Abstract
Fungal infections are a worldwide problem associated with high morbidity and mortality. There are relatively few antifungal agents, and resistance has emerged within these pathogens for the newest antifungal drugs. As the fungal cell wall is critical for growth and development, it is one of the most important targets for drug development. In this review, the currently available cell wall inhibitors and suitable drug candidates for the treatment of fungal infections are explored. Future studies of the fungal cell wall and compounds that have detrimental effects on this important outer structural layer could aid in antifungal drug discovery and lead to the development of alternative cell wall inhibitors to fill gaps in clinical therapies for difficult-to-treat fungal infections.
Keywords: : β (1-3)-glucan, antifungal drug, chitin, fungal cell wall, mannoprotein, unmasking
Fungal infections are a global health problem, where about 20% (325 species where 87 have been described since 1980) of all known human pathogens are fungi [1,2]. The severity of fungal infections can range from superficial, affecting the skin or nails, to severe, causing life-threatening invasive or disseminated infections. The most severe infections occur primarily in patients with an underlying disease or the immunocompromised patient population including cancer patients, hematopoietic stem cell or organ transplant recipients, those on a long-term corticosteroid regime, and HIV patients [3–6]. Some of the most common opportunistic fungal pathogens responsible for infections are Candida, Aspergillus and Cryptococcus, but many other fungi can cause infections including the dimorphic endemic fungi Blastomyces, Histoplasma, Coccidioides, Paracoccidioides, and fungi frequently found in the environment such as Fusarium, Scedosporium, Penicillium, and members of the Mucorales [7–12]. Currently, clinically relevant antifungal agents are divided into five classes based on the structure of the therapeutic compound and the mode of action, and they consist of the polyenes, azoles, allylamines, pyrimidines, and echinocandins [13,14]. Most of these antifungal agents either directly target the membrane (polyenes) or a component of the membrane (allyamines and azoles), while the most recently approved class, the echinocandins, target the fungal cell-wall complex β (1-3)-glucan synthase [15,16]. Although the current antifungal drugs have enabled management of fungal infections, rising drug resistance (azoles, echinocandins), high toxicity (polyenes), and poor oral availability (polyenes, echinocandins), have left an opportunity for improvement in therapeutic antifungal compounds [15,17,18]. In this review, we will cover the available antifungal drugs that target the cell wall and their challenge to eliminate the fungal infection and consider advancements in the identification of new lead compounds and strategies for targeting the fungal cell wall.
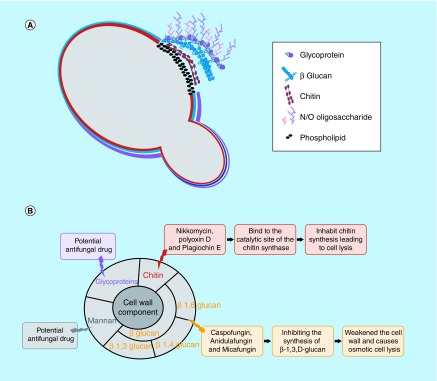
The structure of the cell wall is unique to fungi, and because it is absent in humans, the fungal cell wall is a promising target for antifungal drugs. One of the main roles of the fungal cell wall is to provide protection from environmental stresses including osmotic stress, which can result in cell lysis and provide passive protection against the entrance of potentially harmful macromolecules [19]. The cell wall of most fungi consists of polysaccharides (mainly glucans and chitins) and glycoproteins (Figure 1A). The inner layers of the cell wall include chitin and glucan, where 50–60% of the dry weight of the cell wall is made up of β (1-3)-glucan; therefore, its synthesis is essential for proper cell wall formation and normal development of the cell [20]. Chitin, which is mostly present in filamentous fungi, consists of β-1,4-linked N-acetylglucosamine and is covalently linked to β (1-3)-glucan [21,22]. Chitin synthesis is mediated by chitin synthases (CHS) [23–25]. Chitin provides the structural integrity and physical strength of the cell wall. When the structure of chitin and/or the biosynthesis of the polymer becomes disrupted, the wall becomes disordered and can lead to lysis and cell death [19]. The outer layer of the cell wall consists of homopolymers of mannose, 3–5% protein and 1–2% phosphate [26]. Mannan does not exist as a polysaccharide in the cell wall, but instead is found covalently bound to cell wall-associated proteins forming mannosylated glycoproteins [27–29]. Cell wall glycoproteins modified with N- and O-linked oligosaccharides are attached to the cell membrane. Proteins are firmly attached to the cell wall via covalent bonds between the N- and O-linked sugar moiety or a glycosylphosphatidylinositol (GPI) anchor to the chitin and/or glucan polymers [26,30,31]. Proteins located on the cell wall carry out a wide range of functions and are involved in adhesion to a surface, trafficking of macromolecules, cell-to-cell interactions, and providing protection against xenobiotic toxic substances. Each one of these cell wall components is vital for the normal development of the fungal cell and can be considered a potential target for antifungal agents (Figure 1B).
Figure 1. . Structure of the fungal cell wall and overview of compounds that target components.
The fungal cell wall is composed of layers of chitin and glucan with an outer layer of mannoproteins and other proteins linked by a glycosylphosphatidylinositol anchor to the cell wall (A). An overview of antifungal agents that either directly target an element of the cell wall or impede their biosynthesis (B).
The fungal cell wall
The fungal cell wall can compose approximately 40% of the total cell volume and range in thickness from 0.1 to 1.0 μm [32,33], which can be further extended by the presence of an outer capsule layer in Cryptococcus neoformans and Histoplasma capsulatum. The main components of the cell wall of most fungi, chitin and β (1-3)-glucan, are surrounded by a gel-like substrate comprised primarily of α (1-3)-glucans and galactomannoproteins.
Chitin
An integral component and defining feature of the fungal cell wall, chitin is a polymer of β 1,4-linked N-acetylglucosamine; composing only one to two percent of the total dry weight in Saccharomyces cerevisiae but in considerable quantities in filamentous fungi (10–20%) with important roles in maintaining the mechanical strength and structural integrity of the fungal cell wall [19,21,22,34,35]. Since chitin does not exist in human cells, this component is an attractive target for antifungal therapies with low toxicity for humans. As an indication of the importance of chitin to the fungal cell, inhibition of chitin biosynthesis can result in several detrimental effects ranging from a delay in cell wall maturation and septum and bud ring formation, to a delay or defect in cell division resulting in suboptimal growth of the fungus [36,37].
CHSs are a family of enzymes that are localized to the plasma membrane and are responsible for the biosynthesis of chitin [23]. Filamentous fungi harbor multiple different classes of CHSs within their genomes, while typically three classes of CHSs are present in yeast [38]. Of the three CHS in S. cerevisiae, Chs3p is the primary CHS where it is responsible for generating a majority of the chitin found in the cell wall (approximately 80–90%) [36,38]. Mutants of CHS3 have reduced chitin levels, poor growth and defects in cell wall integrity [36,37]. Aspergillus fumigatus has seven CHSs (CHSA-G), and each of these genes has been systematically disrupted demonstrating none of the CHS genes are essential in this fungus [39]. Single mutants of the CHSA-C, -F and -G genes appear to have no discernible difference from the wild-type fungus [37,39,40]; however, disruption of CHSD and CHSE results in a 20% and a 30% reduction in the production of chitin, respectively [41,42]. Seven CHSs (encoded by CHS1-7) are present in the genome of Neurospora crassa [43–46]. Disruption of CHS6, CHS7, and a double mutant of CHS1;CHS3 results in lower N-acetylglucosamine (0.34, 0.30, 0.35 μg/ml, respectively) [43–47]. Chitin is synthesized in Candida albicans by four CHS genes, where CHS1 is essential for viability. Chs2p is responsible for the production of about 40% of the chitin present in hyphal cells, whereas Chs3p and Chs8p synthesize short- and long-chitin microfibrils, respectively [48–52]. A proportion of chitin is deacetylated to chitosan by chitin deacetylases, and the quantity of chitosan can vary greatly between fungal species. For instance, only a small proportion of chitin is deacetylated to chitosan in C. albicans (<5%); however, a majority of chitin is deacetylated to chitosan in C. neoformans (∼ 65%) [53]. It is postulated that chitin deacetylation may provide the polymer with more flexibility, while decreasing the availability of chitin to be degraded by chitinases [53].
