SUMMARY
Regulation of gene expression is an important aspect of insulin action but in vivo is intertwined with changing levels of glucose and counter-regulatory hormones. Here we demonstrate that under euglycemic clamp conditions, physiological levels of insulin regulate interrelated networks of more than 1,000 transcripts in muscle and liver. These include expected pathways related to glucose and lipid utilization, mitochondrial function, and autophagy, as well as unexpected pathways, such as chromatin remodeling, mRNA splicing, and Notch signaling. These acutely regulated pathways extend beyond those dysregulated in mice with chronic insulin deficiency or insulin resistance and involve a broad network of transcription factors. More than 150 non-coding RNAs were regulated by insulin, many of which also responded to fasting and refeeding. Pathway analysis and RNAi knockdown revealed a role for lncRNA Gm15441 in regulating fatty acid oxidation in hepatocytes. Altogether, these changes in coding and non-coding RNAs provide an integrated transcriptional network underlying the complexity of insulin action.
Graphical Abstract
In Brief
Batista et al. demonstrate potent transcriptional remodeling by physiological insulin action in skeletal muscle and liver, involving interrelated networks of protein-coding genes, transcription factors, and long non-coding RNAs (lncRNAs). From an array of metabolically sensitive lncRNAs, Gm15441 is identified as a regulator of fatty acid oxidation in hepatocytes.
INTRODUCTION
Insulin is the primary hormone controlling the balance between an anabolic and catabolic state. This occurs through regulation of a range of physiological processes involved in metabolism, growth, differentiation, and cell survival (Boucher et al., 2014; Tokarz et al., 2018). At the cellular level, insulin activates the insulin receptor (IR) tyrosine kinase, which triggers two major downstream pathways: the IRS-1/phosphatidylinositol 3-kinase (PI3K)/AKT pathway, which regulates most of insulin’s metabolic actions, and the SHC/RAS/mitogen-activated protein kinase (MAPK) pathway, which regulates growth and differentiation (Haeusler et al., 2018; Taniguchi et al., 2006). In both its metabolic and growth-promoting actions, insulin regulates the expression of many genes, both through direct effects on gene transcription and through indirect effects secondary to changing levels of blood glucose and other hormones.
During fasting, for example, when insulin levels are low and glucagon and glucocorticoid levels are high, the promoter of the phosphoenolpyruvate carboxykinase 1 (Pck1) gene, a key mediator of hepatic gluconeogenesis, is occupied by multiple transcriptional activators, including FOXO1 and FOXO3, FOXA2, SRC-1, CBP/p300, HNF4α, glucocorticoid receptor, and the co-activator PGC1α (Granner, 2015). Feeding, which increases insulin and decreases glucagon levels, induces rapid, phosphorylation-dependent removal of most of these factors from Pck1 promoter, leading to suppression of gluconeogenesis, but to what extent this is the result of increased insulin action versus decreased glucagon action is unclear.
Global mRNA profiling approaches have been useful in identifying insulin-regulated genes in both cell lines and tissues taken from animals before and after insulin administration, during fasting and refeeding, or in the context of insulin resistance (Boucher et al., 2010; Fazakerley et al., 2018; Yang et al., 2016). However, these studies have limitations in the interpretation, because all of these manipulations result in changes of glucose levels, as well as levels of other hormones and metabolites. The gold standard method for assessing insulin action isolated from these other effects is the hyperinsulinemic-euglycemic clamp (Kim, 2009). In this regard, some microarray-based profiling of mRNAs has been performed in skeletal muscle biopsies from healthy and insulin-resistant human subjects undergoing hyperinsulinemiceuglycemic clamps (Coletta et al., 2008; Rome et al., 2003; Sears et al., 2009); however, little has been done using more comprehensive approaches like RNA sequencing (RNA-seq), which allows measurement of all transcripts, including multiple iso-forms due to alternative splicing and the expression of non-coding RNA (ncRNA) species. Likewise, in the human studies, there was no assessment of insulin regulation of gene expression in the liver due to sampling limitations.
To address gene expression changes induced by insulin under constant blood glucose levels, where the effects of counter-regulatory hormones are minimized, we have performed transcriptomic analysis in muscle and liver isolated from mice during a euglycemic clamp. We find that insulin regulates >1,000 transcripts in muscle and liver. These changes involve many well-known pathways related to energy metabolism, development, and auto-phagy, as well as previously unrecognized pathways, including chromatin remodeling and multiple ncRNAs, many of these with unknown function. Knockdown of selected long ncRNAs (lncRNAs) identified by lncRNA-mRNA correlation analysis in hepatocytes reveals a role for Gm15441 in regulation of fatty acid oxidation (FAO) and lipid accumulation. Thus, the response to physiological insulin levels in vivo is associated with a multi-level network of tissue-specific regulation of coding and ncRNAs producing the broad, pleiotropic actions of the hormone.
RESULTS
In vivo Insulin Action and Gene Expression in Skeletal Muscle and Liver
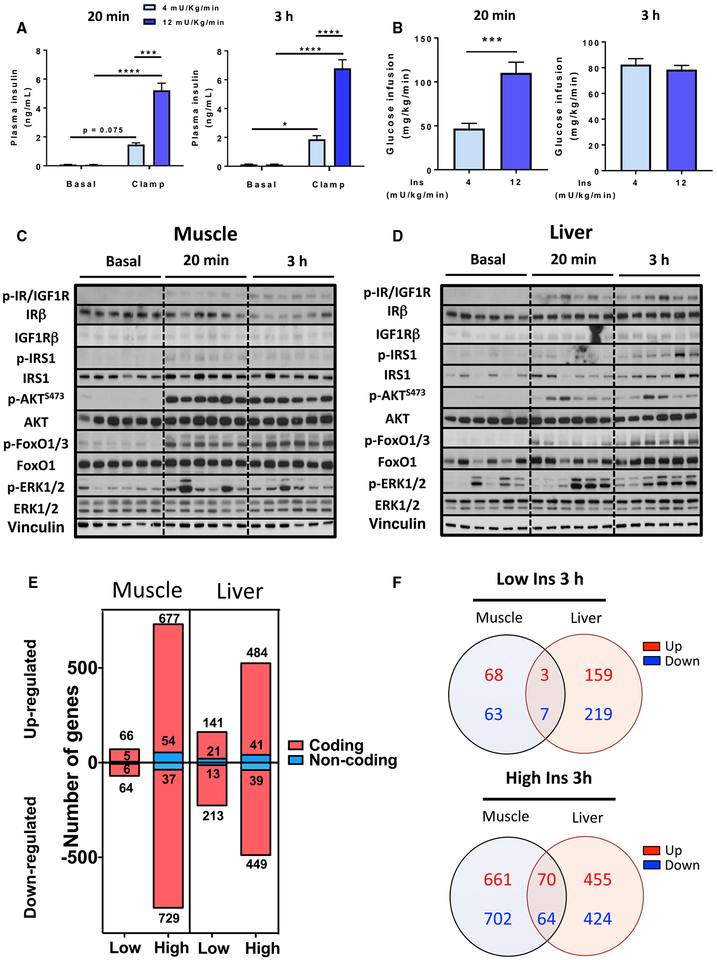
To assess the regulation of gene expression in skeletal muscle and liver in response to physiological insulin levels, without interference of changing glucose levels or counter-regulatory hormones, we performed hyperinsulinemic-euglycemic clamps on conscious mice at low and high physiological (4 or 12 mU/kg/min) insulin infusion rates and collected samples at 20 min and 3 h (Kim, 2009). These doses are within ranges that effectively suppress endogenous glucose production (Ayala et al., 2006; Berglund et al., 2008) while producing half-maximal to maximal effects on glucose uptake (Shen et al., 1999). After some transient, mild increases in glucose in control mice at early time points, blood glucose of all mice remained between 110 and 150 mg/dL throughout the experiment (Figure S1A). At both time points, low and high insulin infusions raised plasma insulin levels to 1.7 ± 0.1 or 5.9 ± 0.5 ng/mL, representing levels seen during refeeding of fasted mice (O’Neill et al., 2015) or after an intraperitoneal glucose tolerance test (ipGTT) (Andrikopoulos et al., 2008), respectively (Figure 1A). In the 20 min clamp and at early time points in the 3 h clamp, the glucose infusion rate (GIR) increased in a dose-dependent manner, but by 3 h, GIR had plateaued and was similar between insulin doses (Figures 1B and S1B).
Figure 1. Euglycemic Clamp, Insulin Signaling, and Gene Expression in Muscle and Liver.
(A) Plasma insulin levels before and after hyperinsulinemic-euglycemic clamp with 4 or 12 mU/kg/min insulin infusion for 20 min or 3 h (n = 2–4). Data are mean ± SEM (n = 6), *p < 0.05, ***p < 0.01, ****p < 0.0001, two-way ANOVA.
(B) Glucose infusion rates indicated as average of t = 10–15 min for the 20 min clamp and t = 120–150 min for the 3 h clamp. ***p < 0.001, Student’s t test. Data are mean ± SEM.
(C and D) Insulin signaling in (C) muscle and (D) liver from mice infused with high insulin for 20 min or 3 h (n = 6). For basal levels, 3 samples from saline-infused mice at each time point were used. See also Figure S1.
(E) Number of regulated genes by low- and high-dose insulin at 3 h (FDR < 0.1).
(F) Venn diagrams of tissue-specific and overlapping genes regulated by low and high insulin. See also Table S1.
Insulin signaling dynamics during the clamp are shown in Figures 1C, 1D, S1C, and S1E. Infusion with high doses of insulin led to IR and/or IGF1 receptor (IGF1R) tyrosine phosphorylation in both muscle and liver by 20 min, which was further increased at 3 h. Total IR protein levels were significantly downregulated at 3 h in muscle but remained stable in liver, indicating differential effects of insulin to downregulate the IR in these two tissues. By contrast, IGF1R remained unaltered in muscle but increased with time in liver. Downstream, tyrosine phosphorylation of IRS-1 in muscle peaked at 20 min and then gradually declined, while in liver, IRS-1 phosphorylation continued to increase to the 3 h point. In both tissues, p-AKT levels were maximal by 20 min, while phosphorylation of its downstream target FOXO1 increased more slowly. Phosphorylation of ERK1/2 was more variable between animals but transiently increased at 20 min and returned to near-basal levels at 3 h.
