Abstract
Although it has been recognized that energy metabolism and mitochondrial structure and functional activity in the immature brain differs from that of the adult, few studies have examined mitochondria specifically at the neuronal synapse during postnatal brain development. In this study, we examined the presynaptic mitochondrial proteome in mice at postnatal day 7 and 42, a period that involves the formation and maturation of synapses. Application of two independent quantitative proteomics approaches – SWATH-MS and super-SILAC – revealed a total of 40 proteins as significantly differentially expressed in the presynaptic mitochondria. In addition to elevated levels of proteins known to be involved in ATP metabolic processes, our results identified increased levels of mitoNEET (Cisd1), an iron-sulfur containing protein that regulates mitochondrial bioenergetics. We found that mitoNEET overexpression plays a cell-type specific role in ATP synthesis and in neuronal cells promotes ATP generation. The elevated ATP levels in SH-SY5Y neuroblastoma cells were associated with increased mitochondrial membrane potential and a fragmented mitochondrial network, further supporting a role for mitoNEET as a key regulator of mitochondrial function.
Keywords: Mitochondria, MitoNEET, Proteomics, Synaptic
INTRODUCTION
The brain, in contrast to many other tissues, is immature at birth and undergoes extensive changes during postnatal development. In particular, neuronal development involves progression through stages of neurogenesis, differentiation, migration, synaptogenesis, and pruning/maturation (Stiles & Jernigan, 2010). Synaptogenesis refers to the biochemical and morphological changes that enable the formation of synapses between neurons to facilitate neurotransmission, which is necessary for normal brain function. In the postnatal mouse brain, synaptogenesis occurs between the first and third weeks after birth, while maturation including pruning and myelination continues until at least four to six weeks, respectively (Semple, Blomgren, Gimlin, Ferriero, & Noble-Haeusslein, 2013). During this critical period of synaptogenesis and maturation of neural circuitries, synaptic transmission increases, cortical electrical activity matures, and energy demands are high to maintain ionic homeostasis (Erecinska, Cherian, & Silver, 2004). This postnatal phase corresponds with higher rates of cerebral blood flow (Kreisman, Olson, Horne, & Holtzman, 1989; Nehlig, Pereira de Vasconcelos, & Boyet, 1989), glucose utilization (Nehlig, de Vasconcelos, & Boyet, 1988), and oxygenation (Kreisman et al., 1989) than found in later life.
In adult humans, the brain, which constitutes 2% of body weight, receives 20% of the blood supply and utilizes high levels of oxygen to generate energy via mitochondrial oxidative phosphorylation. Mitochondrial respiratory chain complexes are enzymatically functional at early stages of human fetal development and after birth their absolute activities are elevated (Minai et al., 2008). Our previous work identified proteomic alterations that occur in brain mitochondria between embryonic day 18 and postnatal day 7 (P7) in line with elevated mitochondrial function to meet energy demands (Villeneuve, Stauch, & Fox, 2014b). Several animal studies also indicate an up-regulation of mitochondrial biogenesis and gene expression in the postnatal period (Izquierdo, Luis, & Cuezva, 1990; Kim, Lecordier, & Bowman, 1995). In the adult as well as in the immature brain, a close correlation between neural electrical activity and brain energy metabolism exists. Presynaptically, neuronal communication requires energy and generates large fluctuations in calcium concentrations. Mitochondria play a key role in supplying energy and buffering calcium at the synapse. In neuronal axons, mitochondria translocate to synaptic areas in situations of increased electric activity and high metabolic demand (Frederick & Shaw, 2007). Consistent with this, the postnatal period of synaptogenesis, when synaptic transmission increases, coincides with increases in mitochondrial number, volume, and cristae content (Gregson & Williams, 1969; Pysh, 1970; Samson, Balfour, & Jacobs, 1960). Further, elevated activities of the electron transport chain (ETC) complexes have been reported in presynaptic mitochondria from rat brain during this postnatal time frame (Bates, Almeida, Heales, & Clark, 1994). Studies of the synaptic proteome during postnatal brain development have revealed dynamic protein expression changes, suggestive of simultaneous increases in metabolism and synaptic transmission (McClatchy, Liao, Lee, Park, & Yates, 2012). While the synaptic proteome has been studied during critical periods of development (Dahlhaus et al., 2011; Gonzalez-Lozano et al., 2016; McClatchy et al., 2012; Moczulska et al., 2014), the presynaptic mitochondrial proteome has not been described during postnatal brain development, specifically at a period that corresponds to synaptogenesis.
Mass spectrometry (MS)-based proteomics is a powerful tool for biological and clinical research and enables relatively comprehensive global analyses. Relative quantitation with stable isotope labels or label-free methods has been widely used to study differential protein expression profiles. For example, stable isotope labeling with amino acids in cell culture (SILAC) is an excellent method for accurate quantitative proteomics and the use of SILAC has been expanded for tissue proteome quantification by using a mix of multiple SILAC-labeled cell lines as internal standards (super-SILAC) (Deeb, D’Souza, Cox, Schmidt-Supprian, & Mann, 2012; Geiger, Cox, Ostasiewicz, Wisniewski, & Mann, 2010; Geiger, Wehner, Schaab, Cox, & Mann, 2012). Alternatively, label-free methods based on a data-independent acquisition (DIA) strategy have emerged as powerful quantification tools because of recent developments in high-resolution quantitative proteomics. Sequential window acquisition of all theoretical fragment-ion spectra (SWATH)-MS is a label-free proteomics strategy where protein quantification is performed using a reference spectral library based on information extracted from the DIA files (Gillet et al., 2012). As every method has its unique advantages, we applied two independent quantitative proteomics approaches – SWATH-MS and super-SILAC – to analyze the proteome of presynaptic mitochondria, in order to explore the alterations that occur during postnatal development. In our study, we compared the proteome of presynaptic mitochondria from mice at two postnatal ages, P7 and P42, a period that involves the formation and maturation of synapses in mice. P7 in mouse corresponds in humans to a 36–40 gestation (term) infant, whereas P42 is teenaged age in humans, between which times significant brain growth, remodeling, and maturation occurs (Semple et al., 2013). After combining the aforementioned analysis datasets, we identified a total of 40 proteins as significantly differentially expressed in the presynaptic mitochondria. In addition to elevated levels of proteins known to be involved in ATP metabolic processes, our results identified mitoNEET (CDGSH iron-sulfur domain-containing protein 1, Cisd1), a mitochondrial outer membrane protein, as a candidate for further study. We found that mitoNEET overexpression plays a cell type specific role in ATP synthesis and in SH-SY5Y neuroblastoma cells promotes ATP synthesis. The elevated ATP levels in SH-SY5Y neuroblastoma cells were associated with increased mitochondrial membrane potential and a fragmented mitochondrial network, further supporting a role for mitoNEET as a key regulator of mitochondrial function.
MATERIALS AND METHODS
Animals
Wild-type (WT) C57BL/6 (substrain C57BL/6NCrl) mice were obtained from Charles River Laboratories International, Inc (Wilmington, MA). The mice were maintained on a 12-h light/dark cycle in a temperature-controlled environment with free access to standard mouse chow and water. All protocols were conducted within National Institutes of Health guidelines with the approval and oversight of the University of Nebraska Medical Center Institutional Animal Care and Use Committee.
Presynaptic Mitochondrial Isolation
Presynaptic mitochondria were isolated from the mice (male, P7 and P42) using our previously published method (Stauch, Purnell, & Fox, 2014a, 2014b). Briefly, mouse fore/midbrains (hindbrain and olfactory bulbs removed) were minced and homogenized (10 strokes in a Dounce homogenizer) in isolation media (225 mM sucrose, 75 mM mannitol, 1 mM EGTA, 5 mM HEPES, and cOmplete Mini, EDTA-free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), pH 7.4). Discontinuous Percoll gradient centrifugation was performed to separate synaptosomes from myelin and free non-synaptic mitochondria (Kristian, 2010). The presynaptic mitochondria were released from the synaptosomes using nitrogen cavitation and further purified using Percoll gradient centrifugation followed by anti-TOM22 immunomagnetic affinity isolation (Miltenyi Biotech, Bergisch Gladbach, Germany). The pure presynaptic mitochondria were lysed in 100 mM Tris-HCl with 4% (w/v) SDS and 0.1 M DTT, pH 7.6 using brief sonication and incubation at 95°C for 5 min. The Pierce 660 nm Protein Assay (Thermo Scientific, Waltham, MA) was used to determine protein concentration.
Isolation of Mouse Cell Line Mitochondria
Mitochondria were isolated from the mouse cell lines C8-D1A, CATH.a, Neuro-2a, and NB41A3 (obtained from American Type Culture Collection (ATCC), Manassas, VA) as previously described (Stauch et al., 2014b). Briefly, the cells were grown in DMEM/F-12 media containing 10% fetal bovine serum, 2 mM L-glutamine, and 1% penicillin/streptomycin. Cells were harvested and the mitochondria were isolated from lysed cells by sequential differential centrifugation using the Mitochondrial Isolation Kit for Cultured Cells (Mitosciences, Eugene, OR) followed by immunomagnetic (anti-TOM22) affinity isolation (Miltenyi Biotech). The cell line mitochondria were lysed and protein concentration was determined as described above for synaptic mitochondria.