Glucans
β (1-3)-glucan is a long, linear polymer of glucose that is important for cell wall structural integrity and is enriched in the inner layer of the two-layered cell wall. β (1-3)-glucan is the major component of cell wall glucan (40–55%); however β (1,6)-, mixed β (1-3)- and β (1-4)-, α (1-3)-, and α (1-4)-linked glucans have also been found in various fungal cell walls [54–56]. β (1-3)-glucan is covalently linked to polymers of β (1-6)-glucan and chitin; this network of covalently bound polymers are able to interact with adjacent polymers in other chains through hydrogen bonds, ultimately forming a complex three-dimensional network of microfibrils [57,58]. Based on the composition of the components in this network, there is a large degree of variation among fungal pathogens; for instance, in A. fumigatus the polysaccharides are primarily galactomannan and β (1-3 and 1-4)-glucans, whereas in C. albicans they are mainly β (1-6)-glucans [59,60]. β (1-3)-glucans are synthesized by the highly conserved β (1-3)-glucan synthase complex, which is composed of a catalytic domain (Fks), responsible for the biosynthesis of the polymer, and a regulatory (Rho1) subunit [55,56,61]. Disruption of the FKS gene yields mutants with cell wall defects and decreased viability [56]. Filamentous fungi encode a single essential FKS gene, while two FKS genes have been identified in S. cerevisiae (FKS1/FKS2) [61]. The predicted membrane topology of Fks1p shows two large hydrophobic domains separated by a large hydrophilic domain, several transmembrane helixes and a cytoplasmic N-terminus [62].
Mannans & mannoprotein
The outer layer of the cell wall consists of heavily mannosylated glycoproteins that are modified with N- and O-linked oligosaccharides [26,30,31], where the structures of the oligosaccharide side chains are diverse between fungi. Some well-characterized examples include the cell walls of the model yeasts S. cerevisiae and C. albicans, which contain proteins that are glycosylated with mannose-rich oligosaccharides (mannans), while the oligosaccharide side chains of N. crassa and A. fumigatus glycoproteins are composed of both mannose and galactose residues, referred to as galactomannan [63,64]. Mannose polymers are linked to the protein through asparagine and threonine (N-glycosidic) or serine (O-glycosidic) residues. Outer chains of branched mannan that contains α-(1-2), α-(1-3), β-(1-2), β-(1-6) and phosphodiester linkages are attached to the N-mannan core through an α-(1-6)-backbone [26,30,31]. The O-mannan consists of a simple, unbranched linear carbohydrate, which is comprised of a series of α-(1-2)-linkages [31]. Mannoproteins are covalently attached to β (1-3)-glucan or β (1-6)-glucan via either their oligosaccharide sidechains or GPI anchors [31,65]. Several mannosyltransferases genes (MNN1-2,4-6,9-12,14) are required for normal cell wall mannan content in C. albicans, and deletion of each gene results in a defect in cell wall function [65,66].
Cell wall proteins
The outer layer of the cell wall contains proteins involved in surface adhesion and plays an important role in colonization and biofilm formation, including the Als (agglutinin-like sequence) and Hwp (hyphal wall protein) proteins [67,68]. One major difference between Candida and Saccharomyces, which are closely related, is that Candida species produce an extensive amount of adhesion proteins that are an important determinant of pathogenicity [69]. In C. albicans, the ALS family consists of eight genes that are engaged in cell adhesion [68,69]. Among them, Als1 and Als3 are considered the most important for biofilm formation [70,71]. The Als proteins have hydrophobic N and C termini, where the N terminus is involved in ligand binding, and the C terminus is modified by the addition of a GPI anchor [70], which is essential for linking the Als protein to the fungal cell wall [69].
As Hwp1 is involved in adhesion, it localizes to the cell surface and undergoes extensive modification followed by covalent attachment via a GPI anchor located at the C-terminus to β-glucan in the cell wall. This protein is involved in controlling hyphal surface gene expression with several other potential functional properties [72]. Hwp1 is rich in the amino acids proline (27%) and glutamine (16%), which may function to maintain the conformation of the polypeptide chains and binding [72,73]. The C terminus of Hwp1 is rich in serine and threonine, which provide O-glycosylation sites predicted to function by extending ligand-binding domains into the extracellular space. Strains without Hwp1 show less adhesion to human epithelial cells and are attenuated in virulence in a murine model of infection [74].
Antifungal agents targeting the cell wall in clinical use
The structure of the cell wall is necessary for the survival and growth of fungal cells; it also provides a barrier from the environmental stress. Damage to the cell wall structure leads to efflux of cytoplasmic molecules, cell membrane disruption, osmotic fragility, and suppression of fungal growth [19]. Efforts have been made to develop a drug to inhibit cell wall activity, but currently available antifungal drugs do not inhibit fungal infection. In this section, we will discuss the currently available antifungal agents that act on the cell wall (Figure 1B).
Inhibitors of β (1-3)-glucan synthase, echinocandins
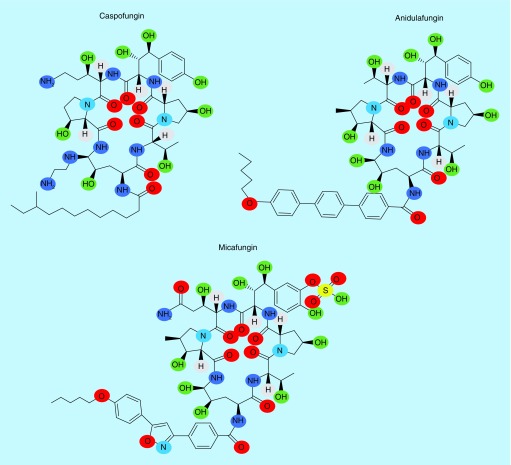
The precursor to the group known as the echinocandins were discovered in the 1970s from Aspergillus nidulans and Aspergillus rugulosus [75]. The production and assembly of glucan requires a series of enzymes and regulatory networks that are mostly specific for fungi, and therefore a prime target for antifungal compounds. The echinocandins are the most recent class of antifungal agents approved by the US FDA for clinical use and consist of caspofungin, anidulafungin, and micafungin (Figure 2 & Table 1). The antifungal activity of echinocandins is derived by their ability to act as a noncompetitive inhibitor of β (1-3)-glucan synthase [76]. The inhibition of β (1-3)-glucan synthesis causes a decrease in the β (1-3)- and β (1-6)-glucan network, leading to a disordered and osmotically unstable cell wall, which results in fungal cell death and/or decreased host tissue damage [77,78]. Furthermore, inhibition of β (1-3)-glucan synthase influences several other key components of both the fungal cell membrane, where there is a reduction in the ergosterol content, and the cell wall, where there is an increase in the chitin content and ultrastructural changes [79]. While the primary in vitro mode of action of echinocandins is understood to be cell wall inhibition, an in vivo mode of action of these compounds is tied to the host immune response [80]. Clearance of a C. albicans systemic infection in a murine model by caspofungin is dependent on the immune receptor, dectin-1, which binds to fungal cell wall β (1-3)-glucan. Candida albicans decreases the efficiency of dectin-1 detection of β (1-3)-glucan by covering it with a layer of mannosylated glycoproteins (mannan), which interfere with the glucan–dectin-1 interaction [80]. This is referred to as masking, and greater exposure of β (1-3)-glucan due to mutations or drug treatment is referred to as unmasking. Studies suggest that the in vivo activity of caspofungin affects the structure of the cell wall and increases β (1-3)-glucan exposure to dectin-1, which increases the response of the immune cells to the fungus [80–82].
Figure 2. . Echinocandins, β (1-3)-glucan synthase inhibitors.
Caspofungin, micafungin, and anidulafungin are inhibitors of fungal β (1-3)-glucan synthesis and are approved by the US FDA.