Despite the rapid signaling events, transcriptomic profiling by RNA-seq 20 min into the clamp revealed no significantly regulated genes in muscle and only one significantly regulated gene in liver, heat shock protein family A member 1B (Hspa1b), which was downregulated by both low and high insulin. By 3 h, however, insulin stimulation produced robust and dose-dependent gene regulation in both tissues. In skeletal muscle, of 13,142 genes detected, using a false discovery rate (FDR) cutoff of 0.1, there were 141 and 1,497 regulated genes with low and high insulin, respectively (Figure 1E, left). In liver, of 13,482 genes detected, we found 388 and 1,013 genes regulated by low and high insulin (Figure 1E, right). For both tissues, significantly changed genes were similarly distributed between upregulated and downregulated. About 9% of the significantly regulated genes in muscle and liver were non-protein coding (Figure 1E). Not surprisingly, the effects of insulin were highly tissue specific, with only about 10% of genes commonly regulated in both tissues (Figure 1F). This interesting subset included genes related to lipid and sterol metabolism and transcriptional regulation (Table S1).
Insulin-Regulated Transcriptional Networks in Skeletal Muscle and Liver
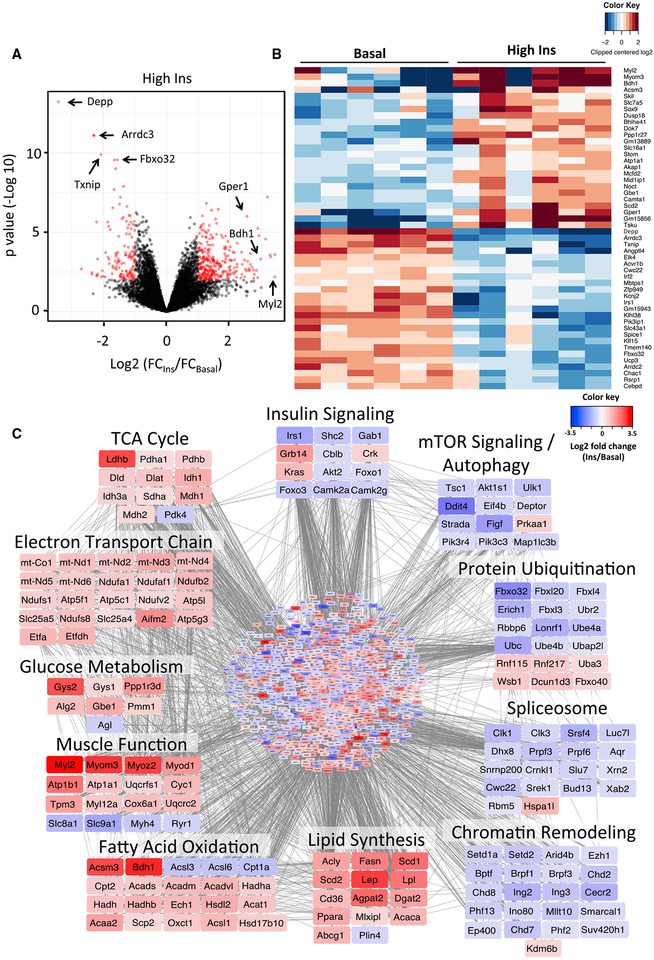
Genes regulated by high-dose insulin in skeletal muscle are shown in a volcano plot in Figure 2A and in heatmap form in Figure 2B. Among the most downregulated genes were two catabolic genes involved in protein ubiquitination (Fbxo32) and autophagy (Depp) and two members of the alpha-arrestin family, thioredoxin-interacting protein (Txnip) and arrestin domain-containing 3 (Arrdc3). Among the most upregulated genes were mediators of myofiber contraction, myosin light chain 2 (Myl2) and myomesin 3 (Myom3), FAO, acyl coenzyme A (CoA) synthetase 3 (Acsm3), and 3-hydroxybutyrate dehydrogenase 1 (Bdh1). Changes in expression of these top-ranking genes were confirmed by qPCR (Figure S2A).
Figure 2. Insulin Regulation of Gene Expression in Skeletal Muscle.
(A) Volcano plot showing distribution of differentially expressed genes (in red) by high insulin at 3 h compared to basal on the log2 scale.
(B) Heatmap showing the top 50 insulin-regulated genes. See also Figure S2.
(C) Network of predicted protein-protein interactions from STRING analysis (Szklarczyk et al., 2017) using insulin-regulated genes in muscle as input. 1,406 nodes and 6,369 interactions were detected (p < 1 × 10−16). Colors of nodes represent log2 fold change values of insulin regulation. See also Figure S3 and Tables S2 and S3.
To better visualize the effects of insulin on skeletal muscle at a system level, we constructed protein-protein interaction networks (Szklarczyk et al., 2017) from gene expression data (Figure 2C; Table S2). This approach identified 11 sets of genes that were highly coordinated in their response to insulin stimulation, with some including as many as 22 genes in a single pathway. Insulin positively regulated multiple members of gene sets involved in the tricarboxylic acid (TCA) cycle, electron transport chain, muscle function, and glucose and lipid metabolism. In addition to these nuclear-encoded genes, insulin increased mRNA levels of 19 genes encoded by mtDNA by ≥1.5-fold, including components of the electron transport chain (mt-Nd1 and mt-Co1) (Figure 2C) and tRNAs (mt-Tl1 and mt-Tp) (Table S3), consistent with a coordinated increase in mitochondrial biogenesis. Conversely, insulin downregulated gene clusters involved in insulin action, components of mTOR signaling and autophagy pathways, and protein ubiquitination. Pathway analysis confirmed enrichment for several of these gene clusters (Figures S3A and S3B). In addition to these classical pathways, insulin downregulated unanticipated gene sets associated with chromatin remodeling and mRNA splicing. The former allows refinement of the transcriptional program by altering chromatin structure, which determines accessibility of transcription factors to gene promoters (Hota and Bruneau, 2016), while the latter allows alternative assembly of the transcribed mRNAs (Chen and Manley, 2009).
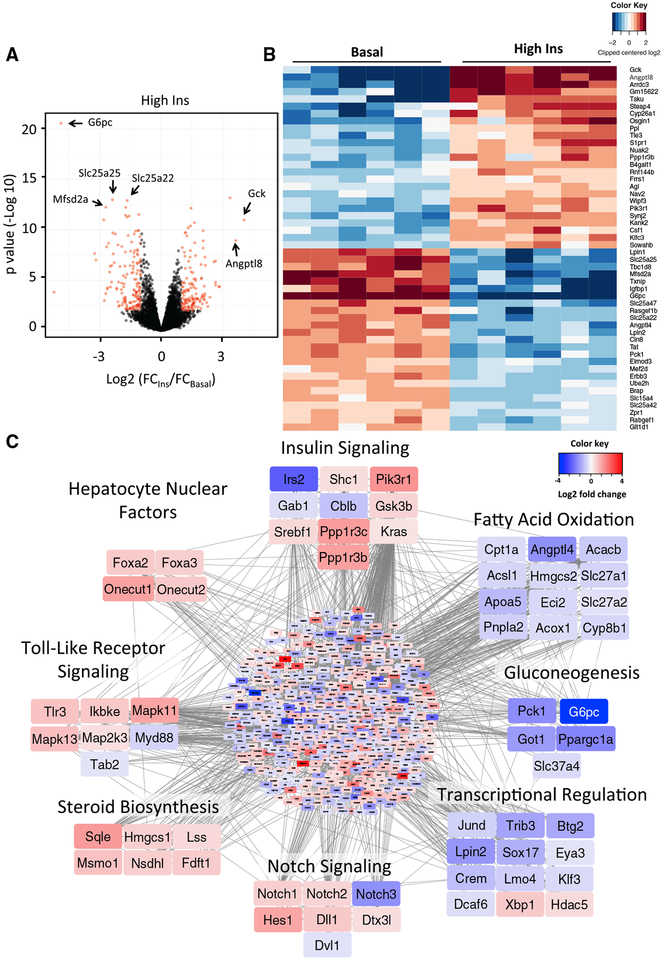
In general, the total number of mRNAs regulated in the liver was less than that in muscle, but the magnitude of regulation was greater, with some mRNAs showing up to 37-fold changes during the clamp. In liver, the top-ranking downregulated genes included the catalytic subunit of glucose-6-phosphatase (G6pc), a rate-limiting enzyme controlling glucose production, and plasma membrane transporters for omega-3 fatty acids (Mfsd2a) and mitochondrial carriers for glutamate (Slc25a22) and ATP/Mg-Pi (Slc25a25) (Figure 3A). Simultaneously, genes involved in glucose utilization (Gck) and lipid storage (Angptl8) were among the most upregulated (Figure 3B). Many of these were confirmed by qPCR (Figure S2B). Analysis of transcriptional networks controlled by insulin in the liver (Figure 3C; Table S2) showed fewer and smaller clusters. Insulin increased expression of mediators of Toll-like receptor and Notch signaling pathways, steroid and cholesterol biosynthetic enzymes, and several members of the hepatocyte nuclear factor family of transcription factors, some of which are defective in maturity onset diabetes of the young (MODY). Gene clusters involved in FAO, gluconeo-genesis, and transcription were predominantly downregulated. These clusters were confirmed by pathway analysis (Figures S3C and S3D).
Figure 3. Insulin Regulation of Gene Expression in Liver.
(A) Volcano plot showing distribution of differentially expressed genes (in red) in liver by high insulin at 3 h compared to basal on the log2 scale.
(B) Heatmap showing the top 50 insulin-regulated genes. See also Figure S2.
