Abstract
3-Hydroxy-3-methylglutaryl-CoA reductase (HMGR) is an essential enzyme in the mevalonate pathway. In higher plants, mevalonate pathway involves in the production of precursor for isoprenoids biosynthesis, including essential components for cell functions. Previously, we confirmed that the Arabidopsis thaliana HMGR1S (AtHMGR1S) is phosphorylated at S577 by the combination of sucrose non-fermenting related kinase-1 (SnRK1) and geminivirus rep-interacting kinase-1 (GRIK1) in vitro. However, even in quantitative phosphoproteomics studies that were directed to find SnRK1 target substrates, AtHMGR1S phosphorylation at S577 has never been detected in planta. In this study, we expressed AtHMGR1S as a C-terminal FLAG-fusion protein in A. thaliana hmg1 mutant to confirm its phosphorylation in planta. Our results provide the first direct evidence that AtHMGR1S is phosphorylated at S577 in planta. Moreover, phosphatase inhibitors treatment to the A. thaliana seedlings induced AtHMGR1S phosphorylation at sites other than S577, suggesting the presence of a novel HMGR regulatory mechanism in planta.
Keywords: Arabidopsis thaliana, HMG-CoA reductase, phosphatase inhibitors, phosphorylation
Introduction
The enzyme 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase catalyzes the NADPH-mediated reaction of HMG-CoA to mevalonic acid as the first committed step of the mevalonate pathway. HMG-CoA reductase (HMGR) is essential in the mevalonate pathway, which is necessary for most eukaryotes to produce some indispensable cell components. In animals, HMGR is encoded by a single gene, and it is highly regulated to control sterol homeostasis for proper cell function (Espenshade and Hughes 2007; Friesen and Rodwell 2004). The molecular mechanisms of mammalian HMGR regulation have been well documented. Mammalian HMGR is regulated at the transcriptional level by binding proteins that bind to the sterol regulatory elements of the HMGR promoter and at the post-translational level by the insulin-induced gene, which leads to HMGR degradation through ubiquitination, and by the AMP-activated protein kinase (AMPK) cascade that leads to the direct inactivation of HMGR via phosphorylation at a conserved serine residue (Espenshade and Hughes 2007). In higher plants, mevalonate pathway involves in the production of precursor for isoprenoids biosynthesis such as sesquiterpenoids and triterpenoids, including essential components for cell functions. Plant HMGR is encoded by multiple genes, with expression induced by developmental and environmental stimuli.
Post-transcriptional feedback regulation of HMGR has been suggested by experiments using chemical inhibitors of the mevalonate pathway (Kobayashi et al. 2007; Nieto et al. 2009; Tang et al. 2010; Wentzinger et al. 2002). HMGR may be post-translationally regulated by interaction with protein phosphatase 2A (PP2A) (Leivar et al. 2011). Moreover, the Arabidopsis thaliana HMGR, which heterologously expressed in bacteria, is inactivated by the protein kinases purified from plant tissues through phosphorylation at the corresponding serine residue of the mammalian HMGR (Dale et al. 1995a; Sugden et al. 1999). Protein kinases were then suggested as the sucrose non-fermenting related kinase-1 (SnRK1), the homolog of mammalian AMPK, by biochemical and immunological experiments (Ball et al. 1995; Barker et al. 1996). In the case of A. thaliana, three different HMGR isoenzymes are encoded by two genes, HMG1 for both AtHMGR1L and AtHMGR1S, and HMG2 for AtHMGR2 (Enjuto et al. 1994; Lumbreras et al. 1995), and it has been reported that HMG1 plays important physiological roles for plant growth and development (Ohyama et al. 2007; Suzuki et al. 2004, 2009). We have also previously elucidated that AtHMGR1S is negatively regulated through phosphorylation by the AKIN10-GRIK1 kinase cascade system in vitro (Robertlee et al. 2017). Because AKIN10 is the ortholog of AMPK and the phosphorylated residue (S577) corresponds to the phosphorylated serine of mammalian HMGR by AMPK, this system resembles the regulatory system of mammal HMGR by the AMPK–AMPKK kinase cascade.