Table 1. . Summary of antifungal drugs in the two major classes targeting cell wall components.
| Target site | Drug | Source | Indication |
|---|---|---|---|
| Inhibitors of β (1,3)-glucan synthase | Caspofungin | Lipopeptide product of Aspergillus nidulans or Aspergillus rugulosus | Candidemia, refractory aspergillosis |
| Anidulafungin | Lipopeptide product of A. nidulans or A. rugulosus and acetylation by Actinoplanes utahensis | Candidiasis, salvage therapy for aspergillosis | |
| Micafungin | Coleophoma empetri | Candidiasis | |
| Inhibitors of chitin synthesis | Polyoxin | Streptomyces cacaoi | |
| Nikkomycin Z | Streptomyces tendae | Coccidioidomycosis | |
All three echinocandins in clinical use are semisynthetic lipopeptide derivatives of natural products and have efficacy against Candida spp. and Aspergillus spp. (Table 1) [81]. The main difference between caspofungin, micafungin, and anidulafungin resides in the side chain, where they possess a fatty acid side chain, a complex aromatic substituent and an alkoxytriphenyl group, respectively (Figure 2) [81]. This side chain modification increases glucan synthase inhibitory activity and decreases their toxicity to the host cells [81]. The long tail fatty acid chain of caspofungin allows interaction of this compound with the fungal cell membrane, leading to disruption of the function of the enzyme Fks1. Mutations in two hot spot regions of the FKS1 gene lead to conformational changes in the enzyme resulting in lower susceptibility to echinocandins [82,84]. These mutations happen in highly conserved regions of the FKS1 gene termed HS1 (positions 641–649) and HS2 (positions 1345–1365) [84,85].
The echinocandins are safer than other classes of antifungal agents; however, a significant drawback is their poor oral bioavailability and high protein binding affinity [86,87]. They are metabolized by the liver and distributed into the lung, liver, and spleen, but not into the CNS and eye [88], and are therefore not recommended for meningeal or eye infections.
Inhibitors of chitin synthase
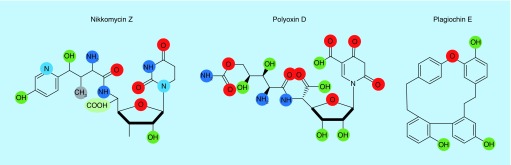
Several compounds that affect chitin synthesis have been identified, primarily the nikkomycins, polyoxins, and plagiochin (Figure 1B). Nikkomycin Z and polyoxins are antifungal drugs that are pyrimidine nucleosides linked to a di- or tripeptide moiety and therefore have structural similarities to UDP-N-acetylglucosamine, the substrate of CHS (Figure 3 & Table 1) [77]. Nikkomycins were first isolated from Streptomyces tendae, and their antifungal activity derives from being competitive inhibitors for the catalytic site of CHS where they bind with higher affinity than N-acetylglucosamine [77,89]. The nucleoside moiety varies in different nikkomycins (nikkomycins X and Z). Studies with S. cerevisiae have demonstrated that this drug can inhibit chitin synthase I and III, but not II, and has various degrees of inhibitory activity to other CHSs [90]. Nikkomycin Z has synergism with other antifungal agents including the Fks inhibiting echinocandins and the ergosterol biosynthesis inhibiting triazoles [91–93]. Nikkomycins have been shown to have clinically relevant antifungal activity in vitro against Candida parapsilosis, Coccidioides immitis, and Blastomyces dermatitidis [94], while in vivo models of fungal infections have indicated that nikkomycin Z may have some efficacy for the treatment of coccidioidomycosis, blastomycosis, and perhaps histoplasmosis [94]. However, other fungi including most of the Candida spp., C. neoformans, Aspergillus spp., Fusarium spp., and other opportunistic fungi have been reported to be resistant to nikkomycins perhaps due to their degradation and difficulty to penetrate the cell membrane [91,94,95]. Combination therapy of the chitin synthase inhibitor nikkomycin Z and an echinocandin enhanced the treatment of A. fumigatus, C. albicans, Rhizopus spp., and C. immitis with resistance to echinocandins [18].
Figure 3. . Chitin synthase inhibitors.
Nikkomycins, polyoxin, and plagiochin are chitin synthase inhibitors and are structurally similar to UDP-N-acetylglucosamine. Their inhibitory activity derive from their ability to bind with a high affinity to the catalytic site of chitin synthases.
Polyoxins are similar to nikkomycins having selective inhibition against CHS in fungi (Figure 3 & Table 1) [94]. First isolated from Streptomyces cacaoi var. asoensis in the 1960s, the polyoxins are a group of 12 structurally related compounds, which have been named alphabetically from A to L. The competition between the nucleoside moieties of the polyoxins and UDP-N-acetylglucosamine for the active site of the enzyme causes inhibition of the chitin synthase. Polyoxins contain two unusual amino acids, carbamoylpolyoxamic and polyoximic acids. In this competition, the carbamoylpolyoxamic acid moiety plays a critical role in the binding of polyoxins to the enzyme [96]. The carbamoylpolyoxamic acid moiety associates with the chitin synthetase due to hydrophobic interactions while the terminal carbamoyloxy group hydrogen bonds with the enzyme through an amide group [97]. Polyoxins are divided into two groups, one having polyoximic acid (III) and a negative rotation, while the other group does not contain polyoximic acid and has a positive rotation [95]. Polyoxins had been used with some success against C. neoformans, C. albicans, Aspergillus flavus and S. cerevisiae, although resistance to polyoxins has been reported in C. albicans mainly due to their poor transport through the cell membrane [98].
Plagiochin E is a phenolic antifungal macrocyclic bisbibenzyl compound that was first identified from liverwort, Marchantia polymorpha L. (Figure 3 & Table 1) [99]. The MIC of plagiochin E was 16 mg/ml against strains of C. albicans. Transmission electron microscopy of C. albicans cells treated with plagiochin E showed the structure of the cell was extremely damaged suggesting the antifungal activity of plagiochin E was associated with its effect on the cell wall [96]. Plagiochin E alters the expression of several chitin synthase genes in C. albicans, where the transcripts of CHS1 were significantly decreased while the expression of CHS2 and CHS3 was upregulated [96]. Candida albicans exposed to plagiochin E displayed the presence of phosphatidylserine at the cytoplasmic membrane, a marker for the early stage of apoptotic events [100]. Plagiochin E acts synergistically with fluconazole on fluconazole-resistant C. albicans strains suggesting it could be used in combinational therapy [99].
Antifungal agents in development
In addition to the antifungal drugs that are currently FDA approved for clinical use, there are several compounds in clinical development. In this section, we summarize and discuss these new compounds in clinical trials, in particular those compounds with a different mode of action.
Rezafungin
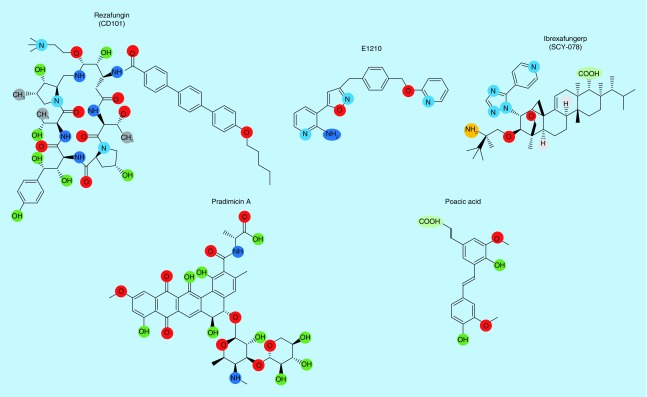
Rezafungin (formerly CD101) is a highly stable echinocandin similar in structure to anidulafungin with efficacy against Aspergillus spp. and Candida spp. (Figure 4) [101,102]. Importantly, CD101 has in vitro antifungal activity against Candida auris and other Candida strains that are resistant to other echinocandins [101,103]. A key characteristic of CD101 is the therapeutically favorable pharmacokinetic properties where the compound displays prolonged plasma stability when compared with anidulafungin and minimal degradation during storage [104]. This compound has a long half-life and very low clearence in animal models when compared with anidulafungin [105]. These advantages allow rezafungin to remain longer inside the patient at a therapeutic level, thereby potentially decreasing the frequency of doses to patients. Rezafungin is currently in Phase III clinical trials for the treatment of candidemia and invasive candidiasis.
