Abstract
Context
Primary ovarian insufficiency (POI) is a cause of female infertility. However, the genetic etiology of this disorder remains unknown in most patients with POI.
Objective
To investigate the genetic etiology of idiopathic POI.
Patients and Methods
We performed whole-exome sequencing of 11 families with idiopathic POI. To gain insights into the potential mechanisms associated with this mutation, we generated two mouse lines via clustered regularly interspaced short palindromic repeats/Cas9 technology.
Results
A pathogenic homozygous missense mutation (c.149A>G; p.Asp50Gly) in the POLR3H gene in two unrelated families was identified. Pathogenic mutations in this subunit have not been associated with human disorders. Loss-of-function Polr3h mutation in mice caused early embryonic lethality. Mice with homozygous point mutation (Polr3hD50G) were viable but showed delayed pubertal development, characterized by late first estrus or preputial separation. The Polr3hD50G female and male mice showed decreased fertility later in life, associated with small litter size and increased time to pregnancy or to impregnate a female. Polr3hD50G mice displayed decreased expression of ovarian Foxo3a and lower numbers of primary follicles.
Conclusion
Our manuscript provides a case of POI caused by missense mutation in POLR3H, expanding the knowledge of molecular pathways of the ovarian function and human infertility. Screening of the POLR3H gene may elucidate POI cases without previously identified genetic causes, supporting approaches of genetic counseling.
This study revealed the first POLR3H mutation in two unrelated families with POI. The animal model showed delayed pubertal development and decreased fertility.
Primary ovarian insufficiency (POI), also known as premature ovarian failure, is a cause of female infertility affecting 1% of women <40 years of age. This disorder is characterized by primary or secondary amenorrhea, hypoestrogenism, and increased gonadotropin levels (FSH >25 IU/L) (1).
The genetic etiology of isolated or syndromic POI is highly heterogeneous, as many genes related to ovarian development and the meiosis pathway have been associated with POI (2). Although several POI cohorts have been analyzed, the genetic etiology of most patients remains unknown (3, 4).
Using exome sequencing, we identified a previously undescribed homozygous mutation in the subunit 3H of RNA polymerase III causing POI in two unrelated families. RNA polymerase III synthesizes several untranslated RNAs and is involved in cell growth, differentiation, and innate immune response (5). However, mutation in this subunit has not been associated with human disorders. To further assess the physiological relevance of this finding, we generated two mouse models by using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology. We found that loss-of-function mutation of Polr3h in mice is embryonically lethal and that mice with the same mutation identified in patients with POI showed impaired reproductive function.
Patients and Methods
Clinical evaluation
Institutional review board approval and written informed consent were obtained from all subjects before blood collection for DNA analysis. The study was approved by the Ethics Committee of Hospital das Clinicas, Faculdade de Medicina, Universidade de São Paulo, Brazil (protocol number 2015/12837/1.015.223).
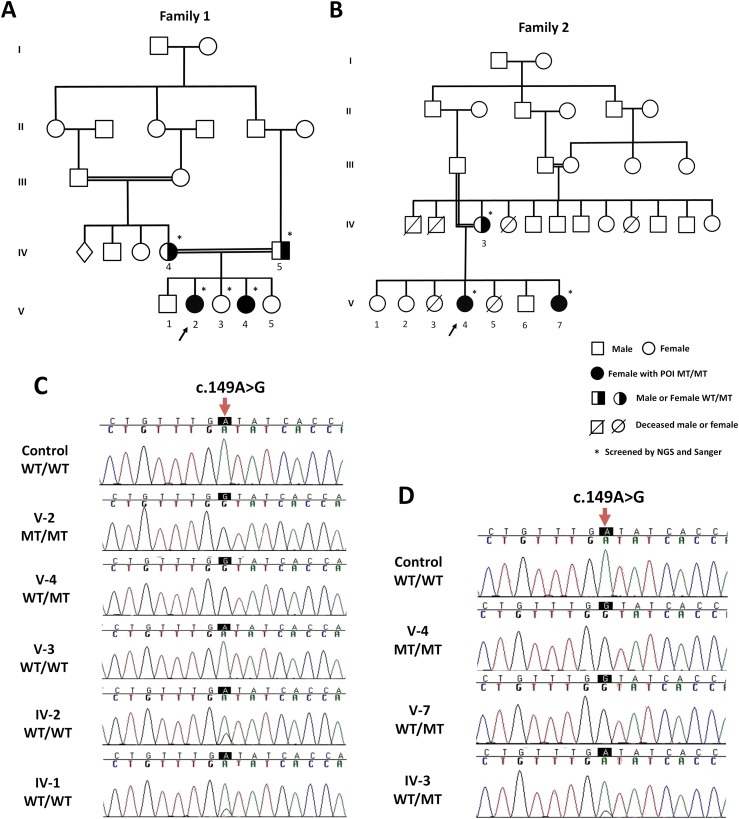
From a cohort of 11 families with POI, we identified two unrelated consanguineous families with two siblings, each clinically classified as displaying primary amenorrhea. The four patients had no breast development and denied menarche. The karyotypes were determined by analysis of 50 metaphases as 46,XX. The gonadotropin levels were elevated, whereas estradiol was undetectable (see all clinical features in Table 1). In family 1, the proband (V-2) presented with obesity over the years (body mass index = 30 kg/m2), and the proband’s brother (V-1) had a diagnosis of 3β-hydroxysteroid dehydrogenase deficiency (6). Other members of families have not presented any phenotype related to infertility or delayed puberty.
Table 1.
Clinical Features of 2 Siblings From 2 Unrelated Families With POI
| Clinical Features | F1 | F1 | F2 | F2 |
|---|---|---|---|---|
| V-2 | V-4 | V-4 | V-7 | |
| Age at diagnosis, y | 17 | 12 | 16 | 14 |
| Weight, kg | 53 | 35 | 41 | 30 |
| Height, cm | 153 | 137 | 153 | 143 |
| Adult height, cm | 157 | 164 | 164 | 158 |
| FSH, U/L | 41 | 36 | 35 | 21 |
| LH, U/L | 16 | 12 | 11 | 4 |
| Estradiol, pg/mL | <13 | <13 | <13 | <13 |
| Breasts/pubic hair | B1/P1 | B1/P1 | B1/P1 | B1/P1 |
| Uterus size at ultrasound | Prepubertal | Prepubertal | Prepubertal | Prepubertal |
| Ovaries at ultrasound | Not visualized | Prepubertal | Not visualized | Prepubertal |
Abbreviations: F1: family 1; F2: family 2.
Adrenal or thyroid autoimmune disorders were excluded from all patients. The treatment with conjugated estrogens followed by progesterone replacement resulted in complete breast development and vaginal bleeding.
Whole-exome sequencing
Genomic DNA was extracted from peripheral blood leukocytes via standard procedures. Exons were captured with a SureSelect Human All Exon Kit (Agilent Technology, Santa Clara, CA) and sequenced on a HiSeq2500 system (Illumina, San Diego, CA). Alignment of raw data and variant calling were performed according to the steps described by França et al. (7, 8). Briefly, the FASTQ files were aligned to human reference genome GRCh37/hg19 with a Burrows-Wheeler alignment tool (9). Variant calling was performed with Freebayes in all the resulting BAM files, and the resulting variants were annotated with ANNOVAR (10–12). The allelic variant was classified according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines (13).
Sanger sequencing
Data from whole-exome sequencing were confirmed by Sanger sequencing in all subjects. As a control, Sanger sequencing was performed to evaluate 200 fertile Brazilian women for the putative damaging variant. Primers flanking the POLR3H variant (exon2: c.149A>G ENS000000100413/NM_001018050) were used for PCR amplification. PCR products were sequenced with a BigDye® Terminator version 3.1 cycle sequencing kit followed by automated sequencing on an ABI PRISM® 3130xl genetic analyzer (Applied Biosystems, Foster City, CA).
