Abstract
Human SCGB1A1 protein has been shown to be significantly reduced in BAL, sputum, and serum from humans with asthma as compared with healthy individuals. However, the mechanism of this reduction and its functional impact have not been entirely elucidated. By mining online datasets, we found that the mRNA of SCGB1A1 was significantly repressed in brushed human airway epithelial cells from individuals with asthma, and this repression appeared to be associated with reduced expression of FOXA2. Consistently, both Scgb1A1 and FoxA2 were downregulated in an ovalbumin-induced mouse model of asthma. Furthermore, compared with wild-type mice, Scgb1a1 knockout mice had increased airway hyperreactivity and inflammation when they were exposed to ovalbumin, confirming the antiinflammatory role of Scgb1a1 in protection against asthma phenotypes. To search for potential asthma-related stimuli of SCGB1A1 repression, we tested T-helper cell type 2 cytokines. Both IL-4 and IL-13 repressed epithelial expression of SCGB1A1 and FOXA2. Importantly, infection of epithelial cells with human rhinovirus similarly reduced expression of these two genes, which suggests that FOXA2 may be the common regulator of SCGB1A1. To establish the causal role of reduced FOXA2 in SCGB1A1 repression, we demonstrated that FOXA2 was required for SCGB1A1 expression at baseline. FOXA2 overexpression was sufficient to drive promoter activity and expression of SCGB1A1 and was also able to restore the repressed SCGB1A1 expression in IL-13–treated or rhinovirus-infected cells. Taken together, these findings suggest that low levels of epithelial SCGB1A1 in asthma are caused by reduced FOXA2 expression.
Keywords: rhinovirus, asthma, CC10, FOXA2, secretoglobin
Clinical Relevance
SCGB1A1 was found to be significantly reduced in asthmatic airways, likely by the action of T-helper cell type 2 cytokines and/or rhinovirus infection. At the molecular level, this reduction was mediated by the downregulation of FOXA2, an essential transcription factor for SCGB1A1 expression.
Club cell secretory protein (CC16, also known as CC10, CCSP, or uteroglobin) is a homodimeric pneumoprotein encoded by SCGB1A1 (secretoglobin family 1A member 1) (1, 2). SCGB1A1 is mainly expressed by nonciliated respiratory epithelial cells and accounts for up to 5% of the total protein in BAL fluid (3, 4). SCGB1A1 can also be detected in sputum and in the systematic circulation (1, 2, 5). Although the precise molecular mechanism through which SCGB1A1 elicits its function is unclear, SCGB1A1 has demonstrated potent antiinflammatory, antitumor, and antitoxicant functions (1, 2, 5) via the inhibition of PLA2 activity (6), proinflammatory prostaglandins (7, 8), chemotaxis (9–13), or cytokine production (14, 15).
Low levels of SCGB1A1 in the circulation and in BAL fluid have been consistently linked to lung function deficits in chronic obstructive pulmonary disease (COPD) (16–25). Along this line, SCGB1A1 has been shown to attenuate airway inflammation and delay lung function decline induced by smoking (26, 27), both of which are major pathogenic markers of COPD. Lower levels of SCGB1A1 were also detected in BAL (28–30), serum (20, 28; 31–33), and urine (34) samples from individuals with asthma as compared with healthy control subjects. Interestingly, a genetic polymorphism, A38G (rs3741240), at 38 bp downstream of the transcription start site of SCGB1A1 has been shown to be a potential risk factor for the development and severity of asthma (35–40). The A allele is also associated with reduced levels of SCGB1A1 in BAL (41) or in the circulation (40), implying that low levels of SCGB1A1 protein in different body compartments of individuals with asthma may originate from decreased cellular SCGB1A1 production. However, the underlying molecular mechanism for this decreased production is unclear.
In this study, we tested the hypothesis that lower levels of SCGB1A1 in individuals with asthma compared with normal subjects are regulated at the transcriptional level by FOXA2. Additionally, we sought to determine whether a reduction in SCGB1A1 exacerbates airway inflammation and hyperreactivity in asthma.
Methods
Gene Expression Omnibus Database and Meta-Analysis
The National Institutes of Health Gene Expression Omnibus database and ArrayExpress program were used for this study, and all available datasets published before November 1, 2017 were included for the initial search. The studies were restricted for samples from adult human bronchial epithelium isolated from both subjects with severe asthma and healthy control subjects. Three studies (GSE43696, GSE63142, and GSE89809) were retrieved (a summary of the sample sizes for these studies is provided in Table 1.
Table 1.
Summary of the GEO Datasets Used for the Meta-Analysis
All programming and data analyses were performed using the R package. Data were preprocessed by the original authors using either the robust multi-array average method or cyclic-loess normalization. The genes that had symbol IDs and were expressed across all three studies were kept for further analyses (17,439 genes). For each gene in each study, a simple linear regression model was fitted and the effect size (log2 difference in expression between subjects with asthma and healthy control subjects) and its SE were calculated. Then a meta-analysis was conducted by using “metaphor” in the R package via a linear mixed-effect model with the previous effect size and corresponding SE obtained from each individual study (42). Finally, the P values were adjusted with the Benjamini-Hochberg procedure (43) to control the false-discovery rate (43).
Ovalbumin-induced Mouse Model of Asthma, Differential Cell Count, and Lung Function Measurement
Scgb1a1 knockout (KO) mice on a C57/B6 background (44) were obtained from the National Institute of Cancer and bred in the University of Arizona animal facility according to an approved institutional animal care and use committee protocol. Wild-type (WT) mice (C57/B6) were purchased from the Jackson Laboratory. All mice were housed in the animal facility at the University of Arizona in an air-conditioned room with a 12-hour light/dark cycle and were used at 8–10 weeks of age. An ovalbumin (OVA)-induced mouse model of asthma was established as previously described (45). Briefly, mice received an intraperitoneal injection of 10 μg of alum-precipitated chicken egg OVA (Sigma) 21 days before inhalational exposure and a booster injection 7 days before being intranasally exposed to OVA solution at 100 μg/ml every other day three times. The control mice were exposed to PBS. One day after exposure, the mice were anesthetized and their tracheas were isolated, cannulated, and connected to a small-animal ventilator. Changes in lung resistance in response to increasing doses of methacholine were directly assessed by using the flexiVent apparatus (SCIREQ) (46). After lung function measurements were obtained, the mice were killed and three successive volumes of 0.55 ml of PBS with 0.1% BSA were then instilled and gently aspirated to obtain BAL. All BAL samples were aliquoted and stored in three bar-coded Eppendorf tubes. The cells in the BAL were cytocentrifuged, air dried, and stained with a HEMA 3 stain set (Thermo Scientific), and the numbers of macrophages, neutrophils, eosinophils, and lymphocytes were then counted in a blinded manner using light microscopy by at least two researchers to ensure an objective evaluation. Differential cell counts (macrophages, neutrophils, eosinophils, and lymphocytes) were presented as lavaged cells/lung.
Cell Culture, Cytokine Treatment, and Rhinovirus Infection
Human bronchial tissues were obtained from the National Disease Research Interchange according to an approved protocol. Primary human bronchial epithelial cells (HBECs) were cultivated as described previously (47) in Ham’s F12:Dulbecco’s modified Eagle medium (1:1) supplemented with eight factors: insulin (5 mg/ml), transferrin (5 mg/ml), epidermal growth factor (10 ng/ml), dexamethasone (0.1 mM), cholera toxin (10 ng/ml), bovine hypothalamus extract (15 mg/ml), BSA (0.5 mg/ml), and all-trans-retinoic acid (30 nM). IL-4 (10 ng/ml), IL-13 (10 ng/ml) (R&D Systems), or PBS (solvent) control was used to treat the cells for 72 hours. Rhinovirus 16 (RV16) stock was amplified and purified based on the previously published protocol (48). The RV infection protocol was performed as described previously (47). Briefly, RV16 was directly diluted into culture media at a multiplicity of infection of 10 and incubated for 24 hours. The control was purified product from the media that was collected from sham-infected cells and subjected to the same purification steps as the RV. After infection, HBECs were washed and incubated in fresh media or conditioned media at 35°C for 24 hours. At the time of sample collection, the cells were washed three times in PBS to remove viral particles in the media. The lack of viruses was confirmed by PCR assay of the final wash as described previously (47).