Despite much biochemical evidence of plant HMGR phosphorylation, plant HMGR phosphorylation at the conserved serine residue (corresponds to S577 in AtHMGR1S) has been undetected in plant phosphoproteomics analysis (Durek et al. 2010; Heazlewood et al. 2008; Heintz et al. 2012; Yao et al. 2012; Zulawski et al. 2013). It has also been undetected in quantitative phosphoproteomics studies that were directed to find SnRK1 kinase target substrates in planta (Cho et al. 2016; Nukarinen et al. 2016). In this study, we expressed AtHMGR1S as a C-terminal FLAG-fusion protein in A. thaliana hmg1 mutant to confirm its phosphorylation in planta. This study provides the first direct evidence that AtHMGR1S is phosphorylated at S577 in planta. In addition, the analysis of the phosphatase inhibitors cocktail treatment to the A. thaliana seedlings induced AtHMGR1S phosphorylation at sites other than S577, suggesting the presence of a novel HMGR regulatory mechanism in planta.
Materials and methods
Generation of 35S::HMG1S-FLAG in hmg1-1 and 35S::HMG1S_S577A-FLAG in hmg1-1 transgenic plants
The coding sequence of HMG1S was amplified from a cDNA clone of Arabidopsis thaliana (pda00683, RIKEN BRC) with modification of stop codon (TGA) to Leu (CTC) using 5′-TGG AGA GAT TAT TCA TTC CCT CCA ATG GA-3′ and 5′-GCT CTC GAG TGT TGT TGT TGT TGT CGT TG-3′. The amplified fragment was cloned into pCR-II TOPO (Invitrogen, Carlsbard, CA, USA). The coding sequence of HMG1S was reamplified with using 5′-TGG AGA GAT TAT TCA TTC CCT CCA ATG GA-3′ and 5′-ACG CCA AGC TAT TTA GGT GAC ACT ATA G-3′ from the resultant plasmid and cloned into pCR-II TOPO (Invitrogen). The resultant plasmid was designated as pCR-II/HMG1S. A FLAG fragment was generated by annealing with 5′-ACT CCC GGG CTC GAG GAT TAC AAG GAT GAT GAT GAT AAG TAA GAG CTC ACC-3′ and 5′-GGT GAG CTC TTA CTT ATC ATC ATC ATC CTT GTA ATC CTC GAG CCC GGG AGT-3′, and the fragment was cloned into pGEM-T easy (Promega, Madison, WI, USA). The resultant plasmid was designated as pGEM-T/FLAG. The FLAG fragment amplified using 5′-CCA GTC ACG ACG TTG TAA AAC GAC-3′ and 5′-TGA CTC TAG AGC TCT CCC ATA TGG TCG AC-3′ from pGEM-T/FLAG as a template and digested with XhoI and XbaI, then the digested fragment was cloned into XhoI and XbaI sites of pCR-II/HMG1S. The AtHMGR1S_S577A-FLAG mutant was prepared by two-step PCR using AtHMGR1S-FLAG as a template and using the forward primer 5′-GAA ATA CAA TAG AGC TAG CCG AGA CAT CTC-3′ and the reverse primer 5′-ATG TCT CGG CTA GCT CTA TTG TAT TTC ATG-3′ to change Ser577 (TCC) to Ala577 (GCT). Both AtHMGR1S-FLAG and AtHMGR1S_S577A-FLAG DNA fragments were cloned into pENTR/D-TOPO (Invitrogen). The resulting entry clones were integrated into the binary vector pBCR-79 (Seki et al. 2008) using the GATEWAY system (Invitrogen). The resulting constructs were introduced into Agrobacterium tumefaciens strain GV3101, followed by introduction into HMG1/hmg1-1 heterozygous plants (WS background) (Suzuki et al. 2009) using the floral dip method (Clough and Bent 1998). The transgenic plants with hmg1-1/ hmg1-1 homozygous background was screened by PCR using the forward primer 5′-ATG AAG AAA AAG CAA GCT GGT CC-3′ and the reverse primer 5′-CAT TTT ATA ATA ACG CTG CGG ACA TCT AC-3′ from genomic extracts as a template.
Plant growth conditions
A. thaliana seeds were surface sterilized (1% SDS and 0.25% NaClO) for 20 min with gentle shaking and then rinsed 5 times with sterilized MiliQ water (Millipore, Billerica, MA, USA). Seeds were placed on 1× concentration of Murashige and Skoog plant salt mixture (Wako, Osaka, Japan) plates containing 3% sucrose and 1.2% agar (Ina Food Industry, Nagano, Japan). After stratification at 4°C for at least 2 days, plates were vertically placed in a growth chamber (Sanyo MLR-351, 23°C) under long-day conditions (16 h under Panasonic FL40S·D fluorescent light with photosynthetic photon flux density 60–80 µmol M−2 s−1, 8 h dark) for 14 days.