Animals
Adult male and female C57BL/6J mice (JAX mice, stock no. 000664) and B6;SJL mice heterozygous for Polr3h loss-of-function mutation (knockout, named Polr3hKO), or heterozygous and homozygous for Polr3h D50G mutation (named Polr3hD50G), were maintained in the University of Michigan animal facility in a light- (12 hours on/off) and temperature-controlled (21°C to 23°C) environment. Mice had free access to water and phytoestrogen-reduced Envigo diet 2916 (16% protein, 4% fat) except during breeding, when they were fed a higher-protein and higher-fat phytoestrogen-reduced Envigo diet 2919 (19% protein, 8% fat). A phytoestrogen-reduced diet was used to avoid the effect of exogenous estrogen on the reproductive physiology. All procedures and experiments were carried out in accordance with the guidelines established by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the University of Michigan Institutional Animal Care and Use Committee (protocol no. 06792).
Histology and in situ hybridization histochemistry
Adult male and female C57BL/6J mice (n = 4 per sex) were transcardially perfused with 10% buffered formalin (Merck KGaA, Darmstadt, Germany), and gonads were dissected out and cryoprotected overnight in 20% sucrose. Single-label in situ hybridization histochemistry was performed as previously described (14–16). Ovary and testis were sectioned in the cryostat (14-μm sections) and directly mounted onto SuperFrost Plus slides. Tissue was dehydrated in increasing concentrations of ethanol, cleared in xylene, rehydrated in decreasing concentrations of ethanol, and placed in prewarmed sodium citrate buffer, pH 6.0. Slides were microwaved for 10 minutes, followed by dehydration in graded ethanol.
The Polr3h cDNA was produced from mouse ovary. The following primers were used to amplify a 578-bp sequence (mRNA, NM_030229.4): F (T3) 5′(CAG AGA TGC AAT TAA CCC TCA CTA AAG GGA GA) GCA TCC CAC ACC AAA GTC CA 3′; R (T7) 5′(CC AAG CCC TCT AAT ACG ACT CAC TAT AGG GAG A) GCT TGG CTC TGG AGA ACA CT 3′. The Polr3h riboprobe was generated by in vitro transcription with 33P-UTP as the radioisotope. 33P-labeled Polr3h probe was diluted in hybridization solution, and slides containing sections of ovary and testis were hybridized overnight at 56°C. The next day, slides were incubated in 0.002% RNAse A followed by stringency washes in sodium citrate–sodium chloride buffer. Sections were dehydrated in increasing concentrations of ethanol, air dried, and placed in X-ray film cassettes with BMR-2 film (Kodak, Rochester, NY) for 2 to 3 days. Slides were then dipped in NTB autoradiographic emulsion (Kodak) and stored in foil-wrapped slide boxes at 4°C for 2 weeks. Slides were developed with Dektel developer (Kodak), counterstained with hematoxylin and eosin (H&E), and coverslipped with distyrene, a plasticizer, and xylene. Sections of gonads were counterstained with H&E.
Ovaries from adult wild type and mutant mice were fixed in 10% buffered formalin for a minimum of 48 hours before standard paraffin-embedded protocol. The entire ovary was sectioned (4-μm sections), and sections 20 μm apart were collected and subjected to standard H&E staining. Quantification of primary, secondary and antral follicles, as well as corpora lutea, was performed in every third section (60 μm apart) in a total of 10 sections per ovary. The area of each section was obtained, and an average area was calculated by dividing the total area by the number of sections.
Generation of Polr3hD50G mice with CRISPR/Cas9 technology
To generate a mouse line with the c.149A>G substitution on Polr3h, we used CRISPR/Cas9 technology (17, 18). The single guide RNA (sgRNA) target and protospacer adjacent motif (PAM) sites in the mouse ch15, nucleotides 81915030 to 81926213 (National Center for Biotechnology Information reference sequence NC_000081.6), were identified on the Web site http://crispor.tefor.net/. Two sgRNAs were selected and tested. The sgRNA1 target and PAM were identified on coding strand 5′ CAC ATA GGC GTC CTC CAG CTT GG 3′, with a predicted cut site 12 bp upstream of the desired point mutation. The sgRNA2 target and PAM were identified on noncoding strand 5′ TCT GTT TGA TAT CAC CAA GCT GG 3′, with a predicted cut site 9 bp upstream of the desired point mutation. The activity of the two sgRNA/Cas9 complexes at the target site was assessed by analysis of genomic DNA extracted from in vitro cultured blastocysts from mouse zygotes that had been microinjected with Cas9 reagents, as described (19). DNA sequence analysis of PCR products obtained by amplification across the Cas9 target showed the presence of insertions/deletions (indels) in the genomic DNA due to nonhomologous end joining repair of double-strand chromosome breaks induced by sgRNA/Cas9 complexes for both sgRNAs. Because it was not possible to know a priori whether the mutations would cause infertility, we chose the sgRNA1 to decrease the chances of producing a homozygous mutation (20). A single-strand donor oligonucleotide (ssODN, 145 bp) that contained the intended mutation site for use as the homology-directed repair (HDR) template was synthesized. Multiple silent coding changes (lowercase letters) were introduced in the Cas9 targets for sgRNAs to block Cas9 recleavage after correct repair of chromosome breaks by the ssODN, as follows: 5′TCT cTT cGg cAT tAC aAA aCT cGA aGA tGC tTA cGT G 3′. For the ease of genotyping of the resulting transgenic mice, we introduced a Taq I restriction enzyme site into the ssODN. The sgRNA target was cloned into the plasmid pX330 (Addgene.org plasmid no. 42230, a kind gift from Feng Zhang) as described (21). The circular pX330 plasmid (5 ng/μL final concentration) (22) and circular donor ssODN (10 ng/μL final concentration) were mixed together for microinjection. Fertilized eggs were produced by mating superovulated B6SJL_F1 with B6SJL_F1 males (JAX® mice, stock no. 100012). Pronuclear microinjection was carried out as described (23, 24). Of 343 zygotes microinjected, 266 surviving zygotes were transferred to B6D2_F1 (JAX® mice, stock no. 100006) pseudopregnant female mice, and 35 potential G0 founders were obtained. Genotyping of genomic DNA extracted from tail tip biopsies (RED Extract-NAmp Tissue PCR Kit XNAT, Merck KGaA, Darmstadt, Germany) was performed by PCR with primers flanking the oligo donor followed by Taq I digestion. Primers sequences were as follows: F: AAC TTC ATG TCC ATC CTA TCC ACT TAA C; and R: CTT CAT CTT TTG TTA CAG AGA AAC GAA CC. Double-distilled water was used as negative control. PCR amplicons were run on a 2% agarose gel with EtBr (Life Technologies, Carlsbad, CA) at 100 mV for 40 minutes and visualized under ultraviolet light. Expected band sizes were wild type: 441 bp; homozygous Polr3h D50G mutation, 347 bp and 94 bp; and heterozygous Polr3h mutation, 441 bp, 347 bp, and 94 bp. Expected mutations were confirmed by DNA sequencing analysis.
To minimize the risk of off-targets by CRISPR/Cas9, selected founders (G0 generation) were backcrossed to C57BL/6J mice for two generations. The Polr3hKO/+ and Polr3hD50G/+ offspring were intercrossed to generate the homozygous Polr3hKO and Polr3hD50G and wild type (wt) control littermates (Polr3h+/+).
Puberty onset and fertility testing
In female mice, pubertal maturation was assessed by age of vaginal opening (puberty onset) and age of first estrus (puberty completion). Mice were monitored daily for vaginal opening starting at postnatal day (PN) 21 and first estrus via vaginal cytology until PN60. Estrous cycles of adult females were then assessed for 6 weeks (PN80 to PN120). Experimental and control female mice were housed with sexually experienced C57BL/6J males for 100 days (PN120 to PN220) to assess fertility, latency to pregnancy, number of litters, and litter size. Two cohorts of six or seven mice per sex and genotype were independently evaluated.
