Abstract
An increase in plant biomass production is desired to reduce emission of carbon dioxide emissions and arrest global climate change because it will provide a more source of energy production than fossil fuels. Recently, we found that forced expression of the rice 45S rRNA gene increased aboveground growth by ca. 2-fold in the transgenic Arabidopsis plants. Here, we created transgenic tobacco plants harboring the rice 45S rRNA driven by the maize ubiquitin promoter (UbiP::Os45SrRNA) or cauliflower mosaic virus 35S promoter (35SP::Os45SrRNA). In 35SP::Os45SrRNA and UbiP::Os45SrRNA transgenic tobacco plants, the leaf length and size were increased compared with control plants, leading to an increase of aboveground growth (dry weight) up to 2-fold at the early stage of seedling development. Conversely, leaf physiological traits, such as photosynthetic capacity, stomatal characteristics, and chlorophylls and RuBisCO protein contents, were similar between the transgenic and control plants. Flow cytometry analysis indicated that the transgenic plants had enhanced cell-proliferation especially in seedling root and leaf primordia. Microarray analysis revealed that genes encoding transcription factors, such as GIGANTEA-like, were more than 2-fold up-regulated in the transgenic plants. Although the mechanism underlying the increased growth has yet to be elucidated, this strategy could be used to increase biomass production in cereals, vegetables, and bio-energy plants.
Keywords: 45S ribosomal RNA, cell proliferation, growth increase, photosynthetic properties, transgenic plants
Introduction
We recently reported that the growth of transgenic Arabidopsis plant was increased by forced expression of the rice 45S rRNA gene (Makabe et al. 2016). The transgenic Arabidopsis plants showed ca. 2-fold increased growth compared with control plants without showing any difference in the size and ploidy level of the leaf cells. Although the mechanism underlying this growth increase is unclear, the results showed that forced expression of the rice, not Arabidopsis, full-length 45S rRNA gene was required to increase the growth of transgenic Arabidopsis. To confirm that this phenomenon was reproducible in another plant, we produced transgenic tobacco plants harboring the rice 45S rRNA gene under the control of the maize ubiquitin promoter (UbiP::Os45SrRNA) and cauliflower mosaic virus (CaMV) 35S promoter (35SP::Os45SrRNA).
Growth increases in organisms can be caused by hybrid vigor (Darwin 1876; Hochholdinger and Hoecker 2007; Lippman and Zamir 2007; Meyer et al. 2004). The hybrid vigor appears only at the early stage of plant development through the enhancement of cell-proliferation, which is probably mediated by circadian rhythms (Chen 2010; Fujimoto et al. 2012; Ni et al. 2009). Polyploidization can also increases plant biomass through the enlargement of cell size (Kondrosi et al. 2000; Sugimoto-Shirasu and Robert 2003). Generally, hybrid vigor and polyploidization cause 1.2–1.5-fold increases in the biomass compared with the original plants (Duvick 1999).
Plant growth is regarded as the product of cell number and cell size if sufficient organic materials are supplied by the photosynthesis. Therefore, photosynthetic capacity is one of the determinants of plant growth. The photosynthetic capacity is mediated by various factors, such as the integrity of the photosynthetic machinery, leaf morphology, and environmental stresses (Saibo et al. 2009). Recently, the physiological state of photosynthesis in intact leaves has been analyzed using pulse amplitude modulation (PAM) (Woo et al. 2008). The PAM analysis can measure chlorophyll fluorescence to estimate a wide range of photosynthetic parameters, such as the photosynthesis rate, non-photochemical quenching (NPQ), stomatal conductance, and the electron transport rate. This method can also generate a two-dimensional image of the photosynthetic rate by scanning chlorophyll fluorescence from photosystem II (φPSII). Simultaneously, because the gas exchange of carbon dioxide, water vapor, and oxygen through stomata significantly affects on the photosynthetic capacity, plants optimize their gas exchange efficiency by regulating stomatal aperture size, stomatal density, stomatal pore openness, and stomatal distribution patterns, all of which affect stomatal conductance (Scheidegger et al. 2000).
Leaf development in plants is an important factor that affects the photosynthetic capacity (Tsukaya 2006). To increase the photosynthetic capacity, plants have to enlarge their leaf-area to the widest extent possible to capture the sun’s energy. In leaf development, cell division first occurs in the primordium and then cell expansion follows to achieve the final leaf size. To date, much knowledge has been accumulated on the genetic regulation of cell-proliferation and cell-expansion in leaves. However, the detailed mechanism underlying the determination of final leaf size has yet to be elucidated. Several Arabidopsis mutants for ribosomal protein genes have shown developmental changes in leaf size (Byrne 2009; Fujikura et al. 2009; Horiguchi et al. 2011; Horiguchi et al. 2012; Ito et al. 2000; Rosado et al. 2012; Van Lijsebettens et al. 1994; Zsogon et al. 2014). Thus, some ribosome-related process might be involved in the co-ordination of cell-proliferation and cell-expansion in the leaf development.
Makabe et al (2016) found that the forced expression of the rice 45S rRNA gene caused a growth increase in transgenic Arabidopsis. The eukaryotic 45S rRNA gene, consisting of the 18S, 5.8S, and 28S rRNAs, is transcribed as a single transcription unit and post-transcriptionally processed into three rRNA molecules (Appels and Dvorak 1982). As the three rRNA sequences within the 45S rRNA transcripts are highly conserved between rice and Arabidopsis, the expression of the two internal transcribed spacers (ITSs), ITS1 between the 18S and 5.8S rRNAs and ITS2 between the 5.8S and 28S rRNAs, might be involved in the growth increase in the transgenic Arabidopsis.
In this study, we produced transgenic tobacco plants with forced expression of the rice 45S rRNA gene and analyzed their leaf photosynthetic and morphological traits in detail.