For stable isotope labeling by amino acids in cell culture (SILAC) experiments, the four cell lines above were SILAC-labeled by culturing in Advanced DMEM/F-12 (Invitrogen, Carlsbad, CA) with heavy isotope-labeled amino acids, (U-13C615N4)-L-arginine (Arg-10) and (U-13C6)-L-lysine (Lys-6) replacing natural lysine and arginine. The media was supplemented with 10% dialyzed fetal bovine serum, SILAC glucose solution, L-glutamine, SILAC phenol red solution, and penicillin/streptomycin. Cells were cultured in the SILAC media until fully labeled (seven generations minimum) as assessed by quantitative mass spectrometry. The mitochondrial super-SILAC mix was prepared by mixing equal amounts of ‘heavy’ labeled mitochondrial lysates from each of the four cell lines as described previously (Stauch et al., 2014b).
Data-Dependent Acquisition (DDA) of Super-SILAC Experimental Samples
Experimental samples (protein lysates from the isolated mouse presynaptic mitochondria) were mixed in a 1:1 ratio with the mitochondrial super-SILAC mix. The filter-aided sample preparation (FASP) technique (Wisniewski, Zougman, Nagaraj, & Mann, 2009) was used for trypsin digestion and peptides were desalted using the Oasis mixed-mode weak cation-exchange (MCX) cartridge protocol (Waters, Milford, MA). Peptides were quantified by absorbance at 205 nm on the NanoDrop 2000 UV-vis Spectrophotometer (Thermo Scientific) using the Scopes method (Scopes, 1974) and the unfractionated samples of peptides were analyzed in quadruplicate (four biological replicates per age group (P7 and P42), n = 4) with two technical replicates by nano-LC-MS/MS in DDA mode on the 5600 TripleTOF instrument (SCIEX, Framingham, MA) and protein identification and quantification was performed using ProteinPilot as previously described (Stauch et al., 2014a, 2014b). Searches were performed against the UniProt Mus Musculus Proteome UP000000589 containing 16,890 reviewed proteins (Swiss-Prot) in ProteinPilot (Version 5.0.1, SCIEX) using the Paragon algorithm and the default settings (Shilov et al., 2007). Exclusion criteria to remove proteins from the analysis were as follows: FDR of 0.05 for both peptides and proteins, peptides must contain at least 6 amino acids, contaminants as identified through the database search, and proteins identified as being in the reverse database. The additional cutoff values of Unused ProtScore ≥ 1.3 and number of unique peptides ≥ 2 were applied to the data. Quantification was performed using the heavy super-SILAC mix as an internal standard and the resulting heavy-to-light (H/L) ratios were normalized to this mix and expressed as light-to-heavy (L/H, sample/super-SILAC internal standards). The L/H expression values were then converted to log2 scale and median normalized so that the total light and heavy intensities in each sample were equivalent since the same amount of light and heavy proteins were mixed. The “ratio of ratio” value was determined, which is the change in protein expression from P7 to P42.
Generating the Mitochondrial SWATH-MS Reference Spectral Library
On the basis of protein quantification, the mitochondrial lysates prepared from the unlabeled C8-D1A, CATH.a, Neuro-2a, and NB41A3 cell lines were mixed in equal amounts. This cell line derived mitochondrial lysate mix was processed using the FASP method (Wisniewski et al., 2009). The peptides were desalted using Oasis MCX cartridges following the manufacturer’s protocols. The resulting peptides were quantified by absorbance at 205 nm (Scopes, 1974). Peptides were fractionated into 12 fractions from pH 3 to 10 (low-resolution kit) by isoelectric focusing using an Agilent 3100 OFFGEL Fractionator (Agilent Technologies, Santa Clara, CA). Fractionated peptides were cleaned and prepared for mass spectrometry using Pierce C-18 PepClean Spin Columns (Thermo Scientific). Samples were dehydrated with a Savant ISS 110 SpeedVac Concentrator (Thermo Scientific) and resuspended in 6 μL of 0.1% FA for LC-MS/MS analysis. The samples (12 fractions of unlabeled cell line mitochondrial peptides) used to generate the SWATH-MS reference spectral library were subjected to traditional DDA as described previously for the generation of our rat SWATH-MS reference spectral library (Villeneuve, Stauch, & Fox, 2014a). Additional samples were added to enrich our library for synaptic proteins as described for our rat SWATH-MS reference spectral library (Villeneuve, Purnell, Boska, & Fox, 2016). Presynaptic mitochondria isolated from WT mouse brain were prepared as described above for the cell line mitochondria and added to the spectral library. For peptide identification, our library was generated in ProteinPilot (Version 5.0.1, SCIEX) using the Paragon algorithm and the default settings (Shilov et al., 2007). All searches were performed against the UniProt Mus Musculus Proteome UP000000589 containing 16,890 reviewed proteins (Swiss-Prot). Combined results yielded a library of 4,302 proteins identified with high confidence (greater than 99%) that passed the global FDR from fit analysis using a critical FDR of 1%. Of note, our library is comparable to results from recently analyzed mitochondrial extracts from mouse tissues where a total of 3,881 proteins were detected (Pagliarini et al., 2008). While these numbers are higher than the commonly quoted ~1500, the high sensitivity of newer generation mass spectrometry instruments may identify proteins not yet known to be mitochondrial as well as co-purifying contaminants. In our analyses, we filtered the identified proteins based on the 1,626 genes listed in the MitoMiner database (Smith, Blackshaw, & Robinson, 2012), as described below.
Data-Independent Acquisition (DIA) of SWATH-MS Experimental Samples.
Experimental samples (protein lysates from the isolated mouse presynaptic mitochondria, the same experimental samples were used for both super-SILAC and SWATH-MS) used for DIA mass spectrometry were digested with trypsin using the FASP method (Wisniewski et al., 2009). The resultant peptides were desalted using Oasis MCX cartridges and then quantified using the Scopes method (Scopes, 1974). The unfractionated samples of peptides from the P7 and P42 mouse synaptic mitochondrial lysates were analyzed in quadruplicate (four biological replicates per age group (P7 and P42, n = 4)) by nano-LC-MS/MS in SWATH-MS mode on the 5600 TripleTOF instrument (SCIEX) and targeted data extraction was performed as previously described (Gillet et al., 2012; Haverland, Fox, & Ciborowski, 2014; Villeneuve et al., 2014a). All of the fragment ion chromatograms were extracted and automatically integrated with PeakView (Version 2.1, SCIEX). In accordance with previously published work (Gillet et al., 2012), we selected five peptides and five transitions option for quantitative analysis and performed targeted data extraction for each peptide. For each peptide the fragment ion chromatograms were extracted using the SWATH isolation window set to a width of 10 min and 50 ppm accuracy (Gillet et al., 2012). To calibrate retention times, synthetic peptides (BiognoSYS, Zurich, Switzerland) were spiked-in to the samples in accordance with manufacturer’s protocol. Samples were normalized to the area counts of the synthetic peptides. Data were normalized to the median peak ratios of common proteins in MarkerView software (Version 1.2.1, SCIEX).
Bioinformatics
The Database for the Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) Bioinformatics Resources 6.8 was used for functional annotation (Huang, 2009). MitoMiner 4.0 (V2018 JUN, http://mitominer.mrc-mbu.cam.ac.uk/release-4.0/begin.do) was used to annotate the proteins identified by SWATH-MS and Super-SILAC as mitochondrial (Smith et al., 2012). The SWATH-MS and Super-SILAC data were uploaded to the Cyber-T Web server (http://cybert.ics.uci.edu/) (Kayala & Baldi, 2012), which implements a t-test using a Bayesian regularization method for quantitative mass spectrometry analysis. Benjamini and Hochberg (BH) multiple-hypothesis testing corrections were performed to obtain q values (q < 0.05 was considered statistically significant). All correlation analysis, linear fits, and frequency distributions were completed using Prism 6 (GraphPad Software, La Jolla, CA).
