Abstract
Balanced development of adaxial and abaxial domains in leaf primordia is critical for the formation of flat symmetric leaf lamina. Arabidopsis ASYMMETRIC LEAVES1 (AS1) and AS2 proteins form a complex (AS1–AS2), which acts as key regulators for the adaxial development by the direct repression of expression of the abaxial gene ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3). Many modifier mutations have been identified, which enhance the defect of as1 and as2 mutations to generate abaxialized filamentous leaves without adaxial traits, suggesting that the development of the adaxial domain is achieved by cooperative repression by AS1–AS2 and the wild-type proteins corresponding to the modifiers. Mutations of several genes for DNA replication-related chromatin remodeling factors such as Chromatin Assembly Factor-1 (CAF-1) have been also identified as modifiers. It is still unknown, however, whether mutations in genes involved in DNA replication themselves might act as modifiers. Here we report that as1 and as2 mutants grown in the presence of hydroxyurea, a known inhibitor of DNA replication, form abaxialized filamentous leaves in a concentration-dependent manner. We further show that a mutation of the INCURVATA2 (ICU2) gene, which encodes the putative catalytic subunit of DNA polymerase α, and a mutation of the Replication Factor C Subunit3 (RFC3) gene, which encodes a protein used in replication as a clamp loader, act as modifiers. In addition, as2-1 icu2-1 double mutants showed increased mRNA levels of the genes for leaf abaxialization. These results suggest a tight link between DNA replication and the function of AS1–AS2 in the development of flat leaves.
Keywords: ASYMMETRIC LEAVES2, DNA replication, ICU2, leaf development, RFC3
Introduction
Leaf primordia that are developed as lateral organs from the shoot apical meristem (SAM) grow up along the proximal–distal, medial–lateral, and adaxial–abaxial axes through repeated cell divisions and cell differentiations. The establishment of adaxial–abaxial polarity at the initial stage of leaf development is crucial for the formation of flat symmetric leaves (Bowman and Floyd 2008; Byrne et al. 2000; Hudson 2000; Nakata and Okada 2013; Semiarti et al. 2001; Steeves and Sussex 1989; Szakonyi and Byrne 2011; Tsukaya 2013; Waites and Hudson 1995). The abaxialization is thought to proceed to adaxialization because abaxial-determining genes were expressed earlier than that of adaxial-determining genes (Eshed et al. 1999, 2001; Sawa et al. 1999) and the defect of the adaxial domains results in the generation of the filamentous leaf that retains only abaxial traits (Li et al. 2005; Ueno et al. 2007; Waites and Hudson 1995). AS1 and AS2 are key regulators of the formation of flat symmetric leaves. AS1 and AS2 encode nuclear proteins and form a complex (designated AS1–AS2) (Guo et al. 2008; Luo et al. 2012; Xu et al. 2003; Yang et al. 2008). Mutations in these genes are associated with pleiotropic abnormalities in leaves along the three developmental axes described above (Byrne et al. 2000; Iwakawa et al. 2002, 2007; Matsumura et al. 2009; Ori et al. 2000; Rédei and Hirono 1964; Semiarti et al. 2001; Tsukaya and Uchimiya 1997), suggesting that AS1–AS2 regulates multiple genes (Iwasaki et al. 2013; Machida et al. 2015; Takahashi et al. 2008; Takahashi et al. 2013) that might be involved in leaf formation along the three axes.
Direct repression by AS1–AS2 of the expression of at least two gene families is critical for leaf development, one of which is the class 1 KNOTTED-like homeobox (KNOX) genes such as BREVIPEDICELLUS (BP), KNAT2 (Guo et al. 2008). The other class of direct targets includes the abaxial-determining gene ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3) (Iwasaki et al. 2013), which is directly repressed by the binding of AS1–AS2 to its promoter region. In addition, AS1–AS2 also indirectly represses the expression of ETT/ARF3 and the functionally redundant gene ARF4 through its positive regulation of the miR390-tasiR-ARF pathway, essential for adaxial development (Iwasaki et al. 2013). AS1–AS2 is also required for maintaining levels of methylated CpGs in the ETT/ARF3 coding region (Iwasaki et al. 2013). The methylation is abolished in the mutant of METHYLTRANSFERASE1 (MET1) gene and the level of ETT/ARF3 transcript increases in shoot apices in met1 (Iwasaki et al. 2013). The observed anti-parallel relationship supports the hypothesis that the CpG methylation in ETT/ARF3 might play a role in repression of ETT/ARF3 expression (Iwasaki et al. 2013).