RNA Extraction, cDNA Synthesis, and Real-Time qPCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen). cDNA was prepared from 2 mg of total RNA with Moloney murine leukemia virus (MoMLV)–reverse transcriptase (Promega, Inc.) by oligo-dT primers for 90 minutes at 42°C in a 20-ml reaction solution, and was then further diluted to 100 μl with water for the following procedures: 2 μl of diluted cDNA was analyzed using the SYBR Green PCR Master Mix in a Veriti Thermal Cycler (Thermo Fisher) according to the manufacturer’s protocol. Primers (Table 2) were used at 0.2 mM. The relative amount of mRNA in each sample was calculated based on the ΔΔCt method using the housekeeping gene GAPDH or actin. The purity of the amplified product was determined from a single peak of a dissociation curve. Results were calculated as fold induction over control (49).
Table 2.
PCR Primers
| Gene | Primer |
|---|---|
| GAPDH, human | Forward CAATGACCCCTTCATTGACC |
| Reverse GACAAGCTTCCCGTTCTCAG | |
| SCGB1A1, human | Forward CAAAAGCCCAGAGAAAGCATC |
| Reverse CAGTTGGGGATCTTCAGCTTC | |
| MUC5AC, human | Forward GCCTTCACTGTACTGGCTGAG |
| Reverse TGGGTGTAGATCTGGTTCAGG | |
| FOXA2, human | Forward CGACTGGAGCAGCTACTATGC |
| Reverse ATGTACGTGTTCATGCCGTTC | |
| FOXA3, human | Forward GGCAAGATGCTGACCTTGAGT |
| Reverse TTGACGAAGCAGTCGTTGAAA | |
| ACTIN, mouse | Forward ACCGTGAAAAGATGACCCAGA |
| Reverse GGAGTCCATCACAATGCCTGT | |
| Scgb1a1, mouse | Forward CAGAGTCTGGTTATGTGGCATC |
| Reverse TAGGATTTTCTCCGTGAGCTTC | |
| Muc5ac, mouse | Forward CCATGCAGAGTCCTCAGAACA |
| Reverse TTACTGGAAAGGCCCAAGCA | |
| Foxa2, mouse | Forward CGACTGGAGCAGCTACTACGC |
| Reverse ATGTGTTCATGCCATTCATCC | |
| Foxa3, mouse | Forward GGCGAGGTGTATTCTCCAGTG |
| Reverse AGGGTAGGGAGAGCTGAGTGG |
Western Blot Analysis
Total cellular protein was collected from HBECs, fractionated on an SDS-PAGE gel, and transferred to a nitrocellulose membrane. The membrane was then incubated with anti-FOXA2 antibody (Santa Cruz Biotechnology) to quantify the protein expression of FOXA2. Equal protein load was confirmed using the staining of anti-actin antibody (Santa Cruz Biotechnology).
Immunofluorescence
For mouse tissue staining, the tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Sections were incubated with anti-MUC5AC antibody (Thermo Scientific) or anti-SCGB1A1 antibody (R&D Systems) overnight at 4°C. For cell staining, HBECs were fixed with 4% paraformaldehyde. Then, the fixed cells were permeated with Triton X-100. Intracellular MUC5AC or SCGB1A1 protein was determined by immunofluorescence staining using anti-MUC5AC or anti-SCGB1A1 antibody, respectively. Then, Alexa 488 (green)– or Alex 594 (red)–conjugated secondary antibody (Thermo Fisher) was used to obtain fluorescence images. The images were acquired by confocal microscopy (LSM 510 Meta; Carl Zeiss). For quantification, images from five random view areas were taken for each biological replicate. Positively stained areas were measured using ImageJ (https://imagej.nih.gov/ij/).
Transfection of HBECs with Electroporation
For transfection of HBECs, 951 bp of WT human SCGB1A1 promoter (−945 to +6) was amplified and cloned into a pGL3 luciferase reporter vector (Promega). A putative FOXA2 binding site (−193 to −157) was predicted based on previously described methods (50, 51) and a mutated SCGB1A1 promoter (mutant) luciferase reporter was created by deleting this site. The entire open reading frame of human FOXA2 was cloned into pcDNA3.1 (Promega). Transfection of human HBECs was performed using the Lonza 4D-Nucleofector system. HBECS were pelleted (200 g, 2 min) and resuspended in 100 μl of prewarmed electroporation buffer. The Lonza P3 Primary Cell 4D-Nucleofector X Kit was used according to the manufacturer’s instructions. For the promoter assay, we used 3 μg of SCGB1A1 promoter (WT or mutant) reporter gene, 3 μg of FOXA2, and 0.6 μg of Renilla luciferase control reporter. For FOXA2 overexpression, we used 3 μg of FOXA2. After transfection, the HBECS were resuspended and seeded at 37°C and 5% CO2 for further analysis.
siRNA
An siRNA sequence (CCATGAACATGTCGTCGTA) against FOXA2 was synthesized by Sigma-Aldrich. siRNA was transfected into the cells using the Lipofectamine 2000 (Invitrogen) according to the manufacturer’s suggested protocol.
Statistical Analysis for Experimental Samples
Experimental groups were compared using a two-sided Student’s t test, with the significance level set at P < 0.05. When the data were not distributed normally, significance was assessed with the Wilcoxon rank-sum test, and P < 0.05 was considered significant.
Results
SCGB1A1 Expression Was Reduced in Epithelial Cells from Subjects with Asthma
We hypothesized that low levels of SCGB1A1 in BAL from individuals with asthma may be caused by reduced transcription of the SCGB1A1 gene. Although no report has been published specifically about SCGB1A1 gene expression in individuals with asthma, the GEO database contains data from several well-controlled studies on epithelial gene profiling in human asthma. Thus, reanalyzing these datasets, instead of directly collecting clinical samples and measuring SCGB1A1 transcripts, may be a more efficient approach. In this analysis, we focused on gene expression profiles from brushed HBECs. Some other studies using samples from BAL cells or biopsies were skipped because of the significant contamination from nonepithelial cell types. Three datasets were identified from the GEO database and subjected to reanalysis and later meta-analysis (with adjusted P < 0.01 as the cutoff). As shown in Table 3, SCGB1A1 mRNA expression was significantly reduced by ∼53% (effect size: −1.10 at log2 scale) in epithelial cells from subjects with asthma as compared with those from healthy control subjects. In contrast, the major airway epithelial mucin gene MUC5AC was highly elevated by approximately twofold (effect size: −1.00 at log2 scale).
Table 3.
Differentially Expressed Genes from the Meta-Analysis
| Gene Symbol | Effect Size* | Adjusted P Value† |
|---|---|---|
| SCGB1A1 | −1.10 | 9.24E−07 |
| MUC5AC | 1.00 | 2.18E−07 |
| FOXA2 | −0.56 | 8.37E−10 |
| FOXA3 | 0.46 | 1.67E−04 |
| FOXA1 | −0.12 | 0.06 |
| CEBPB | −0.14 | 0.07 |
| CEBPA | −0.07 | 0.45 |
| SPDEF | 0.09 | 0.55 |
| CEBPD | 0.03 | 0.76 |
| TTF1 | −0.018 | 0.82 |
Log2 scale.
