Abstract
Lipid remodeling in soybean under phosphorus (P)-limitation stress was investigated via lipidomic analysis. Principle component analysis of lipidome data from plants with 4 unfolded trifoliate leaves revealed that each leaf responded to P-limitation stress differently. Upon P limitation, a substantial decrease in phospholipids was observed particularly in the 1st and 2nd trifoliate leaves, while 3rd, and especially 4th, trifoliate leaves showed lipid profiles similar to those from control plants grown under P sufficiency. Under P-limited conditions, non-phosphorus glycoglycerolipid, glucuronosyldiacylglycerol (GlcADG), significantly increased in the 1st and 2nd trifoliate leaves. The levels of some other non-phosphorus glycoglycerolipids, including monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol (SQDG), were elevated under P-limited growth conditions, while there were only slight changes in the total levels of these lipid classes upon P limitation. These results indicate that the lipid metabolic pathway in tissues of soybean plants does not uniformly respond to P-limitation stress, where lipid remodeling is very active in older leaves and phosphate appears to be preferentially remobilized to the younger tissues under P-limited conditions.
Keywords: glucuronosyldiacylglycerol, lipidome, phosphorus, soybean, sulfoquinovosyldiacylglycerol
Phosphorus (P) is an essential macronutrient for plant growth. Thus, plants have developed various mechanisms to adapt to P-limited environments. One of them is lipid remodeling where a drastic decrease in phospholipids and a complementary increase in non-phosphorus glycolipids are observed (Andersson et al. 2003; Benning and Ohta 2005; Härtel et al. 2000; Jouhet et al. 2004). Phospholipids constitute a major phosphorus pool in plants, accounting for about 30% of the total organic phosphates (Bieleski 1968, 1973; Ergle and Guinn 1959). Thus, remobilization of phosphate from phospholipids or limitation of phosphate supply for phospholipid metabolism is considered to play an essential role in the adaptive response to P-limited growth conditions.
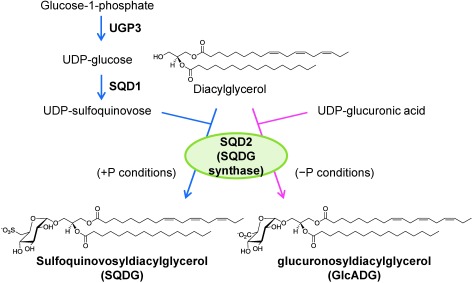
The physiological importance of lipid remodeling in plants under P-deficient growth conditions has been investigated using several loss-of-function mutants, where genes involved in sulfoquinovosyldiacylglycerol (SQDG) biosynthesis (Essigmann et al. 1998; Okazaki et al. 2009; Yu et al. 2002), induced accumulation of galactolipids (Awai et al. 2001; Gaude et al. 2008; Kelly and Dörmann 2002; Kobayashi et al. 2006, 2009) and degradation of phospholipids under P-limited growth conditions (Cheng et al. 2011; Cruz-Ramírez et al. 2006; Gaude et al. 2008; Nakamura et al. 2005, 2009) are disrupted. Some of them showed growth defects under P-limited conditions (Nakamura et al. 2009; Okazaki et al. 2013; Yu et al. 2002). Recently, we have identified a novel plant glycoglycerolipid, glucuronosyldiacylglycerol (GlcADG), which inducibly accumulates in Arabidopsis and rice upon P limitation (Figure 1) (Okazaki et al. 2013). Investigation of various Arabidopsis mutants, deficient in 3 different SQDG-biosynthetic genes (ugp3, sqd1, sqd2), revealed that only sqd2 mutant showed impaired accumulation of GlcADG and severe growth defects under P-limited nutrient conditions as compared to other SQDG-deficient mutants, suggesting that GlcADG contributes to the mitigation of P-limitation stress in plants. Our study also indicates that growth defect of sqd2 mutants of Arabidopsis under P-limited conditions was mainly observed in the older well-developed leaves, the color of which is white due to an induced breakdown of chlorophylls, whereas the changes in leaf color are slight in younger leaves (Okazaki et al. 2013). These results suggest that there are substantial differences between local and systemic reactions to P deficiency, especially in lipid remodeling. Arabidopsis is a good research material for this research purpose, although the young rosette leaves are rather too small for handling. In this study, we investigated the tissue-dependent differences in lipid remodeling upon P limitation using soybean plants because the leaf size of this plant is preferable for our study and each leaf was easily distinguishable compared with Arabidopsis.