Defects in polarity of as1 and as2 leaves are enhanced under certain growth conditions as well as in conjunction with mutations in members of certain groups of genes (Machida et al. 2015), which are designated as modifiers of adaxial–abaxial patterning (Machida et al. 2015; Matsumura et al. 2016; Szakonyi et al. 2010). To date, various modifier mutations that generate filamentous leaves surrounded by abaxialized epidermis in the as1 or as2 mutant backgrounds have been identified and it has been reported that the development of the adaxial domain of leaves is severely defective in those double mutants. The modifier genes include several that mediate the biogenesis of tasiR-ARF (Machida et al. 2015; Yang et al. 2006). Other relevant modifier genes belong to several different groups: those for ribosome biogenesis (Horiguchi et al. 2011a; Matsumura et al. 2016; Pinon et al. 2008; Yao et al. 2008); chromatin modification (Kojima et al. 2011; Ueno et al. 2007); and cell proliferation (Horiguchi et al. 2011b; Ishibashi et al. 2012; Wang et al. 2011; Xu et al. 2012; Yuan et al. 2010). Mutations of genes related to DNA replication-related chromatin remodeling and DNA repair have been identified as modifiers that enhance leaf adaxial–abaxial abnormalities in as1 and as2 (Inagaki et al. 2009; Machida et al. 2015; Xu et al. 2012). Mutations in FASCIATA1 (FAS1) and FAS2 genes that encode Arabidopsis homologs of components of Chromatin Assembly Factor-1 (CAF-1), a histone chaperone essential for chromatin remodeling after DNA replication (Takami et al. 2007), also cause abaxialized filamentous leaves in as1 and as2 (Ishibashi et al. 2013; Xu et al. 2012). Since these factors are not directly involved in reactions of DNA replication, the relationship between AS1–AS2, factors involved in DNA replication and the leaf development is, however, still unknown.
In the present study, we used hydroxyurea, which is an inhibitor of DNA replication (Saban and Bujak 2009), and Arabidopsis mutants of genes for the catalytic subunit of DNA polymerase α and Replication Factor C Subunit3 (RFC3) involved in DNA elongation on a primed DNA template as a clamp loader. The present results show that as1 and as2 mutants grown in the hydroxyurea-containing medium formed abaxialized filamentous leaves in a concentration-dependent manner. The mutations we used also generated abaxialized filamentous leaves in the as1 and as2 backgrounds. These results suggest that the cooperative action of AS1–AS2 and the proper progression of DNA replication are involved in the development of flat leaves in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Col-0 (CS1092), and the mutants as1-1 (CS3374) and as2-1 (CS3117) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA; ABRC). We outcrossed as2-1 with Col-0 three times and as1-1 with Col-0 once, and used the progeny for our experiments (Semiarti et al. 2001). The incurvata2-1 (icu2-1) mutant that has a point mutation in the coding region of INCURVATA2, which encodes the putative catalytic subunit of the DNA polymerase α of Arabidopsis thaliana, was described by Barrero et al. (2007). The rfc3-1 mutant, which has a point mutation in the coding region of a gene encoding a protein with high homology to Replication Factor C Subunit3 of yeast and other eukaryotes, was described by Xia et al. (2009). For analysis of phenotypes, seeds were sown on soil or on agar plates of Murashige and Skoog (MS) medium (Wako Pure Chemical Industries, Ltd., Osaka, Japan) supplemented with 2% (w/v) sucrose and 0.8% agar (Nakagawa et al. 2012). After 2 days at 4°C in darkness, plants were transferred to a daily regimen of 8 h of darkness and 16 h of white light at 50 µmol m−2 s−1 at 22°C, as described previously (Semiarti et al. 2001). Hydroxyurea was purchased from Sigma-Aldrich (St. Louis, Missouri, USA). For chemical treatments, hydroxyurea was dissolved in H2O, mixed with MS agar, and immediately dispensed into plastic dishes. Ages of plants are given in terms of numbers of days after sowing.
Fluorescence microscopy
The as1-1 and as2-1 plants containing the FILAMENTOUS FLOWER (FIL) promoter FILp:GFP were described by Nakagawa et al. (2012). Shoot apices containing leaf primordia were embedded in 5% agar and then the agarose blocks were sliced into sections with a vibratome. Fluorescence was observed with a confocal laser scanning microscope (LSM510 META; Carl Zeiss Inc., Oberkochen, Germany).
Real-time RT-PCR
Shoot apices of mutant and Col-0 (wild-type) plants were harvested 14 days after seeds were sown and immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from the 14-day-old shoot apices with the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. For the analysis of RNA levels in Arabidopsis by real-time qRT-PCR, we prepared 10 µg of total RNA. Reverse transcription was carried out with ReverTra Ace (TOYOBO, Osaka, Japan). Sample volumes were normalized for equal amplification of DNA fragments using primers specific for α-tubulin cDNA. Primer sets are listed in the Supplementary Table. PCR was performed in the presence of the double-stranded DNA-specific dye SYBR Green (Applied Biosystems, Lincoln, CA, USA). Amplification was monitored in real time with the Applied Biosystems StepOnePlus Real-Time PCR system (Applied Biosystems) according to the supplier’s recommendations. The mean value of three technical replicates was normalized by using the ACTIN2 transcripts as a control.