(C) Network of predicted protein-protein interactions from STRING analysis (Szklarczyk et al., 2017) using insulin-regulated genes in liver as input. 933 nodes and 2,895 interactions were detected (p < 1 × 10−16). Colors of nodes represent log2 fold change values of insulin regulation. See also Figure S3 and Table S2.
Identification of Candidate Upstream Regulators by Motif Enrichment Analysis
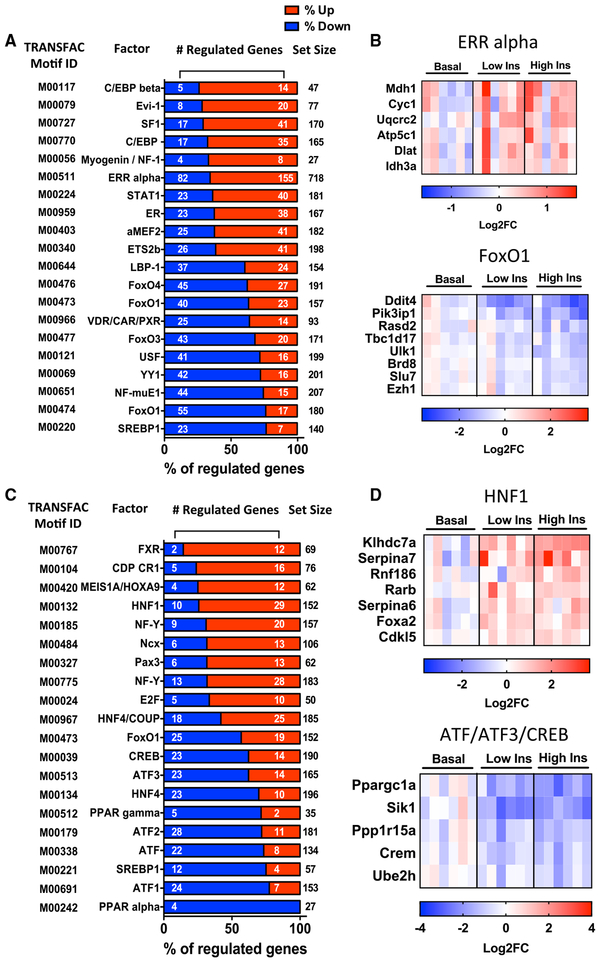
To identify the potential mediators of these changes in gene expression, we analyzed sequences 2 kb upstream or downstream of the transcription start site (TSS) in all regulated genes for transcription factor binding motifs. In muscle, promoter regions of upregulated genes were highly enriched for sites for C/EBPβ, EVI1 (also known as PRDM3), transcription factors involved in muscle development (Myogenin/NF-I and MEF2A), factors previously identified as being involved in insulin action (ETS2), and nuclear receptors, including steroidogenic factor 1 (SF1), estrogen receptor (ER), and estrogen-related receptor alpha (ERRα) (Figure 4A). Of 155 genes upregulated by insulin that showed enrichment for ERRα binding motifs, 51 overlapped with ERRα targets previously identified by chromatin immuno-precipitation sequencing (ChIP-seq) in PGC1α-overexpressing C2C12 myotubes (Salatino et al., 2016) (Figure S4), including genes involved in the TCA cycle (Mdh1 and Idh3a) and oxidative metabolism (Cyc1 and Atp5c1) (Figure 4B, top).
Figure 4. Transcription Factor Motifs Enriched in Insulin-Regulated Genes in Muscle and Liver.
(A and C) Overrepresented transcription factor motifs within 2 kb of TSS in (A) muscle and (C) liver. Plots are percentages of predicted target genes that are significantly up- or downregulated by insulin (FDR < 0.1). Enrichment analysis was performed using data from low and high insulin samples combined (n = 12).
(B) Examples of genes in muscle showing enrichment for ERRα (top) and FOXO1 (bottom) binding motifs.
(D) Examples of genes in liver showing enrichment for HNF1 (top) and ATF protein (BATF, ATF3, and CREB) (bottom) binding motifs. See also Figure S4.
Among downregulated genes, the most enriched transcription factor motifs were several previously shown to be involved in control of metabolism and insulin action (USF and SREBP1) (Mounier and Posner, 2006), mTOR signaling (YY1) (Cunningham et al., 2007), and signaling by vitamin D receptor (VDR) (Wang et al., 2012) (Figure 4A). Consistent with the model of nuclear exclusion of FoxO proteins following insulin-induced phosphor-ylation by AKT (Nakae et al., 2000; O’Neill et al., 2016), a large fraction of the downregulated genes had binding sites for FOXO1, FOXO3, and FOXO4 (Figure 4B, bottom).
In the liver, most insulin-upregulated genes contained transcription factor binding sites for factors related to developmental pathways involving hepatocyte nuclear factor (HNF) 1 and HNF4 (Sheaffer and Kaestner, 2012), homeodomain proteins (NCX, MEIS1/HOXA9, and CDP) (Azcoitia et al., 2005; Borghini et al., 2006; Xu et al., 2010), and the bile acid receptor FXR, as well as for factors related to cell-cycle regulation (NF-Y and E2F) (Di Agostino et al., 2006; Tu et al., 2006) (Figure 4C). Of the 29 upregulated genes that showed HNF1 binding sites, 23 have been identified by ChIP-seq analysis as HNF1α targets in HepG2 hepatocytes (Davis et al., 2018) (Figure 4D, top, and Figure S4). However, genes containing promoter motifs related to lipid metabolism (PPARα, PPARγ, and SREBP1) and cyclic AMP (cAMP) action (B-ATF, ATF1/3, CREB, and CREBP1) were enriched in downregulated genes (Figure 4C). Consistent with the pivotal role of insulin in the transition to the post-absorptive state through suppression of cAMP signaling, several fasting-related genes possessing binding sites for activating transcription factor (ATF) proteins were found to be downregulated (Figure 4D, bottom).
SREBP1 is translated into a 125 kDa precursor protein that, following sterol depletion or insulin action, gets cleaved into an active 68 kDa isoform that translocates to the nucleus and stimulates transcription of several genes involved in lipogenesis (Horton et al., 2002). In the 3 h clamp, SREBP1 activation was confirmed by a 2-fold increase of the cleaved isoform in both muscle and liver (Figures S4C and S4D). Fatty acid synthase (FAS), a well-known target of SREBP1, followed the same increasing trend. In both muscle and liver, we also observed enrichment of SREBP1 motifs, especially in downregulated genes, including those related to transcription, notch, and wnt signaling (Figure S4E), suggesting additional functions of SREBP1 that go beyond regulation of lipogenesis. Thus, multiple general and tissue-specific transcription factors underlie the diverse effects of insulin on gene expression in skeletal muscle and liver.
Identification of Insulin-Regulated Pathways in Diabetic States
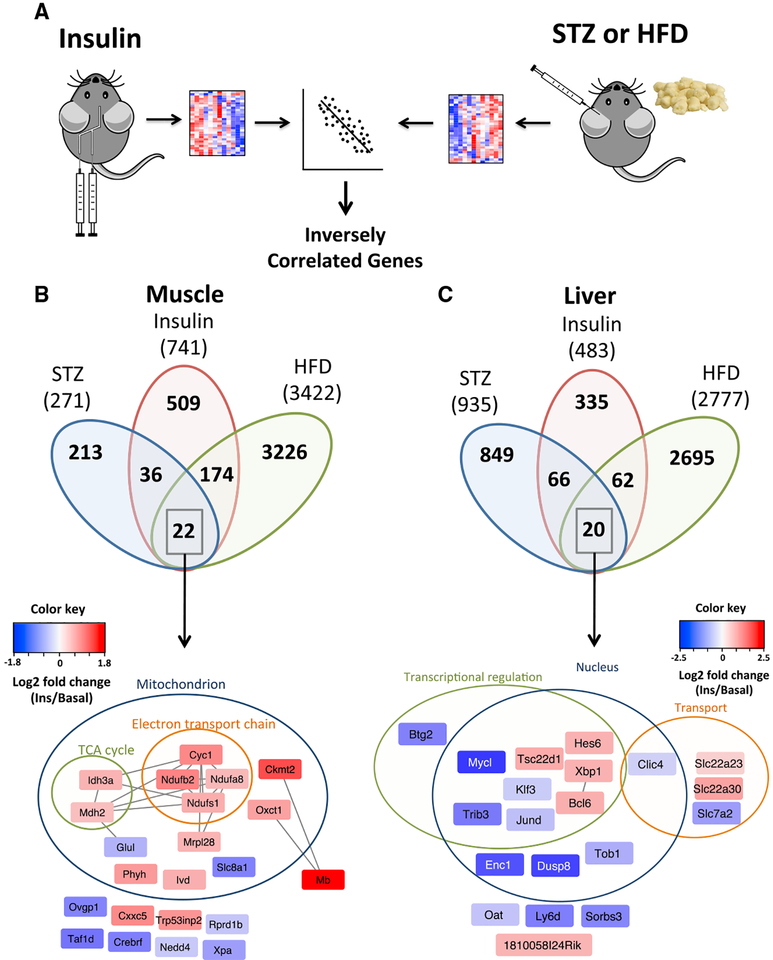
Diabetes is a state of impaired insulin signaling due to either lack of insulin (type 1 diabetes) or insulin resistance (type 2 diabetes). Both of these can lead to altered gene expression due to the loss of insulin signaling, as well as to secondary factors such as hyperglycemia, changes in other hormones, inflammation, and generation of reactive oxygen species (Lan et al., 2003; Yechoor et al., 2004; Yip and Fathman, 2014). To identify pathways directly regulated by insulin action within those dysregulated in diabetes, we intersected our data with gene expression datasets in muscle and livers from insulin-deficient streptozotocin (STZ)-treated mice (Franko et al., 2014; Kivela¨ et al., 2006) and insulin resistance due to high-fat diet (HFD) feeding (Almind and Kahn, 2004), looking for inversely correlated genes (Figure 5A). Of the 741 mRNAs measured in all studies whose expression was regulated by insulin during the clamp in muscle, 58 were inversely regulated in STZ diabetes and 196 were inversely regulated in HFD-induced insulin resistance. Of these reciprocally regulated genes, 22 were common to both patho-physiological conditions (Table S4). These related to mitochondrial function, electron transport chain, and TCA cycle activity (Figure 5B). Of the 483 mRNAs regulated by insulin in liver measured in all studies, 86 were inversely regulated in STZ, 82 were inversely regulated in HFD-induced diabetes, and 20 were common between these diabetic conditions (Figure 5C; Table S4). These were mainly associated with transcriptional regulation and transport of organic anions (Slc22a23 and slc22a30) and cationic amino acids (slc7a2). In both tissues, almost 70% of the genes regulated by insulin during the clamp were not significantly altered by either disease state, suggesting that in these chronic conditions, there are compensatory mechanisms that protect against some of the dysregulated gene expression that would result from a loss of insulin action.