Phosphatase inhibitors treatment
Transgenic A. thaliana seedlings (14 day old, approximately 24 seedlings) were soaked in 1 ml of liquid 1/2×Murashige and Skoog medium (Duchefa, Haarlem, Netherlands) containing 1% sucrose and 2× concentration of phosphatase inhibitors cocktail (PhosSTOP™, Roche, Basel, Switzerland) in the TubeSpin® Bioreactors 50 (TPP, Trasadingen, Switzerland). Plants were vacuum infiltrated (0.6 MPa) for 10 min and then incubated under light in the growth chamber for an additional 6 h.
Protein purification
Frozen tissues were grinded using Multi-Beads Shocker® [model MB1200PT (S), Yasui Kikai, Osaka, Japan]. Each 100 mg (fresh weight) tissue sample was homogenized in 200 µl homogenization buffer (250 mM sucrose, 100 mM tricine, 2 mM EDTA, 50 mM NaCl, pH 7.5) supplemented with 1% TritonX-100, 3× plant protease inhibitor cocktail (P9599, Sigma-Aldrich, St. Louis, MO, USA), 2× PhosSTOP™, and 10 mM DTT, followed by centrifugation at 12000×g for 10 min. FLAG-tagged proteins from the supernatant were purified using anti-FLAG-tag magnetic beads (MBL, Code No. 3343, Nagoya, Japan) according to the manufacturer’s recommendations. Bradford’s assay was used to determine the concentration of the protein using BSA as standard (Quick Start™, Bio-rad, CA, USA).
HMGR dephosphorylation and HMGR activity assay
The HMGR activity assay was performed on the same day as the FLAG-proteins purification. The HMGR dephosphorylation was performed by adding the premix reaction buffer that comprised of 1 mM MgCl2, 40 mM DTT, 1× lambda phosphatase buffer (P0753S, NEB, Ipswich, MA, USA), 1× plant protease inhibitor cocktail (Sigma-Aldrich), and the same volume of either 200 units lambda protein phosphatase (P0753S, NEB) or water, into 40 ng FLAG-purified HMGR. The reaction mixture was incubated at 30°C for 20 min. The HMGR activity assay was started by adding 20 µl of the premix reaction buffer containing 1 µg BSA, 1 mM NADPH, 10 mM DTT, and 1 µl of [glutaryl-3-14C] HMG-CoA (49.2 mCi mmol−1, NEC642010UC, PerkinElmer, Inc., MA, USA) to the whole resulting products of phosphatase assay to reach a final assay volume of 25 µl at 30°C for 25 min. The reaction was stopped, and the resulting mevalonate was lactonized and then extracted with ethyl acetate to measure the radioactivity by a liquid scintillation counter (LSC-5100, Aloka, Tokyo, Japan) as previously described (Robertlee et al. 2017). Two-tailed students t-test was used to analyze all the data using the statistical software StatPlus:mac (AnalystSoft Inc. Version v6. http://www.analystsoft.com/en/).
Phos-tag SDS-PAGE western blotting
We used Mn2+–Phos-tag® SDS-PAGE followed by western blotting with the anti-HMG1 catalytic domain antibody (Tang et al. 2010) to detect the presence of phosphorylated AtHMGR1S. The gel was made according to the Phos-tag manufacturer’s recommendations in a PAGE apparatus (ATTO, model AE-6401, Tokyo, Japan) with 30 µM Phos-tag, 8–9% w/v polyacrylamide separating gel (5.5 ml) and 4% w/v polyacrylamide stacking gel (2.5 ml). The phosphatase treatment for Phos-tag SDS-PAGE was similar as the phosphatase treatment for the HMGR activity assay, excluding DTT and protease inhibitors to avoid protein band distortion during electrophoresis. The phosphatase reaction was stopped by the addition of Laemmli’s sample buffer (Nacalai #30566-22, Kyoto, Japan), and then samples were boiled at 95°C for 4 min. Electrophoresis was performed at a constant current 26 mA h for approximately 2.5 h (ATTO, model AE-6530). The gel was treated using 10 mM EDTA (3×15 min) before transfer to polyvinylidene difluoride membranes (Milipore) using a semi-dry blotting apparatus at a constant voltage, 25 V, for 45 min (ATTO WSE-4020, Tokyo, Japan) with a three transfer buffers system (Kinoshita-Kikuta et al. 2014). The presence of the AtHMGR1S band on the membrane was then analyzed by western blot using anti-AtHMG1 catalytic domain (AtHMG1cd) antibody, as previously described (Robertlee et al. 2017).