Results
Treatments of as1 and as2 plants with hydroxyurea induce filamentous leaves
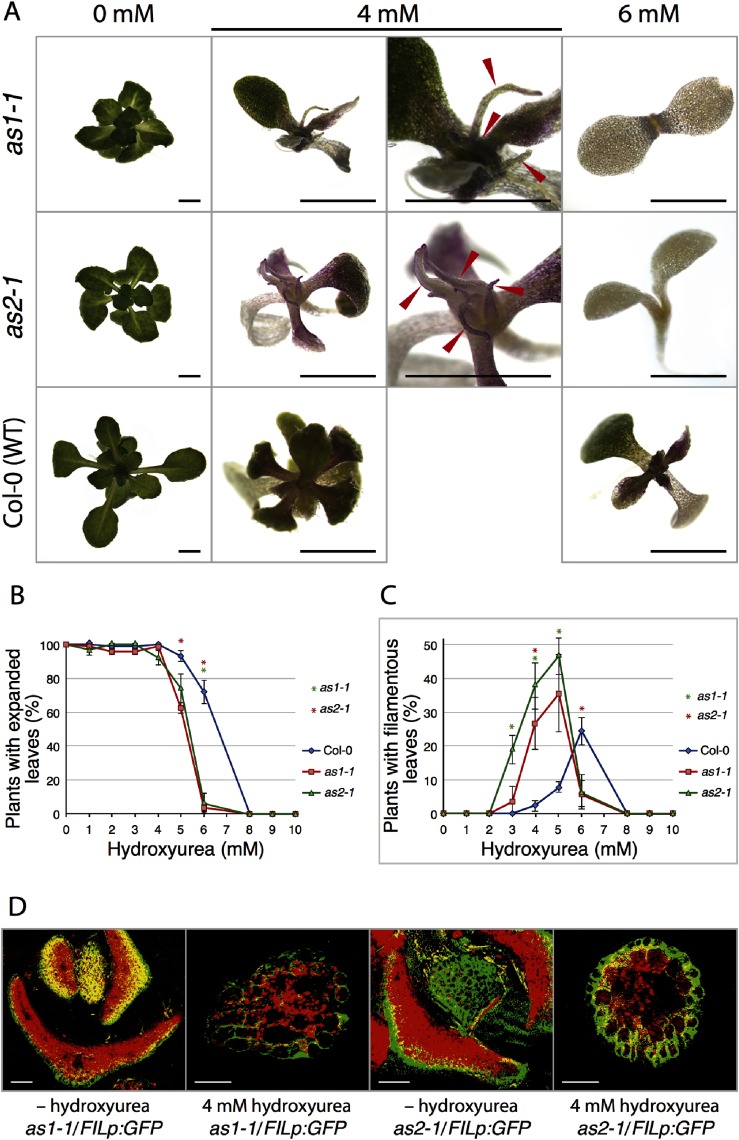
Col-0 (wild-type), as1-1 and as2-1 plants were grown in the presence of hydroxyurea, a known replication inhibitory compound. Using 0 to 10 mM concentrations of hydroxyurea, we examined the dose-dependent effects of this compound on the formation of expanded true leaves and filamentous leaves in these plants (Figure 1A–C). The percentage of plants with expanded true leaves was decreased at higher concentrations of hydroxyurea in as1-1, as2-1, and the wild type (Figure 1B). In the presence of 3 mM hydroxyurea, Col-0, as1-1 and as2-1 plants normally formed expanded true leaves. Mutants, however, exhibited higher sensitivity to hydroxyurea than Col-0 between 4 and 6 mM: all Col-0 and as1-1 plants and 90% of as2-1 formed expanded true leaves at 4 mM; 90% of the wild type and more than 60–75% of mutants formed expanded leaves at 5 mM; 72% of Col-0 and only 7% of mutants formed expanded leaves at 6 mM. As shown in Figure 1C, 8%, 26%, and 34% of as1-1 and 19%, 38%, and 46% of as2-1 plants produced filamentous leaves indicative of a defect in adaxial–abaxial polarity at 3, 4, and 5 mM hydroxyurea, respectively. In contrast, only 1% and 9% of Col-0 plants with filamentous leaves were observed at 4 and 5 mM concentrations of hydroxyurea, respectively. Thus, frequencies of the formation of filamentous leaves in as1-1 and as2-1 increased in a concentration-dependent manner by hydroxyurea. Most of all mutant plants stopped developing true leaves and growing in the presence of 6 mM hydroxyurea (Figure 1A, C), and they eventually exhibited chrolosis (Figure 1A). Seventy-two percent of Col-0 plants generated expanded leaves and 25% generated filamentous leaves under the same growth conditions (Figure 1B, C). Thus, a concentration of 6 mM hydroxyurea should be too strong to use for examining leaf phenotypes in as1-1 and as2-1 mutants, and a concentration of 5 mM or less would be suitable for the examination.
Figure 1. Effects of hydroxyurea on as1-1 and as2-1 mutants. (A) Phenotypes of Col-0, as1-1, and as2-1 plants grown on agar plates in the presence and the absence of 4 mM, 6 mM hydroxyurea. Red arrowheads on as1-1 and as2-1 plants show filamentous leaves. Scale bars=2 mm. (B) Frequencies of plants that have true leaves grown in the presence and in the absence of hydroxyurea. Frequency is defined as the ratio of the number of plants having expanded leaves to the total number (n=30) of plants examined. Plants were grown at 22°C. Observations were performed at 21 days after sowing. Bars represent the s.d. from three biological replicates. (C) Frequencies of plants with filamentous leaves grown in the presence and in the absence of hydroxyurea. Frequency is defined as the ratio of the number of plants with more than one filamentous leaf to the total number (n=30) of plants examined. Plants were grown at 22°C. Observations were performed at 21 days after sowing. Bars represent the s.d. from three biological replicates. (D) as1-1/FILp:GFP and as2-1/FILp:GFP plants were grown on medium with and without 4 mM hydroxyurea for 21 days. Expression patterns of FILp:GFP in transverse sections of leaves are shown. Green signals due to GFP; red, autofluorescence. Yellow signals were due to both GFP and autofluorescence. Scale bars=50 µm. Significant differences from wild type were evaluated by Student’s t-test and are represented by asterisks (* p<0.01) in (B) and (C).