P value adjusted by the Benjamini-Hochberg procedure (false discovery rate), with P < 0.01 considered significant (marked in bold typeface).
Decreased steady-state mRNA levels often indicate reduced transcription. A few transcription factors (TFs) (e.g., FOXA1, A2, A3, SPDEF, TTF1/NKX2-1, CEBPA, CEBPB, and CEBPD) were previously reported to be involved in the transcriptional control of SCGB1A1 in a H441 cell line (52, 53). Thus, we decided to test these TFs by meta-analyses (adjusted P < 0.01 as cutoff) of the same datasets. Only FOXA2 (decreased by ∼32%) and FOXA3 (increased by ∼38%) were differentially expressed between individuals with asthma and healthy control subjects (Table 3). The positive correlation between FOXA3 and MUC5AC is consistent with a previous report that FOXA3 positively regulated mucin and mucous cell metaplasia in response to T-helper cell type 2 (Th2) cytokines (54). Likewise, the positive correlation between FOXA2 and SCGB1A1 implies that FOXA2 may positively regulate SCGB1A1, and the reduction of FOXA2 in asthma may be responsible for decreased SCGB1A1 expression.
Scgb1a1 Was Downregulated in the Mouse Model of Asthma, and Lack of SCGB1A1 Was Associated with an Exacerbated Asthmatic Phenotype
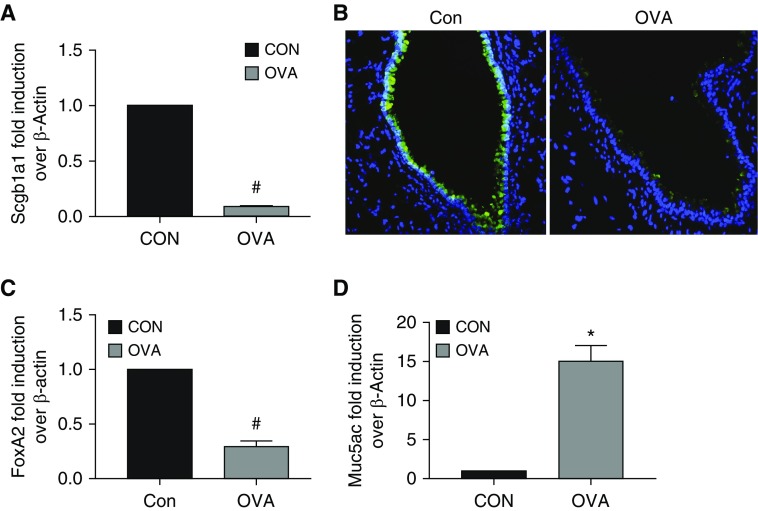
To further demonstrate the causality between asthmatic phenotypes and Scgb1a1 reduction, we used an OVA-induced mouse model of asthma. Indeed, we found that Scgb1a1 mRNA (Figure 1A) and protein (immunofluorescence staining shown in Figure 1B) were significantly reduced in the mice challenged with OVA as compared with the control mice. Additionally, FoxA2 expression was decreased (Figure 1C) and Muc5ac expression was increased (Figure 1D) in OVA-treated mice. These data were consistent with the findings in human asthma (Table 3).
Figure 1.
Differential gene expression in an ovalbumin (OVA)-induced mouse model of asthma. Mice were challenged with OVA or PBS control. As described in Methods, after completion of the experiment, the mice were killed and lung tissues were harvested. Total RNA was extracted and subjected to qPCR analysis. Lung tissues were also preserved, fixed, and subjected to immunofluorescence staining. (A) Scgb1a1 mRNA was measured by qPCR analysis in mouse lung. (B) Scgb1a1 protein was measured by immunofluorescence in mouse airway tissue sections using a specific anti-Scgb1a1 antibody (green). Blue: DAPI staining for the cell nuclei. (C) FoxA2 mRNA was measured by qPCR analysis in mouse lung. (D) Muc5ac mRNA was measured by qPCR analysis in mouse lung. Data shown are mean ± SEM; *P < 0.05 and #P < 0.05; OVA versus control (Con); n = 6.
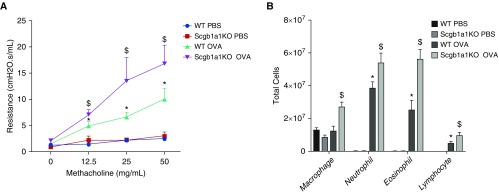
To further characterize the impact of Scgb1a1 deficiency in the lung, we tested Scgb1a1-KO mice. Wang and colleagues previously studied a different line of Scgb1a1-KO mice on a 129J background in an OVA model (55). However, the 129J mouse strain is resistant to OVA challenge and has limited airway hyperreactivity (AHR), which is characteristic of asthma, in response to OVA exposure (56). Accordingly, the AHR in the study by Wang and colleagues was fairly moderate, as the authors pointed out (55). Thus, we decided to retest the OVA challenge in our KO model with the OVA-susceptible C57/B6 strain (56). Indeed, OVA induced robust increases of AHR in response to increasing doses of methacholine, and the lack of Scgb1a1 exacerbated AHR (Figure 2A). Additionally, OVA challenge significantly enhanced airway inflammation, as demonstrated by increased infiltration of neutrophils, eosinophils, and lymphocytes in WT mice, and the number of inflammatory cells was further increased in the KO mice (Figure 2B). Thus, Scgb1a1 appears to play an antiinflammatory role in defending against asthma pathogenesis.
Figure 2.
Exacerbated inflammation and airway hyperreactivity in Scgb1a1 knockout (KO) mice. Both wild-type (WT) and Scgb1a1 KO mice were challenged with OVA or with PBS control. (A) Lung resistance in response to increased doses of methacholine challenge was measured using the flexiVent system. (B) A differential cell count was performed to quantify BAL cells. *P < 0.05 WT OVA versus WT PBS. $P < 0.05 Scgb1a1 KO OVA versus WT OVA. n = 6.