Western Blotting
Isolated presynaptic mitochondria obtained from four biological replicates per age group (n = 4 for P7 and P42, these experimental samples were isolated from a separate set of mice than those used for the super-SILAC and SWATH-MS analyses) were lysed using 4% SDS, 100 mM Tris, 0.1 M DTT. The total protein concentration was determined using the Pierce 660 nm Protein Assay. For each sample, 10 μg of protein was separated by SDS-PAGE on Bolt 4–12% Bis-Tris gels (Thermo Scientific). After electrophoresis, the proteins were transferred onto nitrocellulose membrane using the iBlot Dry Blotting System (Thermo Scientific). After blocking with SuperBlock Blocking Buffer, membranes were probed overnight at 4°C with primary antibodies for Tomm20 (1:2000, Santa Cruz Biotechnology, Dallas, TX, sc-11415), Actin (1:20,000, Abgent, San Diego, CA, AM1829B), Cisd1 (1:2000, ProteinTech, Rosemont, IL, 16006–1-AP), and Atp5a1 (1:2000, Abcam, MS507). The IRDye 680RD goat anti-mouse (LI-COR, Lincoln, NE, 926–68070), IRDye 680RD donkey anti-rabbit (LI-COR, 925–68073), IRDye 800CW donkey anti-goat (LI-COR, 925–32214), and IRDye 800CW goat anti-rabbit (LI-COR, 926–32211) near-infrared fluorescent secondary antibodies were used at a 1:20,000 dilution. Blots were imaged with an Odyssey Fc imaging system and quantified using LI-COR Image Studio software (Version 4.0, LI-COR). Statistical analyses were conducted in Prism using unpaired two-tailed t-tests for immunoblotting. p < 0.05 was considered statistically significant. Data are expressed as mean ± SEM.
MitoNEET Transfection
To study the effects of mitoNEET overexpression on mitochondrial activity, mitoNEET was transiently overexpressed in NIH/3T3 cells and SH-SY5Y cells (both obtained from the ATCC, Manassas, VA). The mitoNEET plasmid was obtained from Origene, Rockville, MD (RC203308). Cotransfection with DsRed-Express (Clontech, Mountain View, CA, 632416) or pMAX GFP (Lonza) was used to identify cells transfected successfully (Supplementary Figures S1A, S2A, and S2B). The empty vector plasmid pcDNA 4/T0 (Thermo Scientific) was used for control transfections. The Amaxa electroporation protocol (Lonza, Allendale, NJ, USA) was used for the mitochondrial membrane potential and ATP experiments, while X-tremeGENE HP DNA Transfection reagent (Roche Diagnostics) was used for mitochondrial morphology experiments. A ratio of 5:1 of mitoNEET:DsRed-Express (or 4:1 of mitoNEET:pMAX GFP) was used with 10 μg of total DNA being utilized in accordance with the protocol. All transfections were conducted in a poly-D-lysine coated, glass-bottom 24 well plate. Each transfection was performed 3 times on 3 separate days. Western blotting was performed to confirm expression of mitoNEET in the NIH/3T3 and SH-SY5Y cells post-transfection (Supplementary Figures S1B and S2C). Mitochondria were isolated and immunoblotting was performed as described above using a primary antibody to mitoNEET (1:2,000, ProteinTech, 16006–1-AP) and Atp5b (1:2000, Abcam, ab14730) or cell lysates were probed for mitoNEET and Actin (1:20,000, Abgent, AM1829B). Subsequent experiments were performed at 48–65 hours post-transfection (as peak expression of mitoNEET was found between 24 and 96 hours post-transfection).
Mitochondrial membrane potential
To determine the effect of mitoNEET overexpression on the mitochondrial membrane potential, the potential was measured in NIH/3T3 and SH-SY5Y cells from control and experimental transfections using live-cell imaging and JC-1 (Molecular Probes, Invitrogen) staining. In JC-1 staining, the JC-1 dye accumulates in the mitochondria of healthy cells as aggregates and are fluorescent red. When the cells are under stress, the mitochondrial potential collapses, and the JC-1 dye can no longer accumulate in the mitochondria. Consequently, the JC-1 aggregates remain in the cytoplasm, in a monomeric form, which is green. In this experiment, the cells were stained for 20 minutes using 2 μM JC-1 at 37°C, then washed 3 times with 1xPBS, and analyzed for mitochondrial membrane potential by measuring the red:green ratio with confocal microscopy (Nikon (Melville, NY) swept-field confocal microscope using a 60x oil-immersion (1.45 NA) objective and a live-cell imaging chamber (Pathology Devices, Inc., Westminster, MD) to maintain appropriate oxygen and carbon dioxide conditions during imaging). Images were acquired using 488 nm excitation and 536 nm and 593 nm emission filters for the green monomers and red dimers, respectively.
To determine membrane polarization, the ratio of red (593 nm) to green (536 nm) fluorescence of JC-1 images was calculated using NIH Image J. The outline of individual cells was circled using the freehand tool to create a region of interest (ROI) and saved in the multimeasure plugin. The average intensity of the same ROI was analyzed in both the red and green channels. The polarization ratio was obtained for each ROI and the average of the ratio was obtained over all ROIs for each transfection condition.
ATP Levels
ATP levels were measured in NIH/3T3 and SH-SY5Y cells after mitoNEET transfection using the ATP determination kit (Molecular Probes, Invitrogen). The bioluminescence assay is based on the reaction of ATP with recombinant firefly luciferase and its substrate luciferin. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to the ATP concentration, which was measured using a luminometer. We measured ATP from the cell pellet lysates (flash freeze fracture method) using a standard curve method.
Mitochondrial Morphology
To determine the effect of mitoNEET overexpression (48 hours) on mitochondrial morphology, the NIH/3T3 and SH-SY5Y cells from control and experimental transfections were stained with MitoTracker Deep Red (Molecular Probes, Invitrogen) prior to fixation using 4% PFA in 1xPHEM buffer. Following fixation, the cells were stained with Wheat Germ Agglutinin 350 (Thermo Scientific) prior to mounting using Fluoromount-G (Thermo Scientific). Imaging was performed on a Zeiss Observer Z1 microscope with the Apotome (at 0.1 μm z-steps, 13–17 slices per image) using Axio vision 4.8.2. The mitochondrial morphology was calculated using MitoGraph v3.0 as described (Harwig et al., 2018).
Statistical Analysis
All statistical analysis was performed using one-way ANOVA with Bonferroni post-hoc tests to compare three samples, or t-test in the cases where only two conditions were studied. For the data other than the proteomics, statistical analyses and linear fits were completed using Prism 6 software (Graphpad Software Inc., La Jolla, CA). For the SH-SY5Y data, linear unmixing was performed before the luciferase assay to account for differences in transfection efficiency.
RESULTS
Mitochondrial protein identification and quantification
In order to explore the alterations that occur during postnatal brain development, we applied two independent quantitative proteomic approaches – SWATH- and super-SILAC-based mass spectrometry (MS) – to analyze the proteome of mitochondria isolated from synaptosomes (Figure 1A), which contain the complete presynaptic nerve terminal contents, including cytoplasm, synaptic vesicles, mitochondria, and cytoskeleton (Dunkley, Jarvie, & Robinson, 2008). The same experimental samples (presynaptic mitochondria from P7 and P42 mice, n = 4) were used for both MS techniques. In the SWATH-MS experiments, a total of 1,293 proteins were identified and quantified across all samples from each experimental age group (Supplementary Table S1), of which 750 proteins (58.0%) were identified as being mitochondrial due to annotation by MitoMiner (Figure 1B, Supplementary Table S2). We next used a Bayesian regularized t-test analysis and multiple testing corrections (Kayala & Baldi, 2012), and 69 mitochondrial proteins were considered significantly differentially expressed (BH q < 0.05) in synaptic mitochondria from P42 mice as compared to those from P7 mice (Supplementary Table S2). In the super-SILAC experiments, a total of 1,032 proteins were quantified across all samples from each experimental age group (Supplementary Table S3). Of these, 667 proteins (64.6%) were annotated as mitochondrial by MitoMiner (Figure 1C, Supplementary Table S4), and 292 mitochondrial proteins were considered significantly differentially expressed (BH q < 0.05) in synaptic mitochondria from P42 mice as compared to those from P7 mice (Supplementary Table S4). Combining the quantified mitochondrial proteins from each approach, a total of 552 mitochondrial proteins were observed to be overlapping between the two approaches (Figure 1D; Supplementary Table S5), and 198 mitochondrial proteins were uniquely quantified in the SWATH-MS proteomic approach, whereas there were 115 unique mitochondrial proteins in the super-SILAC-labeling proteomic approach. We next analyzed the correlation of the expression of the 552 overlapping quantified mitochondrial proteins between the two approaches, and a Pearson correlation analysis revealed a significant correlation (r = 0.4048; p < 0.0001) for SWATH-MS and super-SILAC-labeling proteomics (Figure 1E). We found that both approaches were suitable for proteomic synaptic mitochondrial analysis consistent with our previous work.
Figure 1. Quantitative proteomic analysis of synaptic mitochondria from P7 and P42 mouse brain.