Materials and methods
Plant material
Plantlets of Nicotiana tabaccum ‘Petit Havana’ SR-1 line were maintained in culture bottles under sterile condition and used for the production of transgenic plants.
Production of transgenic tobacco plants
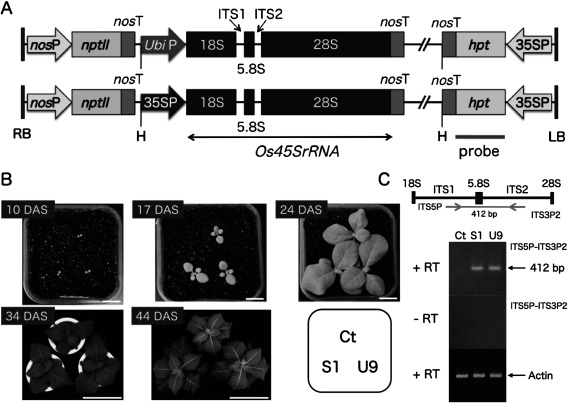
Full-length of 45S rRNA gene (DDBJ Accession No. LC086814) in Oryza sativa ssp. Indica N16 line was amplified (Makabe et al. (2016). The Os45SrRNA fragment (5.8 kb) was ligated between maize ubiquitin promoter (1.0 kb without the first intron) (Christensen and Quail 1996) and nopaline synthase gene terminator (nosT) or between CaMV 35S promoter and nosT to construct UbiP::Os45SrRNA or 35SP::Os45SrRNA chimeric gene, respectively (Makabe et al. 2016). These chimeric genes were inserted into a binary vector pEKH (Takesawa et al. 2002) at HindIII site between kanamycin and hygromycin resistance cassettes (Figure 1B). The binary vector was mobilized to Agrobacterium tumefaciens EHA101 by freeze-thaw method and transformation of tobacco plant was done by leaf-disc method (Horsch et al. 1985). Transgenic plants were selected on 50 mg L−1 kanamycin-containing MS media (Murashige and Skoog 1962).
Figure 1. Production of transgenic tobacco plants with the forced expression of the rice 45S rRNA gene using the maize ubiquitin or CaMV 35S promoter. (A) Schematic representation of transgenes; Full-length 45S rRNA gene (Os45SrRNA, 5.8 kb) of Oryza sativa ecotype Indica cultivar N16 line was linked to the maize ubiquitin promoter (UbiP) or the CaMV 35S promoter (35SP). The chimeric gene was inserted into HindIII (H) site between kanamycin (nosP-nptII-nosT) and hygromycin (35SP-hpt-nosT) resistance cassettes of binary vector pEKH to construct pEKH UbiP::Os45SrRNA or 35SP::Os45SrRNA (Makabe et al. 2016). PCR product of hpt gene was used as probe for Southern blot analysis. (B) Comparison of growth between transgenic (S1, U9) and control (Ct) plants in a growth chamber. Transgenic S1 and U9 plants having a single-copy of transgene were selected by Southern blot analysis for hygromycin resistance gene and segregation analysis for kanamycin resistance gene (Supplementary Figure S1). Photographs were taken at 10, 17, 24, 34, and 44 DAS. 10, 17 and 24 DAS (bar=2 cm), 34 DAS (bar=20 cm), 44 DAS (bar=40 cm). (C) Semi-quantitative RT-PCR was performed to detect the expression of ITS regions within rice 45S rRNA transcripts in transgenic S1 and U9 plants. A pair of primers, ITS5P and ITS3P2, was designed based on the rice ITS sequences. S1 and U9 plants showed similar amount of PCR product, which was absent in Ct plant and without adding reverse transcriptase (−RT). Actin mRNA was also amplified as an internal standard.
Southern blot analysis
Leaf samples were frozen using liquid nitrogen and crushed into fine powder using a Multi-beads Shocker (Yasui Kikai, Kyoto, Japan). Genomic DNA was extracted from 100 mg of leaf tissues using the modified CTAB method (Doyle and Doyle 1987). HindIII-digested genomic DNA (5 µg) was separated through 0.9% agarose gel, blotted to Immobilon-Ny+ membrane (Millipore Corporation, USA), and hybridized with a digoxigenin-labeled probe of hygromycin phosphotransferase (hpt) gene according to the supplier’s instructions (Roche Diagnostics, Mannheim, Germany). Hybridization with the DIG-labeled hpt probe was carried out at 39°C for 16 h. The membrane was treated with anti-DIG alkaline phosphatase and substrate CPD-star (Roche Diagnostics, Mannheim, Germany). Then, the membrane was exposed to Hyperfilm TM-MP X-ray film for 30 min at room temperature.
RT-PCR analysis
Total RNAs were extracted from leaves (100 mg) of transgenic and non-transformation plants using Plant RNA Reagent (Life Technologies, USA). First strand cDNA were synthesized from 1 µg of total RNA in a 20 µl reaction volume using Superscript Transcriptase III (Life Technologies, USA) with oligo dT (20) primer. Two pairs of RT-PCR primers, ITS5P: 5′-CGC GAT ACC ACG AGC T AAA TCC AC-3′-ITS3P2: 5′-GTC CGA GGC GTT CGC TCT CGG TGC-3′ and actin5P: 5′-GAA A ATG GTG AAG GCT GGT TTT G-3′-actin3P: 5′-AGG ATT GAT CCT CCG ATC CAG A-3′ were designed to amplify ITS (ITS1-5.8S-ITS2) region of rice 45S rRNA and actin mRNA (positive control), respectively.