SCGB1A1 Was Downregulated by Treatment with Th2 Cytokines
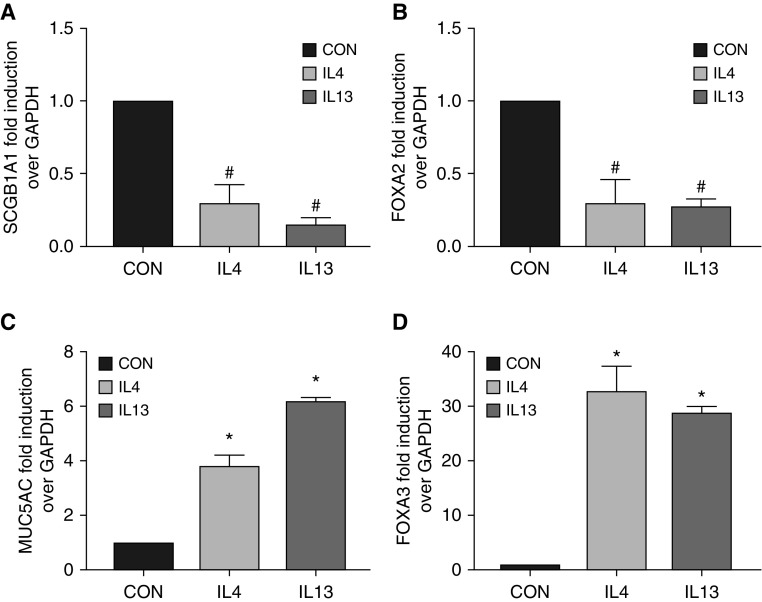
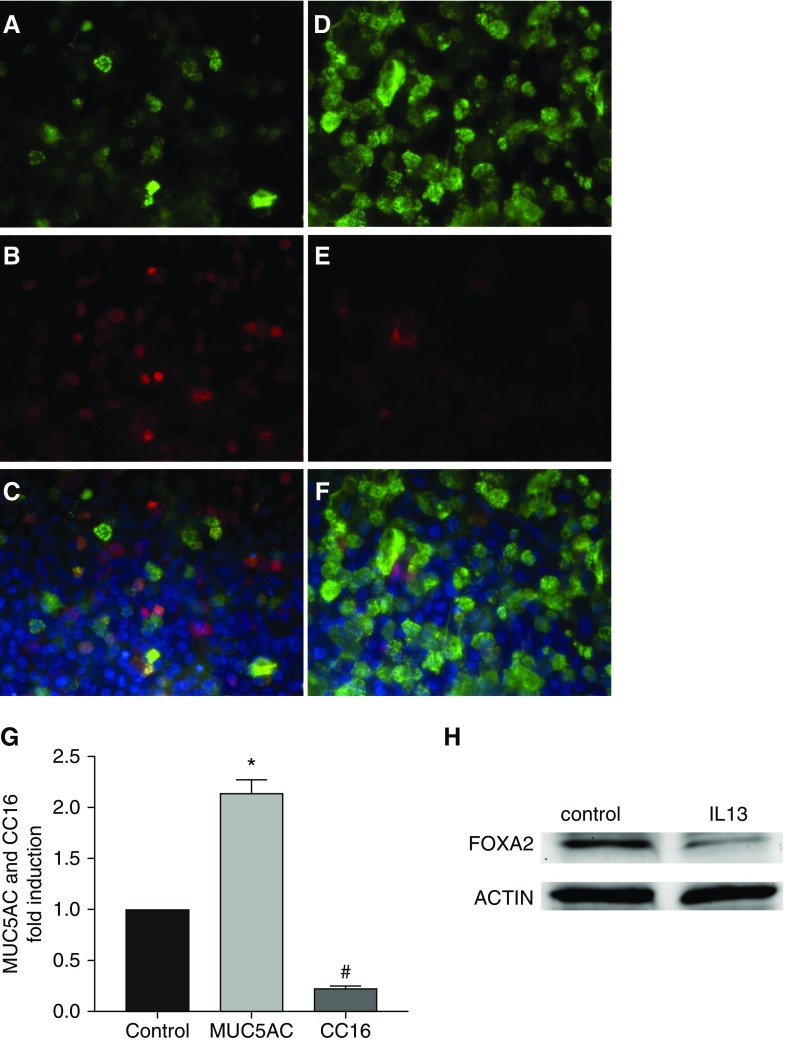
Because one of major characteristics of asthma is a Th2-dominant inflammatory response, we tested whether hallmark Th2 cytokines (e.g., IL-4 and IL-13) could repress SCGB1A1. Indeed, treatment with IL-4 or IL-13 downregulated mRNAs of SCGB1A1 (Figure 3A) and FOXA2 (Figure 3B) in primary HBECs. In contrast, the same treatments significantly elevated the mRNAs of MUC5AC (Figure 3C) and FOXA3 (Figure 3D). These results were consistent with the findings in human asthma (Table 3) and in our mouse model (Figure 1). To confirm the mRNA data in Figure 3, we further measured protein production in HBECs after IL-13 treatment. IL-13 treatment greatly reduced the protein production of SCGB1A1 (immunofluorescence staining shown in Figures 4B, 4E, and 4G) and FOXA2 (Western blot analysis shown in Figure 4H), whereas MUC5AC protein was increased under the same circumstance (Figures 4A, 4D, and 4G). Interestingly, SCGB1A1 and MUC5AC were coexpressed in a small set of epithelial cells (shown as yellow staining in Figures 4C and 4F). These data strongly suggest that Th2 cytokines, which are abundantly present in asthmatic airways, are responsible for reduced expression of SCGB1A1.
Figure 3.
Differential epithelial gene expression in response to IL-4 or IL-13 treatment. Human bronchial epithelial cells (HBECs) were treated with IL-4, IL-13, or solvent Con for 3 days. Total RNA was collected and subjected to qPCR analysis for (A) SCGB1A1, (B) FOXA2, (C) MUC5AC, and (D) FOXA3. Data shown are mean ± SD; *P < 0.05 and #P < 0.05; treatment versus Con; n = 4.
Figure 4.
Differential epithelial protein production in response to IL-13 treatment. (A–F) HBECs were treated with IL-13 (D–F) or solvent control (A–C) for 3 days. Cells were fixed and subjected to immunofluorescence imaging for MUC5AC (A and D, green) and SCGB1A1 (B and E, red). C and F are merged images. Blue: nuclear staining with DAPI. (G) Image quantification from six biological replicates of MUC5AC (A and D) or SCGB1A1 (B and E) expression in response to IL-13 treatment. *P < 0.05 and #P < 0.05; n = 6. (H) Representative image from Western blot analysis of FOXA2 protein after IL-13 treatment. Actin was used as a loading control.
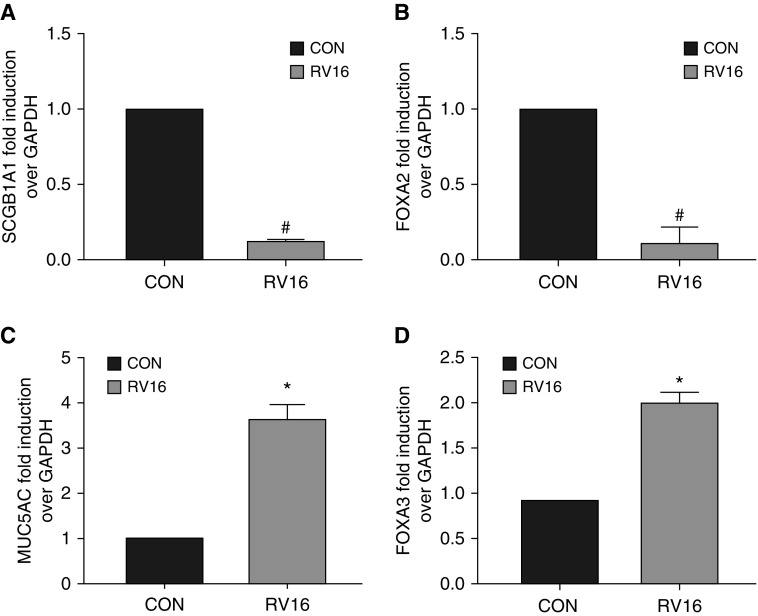
SCGB1A1 Was Downregulated by Human RV
RV infection causes asthma exacerbations and severe asthma. Because the samples in our meta-analysis were collected from subjects with severe asthma, we tested the impact of RV infection on SCGB1A1. RV infection of HBECs caused severe repression of SCGB1A1 (Figure 5A) and FOXA2 expression (Figure 5B), but it increased MUC5AC (Figure 5C) and FOXA3 (Figure 5D). In this system, no Th2 cytokines were detected (data not shown). Thus, RV infection repressed SCGB1A1 expression independently of Th2 cytokines, and likely through a common mechanism with Th2 cytokines by FOXA2 inhibition (Figures 3B and 4H).
Figure 5.
Differential epithelial gene expression in response to rhinovirus 16 (RV16) infection. HBECs were infected with RV16 or sham Con for 24 hours. Total RNA was collected and subjected to qPCR analysis for (A) SCGB1A1, (B) FOXA2, (C) MUC5AC, and (D) FOXA3. Data shown are mean ± SD. *P < 0.05 and #P < 0.05; RV versus Con; n = 4.