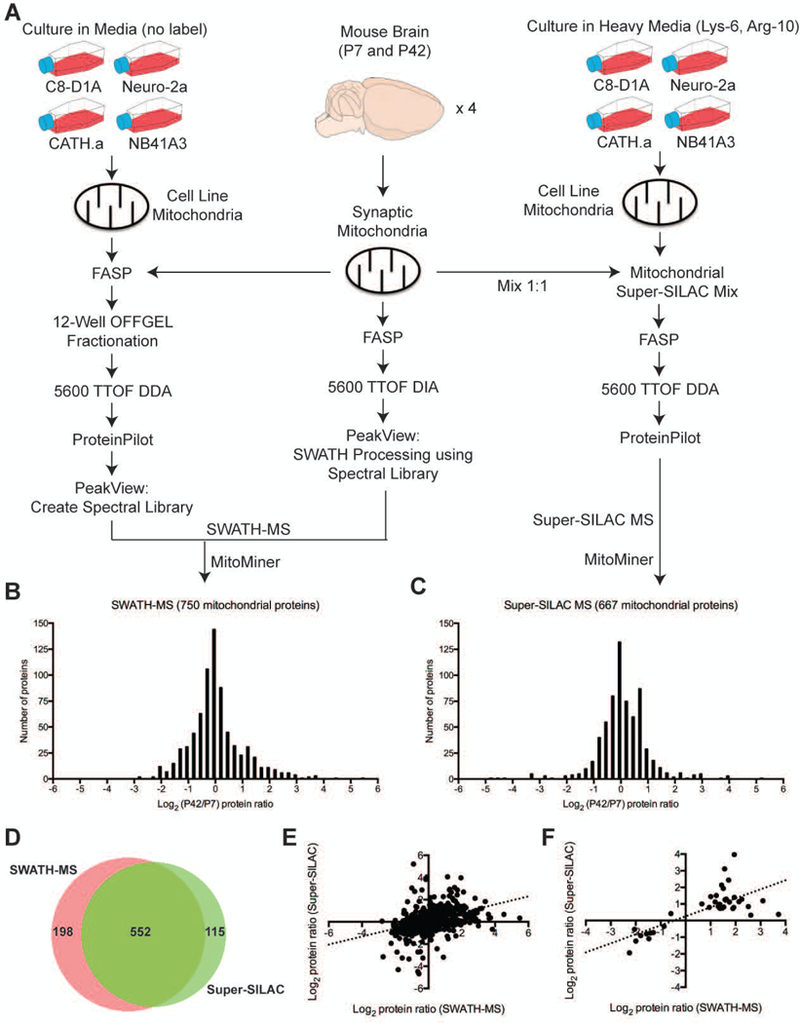
A) Scheme for the synaptic mitochondrial quantitative proteomic analyses using the SWATH-MS and super-SILAC MS strategies. The distribution of the fold changes (log2) in (B) 750 and (C) 667 quantified synaptic mitochondrial proteins (annotated by MitoMiner) between the P7 and P42 mouse groups identified by SWATH-MS and super-SILAC MS, respectively. D) Venn diagram comparison of SWATH- and super-SILAC-based MS quantitative mitochondrial proteins (annotated by MitoMiner). E) Scatter plot of the fold changes (log2) of the 522 overlapping quantified mitochondrial proteins (annotated by MitoMiner) between the two approaches.
Functional annotation of the differentially expressed mitochondrial proteins
Of the 552 mitochondrial proteins quantified by both the SWATH-MS and super-SILAC-labeling experiments, 40 were found to be differentially expressed by both techniques (Supplementary Table S5). We next analyzed the correlation of the expression of the 40 overlapping differentially expressed mitochondrial proteins between the two approaches, and a Pearson correlation analysis revealed a significant correlation (r = 0.7210; p < 0.0001) for SWATH-MS and super-SILAC-labeling proteomics (Figure 1F). While 30 of the 40 overlapping differentially expressed mitochondrial proteins were found to be up-regulated by both techniques, 9 were found to be down-regulated by both techniques. Only one protein, citrate synthase did not show the same change (SWATH predicted down-regulation, while SILAC predicted up-regulation). Bioinformatic analysis of the 30 up-regulated mitochondrial proteins using DAVID functional annotation clustering tool (Huang, 2009) revealed several GO biological process (BP) terms that were altered according to our proteomics results (Supplementary Table S6). The top two terms based on the list of 30 up-regulated mitochondrial proteins in synaptic mitochondria from P42 compared to P7 mice were “ATP metabolic process” and “tricarboxylic acid (TCA) cycle”, which supports the role of mitochondria for energy generation during brain development. The top two BP terms based on functional annotation of the 9 down-regulated mitochondrial proteins in synaptic mitochondria from P42 compared to P7 mice were “protein import into mitochondrial matrix” and “fatty acid beta-oxidation” (Supplementary Table S7).
Confirmation of protein expression changes via Western blotting analysis
To validate the differentially expressed proteins identified from the SWATH-MS and super-SILAC-labeling experiments, we selected two proteins that displayed the same change trend in both methods and applied the classical method for relative protein quantification by Western blotting. These experimental samples (synaptic mitochondria from P7 and P42 mice, n = 4) were from a separate set of mice than those used for the MS analyses. As shown in Figure 2, the relative abundance of the two proteins (i.e. Atp5a1 and Cisd1 (MitoNEET)) was similar to the results from the SWATH-MS and super-SILAC-labeling analyses. Taken together, the data indicates that the results from the validation experiment was consistent with the initial discovery result from the simultaneously application of the two independent quantitative proteomic approaches – SWATH and super-SILAC – to analyze the proteome of synaptic mitochondria.
Figure 2. Validation of two selected proteins in synaptic mitochondria from P7 and P42 mouse brain by western blotting.
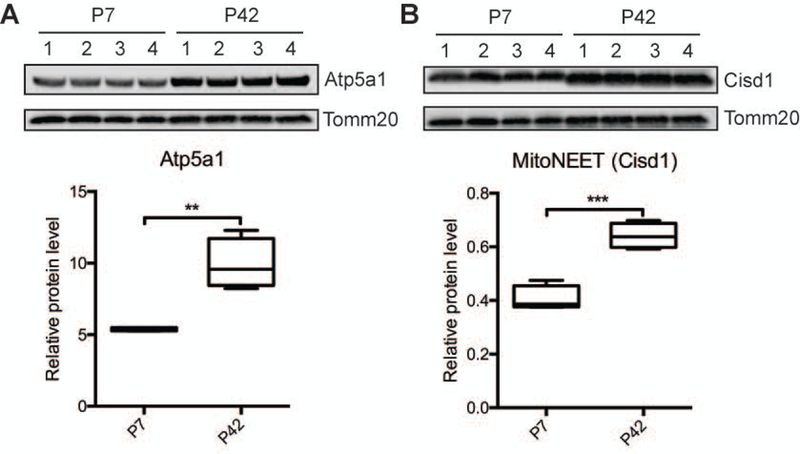
The abundance of Atp5a1 and Cisd1 proteins in synaptic mitochondria from P7 and P42 mouse brains were analyzed by immunoblotting using four animals. Values were normalized to Tomm20. These data were obtained from four independent isolations (n = 4). A box and whisker plot is shown representing the interquartile range (IQR) with median, minimum and maximum values. **p<0.01; ***p<0.0001 compared with P7, unpaired two-tailed t tests.
Overexpression of mitoNEET (Cisd1) increases neuronal ATP levels.
From the identified proteins, we chose to focus on mitoNEET. Since its initial identification as a mitochondrial outer membrane protein mitoNEET which binds the antidiabetic drug pioglitazone (Colca et al., 2004), several compounds have been identified as mitoNEET ligands (Geldenhuys, Leeper, & Carroll, 2014). MitoNEET has been proposed as a mitochondrial protein drug target for several diseases including diabetes, cancer, and neurodegeneration (Geldenhuys et al., 2014). Our proteomic work revealed neuronal mitoNEET, specifically presynaptic mitochondrial levels, increase during postnatal brain development (Table 1). This significant elevation of mitoNEET protein levels was confirmed via immunoblotting (Figure 2) suggesting neuronal mitoNEET function is important during postnatal brain development. Thus, in addition to neurodegenerative diseases, targeting mitoNEET might attenuate mitochondrial abnormalities observed in neurodevelopmental disorders (Falk, 2010; Uittenbogaard & Chiaramello, 2014) as well, particularly when synaptic alterations are implicated.
Table 1.
The average ± standard deviation of the mitochondrial volumes, average width and average length in each cell (n as in Figure Legend 6), as calculated by MitoGraph. The p-values were calculated using unpaired t-tests, n.s. = not significant.
| MitoGraph Output | Control | MitoNEET | p-value |
|---|---|---|---|
| 3T3 Volume from voxels (μm3) | 36.2±15.9 | 33.8±13.9 | n.s. |
| 3T3 Average Width (μm) | 0.19±0.02 | 0.19±0.02 | n.s. |
| 3T3 Total Length (μm) | 420.9±168.8 | 397.4±138.5 | n.s. |
| SH-SY5Y Volume from voxels (μm3) | 11.9±6.9 | 7.6±4.9 | p=0.005 |
| SH-SY5Y Average Width (μm) | 0.19±0.02 | 0.17±0.02 | p<0.001 |
| SH-SY5Y Total Length (μm) | 128.2±56.3 | 91.2±50.7 | p=0.008 |
To determine whether mitoNEET overexpression modulates parameters of mitochondrial function, ATP production and mitochondrial membrane potential were studied in two different cell types, NIH/3T3 (a murine fibroblast line) and SH-SY5Y (a human neuroblastoma line). Initially a western blot time course for expression of the transfected mitoNEET expression plasmid was performed in NIH/3T3 cells (Supplementary Figure S1B). Based on this blot, mitoNEET expression was increased between 24 and 96 hours post-transfection. All subsequent experiments were conducted at 65 hours post-transfection. Overexpression of mitoNEET in NIH/3T3 cells decreased the mitochondrial membrane potential (Figure 3A) and reduced ATP production (Figure 3B), consistent with previous findings in fibroblasts (Kusminski et al., 2012). Conversely, overexpression of mitoNEET in SH-SY5Y cells increased the mitochondrial membrane potential (Figure 4A) and elevated ATP production (Figure 4B). SH-SY5Y cells show many neuronal characteristics, which is important since presynaptic mitochondria would be neuronal in origin. These results suggest that not only is mitoNEET able to alter ATP production and mitochondrial membrane polarization, but also, that the alteration is dependent on the type of cell in which mitoNEET is overexpressed.