Plant growth analysis in a growth chamber
Transgenic (S1, U9) and control (Ct) plants were grown in an environmentally controlled growth chamber (Yamori et al. 2011). The chamber for all the plants was operated with a day/night temperature of 25/20°C, a PPFD of 500 µmolm−2 s−1, a 12h photoperiod and a CO2 concentration of 400 µmol mol−1. Plants were grown in garden mix containing approximately 2 gL−1 of a slow-release fertilizer (Osmocote, Scotts Australia, Castle Hill, Australia). Plant growth analysis was performed at 24, 34, 44, and 50 DAS.
Gas-exchange and chlorophyll fluorescence measurements
CO2 gas exchange and chlorophyll a fluorescence was measured in fully expanded leaves of the transgenic (S1, U9) and control (Ct) plants with a portable gas exchange system (LI-6400, LI-6400-40 leaf chamber fluorometer, LI-COR) (Yamori et al. 2011). The light response of CO2 assimilation rate was measured at a CO2 concentration of 400 µmol mol−1 and 25°C and 65% relative humidity. Non-photochemical quenching (NPQ) was calculated as NPQ=(Fm-Fm′)/Fm′. The quantum yield of photosystem II (φPSII) was calculated as φPSII=(Fm′-F′)/Fm′, and the electron transport rate (ETR) was calculated as ETR=0.5×absI×φPSII, where 0.5 is the fraction of absorbed light reaching PSII and absI is absorbed irradiance taken as 0.84 of incident irradiance. Data represent means±SE
Chlorophyll fluorescence imaging
Chlorophyll fluorescence images were taken in plants of 10-, 17- and 24-DAS with a chlorophyll fluorescence imaging system (IMAGING-PAM, Walz, Effeltrich, Germany) (Yamori et al. 2011). Leaves were dark adapted for 20 min prior to determination of chlorophyll fluorescence. Then, plants were placed at 500 µmol photons m−2s−1, which is similar to the growth light condition, for 20 min. The quantum yield of photosystem II [φPSII=(Fm′-F′)/Fm′], photochemical quenching [qP=(Fm′-F′)/(Fm′-Fo′)], non-photochemical quenching [NPQ=(Fm-Fm′)/Fm′], and the fraction of PSII centers in the open state (with QA oxidized) [qL=qP×(Fo′/F′)] were calculated using the software ImagingWin. Data represent means±SE
Analysis of photosynthetic activity
Seeds of transgenic (S1, U9) and control (Ct) lines were sown in small pots within an environmental controlled growth chamber as described (Yamori et al. 2011). Leaves were exposed to strong light at 2,000 µmol photons m−2s–1 at the corresponding temperature for 90 min. The fraction of active PSII (Fv/Fm) was measured after dark incubation for 30 min. Data represent means±SE, n=5.
Quantifications of photosynthetic components
Immediately after the measurements of gas exchange, leaf samples were taken, immersed in liquid nitrogen and stored at −80°C until determinations of chlorophyll and RuBisCO. Contents of leaf chlorophyll and RuBisCO were quantified according to Yamori et al. (2011). Data represent means±SE, n=5.
Flow cytometric analyses
Relative DNA content per nuclei of somatic cells in cotyledon, root and hypocotyl of seedling at 7, 8, 10, 14, 20 days after sowing (DAS) were measured in triplicate using the laser flow cytometer PAS CA-IV (Partec GmbH, Germany) (Mishiba and Mii 2000). Cell division activity was indicated as the 4C/2C ratio. Mature leaf cells were also analyzed to check polysomaty at 30 DAS. Data represent means±SE
Microarray analysis
Total RNA was isolated from aerial parts of transgenic (S1, U9) and control (Ct) seedlings (12 DAS) using RNeasy Plant Mini Kit (Qiagen, Valencia, CA). We entrusted microarray analysis to DNA Chip Research Institute (Yokohama, Japan) using Agilent tobacco oligo-DNA microarray (Agilent Technologies, Palo Alto, CA).
Results
Production of transgenic tobacco plants expressing the rice 45S rRNA gene
We produced 14 and seven transgenic tobacco plants harboring the UbiP::Os45SrRNA and 35SP::Os45SrRNA transgenes (Figure 1A), respectively. Transgenic lines harboring a single-copy transgene were selected by Southern blot analysis (Supplementary Figure S1) and the homozygous lines for the transgene were obtained by genetic analysis for kanamycin resistance (data not shown).
Comparison of the initial growth of transgenic tobacco in a growth chamber
Seedlings of T2 transgenic lines, which were homozygous for the UbiP::Os45SrRNA (U9) and 35SP::Os45SrRNA (S1) transgenes, and control (Ct) plants were grown together in small pots within a growth chamber. Under these conditions, the transgenic S1 and U9 plants showed similar growth to the control (Ct) plants at 10 DAS (Figure 1B). In contrast, the aboveground growth of the two transgenic S1 and U9 plants was greatly increased compared with the control plants at 17 DAS and later. Semi-quantitative RT-PCR showed that the two transgenic (S1, U9) plants expressed the ITS region of the rice 45S rRNA transcripts at similar and low levels when compared with the actin gene used as an amplification standard (Figure 1C).