Males were assessed for puberty onset by daily analysis of balanopreputial separation (BPS) starting at PN25. A group of adult mice was euthanized at PN60 to assess testis weight. Another group of experimental and control mice were bred with two C57BL/6J females of proven fertility for 100 days (PN70 to PN170). Fertility, latency to impregnate a female, number of litters, and litter size were recorded. Two cohorts of five to eight mice per sex and genotype were independently evaluated.
RNA extraction, cDNA synthesis, and quantitative PCR
Prepubertal (PN15) and adult (PN60 to PN70) Polr3hD50G mice (n = 6 per genotype and age) were euthanized in the morning (females on diestrus) by decapitation under anesthesia (isoflurane). One ovary was removed and immediately frozen in dry ice for mRNA quantification. The other ovary was collected for histological analysis with a standard paraffin-embedded protocol and H&E staining.
Total RNA was extracted with TRIzol reagent (Life Technologies) according to the manufacturer’s protocol. RNA concentration was defined in an Epoch 1000 spectrophotometer (Biotek, Winooski, VT), and an equal amount of RNA (500 ng) from each sample was reverse-transcribed (RT2 First Strand Kit Qiagen, Hilden, Germany). PCR reactions were performed in a CFX384TM Real-Time System (Bio-Rad, Hercules, CA), using SYBR® Green gene expression assays and primer pairs synthesized by Merck KGaA (Darmstadt, Germany) or IDT (Coralville, IA). All samples and standard curves were run in triplicate. Water instead of cDNA was used as a negative control, and Gapdh was used as the housekeeping gene. Determination of gene transcript levels in each sample was obtained by the ΔΔCt method. The threshold cycle (Ct) of mRNA was measured and normalized to the average of the housekeeping genes (ΔCt). The fold change of mRNA in the unknown sample relative to control group was determined by 2−ΔΔCt. Primer sequences are available by request. Results are expressed as fold changes compared with wt.
Data analysis, statistics, and production of photomicrographs
Sections of ovary and testis were analyzed with an Axio Imager M2 microscope (Zeiss, Oberkochen, Germany). Photomicrographs were produced by capturing images with a digital camera (Axiocam, Zeiss, Oberkochen, Germany) mounted directly on the microscope and Zen software. Image-editing software (Adobe Photoshop CS6) was used to combine photomicrographs into plates. Only sharpness, contrast, and brightness were adjusted. Data are expressed as mean ± SEM. Comparisons between two groups were carried out via unpaired two-tailed Student t test. Statistical analyses were performed in GraphPad Prism 7 software, and an α value (P) of 0.05 was considered in all analyses. In multiple t tests, false discovery (q value) was also evaluated. For fertility analysis, we used the linear mixed effects model for litter size, with a fixed effect of mutation genotype and pregnancy number with a random intercept for mice. Model: lmer [Litter.size ≈ Pregnancies + Genotype + (1/Mouse.ID)]. The t tests used the Satterthwaite method (lmerModLmerTest). For latency to pregnancy (or time to impregnate a female) we used multiple t test and survival analyses. The details of statistical analysis are described in the figure legends.
Results
Exome sequencing and Sanger sequencing reveal a recessive mutation in POLR3H
Considering the known consanguinity in both families with two affected siblings and unaffected parents, we assumed that the pathogenic variant would be a rare homozygous variant present in all affected subjects (Fig. 1A and 1B). In each family, the candidate variants were filtered as homozygous in both siblings and heterozygous in their parents, consistent with an autosomal recessive inheritance. Regarding population frequency, we excluded all variants present with a minor allele frequency >0.1% in the 1000 Genomes, Exome Variant Server National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project (ESP), and Exome Aggregation Consortium databases. Of these, only missense, nonsense, frameshift variants in coding regions, and splice sites were included. A single variant met these criteria: the homozygous c.149A>G; p.Asp50Gly (p.D50G) mutation in POLR3H. The mean coverage depth of the exome targeted regions was 88 to 99× and 37 to 51×, with 95% to 96% and 89% to 93% of the target bases covered more than 10-fold in family 1 and family 2, respectively. Sanger sequencing confirmed that the c.149A>G POLR3H mutation was present in a homozygous state in both siblings and in a heterozygous state in both parents (Fig. 1C, 1D). In family one, the heterozygous c.149A>G mutation was identified in the oldest brother (V-1) and in the unaffected sister (V-5).
Figure 1.
Pathogenic homozygous mutation in POLR3H identified in two unrelated families. (A, B) Family pedigrees. The arrows indicate the probands (F1: V-2 and F2: V-4). Pedigree numbers of individuals are indicated above the symbols. The asterisk indicates samples used for whole-exome and Sanger sequencing. (C, D) The electropherogram confirmed homozygous c.149A>G (red arrow on nucleotide peak of interest) in all affected women (F1: V-2 and V-4; F1: V-4 and V-7), heterozygous mutation in their parents (F1: IV-4 and IV-5; F2: IV-3), and absent in unaffected sister (F1: V-3). WT denotes the wild-type allele and MT indicates the c.149A>G mutation.
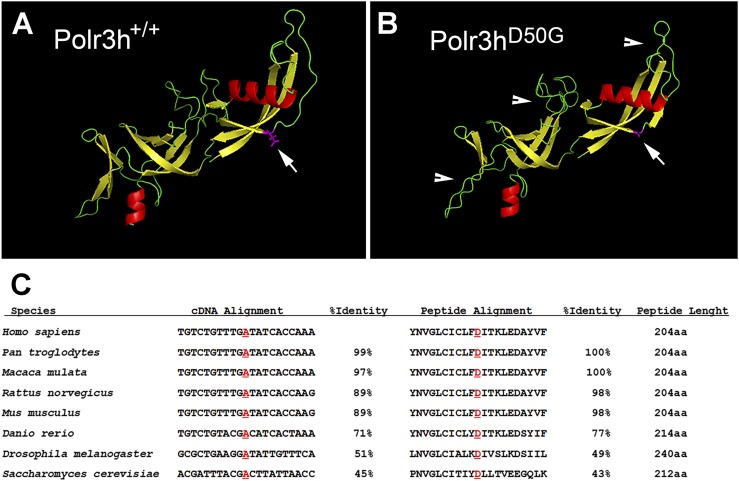
For protein structure prediction, we used the Iterative Threading Assembly Refinement (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) program developed by Yang Zhang’s research group at the University of Michigan (Ann Arbor, MI) (25–27) (Fig. 2A, 2B). The structure change was classified as pathogenic or disease-causing by all base conservation scores and functional prediction tools (SIFT, Polyphen2HVAR, Mutation Taster, Mutation Assessor, RadialSVM, MetaLR, CADD, and GERP).
Figure 2.
POLR3H predicted structure and homology. (A, B) Predicted POLR3H wild type (POLR3H+/+) and mutant (POLR3HD50G) protein structure. Arrows indicate site of point mutation, and arrowheads show additional sites of potential structure disruption. (C) Sequence alignment of POLR3H nucleotides and amino acids among orthologs. Note highly conserved sequence among species.
Regarding the minor allele frequency, the mutation (c.149A>G; p.Asp50Gly) was not present in the 400 alleles from fertile Brazilian women that were analyzed by the Sanger method as well as the 609 elderly Brazilian exomes sequences from the Online Archive of Brazilian Mutations database (28). In addition, the p.Asp50Gly mutation has not been reported in other public databases such as 1000 Genomes and the NHLBI Grand Oppoutunity Exome Sequencing Project (ESP).
Polr3h is conserved in eukaryotes and is highly expressed in mouse gonads
To perform functional studies in mice, we initially compared the sequence homology of Polr3h gene and protein (RPC8) (Fig. 2) and found that the mutated site is highly conserved. According to BioGPS Web site (http://biogps.org), Polr3h is ubiquitously expressed, showing high to moderate levels in adult gonads.
