Abstract
Lignin encrusts lignocellulose polysaccharides, and has long been considered an obstacle for the efficient use of polysaccharides during processes such as pulping and bioethanol fermentation. However, lignin is also a potential feedstock for aromatic products and is an important by-product of polysaccharide utilization. Therefore, producing biomass plant species exhibiting enhanced lignin production is an important breeding objective. Herein, we describe the development of transgenic rice plants with increased lignin content. Five Arabidopsis thaliana (Arabidopsis) and one Oryza sativa (rice) MYB transcription factor genes that were implicated to be involved in lignin biosynthesis were transformed into rice (O. sativa L. ssp. japonica cv. Nipponbare). Among them, three Arabidopsis MYBs (AtMYB55, AtMYB61, and AtMYB63) in transgenic rice T1 lines resulted in culms with lignin content about 1.5-fold higher than that of control plants. Furthermore, lignin structures in AtMYB61-overexpressing rice plants were investigated by wet-chemistry and two-dimensional nuclear magnetic resonance spectroscopy approaches. Our data suggested that heterologous expression of AtMYB61 in rice increased lignin content mainly by enriching syringyl units as well as p-coumarate and tricin moieties in the lignin polymers. We contemplate that this strategy is also applicable to lignin upregulation in large-sized grass biomass plants, such as Sorghum, switchgrass, Miscanthus and Erianthus.
Keywords: AtMYB61, lignin, p-coumarate, tricin, upregulation
Introduction
Lignocellulose biomass accounts for the highest proportion of terrestrial biomass on earth (Yoda 1982), and mainly consists of the secondary cell wall of vascular plants. It represents the most promising natural and renewable resource and, consequently, has attracted increased research interests. Future demands for lignocellulose biomass from the biomass-refinery industry will heighten the need for the methods to increase lignocellulose biomass production. Additionally, improvement in the utilization characteristics of lignocellulose biomass is also a key to boost lignocellulose utilization economy.
Lignocellulose is mainly composed of lignin and polysaccharides, i.e. cellulose and hemicelluloses. Lignin fills the spaces between cell wall polysaccharides and confers mechanical strength and imperviousness to the cell wall of vascular plants (Boerjan et al. 2003). Lignocellulose functions in planta as a structural material to support the plant body, and not as a storage material that is biologically recycled. Therefore, lignocellulose is inherently difficult to exploit by chemical and biochemical conversions. Lignin possesses several properties that interfere with the use of lignocellulosic polysaccharides in processes such as chemical pulping, forage digestion, and enzymatic hydrolysis. It has long been considered that plant materials with low lignin content or easily removed lignin may help to facilitate the polysaccharide utilization (Gressel 2008; Hisano et al. 2009; Vanholme et al. 2008). Lignin is a complex phenylpropanoid polymer that is biosynthesized via oxidative coupling of p-hydroxycinnamyl alcohols (monolignols) and related compounds formed in the cinnamate/monolignol pathway (Umezawa 2010). Numerous transgenic and mutant plants with downregulated expression of genes encoding enzymes of this pathway have been generated. These transgenic plants generally contain lower lignin content, leading to more efficient enzymatic saccharification, forage digestion, and pulping (Chen and Dixon 2007; Chiang 2006; Vanholme et al. 2008; Weng et al. 2008). We also reported transgenic rice (Oryza sativa L. ssp. japonica cv. Nipponbare) lines with decreased lignin content, which exhibited significantly enhanced enzymatic saccharification efficiency (Hattori et al. 2012; Koshiba et al. 2013a, 2013b).
On the other hand, increased lignin content in plants has long been disregarded for lignocellulose utilization even though lignin has valuable attributes related to many aspects of lignocellulose production and utility. First, lignin is a potential feedstock for various bio-based aromatic chemicals and biofuels, and has generated considerable interest as a result (Marshall and Alaimo 2010; Pu et al. 2008; Zakzeski et al. 2010). For example, aromatic components can increase the heat resistance and mechanical strength of engineered plastics, and lignin represents a potential biomass-derived source of these aromatic components (Ishii et al. 2013). Second, because lignin has higher carbon content and heating values than polysaccharides, lignin-derived components in kraft-pulping effluents are being exploited as an important by-product fuel in the pulp mills. Global production of this fuel in 1999 was equivalent to 60 billion liters of crude oil (Yokoyama and Matsumura 2008). Additionally, higher lignin content is beneficial for wood and grass fuels. Third, higher plant lignin content may be correlated with higher biomass production. Relatively large graminaceous plants, such as bamboo (Phyllostachys heterocycla) (approximately 14 m tall) and Erianthus (approximately 4.5 m tall) generally have higher lignin content (approximately 26% and 23–28%, respectively) (Higuchi 1957; Itoh 1990; Yamamura et al. 2013) than smaller graminaceous plants such as rice (O. sativa, approximately 80 cm tall with ca. 10–15% of lignin content) (Suzuki et al. 2009). This difference may be because larger plants require greater mechanical strength than smaller plants, which can be achieved with relatively high lignin content. In addition, excessive reduction in lignin content in breeding programs by cross-fertilization often results in drastic increase in the frequency of lethality (Casler et al. 2002). Therefore, higher lignin content is important for increased lignocellulose production and utility.