Figure 4. . Structure of antifungal agents in various stages of clinical development.
Illustration of chemical structures of rezafungin (CD101), ibrexafungerp (SCY-078), E1210, pradimicin A, and poacic acid.
Ibrexafungerp
Ibrexafungerp (formerly SCY-078) is a glucan synthase inhibitor, which is a semisynthetic derivative of enfumafungin, a triterpene glycoside natural product. While both the echinocandins and ibrexafungerp inhibit glucan synthase, the structure of SCY-078 is significantly different from the echinocandins and has been designated the first member of the triterpene class of antifungal agents (Figure 4). In vitro, SCY-078 has antifungal efficacy (MIC levels below 0.5 μg/ml) against Aspergillus and, importantly, Candida strains that are resistant to the echinocandin glucan synthase inhibitors [106–109]. SCY-078 has good activity against the multidrug-resistant C. auris [110], and is the only β (1-3)-glucan inhibitor that showed some antifungal activity against the filamentous fungus Scedosporium prolificans [111]. This drug is currently in Phase I clinical development for intravenous formulation and initiating Phase III clinical trials as an oral formulation for the treatment of invasive candidiasis.
E1210
The glycophosphatidylinositol anchor is involved in attaching proteins to the fungal cell wall and cell membrane. The compound E1210 is capable of inhibiting inositol acylation via the Gwt1 protein in biosynthesis of the GPI anchor (Figure 4) [112]. The critical role of Gwt1 is to maintain the integrity of the fungal cell wall and enable the fungal cell to adhere to the host mucosal surface. E1210 has broad spectrum in vitro fungicidal activity at 1–2 μg/ml against many clinically important fungi including most Candida spp. (Candida krusei is an exception), Aspergillus spp., Fusarium spp. and Scedosporium spp. [112–114]. In C. albicans, E1210 is able to inhibit germ tube and biofilm formation due to a decrease in the adherence to the surface [112,113]. While E1210 displayed antifungal efficacy in murine models of candidiasis, aspergillosis, and fusariosis, there was no inhibitory activity observed against Pig-W (phosphatidylinositol-glycan biosynthesis, the human ortholog) even at concentrations as high as 100 μM.
Pradimicins
Pradimicins were first reported in 1988 from Actinomadura spp. with broad-spectrum antifungal activities (Figure 4) [115,116]. The mechanism of action of pradimicin is dependent on binding to mannan in the presence of Ca2+ ions, which causes a disruption in the integrity of the cell membrane [115]. These compounds show low toxicity in mammalian cells [115]. A new pradimicin derivative, BMS-181184, has in vitro antifungal activity against many clinically relevant fungi including Candida spp., C. neoformans, Aspergillus spp., dematiaceous molds, and members of the Mucorales [117,118].
Poacic acid
Poacic acid is primarily found in members of the grass family (Poaceae) of plants. This compound has antifungal activity against a broad range of fungi, in particular, against S. cerevisiae (IC50 of 111 μg/ml, 324 μM) (Figure 4) [119]. Poacic acid localizes to the cell wall and causes rapid cell lysis by directly binding to β (1-3)-glucan [119]. In addition to S. cerevisiae, poacic acid showed antifungal activity against Sclerotinia sclerotiorum, Alternaria solani, and Phytophthora sojae [119]. A significant synergistic effect exists between poacic acid and caspofungin, and to a lesser extent between poacic acid and nikkomycin Z [119]. The synergism with caspofungin suggests that poacic acid targets the cell wall via a mechanism that is independent of the mode of action of caspofungin [119]. Caspofungin and poacic acid both cause rapid cell leakage, but in nonactively growing cells, the cell leakage due to treatment with poacic acid is significantly greater than caspofungin [119]. This suggests an alternative mode of action for poacic acid such as a disruption in cell wall integrity rather than enzymatic inhibition by binding directly to β (1-3)-glucan [119]. Poacic acid is localized to the cell surface and targets the β (1-3)-glucan layer leading to turgor pressure and a weakened cell wall [119].
Future discovery & development of potential antifungal drugs
Antifungal resistance is a significant concern, and new treatment strategies are necessary to address this issue. A better understanding of the synthesis of the cell wall precursors, the cell wall assembly, and mechanisms or cellular alterations that perturb this cellular structure, could provide insight into possible targets for the development of new antifungal agents. Due to increased incidence of resistance to currently used antifungal drugs, it is imperative to identify alternative modes of action for new generations of therapeutic agents to be developed.
Facilitate the recognition of fungal pathogens by immune cells
As indicated above, the fungal cell wall is an intricate, ordered cellular structure composed of primarily of mannoproteins, β-glucans, and chitin that protects the fungus from recognition by the innate immune system. Fungal pathogens possess a pathogen-associated molecular pattern (PAMP), which can be recognized by host immune cells such as macrophages, neutrophils, and dendritic cells via a pattern recognition receptor [120].
The dectin-1 receptor, a C-type lectin, recognizes β-glucans with β (1-3)- and β (1-6)-linkages and elicits a proinflammatory response via TNFα to destroy the pathogen [121,122]. The interaction between dectin-1 and β (1-3)-glucans requires a minimum of 10 or 11 glucosyl residues [123–125]. Dectin-1 recognition is very specific and does not recognize monosaccharides or carbohydrates with different linkages. β (1-3)-glucan is enriched in the inner layer of the cell wall, which is covered, or masked, by a layer of heavily glycosylated mannoproteins (mannan), which inhibit binding and recognition by the pattern recognition receptor dectin-1 [126]. Most opportunistic fungal pathogens such as C. albicans mask their PAMPs making it difficult for the immune system to detect [126,127]. The mechanism of PAMP masking and the recognition by the immune system is still unclear, and further understanding of fungal masking and the ability of the cell to escape recognition by the host immune system may aid in the development of an effective antifungal drug. Antifungal agents like caspofungin disrupt the cell wall architecture and cause unmasking of the β (1-3)-glucan layer allowing greater binding by dectin-1. This greater binding is associated with increased recognition of Candida spp. by macrophages, and increased elicitation of cytokines like TNFα. A screen for mutants that exhibit a phenotype similar to the unmasking of the β (1-3)-glucan layer caused by caspofungin could be used to identify potential drug targets in the future.
Targeting the fungal cell wall pathways
Several enzymatic steps are involved in the biosynthetic process of chitin. Chitin synthesis is mediated by PKC, the Ca2+ calcineurin, and the high-osmolarity glycerol (HOG) signaling pathways [128]. Activation of these pathways in the presence of antifungal drugs reduces susceptibility and enables the cells to adapt to the treatment. Additionally, the mannosyltransferases and glycosyltransferases located in the Golgi apparatus, which are responsible for the synthesis of the N-linked and O-linked oligosaccharides that reside on the exterior mannoproteins, are another potential molecular target for the development of antifungal compounds.
Disruption of pathways for expression of fungal adhesion proteins
A variety of adhesion proteins reside on the fungal cell wall and are vital to the infection process where they aid in colonization and adhesion to host tissue. The expression of Als3 and Hwp1 are governed by a network of signal transduction pathways including the cAMP-PKA pathway (Ras1, Tpk1, Tpk2, Efg1) [129]. Each of the components in these pathways is involved in the correct expression of cell surface adhesion factors, and impeding any of the genes in this pathway could reduce fungal cell adhesion and biofilm formation. Hence, a better understanding of the fungal cell wall adhesion proteins and how they are regulated could lead to potential alternative drug targets for the treatment of fungi.