FOXA2 Is the Key TF that Drives SCGB1A1 Expression
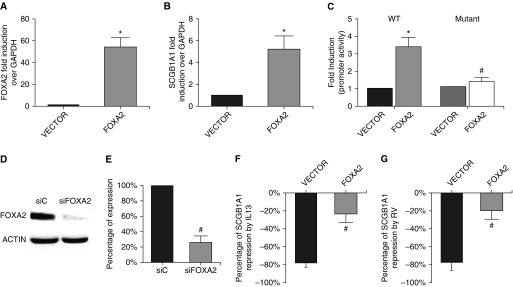
Considering the remarkably correlated findings of FOXA2 and SCGB1A1 expression in three separate models (human asthma, mouse OVA model, and primary cell culture), we suspected that the reduction of FOXA2 in these systems was likely responsible for the decreased expression of SCGB1A1. Because previous studies of transcriptional control of SCGB1A1 were based on artificial promoter constructs in cell lines (52, 53), it is still unknown whether FOXA2 is able to drive SCGB1A1 gene expression in primary human airway epithelial cells. By using a highly efficient electroporation system, we were able to induce high levels of FOXA2 expression in HBECs (Figure 6A). The increase in FOXA2 increased endogenous mRNA abundance by more than fivefold (Figure 6B) and WT SCGB1A1 promoter activity by more than threefold (Figure 6C). Importantly, FOXA2 overexpression failed to stimulate a mutant SCGB1A1 promoter without a FOXA2 binding site (Figure 6C), confirming the role of FOXA2 as a key TF in driving SCGB1A1 gene expression. Because SCGB1A1 is the hallmark of club cells, we tested another club cell marker, CYP2F2, and found that it was not affected by FOXA2 overexpression (data not shown). Furthermore, siRNA knockdown of FOXA2 (Figure 6D) markedly reduced the baseline expression of SCGB1A1 to <26% of that of the control (Figure 6E). Additionally, we performed rescue experiments on IL-13– or RV-induced SCGB1A1 repression. FOXA2 overexpression overrode IL-13–induced repression of SCGB1A1 by lowering the repression percentage from a substantial −78% to a mere −24% (Figure 6F). Likewise, FOXA2 overexpression also rescued RV16-induced repression of SCGB1A1 by lowering the repression percentage from a significant −86% to −20% (Figure 6G). Taken together, these findings indicate that the reduced expression of FOXA2 was causally responsible for the low level of SCGB1A1 in asthma.
Figure 6.
Genetic manipulation of FOXA2 altered SCGB1A1 expression. (A) FOXA2 expression plasmid or its vector control was delivered into HBECs via electroporation, and the cells were then analyzed for FOXA2 expression by qPCR. (B) Endogenous SCGB1A1 expression was measured in the cells with FOXA2 overexpression or with vector control. (C) WT or mutated (mutant) SCGB1A1 promoter luciferase reporter was delivered into HBECs via electroporation with FOXA2 or with its vector control. Promoter activity was analyzed by luciferase assay. *P < 0.05. Cells electroporated with WT promoter + FOXA2 versus cells electroporated with WT promoter + vector. n = 4. #P < 0.05. Cells electroporated with WT promoter + FOXA2 versus cells electroporated with mutant promoter + FOXA2. n = 4. (D) Western blot analysis of FOXA2 expression in the cells transfected with control siRNA (siC) or siRNA against FOXA2 (siFOXA2). (E) Endogenous SCGB1A1 expression was measured by real-time PCR; #P < 0.05; cells transfected with siFOXA2 versus cells transfected with siC; n = 4. (F) HBECs with FOXA2 overexpression or vector control were treated with or without IL-13. The percentage of repression was calculated as the magnitude of IL-13–induced SCGB1A1 repression. (G) HBECs with FOXA2 overexpression or vector control were infected with or without RV. The percentage of repression was calculated as the magnitude of RV-induced SCGB1A1 repression. Data shown are mean ± SD; #P < 0.05; cells electroporated with FOXA2 versus cells electroporated with the vector; n = 4.
Discussion
Circulating SCGB1A1 has frequently been used as a biomarker to monitor lung injury caused by various diseases or environmental exposures (1, 2, 5). Paradoxically, decreases, rather than increases, of SCGB1A1 have been consistently observed to associate with chronic airway diseases such as COPD (16–25) and asthma (20, 28; 31–33). These findings do not support the previous notion that circulating SCGB1A1 is part of leaked proteins from injured lungs, as this scenario predicts increases, rather than decreases, of circulating SCGB1A1. The inverse relationship between the level of SCGB1A1 and COPD severity suggests that SCGB1A1 may be protective against COPD pathogenesis, and this notion was recently supported by two related studies using animal models of smoke-induced COPD (26, 27). In asthmatic airways, the proportion of SCGB1A1-positive epithelial cells was found to be reduced (32, 57). Thus, either outright cell death of SCGB1A1-expressing cells or downregulation of epithelial SCGB1A1 protein, and therefore less cellular staining, may be the culprit in this decrease. Excessive epithelial cell death (e.g., apoptosis and pyroptosis) has been proposed to be responsible for asthma pathogenesis by remodeling the airway and leading to clinical symptoms such as reversible expiratory airflow limitation and AHR (58, 59). However, the nondiscriminatory nature of cell death cannot account for the preferential reduction of SCGB1A1-expressing cells. Thus, cellular SCGB1A1 downregulation is most likely responsible for this reduction in asthmatic airways. In this study, for the first time, we provide evidence that the decrease of SCGB1A1 transcripts (i.e., mRNA) is responsible for its protein reduction. This phenomenon was seen in three independent models (human subjects, an in vivo animal model, and an in vitro primary cell culture model). Steady-state mRNA is usually controlled by a balance of transcription and mRNA stability. We cannot completely exclude the possibility that decreased mRNA stability may also contribute to the decrease of SCGB1A1 transcripts. However, the reduction of transcription is at least partly responsible, as FOXA2, a key TF, was found to be essential for the transcription of SCGB1A1, and FOXA2 expression was also reduced in asthma.
Among numerous pathogenic components for asthma, we tested two major players—Th2 cytokines and RV infection—for their effects on SCGB1A1 reduction. Th2 cytokines are classical players in regulating the “Th2-high” asthma endotype (60, 61), and human RV is the major contributor to asthma exacerbations and severe asthma (62, 63). We have demonstrated the causal link between Th2 cytokines/RV and SCGB1A1 reduction in asthmatic airways. Although the downstream signaling pathways induced by Th2 cytokines (mainly JAK-STAT6) (64) and by RV infection (mainly IFN-STAT1) (65) are largely nonoverlapping, their effects on SCGB1A1 reduction appear to converge on downregulation of the common TF, FOXA2. Our findings are similar to a previous report demonstrating FOXA2 as the common regulator of human mucin gene stimulation by Th2 cytokine treatment or by epidermal growth factor receptor activation (66). The difference between this study and the previous one is that we found that FOXA2 was a negative regulator of human mucin genes but a positive regulator of SCGB1A1.