Figure 3. Decreased mitochondrial membrane potential and ATP levels in NIH/3T3 cells overexpressing mitoNEET.
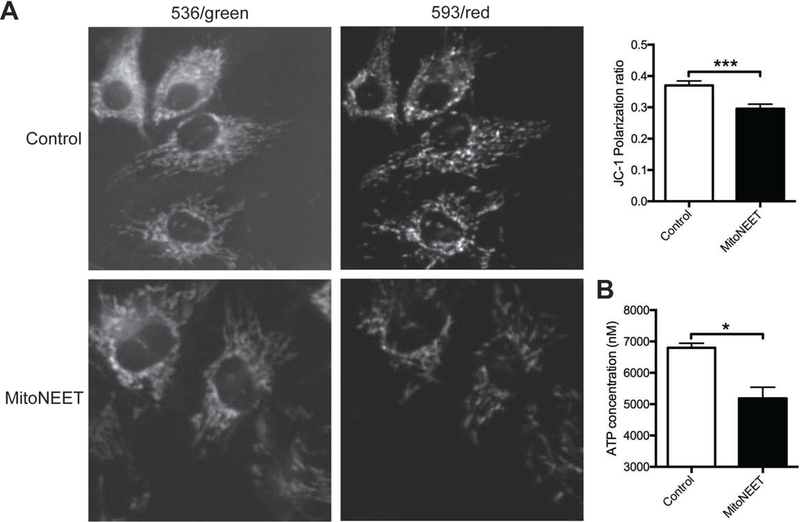
(A) Representative confocal images of JC-1 staining of NIH/3T3 cells 65 h post-transfection with control or mitoNEET overexpression constructs. The loss of red fluorescence (593) and thus increased green (536) mitochondria were observed in transfected mitoNEET overexpressing cells, indicating low mitochondrial membrane potential. (B) Bar graph shows the ratio of red to green fluorescence intensity calculated to characterize mitochondrial membrane potential. Note the decreased mitochondrial membrane potential in mitoNEET overexpressing NIH/3T3 cells. (C) ATP assay in NIH/3T3 cells 65 h post-transfection with control or mitoNEET overexpression constructs. These data were obtained from three independent experiments (n = 3). Error bars indicate ± S.E.M. *p<0.05; ***p<0.001 compared with control transfection, one-way analysis of variance followed by Tukey’s post tests.
Figure 4. Increased mitochondrial membrane potential and ATP levels in SH-SY5Y cells overexpressing mitoNEET.
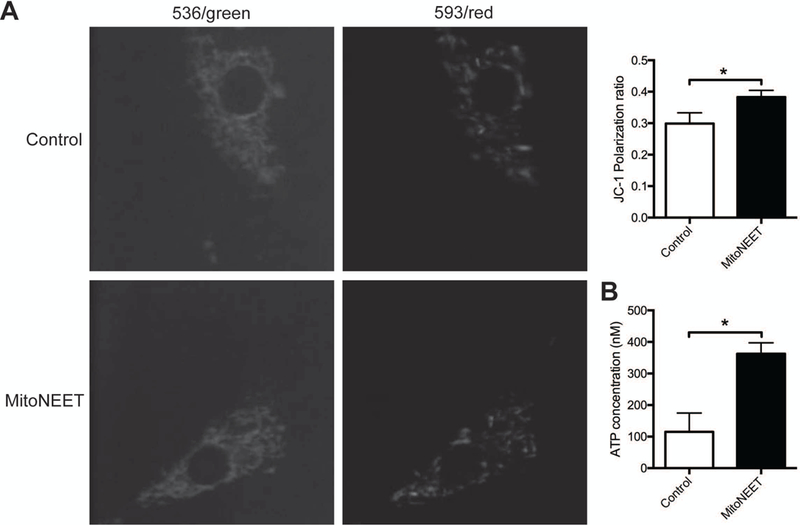
(A) Representative confocal images of JC-1 staining of SH-SY5Y cells 65 h post-transfection with control or mitoNEET overexpression constructs. The increase of red fluorescence (593) and thus decreased green (536) mitochondria were observed in transfected mitoNEET overexpressing cells, indicating high mitochondrial membrane potential. (B) Bar graph shows the ratio of red to green fluorescence intensity calculated to characterize mitochondrial membrane potential. Note the increased mitochondrial membrane potential in mitoNEET overexpressing SH-SY5Y cells. (C) ATP assay in SH-SY5Y cells 65 h post-transfection with control or mitoNEET overexpression constructs. These data were obtained from three independent experiments (n = 3). Error bars indicate ± S.E.M. *p<0.05 compared with control transfection, one-way analysis of variance followed by Tukey’s post tests.
Mitochondrial morphology
Mitochondrial morphology is malleable in cells and such changes can alter function (Westermann, 2012). To assess if mitoNEET overexpression altered the structure of mitochondria, we used MitoTracker Deep Red stained NIH/3T3 and SH-SY5Y cells transfected with mitoNEET compared to cells transfected with the expression vector alone. Images of transfected cells were then analyzed using MitoGraph to characterize the resulting network structure of cellular mitochondria (Figure 5). Figure 6 represents the fold change between the averages of the mitoNEET expressing and control NIH/3T3 and SH-SY5Y cells. No significant changes in any mitochondrial morphology parameters resulting from mitoNEET expression were found in NIH/3T3 cells (Figure 6A and Table 1). As shown in Figure 6B, metrics associated with enhanced fission are increased in mitoNEET overexpressing SH-SY5Y cells (total nodes, edges, and connected components) while metrics associated with enhanced fusion are decreased (PHI, Avg edge length, and Avg degree). This is expressed as the MitoGraph Connectivity Score, which significantly decreased from (mean ± SD) 0.72 ± 0.20 in controls to 0.39 ± 0.15 with mitoNEET overexpression. Furthermore, the total length, average width, and volume of the mitochondria were also impacted by mitoNEET overexpression in the SH-SY5Y cells (Table 1).
Figure 5. MitoGraph analysis of mitoNEET overexpressing NIH/3T3 and SH-SY5Y cells.
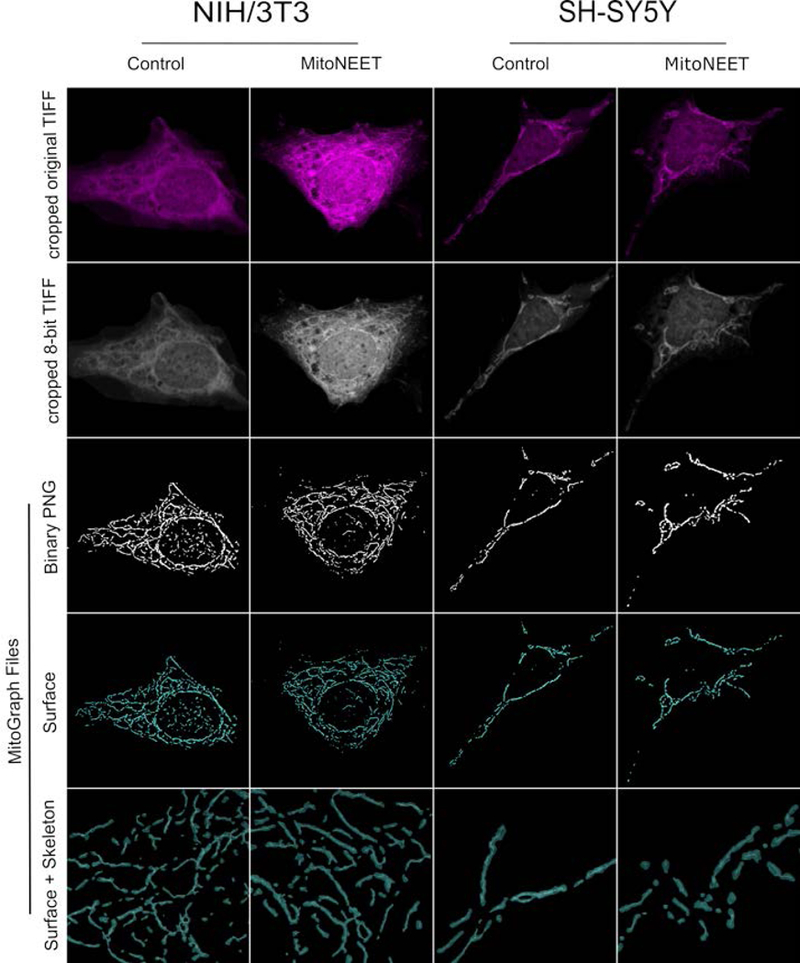
Representative images of MitoTracker Deep Red stained NIH/3T3 and SH-SY5Y cells transfected with mitoNEET or the expression vector alone (control), fixed in 4% PFA, imaged and processed by MitoGraph v3.0 as described previously (Harwig et al., 2018). The top row represents the original cropped TIFF files and the second row represents the cropped 8-bit TIFF files. The bottom 3 rows are output files of MitoGraph (the binary PNG, the 3D surface representation, and a zoom in on the surface with the skeleton of the mitochondrial network).