Results
AtHMGR1S is phosphorylated at S577 in Arabidopsis thaliana under normal growth conditions
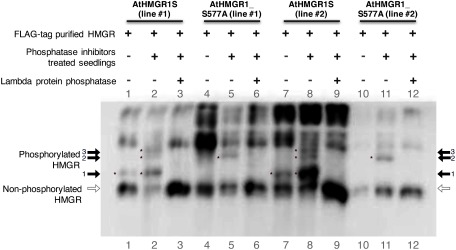
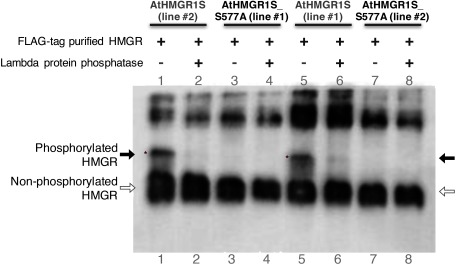
We have previously shown that AtHMGR1S is inactivated through phosphorylation at S577 by the AKIN10-GRIK1 kinase cascade system in vitro (Robertlee et al. 2017). To elucidate whether AtHMGR1S is phosphorylated at S577 in planta, we expressed either AtHMGR1S or AtHMGR1S_S577A as a C-terminal FLAG-fusion protein in the A. thaliana hmg1-1 mutant under 35S promoter. The hmg-1-1 mutant exhibits dwarfing, early senescence, male sterility, and reduced sterol levels under normal growth conditions; moreover, compared to wild type A. thaliana, it exhibits higher sensitivity to lovastatin (HMGR inhibitor) (Suzuki et al. 2004). The expression of either AtHMGR1S or AtHMGR1S_S577A suppressed the dwarf phenotypes of hmg1-1 mutant under normal growth condition (Supplementary Figure S1A–C); suppressed the male sterility phenotype because seeds of these transgenic lines were obtained from hmg1-1 homozygous background; and they also suppressed the lovastatin-sensitive phenotype of hmg1-1 mutant (Supplementary Figure S1D), suggesting that both of them are functional proteins in planta; therefore, these transgenic lines enabled us to confirm the AtHMGR1S phosphorylation at S577 in planta. western blotting analysis using an anti-HMGR1 antibody of crude cellular extracts also demonstrated that the expression of both AtHMGR1S or AtHMGR1S_S577A recovered the HMGR protein level in the hmg-1-1 background mutant (Supplementary Figure S1E). To obtain a clear separation between phosphorylated and non-phosphorylated AtHMGR1S using Phos-tag SDS-PAGE western blotting with an anti-HMGR1 antibody, we examined the phosphorylation level of the FLAG-tag purified proteins in this study. The protein band with reduced electrophoretic mobility was detected from the AtHMGR1S transgenic A. thaliana seedlings (Figure 1: lane 1 and 5). The lambda protein phosphatase treatment abolished the shifted band (Figure 1: lane 2 and 6), suggesting that it was derived from the phosphorylation of AtHMGR1S. There was no protein band with reduced electrophoretic mobility detected from the AtHMGR1S_S577A transgenic A. thaliana seedlings (Figure 1: lane 3 and 7), suggesting no phosphorylated form of AtHMGR1S_S577A was detected.
Figure 1. FLAG-tag purified AtHMGR1S is phosphorylated at S577. The same FLAG-purified samples used for the HMGR activity assay were treated with lambda protein phosphatase and then fractionated with Phos-tag SDS-PAGE, followed by western blotting analysis using an anti-HMG1cd antibody. Phosphorylated HMGR has reduced electrophoretic mobility (indicated by black arrow and asterisks) compared to the non-phosphorylated HMGR (indicated by blank arrow) in Phos-tag SDS-PAGE. The phosphatase treatment abolished the band with reduced electrophoretic mobility. Two transgenic lines are shown for each AtHMGR1S and AtHMGR1S_S577A indicated by line number.
