Abstract
Neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and Parkinson’s disease (PD), cause the loss of hundreds of thousands of lives each year. Effective treatment options able to halt disease progression are lacking. Despite the extensive sequencing efforts in large patient populations, the majority of ALS and PD cases remain unexplained by genetic mutations alone. Epigenetics mechanisms, such as the post-translational modification of histone proteins, may be involved in neurodegenerative disease etiology and progression and lead to new targets for pharmaceutical intervention. Mammalian in vivo and in vitro models of ALS and PD are costly and often require prolonged and laborious experimental protocols. Here, we outline a practical, fast, and cost-effective approach to determining genome-wide alterations in histone modification levels using Saccharomyces cerevisiae as a model system. This protocol allows for comprehensive investigations into epigenetic changes connected to neurodegenerative proteinopathies that corroborate previous findings in different model systems while significantly expanding our knowledge of the neurodegenerative disease epigenome.
Keywords: neurodegeneration, amyotrophic lateral sclerosis, Parkinson’s disease, α-synuclein, fused in sarcoma, TAR DNA-binding protein 43, histone post-translational modifications, epigenetics
SUMMARY:
This protocol outlines experimental procedures to characterize genome-wide changes in the levels of histone post-translational modifications (PTM) occurring in connection with the overexpression of proteins associated with ALS and Parkinson’s disease in Saccharomyces cerevisiae models. After SDS-PAGE separation, individual histone PTM levels are detected with modification-specific antibodies via Western blotting.
INTRODUCTION:
Neurodegenerative diseases are devastating illnesses with little to no treatment options available. Among these, amyotrophic lateral sclerosis (ALS) and Parkinson’s disease (PD) are particularly dreadful. Approximately 90% of ALS and PD cases are considered sporadic, occurring without family history of the disease, while the remaining cases run in families and are generally linked to a specific gene mutation1,2. Interestingly, both of these diseases are associated with protein mislocalization and aggregation3–6. For instance, fused in sarcoma (FUS) and TAR DNA-binding protein 43 (TDP-43) are RNA binding proteins that mislocalize to the cytoplasm and aggregate in ALS7–12, while α-synuclein is the principle component of proteinaceous aggregates termed Lewy bodies in PD5,13–15.
Despite the extensive genome-wide association efforts in large patient populations, the overwhelming majority of ALS and PD cases remain unexplained genetically. Can epigenetics play a role in neurodegenerative disease? Epigenetics comprises changes in gene expression occurring without changes to underlying DNA sequence16. A main epigenetic mechanism involves the post translational modifications (PTMs) of histone proteins16. In eukaryotic cells, genetic material is tightly wrapped into chromatin. The base unit of chromatin is the nucleosome, consisting of 146 base pairs of DNA wrapped around a histone octamer, comprised of four pairs of histones (two copies each of histones H2A, H2B, H3, and H4)17. Each histone has an N-terminal tail that protrudes out of the nucleosome and can be modified by the addition of various chemical moieties, usually on lysine and arginine residues18. These PTMs are dynamic, which means they can be easily added and removed, and include groups such as acetylation, methylation, and phosphorylation. PTMs control the accessibility of DNA to the transcriptional machinery, and thus help control gene expression18. For example, histone acetylation reduces the strength of the electrostatic interaction between the highly basic histone protein and the negatively charged DNA backbone, allowing the genes packed by acetylated histones to be more accessible and thus highly expressed19. More recently, the remarkable biological specificity of particular histone PTMs and their combinations has led to the ‘histone code hypothesis20,21‘ in which proteins that ‘write,’ ‘erase,’ and ‘read’ histone PTMs all act in concert to modulate gene expression.
Yeast is a very useful model to study neurodegeneration. Importantly, many neuronal cellular pathways are conserved from yeast to humans22–24. Yeast recapitulate cytotoxicity phenotypes and protein inclusions upon overexpression of FUS, TDP-43 or α-synuclein22–26. In fact, S. cerevisiae models of ALS have been used to identify genetic risk factors in humans27. Furthermore, yeast overexpressing human α-synuclein allowed for the characterization of the Rsp5 network as a druggable target to ameliorate α-synuclein toxicity in neurons28,29.
Here, we describe a protocol exploiting Saccharomyces cerevisiae to detect genome-wide histone PTM changes associated with neurodegenerative proteinopathies (Figure 1). The use of S. cerevisiae is highly attractive because of its ease of use, low cost, and speed compared to other in vitro and animal models of neurodegeneration. Harnessing previously developed ALS and PD models22,23,25,26, we have overexpressed human FUS, TDP-43, and α-synuclein in yeast and uncovered distinct histone PTM changes occurring in connection with each proteinopathy30. The protocol that we describe here can be completed in less than two weeks from transformation to data analysis.