Figure 1. Biosynthesis of SQDG and GlcADG in Arabidopsis.
SQDG is synthesized in chloroplast by SQDG synthase, which is encoded by SQD2 in Arabidopsis, using diacylglycerol and UDP-sulfoquinovose as substrates. Under P-limited conditions, GlcADG inducibly accumulates in plant leaves. Because biosynthesis of GlcADG requires SQDG synthase, it is proposed that GlcADG is synthesized from diacylglycerol and UDP-glucuronic acid, although the biochemistry of the glycosyltransfer reactions by SQDG synthase remains unclear.
Lipidomic analysis of the 1st trifoliate leaves of soybean grown under P-limited conditions has been previously conducted (Okazaki et al. 2015). The same growing regime was employed in this experiment. Briefly, seeds of soybean (Glycine max (L.) Merr. cv. Enrei) were sown in sterilized wet potting mix. After a 10-day-incubation, the roots were gently washed with water to remove the potting mix, and transplanted in rockwool to which MGRL hydroponic medium (Fujiwara et al. 1992) was supplied. For P-limitation treatment, MGRL without KH2PO4 was supplied to plants. After a 2-week-incubation in rockwool, a central unfolded leaflet of each trifoliate leaf of the soybean plant with 4 trifoliate leaves (Supplementary Figure 1), was cut into small pieces by scissors and mixed well. From the mixture, around 50 mg fresh weight of leaf pieces were randomly collected in a sample tube, and used as one sample batch. These samples were subjected to lipidomic analysis using LC-MS as previously reported (Higashi et al. 2015; Okazaki et al. 2015), to afford a data matrix consisting of about 1,600 metabolite-related signals collected from 3 independent plants grown under P-sufficient or P-limited conditions (Supplementary Dataset).
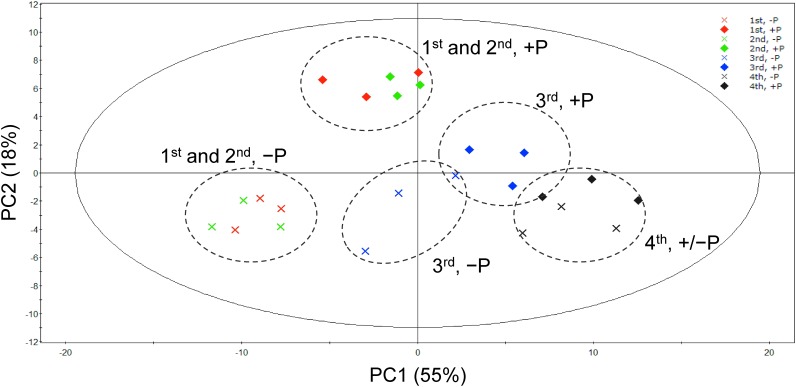
To find the overall changes in the lipid profile of the samples, the dataset was subjected to principle component analysis (PCA), an unsupervised multivariate analysis frequently used to reduce the high-dimensional data and extract the feature of dataset. The score plot of PCA (Figure 2) shows that samples from the 1st, 2nd, 3rd, and 4th trifoliate leaves from plants grown under P-sufficient conditions were scattered from left to right along the PC1 axis in the score plot, and among them, samples of the 1st and 2nd leaves were clustered together. The samples from plants grown under P-limited conditions also exhibited a similar tendency. In addition, samples from 2 different growth conditions were also separated into 2 groups along the PC2 axis (Figure 2). Especially, lipids from the 1st and 2nd trifoliate leaves were clearly separated depending on the P availability.
Figure 2. The score plot of PCA of lipidome data from each individual leaves of soybean grown under P-sufficient or P-limited conditions.
Crude lipid extracts from each trifoliate leaf sample were analyzed by liquid chromatography quadrupole time-of-flight mass spectrometry (Higashi et al. 2015; Okazaki et al. 2015). Each signal detected in the positive ion mode was normalized based on the intensity of the internal standard (PC_10:0/10 : 0). Then the normalized data was pareto-scaled and subjected to PCA.