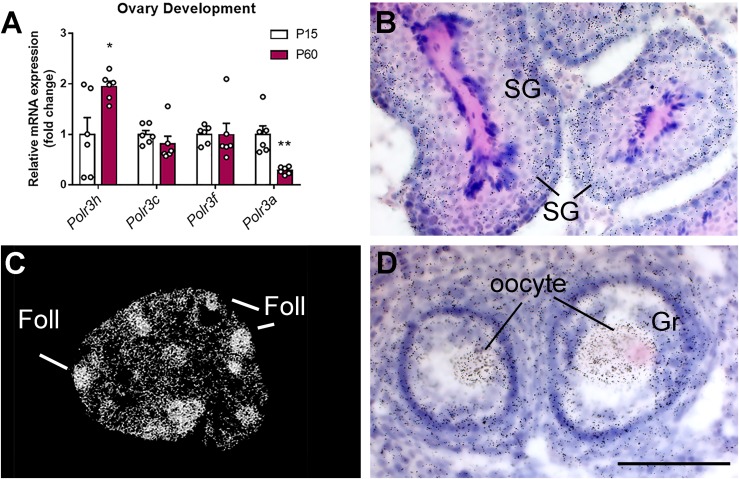
To validate the reported data, we used quantitative PCR and found that ovarian Polr3h expression is higher in adult (PN60 on diestrus) compared with prepubertal (PN15) mice (n = 6 per group) (Fig. 3A). This increase is selective for the subunit 3H (Polr3h), as no changes were observed in Polr3c and Polr3f expression, whereas Polr3a was decreased in ovaries of adult females (PN60).
Figure 3.
Polr3h expression in gonads of mice. (A) Bar graphs showing expression of Pol III subunits in ovaries in prepubertal (PN15) and adult (PN60) mice. Differences were determined by multiple t tests (α < 0.05). Discovery was determined via the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Each Pol III subunit was analyzed individually assuming a consistent SD (n = 6 per group; Polr3h, df = 10, t = 2.7, q = 0.03; Polr3a, df = 10, t = 4.24, q = 0.005). (B–D) Distribution of Polr3h mRNA in the testis (B) and ovary (C, D) of wt mice (silver grains). Higher hybridization signal was observed in developing spermatogonia (SG), granulosa cells (Gr), and oocytes. Scale bar: B, D = 100 μm; C = 1 mm. *P < 0.05; **P < 0.01. Foll, follicles.
To determine the specific cell types that express Polr3h, we performed in situ hybridization in ovary and testis from adult (PN60) mice. High Polr3h expression was observed in developing spermatogonia, oocytes, and ovarian granulosa cells (Fig. 3B–3D). Very low or virtually no expression was detected in mature sperm, Leydig, or theca cells.
Validation of Polr3hD50G mouse model
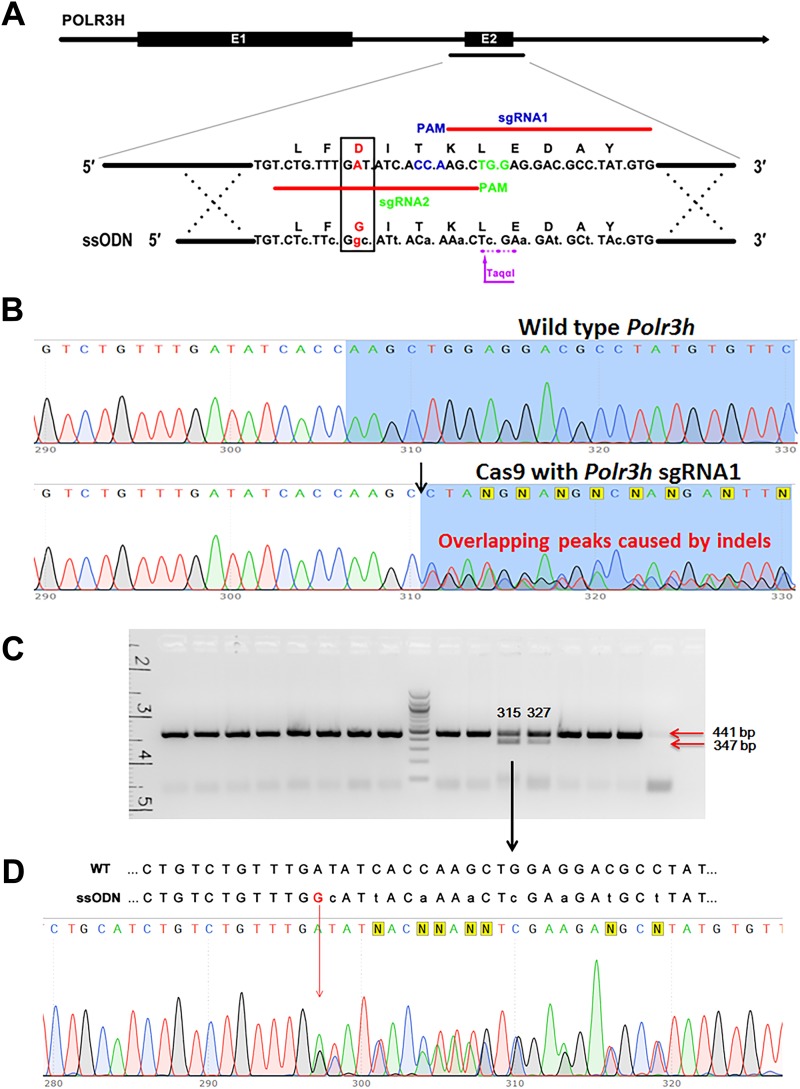
The mouse line with the c.149A>G substitution in the Polr3h gene (Polr3hD50G mice) was generated by CRISPR/Cas9 technology. The sgRNAs, the ssODN (145 bp) with the intended mutation site, and the HDR are shown in Fig. 4A. The activity of each sgRNA was initially assessed by PCR with DNA extracted from blastocysts that developed in in vitro culture from mouse zygotes microinjected with sgRNA/Cas9 constructs. Sanger sequencing further confirmed the activity of two sgRNAs by the presence of indels in the genomic DNA from injected zygotes (Fig. 4B). The efficiency of embryos harboring mutant alleles for the sgRNAs was >80%, indicating that the sgRNAs/Cas9 constructs were able to induce mutation via pronuclear microinjection.
Figure 4.
Generation of a mouse model with introduction of the c.149A>G substitution in Polr3h locus. (A) Schematic diagram of precise editing of Polr3h by CRISPR/Cas 9-induced HDR. Two sgRNAs were designed to match the intended locus (indicated by the two red lines), and the PAM site is indicated in blue and green for sgRNA1 and 2, respectively. The red nucleotide “A” is the target site for the intended mutation, and the red nucleotide “G” is the intended substitution (i.e., c.149A>G; p.Asp50Gly). The amino acids are indicated by capital letters, with the amino acid variation indicated by the red frame. The lowercase letters in the same codon are the silent mutations introduced to prevent recutting after homolog recombination (HR) and to generate a TaqαІ restriction enzyme site. (B) Chromatogram of DNA sequence from PCR products obtained by amplification across the Cas9 target shows the presence of indels in the genomic DNA due to nonhomologous end joining repair of double-strand chromosome breaks induced by sgRNA1/Cas9 (black arrow). (C) Genomic DNA of potential founders (G0) was prepared from tail tips, and a fragment spanning the expected D50G mutation site was PCR amplified and digested by TaqαІ. Polr3h D50G mutations after HDR of Cas9-induced TaqαІ restriction enzyme cutting site produce lower molecular weight fragments (347 bp and 94 bp). Two founders with TaqαІ cutting site were identified, #315 and #327. (D) Further Sanger sequence analysis defined #315 as a founder with the desired c.149A>G; p.Asp50Gly Polr3h mutation and #327 as a Polr3h knockout. E1, exon 1; E2, exon 2.
The sgRNA/Cas9 plasmid and the ssODN were microinjected into mouse zygotes and transferred to pseudopregnant females. Genomic DNA was prepared from tail tips, and a fragment spanning the expected D50G mutation site was PCR amplified. After TaqαІ restriction enzyme digestion and subsequent Sanger sequencing, of 35 potential founders, one (#315; Polr3hD50G_G0) incorporated the ssODN by correct homologous recombination (Fig. 4C–4D). Another founder partially incorporated the ssODN with the TaqαІ recognition site but without the intended mutation site, identified as Polr3h knockout (#327; Polr3hKO_G0). We also obtained 11 founders carrying genomic deletions identified as Polr3h knockouts by Sanger sequencing. However, no incorporation of TaqαІ recognition site was recognized, and therefore they were not used in this study because of the inability to genotype the offspring.
