Abstract
We isolated an ortholog (LjMYB12) of the Arabidopsis R2R3-MYB transcription factor (TF) gene from Lotus japonicus to investigate the regulation of flavonoid biosynthesis, which is driven by many paralogous genes in L. japonicus. We characterized the spatial and temporal expression of LjMYB12 in leaves, stems, roots, flowers, immature seeds, seedling leaves, and seedling roots. Expression was much higher in flowers than in other tissues. To verify the relationship between the expression of LjMYB12 and that of flavonoid biosynthesis genes, we generated transgenic L. japonicus plants overexpressing LjMYB12. Overexpression of LjMYB12 resulted in the upregulation of genes for a chalcone synthase paralog (CHS1), flavanone 3-hydroxylase, and flavonol synthase. Interestingly, LjMYB12 strongly activated CHS1 but did not activate other CHS paralogs. This result suggests differences in the spatial or temporal activation of CHS paralogs by R2R3-MYB TFs. Molecular characterization of R2R3-MYB TFs in L. japonicus will reveal the effects of gene duplication on the regulation of diverse flavonoid biosynthesis.
Keywords: chalcone synthase, flavonol synthase, flavanone 3-hydroxylase, spatial and temporal expression, transgenic Lotus
Leguminous plants use flavonoids and their derivatives in their interactions with other organisms and in response to various environmental stresses (Aoki et al. 2000; Denarie et al. 1992). The regulation of flavonoid biosynthesis in legumes is more complicated than that in other higher plants, because multiple paralogous genes involved in flavonoid biosynthesis exist in the genomes of legumes (Clough et al. 2004; Ryder et al. 1987; Shimada et al. 2003). Spatial and temporal expression of these paralogous genes and the functions of their products are highly diverse (Shimada et al. 2003, 2005). In Arabidopsis thaliana, the expression of flavonoid biosynthesis genes is regulated by several R2R3-MYB transcription factors (TFs) (Borevitz et al. 2000; Nesi et al. 2001). The modular structure of these proteins, which consist of the MYB DNA-binding domain in the N-terminal region and the activation or repression domains in the C-terminal region, places them in one of phylogenetic subgroups 4 to 7 (Kranz et al. 1998; Stracke et al. 2001). In addition, orthologs in subgroups 4 to 7 regulate the expression of flavonoid biosynthesis genes in several other species (Czemmel et al. 2009; Lin-Wang et al. 2010; Paz-Ares et al. 1987). Among R2R3-MYB subgroup 4 to 7, Arabidopsis AtMYB12, which is classified into subgroup 7, is well characterized on the flavonoid biosynthesis (Lewis et al. 2011; Mehrtens et al. 2005; Stracke et al. 2010). Homologs of AtMYB12 also regulated the expression of genes related to flavonoid biosynthesis in tomato (Solanum lycopersicum) and grapevine (Vitis vinifera) (Ballester et al. 2010; Czemmel et al. 2009). Therefore, the molecular characterization of R2R3-MYB subgroup 7 in legumes would reveal the mechanisms that control the expression of many paralogous genes related to flavonoid biosynthesis. In this study, we isolated an ortholog of R2R3-MYB subgroup 7 from Lotus japonicus, a model plant for the study of the molecular genetics of legumes, on the basis of sequence similarity. To understand the regulation of the flavonoid biosynthesis pathways in L. japonicus, we compared spatial and temporal expression patterns between the isolated R2R3-MYB subgroup 7 gene and flavonoid biosynthesis genes. We generated transgenic L. japonicus plants overexpressing the isolated R2R3-MYB subgroup 7 gene and evaluated the expression of flavonoid biosynthesis genes in these plants.