Since filamentous leaves are often generated in plants having defective adaxial–abaxial polarity determination, we examined whether the leaf phenotypes induced by the hydroxyurea treatment are associated with transcript levels of genes that are involved in the polarity development. We observed signals due to green fluorescent protein (GFP), synthesis of which is under the control of the promoter of the abaxial-determining gene FILAMENTOUS FLOWER (FIL) (FILp:GFP) (Watanabe and Okada, 2003) in transgenic as1-1 and as2-1 mutants. As shown in Figure 1D, strong signals were detected in cells located at peripheral positions in filamentous leaves of as1-1 and as2-1 plants grown in the presence of 4 mM hydroxyurea, whereas no signals were detected in cells located on the adaxial side of leaves of as1-1 and as2-1 plants in the absence of hydroxyurea. These results suggested that filamentous leaves in as1-1 and as2-1 treated with hydroxyurea were abaxialized.
Mutation of DNA polymerase α enhanced a leaf adaxial–abaxial polarity defect in as1 and as2
The results in the previous section suggest that inhibition of DNA replication preferentially interfere development of the adaxial domain during formation of leaf primordia on the background of as1-1 and as2-1. We next examined whether the mutation (icu2-1) of the INCURVATA2 gene, which encodes an Arabidopsis homolog of the catalytic subunit of the DNA polymerase α (Barrero et al. 2007), an essential gene for DNA replication, might affect adaxial–abaxial polarity establishment of leaves in the as1 and as2 mutant backgrounds. For this, we generated the double mutants as1-1 icu2-1 and as2-1 icu2-1. As shown in Figure 2 and Table 1, 18.1–21.7% of as1-1 icu2-1 and 4.6–11.7% of as2-1 icu2-1 double mutants showed filamentous leaves, although the wild-type plant and any single mutants of as1-1, as2-1, and icu2-1 did not generate filamentous leaves. We also observed trumpet-like leaves, the weak phenotype of filamentous leaves. Trumpet-like leaves were seen on 16.4–26.1% of as1-1 icu2-1 and 3.0–5.5% of as2-1 icu2-1. These results suggested that the icu2-1 mutation influenced the establishment of leaf adaxial–abaxial polarity in the as1-1 and as2-1 mutant backgrounds.
Figure 2. The inc2-1 mutation enhanced the leaf-phenotype in as1-1 and as2-1 mutants. Gross morphology at 21 days after sowing. Plants indicated below pictures were grown as described in Materials and methods. The as1-1 icu2-1 and as2-1 icu2-1 double mutants exhibited filamentous leaves. Arrowheads indicate filamentous leaves. Scale bars=5 mm.
Table 1. Number of plants with trumpet-like leaves or filamentous leaves.
| Genotype | Experiments | Number of plants examined | Number of plants with trumpet-like leaves (%) | Number of plants with filamentous leaves (%) |
|---|---|---|---|---|
| Col-0 (WT) | 1 | 180 | 0 (0) | 0 (0) |
| 2 | 165 | 0 (0) | 0 (0) | |
| 3 | 173 | 0 (0) | 0 (0) | |
| as1-1 | 1 | 113 | 0 (0) | 0 (0) |
| 2 | 104 | 0 (0) | 0 (0) | |
| 3 | 97 | 0 (0) | 0 (0) | |
| as2-1 | 1 | 273 | 0 (0) | 0 (0) |
| 2 | 193 | 0 (0) | 0 (0) | |
| 3 | 209 | 0 (0) | 0 (0) | |
| icu2-1 | 1 | 251 | 0 (0) | 0 (0) |
| 2 | 186 | 0 (0) | 0 (0) | |
| 3 | 215 | 0 (0) | 0 (0) | |
| as1-1 icu2-1 | 1 | 120 | 23 (19.2) | 21 (17.5) |
| 2 | 138 | 36 (26.1) | 30 (21.7) | |
| 3 | 116 | 19 (16.4) | 21 (18.1) | |
| as2-1 icu2-1 (C2) | 1 | 255 | 11 (4.3) | 19 (7.5) |
| 2 | 231 | 7 (3.0) | 27 (11.7) | |
| 3 | 241 | 10 (4.1) | 24 (10.0) | |
| as2-1 icu2-1 (E7) | 1 | 227 | 8 (3.5) | 18 (7.9) |
| 2 | 238 | 13 (5.5) | 11 (4.6) |
Number of plants with trumpet-like or filamentous leaves is defined as the number of plants with more than one trumpet-like or filamentous leaves, respectively. Percentages in parenthesis indicate the frequency with which trumpet-like or filamentous leaves structure was observed relative to the total number of plants examined. Plants were grown at 22°C for 21–23 days after sowing. C2 and E7 indicate different offspring lines generated by crossing as2-1 with icu2-1.