The objective of this study was to produce transgenic rice plants with elevated lignin content. Various MYB transcription factor genes, comprising a large family of plant transcriptional factors, have been reported to directly or indirectly regulate the expression of lignin biosynthetic genes in many plant species (Nakano et al. 2015; Ye and Zhong 2015; Yoon et al. 2015). Herein, we describe the generation of transgenic rice plants overexpressing five Arabidopsis and one rice MYB genes under the control of the Cauliflower mosaic virus (CaMV) 35S promoter to represent the concept of lignin upregulation. The overexpressions of three Arabidopsis MYBs, i.e., AtMYB55, AtMYB61, and AtMYB63, led to increased lignin content in rice culms of T1 plants. This strategy may be applicable to upregulating lignin production in large graminaceous biomass crops, such as those expected for biomass feedstock production, e.g., Sorghum, switchgrass, Miscanthus, and Erianthus.
Materials and methods
Instrumentation
UV absorbance was measured with a SH-1000 Lab Microplate Reader (Corona Electric Co., Ltd., Ibaraki, Japan). Gas chromatography-mass spectrometry (GC-MS) analysis was performed using Shimadzu GC-MS systems, a QP-5050A for thioacidolysis and a QP-2010 Ultra for nitrobenzene oxidation (Shimadzu Co., Ltd., Kyoto, Japan), respectively. The GC-MS conditions were as follows: Shimadzu HiCap CBP10-M25-025 column (25 m×0.22 mm for QP-5050A, 20 m×0.22 mm for QP-2010 Ultra); carrier gas, helium; injection temperature, 230°C; oven temperature, 40°C at t=0 to 2 min, then to 230°C at 40°C min−1; ionization, electron-impact mode (70 eV).
Nuclear magnetic resonance spectroscopy (NMR) was conducted using a Bruker Biospin Avance III 800 system (800 MHz, Bruker Biospin, Billerica, MA, USA), and the central DMSO solvent peaks (δC/δH: 39.5/2.49 ppm) were used as an internal reference. Adiabatic heteronuclear single-quantum coherence (HSQC) experiments were carried out using a standard implementation (“hsqcgcep.3”) using parameters described previously (Mansfield et al. 2012). Data processing and analysis used Bruker TopSpin 3.1 software (Bruker Biospin, Billerica, MA). HSQC plots were obtained with typical matched Gaussian apodization in F2 and squared cosine-bell apodization and one level of linear prediction (32 coefficients) in F1.
Plant transformation constructs
To overexpress AtMYB55, AtMYB61, AtMYB63, and AtMYB86, we used the previously established Agrobacterium tumefaciens EHA101 strains harboring pGWB2 vectors containing the cDNA inserts of RIKEN Arabidopsis full-length (RAFL) cDNA clones (RIKEN Bioresource Center, Tsukuba, Japan) (Ogawa et al. 2008). To overexpress AtMYB103 and OsMYB55/61, we first amplified the coding sequences (CDS) by polymerase chain reaction (PCR) using KOD Plus DNA polymerase (TOYOBO, Osaka, Japan) or pfu polymerase (Promega KK, Tokyo, Japan), gene specific primer sets, and Arabidopsis cDNAs (Noda et al. 2013) or a cDNA clone (Rice Genome Resource Center, National Institute of Agrobiological Sciences, Tsukuba, Japan) as the templates (Table 1). We also amplified a yellow fluorescent protein (YFP) gene to generate transgenic control plants using pH35YG vector as the template (Table 1). The PCR products were then subcloned into the entry vector pENTR/D-TOPO (Life Technologies Corporation, Tokyo, Japan). The accuracy of the constructs was confirmed by DNA sequencing. The rice transformation vector was produced by an LR Clonase (Life Technologies Corporation) reaction between the entry vector and the pGWB2 destination vector (Nakagawa et al. 2007).
Table 1. Templates and primers used for construction of plant transformation vectors.
| Target gene name | Template | Forward primer | Reverse primer |
|---|---|---|---|
| AtMYB103 | Arabidopsis cDNAs | 5′-CAC CAT GGG TCA TCA CTC ATG CT-3′ | 5′-AAA CGA AGA AGG GAA AGA AGA AGA TAA GGC-3′ |
| OsMYB55/61 | J013087D08 | 5′-CAC CAT GGG GAG ACA TTC CTG-3′ | 5′-GAT ATT CTC AAA AGA CAA GGA CAT CCT TTG-3′ |
| YFP | pH35YG | 5′-CACCATGGTGAGCAAGGGC-3′ | 5′-CTTGTACAGCTCGTCCATGCC-3′ |
Preparation of transgenic rice (T0 generation)
The prepared vectors were introduced into A. tumefaciens strain EHA101 using a freeze-thaw method (Holsters et al. 1978). Primary rice transformants (T0) were generated from O. sativa L. ssp. japonica cv. Nipponbare calli as previously described (Hattori et al. 2012) using the Agrobacterium strains.