Loss-of-function mutation of Polr3h gene causes early embryonic lethality in mice
To evaluate the requirement and potential role of POLR3H in mouse reproductive function, we bred mice heterozygous for Polr3h allele (Polr3hKO/+) to generate the global knockout line. We obtained 12 litters and 57 pups (22 males and 35 females) from six different breeding pairs. After genotyping, 31 mice (54%) were homozygous for the wild type allele, and the remaining (46%) were heterozygous for the Polr3hKO mutation. No mice homozygous for Polr3hKO mutation were obtained. To assess whether Polr3hKO embryos die of developmental abnormalities, a group of pregnant mice (n = 5) was euthanized at midgestation (between embryonic days E10 and E12). We obtained 40 embryos (an average of eight implantations per dam). Of those, 35 embryos were apparently healthy and five were resorbed. After genotyping, we found no mice with the homozygous mutation (Polr3hKO/KO). Of 40 embryos, 22 were heterozygous for the knockout allele (Polr3hKO/+), and the remaining were wt (Polr3h+/+), indicating that Polr3h knockout mice are not viable because of early (preimplantation or germline) defects.
Polr3h D50G mice have delayed puberty
To obtain mice homozygous for the Polr3hD50G mutation and control littermates, we bred mice heterozygous for the Polr3hD50G (Polr3hD50G/+) allele. The distribution of the offspring genotypes followed the Mendelian ratio (i.e., 25% Polr3hD50G/D50G, 50% Polr3hD50G/+, and 25% Polr3h+/+). Only mice homozygous for Polr3hD50G or for the wt allele (controls) were used in this study. Polr3hD50G/+ mice were discarded or used as breeders.
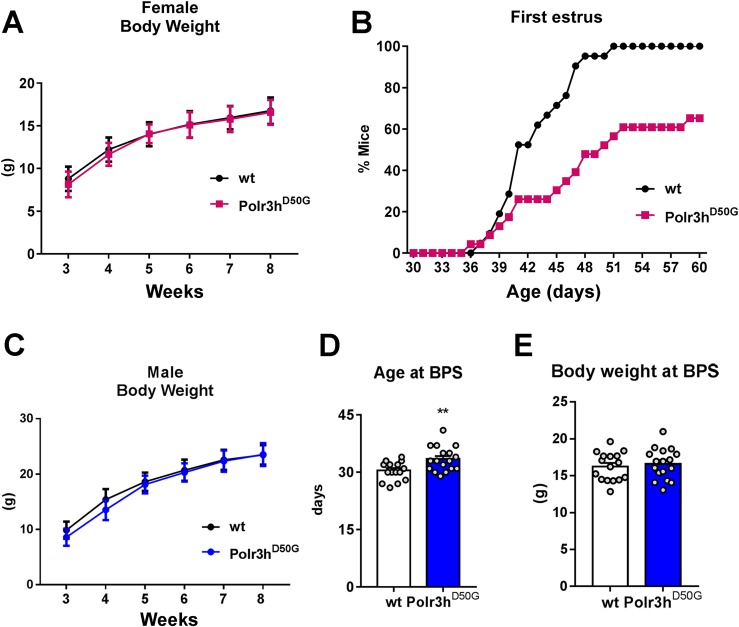
The body weight of female Polr3hD50G mice was similar to that of control littermates (Fig. 5A). No difference in the onset of puberty (age at vaginal opening, 31.4 ± 0.8 days in wt vs. 32 ± 0.9 days in Polr3hD50G) was observed between genotypes (n = 23 wt and n = 21 Polr3hD50G mice). However, puberty completion (age at first estrus) was dramatically delayed in Polr3hD50G mice (Fig. 5B). At PN60, about 35% of Polr3hD50G mice had not shown vaginal cornification. Similarly, Polr3hD50G male mice showed no change in body weight (Fig. 5C) but had delayed pubertal maturation (BPS, n = 17 wt and n = 16 Polr3hD50G mice; Fig. 5D). No difference in body weight at age of BPS was observed (Fig. 5E).
Figure 5.
The Polr3hD50G mice have delayed pubertal maturation. (A) Progression of body weight in females in two cohorts (n = 21 wt and n = 23 Polr3hD50G). (B) Survival graph showing age of first estrus in percentage of mice for two cohorts, as in (A). (C) Progression of body weight in males in two cohorts (n = 10 wt and n = 20 Polr3hD50G). (D) Age at BPS. Data were analyzed by two-tailed t test [n as in (F), df 31, t = 2.9; P = 0.005]. F test to compare variances showed no difference, P = 0.3. (E) Bar graph showing body weight at BPS. Data were analyzed by two-tailed t test [n as in (C), df = 31, t = 0.5; P = 0.60]. F test to compare variances showed no difference, P = 0.76. **P < 0.01.
Polr3h D50G mice show decreased fertility
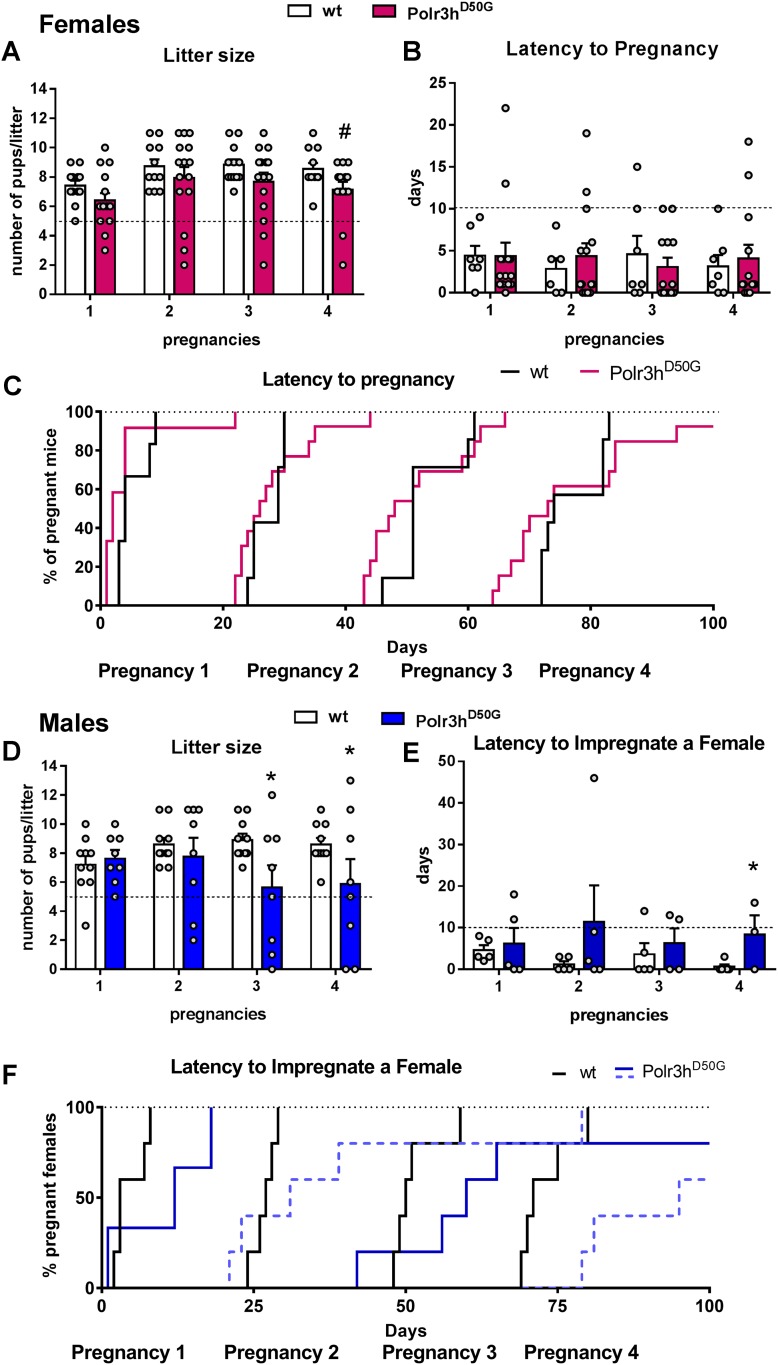
Compared with control littermates, adult Polr3hD50G female mice (PN80 to PN120) showed no differences in the mean number of estrous cycles (5 ± 0.6 in Polr3hD50Gvs. 4.8 ± 0.9 in wt), length of cycles (7.4 ± 1.6 in Polr3hD50Gvs. 7.1 ± 0.9 in wt), and time spent in each cycle phase (n = 5 wt and n = 9 Polr3hD50G mice).