L. japonicus accession Gifu B-129 was obtained from Biological Resource Center in Lotus japonicus and Glycine max of the National BioResource Project (University of Miyazaki). Plants were grown in commercial soil at 26°C in a 16-h light (100–150 µmol s−1 m−2)/8-h dark regime in a growth chamber. Three R2R3-MYB subgroup 7 proteins in Arabidopsis (AtMYB11, AtMYB12, and AtMYB111) contain the R2R3-MYB DNA-binding domain in the N-terminal region and the consensus motif ‘GRTxRSxMK’ (Kranz et al. 1998). The motif of ‘GRTxRSxMK’ serves as a characteristic for distinguishing R2R3-MYB subgroup 7 from the other subgroups. In silico EST sequence analysis was used to search EST clones corresponding to R2R3-MYB subgroup 7 in L. japonicus. We found one EST clone (CN824947) with the motif conserved in R2R3-MYB subgroup 7 among EST clones of L. japonicus. Since the sequence information of the CN824947 clone was partially, the full-length cDNA was synthesized from L. japonicus. Total RNA was extracted from plants with Trizol reagent (Life Technologies, USA). First-strand cDNA was synthesized from total RNA (∼1 µg) with oligo(dT) primer and ReverTra Ace reverse transcriptase (Toyobo, Japan). The full-length cDNA for the R2R3-MYB subgroup 7 gene was synthesized using a 3′-Full RACE Core Set (TaKaRa, Japan) and one specific primer designed on the basis of the sequence information of CN824947 (Supplemental Table). We evaluated the expression levels of LjMYB12, 4-coumarate: CoA ligase (4CL), cinnamic acid 4-hydroxylase (C4H), 3 chalcone isomerases (CHI1–3), 5 chalcone synthases (CHS1, 2, 3, –L1, and –L2), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), 2 isoflavone synthases (IFS1, 2), isoflavone reductase (IFR1), flavonol synthase (FLS), and dihydroflavonol 4-reductase (DFR) by qRT-PCR using SYBR Premix ExTaq II (Tli RNaseH Plus; TaKaRa, Japan) and the primers listed in the Supplemental Table. The reaction was performed in a CFX96 Real-Time System (Bio-Rad Laboratories Inc., Japan) under the following conditions: 40 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 30 s. The plasmid vectors pMDC100-IG and pMDC100-LjMYB12 (Supplemental Figure 1) were constructed by joining the intron-GUS reporter gene (Ohta et al. 1990) or the LjMYB12 gene, respectively, to the cauliflower mosaic virus 35S promoter in the original pMDC100 vector (Curtis and Grossniklaus 2003). Gifu B-129 was transformed and plants were regenerated as described by Stiller et al. (1997) with modifications: Agrobacterium tumefaciens (Rhizobium radiobacter) strain EHA105 was used for transformation; 20 mg l−1 acetosyringone was added into the co-cultivation medium; and the concentration of geneticin for transformant selection was changed from 5 to 10 mg l−1 in the shoot induction medium. Transgenic plants expressing LjMYB12 or intron-GUS were named 12OX or IG, respectively. Total DNA was isolated from fresh leaves of transgenic plants and analyzed by Southern blotting as described by Yamada et al. (2014). A statistical analysis was performed to evaluate the population variances among all samples. The Student’s t test or the Welch’s t test were used to examine whether population means are significantly different from each sample and control. All statistic data was evaluated significantly different from the control when p values were <0.05.