Figure 1:
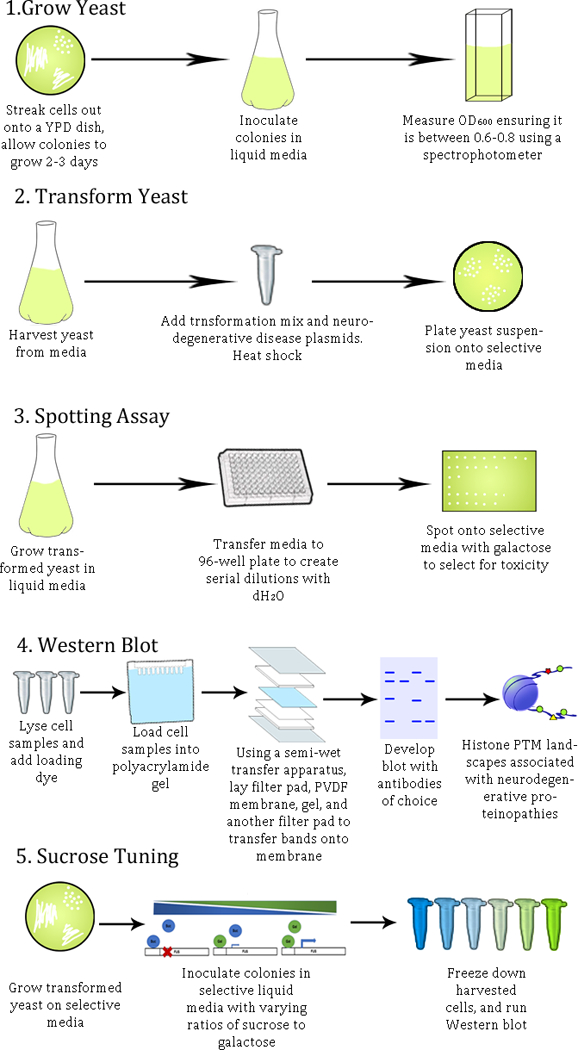
Method overview for the characterization of changes in histone post-translational modifications connected to neurodegenerative disease proteinopathies in yeast models.
PROTOCOL:
1. Transforming S. cerevisiae with neurodegenerative proteinopathy-associated protein constructs
-
1.1
Grow wild type (WT) 303 yeast in yeast extract peptone dextrose (YPD) broth overnight with shaking (200 rpm) at 30 °C.
-
1.2
After 12−16 h of growth, dilute yeast to an optical density at 600 nm (OD600) of 0.25 with YPD. As 10 mL of yeast liquid culture will be needed for each transformation, prepare 50 mL of yeast liquid culture for five transformations corresponding to FUS, TDP-43, α-synuclein, and vector only (ccdB) constructs, as well as a negative control transformation without DNA.
-
1.3
Grow yeast with agitation at 30 °C until an OD600 between 0.60 and 0.80 is reached. This generally takes 4−6 h.
-
1.4
Thirty minutes before yeast growth is complete, linearize plasmid constructs.
NOTE: The plasmid constructs used here must be integrated directly into the genome, and hence linearization is required prior to transformation.
-
1.4.1
Calculate the volume of plasmid stock that amounts to 1 μg. Subtract this volume from 44 μL to calculate the amount of nuclease free H2O needed.
-
1.4.2
Add nuclease free H2O, 5 μL of 10x restriction enzyme buffer (see Table of Materials), 1 μL of NheI restriction enzyme (see Table of Materials), and the appropriate amount of plasmid (calculated in step 1.4.1) to a microcentrifuge tube. Mix thoroughly by pipetting up and down and incubate at 37 °C for 15 min. The plasmid is now linearized and ready for transformation.
-
1.5
After yeast culture reaches an OD600 of 0.6–0.8, harvest cells in a 50 mL conical tube and spin down at 850 x g at 4 °C for 3 min. Discard supernatant.
-
1.6
Wash cell pellet in 10 mL of sterile H2O and centrifuge at 850 x g at 4 °C for 3 min. Discard supernatant. Repeat three more times for a total of four washes.
-
1.7
At the start of the third wash, thaw and boil 10 μL of salmon sperm DNA per transformation reaction for 5 min on a heating block.
-
1.8
Resuspend cell pellet in 100 μL of dH2O per transformation and split into appropriate number of microcentrifuge tubes. For five transformations, resuspend pellet into 500 μL of dH2O and split evenly into five separate microcentrifuge tubes.
-
1.9
Centrifuge for 5 min at 850 x g at room temperature (RT) and remove all remaining water.
-
1.10
Add, in the following order, 50 μL of sterile H2O, 240 μL of 50% polyethylene glycol (PEG), 36 μL of 1 M LiAc, 10 μL of salmon sperm DNA, 20 μL of linearized plasmid DNA (or nuclease free water for no DNA control transformation). Mix thoroughly after each addition and vortex briefly after all transformation components have been added.
-
1.11
Incubate transformation reactions at 42 °C for 20 min.
NOTE: For more consistent temperature control, this incubation should be carried out in a water bath.
-
1.12
Centrifuge at 470 x g at RT for 5 min. Discard supernatant and resuspend each cell pellet in 200 μL of sterile H2O.