Cultivation of rice plants
Transgenic rice plants (T0 generation, approximately 3 months after inoculation with Agrobacterium) were transplanted into 1/10,000a Wagner pots containing a 1 : 1 mixture of vermiculite and “Hanasaki Monogatari” garden soil (Akimoto-Tensanbutsu, Iga, Japan). The transplanted rice plants were grown to maturity at 27°C under a 12-h light/12h-dark photoperiod in a Koitotoron growth chamber (Koito Industries, Yokohama, Japan). The pot bases were constantly immersed in water (a few centimeters deep). The first-generation progeny of the transformants (T1) and wild-type (WT) plants were cultivated as previously described (Koshiba et al. 2013a).
Preparation of dried cell wall samples
Two months after transplanting, the youngest leaves (approximately 30 cm long, including the blade and sheath) were harvested from T0 rice plants and oven-dried overnight at 60°C. About 4 months after transplanted into containers filled with nutrient solution, the aerial parts of WT and T1 rice plants were harvested and dried at room temperature for 1 month. The panicles were removed from the aerial parts, and the remaining plant material was separated into leaves, leaf sheaths, and culms. The separated plant samples were individually cut into pieces with scissors and then pulverized with a TissueLyser (Qiagen, Hilden, Germany), extracted sequentially with methanol, hexane, and distilled water, and then freeze-dried as described previously (Koshiba et al. 2013a, 2013b).
Lignin analysis
Lignin quantitation by the thioglycolic acid method, microscale nitrobenzene oxidation analysis, and thioacidolysis were conducted according to the methods as previously described (Koshiba et al. 2013a, 2013b; Suzuki et al. 2009; Yamamura et al. 2010, 2012). Two-dimensional NMR (2D-NMR) analysis was conducted as previously reported (Mansfield et al. 2012). Briefly, extractive-free and dried cell wall samples were ball-milled using a planetary micro mill Pulverisette 7 (FRITSCH GmbH, Idar-Oberstein, Germany) with ZrO2 vessels containing ZrO2 ball bearings (600 rpm, 10 cycles of 10 min with 5 min interval). The obtained ball-milled cell wall samples (ca. 60 mg) were then dispersed in DMSO-d6/pyridine-d5 (4 : 1, vol/vol) and subjected for NMR measurement.
Quantitative reverse transcription polymerase chain reaction analysis
Total RNAs were extracted from flag leaves at the heading stage of rice T1 and WT rice plants. Quantitative reverse-transcription PCR (qRT-PCR) was conducted as previously described (Koshiba et al. 2013a, 2013b). Each gene was amplified using specific primers: 5′-TTG GTA TGT GGC CAT GTA ACC A-3′ and 5′-GGC CAT ATG CAT GTT GCT GA-3′ for AtMYB55, 5′-AAC ATG GTT GGT TCT GTC CTT CA-3′ and 5′-AAT CGA GGG CTT TAC GCA TAC T-3′ for AtMYB61, 5′-ACA AAC CCG ATC TGC TGG AG-3′ and 5′-TCC AAA TGT CAG GAT CTG AAT CAA-3′ for AtMYB63, respectively. A ubiquitin gene (OsUBQ5; accession no. AK061988) was amplified using specific primers 5′-ACC ACT TCG ACC GCC ACT ACT-3′ and 5′-ACG CCT AAG CCT GCT GGT T-3′ and used as an internal control. The ΔΔCt method was adopted for calculation of gene expression (User Bulletin #2, Applied Biosystems).
Phylogenetic analysis
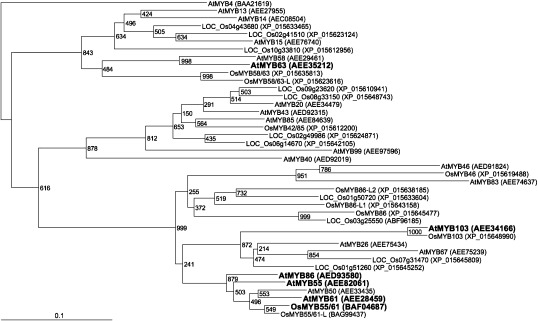
Full-length Arabidopsis and rice R2R3-MYB protein sequences (Hirano et al. 2013a; Zhao and Bartley 2014) were downloaded from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org), the Rice Annotation Project (RAP-DB, http://rapdb.dna.affrc.go.jp/), and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) databases. Multiple alignments were generated from the conserved R2R3 domain (Stracke et al. 2001) with ClustalX (Thompson et al. 1997) under default parameters. Phylogenetic trees (Figure 1) were generated from the alignments using the neighbor-joining method program in ClustalX with the following parameters: “1,000 bootstrap replicates” and “exclude position with gaps”. Then, we specified Arabidopsis AtMYB4 as an outgroup in the phylogenetic tree using NJplot (Perrière and Gouy 1996) and visualized the tree using TreeView (Page 1996).