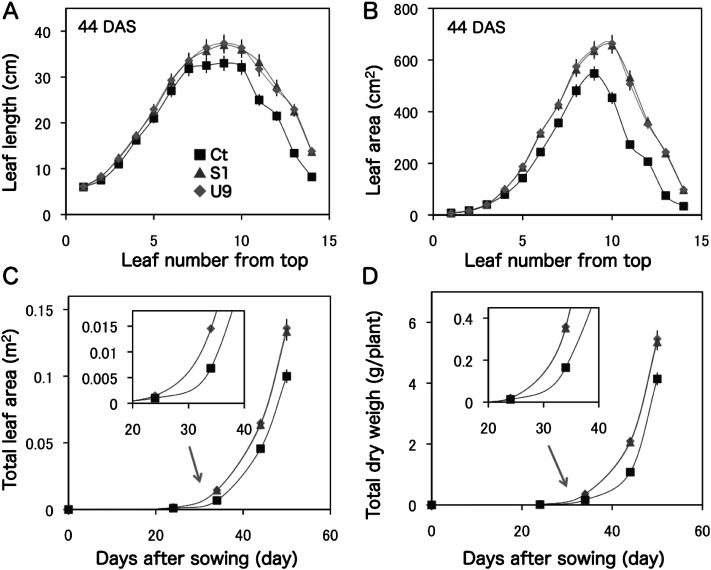
Detailed measurements at 44 DAS showed that leaf-length and leaf-area of the 1st to 3rd leaves from the top were similar between transgenic (S1, U9) and control (Ct) plants, but started to increase from the 4th leaf in the transgenic plants compared with those of the control plants (Figure 2A, B). The total leaf-area and dry-weight of transgenic S1 and U9 plants were maximized up to 2-fold compared with the control plant at 34 DAS (Figure 2C, D). The values of U9 plants were a bit higher than those of S1 plants. Later, these growth differences were reduced to ca. 1.4-fold at 50 DAS in this experiment.
Figure 2. Comparison of aboveground growth between transgenic and control plants. Transgenic (S1, U9) and control (Ct) plants were grown in small pots within a growth chamber and the leaf characters were measured at 44 DAS. (A) Length (cm) of each leaf. (B) Area (cm2) of each leaf. (C) Time-course of total leaf area (cm2). (D) Time-course of total dry weight (g/plant). Transgenic S1 and U9 plants showed similar growth patterns in this condition.
Comparison of plant growth between transgenic and control plants
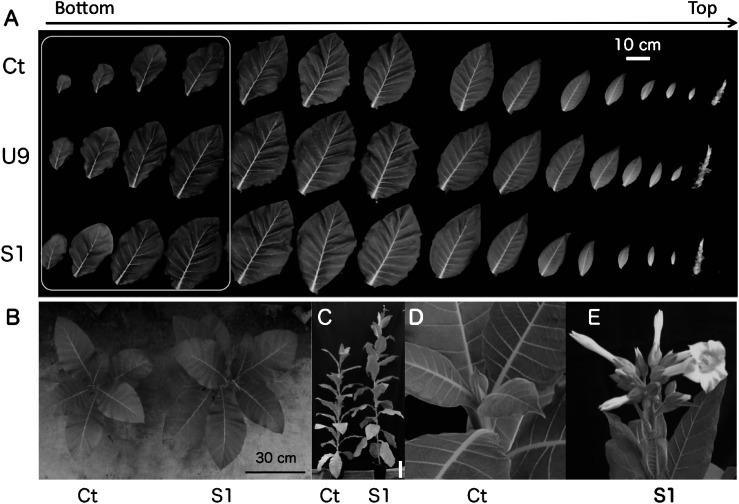
As shown in Figure 3A, transgenic (S1, U9) and control (Ct) plants were grown in growth chamber condition and their leaves were aligned from the bottom to the top at 44 DAS. Although clear growth differences were observed in the first four leaves, these differences became smaller in the later leaves probably because of the limited fertilizer in the small pots. U9 plants increased in size compared with S1 plants when they were planted in a large tray within a growth chamber (Supplementary Figure S2). In the greenhouse conditions, the transgenic S1 and U9 plants showed 1.4 and 2.1-fold increase in the dry weight of aerial tissues, respectively, compared with the Ct plant at 45 DAS (Makabe et al. submitted). The S1 plants grew bigger than the Ct plants at 75 DAS in a greenhouse (Figure 3B). The S1 plants produced their first flowers at 107 DAS (Figure 3C–E). This was much earlier than the flowering of the Ct plants at 152 DAS. Therefore, the fruit number of the S1 plants was ca. 2.8-fold higher than that of the Ct plants. In addition, the 500-seed weights of the S1 and U9 plants were 1.1–1.2-fold greater than that of the Ct plants. In addition, although the nicotine concentration in the leaves, a secondary metabolite of tobacco, was similar between the transgenic and control plants, total nicotine production was increased by 1.4- and 2.1-fold in the transgenic S1 and U9 leaves, respectively, compared with the Ct leaves.
Figure 3. Comparison of growth between transgenic and control plants. (A) Leaves of transgenic (S1, U9) and control Ct plants were aligned from the bottom to the top. The first four leaves of transgenic plants were clearly bigger than those of the control plant (box). (B) Photograph of transgenic S1 and control Ct plants at 75 DAS. (C) Photograph of transgenic S1 and control Ct whole plants at 107 DAS, bar=20 cm. At that time, Ct plant did not have any flower bud (D) while S1 plant started flowering (E).
Comparison of various photosynthetic parameters
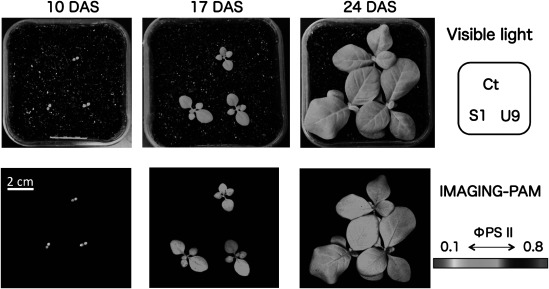
Chlorophyll fluorescence images were analyzed using seedlings at 10-, 17-, and 24-DAS under the growth light intensity. The IMAGING-PAM analysis showed that the fluorescence changed from orange to pale-blue, indicating low to high level of photosynthetic capacities. There were no differences between transgenic (S1, U9) and control (Ct) plants in the two-dimensional fluorescence images (Figure 4) and in the photosynthetic parameters, such as the quantum yield of photosystem II PSII (φPSII), the reduction state of PSII (1-qL), and the non photosynthetic quenching (NPQ) at each DAS (Supplementary Table S1).