Figure 6. Mitochondrial network morphology following mitoNEET overexpression in NIH/3T3 and SH-SY5Y cells.
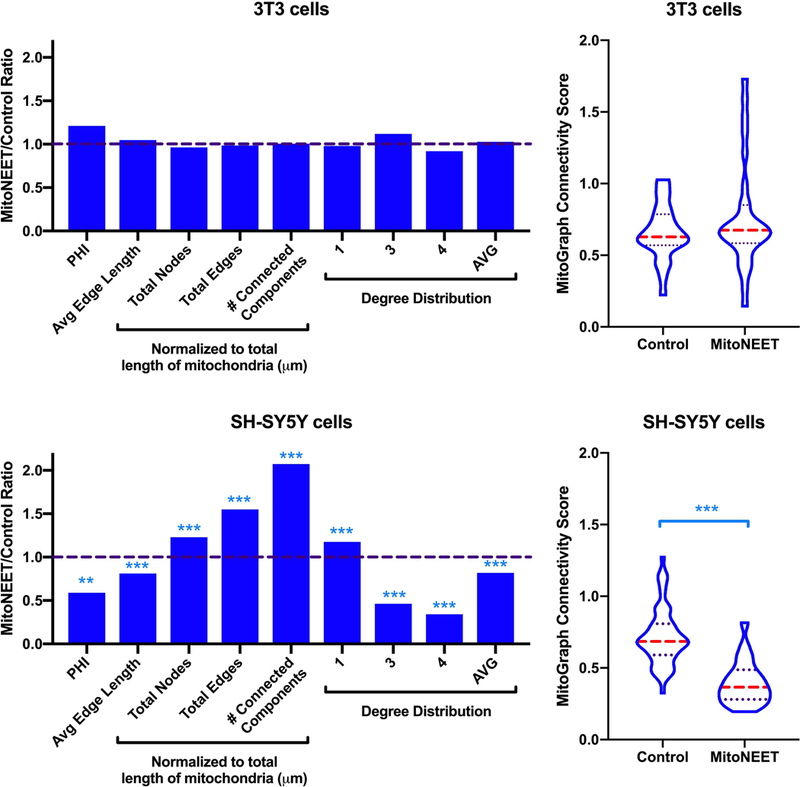
Images from total of 27 (3T3 control), 30 (3T3 mitoNEET), 56 (SH-SY5Y control) and 23 (SH-SY5Y mitoNEET) were analyzed by MitoGraph, which calculated listed measures. These data were obtained from three independent experiments. PHI is the ratio of relative size of the largest connected component to the total mitochondrial size, and the average degree distribution of the nodes is calculated from the relative percentage of free ends (1), 3-way junctions (3), and 4-way junctions (4). Shown on the left are the ratios, for each cell type, of the change resulting from mitoNEET expression relative to control. On the right, the MitoGraph connectivity score is shown (violin plot, dashed line median, dotted lines quartiles). This is calculated from the sum of metrics elevated in a fused state (PHI, Avg Edge Length and Avg Degree) divided by sum of metrics, which are elevated in a fragmented state (total nodes, edges and connected components (normalized)). **p<0.01, ***p<0.001, unpaired t-test.
DISCUSSION
In the present study, our quantitative proteomics results identified differential expression of 40 mitochondrial proteins in mouse synaptosomes during a postnatal development period that coincides with the formation and maturation of synapses. Functional annotation of the 30 up-regulated mitochondrial proteins suggests presynaptic mitochondria undergo proteomic changes important for increased energy generation, specifically ATP synthesis and TCA cycle flux, between postnatal day 7 and 42. Notably, synaptic mitochondrial protein levels of mitoNEET increased during postnatal development and overexpression of mitoNEET in the SH-SY5Y neuroblastoma cell line resulted in elevated ATP levels. Our work implicates a role for mitoNEET in regulating neuronal mitochondrial ATP generation and highlights the importance of ATP provision by mitochondria in the presynaptic compartment during postnatal brain development.
MitoNEET is an iron-sulfur containing protein that was identified as an integral component of the outer mitochondrial membrane and discovered through its ability to bind pioglitazone, an insulin-sensitizing drug used in type 2 diabetes (Colca et al., 2004). Pioglitazone appears to stabilize mitoNEET, slowing the release of iron-sulfur clusters from mitoNEET (Paddock et al., 2007). In fact, the beneficial effects of thiazolidinedione compounds such as pioglitazone seem to be related to mitoNEET and activity on the mitochondrial function. MitoNEET has been described to act as a mitochondrial redox-sensor in several disease models including obesity (Kusminski et al., 2012), cancer (Sohn et al., 2013), cardiac hypoxia and reoxygenation (Habener et al., 2016), and inflammation-induced Parkinson’s disease (Hunter, Choi, Ross, & Bing, 2008).
Previous studies in brain mitochondria isolated from mitoNEET knockout (KO) mice revealed mitochondrial dysfunction, specifically increased production of reactive oxygen species (ROS) and reduced respiration associated with lower ATP generation (Geldenhuys et al., 2017). In fact, mitoNEET KO mice exhibit many of the characteristics of early neurodegeneration in Parkinson’s disease including reduced striatal tyrosine hydroxylase levels, decreased amounts of dopamine, and behavioral impairments (Geldenhuys et al., 2017). In spinal neurons, depletion of mitoNEET results in neuronal loss, and targeting of mitoNEET expression by microRNA-127 regulates neurite outgrowth and apoptosis (He et al., 2016). In addition to a role for MitoNEET in the brain, loss of mitoNEET also leads to mitochondrial dysfunction in other energy demanding tissues, such as the heart. Cardiac mitochondria isolated from mitoNEET deficient mice demonstrate a reduced oxidative capacity (Wiley, Murphy, Ross, van der Geer, & Dixon, 2007). Loss of mitoNEET in mouse embryonic fibroblasts (MEFs) reduces mitochondrial respiration, although this bioenergetics change appeared to be due to lower numbers of mitochondria (Vernay et al., 2017). We found reduced mitochondrial membrane potential in NIH/3T3 fibroblast cells following mitoNEET overexpression, which correlated with lower ATP levels, consistent with previous findings in fibroblasts (Kusminski et al., 2012). In contrast to fibroblasts, in SH-SY5Y cells, a human neuroblastoma line, we found that overexpression of mitoNEET significantly increased ATP production and elevated the mitochondrial membrane potential, a critical parameter that reflects ETC activity.
MitoNEET has been suggested to participate in the control of mitochondrial network morphology. Elevated mitoNEET causes elongated, fused, and dysfunctional mitochondria in white adipose tissue (Kusminski et al., 2012). MitoNEET overexpression in pancreatic β cells leads to collapse of the mitochondrial network (Kusminski et al., 2016). Further, loss of mitoNEET in MEFs decreases intermitochondrial junctions. While reintroducing low levels of mitoNEET expression in these MEFs rescues the mitochondrial network, when mitoNEET is overexpressed at high levels, the mitochondrial network collapses (Vernay et al., 2017). Since previous work revealed that loss or overexpression of mitoNEET alters the mitochondrial network, we chose to assess the effect of mitoNEET overexpression in NIH/3T3 and SH-SY5Y cells. We found that while overexpression of mitoNEET caused no changes to the mitochondrial network in NIH/3T3 cells, it did induce mitochondrial fragmentation in SH-SY5Y cells (Figures 5 and 6). Of note, studies support a role for mitochondrial fission in synaptic health (Knott, Perkins, Schwarzenbacher, & Bossy-Wetzel, 2008). In fact, mitochondrial fission facilitates mitochondrial renewal, redistribution and proliferation into synapses. We observe that in SH-SY5Y cells overexpressing mitoNEET, mitochondrial fragmentation is associated with an elevated mitochondrial membrane potential and increased ATP, suggesting a higher mitochondrial oxidative capacity. While mitochondrial elongation is often thought to promote increased mitochondrial ATP synthesis, in white adipose tissue it was found that the mitochondrial fusion observed with mitoNEET induction is associated with a lower mitochondrial oxidative capacity (Kusminski et al., 2012). Consistent with our findings in mitoNEET overexpressing SH-SY5Y cells, other conditions (i.e. nutrient excess or mitochondrial uncoupling) that result in mitochondrial fragmentation show increased respiration (Liesa & Shirihai, 2013).