Figure 1. Phylogenetic relationships among Arabidopsis and rice MYB proteins. The numbers at nodes represent bootstrap values obtained after 1000 replicates. AtMYB4 is specified as an outgroup. MYB proteins targeted in this study are noted in bold type. The prefixes At and Os indicate Arabidopsis and rice proteins, respectively. Note that not all MYB proteins in the Arabidopsis and rice genomes are shown. The GenBank accession numbers are given in parentheses after gene names. Rice gene names are based on Hirano et al. (2013b), Zhao and Bartley (2014), and Noda et al. (2015).
Results
Lignin upregulation by MYB genes
A number of NAC and MYB transcription factors were proved or suggested to be involved in lignification (Nakano et al. 2015; Umezawa 2010; Ye and Zhong 2015; Yoon et al. 2015). For example, AtNACs such as AtNST1, AtNST2, AtVND6, AtVND7, and AtSND1, and AtMYBs such as AtMYB46 and AtMYB83 were found to be located in upstream steps of transcriptional network for secondary cell wall formation (Nakano et al. 2015; Ye and Zhong 2015; Yoon et al. 2015). Heterologous expression of these upstream regulatory genes in rice affects biosynthesis of all cell wall components, resulting in severe modification of the cell wall formation and eventually growth of transformants. On the other hand, selective upregulation of lignin biosynthesis is probably beneficial to avoid or minimize negative effects of metabolic engineering on the growth. Overexpression of AtMYB61 (Newman et al. 2004), AtMYB85 (Zhong et al. 2008), AtMYB58 (Zhou et al. 2009), and AtMYB63 (Zhou et al. 2009) induced ectopic lignification, while AtMYB103 was reported to activate syringyl (S)-lignin biosynthesis (Öhman et al. 2013). In the present study, as the first preliminary experiment to represent the concept of lignin upregulation for biomass refinery, we selected three of these activator-type MYB genes, AtMYB61, AtMYB63, and AtMYB103. In addition, because Arabidopsis transcription factors may function differently when heterologously expressed in rice, we also included one close rice homolog of AtMYB61, OsMYB55/61 (BAF04687) (Hirano et al. 2013a) (Figure 1), and two Arabidopsis homologs of AtMYB61, namely AtMYB55 and AtMYB86 (Figure 1).
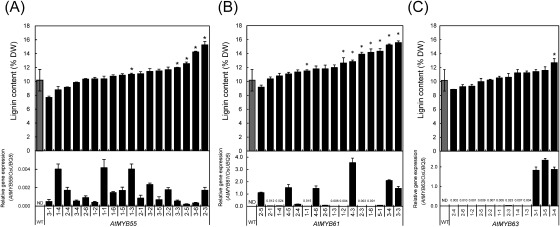
The selected genes were then overexpressed in rice plants under the control of the CaMV 35S promoter. To screen for the MYB genes that are potent for lignin upregulation in rice, we measured the lignin content of ca. 30-cm-long leaves sampled from regenerated T0 plants in the vegetative phase with a plant height of ca. 40 cm. The average lignin content in the transgenic lines is provided in Table 2. The overexpression of AtMYB55, AtMYB61, and AtMYB63 significantly (p<0.05, Table 2) increased lignin content compared with that of the control (Table 2). Therefore, the T1 generation of these transgenic lines was further subjected to more detailed analysis using the plants. The expression of AtMYB55, AtMYB61, and AtMYB63 in the flag leaves of transgenic T1 rice plants was confirmed by qRT-PCR (Figure 2). The lignin content of leaves, leaf sheaths, culms and panicles collected from mature plants was analyzed, and the data for culms are presented in Figure 2. The gene expression and the lignin content seemed inconsistent. In particular, the AtMYB55 transgene expression was as low as less than 0.004 relative to the OsUBQ5 expression. This awaits further studies to elucidate the mechanisms, but might be due to the difference of the organs. We could not obtain both data with the same organ specimens. Because lignin measurement must be done with mature tissues, in which lignification has already ceased, we used alive flag leaves for the gene expression analysis and tried to make sure that the target genes were surely expressed in the plants. The genes were driven by CaMV 35S promoter and therefore the genes should be expressed in the culms as well as the flag leaves. AtMYB55 and AtMYB61 were found to be particularly effective for lignin upregulation. Transgenic rice plants overexpressing these genes contained approximately 1.5-fold lignin than the WT controls (AtMYB55-2-3, 50% increase; AtMYB61-3-3, 53% increase; and AtMYB61-3-4, 50% increase; compared with the WT) (Figure 2). Upregulated lignin production was also observed in an AtMYB63-overexpressing line (AtMYB63-3-4, 25%), albeit at lower levels than the AtMYB55- and AtMYB61-overexpressing lines. There were no noticeable differences in the growth of transgenic lines with increased lignin content and WT lines under the cultivation conditions used in this study.
