | 2716 ± 363 | 2759 ± 393 | 2712 ± 360 | 2772 ± 401 | 2751 ± 384 |
| Breastfeeding status, % | ||||||
| Exclusive | 69.5 | 70.5 | — | — | 0 | 0 |
| Predominant | 7.80 | 9.35 | — | — | 0 | 0 |
| Other | 22.7 | 20.1 | — | — | 57.9 | 51.7 |
| None | 0 | 0 | — | — | 42.1 | 48.3 |
| Plasma retinol,3 µmol/L | 0.629 ± 0.0859 | 0.634 ± 0.0833 | 0.629 ± 0.0639 | 0.636 ± 0.0592 | 0.879 ± 0.139* | 0.913 ± 0.129 |
| Weight, kg | 4.37 ± 0.566 | 4.28 ± 0.547 | — | — | 10.7 ± 1.79 | 11.0 ± 1.65 |
| Length, cm | 54.3 ± 2.29 | 53.8 ± 2.09 | — | — | 83.7 ± 5.01 | 83.5 ± 6.48 |
| Wasting (WHZ <–2), % | 4.96 | 4.34 | — | — | 13.0 | 7.08 |
| Stunting (HAZ <–2), % | 15.6 | 18.8 | — | — | 35.8 | 32.7 |
| Father's educational status, years of school | 8.44 ± 3.72 | 8.68 ± 4.04 | 8.37 ± 3.75 | 9.26 ± 7.70 | 8.49 ± 3.64 | 9.11 ± 8.36 |
| Mother's educational status, years of school | 8.33 ± 3.25 | 8.62 ± 3.46 | 8.36 ± 3.21 | 8.58 ± 3.39 | 8.35 ± 3.13 | 8.56 ± 3.24 |
| Number of people per household | 4.52 ± 1.88 | 4.50 ± 2.01 | 4.54 ± 1.89 | 4.54 ± 2.07 | 4.58 ± 1.96 | 4.34 ± 1.63 |
| Number of live siblings | 0.688 ± 0.855 | 0.669 ± 0.802 | 0.694 ± 0.860 | 0.680 ± 0.822 | 0.625 ± 0.832 | 0.658 ± 0.804 |
| n = 0, % | 53.2 | 51.1 | 53.0 | 51.6 | 57.0 | 51.7 |
| n ≥ 1, % | 46.8 | 48.9 | 47.0 | 48.4 | 43.0 | 48.3 |
| Any children died before (yes), % | 12.1 | 15.8 | 12.7 | 17.2 | 11.7 | 15.0 |
| Access to drinking water, % | ||||||
| Piped water | 90.0 | 88.5 | 90.3 | 89.8 | 88.7 | 89.8 |
| Public tap | 5.0 | 4.3 | 5.22 | 4.69 | 6.45 | 4.23 |
| Tube well | 5.0 | 7.2 | 4.48 | 5.47 | 4.85 | 5.97 |
| Type of toilet | ||||||
| Sanitary latrine | 97.9 | 95.0 | 97.8 | 96.1 | 98.4 | 96.6 |
| Pit latrine | 1.42 | 4.32 | 1.49 | 3.13 | 1.61 | 3.39 |
| Other | 0.68 | 0.68 | 0.71 | 0.77 | 0.0 | 0.0 |
| Floor material, % | ||||||
| Cement or tiles | 95.74 | 94.2 | 95.5 | 95.3 | 96.0 | 94.1 |
| Other | 4.30 | 5.8 | 4.5 | 4.7 | 4.03 | 5.90 |
1Data presented as mean ± SD or frequency as percentage. Difference between treatment groups was determined by Student's t test or chi-squared test. *P < 0.05. VA, neonatal high-dose vitamin A–supplemented group; PL, placebo group.
2Statistics of the 262 infants who have complete set of microbiota data at 6, 11, and 15 wk of age.
3Early-infancy retinol is the average of the 6- and 15-wk plasma retinol levels.
Microbial composition in the study population
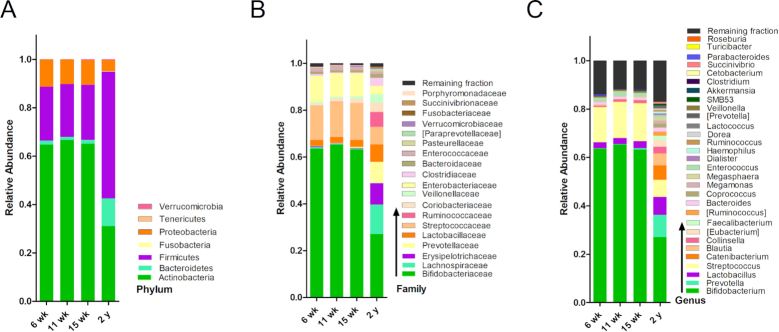
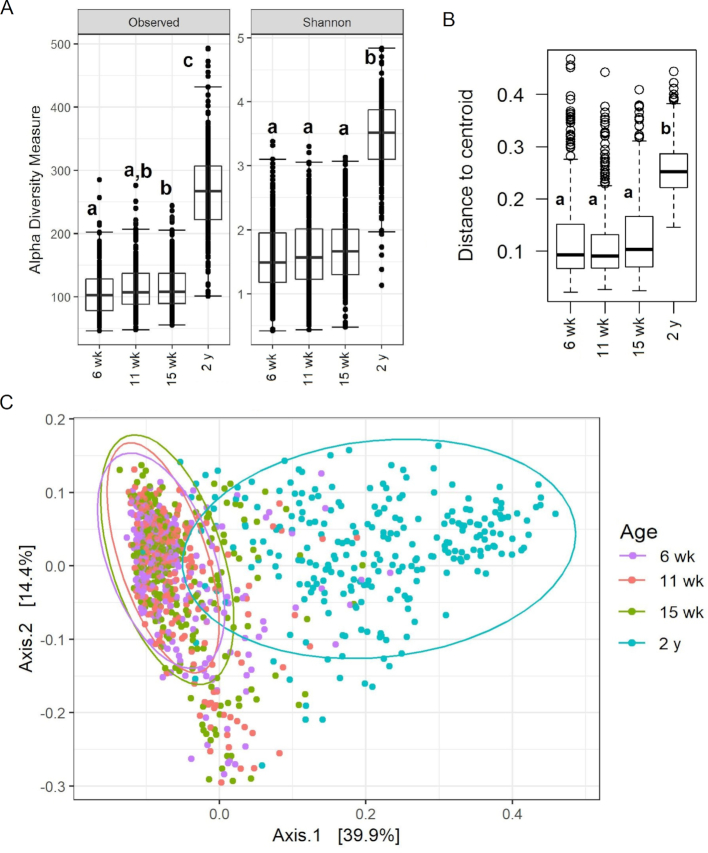
Actinobacteria, predominantly the genus Bifidobacterium, dominated the intestinal flora at 6, 11 and 15 wk of age, with overall composition differing very little at these time points (Figure 1,Supplemental Figure 5). The relative abundance of Bifidobacterium was ∼65% during early infancy and decreased to 28% at 2 y of age, though it still remained the most abundant genus. Firmicutes, rather than Actinobacteria, was the most abundant phylum at 2 y of age. As was seen with bacterial abundance, the 6-, 11-, and 15-wk microbiota had similar α and β diversity measures (Figure 2), although at 15 wk infants had a higher number of observed species than at 6 wk. The microbial composition at 2 y differed from that seen in early infancy.