Figure 5. Genes Inversely Regulated during the Euglycemic Insulin Clamp, STZ Diabetes, and HFD Obesity in Muscle and Liver.
(A) Schematics of insulin clamp and STZ or HFD dataset comparisons.
(B and C) Overlap of inversely correlated genes among insulin, STZ diabetes, and HFD obesity in muscle (B) and liver (C). Genes representing the overlap of all conditions grouped by functional annotation and cellular localization are indicated. Edges represent predicted protein-protein interactions from STRING. Colors of nodes are log2 fold change values of regulation by insulin. See also Table S4.
Insulin Regulation of ncRNA Species
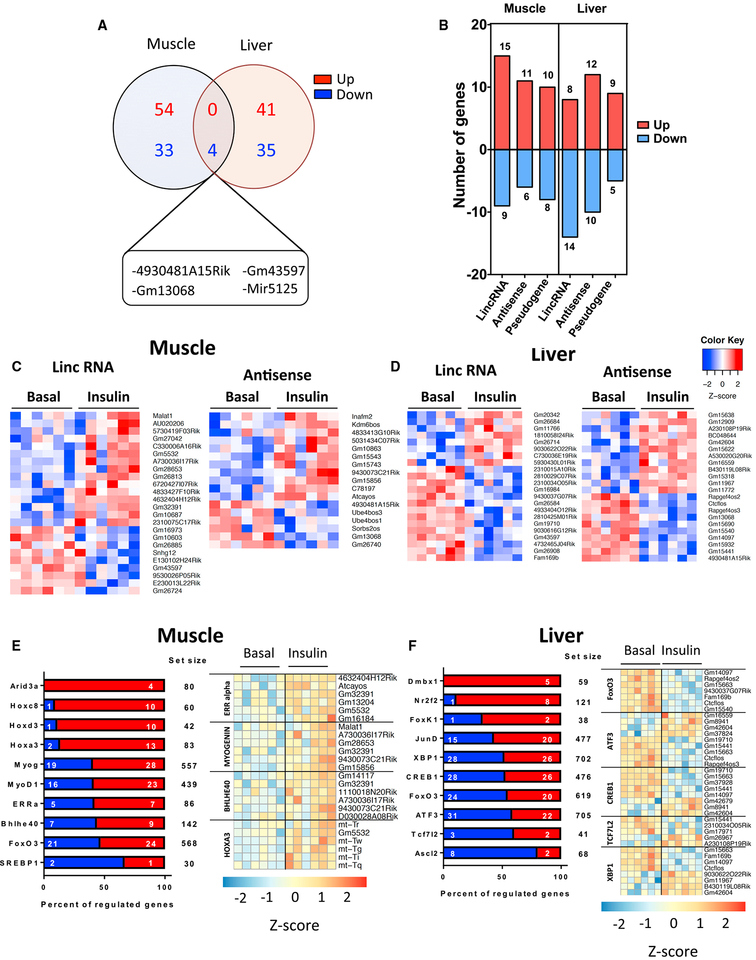
In addition to coding genes, more than 150 ncRNAs were dose-dependently regulated by insulin in muscle and liver (Figure 1E). This is likely an underestimate of the true effect, because library preparation for this study was not designed to capture miRNAs and other very small and non-polyadenylated RNAs. As with coding genes, insulin regulation of ncRNAs at 3 h in response to the higher insulin dose was tissue specific, with only 4 ncRNAs being commonly regulated in muscle and liver, and all of these were downregulated (Figure 6A). More than 60% of the remaining 163 uniquely regulated ncRNAs were lncRNAs, including long intergenic ncRNAs (lincRNAs), antisense RNAs, and pseudogene RNAs (Figure 6B). Top lncRNAs regulated by a high insulin dose in muscle and liver are shown in Figures 6C and 6D, and these were confirmed by qPCR (Figure S5).
Figure 6. Regulation of Non-coding RNA Species by Insulin.
(A) Venn diagrams of tissue-specific and overlapping ncRNAs regulated by high insulin at 3 h.
(B) Number of insulin-regulated lncRNAs across three main categories: lincRNA, antisense RNAs, and pseudogenes in muscle and liver.
(C and D) Heatmaps of top lncRNAs regulated in muscle (C) and liver (D) by high insulin at 3 h. See also Figure S5.
(E and F) Overrepresented transcription factor motifs within 2 kb upstream and 0.2 kb downstream of TSS and example ncRNAs for selected factors in (E) muscle and (F) liver. Plots are percentages of predicted target genes that are significantly up- or downregulated by a high insulin dose (FDR < 0.1).
Binding motif analysis revealed various transcription factors in mediating the effects of insulin on regulation of ncRNAs. In muscle, many upregulated ncRNAs showed enrichment of binding sites for transcription factors involved in development such as AT-rich interaction domain (ARID3A) and homeobox (HOXC8, HOXD3, and HOXA3) proteins (Figure 6E). We also detected binding sites for transcriptional regulators found enriched among coding genes (MYOD1, ERRα, FOXO3, and SREBP1); however, in the case of ncRNAs, the direction of regulation was similar between upregulated and downregulated transcripts. Binding sites for developmental transcription factors (DMBX1 and NR2F2) and transcription factors related to stress response and metabolic adaptation (FOXK1, XBP1, CREB1, FOXO3, and ATF3) were also present among insulin-regulated ncRNAs in liver (Figure 6F).
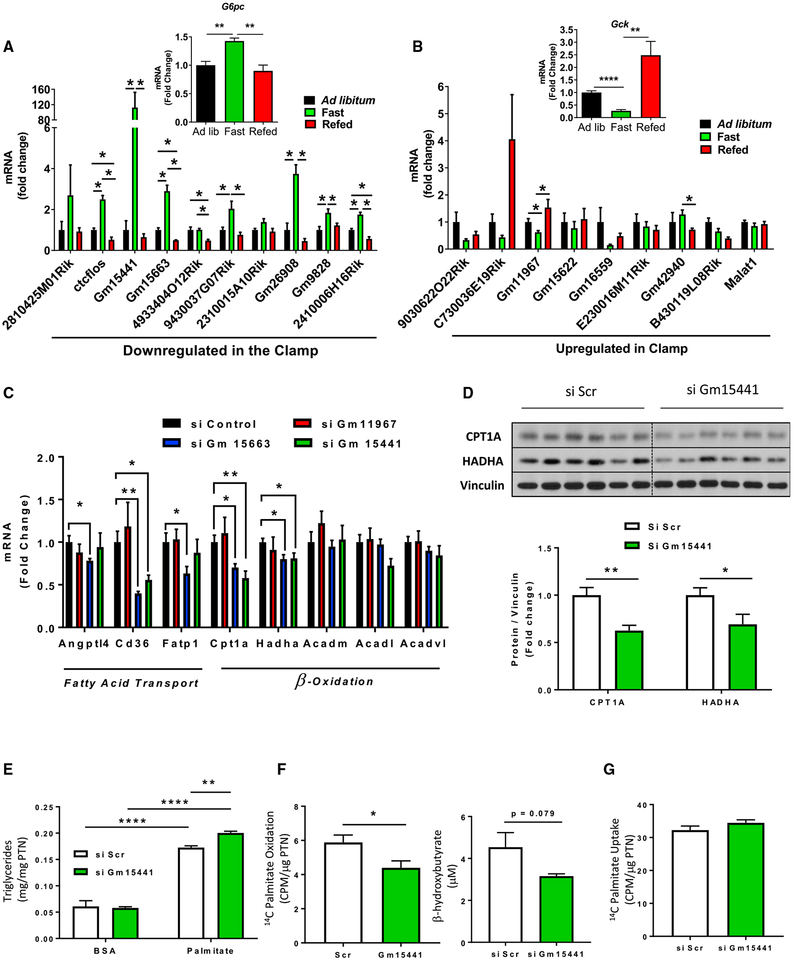
To determine whether the lncRNAs regulated during the clamp are responsive to physiological fluctuations of insulin levels, we subjected mice to a fasting and refeeding paradigm in which samples were collected from three groups of mice: ad libitum fed, overnight fasted, and overnight fasted followed by 8 h of refeeding. As expected, the expression of G6pc and Gck, which are known to respond to feeding cycles (Haeusler et al., 2014) and are major transcriptional targets of insulin in the liver, showed the expected changes, with G6pc expression significantly increased by overnight fasting and returning to ad libitum levels upon refeeding, while Gck was reciprocally regulated with decreased levels after fasting, which returned above ad libitum levels upon refeeding (Figures 7A and 7B, insets). Likewise, many lncRNAs downregulated during the clamp were upregulated by fasting, generally by 2- to 4-fold, although some changed by more than 100-fold, such as Gm15441. Upon refeeding, most of these returned to ad libitum levels or lower, consistent with the pattern of regulation seen during the clamp (Figure 7A). In the case of lncRNAs upregulated by insulin, although there was less robust regulation by fasting and refeeding, lncRNA Gm11967 was significantly downregulated by fasting and increased upon refeeding, and others such as C730036E19Rik, Gm15622, and Gm16559 showed a similar trend (Figure 7B). Thus, the transcriptional remodeling promoted by insulin during the clamp is associated with regulation of multiple lncRNAs. These lncRNAs are also sensitive to fasting and refeeding, indicating a role in metabolic adaptation to nutritional status.