Table 2. Thioglycolic acid lignin content in ca. 30-cm-long leaves of T0 transgenic rice lines.
| Overexpressed genes | Numbers of transgenic lines | Lignin content (%) | p-Value | |
|---|---|---|---|---|
| Average | SD | |||
| YFP (control) | 10 | 5.99 | 1.00 | — |
| AtMYB55 | 16 | 7.31 | 1.37 | 0.005** |
| AtMYB61 | 14 | 7.15 | 1.81 | 0.030* |
| AtMYB63 | 15 | 8.35 | 2.16 | 0.019* |
| AtMYB86 | 9 | 5.54 | 1.06 | 0.177 |
| AtMYB103 | 24 | 6.30 | 1.13 | 0.221 |
| OsMYB55/61 | 14 | 7.09 | 2.40 | 0.072 |
The p-Value was calculated using Student’s t-test to control. SD, standard deviation; *, p<0.05; **, p<0.01 (Student’s t-test).
Figure 2. Relative gene expression rate (lower panels) and lignin content (upper panels) in the culm of wild-type (WT) and AtMYB-overexpressing rice plants. (A), AtMYB55-, (B), AtMYB61-, and (C), AtMYB63-overexpressed rice plants. Each value in the lower panels is the mean of three replicates±SD, and expressed relative to the expression of OsUBQ5. ND, Not Detected; *, Significant difference (Student’s t-test; p<0.05); DW, dry weight.
Lignin analysis of AtMYB61-overexpressing lines
We conducted a series of structural analyses using the extractive-free cell wall samples prepared from AtMYB61-overexpressing lines to obtain structural information regarding the lignins. Nitrobenzene oxidation analysis revealed the syringaldehyde/vanillin (Sa/Va) and p-hydroxybenzaldehyde/vanillin (Ha/Va) ratios were slightly higher for the AtMYB61-overexpressing lines than for the controls (Table 3). This observation suggested that upregulated lignin biosynthesis through the overexpression of AtMYB61 modified the cell wall aromatic composition in addition to increasing the lignin content. However, it is important to note that the degradation of p-coumarates and ferulates in lignin and/or hemicelluloses of grass cell walls also release the same p-hydroxybenzaldehyde derivatives as those released from monolignol-derived typical lignin moieties (Chen 1992). In addition, it is conceivable that syringaldehyde is also formed from the S aromatic ring on tricin units by nitrobenzene oxidation. Hence, we also conducted a thioacidolysis analysis to determine lignin aromatic composition associated with β-O-4 substructures in the lignin polymers. As summarized in Table 4, the syringyl/guaiacyl (S/G) ratios were higher for the three AtMYB61-overexpressing lines than for the controls, implying the abundance of S lignin increased in the cell walls of AtMYB61-overexpressing rice plants.
Table 3. Nitrobenzene oxidation analysis on AtMYB61-overexpressing rice transgenic lines.
| Line | Vanillin (Va) | Syringaldehyde (Sa) | p-Hydroxybenzaldehyde (Ha) | Vanillic acid (VA) | Syringic acid (SA) | p-Hydroxybenzoic acid (HA) | Sa/Va | Ha/Va |
|---|---|---|---|---|---|---|---|---|
| µmol g−1 cell wall residue (CWR) | ||||||||
| WT | 208.69±4.03 | 50.29±0.74 | 52.62±2.37 | 13.67±0.04 | 9.57±0.34 | 15.96±0.10 | 0.24±0.00 | 0.25±0.01 |
| AtMYB61 1–2 | 196.39±6.36* | 50.39±0.21 | 56.00±3.32 | 12.96±0.16** | 9.05±0.27 | 15.72±0.07* | 0.26±0.01* | 0.29±0.03 |
| AtMYB61 3–4 | 164.86±7.96** | 48.52±2.72 | 55.83±4.60 | 12.89±0.05** | 9.07±0.55 | 15.66±0.04** | 0.29±0.00** | 0.34±0.01** |
| AtMYB61 3–3 | 212.51±14.19 | 61.04±0.63** | 65.23±2.23** | 13.67±0.16 | 10.51±0.32* | 16.02±0.11 | 0.29±0.02 | 0.31±0.03* |
Data represent the average±SD (n=3). Asterisks indicate significant differences (Student’s t-test; p<0.05 and 0.01 for * and **, respectively). Sa/Va and Ha/Va: Molar ratios of Sa to Va and Ha to Va, respectively.