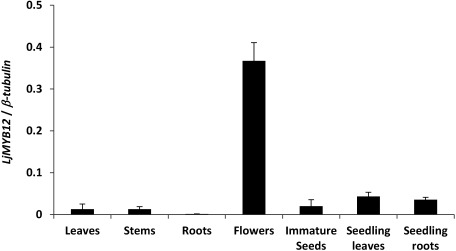
The R2R3-MYB subgroup 7 protein in L. japonicus was predicted to consist of 326 residues (Figure 1). The protein shared the highest sequence identity (83%) with AtMYB12 in the R2R3-MYB DNA-binding domain and the consensus motif (Figure 1). Therefore, the R2R3-MYB subgroup 7 gene isolated in present study was designated as LjMYB12 (AB334529) gene. The nucleotide sequence of LjMYB12 gene was corresponding to that of one gene ID, Lj1g3v4863050, registered on the database of miyakogusa.jp 3.0 (http://www.kazusa.or.jp/lotus/). Subgroup 7 MYBs of Arabidopsis act additively owing to their differential spatial activity in developing seedlings: AtMYB12 controls flavonol biosynthesis mainly in roots, and AtMYB111 controls it mainly in cotyledons (Stracke et al. 2007). The expression of LjMYB12 was much higher in flowers than in leaves, stems, roots, immature seeds, seedling leaves, and seedling roots, its expression did not differ between seedling leaves and roots (Figure 2). Thus, the spatial expression of LjMYB12 differed greatly from that of subgroup 7 MYB in Arabidopsis.
Figure 1. Alignment of amino acid sequences of LjMYB12 (Lj1g3v4863050) with Arabidopsis AtMYB11 (AT3G62610), AtMYB12 (AT2G47460), and AtMYB111 (AT5G49330) in R2R3-MYB subgroup 7 by CLUSTALW. Shaded sequences, conserved residues; bold black underline, R2 and R3 MYB DNA-binding domains; bold gray underline, ‘GRTxRSxMK’ consensus motif of MYB subgroup 7 of Arabidopsis (Kranz et al. 1998).
Figure 2. Expression profile of endogenous LjMYB12 in Gifu B-129. Leaves, stems, roots, and flowers were collected from 14-week-old plants grown at 26°C under a 16/8-h light/dark regime in a growth chamber. Immature seeds were collected from 18-week-old plants. Seedling leaves and roots were collected at 12 days after sowing. Expression levels were determined by normalizing the PCR threshold cycle number of LjMYB12 to that of β-tubulin. Data are means±SD of three biological replicates.
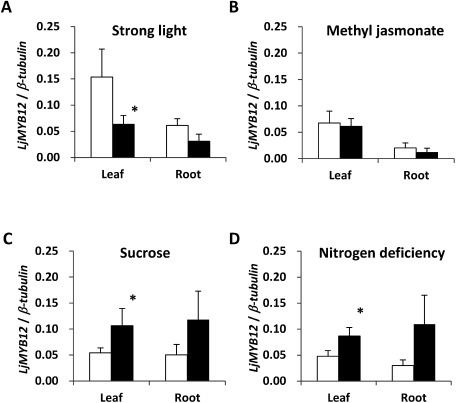
As the expression of MYB genes activated by stressors or inducers such as drought, abscisic acid, sucrose, and ultraviolet light (Abe et al. 1997; Stracke et al. 2010; Teng et al. 2005), we examined the expression profile of LjMYB12 in leaves and roots of seedlings exposed to strong light, methyl jasmonate, sucrose, or nitrogen deficiency. Strong light downregulated LjMYB12 in seedling leaves (Figure 3A). This is a novel finding of a stress response of R2R3-MYB subgroup 7 in higher plants. Methyl jasmonate did not change the expression of LjMYB12 (Figure 3B). Sucrose supply and nitrogen deficiency induced LjMYB12 (Figure 3C and D). Similarly, in Arabidopsis, sucrose supply and nitrogen deficiency weakly induced AtMYB12 in seedlings (Kranz et al. 1998). AtMYB12 controls the expression of flavonoid biosynthesis genes such as CHS, CHI, F3H, and FLS and the activity of UDP-dependent glycosyltransferase in Arabidopsis (Mehrtens et al. 2005; Stracke et al. 2010). To characterize the spatial and temporal expression of flavonoid biosynthesis genes, we evaluated the expression of 14 genes (C4H, 4CL, CHS1, four CHSs [CHS2, CHS3, CHS–L1, and CHS–L2], CHI1, CHI2, CHI3, F3H, F3′H, FLS, IFS1, IFS2, IFR1, and