Figure 4. Comparison of photosynthetic capacity between transgenic and control plants using IMAGING-PAM analysis. Seedlings of transgenic (S1, U9) and control (Ct) plants were grown together in the same pot and images were taken at 10, 17, and 24 DAS. Top: images under visible light. Bottom: Two-dimensional images by the IMAGING-PAM indicated low (orange at 10 DAS) to high (light blue at 24 DAS) photosynthetic capacity corresponding to φPSII indicator bar.
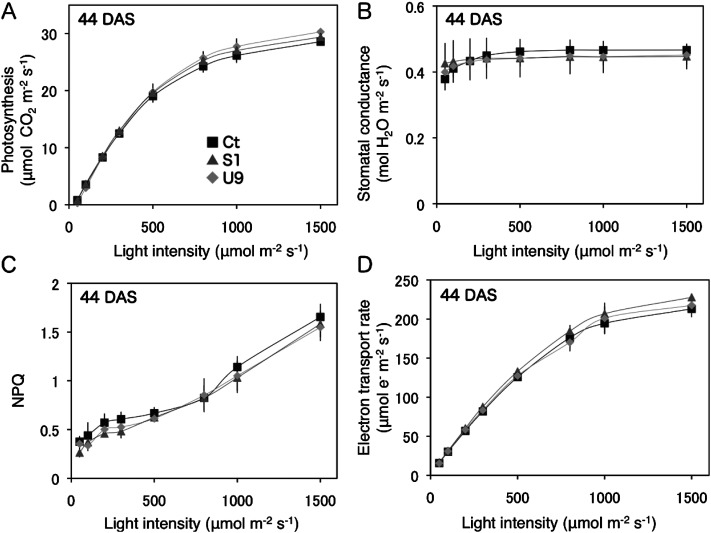
The light-intensity responses of several photosynthetic parameters in leaves were measured at 44 DAS (Figure 5). The photosynthetic rate, stomatal conductance, electron transport rate, and non-photosynthetic quenching (NPQ) at a CO2 concentration of 400 µm mol−1 were all similar between the transgenic and control plants. These data indicate that the transgenic (S1, U9) and control (Ct) plants had the same photosynthetic capacity per unit leaf area.
Figure 5. Comparison of photosynthetic parameters between transgenic and control plants. Four different photosynthetic parameters of transgenic (S1, U9) and control (Ct) leaves were measured under various intensities of light at 44 DAS. (A) Photosynthesis rate (µmol CO2 m−2s−1), (B) Stomatal conductance (µmol H2O m−2s−1), (C) Non photosynthetic quenching (NPQ), (D) Electron transport rate (µmol e−m−2s−1).
Comparison of photosynthetic components
The contents of RuBisCO and chlorophyll per unit leaf area were similar between the transgenic (S1, U9) and control (Ct) plants at 44 DAS (Table 1). All stomatal characteristics, including stomatal size, density and index, were also substantially similar between the transgenic and control plants.
Table 1. Measurements of RuBisCO and chlorophyll contents, and stomatal characteristics in leaves. Transgenic (S1, U9) and control (Ct) leaves at 44 DAS were subjected to measure stomatal characteristics and content of RuBisCO and chlorophyll according to Yamori et al. (2011).
| RuBisCO (g m−2) | Chlorophyll (g m−2) | Stomatal density (mm−2) | Stomatal index | Stomatal length (µm) | Stomatal width (µm) | |
|---|---|---|---|---|---|---|
| Ct | 1.17±0.07 | 0.366±0.015 | 270±12.8 | 0.278±0.009 | 24.7±0.4 | 15.5±0.3 |
| S1 | 1.20±0.06 | 0.370±0.018 | 274±12.5 | 0.254±0.028 | 24.7±0.5 | 15.0±0.4 |
| U9 | 1.24±0.04 | 0.373±0.013 | 278±10.5 | 0.263±0.007 | 24.4±0.6 | 15.1±0.3 |
There were no significant differences between transgenic (S1, U9) and control (Ct) plants. Data represent mean plus standard errors. n=5.
Cell division activities of tobacco seedling
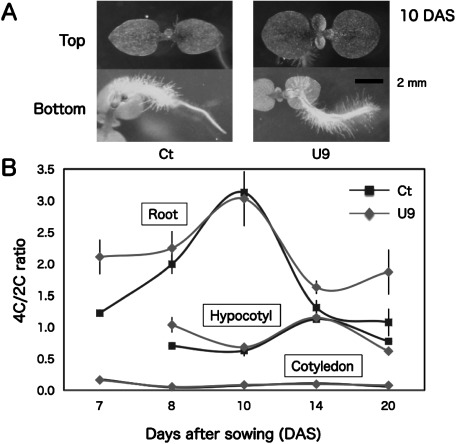
Transgenic U9 seedlings developed true leaves and root hairs earlier than control Ct seedling at 10 DAS (Figure 6A) and their secondary roots started to grow at 14 DAS (data not shown). Flow cytometry analysis showed that cell-division activity, i.e. relative DNA content per nuclei of 4C (G2/M phase) vs. 2C (G1 phase), maximized in roots (10 DAS) and hypocotyls (14 DAS) in transgenic U9 and control Ct seedlings (Figure 6B, raw data was shown in Supplementary Table S2). Because it was difficult to excise hypocotyls from roots at 7 DAS, these tissues were analyzed together as roots. In contrast, cotyledon cells had very low 4C/2C ratios in both seedlings. Transgenic U9 seedlings showed higher 4C/2C ratios in their roots (7 DAS and 14–20 DAS) and hypocotyls (8 DAS) than Ct seedlings.
Figure 6. Comparison of phenotype and relative DNA content per nuclei of somatic cells between transgenic and control plants. (A) Photos of transgenic U9 and control Ct seedlings were taken from top and bottom view at 10 DAS, (B) relative DNA content per nuclei of somatic cells in detached cotyledons, hypocotyls, and roots of U9 and Ct seedlings were measured during 7–20 DAS. The 4C (G2, M phase)/2C (G1 phase) ratio probably corresponded to the activity of cell division.