-
1.13
Plate yeast suspension on selective media plates and spread with rolling beads or spreader. Incubate plates for two to three days at 30 °C.
NOTE: Synthetic-defined (SD)-His plates are used for the plasmids described here.
2. Surveying colony growth suppression and storage in glycerol stocks
-
2.1
Inoculate single colonies from transformation plates in 5 mL of selective media supplemented with 2% raffinose. Grow on a shaker at 30 °C overnight. Select at least 12 colonies per transformation.
NOTE: No colonies are expected in the ‘no DNA’ transformation control plate.
-
2.2
Sterilize pin-frogger with ethanol, followed by flaming.
-
2.3
For each culture, aliquot 100 μL of saturated cell suspension in the first row of a 96-well plate. Add 200 μL of sterile H2O in all wells of the adjacent well columns. For 1:5 serial dilutions, add 50 μL from the first column to the adjacent column behind with multichannel pipet. Mix thoroughly by pipetting. Then add 50 μL from the second column to the third column and mix by pipetting. Repeat this for the rest of the plate.
-
2.4
Plate yeast by placing frogger in a 96-well plate and then stamping by gently and evenly pressing down on selective media plates with and without galactose. Incubate at 30 °C for two to three days.
NOTE: SD-His and SGal-His plates are used for the plasmids described here.
-
2.5
For FUS, TDP-43, and α-synuclein transformations, identify the colony displaying the most growth suppression in the presence of galactose. Select this colony from the SD-His plate and inoculate into 10 mL of selective media supplemented with 2% raffinose for overnight growth. For the vector control, pick a colony displaying no growth suppression in the presence of galactose.
-
2.6
To prepare glycerol stocks, combine 0.5 mL of saturated liquid culture with 0.5 mL of 50% glycerol. Mix by pipetting up and down and freeze at −80 °C.
NOTE: Glycerol stocks can be stored for up to a year at −80 °C.
3. Overexpression of neurodegenerative proteinopathy associated proteins in S. cerevisiae
-
3.1
From frozen glycerol stocks, re-streak yeast on –Histidine, 2% glucose agar selective media plates and incubate at 30 °C for 2–3 days.
-
3.2
Inoculate single colonies for each of overexpression models and control in 5mL of -Histidine selective liquid media supplemented with 2% raffinose. Grow with shaking (200 rpm) at 30 °C.
-
3.3
Grow 100 mL of liquid culture for each of overexpression models and controls (FUS, TDP-43 & control and α-synuclein & control).
-
3.3.1
Prepare 100 mL of culture in selective media supplemented with 2% galactose.
-
3.3.2
Measure the OD600 of overnight cultures and calculate the amount of overnight culture needed to render overexpression cultures at a starting OD600 of 0.3. Typically, overnight cultures will reach an OD600 of 0.9−10, requiring about 5 mL of overnight culture to start 100 mL of fresh culture.
-
3.3.3
Induce protein overexpression by growing yeast on galactose media (described in step 3.3.1) for 5 h (FUS and TDP-43) or 8 h (α-synuclein) with shaking at 30 °C.
-
3.4
Measure OD600 of each culture at the end of the induction period. Standardize all cell counts to the lowest OD600 value. Harvest culture in 50 mL tubes by centrifuging at 850 x g for 5 min at 4 °C. Discard supernatant. For example, if at the end of induction, the measured OD600 for 100 mL of α-synuclein culture is 0.654 and 100 mL of control culture is .984, harvest 95 mL of the α-synuclein culture and 67.1 mL of the control culture. 95 mL of α-synuclein culture to allow for some culture to be used for measurement of the OD600.
-
3.5
Resuspend pellet in 1 mL of sterile dH2O for every 10 mL of culture grown. Split pellet evenly by aliquoting 1 mL of cell resuspension into microcentrifuge tubes. For example, if starting with 100 mL of culture, resuspend pellet in 10 mL of sterile dH2O and divide it into 10 microcentrifuge tubes.
-
3.6
Centrifuge at 850 x g for 5 min at 4 °C, then remove supernatant. Snap freeze cell pellets in liquid nitrogen and store at −80 °C.
NOTE: Cell pellets can be stored at −80 °C for up to one year.
4. Cell lysis and western blotting to detect histone post-translational modifications
-
4.1
Cell lysis
-
4.1.1
Thaw yeast cell pellets on ice and resuspend cell pellets in 100 μL of dH2O.
-
4.1.2
To the cell resuspension, add 300 μL of 0.2 M NaOH and 20 μL of 2-mercaptoethanol. Resuspend by pipetting up and down.
-
4.1.3
Incubate cells on ice for 10 min and then centrifuge at 3,200 x g on a tabletop centrifuge for 30 s at RT. Discard supernatant.
-
4.1.4
Resuspend cell pellet in 100 μL of 1x loading dye and boil for 10 min on a heating block set to 100 °C.
NOTE: The recipe for 6x loading dye can be found in the Table of Materials.