DFR2) in leaves, stems, roots, flowers, immature seeds, seedling leaves, and seedling roots of L. japonicus. Flowers, which showed the highest expression of LjMYB12, showed high expression of C4H, 4CL, CHS1, CHSs, CHI2, F3H, F3′H, FLS, and DFR2 (Supplemental Figure 2). To verify the spatial co-expression of these flavonoid biosynthesis genes and LjMYB12, the expression level was evaluated in transgenic L. japonicus plants overexpressing LjMYB12. Inoculation of explants with A. tumefaciens EHA105 harboring pMDC100-LjMYB12 produced 30 T0 plants, of which 5 showed >9× the expression of LjMYB12 of a control plant which expressed intron-GUS (Supplemental Figure 3). However, these T0 plants did not produce T1 seeds. On the other hand, four T0 plants (12OX-2-1, 12OX-6-3, 12OX-10-2, and 12OX-13-1), which showed moderate expression of LjMYB12, set T1 seeds. Although the remaining 21 plants had seed fertility, the expression level of LjMYB12 was low. High performance liquid chromatography (HPLC) analysis revealed that mature seeds of 12OX10-2 accumulated much more putative flavonoid compounds than control plants (Supplemental Figure 4), indicating that an overexpressing LjMYB12 gene enhances the biosynthesis of flavonoids. These results suggest that the excessive accumulation of flavonoids caused by very high expression of LjMYB12 might reduce seed fertility.
Figure 3. Expression profiles of LjMYB12 in leaves and roots of Gifu B-129 seedlings exposed to (A) strong light, (B) methyl jasmonate, (C) sucrose, or (D) nitrogen deficiency. □ Control plants: seeds were germinated on basal MS medium containing MS vitamins and 0.3% Gelrite for 4 days in the dark at 26°C, and then grown under continuous light (40–60 µmol s−1 m−2) at 26°C. ■ Treated plants: (A) Seedlings were grown at 26°C under continuous light (250–300 µmol s−1 m−2) for 24 h. (B) Seedlings were treated with 500 µM methyl jasmonate and 0.1% Triton X-100 for 24 h. (C) Seedlings were grown on basal MS medium containing 100 mM sucrose for 7 days. (D) Seedlings were grown on basal MS medium without nitrogen for 7 days. All expression levels were determined by normalizing the PCR threshold cycle number of LjMYB12 to that of β-tubulin. Data are means±SD of three experimental replicates. * Significantly different at p<0.05.
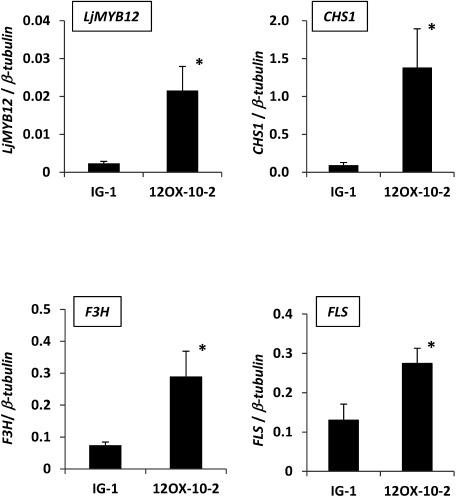
For further analyses we used T2 plants derived from 12OX-10-2, which showed the highest expression level of LjMYB12 among T0 plants with seed fertility and normal morphology (Supplemental Figure 5). At least four bands hybridized with the LjMYB12-specific probe in Southern bolt analysis of the T2 plants, but only one band was detected in Gifu B-129 (Supplemental Figure 6). These results indicate that the T2 plants had multiple copies of LjMYB12. We evaluated the expression of LjMYB12 and flavonoid biosynthesis genes (CHS1, CHSs, CHI1, CHI2, F3H, F3′H, FLS, and DFR2) in leaves of 7-week-old T2 plants. The expression of LjMYB12, CHS1, F3H, and FLS was significantly higher in 12OX-10-2 T2 plants than in the control plants (Figure 4). On the other hand, the expression of the other genes was unchanged (Supplemental Figure 7). These results suggest that CHS1, F3H, and FLS are activated by LjMYB12 in L. japonicus.