Table 4. Changes in lignin β-O-4 substructure composition in the AtMYB61-overexpressed plants.
| Line | Gt | St | Ht | S/G | H/G |
|---|---|---|---|---|---|
| % for total peak area of trithioethylpropane compounds | |||||
| WT | 58.92±0.29 | 31.38±0.39 | 9.70±0.48 | 0.53±0.01 | 0.16±0.01 |
| AtMYB61 1–2 | 57.09±0.68 | 32.56±0.66 | 10.35±0.81 | 0.57±0.01 ** | 0.18±0.02 |
| AtMYB61 3–4 | 52.10±1.12 | 37.16±2.32 | 10.74±1.69 | 0.71±0.06 ** | 0.21±0.03 * |
| AtMYB61 3–3 | 54.11±0.30 | 35.92±1.85 | 9.97±1.61 | 0.66±0.04 ** | 0.18±0.03 |
Data represent the average±SD (n=3). Asterisks indicate significant differences (Student’s t-test; p<0.05 and 0.01 for * and **, respectively. Gt, guaiacyltrithioethylpropane (m/z 269, base-ion); St, syringyltrithioethylpropane (m/z 299, base-ion); Ht, p-hydroxyphenyltrithioethylpropane (m/z 239, base-ion) formed following thioacidolysis of lignin. S/G (syringyl/guaiacyl) and H/G (p-hydroxyphenyl/guaiacyl), peak area ratios of St to Gt and Ht to Gt, respectively.
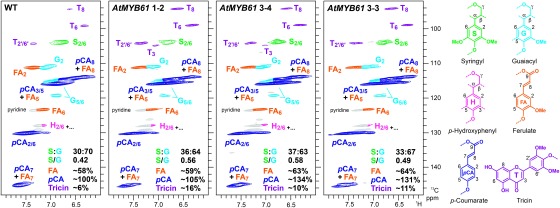
Lastly, 2D-NMR was used for a more in-depth analysis of lignin structures. All HSQC spectra for the cell wall samples of AtMYB61-overexpressing and control lines (Figure 3) displayed typical lignin aromatic signals for G and S units, as well as those from p-hydroxyphenyl (H) unit, albeit at low levels. Aromatic signals from p-coumarate, ferulate and tricin residues in lignin and/or hemicelluloses were clearly observed. A quantitative analysis of the HSQC contour signals revealed that the S/G signal ratio increased in all the three AtMYB61-overexpressing lines. The result was well consistent with the thioacidolysis data (Table 4), and further confirmed that AtMYB61 overexpression increased S lignin content. More significantly, our NMR analysis revealed that the signals from p-coumarate and tricin units were greater in the spectra of AtMYB61-overexpressing lines than in the WT spectrum. Increased p-coumarate and tricin units in the AtMYB61-overexpressing cell wall samples were in line with the increased p-hydroxybenzaldehyde and syringaldehyde yields upon nitrobenzene oxidation (Table 3). Overall, our data suggested that AtMYB61 overexpression increased lignin content in rice cell walls mainly by enriching the abundance of S lignin units as well as p-coumarate and tricin moieties.
Figure 3. Aromatic subregions of short range 1H–13C correlation (HSQC) NMR spectra of cell wall samples isolated from wild-type (WT), and AtMYB61-overexpressing rice plants. Volume integrals are given for the aromatic units that are color-coded to match their assignments in the spectrum. The percentages reported are integrals relative to total of G and S aromatic units.
Discussion
The present study demonstrated that upregulated lignin biosynthesis in rice plants was stimulated by heterologous expression of the genes encoding Arabidopsis transcription factors AtMYB55, AtMYB61, and AtMYB63. Our results indicated that these Arabidopsis MYBs were also functional in rice plants, although, to the best of our knowledge, there are no reports describing a conclusive functional analysis of AtMYB55 in A. thaliana plants. On the other hand, AtMYB61 has been known to play a pleiotropic role (Dubos et al. 2010), affecting lignification (Newman et al. 2004), mucilage production (Penfield et al. 2001) and stomatal aperture (Liang et al. 2005). It was also suggested to potentially control resource acquisition and allocation (Romano et al. 2012). We observed that the overexpression of AtMYB61 in rice plants resulted in increased production of lignin, which is similar to the findings of a previous study in which CaMV 35S-regulated overexpression of AtMYB61 in Arabidopsis plants caused ectopic lignification (Newman et al. 2004). Interestingly, however, AtMYB61 overexpression in rice increased the abundance of S lignin units and also p-coumarate and tricin moieties that are the lignin partial structures characteristic of grasses (Lan et al. 2015; Ralph 2010) (Table 3, 4 and Figure 3). This result may help characterize the transcriptional control for the biosynthesis of grass lignins.
The lignin content in T1 transgenic rice plants was up to 53% higher than that of WT plants (Figure 2), implying it may be possible to produce graminaceous biomass plants with greater lignin content and heating values. The higher heating value (HHV) is highly correlated with lignin content and is calculated for wood using the following equation:
where HHV=higher heating value of extractive-free wood (MJ kg−1) and XL=lignin content (%) (White 1987).
A 53% increase in lignin abundance in lignocellulosic biomass samples originally containing 10% and 20% lignin content gives rise to 15.3% and 30.6% lignin content, resulting in 2.5% and 4.7% increase in HHVs, respectively.