Levels of transcripts of the abaxial determinant genes and class 1 KNOX genes were increased in as2-1 icu2-1
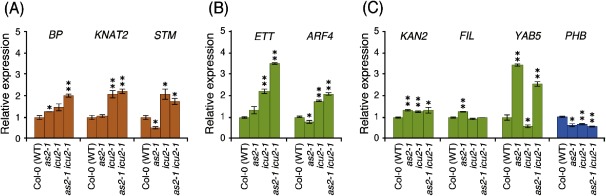
In the previous section, we have shown that the filamentous leaves induced by the hydroxyurea treatment were abaxialized (Figure 1D), suggesting that patterns of expression of genes that control abaxial–adaxial polarity establishment might be altered in the mutants we used. We performed real-time RT-PCR using RNA from the shoot apices of Col-0 (wild-type), as2-1, icu2-1, and as2-1 icu2-1 plants. We quantified transcripts of PHABULOSA (PHB), one of the genes in the HD-ZIP III family that specify the adaxial cell fate; ETT/ARF3, ARF4, KANADI2 (KAN2), FIL, YABBY5 (YAB5) genes that specify the abaxial cell fate; and the BP, KNAT2, SHOOT MERISTEMLESS (STM) genes, which are members of the class 1 KNOX gene family and are expressed in the SAM and its periphery in wild-type plants. As shown in Figure 3, the transcript levels of BP, KAN2, FIL and YAB5 genes were significantly increased in the as2-1 mutant compared with those in Col-0 (wild-type) plant, consistent with our previous report (Kojima et al. 2011; Ishibashi et al. 2013). The transcript levels of KNAT2, STM, ETT/ARF3, ARF4, KAN2 genes were increased in the icu2-1 mutant compared with those in Col-0 (wild-type) plant. The transcript level of the PHB gene was decreased in the as2-1, icu2-1 and as2-1 icu2-1 plants. Furthermore, The accumulated transcript levels of either ETT/ARF3 or ARF4 in the as2-1 icu2-1 double mutant were significantly higher than those levels in both the as2-1 and icu2-1 single mutants.
Figure 3. Transcript levels of genes involved in the determination of leaf polarity and class 1 KNOX genes. Levels of relative expression of (A) class 1 KNOX genes, (B) genes involved in leaf abaxialization (ETT and ARF4), (C) other genes that are involved in leaf abaxialization (KAN2, FIL, YAB5), and a gene that is involved in leaf adaxialization (PHB), respectively, relative to those levels in the wild-type (Col-0) plants. Total RNA was extracted from shoot apices of 14-day-old Col-0, as2-1, icu2-1, and as2-1 icu2-1. Each value was normalized by reference to the level of ACTIN2 (ACT2, at3g18780) transcripts. Light brown, light green and light blue show class 1 KNOX genes, abaxial determinant genes and an adaxial determinant gene, respectively. The values from wild-type plants were arbitrarily set at 1.0. Bars indicate the s.d. among more than three biological replicates. Significant differences from wild type were evaluated by Student’s t-test and are represented by asterisks (* p<0.05 and ** p<0.01).
To date, it has been reported that the formation of filamentous and trumpet-like leaves in several double mutants combined with the as2-1 mutant is, at least partially, due to the increased accumulation of levels of ETT/ARF3 and ARF4 transcripts. These results support the effects of the icu2-1 mutation on the establishment of leaf adaxial–abaxial polarity in the as1-1 and as2-1 mutant background. The increased levels of ETT/ARF3 and ARF4 transcripts in the as2-1 icu2-1 double mutant might be responsible for the formation of the filamentous and trumpet-like leaves.
In the double mutants combined with the as2 mutation that exhibit the formation of filamentous leaves, the accumulated transcript levels of the class 1 KNOX genes BP, KNAT2, KNAT6, and STM have been significantly increased, as compared with those of the wild type (Horiguchi et al. 2011b; Ishibashi et al. 2012; Kojima et al. 2011; Yang et al. 2006). As shown in Figure 3A, in as2-1 icu2-1double mutant, the mRNA levels of BP, KNAT2 and STM genes were markedly increased over those levels in Col-0 (wild-type). These results suggested that leaves in as2-1 icu2-1 double mutants were indeterminate and abaxialized.
Mutation of an Arabidopsis homolog of Replication Factor C subunit 3 (RFC3) enhanced a leaf adaxial–abaxial polarity defect in as1 and as2
To generalize the observation obtained in the above experiment with as2-1 icu2-1, we examined the rfc3-1 mutant for the potential role in leaf development. It is a mutant allele of AtRFC3, which encodes an Arabidopsis homolog of Replication Factor C subunit 3 (Xia et al. 2009; 2010) and is potentially required for stable DNA replication by DNA polymerases (delta and epsilon) (Yin et al. 2009). Leaves of the rfc3-1 mutant were narrow and pointed (Figure 4), which is one of characteristic leaf abnormalities by the modifier mutations to enhance leaf polarity defects in as1 and as2 (Matsumura et al. 2016). To further confirm the involvement of AtRFC3 in the leaf polarity development, we examined leaf phenotypes in the double mutants as1-1 rfc3-1 and as2-1 rfc3-1. As shown in Figure 4 and Table 2, 4.7–7.8% and 11.1–14.5% of the as1-1 rfc3-1 and as2-1 rfc3-1 double mutants showed filamentous leaves, respectively. Trumpet-like leaves were seen on 5.9–8.2% of as1-1 rfc3-1 and 0–2.6% of as2-1 rfc3-1. These results suggested that the rfc3-1 mutation influenced in a certain extent the establishment of leaf adaxial–abaxial polarity in the as1-1 and as2-1 mutant backgrounds.