FIGURE 1.
Mean relative abundance of the gut microbiota at 6 wk (n = 280), 11 wk (n = 281), 15 wk (n = 279), and 2 y (n = 249) of age: (A) phylum, (B) top 20 family, and (C) top 30 genera.
FIGURE 2.
Comparisons of the (A) α diversity measures, (B) β dispersion and (C) β diversity principal coordinate analysis on weighted UniFrac of gut microbiota at 6 wk (n = 280), 11 wk (n = 281), 15 wk (n = 279), and 2 y (n = 249) of age. One-factor ANOVA followed by Tukey highest significant difference post hoc comparisons were used to compare α diversity measures. Box represents the IQR, central line is the median value, and lower and upper whiskers represent the 10th and 90th percentile, respectively. β dispersion measures were compared with the PERMDISP2 permutation test. Different letters on the box represent significant difference and common letters represent no differences. Differences in β diversity were tested by ADONIS (perMANOVA) test (R2 = 0.293, P < 0.001). The ellipse on the principal coordinate analysis plot indicates 95% CI of the clusters by age.
Effect of VAS on Bifidobacterium genus relative abundance at 6, 11, and 15 wk of age
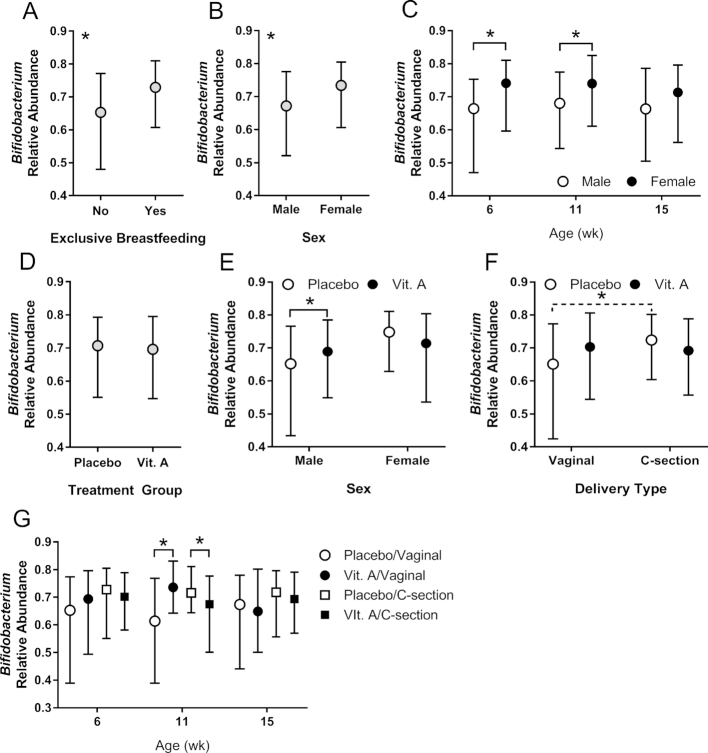
We used a mixed linear model to test our a priori hypothesis that VAS at birth would increase the relative abundance of Bifidobacterium in early infancy. Preplanned interaction terms were included for sex and median birth weight category (above/below) because these factors can affect response to VAS (26). Delivery type (vaginal compared with cesarian) and breastfeeding status (exclusive/not exclusive) were also included in the model with interaction terms. Our primary analysis focused on Bifidobacterium relative abundance determined with 16S analysis at 6, 11, and 15 wk of age (Table 2). In this analysis we found that Bifidobacterium relative abundance was higher in exclusively breastfed infants than in those receiving supplemental feeding (Figure 3A and Table 2). In addition, Bifidobacterium differed by sex, with boys having lower relative abundance than girls (Figure 3B), although this difference was not significant at 15 wk of age (Figure 3C).
TABLE 2.
Results of mixed linear regression analysis (degrees of freedom, F statistic and P value; described in Methods) assessing effect of vitamin A treatment group on Bifidobacterium relative abundance measured in early infancy1
| Effect | F value | P value |
|---|---|---|
| Group | 0.43 | 0.513 |
| Sex | 8.75 | *0.003 |
| BWM | 2.51 | 0.114 |
| Type | 3.28 | *0.071 |
| Age | 1.72 | 0.182 |
| BF | 19.81 | *<0.0001 |
| Group × sex | 5.46 | *0.020 |
| Group × BWM | 0.10 | 0.755 |
| Group × type | 5.43 | *0.021 |
| Group × age | 0.12 | 0.889 |
| Sex × BWM | 0.03 | 0.854 |
| Sex × type | 0.81 | 0.368 |
| Sex × age | 2.44 | *0.089 |
| BWM × type | 0.08 | 0.784 |
| BWM × age | 1.46 | 0.233 |
| Type × age | 1.54 | 0.216 |
| Group × sex × BWM | 0.00 | 0.980 |
| Group × sex × type | 0.00 | 0.971 |
| Group × sex × age | 0.22 | 0.805 |
| Group × BWM × type | 1.58 | 0.209 |
| Group × BWM × age | 0.00 | 0.999 |
| Group × type × age | 2.86 | *0.059 |
| Group × sex × BWM × type | 0.13 | 0.877 |
| Group × sex × BWM × age | 1.34 | 0.256 |
| Group × sex × type × age | 0.60 | 0.664 |
| Group × BWM × type × age | 0.78 | 0.537 |
| Group × sex × BWM × type × age | 1.18 | 0.322 |
1 P values indicated by an asterisk (*) are <0.10. Bifidobacterium relative abundance during early infancy was measured at ages 6 wk (n = 280), 11 wk (n = 281), and 15 wk (n = 279). Group, vitamin A or placebo treatment groups; BWM, birthweight median category (above/below); type, type of delivery (cesarian/vaginal); and BF, breastfeeding status (1 = exclusive/0 = nonexclusive) at 6, 10, and 15 wk of age. The overall model chi-squared value was 41.21 (P < 0.0001).
FIGURE 3.