Drug delivery system for direct targeting to the fungal cells
Although a large number of synthetic and macromolecular antifungal drugs are available, most of them fail to have a sufficient effect on the site of infection. The site-specific release of a drug which directly targets the pathogen, will improve the impact of the drug, even for a drug-resistant pathogen which requires a concentrated dose of the drug. Nanoparticles as drug delivery vehicles were first introduced by Paul Ehrlich in 1954, wherein the drug was directly attached to targeted cells without affecting the healthy ones [130]. Nanoparticles such as liposomes, ethosomes, and niosomes are used as drug delivery systems to precisely target the fungal cell wall. Nanoparticles can be functionalized with materials such as mannan binding protein or Flo1p, which binds to the fungal surface, and could then release the antifungal compound(s) and disrupt the fungal cell wall. This approach could potentially reduce host toxicity while enhancing aqueous drug solubility, bioavailability, and antifungal efficacy [131].
Conclusion
The essential nature of the fungal cell wall makes it a prime target for antifungal agents. While many current therapies in clinical use already target the cell wall, as a better understanding of this structure is revealed, additional therapeutic drugs can potentially be developed for treatment options.
Future perspective
Fungal infections have increased in recent years [132–134], and cause a high degree of morbidity and mortality in patients with weakened immunity. Current available antifungal agents do not meet the expectations for eradicating fungal infections due to the difficulties in selective killing of eukaryotic fungal cells without adverse toxicity to humans. In order to change this situation, a new antifungal drug should possess an extended spectrum of activity against fungi, reduce dosing frequency, oral availability, and have no toxicity to human cells. One of the difficult challenges in antifungal drug development is identifying a suitable target that fulfills all of these criteria. The fungal cell wall is an ideal candidate to develop future antifungal drugs as it is a vital component of the fungus, it is exposed to the external environment, and it is unique to fungi and not present in other eukaryotes [135].
Executive summary.
Currently, five class of antifungal agents are used clinically, where the newest class, the echinocandins, targets the cell wall.
The fungal cell wall is a complex interwoven network composed of primarily of chitin, glucans and various glycoproteins.
Chitin is synthesized from monomers of N-acetylglucosamine by membrane-bound chitin synthases.
β (1-3)-glucan is the major component of cell wall glucans, although multiple other linkages occur in the cell wall. The glucans are synthesized by the glucan synthase complex where the Fks subunit is responsible for the biosynthesis.
The echinocandins are noncompetitive inhibitors of Fks, and there are three currently approved for clinical use that differ in their side chain.
The chitin synthase inhibitor nikkomycin Z is currently in development for coccidioidomycosis.
Multiple other compounds with antifungal activity that target components of the cell wall are in various stages of clinical development, including another echinocandin (rezafungin), a triterpene glucan synthase inhibitor (ibrexafungerp), and a compound that targets the glycosylphosphatidylinositol anchor of cell wall-localized proteins (E1210).
Future approaches to antifungal drug development concerning the cell wall include alteration of the cell wall to facilitate recognition by immune cells, inhibition of pathways regulating cell wall development and targeting critical proteins localized to the cell wall.
Footnotes
Financial & competing interests disclosure
The authors gratefully acknowledge the support of the National Institute of Allergy and Infectious Diseases (K22AI100983), and the article has been deposited to PubMed Central in accordance with NIH guidelines. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Riquelme M, Aguirre J, Bartnicki-Garcia S, et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol. Mol. Biol. Rev. 2018;82(2):e00068-17. doi: 10.1128/MMBR.00068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolhouse M, Gaunt E. Ecological origins of novel human pathogens. Crit. Rev. Microbiol. 2007;33(4):231–242. doi: 10.1080/10408410701647560. [DOI] [PubMed] [Google Scholar]
- 3.Revankar SG, Kirkpatrick WR, Mcatee RK, et al. A randomized trial of continuous or intermittent therapy with fluconazole for oropharyngeal candidiasis in HIV-infected patients: clinical outcomes and development of fluconazole resistance. Am. J. Med. 1998;105(1):7–11. doi: 10.1016/s0002-9343(98)00137-5. [DOI] [PubMed] [Google Scholar]
- 4.Badiee P, Hashemizadeh Z. Opportunistic invasive fungal infections: diagnosis & clinical management. Indian J. Med. Res. 2014;139(2):195–204. [PMC free article] [PubMed] [Google Scholar]
- 5.Rabeneck L, Crane MM, Risser JM, Lacke CE, Wray NP. A simple clinical staging system that predicts progression to AIDS using CD4 count, oral thrush, and night sweats. J. Gen. Intern. Med. 1993;8(1):5–9. doi: 10.1007/BF02600284. [DOI] [PubMed] [Google Scholar]
- 6.Wozniak KL. Interactions of Cryptococcus with dendritic cells. J. Fungi (Basel) 2018;4(1):36. doi: 10.3390/jof4010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler JR, Hube B, Puccia R, Casadevall A, Perfect JR. Fungi that infect humans. Microbiol. Spectrum. 2017;5(3):FUNK-0014–2016. doi: 10.1128/microbiolspec.funk-0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus – what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013;9(12):e1003743. doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt HJ, Blevins A, Sobeck K, Armstrong D. Aspergillus species from hospital air and from patients. Mycoses. 1990;33(11–12):539–541. doi: 10.1111/myc.1990.33.11-12.539. [DOI] [PubMed] [Google Scholar]
- 11.Holbrook ED, Rappleye CA. Histoplasma capsulatum pathogenesis: making a lifestyle switch. Curr. Opin. Microbiol. 2008;11(4):318–324. doi: 10.1016/j.mib.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4(165):165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 13.Perlin DS. Current perspectives on echinocandin class drugs. Future Microbiol. 2011;6(4):441–457. doi: 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker KS, Rogers PD. Recent insights into the mechanisms of antifungal resistance. Curr. Infect. Dis. Rep. 2006;8(6):449–456. doi: 10.1007/s11908-006-0019-3. [DOI] [PubMed] [Google Scholar]
- 15.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999;12(4):501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castelli MV, Butassi E, Monteiro MC, Svetaz LA, Vicente F, Zacchino SA. Novel antifungal agents: a patent review (2011 – present) Expert Opin. Ther. Patents. 2014;24(3):323–338. doi: 10.1517/13543776.2014.876993. [DOI] [PubMed] [Google Scholar]
- 17.Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 2005;3(7):547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- 18.Walker LA, Gow NA, Munro CA. Fungal echinocandin resistance. Fungal Genet. Biol. 2010;47(2):117–126. doi: 10.1016/j.fgb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. BioEssays. 2006;28(8):799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 20.Kapteyn JC, Van Den Ende H, Klis FM. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta. 1999;1426(2):373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- 21.Kollar R, Reinhold BB, Petrakova E, et al. Architecture of the yeast cell wall. β(1-->6)-glucan interconnects mannoprotein, β(1-->)3-glucan, and chitin. J. Biol. Chem. 1997;272(28):17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 22.Kollar R, Petrakova E, Ashwell G, Robbins PW, Cabib E. Architecture of the yeast cell wall. The linkage between chitin and β(1-->3)-glucan. J. Biol. Chem. 1995;270(3):1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 23.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 2002;41(6):367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 24.Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- 25.Munro CA, Gow NA. Chitin synthesis in human pathogenic fungi. Med. Mycol. 2001;39(Suppl. 1):41–53. [PubMed] [Google Scholar]