-
4.2
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and membrane transfer
-
4.2.1
Prepare gel chamber by placing two gels in a gel holder and filling inner chamber to the top with running buffer and filling the outside chamber to the two-gel line mark.
-
4.2.2
Load 15 μL of sample from step 4.1 per well into a 10-well 12% polyacrylamide gel. For the protein ladder lane, load 5 μL.
-
4.2.3
Run gel for approximately 45 min at 150 V, or until loading dye front reaches the bottom of gel.
-
4.2.4
While the gel is running, prepare for membrane transfer by soaking fiber pads (two per gel) in transfer buffer (see Table of Materials) and soaking polyvinylidene fluoride (PVDF) membrane in methanol. The PVDF membrane should be cut to the size of the gel and one PVDF membrane for every gel being transferred. Rinse membrane in transfer buffer before transfer.
-
4.2.5
Assemble, on semi-dry transfer apparatus cell, a transfer ‘sandwich’: from bottom electrode, place (1) presoaked fiber pad, (2) presoaked and rinsed PVDF membrane, (3) gel from step 4.2.3, and (4) a second presoaked fiber pad.
NOTE: Make sure to avoid air bubbles in the transfer ‘sandwich.’ Gently roll ‘sandwich’ with serological pipet to force out bubbles. Once the gel has been placed on top of the PDVF membrane, make sure not to move it.
-
4.2.6
Carry out protein transfer by setting power pack to 150 mA for 1 h. (for one ‘sandwich’).
-
4.3
Detection of histone PTMs with modification specific antibodies
-
4.3.1
Remove membrane from transfer apparatus. Rinse briefly with dH2O.
-
4.3.1.1
Optionally, check transfer of proteins with Ponceau-S stain by pouring enough Ponceau-S stain to cover membrane in a small plastic box and incubating at RT with gentle shaking for 30−60 s. Remove Ponceau-S stain and rinse with dH2O until background stain disappears and protein bands are visible to the naked eye. Continue to rinse with dH2O until all stain is removed from membrane.
-
4.3.2
Block membrane by incubating blot for 1 h at RT with gentle rocking with commercially available tris buffered saline (TBS) blocking buffer (see Table of Materials). Use enough blocking buffer cover blot in small staining box.
NOTE: Be careful to place membrane in staining box upright, so the membrane side on which proteins lie is not facing down.
-
4.3.3
Incubate blot in staining box overnight with a histone modification-specific antibody reactive towards yeast at 4 °C. Dilute the antibody in TBS blocking buffer according to manufacturer specifications. Also include a proper nuclear loading, control such as anti-total H3, raised in a different host species from the modification-specific antibody. For instance, if probing for H3S10ph with an anti-H3S10ph antibody raised in rabbit, use an anti-total H3 antibody raised in mouse as a loading control. Repeat this for every blot as necessary.
NOTE: Antibody dilutions can be reused for a total of three times within a month. Store them at 4 °C.
-
4.3.4
Wash the membrane 4x in house-made TBS + 0.1% polysorbate 20 (TBST) for 5 min with rocking at RT.
-
4.3.5
Incubate blot with fluorescent secondary antibodies at manufacturer-specified dilutions (donkey anti-rabbit 680 and donkey anti-mouse) for 1 h at RT.
NOTE: Fluorescent secondary antibodies must be protected from light during both storage and usage. Carry out incubation in dark plastic boxes or cover with aluminum foil.
-
4.3.6
Wash membrane 4x with TBST for 5 min while rocking at RT.
-
4.3.7
Wash membrane with TBS for 5 min while rocking at RT.
-
4.3.8
Visualize blot on a fluorescent Western blot imaging system for 2 min. Visualize the anti-rabbit 680 and anti-mouse 800 secondary antibodies in the 800 nm and 700 nm channels, respectively.
NOTE: Replicates are independently conducted starting from section 1. Verification that the antibody signal response is within the range over which the signal response is linear is necessary for appropriate data interpretation in these experiments.
5. Data analysis and statistics
-
5.1
Open image of blot in an imaging software (see Table of Materials) and acquire the raw density of modification-specific bands in the vector control, TDP-43 & FUS or vector control & α-synuclein samples. This is done by drawing a rectangle that frames the band in analysis mode.
-
5.2
Repeat step 5.1 for loading control bands. There will be a loading control band and a modification-specific band in each sample.
-
5.3
Calculate the relative density of the modification-specific bands by dividing each band by the density of corresponding band in the vector control sample. This normalizes the data to the vector control.
-
5.4
Calculate the relative density of the loading control band by dividing each band by the density of corresponding loading control band in the vector control sample
-
5.5
Calculate the adjusted relative density of each band by dividing the relative density of the modification-specific band over the relative density of the loading control band for each sample. Now, the data can be visualized in histogram form.
NOTE: For ease of processing, steps 5.3–5.5 can be done in a spreadsheet.
-
5.6
After multiple independent replicates, run Welch’s T-tests between the two control samples and the appropriate proteinopathy model (control versus FUS, control versus TDP-43, and control versus α-synuclein) with p = 0.05 as the cutoff for significance.