After 100 days of continuous fertility testing (two independent cohorts, n = 6 to 8), 10 of 12 wt (83.3%) and 13 of 16 Polr3hD50G (81.25%) female mice had four pregnancies to term. Differences in litter size did not reach statistical difference (P = 0.08). However, the Polr3hD50G, but not the wt mice, delivered less than five pups in nine events (Fig. 6A). On average, Polr3hD50G mice showed a delay of about 10.75 ± 2.0 days for successful pregnancies compared with wt (Fig. 6B, 6C). In males (n = 8 to 10), six of eight (75%) Polr3hD50Gvs. 100% wt mice impregnated the cage mate four times in 100 days. Litter size was lower in the third (P = 0.01) and in the fourth (P = 0.04) pregnancies (Fig. 6D). In eight pregnancies, wt females bred with Polr3hD50G mice gave birth to less than five pups. Latency to impregnate a female was longer (average of 20.25 ± 10.0 days) in Polr3hD50G compared with wt mice (Fig. 6E, 6F). No difference in testis weight (0.092 ± 0.05 vs 0.094 ± 0.002 g) and gross morphology as seen on H&E was detected.
Figure 6.
Polr3h D50G mice show decreased fertility. (A–F) Graphs showing litter size and latency to pregnancy (or to impregnate a female) in 100 days of continuous breeding. For litter size, linear mixed model fit by Restricted Maximum Likelihood was used. The t tests used the Satterthwaite method (lmerModLmerTest). Formula: Litter.size ≈ Pregnancies + Genotype + (1/Mouse.ID). Female: df = 55.09, t = 1.7; #P = 0.08. Male: df = 49.53, Pregnancy 3: t = 2.4, *P = 0.01; Pregnancy 4: t = 2.0, *P = 0.04. Difference in (E) was observed via log-rank (Matel-Cox) test, *P = 0.04. Dashed black lines indicate threshold for comparison (5 pups per litter, 10 days for pregnancy and 100%).
Adult Polr3hD50G mice showed changes in ovarian transcript levels and decreased number of primary follicles
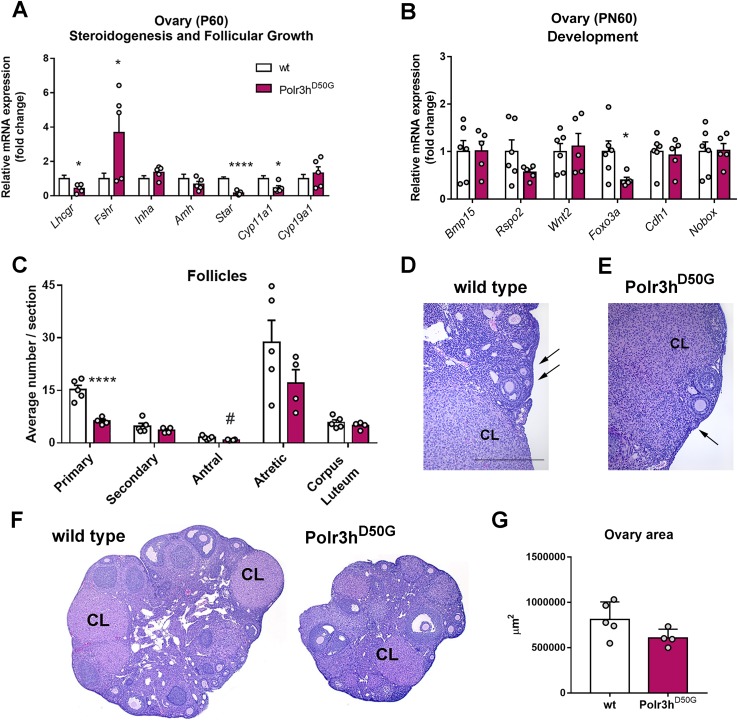
Given the high expression of Polr3h in gonads of control mice, we assessed potential changes in relevant genes: gonadotropin receptors (Lhcgr and Fshr), anti-Müllerian hormone (Amh), inhibin (Inha), and steroidogenesis enzymes (Star, steroidogenic acute regulatory protein and Cyp11a1, cytochrome P450, family 11, subfamily A polypeptide 1). Decreased gonadal Lhcgr, Star, and Cyp11a1 expression and increased Fshr expression were observed in adult Polr3hD50G mice (Fig. 7A).
Figure 7.
Polr3h D50G mice show changes in ovarian transcript levels and decreased number of primary follicles. (A) Relative expression of genes associated with steroidogenesis and follicular growth. Note decreased levels of Lhcgr, Star, and Cyp11A1 and increased Fshr in Polr3hD50G female mice (df = 9; t > 2.4 for differences). (B) Relative expression of genes associated with ovary development. Note decreased levels of Foxo3a in Polr3hD50G female mice (df = 9, t > 2.38 for differences). (C) Average number of follicles per section in different stages of development. Note decreased number of primary follicles (df = 7, t = 5.9). Differences were determined by multiple t test (P < 0.05). Discovery was determined via the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. (D–F) Bright field images showing sections of ovaries. Arrows point to primary follicles (D, E). (G) Graph showing average sectional ovary area for both genotypes. Two-tailed Student t test was used (P = 0.08). *P < 0.05; ****P < 0.001, #P < 0.05 but q > 0.1. Amh, anti-Mullerian hormone; Bmp15, bone morphogenetic protein 15; Cdh1, cadherin 1; CL, corpus luteum; Cyp11a1, cytochrome P450 family 11 subfamily A member 1; Cyp19a1, cytochrome P450 family 19 subfamily A member 1 (aromatase); Foxo3, forkhead box O3; Fshr, follicle stimulating hormone receptor; Inha, inhibin a; Lhcgr, luteinizing hormone/chorionic gonadotropin receptor; Nobox, NOBOX oogenesis homeobox; Rspo2, R-spondin 2; Star, steroidogenic acute regulatory protein; Wnt2, Wnt family member 2.
We next evaluated the expression of genes previously associated with human and mouse ovarian development (3, 29, 30): bone morphogenetic protein 15 (Bmp15), R-spondin 2 (Rspo2), Wnt family member 2 (Wnt2), forkhead box O3 (Foxo3a), cadherin 1 (Cdh1), and NOBOX oogenesis homeobox (Nobox). Decreased expression of Foxo3a was apparent (Fig. 7B).
To assess whether Polr3hD50G mutation promotes depletion of ovarian follicles, we performed histological analysis of ovaries of adult mice. We observed a clear decrease in the average number of primary and antral follicles per section in Polr3hD50G mice (Fig. 7C–7E). We also noticed that some of the individual ovaries were very small, but only a trend for difference in average sectional area was found comparing the genotypes (P = 0.08; Fig. 7F, 7G). Likewise, no change in testis weight was observed (PN80, n = 6 wt and n = 10 Polr3hD50G, 0.091 ± 0.005 g vs. 0.094 ± 0.002 g, t = 0.33, df = 7.3, P = 0.74).