Median relative abundance (error bars indicate 25th/75th percentiles) of Bifidobacterium relative abundance in early infancy (n = 840 observations overall from 280 infants at 6 wk, 281 at 11 wk, and 279 at 15 wk). (A) Overall effect of breastfeeding status (exclusive, n = 474; nonexclusive, n = 366 observations); (B) overall effect of sex (male, n = 415; female, n = 425 observation); (C) age-specific sex differences identified due to significant sex-by-age interaction (male, n = 137, 137, and 141; female, n = 143, 144, and 138 observations at 6, 11, and 15 wk, respectively); (D) overall effect of VA treatment (PL, n = 427; VA n = 413 observations); (E) sex-specific treatment effect identified due to significant group-by-sex interaction (male, n = 208, 207; female, n = 219, 206 observations in PL and VA groups, respectively; (F) delivery type–specific treatment effect identified due to significant group-by-delivery type interaction, with marginal (P = 0.059) treatment effect (vaginal, n = 148, 185; cesarian, n = 279, 228 observations in PL and VA groups, respectively); (G) age– and delivery type–specific treatment effect identified due to significant group-by-age-by-delivery type interaction (PL/vaginal, n = 47, 50, 51; VA/vaginal, n = 63, 61, 61; PL/cesarian, n = 94, 93, 92; VA/cesarian, n = 76, 77, 75 observations at 6, 11, and 15 wk, respectively). Statistical significance was determined with the use of the mixed regression analysis described in Table 2. The asterisks (*) indicate P < 0.05 either as a main effect (panels A and B) or by post hoc comparisons between VA and PL groups (no correction for multiple comparisons); or P < 0.05 adjusted for multiple comparisons in panel F. Panels A, B, D, E, and F show comparisons of mean relative abundance across the 3 ages, whereas panels C and G show age-specific data. PL, placebo; VA, vitamin A.
This analysis also found that Bifidobacterium relative abundance differed by VA treatment group in boys but not girls. Although there was no overall effect of VAS in the model (Figure 3D), there was a significant interaction between treatment group and sex: among boys, the relative abundance of Bifidobacterium was higher in the VA than in the PL group, whereas no difference was seen between the VA and PL groups in girls (Figure 3E). In addition, there was a significant interaction between VAS and type of delivery that was age dependent. Considering the PL group only, infants delivered vaginally had significantly lower Bifidobacterium abundance than infants delivered by cesarian section, whereas no such difference was seen in the infants who received VA (Figure 3F). However, this interaction varied by age: no significant differences were seen at 6 and 15 wk, but at 11 wk of age Bifidobacterium abundance was higher in the VA than in the PL group among infants delivered vaginally, whereas the opposite association was seen in infants delivered by cesarian section (Figure 3G).
Effect of VAS on Proteobacteria relative abundance at 6, 11, and 15 wk of age
We also analyzed the effect of VAS on the phylum Proteobacteria and the family Enterobacteriaceae, according to our original hypothesis, with the same mixed model described for Bifidobacterium in early infancy. No significant effect of treatment was seen (data not shown).
Effect of VAS on B. longum and B. longum subsp. infantis relative abundance at 6 wk of age
This analysis was performed essentially as described above for the genus-level data at 6, 11, and 15 wk, with the use of the same covariates, although a generalized linear model was used rather than a mixed model as only 1 time point, 6 wk, was available for species and subspecies data. The relative abundance of B. longum subsp. infantis was higher in girls than in boys, was higher in exclusively breastfed infants than in those receiving supplemental feeding, was higher in infants delivered by cesarian section than in those delivered vaginally, and was higher in those with birth weights above the median than in those with lower birth weights (Supplemental Figure 6). The same differences were seen at the species level for B. longum (although the difference by delivery type was not statistically significant) and at the genus level for Bifidobacterium (Supplemental Figure 6). VAS did not affect bifidobacterial levels in these analyses, either as a main effect of as an interacting term (data not shown).
VAS and other bacterial taxa
In addition to testing our preplanned hypotheses with regard to Bifidobacterium and Proteobacteria, we analyzed the differential abundance of all taxa between VA and PL groups by fitZIG mixture modeling to accommodate nonnormal distributions and taxa with many zero values. In preliminary analyses we observed interactions of treatment with sex and birth weight. For this reason, and because study subjects were originally randomized to VA and PL groups by sex and birth weight status, we conducted these analyses separately in the 4 randomization groups, i.e., males above or below the male median birth weight (MA and MB, respectively) and females above or below the female median birth weight (FA and FB, respectively), while adjusting for breastfeeding status and delivery type.
Microbiota analysis in early infancy by these methods found no associations for Bifidobacterium (with a sum of counts at 6, 11, and 15 wk of age as the dependent variable) in any of the 4 groups (Tables 3–6), nor were effects seen for other taxa in the MB, FA, or FB groups. However, in the MA infants, the VA group had 2.2-fold higher Corynebacterium (P = 0.0024, adjusted P = 0.11) and 2.2-fold lower Acinetobacter (P = 0.0073, adjusted P = 0.16) than did the PL group (Table 3).
TABLE 3.
Differentially abundant taxa in vitamin A–supplemented group compared with placebo in male infants with birth weight above median1
| Log2 fold change (SE) | P | Adjusted P | |
|---|---|---|---|
| Early infancy | |||
| Phylum | |||
| Proteobacteria | 0.451 (0.334) | 0.18 | 0.51 |
| Family | |||
| Moraxellaceae | −1.18 (0.365) | 0.0023 | 0.067 |
| Corynebacteriaceae | 1.08 (0.415) | 0.011 | 0.16 |
| Genus | |||
| Acinetobacter | −1.10 (0.347) | 0.0024 | 0.11 |
| Corynebacterium | 1.14 (0.413) | 0.0073 | 0.16 |
| 2 y | |||
| Phylum | |||
| Verrucomicrobia | −0.802 (0.605) | 0.178 | 0.42 |
| Family | |||
| Streptococcaceae | −1.21 (0.456) | 0.0092 | 0.16 |
| Comamonadaceae | −0.76 (0.3) | 0.018 | 0.16 |
| Actinomycetaceae | −0.593 (0.244) | 0.019 | 0.16 |
| Eubacteriaceae | −0.343 (0.112) | 0.022 | 0.16 |
| Gemellaceae | −0.437 (0.174) | 0.026 | 0.16 |
| Genus | |||
| Streptococcus | −1.34 (0.437) | 0.0029 | 0.16 |
| Atopobium | −0.576 (0.197) | 0.0090 | 0.16 |
| Megamonas | 1.79 (0.713) | 0.014 | 0.16 |
| Gemella | −0.466 (0.176) | 0.018 | 0.16 |
| Actinomyces | −0.592 (0.243) | 0.019 | 0.16 |
| Comamonas | −0.766 (0.31) | 0.02 | 0.16 |
| Pseudoramibacter_Eubacterium | −0.347 (0.114) | 0.02 | 0.16 |
| Acidaminococcus | −0.541 (0.213) | 0.022 | 0.16 |
| Sarcina | −0.651 (0.294) | 0.036 | 0.23 |
| Dorea | −0.708 (0.352) | 0.047 | 0.28 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 63) and at 2 y of age (n = 65). Taxa with a P value <0.05 or the lowest one were selected for this table. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data at 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
TABLE 6.