6. Sucrose tuning of FUS overexpression
-
6.1.
Overexpress FUS and harvest yeast as described in section 3, but use sucrose to galactose ratios corresponding to: all sucrose; 40 sucrose: 1 galactose; 20 sucrose: 1 galactose; 10 sucrose: 1 galactose; 1 sucrose: 2 galactose; all galactose. The total concentration of sugar in each sample will amount to 2% in all conditions. For example, to make a 100 mL of media containing a 40:1 ratio of sucrose to galactose, add 9.756 mL of 20% sucrose and 244 μL of 20% galactose to 90 mL of selective media.
-
6.2
For data analysis, compare the fold changes in the PTM levels, standardizing to the sucrose condition, and run a one-way ANOVA with a p = 0.05 as a cutoff.
REPRESENTATIVE RESULTS:
To illustrate this method, we will take advantage of recently published results30. WT human FUS and TDP-43 were overexpressed for 5 h, while WT α-synuclein was overexpressed for 8 h. A ccdB construct was used as a vector negative control. Figure 2 shows growth suppression in solid and liquid cultures. Yeast was harvested as described and Western blotting with modification-specific antibodies was performed. Anti-total H3 was used as a loading control. A significant decrease in the levels of H3S10ph and H3K14ac is apparent in the FUS overexpression model (Figure 3a,b). There are significant increases in the levels of acetylation on H4K12 and H4K16 in the TDP-43 overexpression model that are not observed in either the FUS or α-synuclein overexpression model (Figure 3c,d). There are also significant decreases in the levels of H3K36me2 and H2BT129ph in the α-synuclein overexpression model (Figure 3e,f).
Figure 2: Toxicity associated with overexpression of neurodegenerative proteinopathy-related proteins in yeast models.
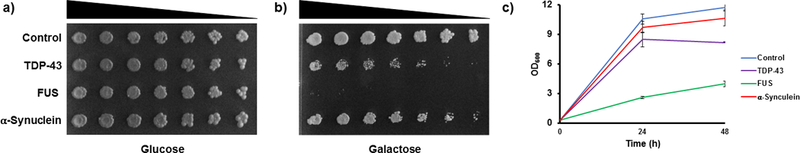
Spotting assays show cell viability for yeast overexpressing vector control, TDP-43, FUS, or α-synuclein in the presence of glucose (a) or galactose (b). (c) Growth curve illustrating cell viability in liquid culture under galactose induction. Error bars indicate the ± SD. n = 3 for each strain. Replicates result from completely independent experiments. Data adapted with permission from Chen, K. et al. Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes. ACS Chem Neurosci. 9 (4), pp 838–848 doi:10.1021/acschemneuro.7b00297. Copyright 2018 American Chemical Society.
Figure 3: Changes in histone post-translational modifications associated with overexpression of neurodegenerative proteinopathy-related proteins in yeast models.
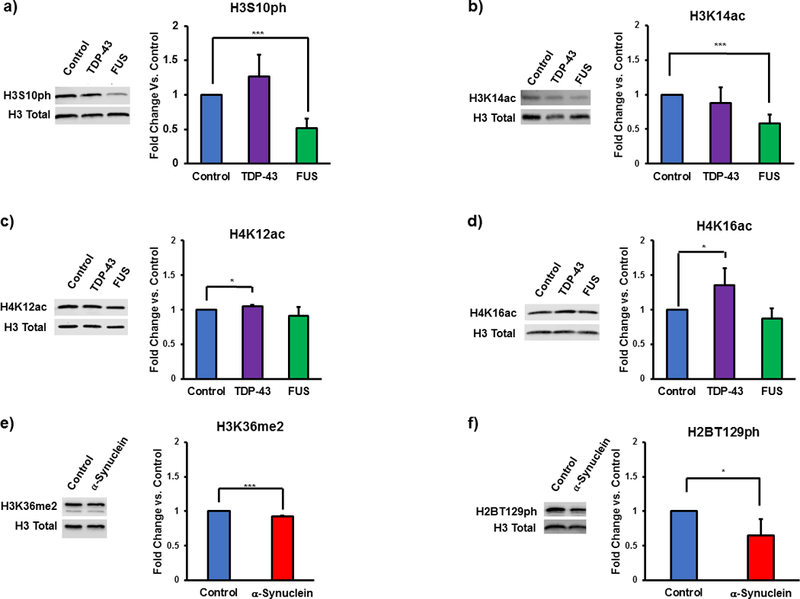
An FUS proteinopathy model shows decreased levels of (a) H3S10ph, n = 6, and (b) H3K14ac, n = 3. Conversely, a TDP-43 proteinopathy model shows increased levels of (c) H4K12ac, n = 3, and (d) H4K16ac, n = 6, while a α-synuclein model shows decreased levels of (e) H3K36me2, n = 3, and (f) H2BT129ph, n= 7. Error bars show + SD. *, p < 0.05, ***, p < 0.001. Replicates result from completely independent experiments. Data adapted with permission from Chen, K. et al. Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes. ACS Chem Neurosci. 9 (4), pp 838–848 doi:10.1021/acschemneuro.7b00297. Copyright 2018 American Chemical Society.