Figure 7. Physiological Regulation and Effect of lncRNA Gm15441 Knockdown on Fatty Acid Oxidation.
(A and B) Expression of (A) downregulated and (B) upregulated lncRNAs by insulin in the clamp in livers of 14- to 16-week-old ad libitum fed, overnight fasted, and 8 h refed mice. Gene expression was normalized to TBP. Data are means ± SEM, n = 4–5, *p < 0.05, **p < 0.01, ****p < 0.0001, one-way ANOVA. (C and D) qPCR analysis of genes involved in lipid transport and oxidation normalized to 18S (C) and CPT1A and HADHA protein levels normalized to vinculin in mouse primary hepatocytes 24 h after transfection with siRNAs against of Gm11967, Gm15663, and Gm15441 (D). Data are means ± SEM. Hepatocyte cultures from 4–5 mice were used. *p < 0.05, **p < 0.01, Student’s t test. See also Tables S5 and S6.
(E) Intracellular triglyceride accumulation in AML-12 hepatocytes treated with 500 μM palmitic acid or vehicle for 8 h.
(F and G) 14C palmitic acid oxidation and β-hydroxybutyrate levels in culture supernatants (F) and 14C palmitic acid uptake in AML-12 hepatocytes (G). Lipid metabolism was assessed 24–36 h after transfection with siRNAs against Gm15441 or scramble controls. Data are means ± SEM, n = 5–6 biological replicates, *p < 0.05, **p < 0.01, ****p < 0.0001, Student’s t test. See also Figures S6 and S7.
Identification of Gm15441 as a Regulator of Lipid Metabolism
The regulatory roles exerted by lncRNAs extend to virtually every step of the transmission of genetic information, including transcription, mRNA processing, translation, and degradation (Villegas and Zaphiropoulos, 2015). Because most lncRNAs are not annotated on pathway databases, we performed correlation analysis of lncRNA-mRNA expression changes in muscle and liver (Table S5) and, from these sets of coding genes, analyzed the associated biological processes. This approach has been previously employed to identify metabolic roles of lncRNAs in liver (Li et al., 2015; Yang et al., 2016).
Based on gene expression changes in liver seen in the clamp and fasting and refeeding experiments, we selected the downregulated lncRNAs Gm15663 and Gm15441 and upregulated lncRNA Gm11967 for follow-up studies. These were also of interest, because Gm11967 and Gm15441 are close to two other insulin-regulated genes, Gck and Txnip genes (Figure S6A), which are well-established regulators of glucose homeostasis in mice and humans (Chutkow et al., 2010; Parikh et al., 2007; Postic et al., 1999; Steele et al., 2014). Gene ontology (GO) analysis of the correlated mRNAs indicated an association of Gm11967 expression with pathways related to differentiation and development, while Gm15663 showed enrichment with mineral transport (iron, copper, and sulfur), the mitochondrial genome, and stem cell maintenance, as well as fatty acid transport and oxidation (Figure S6B; Table S6). For Gm15441, however, eight of the top ten correlated pathways were related to lipid metabolism.
Using RNAi, we knocked down Gm11967, Gm15663, and Gm15441 in mouse primary hepatocytes with an efficiency of 55% to 89%. This occurred without changes in expression of neighboring genes (Figure S6C), confirming the specificity of the knockdown and suggesting that these lncRNAs are more likely to function in trans on more distant genomic regions. Consistent with our initial prediction, knockdown of both Gm15663 and Gm15441 resulted in downregulation of genes involved in lipid transport (Cd36) and β-oxidation (Cpt1a and Hadha), while knockdown of Gm11967 had no effects on genes involved in lipid metabolism (Figure 7C). Changes in CPT1A and HADHA following Gm15441 knockdown were confirmed at the protein level (Figure 7D). These effects were specific to fatty acid metabolism, because genes involved in gluconeogenesis and cholesterol and steroid synthesis remained largely unaffected by knockdown of either of these lncRNAs (Figure S7A). Knockdown of Gm15663 and Gm15441 also downregulated the major transcriptional regulator of lipid metabolism Ppar gamma by 50%–60%, while Ppar alpha expression was reduced by Gm11967 knockdown and Ppargc1a (PGC1α), which co-activates both of these nuclear receptors, was specifically downregulated by Gm15441 knockdown (Figure S7B).
Altogether, the pathway analysis and gene expression studies indicated that Gm15441 was the lncRNA most closely related to metabolic adaptation and specifically to lipid metabolism. To further explore this link at a functional level, we knocked down Gm15441 in the mouse hepatocyte cell line AML-12. This occurred with an efficiency of 70% (Figure S7C), similar to that in primary hepatocytes, and was accompanied by downregulation of Cpt1a plus a reduction of other β-oxidation genes (Acadm and Acadl) (Figure S7D) and the transcriptional regulators of lipid metabolism, Ppar gamma and Ppargc1a (Figure S7E). In a lipid accumulation assay using palmitic acid as substrate, control cells showed a 3-fold increase in intracellular triglyceride levels, and this was enhanced by Gm15441 knockdown (Figure 7E). The latter was associated with a 25% reduction in FAO rates and a nearly significant decrease of β-hydroxybutyrate, a ketone byproduct of fatty acid catabolism, in culture supernatants (Figure 7F). These changes were not attributable to differences in fatty acid uptake (Figure 7G). Collectively, these data indicate that Gm15441 is an insulin-sensitive lncRNA that contributes to regulation of the FAO program.
DISCUSSION
Loss of transcriptional integrity in response to insulin deficiency is linked to many features of uncontrolled diabetes (Patti, 2004; Sears et al., 2009). While much is known from In vitro studies about insulin regulation of gene expression, establishing a comprehensive view of insulin-regulated transcriptional networks in vivo in a tissue-specific manner is more difficult, because glucose homeostasis involves a complex metabolic response of insulin, multiple counter-regulatory hormones, and other metabolites. In the present study, we have assessed the effects of insulin on gene expression at constant blood glucose levels by performing euglycemic clamps at low and high physiological levels of insulin comparable to those observed after feeding or during an oral glucose challenge. We find that in both muscle and liver, insulin acutely regulates a broad and multi-dimensional network of gene expression.
Although the euglycemic clamp is the best available method to assess insulin action at constant blood glucose levels and with minimal counter-regulation (Kim, 2009), there are methodolog ical considerations. First, constant insulin infusion does not recapitulate the dynamic nature of insulin secretion, which may be more efficient in regulation of insulin action (Matveyenko et al., 2012). Second, by its nature, a clamp is based on peripheral insulin delivery, whereas physiologically insulin is secreted into the portal vein, thus changing the balance of effects in muscle compared to liver (Farmer et al., 2015). Third, repeated tail tip blood sampling may have an impact on catecholamine levels, although this would also affect the control mice receiving saline (Ayala et al., 2006).
In muscle, mitochondria are a major target of insulin’s transcriptional actions, with coordinate regulation of both nuclear and mtDNA transcripts linked to glucose utilization through the TCA cycle and oxidative phosphorylation, as well as recruitment of genes involved in glycogenesis and triglyceride synthesis for glucose storage. This transcriptional response is consistent with the role of insulin in muscle as the major site of glucose disposal during the clamp (Thiebaud et al., 1982). The nuclear receptor ERRα is a likely a major upstream regulator of the transcriptional response to insulin associated with mitochondrial metabolism, as indicated by enrichment of ERRα binding sites within promoters of many insulin-regulated genes involved with oxidative metabolism and by increased mRNA levels of the transcriptional co-activator Ppargc1b (PGC1β), which is essential for ERRα activity (Mootha et al., 2004), as well as increased levels of the ERRα downstream target Perm1 (Cho et al., 2013) in the insulin-stimulated state. In addition to control of glucose utilization, a major function of insulin in muscle is prevention of protein breakdown (Abdulla et al., 2016). Consistent with this anticatabolic function, during the clamp there is suppression of auto-phagy genes and E3-ubiquitin ligases. These major pathways regulating muscle proteostasis have been shown to be controlled by FoxO transcription factors (O’Neill et al., 2019; Sandri et al., 2004). Several insulin-regulated autophagy genes (including Ulk1 and Tbc1d17) showed enrichment for FOXO1, FOXO3, and FOXO4 binding sites on promoter regions, consistent with highest FOXO1/3 phosphorylation at 3 h.
Impaired expression of genes involved in oxidative metabolism is observed in muscle from subjects with type 1 diabetes (Karakelides et al., 2007) and type 2 diabetes (T2D) (Mootha et al., 2003; Patti et al., 2003). Intersection of genes regulated by insulin in muscle during the clamp with data from STZ and HFD models reveals that downregulation of mitochondrial function genes related to TCA cycle and oxidative phosphorylation in diabetes is linked to impaired insulin action. The relationship between insulin resistance and diabetic state on gene expression is illustrated by comparing mice with IR knockout in muscle (MIRKO) and mice made diabetic by STZ treatment (Yechoor et al., 2004). In this comparison, genes of oxidative phosphor-ylation are downregulated by STZ diabetes but remain unchanged upon muscle-specific deletion of the IR, indicating that metabolic factors other than insulin resistance are involved in the downregulation of mitochondrial genes. Insulin replacement restores gene expression in lox-STZ mice, but not in MIRKO-STZ mice, indicating that insulin action provides an initiating force in this gene regulation.