Studies suggest that mitochondrial fragmentation may be beneficial for systems working at maximal respiratory capacity, and although the mechanisms by which mitochondrial fragmentation is beneficial is not yet fully understood, it is hypothesized that fragmentation might alter cristae membrane structure allowing for increased nutrient import (Liesa & Shirihai, 2013). Overexpression of mitoNEET in cells has been shown to lead to an increase in the protein levels of mitochondrial respiratory complex subunits, which reside in the cristae membrane (Salem, Whitaker-Menezes, Howell, Sotgia, & Lisanti, 2012). Consistent with this, our proteomic findings revealed elevated expression of ETC proteins coinciding with increased mitoNEET levels in synaptic mitochondria during early postnatal brain development in mice. Similarly, previous synaptic proteome work showed upregulation of several mitochondrial proteins involved in energy generation during brain development, particularly subunits of the ETC (McClatchy et al., 2012). In rat brain, significant alterations in the activities of the complexes of the ETC in presynaptic mitochondria have been reported, in particular, elevated activities of complex II-III (from P10 to P15), complex IV (from P10 to P21), and complex V (from P10 to P60) were found (Bates et al., 1994). Thus, taken with our findings that mitoNEET levels are elevated in mitochondria of neuronal origin during postnatal brain development, when energy demand is increased, and that mitoNEET overexpression increases ATP levels in cells of neuronal origin, supports an important role for mitoNEET in regulating the bioenergetics of neuronal mitochondria. Indeed mitoNEET plays an important role in mitochondrial energy function, because in the absence of mitoNEET, the regulation of oxidative phosphorylation and electron transport in mitochondria is significantly diminished.
In addition to uncovering a role for elevated mitoNEET as a potential regulator of mitochondrial bioenergetics at the synapse in response to energy demands during early postnatal brain development. Elevated mitoNEET levels may also protect the immature brain from oxidative stress, which occurs under conditions of hypoxic-ischemic injury found in preterm neonates and interm infants suffering from birth asphyxia. The developing brain is particularly susceptible to free radical injury due to its high iron levels, which facilitate the catalytic formation of free radicals, and its lower antioxidant system capacity (Blomgren & Hagberg, 2006; Hagberg, Mallard, Rousset, & Thornton, 2014). MitoNEET protects cells from oxidative stress (Habener et al., 2016) and regulates mitochondrial iron levels (Kusminski et al., 2012), thus therapeutics targeting mitoNEET may be useful for injury in the developing brain in addition to age-related neurodegeneration (Carroll, Brilhante, & Suomalainen, 2014).
In summary, our quantitative analysis of the presynaptic mitochondrial proteome shows that proteins important for ATP synthesis, including subunits of the ETC complexes, are upregulated during a period of postnatal brain development, consistent with higher energy demands during synaptogenesis. Further, we uncovered elevated presynaptic mitochondrial protein levels of mitoNEET in P42 compared to P7 mice. Overexpression of mitoNEET resulted in increased ATP production, elevated mitochondrial membrane potential, and a fragmented mitochondrial network with decreased mitochondrial biomass in cells of neuronal origin. Taken together these findings suggest that mitoNEET may play a role in the increase of the mitochondrial respiratory chain activity that is observed during brain development, particularly in the neuron during synaptogenesis.
Supplementary Material
Supplementary Figure S1. Confirmation of cell transfection and mitoNEET overexpression.
(A) Control and mitoNEET overexpression constructs were transfected into NIH/3T3 and SH-SY5Y cells. The DsRed fluorescence (568) was imaged 65 h after transfection for direct observation of cellular uptake. (B) A representative western blot is shown demonstrating mitoNEET expression in mitochondria isolated from transfected NIH/3T3 cells. Atp5b is shown as a loading control.
Supplementary Figure S2. Confirmation of cell transfection and mitoNEET overexpression for morphology experiments.
Control and mitoNEET overexpression constructs were transfected into (A) NIH/3T3 and (B) SH-SY5Y cells. MitoTracker Deep Red fluorescence was imaged for mitochondrial morphology assessments by MitoGraph. The GFP fluorescence was imaged 48 hours after transfection for direct observation of cellular uptake. The WGA fluorescence was imaged for delineating cell membranes. Combined image. (C) Western blot.
Acknowledgements
This work was supported by the National Institutes of Health grant P30 MH062261 and the Michael J. Fox Foundation grant 9524.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Bates TE, Almeida A, Heales SJ, & Clark JB (1994). Postnatal development of the complexes of the electron transport chain in isolated rat brain mitochondria. Dev Neurosci, 16(5–6), 321–327. [DOI] [PubMed] [Google Scholar]
- Blomgren K, & Hagberg H (2006). Free radicals, mitochondria, and hypoxia-ischemia in the developing brain. Free Radic Biol Med, 40(3), 388–397. doi: 10.1016/j.freeradbiomed.2005.08.040 [DOI] [PubMed] [Google Scholar]
- Carroll CJ, Brilhante V, & Suomalainen A (2014). Next-generation sequencing for mitochondrial disorders. Br J Pharmacol, 171(8), 1837–1853. doi: 10.1111/bph.12469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, … Mathews WR (2004). Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab, 286(2), E252–260. doi: 10.1152/ajpendo.00424.2003 [DOI] [PubMed] [Google Scholar]
- Dahlhaus M, Li KW, van der Schors RC, Saiepour MH, van Nierop P, Heimel JA, … Levelt CN (2011). The synaptic proteome during development and plasticity of the mouse visual cortex. Mol Cell Proteomics, 10(5), M110 005413. doi: 10.1074/mcp.M110.005413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb SJ, D’Souza RC, Cox J, Schmidt-Supprian M, & Mann M (2012). Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics, 11(5), 77–89. doi: 10.1074/mcp.M111.015362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley PR, Jarvie PE, & Robinson PJ (2008). A rapid Percoll gradient procedure for preparation of synaptosomes. Nat Protoc, 3(11), 1718–1728. doi: 10.1038/nprot.2008.171 [DOI] [PubMed] [Google Scholar]
- Erecinska M, Cherian S, & Silver IA (2004). Energy metabolism in mammalian brain during development. Prog Neurobiol, 73(6), 397–445. doi: 10.1016/j.pneurobio.2004.06.003 [DOI] [PubMed] [Google Scholar]
- Falk MJ (2010). Neurodevelopmental manifestations of mitochondrial disease. J Dev Behav Pediatr, 31(7), 610–621. doi: 10.1097/DBP.0b013e3181ef42c1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, & Shaw JM (2007). Moving mitochondria: establishing distribution of an essential organelle. Traffic, 8(12), 1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, & Mann M (2010). Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods, 7(5), 383–385. doi: 10.1038/nmeth.1446 [DOI] [PubMed] [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, & Mann M (2012). Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics, 11(3), M111 014050. doi: 10.1074/mcp.M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys WJ, Benkovic SA, Lin L, Yonutas HM, Crish SD, Sullivan PG, … Richardson JR (2017). MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson’s Disease Phenotype. ACS Chem Neurosci. doi: 10.1021/acschemneuro.7b00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys WJ, Leeper TC, & Carroll RT (2014). mitoNEET as a novel drug target for mitochondrial dysfunction. Drug Discov Today, 19(10), 1601–1606. doi: 10.1016/j.drudis.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, … Aebersold R (2012). Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics, 11(6), O111 016717. doi: 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lozano MA, Klemmer P, Gebuis T, Hassan C, van Nierop P, van Kesteren RE, … Li KW (2016). Dynamics of the mouse brain cortical synaptic proteome during postnatal brain development. Sci Rep, 6, 35456. doi: 10.1038/srep35456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson NA, & Williams PL (1969). A comparative study of brain and liver mitochondria from new-born and adult rats. J Neurochem, 16(4), 617–626. [DOI] [PubMed] [Google Scholar]
- Habener A, Chowdhury A, Echtermeyer F, Lichtinghagen R, Theilmeier G, & Herzog C (2016). MitoNEET Protects HL-1 Cardiomyocytes from Oxidative Stress Mediated Apoptosis in an In Vitro Model of Hypoxia and Reoxygenation. PLoS One, 11(5), e0156054. doi: 10.1371/journal.pone.0156054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg H, Mallard C, Rousset CI, & Thornton C (2014). Mitochondria: hub of injury responses in the developing brain. Lancet Neurol, 13(2), 217–232. doi: 10.1016/S1474-4422(13)70261-8 [DOI] [PubMed] [Google Scholar]