To identify the major difference in the lipid profile in the 1st and 2nd leaves grown under different P conditions, the dataset of these 2 tissues were further subjected to orthogonal projection to a latent structure-discriminant analysis (OPLS-DA) (Supplementary Figure 2A) (Wiklund et al. 2008), a supervised multivariate analysis method, which maximizes variance contributing to group separation. The discriminative metabolite ions whose levels from P-starved plants were higher than those from P-sufficient plants are shown in the upper-right region of the S-plot of OPLS-DA (Supplementary Figure 2B and Table 1), while the discriminative ions whose levels from plants grown under P-limited conditions were lower than those of plants grown under P-sufficiency are shown in the lower-left region (Supplementary Figure 2B and Table 2). Table 1 shows that the levels of several GlcADG species were significantly elevated upon P limitation. For example, the level of GlcADG_34:2 in the 1st and 2nd trifoliate leaves under P-limited conditions was about 11- and 14-fold higher than that under P-sufficient conditions, respectively. These results are fundamentally in accordance with the results of our recent report (Okazaki et al. 2015). Similarly, levels of some molecular species, such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and triacylglycerol (TAG), increased upon P deficiency. These results suggest that P deficiency preferentially induces the accumulation of non-phosphorus glycerolipid species. In contrast, Table 2 shows the representative lipids whose levels were lower in the 1st and 2nd leaves in the plants grown under P-limited conditions than in those grown under P-sufficient conditions, including several species of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and pheophytin a which is a chlorophyll a lacking a central Mg2+ ion. These results suggest that phospholipids decrease under P-limited conditions as observed in other plants, such as Arabidopsis, rice, and oat (Andersson et al. 2003; Benning and Ohta 2005; Jouhet et al. 2004; Härtel et al. 2000; Okazaki et al. 2013), and phosphorus limitation could affect the chlorophyll-related metabolism.
Table 1. Discriminative metabolites whose detected levels increased upon P depletion, predicted by OPLS-DA.
| Retention time (min) | m/z | Annotation | Fold change in 1st trifoliate leaf (−P/+P) | Fold change in 2nd trifoliate leaf (−P/+P) |
|---|---|---|---|---|
| 4.15 | 786.5728 | GlcADG_34.2 ([M+NH4]+) | 11.30 | 13.50 |
| 3.79 | 808.5561 | GlcADG_36.5 ([M+NH4]+) | 7.69 | 9.73 |
| 4.31 | 762.5722 | GlcADG_32.0 ([M+NH4]+) | 6.00 | 10.90 |
| 3.95 | 784.5572 | GlcADG_34.3 ([M+NH4]+) | 5.43 | 6.10 |
| 8.40 | 846.7548 | TAG_50.3 ([M+NH4]+) | 4.48 | 14.10 |
| 4.30 | 812.5900 | GlcADG_36.3 ([M+NH4]+) | 4.19 | 4.69 |
| 8.71 | 848.7697 | TAG_50.2 ([M+NH4]+) | 3.91 | 7.73 |
| 4.17 | 934.6459 | DGDG_34.2 ([M+NH4]+) | 3.30 | 3.88 |
| 4.52 | 772.5929 | MGDG_34.2 ([M+NH4]+) | 3.01 | 3.63 |
| 7.85 | 868.7392 | TAG_52.6 ([M+NH4]+) | 2.94 | 6.77 |
| 8.13 | 870.7542 | TAG_52.5 ([M+NH4]+) | 2.56 | 4.73 |
| 4.56 | 962.6770 | DGDG_36.2 ([M+NH4]+) | 2.29 | 2.76 |
| 3.80 | 956.6295 | DGDG_36.5 ([M+NH4]+) | 1.49 | 1.65 |
| 4.33 | 960.6608 | DGDG_36.3 ([M+NH4]+) | 1.27 | 1.40 |
| 3.96 | 932.6298 | DGDG_34.3 ([M+NH4]+) | 1.18 | 1.16 |
Table 2. Discriminative metabolites whose detected levels decreased upon P depletion, predicted by OPLS-DA.