- 26.Kogan G, Pavliak V, Masler L. Structural studies of mannans from the cell walls of the pathogenic yeasts Candida albicans serotypes A and B and Candida parapsilosis . Carbohydr. Res. 1988;172(2):243–253. doi: 10.1016/s0008-6215(00)90858-9. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd MG. Cell envelope of Candida albicans . Crit. Rev. Microbiol. 1987;15(1):7–25. doi: 10.3109/10408418709104445. [DOI] [PubMed] [Google Scholar]
- 28.Shepherd MG, Poulter RT, Sullivan PA. Candida albicans: biology, genetics, and pathogenicity. Annu. Rev. Microbiol. 1985;39:579–614. doi: 10.1146/annurev.mi.39.100185.003051. [DOI] [PubMed] [Google Scholar]
- 29.Cassone A. Cell wall of Candida albicans: its functions and its impact on the host. Curr. Top. Med. Mycol. 1989;3:248–314. doi: 10.1007/978-1-4612-3624-5_10. [DOI] [PubMed] [Google Scholar]
- 30.Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 1998;62(1):130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata N, Ikuta K, Imai T, et al. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. Structure–antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis . J. Biol. Chem. 1995;270(3):1113–1122. doi: 10.1074/jbc.270.3.1113. [DOI] [PubMed] [Google Scholar]
- 32.Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10(7):851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 33.Klis FM, Mol P, Hellingwerf K, Brul S. Dynamics of cell wall structure in Saccharomyces cerevisiae . FEMS Microbiol. Rev. 2002;26(3):239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 34.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 35.Morozov AA, Likhoshway YV. Evolutionary history of the chitin synthases of eukaryotes. Glycobiology. 2016;26(6):635–639. doi: 10.1093/glycob/cww018. [DOI] [PubMed] [Google Scholar]
- 36.Bulawa CE. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis . Mol. Cell. Biol. 1992;12(4):1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw JA, Mol PC, Bowers B, et al. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1991;114(1):111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdivieso MH, Mol PC, Shaw JA, Cabib E, Duran A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae . J. Cell Biol. 1991;114(1):101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellado E, Aufauvre-Brown A, Gow NA, Holden DW. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 1996;20(3):667–679. doi: 10.1046/j.1365-2958.1996.5571084.x. [DOI] [PubMed] [Google Scholar]
- 40.Walsh TJ, Anaissie EJ, Denning DW, et al. Treatment of Aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 2008;46(3):327–360. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 41.Mellado E, Specht CA, Robbins PW, Holden DW. Cloning and characterization of chsD, a chitin synthase-like gene of Aspergillus fumigatus . FEMS Microbiol. Lett. 1996;143(1):69–76. doi: 10.1111/j.1574-6968.1996.tb08463.x. [DOI] [PubMed] [Google Scholar]
- 42.Aufauvre-Brown A, Mellado E, Gow NA, Holden DW. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet. Biol. 1997;21(1):141–152. doi: 10.1006/fgbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 43.Yarden O, Yanofsky C. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa . Genes Dev. 1991;5(12b):2420–2430. doi: 10.1101/gad.5.12b.2420. [DOI] [PubMed] [Google Scholar]
- 44.Din AB, Yarden O. The Neurospora crassa chs-2 gene encodes a non-essential chitin synthase. Microbiology. 1994;140(Pt 9):2189–2197. doi: 10.1099/13500872-140-9-2189. [DOI] [PubMed] [Google Scholar]
- 45.Din AB, Specht CA, Robbins PW, Yarden O. chs-4, a class IV chitin synthase gene from Neurospora crassa . Mol. Gen. Genet. 1996;250(2):214–222. doi: 10.1007/BF02174181. [DOI] [PubMed] [Google Scholar]
- 46.Borkovich KA, Alex LA, Yarden O, et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68(1):1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fajardo-Somera RA, Johnk B, Bayram O, Valerius O, Braus GH, Riquelme M. Dissecting the function of the different chitin synthases in vegetative growth and sexual development in Neurospora crassa . Fungal Genet. Biol. 2015;75:30–45. doi: 10.1016/j.fgb.2015.01.002. [DOI] [PubMed] [Google Scholar]; • Investigate the high degree of functional redundancy in a fungus with multiple chitin synthases. Interestingly, orthologous chitin synthases may have different roles in development in various fungi.
- 48.Bulawa CE, Miller DW, Henry LK, Becker JM. Attenuated virulence of chitin-deficient mutants of Candida albicans . Proc. Natl Acad. Sci. USA. 1995;92(23):10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gow NA, Robbins PW, Lester JW, et al. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans . Proc. Natl Acad. Sci. USA. 1994;91(13):6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mio T, Yabe T, Sudoh M, et al. Role of three chitin synthase genes in the growth of Candida albicans . J. Bacteriol. 1996;178(8):2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munro CA, Whitton RK, Hughes HB, Rella M, Selvaggini S, Gow NA. CHS8-a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet. Biol. 2003;40(2):146–158. doi: 10.1016/s1087-1845(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 52.Munro CA, Winter K, Buchan A, et al. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 2001;39(5):1414–1426. doi: 10.1046/j.1365-2958.2001.02347.x. [DOI] [PubMed] [Google Scholar]; • Through use of a conditional mutant, the study shows that Chs1 of the most common fungal pathogen Candida albicans is responsible for septum formation of yeast and hyphal cells, and is required for cell wall integrity. As Chs1 was essential, it is an attractive target for development of cell wall targeting antifungal agents.
- 53.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans . Eukaryot. Cell. 2007;6(5):855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernard M, Latge JP. Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 2001;39(Suppl. 1):9–17. [PubMed] [Google Scholar]
- 55.Klis FM, De Groot P, Hellingwerf K. Molecular organization of the cell wall of Candida albicans . Med. Mycol. 2001;39(Suppl. 1):1–8. [PubMed] [Google Scholar]
- 56.Grun CH, Hochstenbach F, Humbel BM, et al. The structure of cell wall α-glucan from fission yeast. Glycobiology. 2005;15(3):245–257. doi: 10.1093/glycob/cwi002. [DOI] [PubMed] [Google Scholar]
- 57.Fontaine T, Mouyna I, Hartland RP, Paris S, Latge JP. From the surface to the inner layer of the fungal cell wall. Biochem. Soc. Trans. 1997;25(1):194–199. doi: 10.1042/bst0250194. [DOI] [PubMed] [Google Scholar]
- 58.Latge JP. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 2007;66(2):279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 59.Fontaine T, Simenel C, Dubreucq G, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000;275(36):27594–27607. doi: 10.1074/jbc.M909975199. [DOI] [PubMed] [Google Scholar]
- 60.Aimanianda V, Clavaud C, Simenel C, Fontaine T, Delepierre M, Latge JP. Cell wall β-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis. J. Biol. Chem. 2009;284(20):13401–13412. doi: 10.1074/jbc.M807667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qadota H, Python CP, Inoue SB, et al. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272(5259):279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 62.Douglas CM, Foor F, Marrinan JA, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-D-glucan synthase. Proc. Natl Acad. Sci. USA. 1994;91(26):12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First description of FKS1 being a transmembrane protein and a subunit of the fungal glucan synthase complex.