In the liver, insulin upregulates genes involved in steroid and cholesterol biosynthesis and downregulates pathways related to glucose production and fatty acid catabolism. In terms of suppression of endogenous production in liver, insulin action to phosphorylate FOXO1 plays a central role in increasing Gck transcription while suppressing G6pc levels (Haeusler et al., 2014). Insulin resistance in liver due to deletion of the IR or its downstream partners, IRS proteins or AKT1/AKT2, is characterized by unrestrained gluconeogenesis, and this can be reversed by liver-specific deletion of FOXO1 (Lu et al., 2012; Matsumoto et al., 2007; O-Sullivan et al., 2015). In our study, the involvement of FOXOs in the transcriptional response to insulin in the liver is indicated by increase of FOXO1 and FOXO3 phosphorylation in association with coordinate upregulation of Gck and downregulation of G6pc and other canonical FOXO targets in the liver, such as Pck1, Igfbp1, and Ppargc1a.
HNFs also appear to mediate part of the transcriptional effects of insulin in liver metabolism. During the 3 h clamp, insulin increases the expression of Foxa2 (HNF3β), Foxa3 (HNF3γ), Onecut1 (HNF6α), and Onecut2 (HNF6β) in the liver, and promoters of several regulated genes are enriched for HNF1 and HNF4 binding motifs. Insulin has been shown to also regulate liver transcription factors at the post-translational level, such as by promoting the inhibitory phosphorylation of FOXA2 at Thr156 by AKT, resulting in nuclear exclusion and suppression of gluconeo-genesis genes (Wolfrum et al., 2003), and through FOXO1 phosphorylation, which disrupts an inhibitory interaction with HNF4α, thus promoting its transcriptional activity (Hirota et al., 2008). Thus, HNFs, in combination with FOXOs, play important roles in determining the transcriptional response to insulin in the liver.
Control of lipid metabolism is a central metabolic function of insulin and occurs with different transcriptional profiles in muscle and liver. In liver, 3 h insulin stimulation downregulates the rate-limiting FAO enzymes Cpt1a and Cpt2 and increases genes involved in steroid and cholesterol biosynthesis (Hmgcs1 and Sqle), while mRNA levels of the major lipogenic genes remain unchanged at this time point. In muscle, insulin increases expression of enzymes of triglyceride synthesis (Gk and Dgat2), as well as genes involved in de novo lipogenesis (Fasn and Acly). Most lipogenic actions of insulin are controlled by the sterol response element binding proteins (SREBPs), with SREBP1c being more selective toward fatty acid and triglyceride synthesis and SREBP2 stimulation of cholesterol synthesis (Horton et al., 2002). Although the active SREBP1 isoform is induced by insulin in both tissues, the transcriptional profile from liver is most consistent with SREBP2 activation, whereas in muscle, SREBP1c is likely the dominant isoform. This agrees with previous observations of impaired SREBP2-mediated cholesterogenic gene expression in livers lacking IR expression specifically in hepatocytes (Miao et al., 2014). Motif analysis also indicate potential SREBP1 target genes involved in functions other than lipid metabolism, including transcription, chromatin modification, and notch and wnt signaling that are suppressed during the clamp. This is in line with previous reports of downregulation of genes involved in gluconeogenesis, cytochrome P450, and IRS-2-mediated signaling by SREBPs (Ide et al., 2004; Jang et al., 2016; Yamamoto et al., 2004).
Another aspect of metabolic regulation revealed by this study is the important role of insulin in regulation of ncRNAs. ncRNAs have gained increased attention in genetic research, because more than 85% of SNPs associated with T2D occur in non-coding intronic or intergenic regions of the genome (Jenkinson et al., 2015). In addition, dysregulated expression of lncRNAs has been demonstrated under diabetic conditions (He et al., 2017). However, a comprehensive understanding of lncRNA regulation by metabolic signals and downstream functions is lacking. We find that lncRNA regulation accounts for a significant part of the transcriptional response to insulin in muscle and liver. This is not limited to the clamp but is also observed by physiological insulin oscillations induced by fasting and refeeding. One such example is Gm15441, which is upregulated >100-fold by fasting and promptly returns to basal levels after refeeding, consistent with its downregulation during the clamp. This is in line with an upregulation of this lncRNA in livers of mice on a ketogenic diet, which lowers insulin levels (Newman et al., 2017).
Correlation between levels of Gm15441 with expression of FAO genes suggested a role for this lncRNA in regulating lipid metabolism. Knockdown of Gm15441 lowered palmitic acid-derived CO2 and ketone body formation. Further studies are needed to determine precisely how Gm15441 regulates lipid metabolism, but our study provides two important facets of this mechanism. First, PPAR nuclear receptors are likely to be involved, because the expression of Ppar gamma and its co-activator PGC1α are downregulated by more than 50% upon knockdown of Gm15441. Although FAO is linked to Ppar alpha, whose expression was unchanged in our knockdown experiments, previous studies have shown that lower levels of PGC1 in the liver can also affect FAO (Chambers et al., 2012; Estall et al., 2009). Second, the lack of regulation of neighboring genes upon Gm15441 and Gm15663 knockdown and the different chromosomal location of potential targets PPARγ and PGC1α from these lncRNAs suggest a long-range trans acting mechanism.
In conclusion, insulin promotes rapid and tissue-specific remodeling of gene transcription in vivo involving multiple layers of regulation. These changes extend include aspects not previously linked to diabetes, such as transcriptional regulation of lipid metabolism by ncRNAs. This network of gene regulation provides potential targets for therapeutic approaches involving insulin action in muscle and liver.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, C. Ronald Kahn (c.ronald.kahn@joslin.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Experiments were performed on three month-old male C57BL/6J mice (Jackson Laboratories). Mice were housed in standard conditions under 12-hour light/12-hour dark cycle and fed a 22% fat chow diet (Mouse Diet 9F, LabDiet). All procedures described were approved by the IACUC of the Joslin Diabetes Center, Boston, MA 02215 and University of Massachusetts Medical School, Worcester, MA 01655, and were in accordance with NIH guidelines.
In vitro cell models
For primary hepatocyte isolation, mouse livers were perfused through the inferior cava vein with PBS-EDTA, followed by perfusion with a pre-warmed type I collagenase solution (GIBCO, catalog 17100017). Hepatocytes were released from excised livers in a 10 cm Petri dish, following two cycles of filtering through a 100 μm strainer and centrifugation at 50 g for 90 s at 4°C. Hepatocytes from preparations with 85% viability or higher, determined by trypan blue staining, were plated in 6-well plates pre-coated with collagen I (Sigma, catalog C8919). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 units of penicillin-streptomycin.
Mouse TGFα-immortalized AML-12 hepatocytes were maintained in DMEM/F12 medium containing 10% FBS, 10 μg/mL insulin, 5.5 μg/mL transferrin, 5 ng/mL selenium, 40 ng/mL dexamethasone, 11 mg/mL sodium pyruvate and 100 units of penicillin-streptomycin. All cells were maintained in a humidified incubator at 37°C and 5% CO2.
METHOD DETAILS
Non-labeled Hyperinsulinemic-Euglycemic Clamp
Mice (n = 6 per condition) were anestethized with a mixture of ketamine/xylazine and had an indwelling catheter placed in the right internal jugular vein. After recovery (4–5 days) mice were fasted overnight (~16 hr) and placed on rat-sized restrainers. After acclimation period, mice received an insulin bolus of 16 or 48 mU/kg and were continuously perfused with either 4 or 12 mU/kg/min insulin or saline as control. Blood samples were collected from the tail tip at 5 min intervals (for the 20 min clamp) or 10–30 min intervals (for the 3 h clamp), and a 20% glucose solution was infused to maintain blood glucose levels at euglycemia (110 to 150 mg/dL). At 20 min or 3 h of clamp, mice were euthanized by cervical dislocation, and tissues were harvested, snap frozen in liquid nitrogen and kept at −80°C until analysis. These in vivo experiments were conducted at the National Mouse Metabolic Phenotyping Center (MMPC) at UMass Medical School.
mRNA isolation, RNA-sequencing and qPCR
Total RNA was isolated from quadriceps muscle and liver fragments homogenized in QIAzol reagent (QIAGEN), followed by chlorophorm/isopropanol/ethanol extraction. RNA quality and quantification was verified at the Joslin Genomics Core using a 2100 Agilent Bioanalyzer instrument. Total RNA samples (2 μg) that passed quality test (RNA integrity score > 7), were submitted to the Biopolymers Facility at Harvard Medical School. Stranded cDNA libraries with unique index tags for each sample (48 multiplexed) were made using a directional RNA-seq kit (Wafergen). Sequencing was performed on a Illumina HiSeq 2500 run on rapid mode (2×50). qPCR reactions were prepared using iQ SybrGreen Supermix (Bio-Rad, catalog 1708884) and run on a C1000 Thermal Cycler (BioRad, catalog CFX384) using TATA box binding protein (Tbp) or 18S ribosomal RNA as internal controls as indicated. Primer sequences used are listed in Table S7.
Bioinformatics analysis
Reads were aligned to the mouse genome from Gencode (GRCm38.p4) using STAR (Dobin et al., 2013) and counted with feature-Counts (Liao et al., 2014). Read counts were transformed to log2-counts per million (LogCPM), their mean-variance relationship was estimated, their weights were computed with voom (Law et al., 2014), and their differential expression was assessed using linear modeling with the R package limma (Ritchie et al., 2015). P values were corrected using the Benjamini-Hochberg false discovery rate (FDR), and FDR < 0.1 was considered statistically significant. Gene sets based on transcription factor targets, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology Biological Process (BP) databases were tested using the limma Roast method (Wu et al., 2010).
Gene expression networks were designed by uploading differentially expressed genes to STRING database (Szklarczyk et al., 2017) and were further modeled in Cytoscape (Shannon et al., 2003). Gene clusters were identified using KEGG gene sets and ClusterOne function (Nepusz et al., 2012). Functional annotation analysis was perfomed using the DAVID platform V6.8 (Huang et al., 2009).