Lambda phosphatase treatment increased the HMGR activity of the FLAG-tag purified AtHMGR1S
To confirm whether S577 phosphorylation of AtHMGR1S in planta leads to HMGR inactivation, we examined the HMGR activity assay of the FLAG-tag purified proteins after the lambda phosphatase assay. The phosphatase treatment of the AtHMGR1S increased the HMGR activity approximately 10.7% relative to the non-phosphatase treated control. On the contrary, the HMGR activity of the AtHMGR1S_S577A was not altered by phosphatase treatment (Figure 2).
Figure 2. Lambda phosphatase treatment increased the activity of the FLAG-tag purified AtHMGR1S. Transgenic A. thaliana seedlings (14 day old, 96 seedlings) harboring AtHMGR1S or AtHMGR1S_S577A were used for FLAG-tag protein purification. The HMGR activity of FLAG-tag purified proteins is shown relative to the control (not treated with lambda phosphatase) which indicated 100% for each of the AtHMGR1S_S577A and AtHMGR1S FLAG-tag purified proteins. Two transgenic lines are shown for each AtHMGR1S and AtHMGR1S_S577A indicated by line number. [A] The activity being 19.49 and 16.44 nmol mevalonate min−1 mg−1 protein for AtHMGR1S (line #1) and AtHMGR1S_S577A (line #1) respectively. The mean of relative activities and standard error of mean for at least five measurements, in two experiments, are shown. [B] The activity being 35.68 and 13.33 nmol mevalonate min−1 mg−1 protein for AtHMGR1S (line #2) and AtHMGR1S_S577A (line #2) respectively. The mean of relative activities and standard error of mean for three experiments by using one FLAG-tag purified protein are shown. Asterisks indicates significant difference (t-test, * p<0.005) compared to corresponding controls.
Phosphatase inhibitors treatment to A. thaliana seedlings induced the phosphorylation of AtHMGR1S at sites other than S577
We have previously shown that the combination of SnRK1 and GRIK1 phosphorylated AtHMGR1S at sites other than S577 in vitro (Robertlee et al. 2017). To determine whether HMGR can be phosphorylated at sites other than S577 in A. thaliana, we examined AtHMGR1S proteins in transgenic A. thaliana seedlings after treatment with the phosphatase inhibitors cocktail. Phos-tag SDS-PAGE western blot analysis was used with the anti-HMGR1 antibody to assess the phosphorylation status of the FLAG-tag purified proteins. A protein band with reduced electrophoretic mobility was detected from the AtHMGR1S transgenic seedlings untreated with phosphatase inhibitors (Figure 3: arrow 1, lane 1 and 7). However, two additional protein bands were detected from the AtHMGR1S transgenic seedlings treated with phosphatase inhibitors (Figure 3: arrows 2 and 3, lane 2 and 8). There was no band with reduced electrophoretic mobility detected from the untreated AtHMGR1S_S577A transgenic seedlings (Figure 3: lane 4 and 10). But an additional band was detected from AtHMGR1S_S577A transgenic seedlings treated with phosphatase inhibitor (Figure 3: arrow 2, lane. 5 and 11). Lambda phosphatase treatment of the FLAG-tag purified proteins abolished these shifted bands, resulting higher signal intensity of the dephosphorylated proteins (Figure 3: blank arrow, lanes 3, 6, 9, 12).
Figure 3. Phosphatase inhibitors treatment induced the phosphorylation of AtHMGR1S at sites other than S577. Transgenic A. thaliana seedlings (14 day old) were treated with phosphatase inhibitors cocktail and incubated for an additional 6 h. The seedlings were then used for FLAG-tag protein purification. FLAG-tag purified proteins were then fractionated with Phos-tag SDS-PAGE, followed by western blotting analysis using an anti-HMG1cd antibody. The phosphorylated HMGR has slower electrophoretic mobility (indicated by black arrow and asterisks) compared to the non-phosphorylated HMGR (indicated by blank arrow) in Phos-tag SDS-PAGE. There are three additional bands from AtHMGR1S (black arrow-1, 2, 3) and one additional band from AtHMGR1S_S577A FLAG-tag purified proteins (black arrow-2) from the seedlings treated with phosphatase inhibitors. The lambda phosphatase treatment abolished the additional bands of the FLAG-purified proteins. The Phos-tag SDS-PAGE of samples in lanes 1, 2, 10, and 11 was repeated without the addition of lambda phosphatase buffer to get sharper separation of protein bands and shown in Supplementary Figure S2.