FOXA2 belongs to the forkhead box family of TFs, the dysregulation of which is associated with respiratory illnesses, congenital disorders, diabetes mellitus, and carcinogenesis (67). FOXA1 and A3 were responsible for airway mucous cell development in the healthy lung, and for mucous cell metaplasia in chronic airway diseases. In all of our models, FOXA3 was associated with MUC5AC gene expression, a hallmark mucin gene of mucous cells. FOXA2 was shown to repress mucin gene expression (68). Thus, FOXA2 may behave as a molecular switch to turn on SCGB1A1 but turn off mucin genes such as MUC5AC. In a previous study using an OVA model with a short exposure protocol, Muc5ac expression was found in a subset of SCGB1A1-expressing cells (69). In that study, SCGB1A1-expressing cells were not reduced. Because the duration and frequency of OVA exposure determine the strength of asthmatic phenotypes, the lack of a SCGB1A1 reduction might be associated with the mild nature of such a short exposure. However, this model provided a unique window into an intermediate state in which epithelial cells transitioned from SCGB1A1 expression to MUC5AC expression without an overall reduction in SCGB1A1. The longer exposure in our model may represent the severe form of the asthmatic phenotype, which is associated with a significant SCGB1A1 reduction. This is consistent with our meta-analysis of human data, in which a significant reduction of SCGB1A1 was only found in samples from subjects with severe asthma. However, further studies are needed to elucidate the molecular and cellular mechanisms of this switch.
Because SCGB1A1 is increasingly emerging as a protector against various chronic airway diseases, our finding suggests that SCGB1A1 replacement/augmentation may be an effective therapy for asthma, particularly severe asthma. Interestingly, in a previous study, Scgb1a1 restoration in KO mice was shown to repress expression of Th2 cytokines (e.g., IL-4, IL-5, and IL-13) without affecting the levels of Th1 cytokines (e.g., IFN-γ) (15), supporting the potential therapeutic application of SCGB1A1 in asthma. Unfortunately, a gene therapy approach using liposome-delivered SCGB1A1 DNA, which is not feasible as an asthma therapy, was used in that study. A medical-grade recombinant SCGB1A1 has been used in a clinical trial in premature infants with respiratory distress syndrome (70), and it will be interesting to examine its effect in asthma. An alternative approach to enhance SCGB1A1 may be to modulate its native production. We recently reported that retinoids were able to significantly upregulate SCGB1A1 both in vitro and in human subjects (71). In the present study, we further show that FOXA2 is required for SCGB1A1 transcription and its reduction is responsible for the decrease of SCGB1A1 in asthma. Interestingly, retinoic acid–binding elements were previously predicted in the promoter of FOXA2 (72). We speculate that retinoids may regulate SCGB1A1 indirectly through FOXA2, providing a novel approach to treat asthma by indirectly elevating SCGB1A1. This, in turn, could potentially boost antiinflammatory effects in the airway.
One limitation of this study is that our search was not exhaustive. We do not know all the components in asthmatic airways that are responsible for an SCGB1A1 reduction. The existence of positive factors that increase SCGB1A1 production is also possible. The strength and duration of net negative factors likely dominate in asthmatic airways over time. On the other hand, whether there are other TFs in addition to FOXA2, and whether a post-transcriptional mechanism is involved remain unclear. Furthermore, the downstream pathways induced by IL-13 treatment or by RV infection that converge on FOXA2 downregulation are also unclear. Future studies in these areas are urgently needed to completely elucidate the molecular mechanism underlying the SCGB1A1 reduction in asthma.
In conclusion, SCGB1A1 was found to be significantly reduced in asthmatic airways, likely by the action of Th2 cytokines and/or RV infection. At the molecular level, this reduction was mediated by the downregulation of FOXA2, an essential TF for SCGB1A1 expression.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants ES027013, ES028889, AI39439, and AI113526; Clinical Innovator Award 123055 from the Flight Attendant Medical Research Institute; and Arizona Biomedical Investigator Award BIG-3064.
Author Contributions: L.Z., L.A., R.W.H., and Y.C. conceived or designed the study. L.Z., D.R., R.L., C.B., X.Z., and S.-Y.L. acquired, analyzed, or interpreted data. L.Z., L.A., R.W.H., and Y.C. drafted the manuscript and revised it critically for important intellectual content. L.A., R.W.H., and Y.C. approved the final version for publication, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0199OC on December 21, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Broeckaert F, Bernard A. Clara cell secretory protein (CC16): characteristics and perspectives as lung peripheral biomarker. Clin Exp Allergy. 2000;30:469–475. doi: 10.1046/j.1365-2222.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocr Rev. 2007;28:707–725. doi: 10.1210/er.2007-0018. [DOI] [PubMed] [Google Scholar]
- 3.Mansur AH. Secretoglobin 1A1 gene and asthma pre-disposition: what is the evidence? Clin Exp Allergy. 2009;39:8–11. doi: 10.1111/j.1365-2222.2008.03161.x. [DOI] [PubMed] [Google Scholar]
- 4.Singh G, Katyal SL. Clara cell proteins. Ann N Y Acad Sci. 2000;923:43–58. doi: 10.1111/j.1749-6632.2000.tb05518.x. [DOI] [PubMed] [Google Scholar]
- 5.Lakind JS, Holgate ST, Ownby DR, Mansur AH, Helms PJ, Pyatt D, et al. A critical review of the use of Clara cell secretory protein (CC16) as a biomarker of acute or chronic pulmonary effects. Biomarkers. 2007;12:445–467. doi: 10.1080/13547500701359327. [DOI] [PubMed] [Google Scholar]
- 6.Levin SW, Butler JD, Schumacher UK, Wightman PD, Mukherjee AB. Uteroglobin inhibits phospholipase A2 activity. Life Sci. 1986;38:1813–1819. doi: 10.1016/0024-3205(86)90135-9. [DOI] [PubMed] [Google Scholar]
- 7.Mandal AK, Ray R, Zhang Z, Chowdhury B, Pattabiraman N, Mukherjee AB. Uteroglobin inhibits prostaglandin F2α receptor-mediated expression of genes critical for the production of pro-inflammatory lipid mediators. J Biol Chem. 2005;280:32897–32904. doi: 10.1074/jbc.M502375200. [DOI] [PubMed] [Google Scholar]
- 8.Mandal AK, Zhang Z, Ray R, Choi MS, Chowdhury B, Pattabiraman N, et al. Uteroglobin represses allergen-induced inflammatory response by blocking PGD2 receptor-mediated functions. J Exp Med. 2004;199:1317–1330. doi: 10.1084/jem.20031666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antico G, Lingen MW, Sassano A, Melby J, Welch RW, Fiore S, et al. Recombinant human uteroglobin/CC10 inhibits the adhesion and migration of primary human endothelial cells via specific and saturable binding to fibronectin. J Cell Physiol. 2006;207:553–561. doi: 10.1002/jcp.20604. [DOI] [PubMed] [Google Scholar]
- 10.Geerts L, Jorens PG, Willems J, De Ley M, Slegers H. Natural inhibitors of neutrophil function in acute respiratory distress syndrome. Crit Care Med. 2001;29:1920–1924. doi: 10.1097/00003246-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Lesur O, Bernard A, Arsalane K, Lauwerys R, Bégin R, Cantin A, et al. Clara cell protein (CC-16) induces a phospholipase A2-mediated inhibition of fibroblast migration in vitro. Am J Respir Crit Care Med. 1995;152:290–297. doi: 10.1164/ajrccm.152.1.7541278. [DOI] [PubMed] [Google Scholar]