Figure 4. The rfc3-1 mutation enhanced the leaf-phenotype in as1-1 and as2-1 mutants. The as1-1 rfc3-1 double mutants exhibited trumpet-like leaves, while the as2-1 rfc3-1 double mutants exhibited filamentous leaves. Gross morphology at 21 days after sowing. Arrowheads indicate higher magnification views of trumpet-like leaves or filamentous leaves. Scale bars=5 mm.
Table 2. Number of plants with trumpet-like leaves or filamentous leaves.
| Genotype | Experiments | Number of plants examined | Number of plants with trumpet-like leaves | Number of plants with filamentous leaves |
|---|---|---|---|---|
| Col-0 (WT) | 1 | 90 | 0 (0) | 0 (0) |
| 2 | 78 | 0 (0) | 0 (0) | |
| 3 | 85 | 0 (0) | 0 (0) | |
| as1-1 | 1 | 93 | 0 (0) | 0 (0) |
| 2 | 90 | 0 (0) | 0 (0) | |
| 3 | 71 | 0 (0) | 0 (0) | |
| as2-1 | 1 | 68 | 0 (0) | 0 (0) |
| 2 | 76 | 0 (0) | 0 (0) | |
| 3 | 81 | 0 (0) | 0 (0) | |
| rfc3-1 | 1 | 86 | 0 (0) | 0 (0) |
| 2 | 85 | 0 (0) | 0 (0) | |
| 3 | 80 | 0 (0) | 0 (0) | |
| as1-1 rfc3-1 | 1 | 102 | 7 (6.9) | 8 (7.8) |
| 2 | 85 | 5 (5.9) | 4 (4.7) | |
| 3 | 85 | 7 (8.2) | 6 (7.1) | |
| as2-1 rfc3-1 | 1 | 63 | 0 (0) | 8 (12.7) |
| 2 | 76 | 2 (2.6) | 11 (14.5) | |
| 3 | 81 | 1 (1.2) | 9 (11.1) |
Number of plants with trumpet-like or filamentous leaves is defined as the number of plants with more than one trumpet-like or filamentous leaves, respectively. Percentages in parenthesis indicate the frequency with which trumpet-like or filamentous leaves structure was observed relative to the total number of plants examined. Plants were grown at 22°C for 21–23 days after sowing.
Discussion
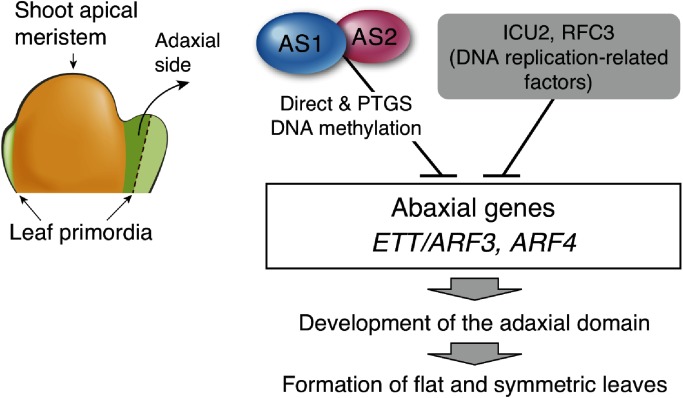
The results in the present study showed that hydroxyurea, which interferes DNA synthesis, disrupted the establishment of the leaf adaxial–abaxial polarity in the as1 or as2 mutant, which resulted in the formation of the abaxialized filamentous leaves (Figure 1A, C, D). Hydroxyurea, a known replication inhibitory compound, inhibits ribonucleotide reductase enzyme known to be crucial in the conversion of ribonucleotides into deoxyribonucleotides and prevents cells from leaving the G1/ S phase of the cell cycle (Saban and Bujak 2009). Our results imply that normal progression of the replication step might be required for adaxial–abaxial establishment of leaf development. In addition, mutations of ICU2 and RFC3 genes that are required for DNA replication also induced the formation of filamentous leaves in the as1 or as2 genetic background (Figures 2 and 4). These results suggested that a proper progression of DNA replication is critical, at least in part, for the development of the adaxial domain in as1 and as2 mutants. Since the frequencies of formation of filamentous leaves were increased in the as1 or as2 mutant treated with hydroxyurea or in the double mutants, as1-1 icu2-1, as2-1 icu2-1, as1-1 rfc3-1, as2-1 rfc3-1, the AS1–AS2-mediated pathway and the progression of DNA replication might be independently, but cooperatively, involved in the development of the adaxial domain of leaves in the wild-type Arabidopsis (Figure 5). Such cooperative actions in the leaf development are also observed in combinations of as mutations with other modifiers (Machida et al. 2015).