Differentially abundant taxa in vitamin A–supplemented group compared with placebo during early infancy and at 2 y of age in female infants with birth weight below medianwith placebo during early infancy and at 2 y of age in female infants with birth weight above median1
| Log2 fold change (SE) | P | Adjusted P | |
|---|---|---|---|
| Early infancy | |||
| Phylum | |||
| Actinobacteria | −0.192 (0.119) | 0.12 | 0.43 |
| Family | |||
| Gemellaceae | −0.739 (0.326) | 0.027 | 0.44 |
| Peptostreptococcaceae | −0.423 (0.148) | 0.030 | 0.44 |
| Genus | |||
| Gemella | −0.725 (0.325) | 0.030 | 0.44 |
| Oscillospira | −0.634 (0.271) | 0.030 | 0.44 |
| Peptostreptococcus | −0.414 (0.142) | 0.034 | 0.44 |
| 2 y | |||
| Phylum | |||
| Firmicutes | −0.274 (0.152) | 0.098 | 0.332 |
| Family | |||
| Neisseriaceae | 0.566 (0.196) | 0.010 | 0.32 |
| Genus | |||
| Neisseria | 0.556 (0.201) | 0.0096 | 0.46 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 66) and at 2 y of age (n = 63). Taxa with a P-value <0.05 or the lowest one were selected for this table. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data at 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
Taxa at 2 y of age were also examined for association with VA treatment at birth. For the FA infants, the VA group had a 1.5-fold higher relative abundance of the phylum Actinobacteria (primarily comprised of Bifidobacterium) than the PL group (P = 0.049, adjusted P = 0.15; Table 4). With regard to other taxa, multiple effects were seen in the MA infants (Table 3) with the lowest P value seen for a lower relative abundance of Streptococcus in the VA than in the PL group (P = 0.0029; adjusted P = 0.16). In the MB infants, Parabacteroides relative abundance was lower in the VA than in the PL group (Table 4). In the FA infants, Megasphaera relative abundance was higher in the VA than in the PL group (Table 5).
TABLE 4.
Differentially abundant taxa in vitamin A–supplemented group compared with placebo during early infancy and at 2 y of age in male infants with birth weight below median1
| Log2 fold change (SE) | P | Adjusted P | |
|---|---|---|---|
| Early infancy | |||
| Phylum | |||
| Firmicutes | −0.261 (0.23) | 0.26 | 0.96 |
| Family | |||
| Gemellaceae | −0.646 (0.293) | 0.037 | 0.86 |
| Genus | |||
| Catenibacterium | 0.714 (0.378) | 0.062 | 0.81 |
| 2 y | |||
| Phylum | |||
| Bacteroidetes | 0.485 (0.462) | 0.29 | 0.83 |
| Family | |||
| Porphyromonadaceae | −1.45 (0.466) | 0.0031 | 0.095 |
| Bacteroidaceae | −1.56 (0.644) | 0.017 | 0.25 |
| Eubacteriaceae | −0.6 (0.245) | 0.025 | 0.25 |
| Genus | |||
| Parabacteroides | −1.47 (0.458) | 0.0023 | 0.14 |
| Bacteroides | −1.57 (0.655) | 0.019 | 0.43 |
| Anaerostipes | −0.467 (0.193) | 0.026 | 0.43 |
| Faecalibacterium | 1.16 (0.543) | 0.035 | 0.43 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 66) and at 2 y of age (n = 57). Taxa with a P value <0.05 or the lowest one were selected for this table. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data at 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
TABLE 5.
Differentially abundant taxa in vitamin A–supplemented group compared with placebo during early infancy and at 2 y of age in female infants with birth weight above median1
| Log2 fold change (SE) | P | Adjusted P | |
|---|---|---|---|
| Early infancy | |||
| Phylum | |||
| Fusobacteria | 0.355 (0.27) | 0.20 | 0.77 |
| Family | |||
| Corynebacteriaceae | 0.721 (0.357) | 0.046 | 0.98 |
| Genus | |||
| Corynebacterium | 0.708 (0.361) | 0.052 | 0.96 |
| 2 y | |||
| Phylum | |||
| Actinobacteria | 9.55 (0.316) | 0.049 | 0.15 |
| Bacteroidetes | 4.88 (0.336) | 0.050 | 0.15 |
| Family | |||
| Carnobacteriaceae | −0.381 (0.232) | 0.038 | 0.92 |
| Genus | |||
| Megasphaera | 2.33 (0.639) | <0.001 | 0.031 |
| Faecalibacterium | −1.31 (0.558) | 0.020 | 0.56 |
| Slackia | −0.63 (0.294) | 0.038 | 0.56 |
| Granulicatella | −0.511 (0.233) | 0.038 | 0.56 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 67) and at 2 y of age (n = 64). Taxa with a P value <0.05 or the lowest one were selected for this table. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data at 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
VA status is associated with gut microbial composition
We measured plasma retinol at 6 and 15 wk of age and used the mean of these 2 values for each infant to determine the association between VA status in early infancy and the composition of the gut microbiome at that time and at 2 y of age through the use of the fitZIG approach described above. Initial analyses of the association between microbiota and serum retinol with all infants found a significant interaction between serum retinol and sex for some taxa, and thus we analyzed boys and girls separately. In these analyses for early infancy, the family Leuconostocaceae was negatively associated with plasma retinol but only in boys (Table 7), whereas no associations were seen in girls (Table 8). When 2-y microbiota data were analyzed, the relative abundance of the genus Cetobacterium [also at family (Fusobacteriaceae) and phylum (Fusobacteria) levels] was ∼200-fold higher for every unit (µmol/L) increase in early-life plasma retinol for boys (Table 7). On the other hand, in girls, plasma retinol in early infancy was positively associated with Acidaminococcus, Comamonas, and Eggerthella abundance at 2 y of age, whereas the relative abundance of Mitsuokella was negatively associated with early-life plasma retinol in girls (Table 8).
TABLE 7.