| Retention time (min) | m/z | Lipid | Fold change in 1st trifoliate leaf (−P/+P) | Fold change in 2nd trifoliate leaf (−P/+P) |
|---|---|---|---|---|
| 4.58 | 786.6006 | PC_36:2 ([M+H]+) | 0.375 | 0.283 |
| 4.87 | 788.6161 | PC_36:1 ([M+H]+) | 0.396 | 0.325 |
| 5.03 | 814.6317 | PC_38:2 ([M+H]+) | 0.397 | 0.338 |
| 4.70 | 744.5537 | PE_36:2 ([M+H]+) | 0.400 | 0.352 |
| 4.45 | 760.5849 | PC_34:1 ([M+H]+) | 0.404 | 0.380 |
| 4.29 | 716.5227 | PE_34:2 ([M+H]+) | 0.447 | 0.401 |
| 4.02 | 782.5691 | PC_36:4 ([M+H]+) | 0.533 | 0.408 |
| 4.07 | 714.5070 | PE_34:3 ([M+H]+) | 0.561 | 0.622 |
| 4.18 | 758.5695 | PC_34:2 ([M+H]+) | 0.564 | 0.504 |
| 3.62 | 778.5381 | PC_36:6 ([M+H]+) | 0.607 | 0.612 |
| 3.80 | 780.5537 | PC_36:5 ([M+H]+) | 0.611 | 0.541 |
| 4.11 | 740.5237 | PE_36:4 ([M+H]+) | 0.626 | 0.524 |
| 3.97 | 756.5535 | PC_34:3 ([M+H]+) | 0.651 | 0.713 |
| 6.23 | 871.5726 | pheophytin a ([M+H]+) | 0.839 | 0.833 |
| 3.99 | 549.4887 | unknown | 0.576 | 0.554 |
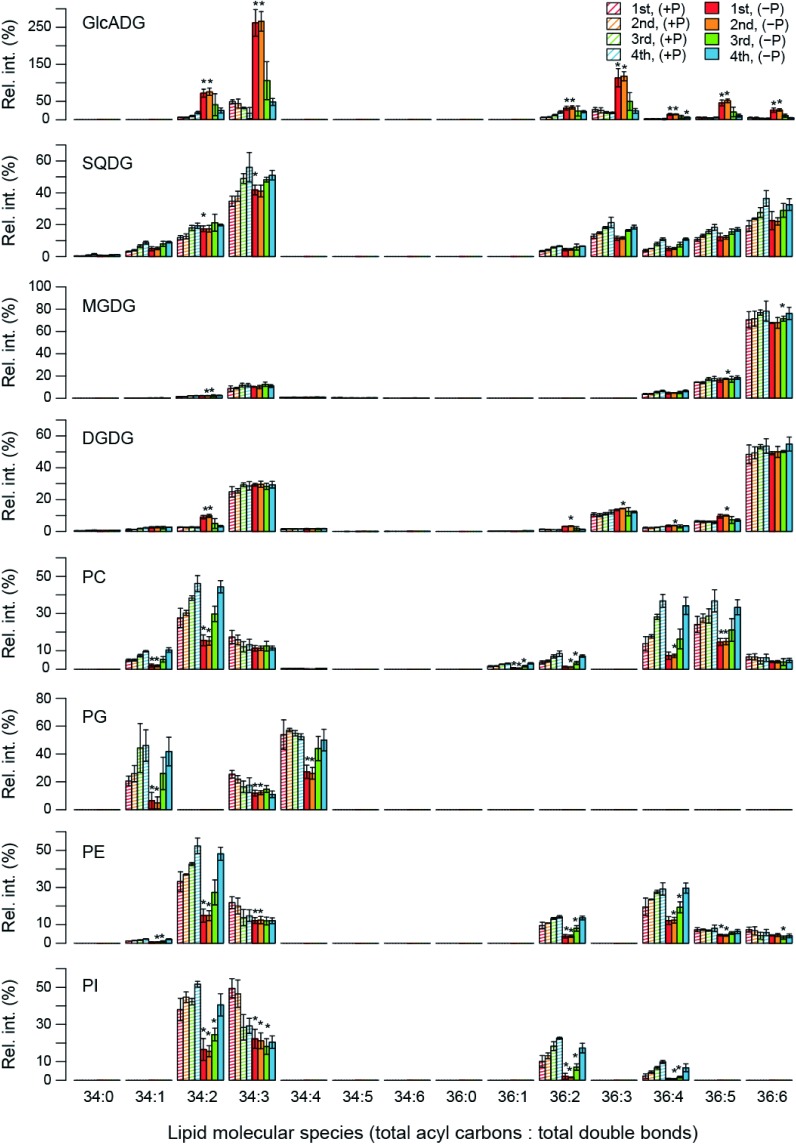
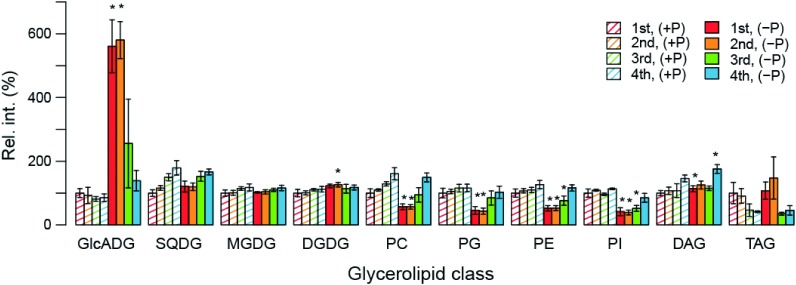
Since the above-mentioned analyses showed that P limitation significantly affected the metabolism of glycerolipids, changes in the level of each lipid molecule upon P limitation were investigated. In Figure 3, the levels of individual lipid molecules are expressed as the relative value against the levels of total lipids of the same class in the 1st trifoliate leaf grown under P-sufficient conditions. The total of the relative values of individual lipid molecules of the same lipid class in Figure 3 were also calculated and represented in Figure 4 to examine the effect of P limitation on the total level of each lipid class.