Discussion
Plants produce diverse structures of isoprenoids through the mevalonate and methylerythritol 4-phosphate pathways. Enhancing the mevalonate pathway by increasing the mRNA level of HMGR has been one strategy to increase the production of specialized metabolites from economically important plants (Liao et al. 2016). The enhancement of downstream production by the mevalonate pathway such as phytosterols by removing the potential phosphorylation site to avoid negative regulation of the enzyme activity has also been an emerging strategy (Hey et al. 2006; Pütter et al. 2017). Although the negative regulation of HMGR through phosphorylation is likely to occur in planta, it has not been experimentally verified.
The data presented in this study strongly suggest that A. thaliana HMGR1 is phosphorylated in planta. First, there was a protein band in the FLAG-tag purified AtHMGR1S proteins with reduced electrophoretic mobility in the Phos-tag SDS-PAGE western blotting that was absent in the AtHMGR1S_S577A proteins, and this band disappeared after phosphatase treatment. Second, lambda phosphatase treatment of the FLAG-tag-purified AtHMGR1S proteins increased the HMGR activity (10.7%), but lambda phosphatase treatment did not affect the HMGR activity of the AtHMGR1S_S577A proteins. A plausible explanation for the slight increase in HMGR activity after lambda phosphatase treatment is the low concentration of phosphorylated AtHMGR1S compared to the non-phosphorylated counterpart (Figure 1). Our results confirmed that HMGR phosphorylation at S577 reduced the enzyme activity in vitro (Robertlee et al. 2017) and in planta. These data indicated that phosphorylation at S577 negatively regulates HMGR1 activity in A. thaliana. Third, AtHMGR1S is phosphorylated at sites other than S577 when the A. thaliana seedlings were treated with the phosphatase inhibitors cocktail, suggesting the presence of a novel HMGR regulatory mechanism in planta.
We have previously shown that AtHMGR1S_S577A is also phosphorylated by the combination of SnRK1 and GRIK1 in vitro (Robertlee et al. 2017). In an attempt to confirm AtHMGR1S phosphorylation at sites other than S577 in planta, we treated the transgenic A. thaliana seedlings with a phosphatase inhibitors cocktail. The Phos-tag SDS-PAGE western blotting results show three additional bands from AtHMGR1S and one additional band from the AtHMGR1S_S577A FLAG-tag purified proteins from phosphatase inhibitors treated seedlings. The lambda protein phosphatase treatment of the FLAG-tag purified AtHMGR1S and AtHMGR1S_S577A abolished these additional bands and increased the unshifted protein band signal intensity (Figure 3: blank arrow, lanes 3, 6, 9, and 12) compared to the non-phosphatase treated sample, suggesting that they were derived from the phosphorylated form of the FLAG-tag purified proteins. The band with reduced electrophoretic mobility from the untreated AtHMGR1S seedlings (Figure 3: arrow 1, lanes 1 and 7) was most probably phosphorylated at S577 because there was no detected protein band with a similar relative distance from the untreated AtHMGR1S_S577A seedlings (Figure 3: arrow 1, lanes 4 and 10). This protein band has a higher intensity in the sample treated with the phosphatase inhibitors (Figure 3: arrow 1, lanes 2 and 8) compared to that in the untreated (Figure 3: arrow-1 lane 1 and 7) seedlings, suggesting that S577 phosphorylation was highly accumulated in the phosphatase inhibitors treatment.