Protein extraction and immunoblotting
Tissue fragments and cells were homogenized in RIPA buffer (EMD Millipore) supplemented with protease and phosphatase inhibitors (Biotool). Lysates were separated from insoluble material by centrifugation (12,000 RPM, 15 min, 4°C) and total protein content was determined by BCA assay (ThermoFisher). Equal protein ammounts (~10 μg) were resolved by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore). Membranes were immunoblotted with the indicated antibodies: p-IR/IGF1R (#3024, Cell Signaling), IRβ (sc-711, Santa Cruz), IGF1Rβ (#3027, Cell Signaling), p-IRS1 (09–432, Millipore), IRS1 (611394, BD), p-AktS473 (#9271, Cel Signaling), Akt pan (#4685, Cell Signaling), p-FOXO1/3 (#9464, Cell Signaling), FoxO1 (#9454, Cell Signaling), p-ERK1/2 (#9101, Cell Signaling), ERK1/2 (#9102, Cell Signaling), CPT1A (ab128568, Abcam), HADHA (ab54477, Abcam), SREBP1 (sc-8984, Santa Cruz), Fatty Acid Synthase (ab22759, Abcam), Vinculin (#3574, Chemicon).
LncRNA knockdown
Twenty four hours after isolation, primary hepatocytes were transfected with 75 pmoles of custom-made siRNAs (Dharmacon/GE Lifesciences) against Gm11967, Gm15663, Gm15441 or non-targeting controls using Lipofectamine RNAiMAX (Invitrogen). Twenty four hours post transfection, cells were washed with PBS and collected with 1 mL TRIzol reagent (Invitrogen) followed by RNA extraction according to manufacturer’s instructions. Protein extracts were prepared as described above. Mouse AML-12 hepatocytes were transfected with either 60 pmoles (12-well) or 30 pmoles (24-well) at 70%–80% confluency. All assays were performed within 24–36 h post-transfection. siRNA sequences are as follows: Gm11967 sense 5′ CAGAAGAUGAUGAUCGGAUUU 3′, antisense 5′ AUCCGAUCAUCAUCUUCUGUU 3′; Gm15663 sense: 5′ GAAAUACGGUGCAGAAGAAUU 3′, antisense: 0035′ UUCUUCUGCACCGU AUUUCUU 3′, Gm15441 sense: 5′ ACAUAAGACUUCAGGAGAAUU 3′, antisense: 5′ UUCUCCUGAAGUCUUAUGUUU 3′.
Fatty acid accumulation, oxidation and uptake
For lipid accumulation, cells were pre-incubated overnight (~16 h) in DMEM/F12 + 2.5% FBS (no supplements) followed by incubatition with 500 μM palmitic acid (Sigma, catalog P5585) pre-complexed with fatty acid-free bovine serum albumin (FAF-BSA) or BSA-only as controls for 8 h. Total intracellular triglycerides were measured by an enzymatic plate-based assay (Pointe Scientific) and normalized to protein content. For FAO, cells were serum-starved in the presence of 0.1% FAF-BSA for 3 h, followed by 1 h with fatty acid incubation media (FAIM) containing 20 μM palmitic acid conjugated with FAF-BSA plus 1 h in the presence of 0.1 μCi 14C palmitic acid (Perkin Elmer). FAO was determined as the fraction of 14C palmitic acid-derived CO2 trapped in filter papers normalized to protein content as previously described (Akie and Cooper, 2015). Before addition of 14C palmitic acid, an aliquot of FAIM was collected and assayed for β-hydroxybutyrate levels using a fluorimetric method (Cayman Chemical) according to manufacturer’s instructions. For fatty acid uptake, cells were serum-starved for 3 h and incubated with FAIM containing 200 μM palmitic acid conjugated with FAF-BSA for 30 min plus another 30 min in the presence of 0.1 μCi 14C palmitic acid. Fatty acid uptake was interpreted as protein-normalized counts from cell lysates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as means ± SEM. Comparisons between two groups was performed using Student’s t test. Comparisons between more than two groups was performed using One-way ANOVA followed by post hoc t tests. Comparisons between two groups and two nominal variables (e.g., basal versus clamp) was performed using Two-way ANOVA followed by Holm-Sidak’s post hoc test. Statistical analysis was performed using GraphPad Prism (Version 7.02). Significance level was set at p < 0.05.
DATA AND SOFTWARE AVAILABILITY
RNA-seq data described here is deposited in Gene Expression Omnibus (GEO) under accession number GEO: GSE117741. Expression data from muscle and liver microarrays of HFD C57BL/6 mice are under accession number GEO: GSE123394. The metabolic phenotype of these mice has been described (Almind and Kahn, 2004). Data-sets from STZ-treatment in mouse muscle GEO: GSE1659 (Kivela¨ et al., 2006) and liver GEO: GSE39752 (Franko et al., 2014) were downloaded from GEO.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-IGF-I Receptor β (Tyr21135/1136)/lnsulin Receptor β (Tyr1150/1151) (19H7) | Cell Signaling Technology | Cat# 3024; RRID:AB_331253 |
| IGF-I Receptor β | Cell Signaling Technology | Cat# 3027; RRID:AB_2122378 |
| Phospho-AKT (Ser473) | Cell Signaling Technology | Cat# 9271; RRID:AB_329825 |
| Akt (pan) (11E7) | Cell Signaling Technology | Cat# 4685; RRID:AB_2225340 |
| Phospho-FOXO1 (Thr24)/FOXO3A (Thr32) | Cell Signaling Technology | Cat# 9464; RRID:AB_329842 |
| FOXO1 | Cell Signaling Technology | Cat# 9454; RRID:AB_823503 |
| p44/42 MAP kinase (phosphorylated Erk1/2) | Cell Signaling Technology | Cat# 9101; RRID:AB_331646 |
| p44/42MAPK(Erk1/2) | Cell Signaling Technology | Cat# 9102; RRID:AB_330744 |
| Insulin Receptor beta (C-19) | Santa Cruz Biotechnology | Cat# sc-711; RRID:AB_631835 |
| Phospho-IRS1 (Tyr608) mouse/ (Tyr612) human | Millipore | Cat# 09-432; RRID:AB_1163457 |
| IRS-1 | BD Biosciences | Cat# 611394; RRID:AB_398916 |
| Vinculin, clone VIIF9 (7F9) | Millipore | Cat# MAB3574; RRID:AB_2304338 |
| CPT1A[8F6AE9] | Abeam | Cat# ab128568; RRID:AB_11141632 |
| HADHA | Abeam | Cat# ab54477; RRID:AB_2263836 |
| SREBP1 | Santa Cruz Biotechnology | Cat# sc-8984; RRID:AB_2194223 |
| Fatty Acid Synthase | Abeam | Cat# ab22759; RRID:AB_732316 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RIPA Lysis Buffer, 10X for Immunoprecipitation & Western Blotting | Millipore | Cat# 20-188 |
| Collagenase, Type I from Clostridium histolyticum | GIBCO | Cat# 17100017 |
| Collagen from calf skin | Sigma-Aldrich | Cat# C8919; CAS: 9007-34-5 |
| Lipofectamine RNAiMAXTransfection Reagent | Invitrogen | Cat# 13778030 |
| PALMITIC ACID, [1–14C]-, 50 μCi | Perkin Elmer | Cat# NEC075H050UC |
| Critical Commercial Assays | ||
| β-Hydroxybutyrate (Ketone Body) Fluorometric Assay Kit | Cayman Chemical | Cat# 700740 |
| Triglyceride Enzymatic Assay | Pointe Scientific | Cat# T7532 |
| Deposited Data | ||
| Raw sequence data (muscle and liver) | This paper | GEO: GSE117741 |
| Expression profiling by array | Almind and Kahn, 2004 | GEO: GSE123394 |
| Experimental Models: Cell Lines | ||
| Mouse: AML12 hepatocytes | ATCC | Cat# CRL-2254; RRID:CVCL_0140 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat# 000664 |
| Oligonucleotides | ||
| siRNA against Gm11967 (sense): 5’ CAGAAGAUGAUGAUCGGAUUU 3’ | This paper | N/A |
| siRNA against Gm11967 (antisense): 5’ AUCCGAUCAUCAUCUUCUGUU 3’ | This paper | N/A |
| siRNA against Gm15663 (sense): 5’ GAAAUACGGUGCAGAAGAAUU 3’, | This paper | N/A |
| siRNA against Gm15663 (antisense): 5’ UUCUUCUGCACCGUAUUUCUU 3’ | This paper | N/A |
| siRNA against Gm15441 (sense): 5’ ACAUAAGACUUCAGGAGAAUU 3’, | This paper | N/A |
| siRNA against Gm15441 (antisense): 5’ UUCUCCUGAAGUCUUAUGUUU 3’ | This paper | N/A |
| Software and Algorithms | ||
| STAR | Dobin et al., 2013 | http://code.google.eom/p/ma-star/ |
| The R Project for Statistical Computing | Freely available online | https://www.r-project.org/ |
| STRING | Szklarczyk et al., 2017 | https://string-db.org/ |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org |
| ClusterOne | Nepusz et al., 2012 | http://www.paccanarolab.org/cluster-one/ |
| DAVID | Huang et al., 2009 | https://david.ncifcrf.gov/ |
Highlights.