To show that the change in histone PTMs correlates with the amount of expression of the neurodegenerative proteinopathy protein, the expression of FUS can be ‘tuned’ by inducing protein expression in varying levels of galactose (Figure 4a). Galactose is mixed with sucrose in changing ratios such that the total concentration of sugar is constant at 2%, but the amount of galactose is modified resulting in a range of protein expression levels. In S. cerevisiae, sucrose is slowly metabolized to glucose, which suppresses galactose induction31. Hence, sucrose neither activates nor suppresses galactose induction. The lower the amount of galactose used to induce FUS overexpression, the less toxicity was observed in both solid and liquid culture (Figure 4b,c). Importantly, the lower the level of FUS overexpression, the smaller the magnitude of the reduction in H3S10ph levels (Figure 4d–f).
Figure 4: Extent of decrease in H3S10ph levels is tied to the level of FUS overexpression.
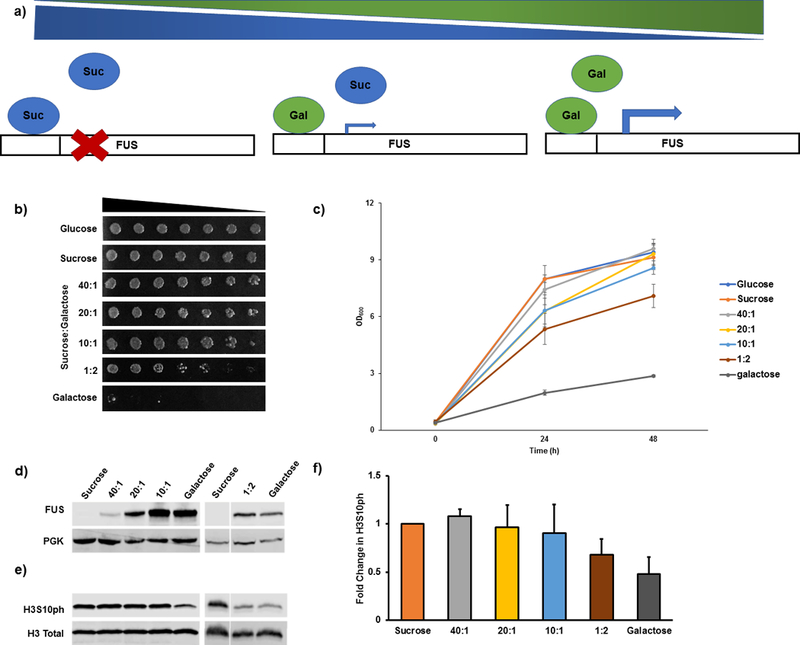
(a) Cartoon illustration of the use of sucrose and galactose ratios to ‘tune’ the amount of FUS overexpression. (b) Spotting assays showing cell viability in the presence of varying levels of galactose. (c) Growth curve in liquid culture showing cell viability in the presence of varying levels of galactose. Error bars indicate ± SD. (d) Representative immunoblots showing that the FUS protein levels rise as cells are exposed to increasing ratios of galactose. Phosphoglycerate kinase (PGK) was used as a loading control. (e) Representative immunoblots showing corresponding decreases in H3S10ph levels with increasing ratios of galactose. (f) Quantitation histogram of (e). n = 3 for each condition. Error bars indicate + SD for quantification chart. Replicates result from completely independent experiments. Data adapted with permission from Chen, K. et al. Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes. ACS Chem Neurosci. 9 (4), pp 838–848 doi:10.1021/acschemneuro.7b00297. Copyright 2018 American Chemical Society.
DISCUSSION:
The protocol described here provides a straightforward, expedient, and cost-effective way of categorizing genome-wide histone PTM changes correlated with neurodegenerative proteinopathies. While there are other models of ALS and PD, such as in vitro human cell lines and murine models32, S. cerevisiae remains attractive because of its ease of use. For instance, yeast models do not require use of a sterile hood, nor do they require the intensive training that goes along with cell culture work. Furthermore, the reagents for culturing yeast are also much more affordable than mammalian cell culture supplies. Yeast models have been exploited to not only reveal the domain determinants of protein aggregation22–26, but also have led to uncovering genetic risk factors in ALS27, as well as modifiers of protein aggregation toxicity22. Furthermore, the deletion of Set3, a member of the histone deacetylase complex, has been found to reduce TDP-43 toxicity in yeast33. Interestingly, our findings in yeast are in agreement with recent reports on histone modification alterations in both an FUS mouse model and in a human SH-SY5Y FUS overexpression model34,35. Furthermore, recently discovered alterations in DNA methylation patterns around key PD genes, such as SNCA and PARK2, support a role for epigenetics in PD36,37.