- 63.Latge JP, Kobayashi H, Debeaupuis JP, et al. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus . Infect. Immun. 1994;62(12):5424–5433. doi: 10.1128/iai.62.12.5424-5433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakajima T, Yoshida M, Nakamura M, Hiura N, Matsuda K. Structure of the cell wall proteogalactomannan from Neurospora crassa. II. Structural analysis of the polysaccharide part. J. Biochem. 1984;96(4):1013–1020. doi: 10.1093/oxfordjournals.jbchem.a134917. [DOI] [PubMed] [Google Scholar]
- 65.Gentzsch M, Tanner W. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology. 1997;7(4):481–486. doi: 10.1093/glycob/7.4.481. [DOI] [PubMed] [Google Scholar]
- 66.Hall RA, Bates S, Lenardon MD, et al. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans . PLOS Pathog. 2013;9(4):e1003276. doi: 10.1371/journal.ppat.1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Munro CA, Bates S, Buurman ET, et al. Mnt1p and Mnt2p of Candida albicans are partially redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005;280(2):1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alberti-Segui C, Morales AJ, Xing H, et al. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast. 2004;21(4):285–302. doi: 10.1002/yea.1061. [DOI] [PubMed] [Google Scholar]
- 69.Lu CF, Montijn RC, Brown JL, et al. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β 1,6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1995;128(3):333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoyer LL, Cota E. Candida albicans agglutinin-like sequence (Als) family vignettes: a review of Als protein structure and function. Front. Microbiol. 2016;7:280. doi: 10.3389/fmicb.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Araujo D, Henriques M, Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017;25(1):62–75. doi: 10.1016/j.tim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 72.Staab JF, Ferrer CA, Sundstrom P. Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans . J. Biol. Chem. 1996;271(11):6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 73.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem. J. 1994;297(Pt 2):249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283(5407):1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 75.Nyfeler R, Keller-Schierlein W. [Metabolites of microorganisms. 143. Echinocandin B, a novel polypeptide-antibiotic from Aspergillus nidulans var. echinulatus: isolation and structural components] Helv. Chim. Acta. 1974;57(8):2459–2477. doi: 10.1002/hlca.19740570818. [DOI] [PubMed] [Google Scholar]
- 76.Odds FC, Brown AJ, Gow NA. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11(6):272–279. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 77.Kathiravan MK, Salake AB, Chothe AS, et al. The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 2012;20(19):5678–5698. doi: 10.1016/j.bmc.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 78.Song JC, Stevens DA. Caspofungin: pharmacodynamics, pharmacokinetics, clinical uses and treatment outcomes. Crit. Rev. Microbiol. 2016;42(5):813–846. doi: 10.3109/1040841X.2015.1068271. [DOI] [PubMed] [Google Scholar]
- 79.Pfaller M, Riley J, Koerner T. Effects of cilofungin (LY121019) on carbohydrate and sterol composition of Candida albicans . Eur. J. Clin. Microbiol. Infect. Dis. 1989;8(12):1067–1070. doi: 10.1007/BF01975172. [DOI] [PubMed] [Google Scholar]
- 80.Marakalala MJ, Vautier S, Potrykus J, et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013;9(4):e1003315. doi: 10.1371/journal.ppat.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eschenauer G, Depestel DD, Carver PL. Comparison of echinocandin antifungals. Ther. Clin. Risk Manag. 2007;3(1):71–97. doi: 10.2147/tcrm.2007.3.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goncalves SS, Souza ACR, Chowdhary A, Meis JF, Colombo AL. Epidemiology and molecular mechanisms of antifungal resistance in Candida and Aspergillus . Mycoses. 2016;59(4):198–219. doi: 10.1111/myc.12469. [DOI] [PubMed] [Google Scholar]
- 83.Pianalto KM, Alspaugh JA. New horizons in antifungal therapy. J. Fungi. 2016;2(4):26. doi: 10.3390/jof2040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balashov SV, Park S, Perlin DS. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 2006;50(6):2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 2007;10(3):121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiederhold NP, Lewis RE. The echinocandin antifungals: an overview of the pharmacology, spectrum and clinical efficacy. Expert Opin. Investig. Drugs. 2003;12(8):1313–1333. doi: 10.1517/13543784.12.8.1313. [DOI] [PubMed] [Google Scholar]
- 87.Zaas AK, Alexander BD. Echinocandins: role in antifungal therapy, 2005. Expert Opin. Pharmacother. 2005;6(10):1657–1668. doi: 10.1517/14656566.6.10.1657. [DOI] [PubMed] [Google Scholar]
- 88.Prabhu RM, Orenstein R. Failure of caspofungin to treat brain abscesses secondary to Candida albicans prosthetic valve endocarditis. Clin. Infect. Dis. 2004;39(8):1253–1254. doi: 10.1086/424449. [DOI] [PubMed] [Google Scholar]
- 89.Behr JB, Gautier-Lefebvre I, Mvondo-Evina C, Guillerm G, Ryder NS. Inhibition of chitin synthetase from Saccharomyces cerevisiae by a new UDP-GlcNAc analogue. J. Enzyme Inhib. 2001;16(2):107–112. doi: 10.1080/14756360109162360. [DOI] [PubMed] [Google Scholar]
- 90.Gaughran JP, Lai MH, Kirsch DR, Silverman SJ. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo . J. Bacteriol. 1994;176(18):5857–5860. doi: 10.1128/jb.176.18.5857-5860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li RK, Rinaldi MG. In vitro antifungal activity of nikkomycin Z in combination with fluconazole or itraconazole. Antimicrob. Agents Chemother. 1999;43(6):1401–1405. doi: 10.1128/aac.43.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiou CC, Mavrogiorgos N, Tillem E, Hector R, Walsh TJ. Synergy, pharmacodynamics, and time-sequenced ultrastructural changes of the interaction between nikkomycin Z and the echinocandin FK463 against Aspergillus fumigatus . Antimicrob. Agents Chemother. 2001;45(12):3310–3321. doi: 10.1128/AAC.45.12.3310-3321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luque JC, Clemons KV, Stevens DA. Efficacy of micafungin alone or in combination against systemic murine aspergillosis. Antimicrob. Agents Chemother. 2003;47(4):1452–1455. doi: 10.1128/AAC.47.4.1452-1455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hector RF, Zimmer BL, Pappagianis D. Evaluation of nikkomycins X and Z in murine models of coccidioidomycosis, histoplasmosis, and blastomycosis. Antimicrob. Agents Chemother. 1990;34(4):587–593. doi: 10.1128/aac.34.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tariq VN, Devlin PL. Sensitivity of fungi to nikkomycin Z. Fungal Genet. Biol. 1996;20(1):4–11. doi: 10.1006/fgbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 96.Wu XZ, Cheng AX, Sun LM, Lou HX. Effect of plagiochin E, an antifungal macrocyclic bis(bibenzyl), on cell wall chitin synthesis in Candida albicans . Acta Pharmacol. Sin. 2008;29(12):1478–1485. doi: 10.1111/j.1745-7254.2008.00900.x. [DOI] [PubMed] [Google Scholar]
- 97.Hori M, Kakiki K, Misato T. Further study on the relation of polyoxin structure to chitin synthetase inhibition. Agricultural Biol. Chem. 1974;38(4):691–698. [Google Scholar]
- 98.Mehta RJ, Kingsbury WD, Valenta J, Actor P. Anti-Candida activity of polyoxin: example of peptide transport in yeasts. Antimicrob. Agents Chemother. 1984;25(3):373–374. doi: 10.1128/aac.25.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guo XL, Leng P, Yang Y, Yu LG, Lou HX. Plagiochin E, a botanic-derived phenolic compound, reverses fungal resistance to fluconazole relating to the efflux pump. J. Appl. Microbiol. 2008;104(3):831–838. doi: 10.1111/j.1365-2672.2007.03617.x. [DOI] [PubMed] [Google Scholar]
- 100.Wu XZ, Chang WQ, Cheng AX, Sun LM, Lou HX. Plagiochin E, an antifungal active macrocyclic bis(bibenzyl), induced apoptosis in Candida albicans through a metacaspase-dependent apoptotic pathway. Biochim. Biophys. Acta. 2010;1800(4):439–447. doi: 10.1016/j.bbagen.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Y, Perez WB, Jiménez-Ortigosa C, et al. CD101: a novel long-acting echinocandin. Cell. Microbiol. 2016;18(9):1308–1316. doi: 10.1111/cmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first paper describing the antifungal efficacy of the echinocandin rezafungin against many clinically relevant fungi and an increased pharmacokinetic profile in vivo.