Discussion
In this study, we identified a homozygous missense mutation (c.149A>G; p.Asp50Gly) in POLR3H, the gene encoding the Pol III polypeptide H, in four patients (two pairs of sisters) from two unrelated consanguineous families diagnosed with idiopathic POI. In addition, we demonstrated that Polr3hD50G mice have delayed puberty and decreased fertility associated with decreased expression of ovarian Foxo3a and lower primary follicle counts.
The Pol Ш is a DNA-dependent RNA polymerase that catalyzes the transcription of DNA into RNA to generate small RNAs, such as 5S rRNA, tRNA, 7SL RNA, 7SK RNA, MRP, U6, and H1 RNA (5, 31). It contains 17 different subunits and plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses, acting as both nuclear and cytosolic DNA sensors involved in innate immune responses (31). Pol Ш activity is also crucial for the regulation of cell cycle, cell growth, and differentiation (31).
The POLR3H gene is located in chromosome 22, position 22.q13.2. The coding base pair c.149A>G resulted in a change from aspartic acid to glycine. According to ClustalW/Ensembl tools, aspartic acid is highly conserved on sequence alignment from humans to primitive eukaryotes as Sacharomyces cerevisiae. The mutation (c.149A>G; p.Asp50Gly) was absent from matched Brazilian controls and from the Online Archive of Brazilian Mutations database (28). In addition, the p.Asp50Gly mutation has not been reported in other public databases such as 1000 Genomes and the NHLBI ESP Exome Variant Server. This mutation is present in the gnomAD database with five-allele counts among 277,204 alleles analyzed, which represents 0.00001804 of allele frequency. However, no homozygous alleles were previously identified.
Available databases have documented that POLR3H is ubiquitously expressed. In mice gonads, Polr3h is preferentially expressed in oocytes, ovarian granulosa cells, and developing spermatogonia. This finding is highly relevant because the evaluated patients had isolated POI with no other identified clinical manifestations. However, one proband of family 1 developed class I obesity over the years. Whether this metabolic dysfunction was induced or facilitated by POLR3H mutation is unknown. In our mouse model (Polr3hD50G), no evidence of changes in body weight regulation was noticed during the experimental period.
Moreover, the advantages of early POI diagnosis could improve the clinical management approach, such as better treatment options for bone health, cardiovascular issues, psychosexual and psychological function, and implications for relatives of women with POI (1). Because diagnoses of primary amenorrhea are based on the absence of menarche and breast development after biochemical confirmation, we could provide personal care to address all clinical needs at a precocious age, as recommended by POI guidelines (1, 2). Furthermore, because of the early manifestations of POI in these families, we were able to reach younger sisters, providing clinical management at an early age and thus avoiding consequences of untreated POI.
Although the POLR3H subunit has not been associated with any disorder in mammals, missense mutations in Pol III polypeptides A and B (POLR3A and POLR3B) have been implicated in autosomal recessive 4H syndrome (i.e., hypomyelinating, hypodontia, hypogonadotropic hypogonadism, and leukodystrophy syndrome) (32–34). A POLR3B mutation was also reported in patients with isolated hypogonadotropic hypogonadism without neurologic or dental anomalies (35). In transgenic mice with the homozygous Polr3a p.Gly672Glu variant, which causes hypomyelinating leukodystrophy in humans, no neurologic abnormalities were observed (36). These studies emphasize the wide spectrum of phenotypes associated with POL III disorders in humans and animal models. Our findings add another layer of complexity for the role of POL III in human physiology and establish the POLR3H as a key component for typical follicle development and ovary function.
The genetic basis of POI has comprised ≥60 genes classifying POI as a highly heterogenous disorder that presents a diverse genetic background (2). Although these genes were previously identified in familial and sporadic POI cases belonging to larger POI populations, the genetic etiology of most POI cases remains unclear (3, 4).
The described genes are implicated in several processes such as gonadal development, hormonal signaling, DNA replication and meioses and DNA repair, immune function, and metabolism (3). Considering that mouse genetic model has become an important tool to prove the functional relevance of genetic mutations identified in humans (37), we generated two mouse lines with Polr3h mutations to gain insights into the mechanism associated with POLR3H dysfunction and POI. Mice with loss-of-function mutation in Polr3h gene (Polr3hKO) are not viable. In this study, we were unable to determine the causes of lethality and the developmental abnormalities, but because no homozygous knockout mice were identified at embryonic days E10 to E12, our findings suggest that POLR3H is necessary for germline differentiation, for early embryonic development and/or implantation.
Although the Polr3hKO mutation is embryonically lethal, the Polr3hD50G mouse line is viable. The sgRNA used to target Porl3h with CRISPR has a high specificity score of 75. This score exceeds recommendations for specificity and minimizes potential off-target hits (38). Moreover, predictions of possible off-target hits in exons in the mouse genome show that off-target hits will not occur on mouse chromosome 15. Thus, linked mutations on chromosome 15 that will cosegregate with the Polr3h gene are not predicted to occur. The founder mice were also mated with wt C57BL/6 mice to move the Polr3h mutations onto the C57BL/6 background over several generations. Because offspring positive for Polr3h mutation are used for subsequent breeding, any chromosome with an off-target hit from the founder mutant mouse will be diluted out and potentially removed by independent assortment during breeding. Therefore, the lethality of the Polr3hKO mutation and the phenotype of the Polr3hD50G mouse line is unlikely to be due to off-target effects. Female and male Polr3hD50G mice showed a delay for pubertal completion, the same phenotype we observed in the patients with POI. Indeed, ≥70% of patients presenting with primary amenorrhea also have delay of puberty and absence of breast development (3). Moreover, Polr3hD50G mice demonstrated signs of progressive gonadal insufficiency associated with smaller litter size, longer time for successful pregnancies, and fewer primary follicles. The ovaries of Polr3hD50G mice also showed decreased Star and Cyp11a1 gene expression, suggesting that POLR3H has an effect in ovarian steroidogenesis, in agreement with the hypoestrogenism observed in patients with POI. Additional studies to assess the role of POLR3H in sex steroid production are warranted.
Follicular development depends on endocrine and intraovarian regulators, such as growth factors and gonadal steroids (39). Disruption of ovarian physiology may be a direct effect of follicular atresia driven by apoptosis of granulosa cell layer, followed by apoptosis of the theca cells (40). Our findings indicate that downregulation of POLR3H promotes early depletion of ovarian function, resulting in the POI phenotype. Other evidence reinforcing this argument is the decreased expression of ovarian Foxo3a gene observed in Polr3hD50G mice. Several oocyte transcriptional regulators play key roles in the control of primordial follicle formation and follicle maintenance (41). FOXO3A is one of them, and it has already been associated with POI (2). Foxo3a−/− mice exhibited early depletion of functional ovarian follicles, which resulted in a marked decline in fertility (42). In mammals, FOXO factors play a key role in cell cycle arrest and apoptosis. Specifically, FOXO3A plays an intraoocyte role controlling follicular activation and early development (43). Moreover, studies have shown that phosphorylation of FOXO3A leads to its nuclear export and changes in apoptotic transcriptional activities (44). Our data suggest that disruption of POLR3H alters Foxo3a expression and intracellular dynamics inducing early depletion of ovarian follicles. Additional studies on the molecular mechanisms are warranted, but our findings in human and mouse genetics define POLR3H as a genetic cause of POI, expanding the knowledge of molecular pathways associated with control of ovarian function.
Acknowledgments
Financial Support: This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant 2014/14231-0 (to M.M.F.); FAPESP grant 2013/02162-8, Nucleo de Estudos e Terapia Celular e Molecular, and Conselho Nacional de Desenvolvimento Científico e Tecnológico grant 303002/2016-6 (to B.B.M.); and FAPESP grant 2014/50137-5 (to Laboratorio de Sequenciamento em Larga Escala). C.F.E. is funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health (NIH) (grants HD069702 and HD090567). The Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities under the direction of Dr. Thom Saunders is supported by NIH grants P30CA046592, DK34933 (University of Michigan Gut Peptide Research Center), and P30DK08194 (University of Michigan George M. O’Brien Renal Core Center).