- 12.Ray R, Zhang Z, Lee YC, Gao JL, Mukherjee AB. Uteroglobin suppresses allergen-induced TH2 differentiation by down-regulating the expression of serum amyloid A and SOCS-3 genes. FEBS Lett. 2006;580:6022–6026. doi: 10.1016/j.febslet.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasanthakumar G, Manjunath R, Mukherjee AB, Warabi H, Schiffmann E. Inhibition of phagocyte chemotaxis by uteroglobin, an inhibitor of blastocyst rejection. Biochem Pharmacol. 1988;37:389–394. doi: 10.1016/0006-2952(88)90204-3. [DOI] [PubMed] [Google Scholar]
- 14.Dierynck I, Bernard A, Roels H, De Ley M. Potent inhibition of both human interferon-gamma production and biologic activity by the Clara cell protein CC16. Am J Respir Cell Mol Biol. 1995;12:205–210. doi: 10.1165/ajrcmb.12.2.7865218. [DOI] [PubMed] [Google Scholar]
- 15.Hung CH, Chen LC, Zhang Z, Chowdhury B, Lee WL, Plunkett B, et al. Regulation of TH2 responses by the pulmonary Clara cell secretory 10-kd protein. J Allergy Clin Immunol. 2004;114:664–670. doi: 10.1016/j.jaci.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 16.Lomas DA, Silverman EK, Edwards LD, Miller BE, Coxson HO, Tal-Singer REvaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Investigators Evaluation of serum CC-16 as a biomarker for COPD in the ECLIPSE cohort Thorax 2008631058–1063.18757456 [Google Scholar]
- 17.Bernard A, Marchandise FX, Depelchin S, Lauwerys R, Sibille Y. Clara cell protein in serum and bronchoalveolar lavage. Eur Respir J. 1992;5:1231–1238. [PubMed] [Google Scholar]
- 18.Pilette C, Godding V, Kiss R, Delos M, Verbeken E, Decaestecker C, et al. Reduced epithelial expression of secretory component in small airways correlates with airflow obstruction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163:185–194. doi: 10.1164/ajrccm.163.1.9912137. [DOI] [PubMed] [Google Scholar]
- 19.Braido F, Riccio AM, Guerra L, Gamalero C, Zolezzi A, Tarantini F, et al. Clara cell 16 protein in COPD sputum: a marker of small airways damage? Respir Med. 2007;101:2119–2124. doi: 10.1016/j.rmed.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Ye Q, Fujita M, Ouchi H, Inoshima I, Maeyama T, Kuwano K, et al. Serum CC-10 in inflammatory lung diseases. Respiration. 2004;71:505–510. doi: 10.1159/000080636. [DOI] [PubMed] [Google Scholar]
- 21.Sin DD, Leung R, Gan WQ, Man SP. Circulating surfactant protein D as a potential lung-specific biomarker of health outcomes in COPD: a pilot study. BMC Pulm Med. 2007;7:13. doi: 10.1186/1471-2466-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsoumakidou M, Bouloukaki I, Thimaki K, Tzanakis N, Siafakas NM. Innate immunity proteins in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Exp Lung Res. 2010;36:373–380. doi: 10.3109/01902141003690389. [DOI] [PubMed] [Google Scholar]
- 23.Vestbo J, Edwards LD, Scanlon PD, Yates JC, Agusti A, Bakke P, et al. ECLIPSE Investigators. Changes in forced expiratory volume in 1 second over time in COPD. N Engl J Med. 2011;365:1184–1192. doi: 10.1056/NEJMoa1105482. [DOI] [PubMed] [Google Scholar]
- 24.Park HY, Churg A, Wright JL, Li Y, Tam S, Man SF, et al. Club cell protein 16 and disease progression in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:1413–1419. doi: 10.1164/rccm.201305-0892OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra S, Halonen M, Vasquez MM, Spangenberg A, Stern DA, Morgan WJ, et al. Relation between circulating CC16 concentrations, lung function, and development of chronic obstructive pulmonary disease across the lifespan: a prospective study. Lancet Respir Med. 2015;3:613–620. doi: 10.1016/S2213-2600(15)00196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laucho-Contreras ME, Polverino F, Gupta K, Taylor KL, Kelly E, Pinto-Plata V, et al. Protective role for club cell secretory protein-16 (CC16) in the development of COPD. Eur Respir J. 2015;45:1544–1556. doi: 10.1183/09031936.00134214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu L, Di PY, Wu R, Pinkerton KE, Chen Y. Repression of CC16 by cigarette smoke (CS) exposure. PLoS One. 2015;10:e0116159. doi: 10.1371/journal.pone.0116159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerra S, Vasquez MM, Spangenberg A, Halonen M, Martin RJ. Club cell secretory protein in serum and bronchoalveolar lavage of patients with asthma. J Allergy Clin Immunol. 2016;138:932–934.e1. doi: 10.1016/j.jaci.2016.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lensmar C, Nord M, Gudmundsson GH, Roquet A, Andersson O, Jörnvall H, et al. Decreased pulmonary levels of the anti-inflammatory Clara cell 16 kDa protein after induction of airway inflammation in asthmatics. Cell Mol Life Sci. 2000;57:976–981. doi: 10.1007/PL00000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Vyve T, Chanez P, Bernard A, Bousquet J, Godard P, Lauwerijs R, et al. Protein content in bronchoalveolar lavage fluid of patients with asthma and control subjects. J Allergy Clin Immunol. 1995;95:60–68. doi: 10.1016/s0091-6749(95)70153-2. [DOI] [PubMed] [Google Scholar]
- 31.Gioldassi XM, Papadimitriou H, Mikraki V, Karamanos NK. Clara cell secretory protein: determination of serum levels by an enzyme immunoassay and its importance as an indicator of bronchial asthma in children. J Pharm Biomed Anal. 2004;34:823–826. doi: 10.1016/S0731-7085(03)00570-3. [DOI] [PubMed] [Google Scholar]
- 32.Shijubo N, Itoh Y, Yamaguchi T, Abe S. Development of an enzyme-linked immunosorbent assay for Clara cell 10-kDa protein: in pursuit of clinical significance of sera in patients with asthma and sarcoidosis. Ann N Y Acad Sci. 2000;923:268–279. doi: 10.1111/j.1749-6632.2000.tb05535.x. [DOI] [PubMed] [Google Scholar]
- 33.Shijubo N, Itoh Y, Yamaguchi T, Sugaya F, Hirasawa M, Yamada T, et al. Serum levels of Clara cell 10-kDa protein are decreased in patients with asthma. Hai. 1999;177:45–52. doi: 10.1007/pl00007626. [DOI] [PubMed] [Google Scholar]
- 34.Ma YN, Wang J, Lee YL, Ren WH, Lv XF, He QC, et al. Association of urine CC16 and lung function and asthma in Chinese children. Allergy Asthma Proc. 2015;36:59–64. doi: 10.2500/aap.2015.36.3853. [DOI] [PubMed] [Google Scholar]
- 35.Candelaria PV, Backer V, Laing IA, Porsbjerg C, Nepper-Christensen S, de Klerk N, et al. Association between asthma-related phenotypes and the CC16 A38G polymorphism in an unselected population of young adult Danes. Immunogenetics. 2005;57:25–32. doi: 10.1007/s00251-005-0778-2. [DOI] [PubMed] [Google Scholar]
- 36.Laing IA, Goldblatt J, Eber E, Hayden CM, Rye PJ, Gibson NA, et al. A polymorphism of the CC16 gene is associated with an increased risk of asthma. J Med Genet. 1998;35:463–467. doi: 10.1136/jmg.35.6.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laing IA, Hermans C, Bernard A, Burton PR, Goldblatt J, Le Souëf PN. Association between plasma CC16 levels, the A38G polymorphism, and asthma. Am J Respir Crit Care Med. 2000;161:124–127. doi: 10.1164/ajrccm.161.1.9904073. [DOI] [PubMed] [Google Scholar]
- 38.Saadat M, Saadat I, Saboori Z, Emad A. Combination of CC16, GSTM1, and GSTT1 genetic polymorphisms is associated with asthma. J Allergy Clin Immunol. 2004;113:996–998. doi: 10.1016/j.jaci.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Sengler C, Heinzmann A, Jerkic SP, Haider A, Sommerfeld C, Niggemann B, et al. Clara cell protein 16 (CC16) gene polymorphism influences the degree of airway responsiveness in asthmatic children. J Allergy Clin Immunol. 2003;111:515–519. doi: 10.1067/mai.2003.180. [DOI] [PubMed] [Google Scholar]