Considering the relative distance of the protein bands from the seedlings treated with phosphatase inhibitors, one additional shifted band of AtHMGR1S_S577A (Figure 3: arrow 2, lanes 5 and 11) corresponds to the same band in AtHMGR1S FLAG-tag purified proteins (Figure 3: arrow 2, lanes 2 and 8). Interestingly, the other band in AtHMGR1S (Figure 3: arrow 3, lanes 2 and 8) did not exist in AtHMGR1S_S577A. A protein containing multiple phosphorylation sites has slower electrophoresis mobility compared to the same protein containing a single phosphorylation site in Phos-tag SDS-PAGE. However, the same protein containing a single phosphorylation site at different sites may also demonstrate differential electrophoresis mobility in Phos-tag SDS-PAGE (Hosokawa et al. 2010). Taken together, our results offer two possibilities for AtHMGR1S phosphorylated isoforms. First, AtHMGR1S has an additional phosphorylation site in addition to S577. For this possibility, three bands correspond to the following state: 1) phosphorylated at S577 only (Figure 3: arrow 1), 2) phosphorylated at an unidentified phosphorylation site designated as X only (Figure 3: arrow 2), and 3) simultaneously phosphorylated at two phosphorylation sites (S577 and X; as shown in Figure 3: arrow 3). Therefore, for the state 3), AtHMGR1S_S577A was only phosphorylated at site X (Figure 3: arrow 2). The second possibility is that AtHMGR1S has three different phosphorylation sites, S577 and two other unidentified phosphorylation sites, designated as X and Y. In this case, three shifted bands may correspond to single, double, and triple phosphorylated AtHMGR1S or single phosphorylated AtHMGR1S at each different site. We could not exclude the possibility that these additional phosphorylation isoforms might be artifacts caused by the phosphatase inhibitors treatment, which does not mimic natural physiological conditions. However, these data suggested that HMGR is phosphorylated at sites other than S577 in planta. Other studies have suggested the presence of HMGR phosphorylation at sites other than S577 by showing the interaction of PP2A with the N-terminal region of HMGR in experiments using A. thaliana (Leivar et al. 2011). Moreover, AtHMGR1 and AtHMGR2 phosphorylation at two sites other than S577 were found in experiments, as annotated in PhosPhat 4.0 database (Durek et al. 2010; Heazlewood et al. 2008; Zulawski et al. 2013). AtHMGR1S phosphorylation at sites other than S577 might occur under specific circumstances. We would like to verify our hypothesis by utilizing other growth conditions in the near future.
We could also not confirm the protein kinases responsible for AtHMGR1S phosphorylation in this study. But our study and other studies have previously shown that plant HMGR isoforms are most probably regulated by S577 phosphorylation by a protein kinase cascade that resembles mammalian HMGR by in vitro experiments (Dale et al. 1995a, b; Robertlee et al. 2017; Sugden et al. 1999). Elucidation of the regulatory elements involved in the regulation of the mevalonate pathway is a complicated but a challenging research topic to increase the productivity of economically important isoprenoids in plants.
Acknowledgments
We are grateful to Dr. Hikaru Seki, Dr. Shuhei Yasumoto, and Ms. Pisanee Srisawat for helpful discussions. This study was supported by the Grant-in-Aid for Scientific Research no. JP15H04485 awarded to TM; and the Monbukagakusho Scholarship awarded to JR from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT).
Abbreviations
- HMG-CoA
3-hydroxy-3-methylglutaryl-CoA
- HMGR
HMG-CoA reductase
- AtHMGR1S
Arabidopsis thaliana HMGR1S
- AMPK
AMP-activated protein kinase
- AMPKK
AMPK kinase
- SNF1
sucrose non-fermenting-1
- SnRK1
sucrose non-fermenting related kinase-1
- GRIK1
geminivirus Rep-interacting kinase-1
- PP2A
protein phosphatase 2A
Supplementary Data
References
- Ball KL, Barker J, Halford NG, Hardie DG (1995) Immunological evidence that HMG-CoA reductase kinase-A is the cauliflower homologue of the RKIN1 subfamily of plant protein kinases. FEBS Lett 377: 189–192 [DOI] [PubMed] [Google Scholar]
- Barker JH, Slocombe SP, Ball KL, Hardie DG, Shewry PR, Halford NG (1996) Evidence that barley 3-hydroxy-3-methylglutaryl-coenzyme A reductase kinase is a member of the sucrose nonfermenting-1-related protein kinase family. Plant Physiol 112: 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Wen TN, Wang YT, Shih MC (2016) Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J Exp Bot 67: 2745–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dale S, Arró M, Becerra B, Morrice, NG, Boronat A, Hardie DG, Ferrer A (1995a) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 233: 506–513 [DOI] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG (1995b) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett 361: 191–195 [DOI] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX (2010) PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38(suppl_1): D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M, Balcells L, Campos N, Caelles C, Arro M, Boronat A (1994) Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA 91: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL (2007) Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 41: 401–427 [DOI] [PubMed] [Google Scholar]