Microarray analysis
Microarray analysis of aerial parts of 12 DAS seedlings revealed that 37 and 45 genes were more than 2-fold up- or down-regulated, respectively, in both transgenic S1 and U9 plants compared with Ct plants (Supplementary Tables S3, S4). Of the 37 up-regulated genes, most genes encoded functional and structural proteins, such as 5-epi-aristolochene synthase, P-rich protein NtEIG-C29, and glutathione S-transferase. Genes involved in transcription, translation, and signal transduction are listed in Table 2. These 23 genes encoded a transformer SR ribonucleoprotein, 60S ribosomal protein L30-like, blue light photoreceptor PHR2, translation initiation factor 5A1, MOB kinase activator like 1, two receptor kinases, four transcription factors; 2 GIGANTEA-like, homeobox leucine-zipper HAT7-like, Lateral Organ Boundaries (LOB) domain-containing protein 41-like, and 12 auxin repressed protein (ARP)-like proteins. Of 45 down-regulated genes, eight genes encoding two mitochondrial 39S ribosomal protein L41A-like proteins, a splicing specificity factor, three signal transduction related proteins, and two transcription factors (TGA10 and LEUNIG-like corepressor).
Table 2. More than 2-fold up- or down-regulated genes in both S1 and U9 transgenic plants.
| Probe Name | Description | Fold change | |
|---|---|---|---|
| S1 vs Ct | U9 vs Ct | ||
| A_95_P005211 | Transformer-SR ribonucleoprotein | 5.67 | 5.82 |
| A_95_P225937 | Blue-light photoreceptor PHR2 (LOC104104471 | 4.54 | 6.29 |
| A_95_P093968 | 60S ribosomal protein L30-like (LOC104229770) | 4.38 | 4.29 |
| A_95_P091298 | Eukaryotic translation initiation factor 5A-1 (LOC104242538) | 4.64 | 2.77 |
| A_95_P258451 | LOB domain-containing protein 41-like (LOC104239409) | 4.22 | 2.72 |
| A_95_P297428 | GIGANTEA-like (LOC104104191) | 2.45 | 3.97 |
| A_95_P108877 | Auxin-repressed protein (ARP1)-like | 3.17 | 3.20 |
| A_95_P114717 | Auxin-repressed protein (ARP1)-like | 2.92 | 3.08 |
| A_95_P105487 | Auxin-repressed protein (ARP1)-like | 2.58 | 3.38 |
| A_95_P163447 | Membrane located receptor kinase-like protein (NtC7) | 2.04 | 3.88 |
| A_95_P105232 | Auxin-repressed protein (ARP1)-like | 2.67 | 3.24 |
| A_95_P176997 | Auxin-repressed protein (ARP1)-like | 2.48 | 3.37 |
| A_95_P110457 | Auxin-repressed protein (ARP1)-like | 2.71 | 2.87 |
| A_95_P106487 | Auxin-repressed protein (ARP1)-like | 2.69 | 2.87 |
| A_95_P114372 | Auxin-repressed protein (ARP1)-like | 2.71 | 2.74 |
| A_95_P177002 | Auxin-repressed protein (ARP1)-like | 2.59 | 2.81 |
| A_95_P310088 | G-type lectin S-receptor-like serine/threonine-protein kinase | 2.63 | 2.71 |
| A_95_P106782 | Auxin-repressed protein (ARP1)-like | 2.42 | 2.84 |
| A_95_P107032 | Auxin-repressed protein (ARP1)-like | 2.16 | 2.67 |
| A_95_P092983 | Homeobox-leucine zipper protein HAT7-like (LOC104232387) | 2.46 | 2.32 |
| A_95_P003171 | Auxin-repressed protein (ARP1)-like | 2.06 | 2.63 |
| A_95_P094463 | MOB kinase activator-like 1 (LOC104233287) | 2.18 | 2.29 |
| A_95_P025081 | GIGANTEA-like (LOC104222517) | 2.04 | 2.07 |
| A_95_P125607 | Splicing specificity factor subunit 3-I-like (LOC104216883) | −25.23 | −2.05 |
| A_95_P233824 | Mitochondrial 39S ribosomal protein L41A-like (LOC104224887) | −4.26 | −4.39 |
| A_95_P014791 | Mitochondrial 39S ribosomal protein L41A-like (LOC104224887) | −4.19 | −4.34 |
| A_95_P131377 | T-complex protein 1 subunit eta (LOC104236456) | −5.97 | −2.54 |
| A_95_P239499 | Putative GEM-like protein 8 (LOC104228486) | −3.35 | −2.44 |
| A_95_P299943 | Putative virus-specific-signaling-pathway regulated protein | −2.10 | −3.12 |
| A_95_P034838 | TGA10 transcription factor | −2.39 | −2.62 |
| A_95_P065840 | Transcriptional corepressor LEUNIG-like (LOC104246300) | −2.07 | −2.13 |
Probe name: based on Agilent tobacco oligo-DNA microarray
Discussion
Transgenic tobacco seedlings harboring a single-copy of the 35SP::Os45SrRNA (S1) or UbiP::Os45SrRNA (U9) transgene (Figure 1A) showed increased growth at 17 DAS compared with the control (Ct) plants in growth chamber conditions (Figure 1B). As shown in Figures 2A and 2B, the enlargement of transgenic (S1, U9) leaves was started at the 4–5th leaf from the top. At this leaf stage, because cell expansion becomes more prominent than cell proliferation, the proliferation of leaf cells might have occurred in the transgenic seedlings at an earlier stage. Although the total leaf-area and dry-weight in S1 and U9 plants were increased by up to 2-fold compared with those of the Ct plants at 34 DAS, the differences in the growth was reduced to ca. 1.4-fold at 50 DAS (Figure 2C, D). This growth retardation during the late stage was considered to be due to fertilizer deficiency and/or limited growth of the root system because the plants were grown in small pots within a growth chamber. In fact, U9 plants seemed to grow bigger than S1 plants when they were grown in a larger tray (Supplementary Figure S2).