Current Affiliation: M.M. Franca’s current affiliation is the Department of Medicine, Section of Endocrinology, The University of Chicago, Chicago, IL 60637.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- BPS
balanopreputial separation
- CRISPR
clustered regularly interspaced short palindromic repeats
- Ct
threshold cycle
- E
embryonic day
- ESP
Exome Sequencing Project
- HDR
homology-directed repair
- H&E
hematoxylin and eosin
- indel
insertion/deletion
- NHLBI
National Heart, Lung, and Blood Institute
- PAM
protospacer adjacent motif
- PN
postnatal day
- POI
primary ovarian insufficiency
- sgRNA
single guide RNA
- ssODN
single-strand donor oligonucleotide
- wt
wild type
References
- 1. Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, Cifkova R, de Muinck Keizer-Schrama S, Hogervorst E, Janse F, Liao L, Vlaisavljevic V, Zillikens C, Vermeulen N; European Society for Human Reproduction and Embryology (ESHRE) Guideline Group on POI. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31(5):926–937. [DOI] [PubMed] [Google Scholar]
- 2. Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, Heddar A, Jarzabek K, Laisk-Podar T, Salumets A, Tapanainen JS, Veitia RA, Visser JA, Wieacker P, Wolczynski S, Misrahi M. Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol Metab. 2018;29(6):400–419. [DOI] [PubMed] [Google Scholar]
- 3. Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37(6):609–635. [DOI] [PubMed] [Google Scholar]
- 4. Laissue P. The molecular complexity of primary ovarian insufficiency aetiology and the use of massively parallel sequencing. Mol Cell Endocrinol. 2018;460:170–180. [DOI] [PubMed] [Google Scholar]
- 5. White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6(1):69–78. [DOI] [PubMed] [Google Scholar]
- 6. Mendonça BB, Russell AJ, Vasconcelos-Leite M, Arnhold IJ, Bloise W, Wajchenberg BL, Nicolau W, Sutcliffe RG, Wallace AM. Mutation in 3 beta-hydroxysteroid dehydrogenase type II associated with pseudohermaphroditism in males and premature pubarche or cryptic expression in females. J Mol Endocrinol. 1994;12(1):119–122. [DOI] [PubMed] [Google Scholar]
- 7. França MM, Lerario AM, Funari MFA, Nishi MY, Narcizo AM, de Mello MP, Guerra-Junior G, Maciel-Guerra AT, Mendonça BB. A novel homozygous missense FSHR variant associated with hypergonadotropic hypogonadism in two siblings from a Brazilian family. Sex Dev. 2017;11(3):137–142. [DOI] [PubMed] [Google Scholar]
- 8. França MM, Funari MFA, Lerario AM, Nishi MY, Pita CC, Fontenele EGP, Mendonca BB. A novel homozygous 1-bp deletion in the NOBOX gene in two Brazilian sisters with primary ovarian failure. Endocrine. 2017;58(3):442–447. [DOI] [PubMed] [Google Scholar]
- 9. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garrison A, Marth G. Haplotype-based variant detection from short-read sequencing. 2012.http://arxiv.org/abs/1207.3907. Last revised 20 July 2012. Accessed April 2016.
- 11. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Álvarez-Castro JM, Yang RC. Multiallelic models of genetic effects and variance decomposition in non-equilibrium populations. Genetica. 2011;139(9):1119–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494(3):528–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott MM, Lachey JL, Sternson SM, Lee CE, Elias CF, Friedman JM, Elmquist JK. Leptin targets in the mouse brain. J Comp Neurol. 2009;514(5):518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donato J Jr, Lee C, Ratra DV, Franci CR, Canteras NS, Elias CF. Lesions of the ventral premammillary nucleus disrupt the dynamic changes in Kiss1 and GnRH expression characteristic of the proestrus-estrus transition. Neuroscience. 2013;241:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 2014;14(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. [DOI] [PubMed] [Google Scholar]
- 21. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3(1):3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Becker K, Jechow B. Generation of transgenic mice by pronuclear microinjection. In: Pease S, Saunders TL, eds. Advanced Protocols for Animal Transgenesis: An ISTT Manual. Heidelberg, Germany: Springer Berlin; 2011:99–115. [Google Scholar]
- 24. Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG, Saunders TL. Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 2009;18(5):769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naslavsky MS, Yamamoto GL, de Almeida TF, Ezquina SAM, Sunaga DY, Pho N, Bozoklian D, Sandberg TOM, Brito LA, Lazar M, Bernardo DV, Amaro E Jr, Duarte YAO, Lebrão ML, Passos-Bueno MR, Zatz M. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum Mutat. 2017;38(7):751–763. [DOI] [PubMed] [Google Scholar]
- 29. Cheng Y, Kawamura K, Takae S, Deguchi M, Yang Q, Kuo C, Hsueh AJ. Oocyte-derived R-spondin2 promotes ovarian follicle development. FASEB J. 2013;27(6):2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ricken A, Lochhead P, Kontogiannea M, Farookhi R. Wnt signaling in the ovary: identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology. 2002;143(7):2741–2749. [DOI] [PubMed] [Google Scholar]
- 31. Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9(18):3687–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bernard G, Chouery E, Putorti ML, Tétreault M, Takanohashi A, Carosso G, Clément I, Boespflug-Tanguy O, Rodriguez D, Delague V, Abou Ghoch J, Jalkh N, Dorboz I, Fribourg S, Teichmann M, Megarbane A, Schiffmann R, Vanderver A, Brais B. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(3):415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saitsu H, Osaka H, Sasaki M, Takanashi J, Hamada K, Yamashita A, Shibayama H, Shiina M, Kondo Y, Nishiyama K, Tsurusaki Y, Miyake N, Doi H, Ogata K, Inoue K, Matsumoto N. Mutations in POLR3A and POLR3B encoding RNA polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet. 2011;89(5):644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tétreault M, Choquet K, Orcesi S, Tonduti D, Balottin U, Teichmann M, Fribourg S, Schiffmann R, Brais B, Vanderver A, Bernard G. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(5):652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richards MR, Plummer L, Chan YM, Lippincott MF, Quinton R, Kumanov P, Seminara SB. Phenotypic spectrum of POLR3B mutations: isolated hypogonadotropic hypogonadism without neurological or dental anomalies. J Med Genet. 2017;54(1):19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choquet K, Yang S, Moir RD, Forget D, Larivière R, Bouchard A, Poitras C, Sgarioto N, Dicaire MJ, Noohi F, Kennedy TE, Rochford J, Bernard G, Teichmann M, Coulombe B, Willis IM, Kleinman CL, Brais B. Absence of neurological abnormalities in mice homozygous for the Polr3a G672E hypomyelinating leukodystrophy mutation. Mol Brain. 2017;10(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen D, Xu T. The expanding role of mouse genetics for understanding human biology and disease. Dis Model Mech. 2008;1(1):56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Drummond AE. The role of steroids in follicular growth. Reprod Biol Endocrinol. 2006;4(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Craig J, Orisaka M, Wang H, Orisaka S, Thompson W, Zhu C, Kotsuji F, Tsang BK. Gonadotropin and intra-ovarian signals regulating follicle development and atresia: the delicate balance between life and death. Front Biosci. 2007;12(8–12):3628–3639. [DOI] [PubMed] [Google Scholar]
- 41. Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301(5630):215–218. [DOI] [PubMed] [Google Scholar]
- 43. Liu L, Rajareddy S, Reddy P, Du C, Jagarlamudi K, Shen Y, Gunnarsson D, Selstam G, Boman K, Liu K. Infertility caused by retardation of follicular development in mice with oocyte-specific expression of Foxo3a. Development. 2007;134(1):199–209. [DOI] [PubMed] [Google Scholar]
- 44. Liu K, Rajareddy S, Liu L, Jagarlamudi K, Boman K, Selstam G, Reddy P. Control of mammalian oocyte growth and early follicular development by the oocyte PI3 kinase pathway: new roles for an old timer. Dev Biol. 2006;299(1):1–11. [DOI] [PubMed] [Google Scholar]