Figure 3. Profiles of polar glycerolipids in each trifoliate leaf of soybean grown under P-sufficient or P-limited conditions.
Intensity of each lipid signal was normalized based on that of the internal standard. Then, the normalized peak intensity of each lipid class of the 1st trifoliate leaf grown under P-sufficient growth conditions were summed, and the mean value of 3 biological replicates were calculated. Using this value (=100% of y-axis of this figure), relative intensity (Rel. int.) of each lipid signals was calculated and shown in this figure. Each data point represents the mean±standard deviation (SD) value of 3 biological replicates. Statistical significance in the changes in the levels of individual lipids upon P deficiency were examined by Welch’s t-test using the function (t.test) in the statistical software R version 3.2.2 (http://www.r-project.org/).
Figure 4. Changes in the total level of each lipid class upon P limitation.
To investigate the effect of P limitation on the total level of each lipid class in the leaves of the same growth stage, relative intensity of each lipid signal of the same lipid class in Figure 3 was summed and represented in this figure. Each data point represents the mean±SD value of 3 biological replicates. Statistical significance in the changes in the total levels of each lipid class upon P deficiency was examined by Welch’s t-test.
Figures 3 and 4 indicate that levels of all the detected GlcADG molecules, especially in the 1st and 2nd trifoliate leaves, were elevated upon P limitation as shown in the Table 1. GlcADG levels were also likely to be elevated in the 3rd and 4th leaves grown under P-limited conditions (Figure 4). Although the results given in Table 1 suggest the induction of accumulation of some species of DGDG and TAG upon P limitation (Supplementary Figure 3), there were only slight changes in the total levels of these lipid classes (Figure 4). A similar tendency was also observed in DAG, which is the major intermediate of glycerolipid metabolic pathway (Supplementary Figure 3 and Figure 4). During the lipid remodeling process accompanying lipid decomposition, free fatty acid can be produced. Since the increase of free fatty acid is reported to be show the toxicity in yeast (Fakas et al. 2011), accumulation of some TAG molecules under phosphorus limitation might be an adaptive response in plants to remove free fatty acids in some cellular spaces where this mechanism is required. In addition, it should be noted that accumulation of SQDG upon P limitation was not significantly induced here unlike in Arabidopsis and rice (Essigmann et al. 1998; Okazaki et al. 2009, 2013; Yu et al. 2002).
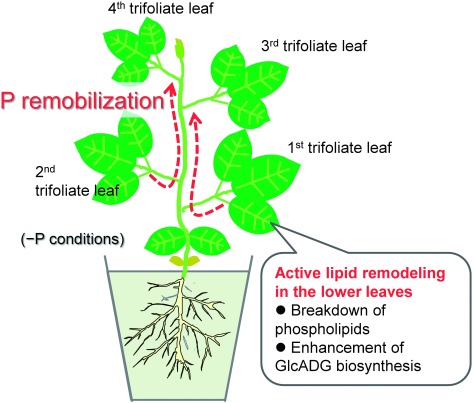
Figures 3 and 4 also display the significant decrease in all the phospholipid classes including PC, PG, PE, and PI in the 1st and 2nd trifoliate leaves. Their content in plants grown under P limitation are also likely to be slightly lower in the 3rd leaf, whereas in the 4th leaf, the total amounts of each lipid class in plants grown under P-limited conditions were almost similar to those in plants grown under P-sufficient conditions. These results collectively suggest that lipid remodeling upon P limitation occurs mainly in the older leaves where phospholipids are reduced along with an increase in GlcADG levels. Such a tissue-dependent difference in lipid remodeling probably contributes to the preferential transfer of phosphorus to the relatively juvenile tissues where cell division occurs actively (Figure 5).