In addition to transcriptional activators, several transcriptional suppressors (Nakano et al. 2015; Xu et al. 2014; Ye and Zhong 2015; Yoon et al. 2015) are known to regulate lignin biosynthesis. Downregulation of these suppressors is likely to induce upregulation of lignin biosynthesis. Knockout of the suppressor genes may be achieved by non-transgenic technologies such as mutagenesis using chemicals or ion beam irradiation.
These strategies describe herein for lignin upregulation may be applicable to large-sized graminaceous plants such as Sorghum, Erianthus, napier grass, switchgrass, and Miscanthus. The increased lignin content in these plants may be beneficial for fuel use and aromatic feedstock production, and ultimately lead to greater biomass productivity, which will help decrease the land area required for biomass production.
In conclusion, we have generated transgenic rice lines with lignin content approximately 1.5-fold higher than that of control plants by upregulation of MYB transcription factor genes. This approach may be applicable for lignin upregulation in large graminaceous plants, which are biomass resources potentially useful for the production of fuels and aromatic industrial feedstock production.
Acknowledgments
The authors thank Prof. Tsuyoshi Nakagawa, Shimane University, for providing the pGWB2 vector, Prof. Taku Demura, Nara Institute for Science and Technology, for providing pH35YG vector, and Prof. Hironori Kaji and Ms. Ayaka Maeno, the collaborative research facility at the Institute for Chemical Research, Kyoto University, for their assistance in NMR analysis. The authors also thank Mr. Atsushi Hosouchi, Dr. Nozomu Sakurai and Dr. Koei Okazaki, Kazusa DNA Research Institute, for technical advice regarding the construction of pGWB2 vector, and Ms. Aiko Morita, Ms. Kumiko Murata, Ms. Mayumi Inutsuka, Ms. Megumi Ozaki, and Ms. Keiko Tsuchida for technical assistance. This research was partly supported by the grant “Genetic Modification of Rice Cell Wall for Efficient Saccharification” from the New Energy and Industrial Technology Development Organization (NEDO) and by the grant “Molecular Breeding of Lignocellulose with Greater Heating Values” from the Advanced Low Carbon Technology Research and Development Program of the Japan Science and Technology Agency (JST). This research was also supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (nos. 20380102 and 25292104) and by the Science and Technology Research Partnership for Sustainable Development (SATREPS), JST/the Japan International Cooperation Agency (JICA). A part of this study was conducted using the Development and Assessment of Sustainable Humanosphere/Forest Biomass Analytical System at the Research Institute for Sustainable Humanosphere, Kyoto University, Japan.
References
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Casler MD, Buxton DR, Vogel KP (2002) Genetic modification of lignin concentration affects fitness of perennial herbaceous plants. Theor Appl Genet 104: 127–131 [DOI] [PubMed] [Google Scholar]
- Chen CL (1992) Nitrobenzene and cupric oxide oxidations. In: Lin SY, Dence CW (eds) Methods in Lignin Chemistry. Springer, Berlin, pp 301–321
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nature Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chiang VL (2006) Monolignol biosynthesis and genetic engineering of lignin in trees, a review. Environ Chem Lett 4: 143–146 [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Gressel J (2008) Transgenics are imperative for biofuel crops. Plant Sci 174: 246–263 [Google Scholar]
- Hattori T, Murakami S, Mukai M, Yamada T, Hirochika H, Ike M, Tokuyasu K, Suzuki S, Sakamoto M, Umezawa T (2012) Rapid analysis of transgenic rice straw using near-infrared spectroscopy. Plant Biotechnol 29: 359–366 [Google Scholar]
- Higuchi T (1957) Biochemical studies of lignin formation. III. Physiol Plant 10: 633–648 [Google Scholar]
- Hirano K, Aya K, Morinaka Y, Nagamatsu S, Sato S, Antonio BA, Namiki N, Nagamura Y, Matsuoka M (2013a) Survey of genes involved in rice secondary cell wall formation through a co-expression network. Plant Cell Physiol 54: 1803–1821 [DOI] [PubMed] [Google Scholar]
- Hirano K, Kondo M, Aya K, Miyao A, Sato S, Antonio BA, Namiki N, Nagamura Y, Matsuoka M (2013b) Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol 54: 1791–1802 [DOI] [PubMed] [Google Scholar]
- Hisano H, Nandakumar R, Wang Z-Y (2009) Genetic modication of lignin biosynthesis for improved biofuel production. In Vitro Cell Dev Biol Plant 45: 306–313 [Google Scholar]
- Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Ishii D, Maeda H, Hayashi H, Mitani T, Shinohara N, Yoshioka K, Watanabe T (2013) Effect of polycondensation conditions on structure and thermal properties of poly(caffeic acid). In Cheng HN, Gross RA, Smith PB (eds) Green Polymer Chemistry: Biocatalysis and Materials II, ACS Symposium Series, Vol. 1144, American Chemical Society, Washington, DC, pp 237–249