In the greenhouse, transgenic tobacco plants reached the flowering stage much earlier than control plants (Figure 3C–E). Therefore, it is possible to produce a 2–3-fold seed yield increase in the transgenic tobacco plants because the fruit number in S1 plants was increased by 2.8-fold compared with Ct plants. Although the 500-seed weight of the U9 plants was 17% heavier than the Ct plants, this difference cannot account for the 2-fold growth increase of the transgenic seedlings. In addition, transgenic (S1, U9) leaves had a 14% higher nicotine content than Ct leaves at 30 DAS. As older leaves have a higher nicotine content than younger leaves in tobacco plants (Igaki 1929), this probably reflects a difference in substantial leaf-age between the transgenic and control plants (Figure 3C).
In the IMAGING-PAM analysis, transgenic (S1, U9) and control Ct plants showed the same photosynthetic capacity (Figure 4). Four different parameters affecting the photosynthetic capacity, including the photosynthesis rate, stomatal conductance, non-photochemical quenching (NPQ), and the electron transporter rate under various light intensities, showed the same values in transgenic and control plants (Figure 5A–D). In addition, the RuBisCO and chlorophyll contents and all stomatal characteristics were similar level between the transgenic and control plants (Table 1). These data indicate that forced expression of the rice 45S rRNA gene promotes up to 2-fold increased aboveground growth without changing the photosynthetic and stomatal characteristics of the transgenic plants.
Because tobacco plants do not show much polysomaty like Arabidopsis thaliana, the cell division activity was inferred from the 4C/2C ratio using a flow cytometer. Interestingly, the maximum peaks of cell division activity differed between (secondary) roots (10 DAS) and hypocotyls (14 DAS) in both U9 and Ct seedlings (Figure 6B). These data suggest that root system development might occur faster than aerial tissues development in tobacco plants. When compared with Ct seedlings, U9 seedlings showed higher cell division activity in 7 DAS roots and 8 DAS hypocotyls. Similarly, transgenic Arabidopsis seedlings showed a well-developed root system compared with control seedlings (Makabe et al. 2016). Therefore, higher cell division activity in the root and leaf primordia at the early seedling stage is probably important for the enhanced growth in the transgenic plants.
Microarray analysis was performed using mRNAs extracted from the aerial parts of 12 DAS seedlings because phenotypic growth differences between the transgenic (S1, U9) and control (Ct) seedlings were found at 10 DAS (Figure 6A). Among the more than 2-fold up- and down-regulated genes, 23 (of 37) and 8 (of 45) genes are listed in Table 2, respectively. The up-regulation of two GIGANTEA (GI)-like transcription factor genes is interesting because Arabidopsis GI genes are controlled by the circadian rhythm and regulate flowering time genes (Fowler et al. 1999). Because Ni et al. (2009) and Chen (2010) suggested that altered circadian rhythms promote growth in the hybrid vigor, circadian genes are probably responsible for the cell proliferation in the hybrid vigor and growth increase found in this study. Thus, the analysis of the circadian genes controlling the expression of tobacco GI genes will be necessary to reveal the mechanism of the growth increase.
The up-regulated ribosomal L30 and down-regulated mitochondrial S41A-like genes were also interesting because mutations of several ribosomal protein genes affect the regulation of cell proliferation and expansion in Arabidopsis leaves (Tsukaya 2006). Although 12 auxin repressed protein ARP1-like genes were up-regulated, they were probably induced to suppress overgrowth of organs in the transgenic plant because over-expression of the ARP1 gene represses plant growth (Zhao et al. 2014). There is no down-regulated gene that is responsible for the control of cell cycle.
In case of the transgenic Arabidopsis, several ethylene responsive transcription factor genes were up-regulated in 12–14 DAS seedlings (Makabe et al. 2016). However, such genes, up- or down- regulated in the transgenic Arabidopsis, were not detected in the microarray analysis of transgenic tobacco seedlings (12 DAS). Although the reasons for the differences in gene expression between Arabidopsis and tobacco are unclear, microarray analysis of seedlings at 12–14 DAS was too late to resolve the genes that were responsible for the growth increase in the transgenic Arabidopsis. The flow cytometry analysis in this study suggests that the transcriptomes in the root and leaf primordia of tobacco seedlings need to be analyzed before 7 DAS.
Semi-quantitative PCR showed that the S1 and U9 transgenic plants expressed the rice 45S rRNA at a similar level (Figure 1C). However, the expressed rice 45S rRNA transcripts might not play roles as rRNA molecules because they were expressed at a quite low level. Because the sequences of the 18S, 5.8S, and 28S rRNAs within the 45S rRNA transcripts are highly homologous between rice and tobacco, the expression of species-specific ITS sequences might be responsible for the growth increase found in the transgenic tobacco and Arabidopsis (Makabe et al. 2016).
Plant leaf development is governed through the mechanisms that regulate the number and size of leaf cells (Palatnik et al. 2003). Therefore, co-ordination between cell proliferation and post-mitotic cell expansion mediates the final leaf size (Gonzalez et al. 2010). Several Arabidopsis mutants with defective genes for cell proliferation show increased cell expansion in their leaves (Horiguchi et al. 2005). Polyploidization can also cause a growth increase in plants through cell expansion (Miller et al. 2012). The aboveground growth increase in the transgenic tobacco plants was not caused by cell expansion because the size and number of stomatal guard cells (Table 1) and the ploidy level of leaf cells (Supplementary Figure S3) were similar between the transgenic and control leaves. Stomatal size is considered as good indicators of the ploidy level in plant cells (Wood et al. 2009), and only the total leaf-area and dry weight of transgenic tobacco plants were increased by ca. 2-fold compared with those of control plants. Taking previous findings together with the results of this study, forced expression of the rice 45S rRNA accelerates cell proliferation without changing the morphological and physiological traits of somatic cells in the transgenic plants.