Figure 5. P remobilization in soybean plants through tissue-dependent lipid remodeling proposed by current study.
Induced accumulation of SQDG upon P limitation has been reported from various organisms including Arabidopsis (Benning and Ohta 2005; Härtel et al. 2000; Jouhet et al. 2004), rice (Okazaki et al. 2013), Chlamydomonas (Riekhof et al. 2003), and bacteria (Benning et al. 1993; Güler et al. 1996). The current study revealed that in soybean leaves, the SQDG content was not significantly affected by P deficiency as compared to that of GlcADG, which was also synthesized by SQDG synthase (Figure 1). The transcript levels of SQDG synthase homologs are known to be elevated in soybean leaves under P-limited conditions (Okazaki et al. 2015). Thus, the upregulation of SQDG synthase homologs in soybean leaves upon P limitation presumably mainly contributes to the induced accumulation of GlcADG rather than of SQDG. In addition, these results suggest the possibility of GlcADG being selected as an alternative of SQDG in some plant species, such as soybean. Further studies on gene expressions of SQD1 and UGP3 homologs in each soybean leaf may reveal the state of metabolic pathways involved in the supply of sugar donors for SQDG and GlcADG biosynthesis, which will provide us more comprehensive understanding of GlcADG-biased synthesis by SQDG synthase in lower leaves of soybean under P-limited conditions. Activation of SQDG synthesis requires activation of the sulfur assimilation pathway, which probably requires substantial energy. Thus, elevation of GlcADG levels, which is the sulfur-free SQDG analog under P-limited conditions, might be suited for energy conservation especially under severe stressed conditions. Studies on GlcADG are still limited. Further studies on lipid remodeling in other plant species will reveal the functional relationship of these 2 anionic glycolipids synthesized by SQDG synthase.
Acknowledgments
This work was supported in part by grant aid from the Strategic International Collaborative Research Program of Japan Science and Technology Agency (Metabolomics for a Low Carbon Society, JST-NSF) and Competitive Program for Creative Science and Technology of RIKEN (Integrated Lipidology). This work was also supported by the Japan Advanced Plant Science Network.
Abbreviations
- GlcADG
glucuronosyldiacylglycerol
- SQDG
sulfoquinovosyldiacylglycerol
- LC-MS
liquid chromatography mass spectrometry
Supplementary Data
References
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Awai K, Marechal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J (2001) Two types of MGDG synthase genes, found widely in both 16 : 3 and 18 : 3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Beatty JT, Prince RC, Somerville CR (1993) The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc Natl Acad Sci USA 90: 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bieleski RL (1968) Levels of phosphate esters in Spirodela. Plant Physiol 43: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol Plant Mol Biol 24: 225–252 [Google Scholar]
- Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernandez F, Ramirez-Chavez E, Herrera-Estrella L (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergle DR, Guinn G (1959) Phosphorus compounds of cotton embryos and their changes during germination. Plant Physiol 34: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakas S, Qiu Y, Dixon JL, Han GS, Ruggles KV, Garbarino J, Sturley SL, Carman GM (2011) Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J Biol Chem 286: 29074–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S (1992) Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler S, Seeliger A, Härtel H, Renger G, Benning C (1996) A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J Biol Chem 271: 7501–7507 [DOI] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Okazaki Y, Myouga F, Shinozaki K, Saito K (2015) Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci Rep 5: 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Marechal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Dörmann P (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya K, Ohta H (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47: 238–248 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci USA 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Nishizawa T, Takano K, Ohnishi M, Mimura T, Saito K (2015) Induced accumulation of glucuronosyldiacylglycerol in tomato and soybean under phosphorus deprivation. Physiol Plant 155: 33–42 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K (2013) A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun 4: 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, Tanaka H, Matsuda F, Hirai A, Hirai MY, et al. (2009) A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21: 892–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Ruckle ME, Lydic TA, Sears BB, Benning C (2003) The sulfolipids 2′-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol 133: 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J (2008) Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem 80: 115–122 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.