- 40.Taniguchi N, Konno S, Hattori T, Isada A, Shimizu K, Shimizu K, et al. The CC16 A38G polymorphism is associated with asymptomatic airway hyper-responsiveness and development of late-onset asthma. Ann Allergy Asthma Immunol. 2013;111:376–381.e1. doi: 10.1016/j.anai.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Ohchi T, Shijubo N, Kawabata I, Ichimiya S, Inomata S, Yamaguchi A, et al. Polymorphism of Clara cell 10-kD protein gene of sarcoidosis. Am J Respir Crit Care Med. 2004;169:180–186. doi: 10.1164/rccm.200304-559OC. [DOI] [PubMed] [Google Scholar]
- 42.Konstantopoulos S. Fixed effects and variance components estimation in three-level meta-analysis. Res Synth Methods. 2011;2:61–76. doi: 10.1002/jrsm.35. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 44.Zhang Z, Kundu GC, Yuan CJ, Ward JM, Lee EJ, DeMayo F, et al. Severe fibronectin-deposit renal glomerular disease in mice lacking uteroglobin. Science. 1997;276:1408–1412. doi: 10.1126/science.276.5317.1408. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Zhao YH, Wu R. In silico cloning of mouse Muc5b gene and upregulation of its expression in mouse asthma model. Am J Respir Crit Care Med. 2001;164:1059–1066. doi: 10.1164/ajrccm.164.6.2012114. [DOI] [PubMed] [Google Scholar]
- 46.Bates JH, Rincon M, Irvin CG. Animal models of asthma. Am J Physiol Lung Cell Mol Physiol. 2009;297:L401–L410. doi: 10.1152/ajplung.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Hamati E, Lee PK, Lee WM, Wachi S, Schnurr D, et al. Rhinovirus induces airway epithelial gene expression through double-stranded RNA and IFN-dependent pathways. Am J Respir Cell Mol Biol. 2006;34:192–203. doi: 10.1165/rcmb.2004-0417OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Lee WM, Mosser AG, Rueckert RR. WIN 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J Virol. 1998;72:1210–1218. doi: 10.1128/jvi.72.2.1210-1218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oslund KL, Zhou X, Lee B, Zhu L, Duong T, Shih R, et al. Synergistic up-regulation of CXCL10 by virus and IFN γ in human airway epithelial cells. PLoS One. 2014;9:e100978. doi: 10.1371/journal.pone.0100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 52.Nord M, Cassel TN, Braun H, Suske G. Regulation of the Clara cell secretory protein/uteroglobin promoter in lung. Ann N Y Acad Sci. 2000;923:154–165. doi: 10.1111/j.1749-6632.2000.tb05527.x. [DOI] [PubMed] [Google Scholar]
- 53.Sawaya PL, Luse DS. Two members of the HNF-3 family have opposite effects on a lung transcriptional element; HNF-3 alpha stimulates and HNF-3 beta inhibits activity of region I from the Clara cell secretory protein (CCSP) promoter. J Biol Chem. 1994;269:22211–22216. [PubMed] [Google Scholar]
- 54.Chen G, Korfhagen TR, Karp CL, Impey S, Xu Y, Randell SH, et al. Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am J Respir Crit Care Med. 2014;189:301–313. doi: 10.1164/rccm.201306-1181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang SZ, Rosenberger CL, Espindola TM, Barrett EG, Tesfaigzi Y, Bice DE, et al. CCSP modulates airway dysfunction and host responses in an OVA-challenged mouse model. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1303–L1311. doi: 10.1152/ajplung.2001.281.5.L1303. [DOI] [PubMed] [Google Scholar]
- 56.Brewer JP, Kisselgof AB, Martin TR. Genetic variability in pulmonary physiological, cellular, and antibody responses to antigen in mice. Am J Respir Crit Care Med. 1999;160:1150–1156. doi: 10.1164/ajrccm.160.4.9806034. [DOI] [PubMed] [Google Scholar]
- 57.Shijubo N, Itoh Y, Yamaguchi T, Imada A, Hirasawa M, Yamada T, et al. Clara cell protein-positive epithelial cells are reduced in small airways of asthmatics. Am J Respir Crit Care Med. 1999;160:930–933. doi: 10.1164/ajrccm.160.3.9803113. [DOI] [PubMed] [Google Scholar]
- 58.Panganiban RA, Sun M, Dahlin A, Park HR, Kan M, Himes BE, et al. A functional splice variant associated with decreased asthma risk abolishes the ability of gasdermin B to induce epithelial cell pyroptosis. J Allergy Clin Immunol. 2018;142:1469–1478, e2. doi: 10.1016/j.jaci.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian BP, Zhou HB, Xia LX, Shen HH, Ying S. Balance of apoptotic cell death and survival in allergic diseases. Microbes Infect. 2014;16:811–821. doi: 10.1016/j.micinf.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Stokes JR, Casale TB. Characterization of asthma endotypes: implications for therapy. Ann Allergy Asthma Immunol. 2016;117:121–125. doi: 10.1016/j.anai.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 61.Brasier AR. Identification of innate immune response endotypes in asthma: implications for personalized medicine. Curr Allergy Asthma Rep. 2013;13:462–468. doi: 10.1007/s11882-013-0363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140:895–906. doi: 10.1016/j.jaci.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson WC, III, Apter AJ, Dutmer CM, Searing DA, Szefler SJ. Advances in asthma in 2016: designing individualized approaches to management. J Allergy Clin Immunol. 2017;140:671–680. doi: 10.1016/j.jaci.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 64.Vale K. Targeting the JAK-STAT pathway in the treatment of ‘Th2-high’ severe asthma. Future Med Chem. 2016;8:405–419. doi: 10.4155/fmc.16.4. [DOI] [PubMed] [Google Scholar]
- 65.Dotzauer A, Kraemer L. Innate and adaptive immune responses against picornaviruses and their counteractions: an overview. World J Virol. 2012;1:91–107. doi: 10.5501/wjv.v1.i3.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, Paquet AC, et al. IL-13 and epidermal growth factor receptor have critical but distinct roles in epithelial cell mucin production. Am J Respir Cell Mol Biol. 2007;36:244–253. doi: 10.1165/rcmb.2006-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katoh M, Katoh M. Human FOX gene family (review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 68.Lai H, Rogers DF. New pharmacotherapy for airway mucus hypersecretion in asthma and COPD: targeting intracellular signaling pathways. J Aerosol Med Pulm Drug Deliv. 2010;23:219–231. doi: 10.1089/jamp.2009.0802. [DOI] [PubMed] [Google Scholar]
- 69.Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, et al. Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol. 2004;31:382–394. doi: 10.1165/rcmb.2004-0060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine CR, Gewolb IH, Allen K, Welch RW, Melby JM, Pollack S, et al. The safety, pharmacokinetics, and anti-inflammatory effects of intratracheal recombinant human Clara cell protein in premature infants with respiratory distress syndrome. Pediatr Res. 2005;58:15–21. doi: 10.1203/01.PDR.0000156371.89952.35. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y, Vasquez MM, Zhu L, Lizarraga RE, Krutzsch M, Einspahr J, et al. Effects of retinoids on augmentation of club cell secretory protein. Am J Respir Crit Care Med. 2017;196:928–931. doi: 10.1164/rccm.201608-1611LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lalevée S, Anno YN, Chatagnon A, Samarut E, Poch O, Laudet V, et al. Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp) J Biol Chem. 2011;286:33322–33334. doi: 10.1074/jbc.M111.263681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.