- Itoh T (1990) Lignification of bamboo (Phyllostachys heterocycla Mitf.) during its growth. Holzforschung 44: 191–200 [Google Scholar]
- Koshiba T, Hirose N, Mukai M, Yamamura M, Hattori T, Suzuki S, Sakamoto M, Umezawa T (2013a) Characterization of 5-Hydroxyconiferaldehyde O-Methyltransferase in Oryza sativa. Plant Biotechnol 30: 157–167 [Google Scholar]
- Koshiba T, Murakami S, Hattori T, Mukai M, Takahashi A, Miyao A, Hirochika H, Suzuki S, Sakamoto M, Umezawa T (2013b) CAD2 deficiency causes both brown midrib and gold hull and internode phenotypes in Oryza sativa L. cv. Nipponbare. Plant Biotechnol 30: 365–373 [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y-K, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15: 1201–1206 [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Marshall A-L, Alaimo PJ (2010) Useful products from complex starting materials: Common chemicals from biomass feedstocks. Chem Eur J 16: 4970–4980 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM (2004) Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 37: 239–250 [DOI] [PubMed] [Google Scholar]
- Noda S, Koshiba T, Hattori T, Yamaguchi M, Suzuki S, Umezawa T (2015) The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta 242: 589–600 [DOI] [PubMed] [Google Scholar]
- Noda S, Takahashi Y, Tsurumaki Y, Yamamura M, Nishikubo N, Yamaguchi M, Sakurai N, Hattori T, Suzuki H, Demura T, et al. (2013) ATL54, a RING-H2 domain protein selected by a gene co-expression network analysis, is associated with secondary cell wall formation in Arabidopsis. Plant Biotechnol 30: 169–177 [Google Scholar]
- Ogawa Y, Dansako T, Yano K, Sakurai N, Suzuki S, Aoki K, Noji M, Saito K, Shibata D (2008) Efficient and high-throughput vector construction and Agrobacterium-mediated transformation of Arabidopsis thaliana suspension-cultured cells for functional genomics. Plant Cell Physiol 49: 242–250 [DOI] [PubMed] [Google Scholar]
- Öhman D, Demedts B, Kumar M, Gerber L, Gorzsás A, Goeminne G, Hedenström M, Ellis B, Boerjan W, Sundberg B (2013) MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73: 63–76 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M (1996) WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Pu Y, Zhang D, Singh PM, Ragauskas AJ (2008) The new forestry biofuels sector. Biofuels Bioprod Bioref 2: 58–73 [Google Scholar]
- Ralph J (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang K-Y, Li E, Douglas CJ, Western TL, et al. (2012) AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol 195: 774–786 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Suzuki Y, Yamamoto N, Hattori T, Sakamoto M, Umezawa T (2009) High-throughput determination of thioglycolic acid lignin from rice. Plant Biotechnol 26: 337–340 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T (2010) The cinnamate/monolignol pathway. Phytochem Rev 9: 1–17 [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Weng J-K, Li X, Bonawitz ND, Chapple C (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol 19: 166–172 [DOI] [PubMed] [Google Scholar]
- White RH (1987) Effect of lignin content and extractives on the higher heating value of wood. Wood Fiber Sci 19: 446–452 [Google Scholar]
- Xu Q, Yin X, Zeng J, Ge H, Song M, Xu C, Li X, Ferguson IB, Chen K (2014) Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J Exp Bot 65: 4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura M, Hattori T, Suzuki S, Shibata D, Umezawa T (2010) Microscale alkaline nitrobenzene oxidation method for high-throughput determination of lignin aromatic components. Plant Biotechnol 27: 305–310 [Google Scholar]
- Yamamura M, Hattori T, Suzuki S, Shibata D, Umezawa T (2012) Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol 29: 419–423 [Google Scholar]
- Yamamura M, Noda S, Hattori T, Shino A, Kikuchi J, Takabe K, Tagane S, Gau M, Uwatoko N, Mii M, et al. (2013) Characterization of lignocellulose of Erianthus arundinaceus in relation to enzymatic saccharification efficiency. Plant Biotechnol 30: 25–35 [Google Scholar]
- Ye ZH, Zhong R (2015) Molecular control of wood formation in trees. J Exp Bot 66: 4119–4131 [DOI] [PubMed] [Google Scholar]
- Yoda K (1982) Effects of terrestrial ecosystems on the carbon dioxide concentration in the atmosphere. Chikyukagaku (Geochemistry) 16: 78–85 [Google Scholar]
- Yokoyama S, Matsumura Y (eds) (2008) The Asian Biomass Handbook: A Guide for Biomass Production and Utilization. http://www.jie.or.jp/biomass/AsiaBiomassHandbook/English/All_E-080917.pdf (accessed on July 4, 2016)
- Yoon J, Choi H, An G (2015) Roles of lignin biosynthesis and regulatory genes in plant development. J Integr Plant Biol 57: 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakzeski J, Bruijnincx PCA, Jongerius AL, Weckhuysen BM (2010) The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110: 3552–3599 [DOI] [PubMed] [Google Scholar]
- Zhao K, Bartley LE (2014) Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol 14: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye Z-H (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]