Figure 4. Expression profiles of LjMYB12, CHS1, F3H, and FLS in leaves of IG-1 and 12OX-10-2 T2 plants 7 weeks after sowing. Expression levels were determined by normalizing the PCR threshold cycle number of each gene to that of β-tubulin. Data are means±SD of three biological replicates. * Significantly different at p<0.05.
MYB12 ortholog activity differed between L. japonicus and Arabidopsis. AtMYB12 activated the transcription of CHI in addition to CHS, F3H, and FLS in Arabidopsis (Mehrtens et al. 2005), but overexpressed LjMYB12 did not increase the expression of CHI paralogs in transgenic L. japonicus plants. Interestingly, LjMYB12 strongly activated CHS1 in L. japonicus, but did not activate other CHSs (Figure 4 and Supplemental Figure 7). Shimada et al. (2007) classified CHS1 into a different clade from the other CHS orthologs in L. japonicus. Since the expression of flavonoid biosynthesis genes is also regulated by other R2R3-MYB TFs in addition to the subgroup 7 (Borevitz et al. 2000; Kranz et al. 1998; Nesi et al. 2001; Stracke et al. 2001), molecular characterization of MYB TFs in legumes will reveal how gene duplication affects the regulation of flavonoid biosynthesis.
Acknowledgments
We thank T. Miyakawa and M. Suzuki for technical assistance. This work supported in part by Grants-in-Aid for Scientific Research form the Ministry of Education, Culture, Sports, Science and Technology of Japan (20580003 and 23580002).
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Akashi T, Ayabe S (2000) Flavonoids of leguminous plants: Structure, biological activity, and biosynthesis. J Plant Res 113: 475–488 [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, Hekkert BTL, Orzaez D, Fernandez-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor S1MYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Tuteja JH, Li M, Marek LF, Shoemaker RC, Vodkin LO (2004) Features of a 103-kb gene-rich region in soybean include an inverted perfect repeat cluster of CHS genes comprising the I locus. Genome 47: 819–831 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J, Debelle F, Rosenberg C (1992) Signaling and host range variation in nodulation. Annu Rev Microbiol 46: 497–531 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BSJ, Muday GK (2011) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol 156: 144–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31: 805–813 [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory C1 locus of Zea mays encode a protein with homology to MYB protooncogene product and with structural similarities to transcriptional activators. EMBO J 6: 3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder TB, Hedrick SA, Bell JN, Liang XW, Clouse SD, Lamb CJ (1987) Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris. Mol Gen Genet 210: 219–233 [DOI] [PubMed] [Google Scholar]
- Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol 131: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada N, Sasaki R, Sato S, Kaneko T, Tabata S, Aoki T, Ayabe S (2005) A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J Exp Bot 56: 2573–2585 [DOI] [PubMed] [Google Scholar]
- Shimada N, Sato S, Akashi T, Nakamura Y, Tabata S, Ayabe S, Aoki T (2007) Genome-wide analyses of the structural gene families involved in the legume-specific 5-deoxyisoflavonoid biosynthesis of Lotus japonicus. DNA Res 14: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J, Martirani L, Tuppale S, Chian RJ, Chiurazzi M, Gresshoff PM (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48: 1357–1365 [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol 188: 985–1000 [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Mori Y, Yasue K, Maruyama N, Kitamura K, Abe J (2014) Knockdown of the 7S globulin subunits shifts distribution of nitrogen sources to the residual protein fraction in transgenic soybean seeds. Plant Cell Rep 33: 1963–1976 [DOI] [PubMed] [Google Scholar]