Differentially abundant taxa for every unit increase of plasma retinol during early infancy and at 2 y of age in male subjects1
| Log2 fold difference (SE) | P | Adjusted P | |
|---|---|---|---|
| Plasma retinol at early infancy | |||
| Microbiota in early infancy | |||
| Phylum | |||
| Verrucomicrobia | 6.24 (3.05) | 0.04 | 0.25 |
| Family | |||
| Leuconostocaceae | −8.03 (2.84) | 0.0054 | 0.16 |
| Enterobacteriaceae | −4.91 (2.06) | 0.018 | 0.27 |
| Genus | |||
| Leuconostoc | −7.72 (2.77) | 0.0059 | 0.26 |
| f_Enterobacteriaceae_g_NA | −5.10 (2.02) | 0.012 | 0.27 |
| Microbiota at 2 y | |||
| Phylum | |||
| Fusobacteria | 7.38 (1.88) | <0.001 | 0.0011 |
| Family | |||
| Fusobacteriaceae | 7.49 (1.87) | <0.001 | 0.0043 |
| Comamonadaceae | −2.43 (1.12) | 0.037 | 0.57 |
| Genus | |||
| Cetobacterium | 7.80 (1.76) | <0.001 | <0.001 |
| Mitsuokella | −3.06 (1.08) | 0.010 | 0.21 |
| Comamonas | −2.64 (1.12) | 0.020 | 0.47 |
| Plasma retinol at 2 y | |||
| Microbiota at 2 y | |||
| Phylum | |||
| Verrucomicrobia | 3.43 (1.30) | 0.0088 | 0.053 |
| Family | |||
| Verrucomicrobiaceae | 3.59 (1.31) | 0.007 | 0.21 |
| Eubacteriaceae | 1.02 (0.45) | 0.033 | 0.43 |
| Genus | |||
| Akkermansia | 3.77 (1.3) | 0.0050 | 0.27 |
| Lactococcus | −3.38 (1.47) | 0.023 | 0.42 |
| Pseudoramibacter_Eubacterium | 1.05 (0.46) | 0.03 | 0.42 |
| Cetobacterium | 1.72 (0.79) | 0.032 | 0.42 |
| Mitsuokella | −1.23 (0.6) | 0.047 | 0.42 |
| Gemella | −0.85 (0.41) | 0.050 | 0.42 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 129) and at 2 y of age (n = 122). Taxa with a P value <0.05 or the lowest one were selected for this table. Plasma retinol concentration is the mean of the 6- and 15-wk values. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data at 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
TABLE 8.
Differentially abundant taxa for every unit increase of plasma retinol during early infancy and at 2 y of age in female subjects1
| Log2 fold difference (SE) | P | Adjusted P | |
|---|---|---|---|
| Plasma retinol at early infancy | |||
| Microbiota in early infancy | |||
| Phylum | |||
| Proteobacteria | −3.75 (2.09) | 0.07 | 0.40 |
| Family | |||
| Prevotellaceae | −7.51 (3.07) | 0.015 | 0.37 |
| Coriobacteriaceae | −9.33 (4.17) | 0.026 | 0.37 |
| Lactobacillaceae | −7.24 (3.65) | 0.048 | 0.46 |
| Genus | |||
| Prevotella | −8.2 (2.99) | 0.0065 | 0.29 |
| Lactobacillus | −7.81 (3.7) | 0.035 | 0.59 |
| Collinsella | −9.26 (4.53) | 0.041 | 0.59 |
| Microbiota at 2 y | |||
| Phylum | |||
| Verrucomicrobia | −1.69 (2.61) | 0.52 | 0.86 |
| Family | |||
| Comamonadaceae | 2.95 (0.898) | 0.0030 | 0.089 |
| Alcaligenaceae | 4.20 (2.09) | 0.046 | 0.72 |
| Genus | |||
| Acidaminococcus | 2.45 (0.588) | <0.001 | 0.023 |
| Comamonas | 2.92 (0.896) | 0.0027 | 0.079 |
| Mitsuokella | −5.89 (2.04) | 0.0051 | 0.101 |
| Eggerthella | 4.55 (1.64) | 0.0069 | 0.101 |
| Sutterella | 4.68 (2.05) | 0.024 | 0.28 |
| Oscillospira | 3.63 (1.80) | 0.046 | 0.45 |
| Plasma retinol at 2 y | |||
| Microbiota at 2 y | |||
| Phylum | |||
| Verrucomicrobia | 4.26 (1.10) | <0.001 | 0.0011 |
| Actinobacteria | 1.85 (0.875) | 0.036 | 0.11 |
| Family | |||
| Verrucomicrobiaceae | 4.22 (1.08) | <0.001 | 0.0061 |
| Turicibacteraceae | 3.06 (1.09) | 0.0058 | 0.091 |
| Carnobacteriaceae | −1.74 (0.64) | 0.0097 | 0.101 |
| Bifidobacteriaceae | 1.94 (0.92) | 0.037 | 0.29 |
| Genus | |||
| Akkermansia | 4.29 (1.10) | <0.001 | 0.011 |
| Turicibacter | 3.325 (1.06) | 0.0023 | 0.067 |
| Eggerthella | 2.14 (0.75) | 0.0054 | 0.11 |
| Granulicatella | −1.74 (0.645) | 0.0096 | 0.14 |
| Slackia | −1.65 (0.73) | 0.027 | 0.32 |
| Actinobacillus | −1.55 (0.748) | 0.043 | 0.42 |
1Differential taxa abundance at phylum, family, and genus levels was determined by fitZIG mixture modeling during early infancy (average of 6, 11, and 15 wk; n = 133) and at 2 y of age (n = 127). Taxa with a P value <0.05 or the lowest one were selected for this table. Plasma retinol concentration is the mean of the 6- and 15-wk values. Early-infancy microbiota data were adjusted for type of delivery and breastfeeding status. Microbiota data 2 y were additionally adjusted for age group at 2 y (older and younger). False discovery rate–adjusted P value (adjusted P) was calculated by the Benjamin-Hochberg method.
Plasma retinol measured at 2 y of age was also used to identify associations with microbiota at that age in boys and girls separately. In this analysis, Actinobacteria relative abundance was positively associated with plasma retinol in girls but not boys (Table 8). For every 1 µmol/L increase of plasma retinol at 2 y a 3.6-fold increase in Actinobacteria relative abundance was seen. At 2 y, 30.4% of the total bacteria belonged to the phylum Actinobacteria and 86.7% of these were the genus Bifidobacterium, which made it the most abundant genus. Interestingly, the relative abundance of another mucosa-associated bacterium, Akkermansia, was also positively associated with plasma retinol in girls (Table 8). Every unit increase of plasma retinol was associated with 20-fold higher Akkermansia abundance regardless of breastfeeding status and type of delivery. This association was consistent from the phylum (Verrucomicrobia) through the genus (Akkermansia) level. In boys, a positive association of similar magnitude was also seen, but was statistically significant only at the level of the phylum Verrucomicrobia (Table 7). Consistent with plasma retinol in early infancy, 2-y plasma retinol was also positively associated with 2-y Eggerthella relative abundance in girls (Table 8). Also, in girls the abundance of 2 other bacteria, Turicibacter and Granulicatella, was positively and negatively associated with plasma retinol levels, respectively (Table 8).