- Friesen JA, Rodwell VW (2004) The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol 5: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX (2008) PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36(Database): D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz D, Gallien S, Compagnon V, Berna A, Suzuki M, Yoshida S, Muranaka T, Van Dorsselaer A, Schaeffer C, Bach TJ, et al. (2012) Phosphoproteome exploration reveals a reformatting of cellular processes in response to low sterol biosynthetic capacity in Arabidopsis. J Proteome Res 11: 1228–1239 [DOI] [PubMed] [Google Scholar]
- Hey SJ, Powers SJ, Beale MH, Hawkins ND, Ward JL, Halford NG (2006) Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol J 4: 219–229 [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Saito T, Asada A, Fukunaga K, Hisanaga S (2010) Quantitative measurement of in vivo phosphorylation states of Cdk5 activator p35 by Phos-tag SDS-PAGE. Mol Cell Proteomics 9: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E, Kinoshita E, Matsuda A, Koike T (2014) Tips on improving the efficiency of electrotransfer of target proteins from Phos-tag SDS-PAGE gel. Proteomics 14: 2437–2442 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki M, Tang J, Nagata N, Ohyama K, Seki H, Kiuchi R, Kaneko Y, Nakazawa M, Matsui M, et al. (2007) Lovastatin insensitive 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol 48: 322–331 [DOI] [PubMed] [Google Scholar]
- Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, Boronat A, Campos N (2011) Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P, Hemmerlin A, Bach TJ, Chye ML (2016) The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol Adv 34: 697–713 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Campos N, Boronat A (1995) The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J 8: 541–549 [DOI] [PubMed] [Google Scholar]
- Nieto B, Forés O, Arró M, Ferrer A (2009) Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry 70: 53–59 [DOI] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Suzuki M, Masuda K, Yoshida S, Muranaka T (2007) Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem Pharm Bull 55: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Pütter KM, van Deenen N, Unland K, Prüfer D, Schulze Gronover C (2017) Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biol 17: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertlee J, Kobayashi K, Suzuki M, Muranaka T (2017) AKIN10, a representative Arabidopsis SNF1-related protein kinase 1 (SnRK1), phosphorylates and downregulates plant HMG-CoA reductase. FEBS Lett 591: 1159–1166 [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice B-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kamide Y, Nagata N, Seki H, Ohyama K, Kato H, Masuda K, Sato S, Kato T, Tabata S, et al. (2004) Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J 37: 750–761 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nakagawa S, Kamide Y, Kobayashi K, Ohyama K, Hashinokuchi H, Kiuchi R, Saito K, Muranaka T, Nagata N (2009) Complete blockage of the mevalonate pathway results in male gametophyte lethality. J Exp Bot 60: 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Kobayashi K, Suzuki M, Matsumoto S, Muranaka T (2010) The mitochondrial PPR protein LOVASTATIN INSENSITIVE 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through RNA editing. Plant J 61: 456–466 [DOI] [PubMed] [Google Scholar]
- Wentzinger LF, Bach TJ, Hartmann M-A (2002) Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiol 130: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Bollinger C, Gao J, Xu D, Thelen JJ (2012) P3DB: An integrated database for plant protein phosphorylation. Front Plant Sci 3: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulawski M, Braginets R, Schulze W (2013) PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res 41(D1): D1176–D1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 2. Lambda phosphatase treatment increased the activity of the FLAG-tag purified AtHMGR1S. Transgenic A. thaliana seedlings (14 day old, 96 seedlings) harboring AtHMGR1S or AtHMGR1S_S577A were used for FLAG-tag protein purification. The HMGR activity of FLAG-tag purified proteins is shown relative to the control (not treated with lambda phosphatase) which indicated 100% for each of the AtHMGR1S_S577A and AtHMGR1S FLAG-tag purified proteins. Two transgenic lines are shown for each AtHMGR1S and AtHMGR1S_S577A indicated by line number. [A] The activity being 19.49 and 16.44 nmol mevalonate min−1 mg−1 protein for AtHMGR1S (line #1) and AtHMGR1S_S577A (line #1) respectively. The mean of relative activities and standard error of mean for at least five measurements, in two experiments, are shown. [B] The activity being 35.68 and 13.33 nmol mevalonate min−1 mg−1 protein for AtHMGR1S (line #2) and AtHMGR1S_S577A (line #2) respectively. The mean of relative activities and standard error of mean for three experiments by using one FLAG-tag purified protein are shown. Asterisks indicates significant difference (t-test, * p<0.005) compared to corresponding controls.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/6c23/6543733/1c7c37bd2ed6/plantbiotechnology-35-1-17.1208a-figure02.jpg)