- Harwig MC, Viana MP, Egner JM, Harwig JJ, Widlansky ME, Rafelski SM, & Hill RB (2018). Methods for imaging mammalian mitochondrial morphology: A prospective on MitoGraph. Anal Biochem, 552, 81–99. doi: 10.1016/j.ab.2018.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverland NA, Fox HS, & Ciborowski P (2014). Quantitative proteomics by SWATH-MS reveals altered expression of nucleic acid binding and regulatory proteins in HIV-1-infected macrophages. J Proteome Res, 13(4), 2109–2119. doi: 10.1021/pr4012602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QQ, Xiong LL, Liu F, He X, Feng GY, Shang FF, … Wang TH (2016). MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection. Sci Rep, 6, 35205. doi: 10.1038/srep35205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Sherman BT, Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc, 4(1), 44–57. [DOI] [PubMed] [Google Scholar]
- Hunter RL, Choi DY, Ross SA, & Bing G (2008). Protective properties afforded by pioglitazone against intrastriatal LPS in Sprague-Dawley rats. Neurosci Lett, 432(3), 198–201. doi: 10.1016/j.neulet.2007.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Luis AM, & Cuezva JM (1990). Postnatal mitochondrial differentiation in rat liver. Regulation by thyroid hormones of the beta-subunit of the mitochondrial F1-ATPase complex. J Biol Chem, 265(16), 9090–9097. [PubMed] [Google Scholar]
- Kayala MA, & Baldi P (2012). Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res, 40(Web Server issue), W553–559. doi: 10.1093/nar/gks420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lecordier A, & Bowman LH (1995). Both nuclear and mitochondrial cytochrome c oxidase mRNA levels increase dramatically during mouse postnatal development. Biochem J, 306 ( Pt 2), 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, & Bossy-Wetzel E (2008). Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci, 9(7), 505–518. doi: 10.1038/nrn2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisman NR, Olson JE, Horne DS, & Holtzman D (1989). Cerebral oxygenation and blood flow in infant and young adult rats. Am J Physiol, 256(1 Pt 2), R78–85. doi: 10.1152/ajpregu.1989.256.1.R78 [DOI] [PubMed] [Google Scholar]
- Kristian T (2010). Isolation of mitochondria from the CNS. Curr Protoc Neurosci, Chapter 7, Unit 7 22. doi: 10.1002/0471142301.ns0722s52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Chen S, Ye R, Sun K, Wang QA, Spurgin SB, … Scherer PE (2016). MitoNEET-Parkin Effects in Pancreatic alpha- and beta-Cells, Cellular Survival, and Intrainsular Cross Talk. Diabetes, 65(6), 1534–1555. doi: 10.2337/db15-1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, … Scherer PE (2012). MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med, 18(10), 1539–1549. doi: 10.1038/nm.2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, & Shirihai OS (2013). Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab, 17(4), 491–506. doi: 10.1016/j.cmet.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy DB, Liao L, Lee JH, Park SK, & Yates JR 3rd. (2012). Dynamics of subcellular proteomes during brain development. J Proteome Res, 11(4), 2467–2479. doi: 10.1021/pr201176v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Martinovic J, Chretien D, Dumez F, Razavi F, Munnich A, & Rotig A (2008). Mitochondrial respiratory chain complex assembly and function during human fetal development. Mol Genet Metab, 94(1), 120–126. doi: 10.1016/j.ymgme.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Moczulska KE, Pichler P, Schutzbier M, Schleiffer A, Rumpel S, & Mechtler K (2014). Deep and precise quantification of the mouse synaptosomal proteome reveals substantial remodeling during postnatal maturation. J Proteome Res, 13(10), 4310–4324. doi: 10.1021/pr500456t [DOI] [PubMed] [Google Scholar]
- Nehlig A, de Vasconcelos AP, & Boyet S (1988). Quantitative autoradiographic measurement of local cerebral glucose utilization in freely moving rats during postnatal development. J Neurosci, 8(7), 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlig A, Pereira de Vasconcelos A, & Boyet S (1989). Postnatal changes in local cerebral blood flow measured by the quantitative autoradiographic [14C]iodoantipyrine technique in freely moving rats. J Cereb Blood Flow Metab, 9(5), 579–588. doi: 10.1038/jcbfm.1989.83 [DOI] [PubMed] [Google Scholar]
- Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, … Jennings PA (2007). MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci U S A, 104(36), 14342–14347. doi: 10.1073/pnas.0707189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, … Mootha VK (2008). A mitochondrial protein compendium elucidates complex I disease biology. Cell, 134(1), 112–123. doi: 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh JJ (1970). Mitochondrial changes in rat inferior colliculus during postnatal development: an electron microscopic study. Brain Res, 18(2), 325–342. [DOI] [PubMed] [Google Scholar]
- Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, & Lisanti MP (2012). Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle, 11(22), 4174–4180. doi: 10.4161/cc.22376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson FE Jr., Balfour WM, & Jacobs RJ (1960). Mitochondrial changes in developing rat brain. Am J Physiol, 199, 693–696. doi: 10.1152/ajplegacy.1960.199.4.693 [DOI] [PubMed] [Google Scholar]
- Scopes RK (1974). Measurement of protein by spectrophotometry at 205 nm. Anal Biochem, 59(1), 277–282. [DOI] [PubMed] [Google Scholar]
- Semple BD, Blomgren K, Gimlin K, Ferriero DM, & Noble-Haeusslein LJ (2013). Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol, 106–107, 1–16. doi: 10.1016/j.pneurobio.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, … Schaeffer DA (2007). The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics, 6(9), 1638–1655. doi: 10.1074/mcp.T600050-MCP200 [DOI] [PubMed] [Google Scholar]
- Smith AC, Blackshaw JA, & Robinson AJ (2012). MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res, 40(Database issue), D1160–1167. doi: 10.1093/nar/gkr1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, … Mittler R (2013). NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci U S A, 110(36), 14676–14681. doi: 10.1073/pnas.1313198110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauch KL, Purnell PR, & Fox HS (2014a). Aging synaptic mitochondria exhibit dynamic proteomic changes while maintaining bioenergetic function. Aging (Albany NY), 6(4), 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauch KL, Purnell PR, & Fox HS (2014b). Quantitative proteomics of synaptic and nonsynaptic mitochondria: insights for synaptic mitochondrial vulnerability. J Proteome Res, 13(5), 2620–2636. doi: 10.1021/pr500295n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J, & Jernigan TL (2010). The basics of brain development. Neuropsychol Rev, 20(4), 327–348. doi: 10.1007/s11065-010-9148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, & Chiaramello A (2014). Mitochondrial biogenesis: a therapeutic target for neurodevelopmental disorders and neurodegenerative diseases. Curr Pharm Des, 20(35), 5574–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernay A, Marchetti A, Sabra A, Jauslin TN, Rosselin M, Scherer PE, … Cosson P (2017). MitoNEET-dependent formation of intermitochondrial junctions. Proc Natl Acad Sci U S A, 114(31), 8277–8282. doi: 10.1073/pnas.1706643114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve LM, Purnell PR, Boska MD, & Fox HS (2016). Early Expression of Parkinson’s Disease-Related Mitochondrial Abnormalities in PINK1 Knockout Rats. Mol Neurobiol, 53(1), 171–186. doi: 10.1007/s12035-014-8927-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve LM, Stauch KL, & Fox HS (2014a). Proteomic analysis of the mitochondria from embryonic and postnatal rat brains reveals response to developmental changes in energy demands. J Proteomics. doi: 10.1016/j.jprot.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve LM, Stauch KL, & Fox HS (2014b). Proteomic analysis of the mitochondria from embryonic and postnatal rat brains reveals response to developmental changes in energy demands. J Proteomics, 109, 228–239. doi: 10.1016/j.jprot.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B (2012). Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta, 1817(10), 1833–1838. doi: 10.1016/j.bbabio.2012.02.033 [DOI] [PubMed] [Google Scholar]
- Wiley SE, Murphy AN, Ross SA, van der Geer P, & Dixon JE (2007). MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci U S A, 104(13), 5318–5323. doi: 10.1073/pnas.0701078104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, & Mann M (2009). Universal sample preparation method for proteome analysis. Nat Methods, 6(5), 359–362. doi: 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Confirmation of cell transfection and mitoNEET overexpression.
(A) Control and mitoNEET overexpression constructs were transfected into NIH/3T3 and SH-SY5Y cells. The DsRed fluorescence (568) was imaged 65 h after transfection for direct observation of cellular uptake. (B) A representative western blot is shown demonstrating mitoNEET expression in mitochondria isolated from transfected NIH/3T3 cells. Atp5b is shown as a loading control.
Supplementary Figure S2. Confirmation of cell transfection and mitoNEET overexpression for morphology experiments.
Control and mitoNEET overexpression constructs were transfected into (A) NIH/3T3 and (B) SH-SY5Y cells. MitoTracker Deep Red fluorescence was imaged for mitochondrial morphology assessments by MitoGraph. The GFP fluorescence was imaged 48 hours after transfection for direct observation of cellular uptake. The WGA fluorescence was imaged for delineating cell membranes. Combined image. (C) Western blot.