Discussion
Colonization of the gut with commensal microbiota during early infancy has both short-term and long-lasting benefits for infant health that result from the direct effects of commensal flora on the development of the infant's immune system (41). Because VA is also required for development of the mucosal immune system (42), as well as for integrity of the mucosal epithelium (43), VAD in early infancy may thus disrupt normal development of the commensal microbiota, whereas VAS during this period for infants at risk of VAD may restore this process. We tested this hypothesis in the present study and found that VAS at birth increased the relative abundance of Bifidobacterium in early infancy. This bifidogenic effect was seen in boys but not girls. Boys had lower Bifidobacterium abundance than girls and VAS increased the levels in boys to approximately that seen in girls. It is possible that boys benefited because they had lower VA status at birth, although when we measured serum retinol at 6 wk of age no difference was seen. Observational data in humans support a sex-specific effect of diet on intestinal microbiota (44), and such differences are also seen in mice and have been attributed to the effects of sex hormones through experiments involving ovariectomy and gonadectomy (45). Sex differences in the microbiome have also been reported from human neonates in 2 recent studies (although not involving Bifidobacterium) (46, 47), a period when testosterone levels are higher in boys than girls (48). Interestingly, a study of New Hampshire infants (49) has reported a sex difference in the microbiome related to environmental exposure to arsenic. In that study, higher arsenic exposure was associated with lower Bifidobacterium levels in boys but not girls. Arsenic exposure is common in some rural areas of Bangladesh due to high levels in groundwater (50). Arsenic exposure (as determined by urinary arsenic) is much higher in neonates from 1 affected rural area of Bangladesh than in the New Hampshire study with affected Bifidobacterium abundance, although breast milk levels were quite low in the Bangladeshi mothers (51). However, arsenic exposure in the city of Dhaka, where our study was completed, is expected to be much lower than in rural areas with high arsenic concentrations in groundwater (50). Thus we cannot attribute the lower Bifidobacterium abundance in boys than girls in our study to arsenic exposure due to this lower risk and our lack of exposure data for this study, although we felt it important to note the possibility. Finally, we feel that this sex difference in the microbiome response to VAS is consistent with other studies in infants. In particular, mortality rates have been seen to decrease to a greater degree in boys than girls (52, 53), an effect that is plausibly linked to effects on the intestinal microbiome.
To our knowledge this is the first controlled intervention trial that has examined the effect of VAS on the infant microbiome. The gut microbiome of infants <1 y of age hospitalized with diarrhea was examined by VA status through the use of serum retinol measurements, but Bifidobacterium levels did not differ between these groups (54). A study of children with autism spectrum disorder who were 1–8 y of age found a decrease in Bifidobacterium over a 6-mo period of VAS but a parallel control group was not used and thus a causal inference cannot be drawn from this study (55). Two studies in mice have shown that retinoic acid treatment can increase gut Bifidobacterium abundance (56, 57), supporting the observation in the present report, although another recent study in VAD and control have not reported effects on bifidobacteria (58).
Although we did not examine mechanisms in this study, we presume that VA could increase Bifidobacterium abundance by affecting the epithelial or immunologic functions of the mucosal barrier. It is possible that VA enhanced regulatory immunity to directly facilitate colonization by commensal bacteria, or protective immunity to decrease competition from enteric pathogens that might disrupt this process, because VA has many effects on the mucosal immune system (42). It is also possible that VAS could promote normal development of the colonic epithelial barrier (9–12), or enhance its repair after damage (14–16), to maintain an appropriate, low oxygen tension in the colonic lumen (59). Because VAS occurred within 48 h of birth, it is likely that VA did not act directly on intestinal bacteria. Studies in model systems would be required to identify the specific mechanism involved.
At 6 wk of age, both Bifidobacterium and B. longum subsp. infantis relative abundance were higher in infants delivered by cesarian than in those delivered vaginally (Supplemental Figure 6). However, this difference by delivery type was statistically significant only in infants in the PL group in our model examining Bifidobacterium at 6, 11, and 15 wk of age (Figure 3), suggesting that the association changed over time in early infancy. Previous studies have reported a lower abundance of Bifidobacterium in cesarian delivery infants, as recently reviewed (2). These previous studies were largely in Western settings and our data are from a maternity hospital in Bangladesh, and it is possible that, in addition to VAS, practices in that hospital may mitigate the detrimental effect of cesarian delivery on Bifidobacterium colonization.
In our study population we found that the genus Akkermansia was positively associated with 2-y plasma retinol levels, suggesting that VA may promote its colonization of the infant gut. Akkermansia muciniphila is a mucin-utilizing bacterium identifed in the human gut relatively recently (60) and, as recently reviewed, is a common commensal in both infants and adults which associates with the colonic mucosa (61). Because VAD decreases goblet cell levels in the rat small and large intestines (9, 10, 12) it is tempting to speculate that better VA status may help provide a niche for this organism to thrive. The observed lower abundance of Akkermansia in association with metabolic disease (62, 63) has stimulated an interest in understanding its role in metabolic health, and the present observation raises the interesting possibility that its niche in the colon might be affected by VA status.
In the present study we found that VAS was associated with increased Bacteroidetes abundance at 2 y in female infants above the median birthweight. In these subjects, 67% of the reads within the phylum Bacteroidetes at 2 y consisted of the genus Prevotella. Similarly, the VA intervention trial in children with autism spectrum of Liu et al. (55) found an increase in Prevotella abundance, although the study did not have a parallel control group, as noted previously.
Microbial diversity and composition at 6, 11, and 15 wk were similar except the observed number of species at 15 wk was higher than at earlier ages. The higher observed species at 15 wk is probably a result of the early introduction of complementary food and decreased exclusive breastfeeding rate compared with 6 and 11 wk. The 2-y microbiota was more diverse, with a number of new genera that were not seen in early infancy. The introduction of solid foods is known to be associated with the development of a more diverse microbiota (46). At 2 y of age, infants had more Firmicutes than in early infancy, indicating the development of a more adult-like microbiota as they increased consumption of solid and semisolid food.
Our study has several strengths, including a large sample size, its placebo-controlled intervention format, the early intervention to affect VA status at the onset of microbial colonization, the follow-up of infants at 2 y of age who were recruited at birth, recruitment of infants from a single institution which could minimize variability in results, and the repeated sampling early in infancy, which may have facilitated identification of the difference between intervention groups, as well as other differences due to sex, breastfeeding, and delivery status. The high abundance of Bifidobacterium during early infancy enabled us to use a standard mixed-model, repeated-measures approach to statistical analysis that allowed identification of the VA treatment effect in early infancy. Limitations to the study include the lack of stool sampling prior to 6 wk and between 15 wk and 2 y of age. Recruitment from a single maternity hospital where cesarian delivery rates were high and maternal and infant care was good may have limited the heterogeneity of the study population and thus limited generalizability to populations with different levels of care. Our study population was urban, where VA status is somewhat better than in rural areas of Bangladesh according to recent survey data (64), which may also limit the generalizability of our results to populations with higher rates of deficiency in rural areas of south Asia.