Figure 5. Roles of AS1–AS2, ICU2, and RFC3 in leaf development in A. thaliana. The AS1–AS2 complex and DNA replication-related factors ICU2 (or RFC3) act cooperatively to repress expression of the leaf abaxial determinant gene ETT/ARF3 and ARF4. Repression of ARFs is crucial for development of the adaxial domain of leaves and then formation of flat and symmetric leaves.
The transcript level of ETT/ARF3 gene, which is a direct target of AS1–AS2 (Iwasaki et al. 2013), was increased in the icu2-1 mutant. Furthermore, transcript levels of ETT/ARF3 and ARF4 in the as2-1 icu2-1 double mutant were higher than those in both the as2-1 and icu2-1 single mutants (Figure 3). Since we have already shown that repression of expression of the abaxial genes ETT/ARF3 and ARF4 by AS1–AS2 and modifiers is important to form flat leaves (Iwasaki et al. 2013; Matsumura et al. 2016; Takahashi et al. 2013), phenotypes of the filamentous and trumpet-like leaves observed in as1-1 icu2-1 and as2-1 icu2-1 might be caused similarly by the up-regulation of ETT/ARF3 during leaf development. It has been shown that ICU2 genetically interacts with TERMINAL FLOWER2, which encodes an ortholog of HETEROCHROMATIN PROTEIN1 of animals and yeasts, and CURLY LEAF, the Polycomb group (PcG) gene (Barrero et al. 2007). Another study of the icu2-1 mutant revealed that ICU2 is required for ensuring the stable maintenance of repressive histone modifications, the H3K27me3 level at the FLOWERING LOCUS C region, and other polycomb repressive complex 2 targets as well as at heterochromatic retroelements (Hyun et al. 2013). Therefore, the maintenance of repressive epigenetic marks in ETT/ARF3 might be disrupted in the icu2-1 mutant. No enrichment of H3K27me3 was detected, however, at the ETT/ARF3 promoter regions, the AS1–AS2 binding site, or within the ETT/ARF3 coding sequence (Husbands et al. 2015; Iwasaki et al. 2013; Roudier et al. 2011; Zhang et al. 2007).
AS1–AS2 is involved in maintaining levels of CpG methylation in the ETT/ARF3 coding region, suggesting that AS1–AS2 plays a key role in epigenetic repression of ETT/ARF3 (Iwasaki et al. 2013). Methylation at these CpG sites were abolished in the mutant of MET1 and the transcript level of ETT/ARF3 in shoot apices was increased (Iwasaki et al. 2013). MET1 is an Arabidopsis homolog of vertebrate DNA methyltransferase1 (DNMT1) (Finnegan et al. 1996), which is responsible for maintaining methylated CpGs during DNA replication (Long et al. 2013). Molecular mechanisms of the DNA replication-coupled maintenance of methylated CpGs in mammalian cells have recently been reported (Ferry et al. 2017; Nishiyama et al. 2013). Mechanisms involving the cooperative action of AS1–AS2 and the replication machinery including ICU2, RFC3 and newly identified factors might be involved in epigenetic repression of the ETT/ARF3 gene that is critical for the formation of flat and symmetric leaves. Recently, we have shown that some modifier genes for the AS2 function are implicated in maintaining CpG methylation in the ETT/ARF3 gene. It should be intriguing to investigate the molecular mechanism of epigenetic regulation of the ETT/ARF3 gene by the cooperative action AS1–AS2 and replication factors such as ICU2 and RFC3 to form flat symmetric leaves.
Acknowledgments
The authors are grateful to Ms. Funahashi and Mr. Ito for their helpful technical support. This work was supported by a Grant-in Aid for Chubu University Grant, D, 2014–2015 [no. 26IM03D to T.Q.L.]; Japan Society for the Promotion of Science (JSPS) KAKENHI [grant numbers JP26291056, JP25650094, JP15K07116]; The Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI [grant numbers JP19060015, JP16H01246].