- 102.Pfaller MA, Messer SA, Rhomberg PR, Castanheira M. CD101, a long-acting echinocandin, and comparator antifungal agents tested against a global collection of invasive fungal isolates in the SENTRY 2015 Antifungal Surveillance Program. Int. J. Antimicrob. Agents. 2017;50(3):352–358. doi: 10.1016/j.ijantimicag.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 103.Berkow EL, Lockhart SR. Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris . Diagn. Microbiol. Infect. Dis. 2018;90(3):196–197. doi: 10.1016/j.diagmicrobio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 104.Krishnan BR, James KD, Polowy K, et al. CD101, a novel echinocandin with exceptional stability properties and enhanced aqueous solubility. J. Antibiot. 2016;70:130. doi: 10.1038/ja.2016.89. [DOI] [PubMed] [Google Scholar]
- 105.Ong V, James KD, Smith S, Krishnan BR. Pharmacokinetics of the novel echinocandin CD101 in multiple animal species. Antimicrob. Agents Chemother. 2017;61(4):e01626-16. doi: 10.1128/AAC.01626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pfaller MA, Messer SA, Motyl MR, Jones RN, Castanheira M. In vitro activity of a new oral glucan synthase inhibitor (MK-3118) tested against Aspergillus spp. by CLSI and EUCAST broth microdilution methods. Antimicrob. Agents Chemother. 2013;57(2):1065–1068. doi: 10.1128/AAC.01588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lepak AJ, Marchillo K, Andes DR. Pharmacodynamic target evaluation of a novel oral glucan synthase inhibitor, SCY-078 (MK-3118), using an in vivo murine invasive candidiasis model. Antimicrob. Agents Chemother. 2015;59(2):1265–1272. doi: 10.1128/AAC.04445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pfaller MA, Messer SA, Motyl MR, Jones RN, Castanheira M. Activity of MK-3118, a new oral glucan synthase inhibitor, tested against Candida spp. by two international methods (CLSI and EUCAST) J. Antimicrob. Chemother. 2013;68(4):858–863. doi: 10.1093/jac/dks466. [DOI] [PubMed] [Google Scholar]
- 109.Jiménez-Ortigosa C, Paderu P, Motyl MR, Perlin DS. Enfumafungin derivative MK-3118 shows increased in vitro potency against clinical echinocandin-resistant Candida species and Aspergillus species isolates. Antimicrob. Agents Chemother. 2014;58(2):1248–1251. doi: 10.1128/AAC.02145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that the β-D-glucan synthase inhibitor ibrexafungerp (formerly SCY-078) has antifungal efficacy against clinically relevant fungi that are resistant to echinocandins.
- 110.Berkow EL, Angulo D, Lockhart SR. In vitro activity of a novel glucan synthase inhibitor, SCY-078, against clinical isolates of Candida auris . Antimicrob. Agents Chemother. 2017;61(7) doi: 10.1128/AAC.00435-17. pii:e00435-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lamoth F, Alexander BD. Antifungal activities of SCY-078 (MK-3118) and standard antifungal agents against clinical non-Aspergillus mold isolates. Antimicrob. Agents Chemother. 2015;59(7):4308–4311. doi: 10.1128/AAC.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watanabe NA, Miyazaki M, Horii T, Sagane K, Tsukahara K, Hata K. E1210, a new broad-spectrum antifungal, suppresses Candida albicans hyphal growth through inhibition of glycosylphosphatidylinositol biosynthesis. Antimicrob. Agents Chemother. 2012;56(2):960–971. doi: 10.1128/AAC.00731-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the mode of action of E1210 where it inhibited inositol acylation of glycosylphosphatidylinositol-anchored proteins, resulting in a reduction of germ tube formation, adhesion and biofilm formation in C. albicans.
- 113.Miyazaki M, Horii T, Hata K, et al. In vitro activity of E1210, a novel antifungal, against clinically important yeasts and molds. Antimicrob. Agents Chemother. 2011;55(10):4652–4658. doi: 10.1128/AAC.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castanheira M, Duncanson FP, Diekema DJ, Guarro J, Jones RN, Pfaller MA. Activities of E1210 and comparator agents tested by CLSI and EUCAST broth microdilution methods against Fusarium and Scedosporium species identified using molecular methods. Antimicrob. Agents Chemother. 2012;56(1):352–357. doi: 10.1128/AAC.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walsh TJ, Giri N. Pradimicins: a novel class of broad-spectrum antifungal compounds. Eur. J. Clin. Microbiol. Infect. Dis. 1997;16(1):93–97. doi: 10.1007/BF01575126. [DOI] [PubMed] [Google Scholar]
- 116.Paudel S, Lee HC, Kim BS, Sohng JK. Enhancement of pradimicin production in Actinomadura hibisca P157-2 by metabolic engineering. Microbiol. Res. 2011;167(1):32–39. doi: 10.1016/j.micres.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 117.Fung-Tomc JC, Minassian B, Huczko E, Kolek B, Bonner DP, Kessler RE. In vitro antifungal and fungicidal spectra of a new pradimicin derivative, BMS-181184. Antimicrob. Agents Chemother. 1995;39(2):295–300. doi: 10.1128/aac.39.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oakley KL, Moore CB, Denning DW. Activity of pradimicin BMS-181184 against Aspergillus spp. Int. J. Antimicrob. Agents. 1999;12(3):267–269. doi: 10.1016/s0924-8579(99)00072-2. [DOI] [PubMed] [Google Scholar]
- 119.Piotrowski JS, Okada H, Lu F, et al. Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc. Natl Acad. Sci. USA. 2015;112(12):e1490–e1497. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brown GD, Taylor PR, Reid DM, et al. Dectin-1 is a major β-glucan receptor on macrophages. J. Exp. Med. 2002;196(3):407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003;197(9):1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kerrigan AM, Brown GD. Syk-coupled C-type lectin receptors that mediate cellular activation via single tyrosine based activation motifs. Immunol. Rev. 2010;234(1):335–352. doi: 10.1111/j.0105-2896.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- 123.Palma AS, Feizi T, Zhang Y, et al. Ligands for the β-glucan receptor, Dectin-1, assigned using ‘designer’ microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J. Biol. Chem. 2006;281(9):5771–5779. doi: 10.1074/jbc.M511461200. [DOI] [PubMed] [Google Scholar]
- 124.Willment JA, Gordon S, Brown GD. Characterization of the human β-glucan receptor and its alternatively spliced isoforms. J. Biol. Chem. 2001;276(47):43818–43823. doi: 10.1074/jbc.M107715200. [DOI] [PubMed] [Google Scholar]
- 125.Legentil L, Paris F, Ballet C, et al. Molecular interactions of β-(1-->3)-glucans with their receptors. Molecules. 2015;20(6):9745–9766. doi: 10.3390/molecules20069745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hasim S, Allison DP, Retterer ST, et al. β-(1,3)-glucan unmasking in some Candida albicans mutants correlates with increases in cell wall surface roughness and decreases in cell wall elasticity. Infect. Immun. 2017;85(1):e00601-16. doi: 10.1128/IAI.00601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bain JM, Louw J, Lewis LE, et al. Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio. 2014;5(6):e01874. doi: 10.1128/mBio.01874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Munro CA, Selvaggini S, De Bruijn I, et al. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans . Mol. Microbiol. 2007;63(5):1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fan Y, He H, Dong Y, Pan H. Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans . Mycopathologia. 2013;176(5):329–335. doi: 10.1007/s11046-013-9684-6. [DOI] [PubMed] [Google Scholar]
- 130.Iqbal J, Anwar F, Afridi S. Targeted drug delivery systems and their therapeutic applications in cancer and immune pathological conditions. Infect. Disord. Drug Targets. 2017;17(3):149–159. doi: 10.2174/1871526517666170606102623. [DOI] [PubMed] [Google Scholar]
- 131.Soliman GM. Nanoparticles as safe and effective delivery systems of antifungal agents: achievements and challenges. Int. J. Pharm. 2017;523(1):15–32. doi: 10.1016/j.ijpharm.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 132.Binder U, Lass-Florl C. Epidemiology of invasive fungal infections in the mediterranean area. Mediterr. J. Hematol. Infect. Dis. 2011;3(1):e20110016. doi: 10.4084/MJHID.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanz Alonso MA, Jarque Ramos I, Salavert Lletí M, Pemán J. Epidemiology of invasive fungal infections due to Aspergillus spp. and Zygomycetes. Clin. Microbiol. Infect. 2006;12:2–6. [Google Scholar]
- 134.Chen M, Xu Y, Hong N, et al. Epidemiology of fungal infections in China. Front. Med. 2018;12(1):58–75. doi: 10.1007/s11684-017-0601-0. [DOI] [PubMed] [Google Scholar]
- 135.De Nobel H, Van Den Ende H, Klis FM. Cell wall maintenance in fungi. Trends Microbiol. 2000;8(8):344–345. doi: 10.1016/s0966-842x(00)01805-9. [DOI] [PubMed] [Google Scholar]