In conclusion, VA supplementation was associated with a higher relative abundance of Bifidobacterium and better VA status was associated with higher abundance of another mucosa-associated genus, Akkermansia. Given the role of VA in promoting intestinal immunity and epithelial integrity, as discussed above, VA appears to also promote healthy colonization of the intestinal mucosa with commensal bacteria, a role that has not been previously appreciated. Increasing the abundance of Bifidobacterium may modulate immune development to promote health in infancy as well as later in life, including the promotion of more robust vaccine responses (27) as well as decreasing the risk of immune-mediated diseases such as atopic asthma (41, 65), whereas increasing the abundance of Akkermansia could decrease the risk of metabolic disease (61). The lower levels of a Proteobacteria genus containing invasive enteric pathogens, Acinetobacter (66), also suggests a more traditional role for VAS in this study, decreasing the risk or severity of enteric infection (43). Future mechanistic work would help identify mechanisms underlying these associations.
Supplementary Material
Acknowledgments
We thank Janet M Peerson, Senior Statistician, WHNRC, for her assistance and Dr Sherry A Tanumihardjo of the University of Wisconsin, Madison, for analysis of plasma retinol at 15 wk of age. The authors’ responsibilities were as follows: CS: was the PI responsible for the overall design of these studies, conducted some statistical analysis, and wrote portions of the manuscript; DM and MU: designed the aspects of the study involving microbiome analysis (along with CS) and participated in writing and revising the manuscript; DM: oversaw the microbiome laboratory work; MNH: participated in the design and analysis of the microbiome studies, carried out laboratory work for sample preparation and microbiome analysis, conducted statistical analysis, and wrote the first draft of the manuscript; SA and RR: were coinvestigators involved in the design and implementation of these studies; MA and AK: conducted laboratory work for preparation of samples and DNA isolation for microbiome analysis: DT and KK: performed laboratory work on microbiome analysis and contributed to the writing of the manuscript; and all authors: have read and approved the manuscript.
Notes
Funding was received from the WHO via a Bill and Melinda Gates Foundation grant (CBS), from the Thrasher Research Fund (CBS) and from National Institutes of Health awards F32HD093185 (DHT), AT007079 and AT008759 (DAM), and from the Peter J Shields Endowed Chair in Dairy Food Science (DAM).
Author disclosures: MNH, SMA, KMK, DHT, MJA, AK, RR, MAU, DAM, and CBS, no conflicts of interest.
Supplemental Table 1 and Supplemental Figures 1–6 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: BWM, birth weight median; FA, females above birth weight; FB, females below birth weight median; FDR, false discovery rate; MA, males above birth weight; MB, males below birth weight median; PL, placebo; RBP, retinol-binding protein; VA, vitamin A; VAD, vitamin A deficiency; VAS, vitamin A supplementation; ZIG, zero-inflated Gaussian.
References
- 1. Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8:143–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L et al.. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81:(4):e00036–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis ZT, Mills DA. Differential establishment of Bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. 2017;88:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. 2015;77:229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362:eaat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherwin JC, Reacher MH, Dean WH, Ngondi J. Epidemiology of vitamin A deficiency and xerophthalmia in at-risk populations. Trans R Soc Trop Med Hyg. 2012;106:205–14. [DOI] [PubMed] [Google Scholar]
- 7. Olson JA, Gunning DB, Tilton RA. Liver concentrations of vitamin A and carotenoids, as a function of age and other parameters, of American children who died of various causes. Am J Clin Nutr. 1984;39:903–10. [DOI] [PubMed] [Google Scholar]
- 8. Montreewasuwat N, Olson JA. Serum and liver concentrations of vitamin A in Thai fetuses as a function of gestational age. Am J Clin Nutr. 1979;32:601–6. [DOI] [PubMed] [Google Scholar]
- 9. Zile M, Bunge C, Deluca HF. Effect of vitamin A deficiency on intestinal cell proliferation in the rat. J Nutr. 1977;107:552–60. [DOI] [PubMed] [Google Scholar]
- 10. Olson JA, Rojanapo W, Lamb AJ. The effect of vitamin A status on the differentiation and function of goblet cells in the rat intestine. Ann N Y Acad Sci. 1981;359:181–91. [DOI] [PubMed] [Google Scholar]
- 11. Warden RA, Strazzari MJ, Dunkley PR, O'Loughlin EV. Vitamin A-deficient rats have only mild changes in jejunal structure and function. J Nutr. 1996;126:1817–26. [DOI] [PubMed] [Google Scholar]
- 12. Reifen R, Nyska A, Koperstein L, Zusman I. Intestinal and hepatic cell kinetics and mucous changes in vitamin-A-deficient rats. Int J Mol Med. 1998;1:579–82. [DOI] [PubMed] [Google Scholar]
- 13. Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed F, Jones DB, Jackson AA. The interaction of vitamin A deficiency and rotavirus infection in the mouse. Br J Nutr. 1990;63:363–73. [DOI] [PubMed] [Google Scholar]
- 15. Warden RA, Noltorp RS, Francis JL, Dunkley PR, O'Loughlin EV. Vitamin A deficiency exacerbates methotrexate-induced jejunal injury in rats. J Nutr. 1997;127:770–6. [DOI] [PubMed] [Google Scholar]
- 16. Maciel AA, Oria RB, Braga-Neto MB, Braga AB, Carvalho EB, Lucena HB, Brito GA, Guerrant RL, Lima AA. Role of retinol in protecting epithelial cell damage induced by Clostridium difficile toxin A. Toxicon. 2007;50:1027–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pantazi E, Marks E, Stolarczyk E, Lycke N, Noelle RJ, Elgueta R. Cutting edge: retinoic acid signaling in B cells is essential for oral immunization and microflora composition. J Immunol. 2015;195:1368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–38. [DOI] [PubMed] [Google Scholar]
- 20. Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J Pediatr. 2010;156:S8–15. [DOI] [PubMed] [Google Scholar]
- 21. Kozakova H, Hanson LA, Stepankova R, Kahu H, Dahlgren UI, Wiedermann U. Vitamin A deficiency leads to severe functional disturbance of the intestinal epithelium enzymes associated with diarrhoea and increased bacterial translocation in gnotobiotic rats. Microbes Infect. 2003;5:405–11. [DOI] [PubMed] [Google Scholar]
- 22. Hong CP, Park A, Yang BG, Yun CH, Kwak MJ, Lee GW, Kim JH, Jang MS, Lee EJ, Jeun EJ et al.. Gut-specific delivery of T-helper 17 cells reduces obesity and insulin resistance in mice. Gastroenterology. 2017;152:1998–2010. [DOI] [PubMed] [Google Scholar]
- 23. Sutherland DB, Suzuki K, Fagarasan S. Fostering of advanced mutualism with gut microbiota by immunoglobulin A. Immunol Rev. 2016;270:20–31. [DOI] [PubMed] [Google Scholar]