Here, we show that these yeast models can also be used to discover how neurodegenerative proteinopathies interact with the epigenome30. We find that distinct changes in histone modifications are associated with each proteinopathy model. The protocol outlined here allows us to corroborate previous findings in other model systems. Most remarkably, this method has heightened our understanding of the epigenomic panorama of neurodegenerative disease. The expanded set of modifications presented here could uncover novel histone “writer” and “eraser” targets for pharmacological intervention. These results highlight the possible contribution of histones PTMs and epigenetics in the pathology of ALS, PD, and other neurodegenerative diseases and may provide novel avenues for treatment of these diseases.
Particular care needs to be taken in steps 1.9 and 1.10 when transforming yeast to overexpress neurodegenerative proteinopathies. Specifically adding the reagents in the proper order during step 1.10 is necessary to ensure high transformation efficiency. Furthermore, it is very important to pick the colony displaying the most growth suppression in galactose. Care should also be taken when assembling the Western ‘sandwich’. Accidental addition of bubbles between the gel and membrane will block transfer of the protein and provide block spots on the blot that can hinder data analysis.
One limitation of this protocol is that antibodies are restricted to only binding and detecting one or two histone modifications at a time. It is also crucial to properly standardize samples after protein overexpression (steps 3.4−3.6). Completely removing all growth media will ensure that each cell pellet winds up resuspended in the same volume. Similarly, it is important to carefully aliquot the same amount of cell suspension into the microcentrifuge tubes (step 3.4). If the samples are not properly standardized, it is difficult to draw any conclusions from any changes on histone PTM levels. Such a problem would become apparent in discrepancies on the levels of the loading control of the Western blot (step 4.2). To remedy this, an SDS-PAGE gel loaded with relevant samples can be Coomassie stained, allowing for protein band quantification of the samples (section 5) and subsequent re-standardization of samples based on the amount of protein present on each sample.
In conclusion, this protocol allows for the analysis of histone PTM changes associated with the overexpression of proteins associated with neurodegenerative proteinopathies in just less than two weeks, from transformation to statistical analysis. Aside from enabling the study of proteinopathies associated with neurodegenerative diseases, this general protocol can be used to study histone PTM alterations in any yeast overexpression model.
Supplementary Material
ACKNOWLEDGMENTS:
We thank Royena Tanaz, Huda Yousuf, and Sadiqa Taasen for technical help. We are very grateful to Prof. James Shorter for the generous provision of reagents and intellectual assistance in the design of sucrose tuning experiments. Yeast plasmids were a generous gift from Prof. Aaron Gitler (including 303Gal-FUS; Addgene plasmid # 29614). Brooklyn College and the Advanced Science Research Center (CUNY) as well as an NIH NINDS Advanced Postdoctoral Transition Award (K22NS09131401) supported M.P.T.
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES:
- 1.Landgrave-Gómez J, Mercado-Gómez O, Guevara-Guzmán R Epigenetic mechanisms in neurological and neurodegenerative diseases. Frontiers in Cellular Neuroscience. 9, 58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, Goutman SA, Feldman EL Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nature Reviews Neurology. 11 (5), 266–279, doi: 10.1038/nrneurol.2015.57, (2015). [DOI] [PubMed] [Google Scholar]
- 3.Beitz JM Parkinson’s Disease: a Review. Frontiers in bioscience (6), 65–74, doi: 10.2741/S415 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495, 467, doi:10.1038/nature1192210.1038/nature11922https://www.nature.com/articles/nature11922-supplementary-informationhttps://www.nature.com/articles/nature11922-supplementary-information, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poewe W et al. Parkinson disease. Nature Reviews Disease Primers. 3, 17013, doi:10.1038/nrdp.2017.1310.1038/nrdp.2017.13https://www.nature.com/articles/nrdp201713-supplementary-informationhttps://www.nature.com/articles/nrdp201713-supplementary-information, (2017). [DOI] [PubMed] [Google Scholar]
- 6.Robberecht W & Philips T The changing scene of amyotrophic lateral sclerosis. Nature Reviews Neuroscience. 14 248, doi:10.1038/nrn343010.1038/nrn3430https://www.nature.com/articles/nrn3430-supplementary-informationhttps://www.nature.com/articles/nrn3430-supplementary-information, (2013). [DOI] [PubMed] [Google Scholar]
- 7.Chen-Plotkin AS, Lee VMY & Trojanowski JQ TAR DNA-binding protein 43 in neurodegenerative disease. Nature Reviews Neurology. 6 211, doi: 10.1038/nrneurol.2010.18, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couthouis J et al. A yeast functional screen predicts new candidate ALS disease genes. Proceedings of the National Academy of Sciences. 108 (52), 20881 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Da Cruz S & Cleveland DW Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Current Opinion in Neurobiology. 21 (6), 904–919, doi: 10.1016/j.conb.2011.05.029, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King OD, Gitler AD & Shorter J The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Research. 1462 61–80, doi: 10.1016/j.brainres.2012.01.016, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann M et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 134 (9), 2595–2609, doi: 10.1093/brain/awr201, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann M et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 314 (5796), 130 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Baba M et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. The American Journal of Pathology. 152 (4), 879–884 (1998). [PMC free article] [PubMed] [Google Scholar]