The forced expression of exogenous 45S rRNA (FEE45) is a simple technology that will contribute to increasing the growth of transgenic plants. Unlike hybrid vigor, the growth increase by FEE45 can be fixed as a homozygous allele in practical cultivars. The mechanism of the growth increase through the enhancement of cell proliferation at the early seedling stage might be related to each other between hybrid vigor and FEE45. The FEE45 technology could be applied to increase the production of secondary metabolites in medicinal plants and to breed high yielding cultivars of cereals, vegetables, trees, and especially biomass plants for bio-energy production.
Abbreviations
- 35SP
cauliflower mosaic virus 35S promoter
- ARP
auxin repressed protein
- DAS
days after sowing
- ETR
electron transport rate
- φPSII
quantum yield of photosystem II
- FEE45
forced expression of exogenous 45S rRNA
- GI
GIGANTEA
- hpt
hygromycin phosphotransferase
- ITS
internal transcribed spacers
- MS
Murashige and Skoog
- NPQ
non-photosynthetic quenching
- nosT
nopaline synthase gene terminator
- PAM
pulse amplitude modulation
- PAM
pulse amplitude modulation
- rRNA
ribosomal RNA
- UbiP
maize ubiquitin promoter
Supplementary Data
References
- Appels R, Dvorak J (1982) Relative rates of divergence of spacer and gene sequences within the rDNA region of species in the Triticeae: Implications for the maintenance of homogeneity of a repeated gene family. Theor Appl Genet 63: 361–365 [DOI] [PubMed] [Google Scholar]
- Byrne ME (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2010) Molecular mechanism of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Darwin CR (1876) The Effects of Cross- and Self-fertilization in the Vegetable Kingdom, John Murry
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Duvick DN (1999) Heterosis: Feeding people and natural resources. In: Coors JG, Pandey S (eds) The Genetics and Exploitation of Heterosis in Crops. Amer Soc of Agron, Crop Sci Soc of Amer, and Soil Sci Soc of Amer, pp 19–29
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H (2009) Coordination of cell proliferation and cell expansion mediated by ribosome related processes in the leaves of Arabidopsis thaliana. Plant J 59: 499–508 [DOI] [PubMed] [Google Scholar]
- Fujimoto R, Taylora JM, Shirasawab S, Peacocka WJ, Dennisa ES (2012) Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA 109: 7109–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Wiegel D, Kamiya Y, et al. (2010) Increased leaf size: Different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Hoecker N (2007) Towards the molecular basis of heterosis. Trends Plant Sci 12: 427–432 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 122–133 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65: 724–736 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H (2012) Ribosomes and translation in plant developmental control. Plant Sci 191–192: 24–34 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Roger SG, Fraley RT (1985) Simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Igaki S (1929) Quantitative changes of the constituents of each part of tobacco during growth. Bult Sci Fakult Terk Kjusu Imp Univ 3: 317–326 (in Japanese) [Google Scholar]
- Ito T, Kim GT, Shinozaki K (2000) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22: 257–264 [DOI] [PubMed] [Google Scholar]
- Kondrosi E, Roundier F, Gendreau E (2000) Plant cell-size control growing by ploidy? Curr Opin Plant Biol 3: 488–492 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D (2007) Heterosis: Revisiting the magic. Trends Genet 23: 60–66 [DOI] [PubMed] [Google Scholar]
- Makabe S, Motohashi R, Nakamura I (2016) Growth increase of Arabidopsis by forced expression of rice 45S rRNA gene. Plant Cell Rep doi: 10.1007/s00299-016-2075-y [DOI] [PubMed] [Google Scholar]
- Meyer RC, Törjék O, Becher M, Altmann T (2004) Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol 134: 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Zhang C, Chen ZJ (2012) Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K, Mii M (2000) Polysomaty analysis in diploid and tetraploid Portulaca grandiflora. Plant Sci 156: 213–219 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Rosado A, Li R, Van de Ven W, Hsu E, Raikhel NV (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA 109: 19537–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo NJ, Lourenço T, Oliveira MM (2009) Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot (Lond) 103: 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidegger Y, Saurer M, Bahn M, Siegwolf R (2000) Linking stable oxygen and carbon isotopes with stomatal conductance and photosynthetic capacity: A conceptual model. Oecologia 125: 350–357 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: Endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Takesawa T, Ito M, Kanzaki H, Kameya N, Nakamura I (2002) Over-expression of ζ glutathione S -transferase in transgenic rice enhances germination and growth at low temperature. Mol Breed 9: 93–101 [Google Scholar]
- Tsukaya H (2006) Mechanism of leaf shape determination. Annu Rev Plant Biol 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo NS, Badger MR, Pogson BJ (2008) A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Nagai T, Makino A (2011) The rate-limiting step for CO2 assimilation at different temperatures is influenced by the leaf nitrogen content in several C3 crop species. Plant Cell Environ 34: 764–777 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li C, Ge J, Xu M, Zhu Q, Wu T, Guo A, Xie J, Dong H (2014) Recessive mutation identifies auxin-repressed protein ARP1, which regulates growth and disease resistance in tobacco. Mol Plant Microbe Interact 27: 638–654 [DOI] [PubMed] [Google Scholar]
- Zsogon A, Szakony D, Shi X, Byrne ME (2014) Ribosomal protein RPL27a promotes female gametophyte development in a dose-dependent manner. Plant Physiol 165: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.