- 24. Bunker JJ, Bendelac A. IgA responses to microbiota. Immunity. 2018;49:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imdad A, Mayo-Wilson E, Herzer K, Bhutta ZA. Vitamin A supplementation for preventing morbidity and mortality in children from six months to five years of age. Cochrane Database Syst Rev. 2017;3:CD008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahmad SM, Raqib R, Qadri F, Stephensen CB. The effect of newborn vitamin A supplementation on infant immune functions: trial design, interventions, and baseline data. Contemp Clin Trials. 2014;39:269–79. [DOI] [PubMed] [Google Scholar]
- 27. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanumihardjo SA, Russell RM, Stephensen CB, Gannon BM, Craft NE, Haskell MJ, Lietz G, Schulze K, Raiten DJ. Biomarkers of Nutrition for Development (BOND)—vitamin A review. J Nutr. 2016;146:S1816–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. R Core Team. A language and environment for statistical computing. [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2017. Available from: http://www.R-project.org. [Google Scholar]
- 31. Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ, Suggests M. The vegan package. Community ecology package. 2007;10:631–7. [Google Scholar]
- 32. McMurdie PJ, Holmes S.. PhyloSeq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wickham H. GGplot2: Elegant Graphics for Data Analysis. Springer Science & Business Media; 2009. [Google Scholar]
- 34. Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poole AC, Pischel L, Ley C, Suh G, Goodrich JK, Haggerty TD, Ley RE, Parsonnet J. Crossover control study of the effect of personal care products containing triclosan on the microbiome. mSphere. 2016;1:(3):e00056–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lim MY, Yoon HS, Rho M, Sung J, Song Y-M, Lee K, Ko G. Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci Rep. 2016;6:23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB et al.. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J et al.. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM et al.. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, Li X, Graeber TG, Schwager E, Huttenhower C et al.. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 2014;8:2193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sirisinha S. The pleiotropic role of vitamin A in regulating mucosal immunity. Asian Pac J Allergy Immunol. 2015;33:71–89. [PubMed] [Google Scholar]
- 43. Stephensen CB. Vitamin A, infection, and immune function. Annu Rev Nutr. 2001;21:167–92. [DOI] [PubMed] [Google Scholar]
- 44. Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, Lusis AJ, Knight R, Caporaso JG, Svanback R. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, Lusis AJ. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7:313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K et al.. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11:e0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, Maas K, Graf J. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One. 2016;11:e0152751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hines M, Spencer D, Kung KT, Browne WV, Constantinescu M, Noorderhaven RM. The early postnatal period, mini-puberty, provides a window on the role of testosterone in human neurobehavioural development. Curr Opin Neurobiol. 2016;38:69–73. [DOI] [PubMed] [Google Scholar]
- 49. Hoen AG, Madan JC, Li Z, Coker M, Lundgren SN, Morrison HG, Palys T, Jackson BP, Sogin ML, Cottingham KL et al.. Sex-specific associations of infants' gut microbiome with arsenic exposure in a US population. Sci Rep. 2018;8:12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yunus FM, Khan S, Chowdhury P, Milton AH, Hussain S, Rahman M. A review of groundwater arsenic contamination in Bangladesh: the Millennium Development Goal era and beyond. Int J Environ Res Public Health. 2016;13:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Islam MR, Attia J, Alauddin M, McEvoy M, McElduff P, Slater C, Islam MM, Akhter A, d'Este C, Peel R et al.. Availability of arsenic in human milk in women and its correlation with arsenic in urine of breastfed children living in arsenic contaminated areas in Bangladesh. Environ Health. 2014;13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Benn CS, Fisker AB, Napirna BM, Roth A, Diness BR, Lausch KR, Ravn H, Yazdanbakhsh M, Rodrigues A, Whittle H et al.. Vitamin A supplementation and BCG vaccination at birth in low birthweight neonates: two by two factorial randomised controlled trial. BMJ. 2010;340:c1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sommer A, Tarwotjo I, Djunaedi E, West KP Jr., Loeden AA, Tilden R, Mele L. Impact of vitamin A supplementation on childhood mortality. A randomised controlled community trial. Lancet. 1986;1:1169–73. [DOI] [PubMed] [Google Scholar]
- 54. Lv Z, Wang Y, Yang T, Zhan X, Li Z, Hu H, Li T, Chen J. Vitamin A deficiency impacts the structural segregation of gut microbiota in children with persistent diarrhea. J Clin Biochem Nutr. 2016;59:113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu J, Liu X, Xiong XQ, Yang T, Cui T, Hou NL, Lai X, Liu S, Guo M, Liang XH et al.. Effect of vitamin A supplementation on gut microbiota in children with autism spectrum disorders—a pilot study. BMC Microbiol. 2017;17:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu HX, Hu Y, Wan YJ. Microbiota and bile acid profiles in retinoic acid-primed mice that exhibit accelerated liver regeneration. Oncotarget. 2016;7:1096–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee H, Ko G. Antiviral effect of vitamin A on norovirus infection via modulation of the gut microbiome. Sci Rep. 2016;6:25835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tian Y, Nichols RG, Cai J, Patterson AD, Cantorna MT. Vitamin A deficiency in mice alters host and gut microbial metabolism leading to altered energy homeostasis. J Nutr Biochem. 2018;54:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Byndloss MX, Pernitzsch SR, Baumler AJ. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol. 2018;11:1299–305. [DOI] [PubMed] [Google Scholar]
- 60. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- 61. Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–81. [DOI] [PubMed] [Google Scholar]
- 62. Schneeberger M, Everard A, Gomez-Valades AG, Matamoros S, Ramirez S, Delzenne NM, Gomis R, Claret M, Cani PD. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cani PD, de Vos WM. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rahman S, Rahman AS, Alam N, Ahmed AS, Ireen S, Chowdhury IA, Chowdhury FP, Rahman SM, Ahmed T. Vitamin A deficiency and determinants of vitamin A status in Bangladeshi children and women: findings of a national survey. Public Health Nutr. 2017;20:1114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW et al.. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marchaim D, Levit D, Zigron R, Gordon M, Lazarovitch T, Carrico JA, Chalifa-Caspi V, Moran-Gilad J. Clinical and molecular epidemiology of Acinetobacter baumannii bloodstream infections in an endemic setting. Future Microbiol. 2017;12:271–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.