- 14.Lotharius J & Brundin P Pathogenesis of parkinson’s disease: dopamine, vesicles and α-synuclein. Nature Reviews Neuroscience. 3 932, doi: 10.1038/nrn983, (2002). [DOI] [PubMed] [Google Scholar]
- 15.Spillantini MG et al. α-Synuclein in Lewy bodies. Nature. 388 839, doi: 10.1038/42166, (1997). [DOI] [PubMed] [Google Scholar]
- 16.Probst AV, Dunleavy E & Almouzni G Epigenetic inheritance during the cell cycle. Nature Reviews Molecular Cell Biology. 10 192, doi: 10.1038/nrm2640, (2009). [DOI] [PubMed] [Google Scholar]
- 17.Luger K, Mäder AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 389 251, doi: 10.1038/38444, (1997). [DOI] [PubMed] [Google Scholar]
- 18.Mazzio EA & Soliman KFA Basic concepts of epigenetics. Epigenetics. 7 (2), 119–130, doi: 10.4161/epi.7.2.18764, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struhl K Histone acetylation and transcriptional regulatory mechanisms. Genes & Development. 12 (5), 599–606 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Garcia BA, Shabanowitz J & Hunt DF Characterization of histones and their post-translational modifications by mass spectrometry. Current Opinion in Chemical Biology. 11 (1), 66–73, doi: 10.1016/j.cbpa.2006.11.022, (2007). [DOI] [PubMed] [Google Scholar]
- 21.Strahl BD & Allis CD The language of covalent histone modifications. Nature. 403 41, doi: 10.1038/47412, (2000). [DOI] [PubMed] [Google Scholar]
- 22.Jovicic A et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nature Neuroscience. 18 (9), 1226–1229, doi: 10.1038/nn.4085, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez Y & Lindquist SL HSP104 required for induced thermotolerance. Science. 248 (4959), 1112 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Outeiro TF & Lindquist S Yeast Cells Provide Insight into Alpha-Synuclein Biology and Pathobiology. Science. 302 (5651), 1772 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson BS et al. TDP-43 Is Intrinsically Aggregation-prone, and Amyotrophic Lateral Sclerosis-linked Mutations Accelerate Aggregation and Increase Toxicity. Journal of Biological Chemistry. 284 (30), 20329–20339, doi: 10.1074/jbc.M109.010264, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Z et al. Molecular Determinants and Genetic Modifiers of Aggregation and Toxicity for the ALS Disease Protein FUS/TLS. PLOS Biology. 9 (4), e1000614, doi: 10.1371/journal.pbio.1000614, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elden AC et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 466 1069, doi:10.1038/nature0932010.1038/nature09320https://www.nature.com/articles/nature09320-supplementary-informationhttps://www.nature.com/articles/nature09320-supplementary-information, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijayanti I, Watanabe D, Oshiro S & Takagi H Isolation and functional analysis of yeast ubiquitin ligase Rsp5 variants that alleviate the toxicity of human α-synuclein. The Journal of Biochemistry. 157 (4), 251–260, doi: 10.1093/jb/mvu069, (2015). [DOI] [PubMed] [Google Scholar]
- 29.Tardiff DF et al. Yeast Reveal a “Druggable” Rsp5/Nedd4 Network that Ameliorates α-Synuclein Toxicity in Neurons. Science. 342 (6161), 979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K et al. Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes. ACS Chemical Neuroscience. 9 (4), 838–848, doi: 10.1021/acschemneuro.7b00297, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flick JS & Johnston M Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Molecular and Cellular Biology. 10 (9), 4757 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Santiago R & Ezquerra M Epigenetic Research of Neurodegenerative Disorders Using Patient iPSC-Based Models. Stem Cells International. 2016 16, doi: 10.1155/2016/9464591, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armakola M et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nature genetics. 44 (12), 1302–1309, doi: 10.1038/ng.2434, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani M et al. Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1. Human Molecular Genetics. 24 (3), 773–786, doi: 10.1093/hmg/ddu494, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masala A et al. Epigenetic Changes Associated with the Expression of Amyotrophic Lateral Sclerosis (ALS) Causing Genes. Neuroscience. 390 1–11, doi: 10.1016/j.neuroscience.2018.08.009, (2018). [DOI] [PubMed] [Google Scholar]
- 36.Eryilmaz IE et al. Epigenetic approach to early-onset Parkinson’s disease: low methylation status of SNCA and PARK2 promoter regions. Neurological Research. 39 (11), 965–972, doi: 10.1080/01616412.2017.1368141, (2017). [DOI] [PubMed] [Google Scholar]
- 37.Daniele S et al. Epigenetic Modifications of the α-Synuclein Gene and Relative Protein Content Are Affected by Ageing and Physical Exercise in Blood from Healthy Subjects. Oxidative Medicine and Cellular Longevity. 2018 16, doi: 10.1155/2018/3740345, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