Supplementary Data
References
- Barrero JM, Gonzalez-Bayon R, del Pozo JC, Ponce MR, Micol JL (2007) INCURVATA2 encodes the catalytic subunit of DNA polymerase α and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19: 2822–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK (2008) Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 59: 67–88 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, et al. (2017) Methylation of DNA Ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell 67: 550–565.e5 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H (2011a) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65: 724–736 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H (2011b) ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52: 112–124 [DOI] [PubMed] [Google Scholar]
- Hudson A (2000) Development of symmetry in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 349–370 [DOI] [PubMed] [Google Scholar]
- Husbands AY, Benkovics AH, Nogueira FTS, Lodha M, Timmermans MCP (2015) The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial–abaxial patterning and leaf complexity. Plant Cell 27: 3321–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y (2013) The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140: 156–166 [DOI] [PubMed] [Google Scholar]
- Inagaki S, Nakamura K, Morikami A (2009) A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLoS Genet 5: e1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi N, Kanamaru K, Ueno Y, Kojima S, Kobayashi T, Machida C, Machida Y (2012) ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol Open 1: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi N, Machida C, Machida Y (2013) ASYMMETRIC LEAVES2 and FASCIATA2 cooperatively regulate the formation of leaf adaxial–abaxial polarity in Arabidopsis thaliana. Plant Biotechnol 30: 411–415 [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C (2007) Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Takahashi H, Iwakawa H, Nakagawa A, Ishikawa T, Tanaka H, Matsumura Y, Pekker I, Eshed Y, Vial-Pradel S, et al. (2013) Dual regulation of ETTIN (ARF3) gene expression by AS1–AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial–abaxial partitioning in Arabidopsis. Development 140: 1958–1969 [DOI] [PubMed] [Google Scholar]
- Kojima S, Iwasaki M, Takahashi H, Imai T, Matsumura Y, Fleury D, Van Lijsebettens M, Machida Y, Machida C (2011) Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol 52: 1259–1273 [DOI] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H (2005) The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Blackledge NP, Klose RJ (2013) 1ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans 41: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Ando S, Sasabe M, Machida C, Kurihara D, Higashiyama T, Machida Y (2012) Arabidopsis ASYMMETRIC LEAVES2 protein required for leaf morphogenesis consistently forms speckles during mitosis of tobacco BY-2 cell via signals in its specific sequence. J Plant Res 125: 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C, Nakagawa A, Kojima S, Takahashi H, Machida Y (2015) The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. WIREs Dev Biol 4: 655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Iwakawa H, Machida Y, Machida C (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Ohbayashi I, Takahashi H, Kojima S, Ishibashi N, Keta S, Nakagawa A, Hayashi R, Saez-Vasquez J, Echeverria M, et al. (2016) A genetic link between epigenetic repressor AS1–AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol Open 5: 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Takahashi H, Kojima S, Sato N, Ohga K, Cha BY, Woo JT, Nagai K, Horiguchi G, Tsukaya H, et al. (2012) Berberine enhances defects in the establishment of leaf polarity in asymmetric leaves1 and asymmetric leaves2 of Arabidopsis thaliana. Plant Mol Biol 79: 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Okada K (2013) The leaf adaxial–abaxial boundary and lamina growth. Plants 2: 174–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502: 249–253 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324 [DOI] [PubMed] [Google Scholar]
- Rédei GP, Hirono Y (1964) Linkage studies. Arabidopsis Inf Serv 1: 9 [Google Scholar]
- Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban N, Bujak M (2009) Hydroxyurea and hydroxamic acid derivatives as antitumor drugs. Cancer Chemother Pharmacol 64: 213–221 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM (1989) Patterns in Plant Development 2nd ed. Cambridge University Press, Cambridge
- Szakonyi D, Byrne ME (2011) Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65: 269–281 [DOI] [PubMed] [Google Scholar]
- Szakonyi D, Moschopoulos A, Byrne ME (2010) Perspectives on leaf dorsoventral polarity. J Plant Res 123: 281–290 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Iwakawa H, Ishibashi N, Kojima S, Matsumura Y, Prananingrum P, Iwasaki M, Takahashi A, Ikezaki M, Luo L, et al. (2013) Meta-analyses of microarrays of Arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial–abaxial patterning and cell division during leaf development. Plant Cell Physiol 54: 418–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwakawa H, Nakao S, Ojio T, Morishita R, Morikawa S, Machida Y, Machida C, Kobayashi T (2008) Knowledge-based fuzzy adaptive resonance theory and its application to the analysis of gene expression in plants. J Biosci Bioeng 106: 587–593 [DOI] [PubMed] [Google Scholar]
- Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T (2007) Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell 18: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H (2013) Leaf development. Arabodopsis Book 11: e0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Uchimiya H (1997) Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol Gen Genet 256: 231–238 [DOI] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Hudson A (1995) phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Wang W, Xu B, Wang H, Li J, Huang H, Xu L (2011) YUCCA genes are expressed in response to leaf adaxial–abaxial juxtaposition and are required for leaf margin development. Plant Physiol 157: 1805–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Okada K (2003) Two discrete cis elements control the Abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Xiao L, Gannon P, Li X (2010) RFC3 regulates cell proliferation and pathogen resistance in Arabidopsis. Plant Signal Behav 5: 168–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Zhu Z, Hao L, Chen JG, Xiao L, Zhang Y, Li X (2009) Negative regulation of systemic acquired resistance by replication factor C subunit 3 in Arabidopsis. Plant Physiol 150: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Huang W, Li Y, Wang H, Huang H, Cui X (2012) Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J 69: 792–808 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H (2003) Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH (2008) betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang W, Wang H, Cai R, Xu Y, Huang H (2006) Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol Biol 61: 63–78 [DOI] [PubMed] [Google Scholar]
- Yao Y, Ling Q, Wang H, Huang H (2008) Ribosomal proteins promote leaf adaxial identity. Development 135: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Yin H, Zhang X, Liu J, Wang Y, He J, Yang T, Hong X, Yang Q, Gong Z (2009) Epigenetic regulation, somatic homologous recombination, and abscisis acid signaling are influenced by DNA Polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X (2010) Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J 64: 331–342 [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.