Physiological insulin regulates a broad transcriptional network in muscle and liver
In addition to mRNA of coding genes, insulin regulates mRNA splicing and lncRNAs
Insulin-regulated gene expression involves multiple transcriptional regulators
The insulin-suppressed lncRNA Gm15441 regulates fatty acid oxidation in hepatocytes
ACKNOWLEDGMENTS
This work was supported by NIH grants R37DK031036, R01DK033201 (to C.R.K.), P30DK036836 (to Joslin Diabetes Center), and 5U2C-DK093000 (to J.K.K.) and the Mary K. Iacocca Professorship (to C.R.K.). T.M.B. was partially supported by a grant from Sao Paulo Research Foundation (2014/25370–8). R.G.M. was supported by a Deutsche Forschungsgemeinschaft (DFG) fellowship (GA 2426/1–1). B.T.O. was funded by a K08 training award (K08DK100543) and an R03 award (R03DK112003) from the NIDDK of the NIH. We thank Jonathan M. Dreyfuss and Hui Pan from Joslin Diabetes Center DRC Genomics and Bioinformatics Core for assistance with data analysis.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.celrep.2019.02.081.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abdulla H, Smith K, Atherton PJ, and Idris I (2016). Role of insulin in the regulation of human skeletal muscle protein synthesis and breakdown: a systematic review and meta-analysis. Diabetologia 59, 44–55. [DOI] [PubMed] [Google Scholar]
- Akie TE, and Cooper MP (2015). Determination of Fatty Acid Oxidation and Lipogenesis in Mouse Primary Hepatocytes. J. Vis. Exp 102, e52982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almind K, and Kahn CR (2004). Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes 53, 3274–3285. [DOI] [PubMed] [Google Scholar]
- Andrikopoulos S, Blair AR, Deluca N, Fam BC, and Proietto J (2008). Evaluating the glucose tolerance test in mice. Am. J. Physiol. Endocrinol. Metab 295, E1323–E1332. [DOI] [PubMed] [Google Scholar]
- Ayala JE, Bracy DP, McGuinness OP, and Wasserman DH (2006). Considerations in the design of hyperinsulinemic-euglycemic clamps in the conscious mouse. Diabetes 55, 390–397. [DOI] [PubMed] [Google Scholar]
- Azcoitia V, Aracil M, Martínez-A C, and Torres M (2005). The homeodo-main protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol 280, 307–320. [DOI] [PubMed] [Google Scholar]
- Berglund ED, Li CY, Poffenberger G, Ayala JE, Fueger PT, Willis SE, Jewell MM, Powers AC, and Wasserman DH (2008). Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes 57, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghini S, Bachetti T, Fava M, Di Duca M, Cargnin F, Fornasari D, Ravazzolo R, and Ceccherini I (2006). The TLX2 homeobox gene is a transcriptional target of PHOX2B in neural-crest-derived cells. Biochem. J 395, 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher J, Tseng YH, and Kahn CR (2010). Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J. Biol. Chem 285, 17235–17245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher J, Kleinridders A, and Kahn CR (2014). Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol 6, a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers KT, Chen Z, Crawford PA, Fu X, Burgess SC, Lai L, Leone TC, Kelly DP, and Finck BN (2012). Liver-specific PGC-1beta deficiency leads to impaired mitochondrial function and lipogenic response to fasting-re-feeding. PLoS ONE 7, e52645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, and Manley JL (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Hazen BC, Russell AP, and Kralli A (2013). Peroxisome proliferator-activated receptor g coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J. Biol. Chem 288, 25207–25218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Birkenfeld AL, Brown JD, Lee HY, Frederick DW, Yoshioka J, Patwari P, Kursawe R, Cushman SW, Plutzky J, et al. (2010). Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes 59, 1424–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta DK, Balas B, Chavez AO, Baig M, Abdul-Ghani M, Kashyap SR, Folli F, Tripathy D, Mandarino LJ, Cornell JE, et al. (2008). Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am. J. Physiol. Endocrinol. Metab 294, E910–E917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, and Puigserver P (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740. [DOI] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46 (D1), D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, and Piaggio G (2006). Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10, 191–202. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estall JL, Kahn M, Cooper MP, Fisher FM, Wu MK, Laznik D, Qu L, Cohen DE, Shulman GI, and Spiegelman BM (2009). Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 58, 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer TD, Jenkins EC, O’Brien TP, McCoy GA, Havlik AE, Nass ER, Nicholson WE, Printz RL, and Shiota M (2015). Comparison of the physiological relevance of systemic vs. portal insulin delivery to evaluate whole body glucose flux during an insulin clamp. Am. J. Physiol. Endocrinol. Metab 308, E206–E222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley DJ, Chaudhuri R, Yang P, Maghzal GJ, Thomas KC, Krycer JR, Humphrey SJ, Parker BL, Fisher-Wellman KH, Meoli CC, et al. (2018). Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. eLife 7, e32111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko A, von Kleist-Retzow JC, Neschen S, Wu M, Schommers P, Böse M, Kunze A, Hartmann U, Sanchez-Lasheras C, Stoehr O, et al. (2014). Liver adapts mitochondrial function to insulin resistant and diabetic states in mice. J. Hepatol 60, 816–823. [DOI] [PubMed] [Google Scholar]
- Granner DK (2015). In pursuit of genes of glucose metabolism. J. Biol. Chem 290, 22312–22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Hartil K, Vaitheesvaran B, Arrieta-Cruz I, Knight CM, Cook JR, Kammoun HL, Febbraio MA, Gutierrez-Juarez R, Kurland IJ, and Accili D (2014). Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat. Commun 5, 5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, McGraw TE, and Accili D (2018). Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol 19, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ou C, Xiao Y, Han Q, Li H, and Zhou S (2017). LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget 8, 71325–71341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Sakamaki J, Ishida J, Shimamoto Y, Nishihara S, Kodama N, Ohta K, Yamamoto M, Tanimoto K, and Fukamizu A (2008). A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J. Biol. Chem 283, 32432–32441. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, and Brown MS (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, and Bruneau BG (2016). ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, et al. (2004). SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol 6, 351–357. [DOI] [PubMed] [Google Scholar]
- Jang H, Lee GY, Selby CP, Lee G, Jeon YG, Lee JH, Cheng KK, Titchenell P, Birnbaum MJ, Xu A, et al. (2016). SREBP1c-CRY1 signalling represses hepatic glucose production by promoting FOXO1 degradation during refeeding. Nat. Commun 7, 12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson CP, Göring HH, Arya R, Blangero J, Duggirala R, and DeFronzo RA (2015). Transcriptomics in type 2 diabetes: Bridging the gap between genotype and phenotype. Genom. Data 8, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakelides H, Asmann YW, Bigelow ML, Short KR, Dhatariya K, Coenen-Schimke J, Kahl J, Mukhopadhyay D, and Nair KS (2007). Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects. Diabetes 56, 2683–2689. [DOI] [PubMed] [Google Scholar]
- Kim JK (2009). Hyperinsulinemic-euglycemic clamp to assess insulin sensitivity in vivo. Methods Mol. Biol 560, 221–238. [DOI] [PubMed] [Google Scholar]
- Kivelä R, Silvennoinen M, Touvra AM, Lehti TM, Kainulainen H, and Vihko V (2006). Effects of experimental type 1 diabetes and exercise training on angiogenic gene expression and capillarization in skeletal muscle. FASEB J 20, 1570–1572. [DOI] [PubMed] [Google Scholar]
- Lan H, Rabaglia ME, Stoehr JP, Nadler ST, Schueler KL, Zou F, Yan-dell BS, and Attie AD (2003). Gene expression profiles of nondiabetic and diabetic obese mice suggest a role of hepatic lipogenic capacity in diabetes susceptibility. Diabetes 52, 688–700. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, and Cao H (2015). A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 21, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lu M, Wan M, Leavens KF, Chu Q, Monks BR, Fernandez S, Ahima RS, Ueki K, Kahn CR, and Birnbaum MJ (2012). Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med 18, 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, and Accili D (2007). Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6, 208–216. [DOI] [PubMed] [Google Scholar]
- Matveyenko AV, Liuwantara D, Gurlo T, Kirakossian D, Dalla Man C, Cobelli C, White MF, Copps KD, Volpi E, Fujita S, and Butler PC (2012). Pulsatile portal vein insulin delivery enhances hepatic insulin action and signaling. Diabetes 61, 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Haas JT, Manthena P, Wang Y, Zhao E, Vaitheesvaran B, Kur-land IJ, and Biddinger SB (2014). Hepatic insulin receptor deficiency impairs the SREBP-2 response to feeding and statins. J. Lipid Res 55, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. (2004). Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101, 6570–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier C, and Posner BI (2006). Transcriptional regulation by insulin: from the receptor to the gene. Can. J. Physiol. Pharmacol 84, 713–724. [DOI] [PubMed] [Google Scholar]
- Nakae J, Barr V, and Accili D (2000). Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J 19, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepusz T, Yu H, and Paccanaro A (2012). Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 9, 471–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Covarrubias AJ, Zhao M, Yu X, Gut P, Ng CP, Huang Y, Haldar S, and Verdin E (2017). Ketogenic Diet Reduces Midlife Mortality and Improves Memory in Aging Mice. Cell Metab 26, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O-Sullivan I, Zhang W, Wasserman DH, Liew CW, Liu J, Paik J, DePinho RA, Stolz DB, Kahn CR, Schwartz MW, and Unterman TG (2015). FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun 6, 7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, and Kahn CR (2015). Differential Role of Insulin/IGF-1 Receptor Signaling in Muscle Growth and Glucose Homeostasis. Cell Rep 11, 1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill BT, Lee KY, Klaus K, Softic S, Krumpoch MT, Fentz J, Stanford KI, Robinson MM, Cai W, Kleinridders A, et al. (2016). Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J. Clin. Invest 126, 3433–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill BT, Bhardwaj G, Penniman CM, Krumpoch MT, Suarez Beltran PA, Klaus K, Poro K, Li M, Pan H, Dreyfuss JM, et al. (2019). FoxO Transcription Factors are Critical Regulators of Diabetes-Related Muscle Atrophy. Diabetes 68, 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, et al. (2007). TXNIP regulates peripheral glucose metabolism in humans. PLoS Med 4, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME (2004). Gene expression in the pathophysiology of type 2 diabetes mellitus. Curr. Diab. Rep 4, 176–181. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. (2003). Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 100, 8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, and Magnuson MA (1999). Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem 274, 305–315. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S, Clément K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS, Riou JP, Laville M, and Vidal H (2003). Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperinsulinemic clamp. J. Biol. Chem 278, 18063–18068. [DOI] [PubMed] [Google Scholar]
- Salatino S, Kupr B, Baresic M, Omidi S, van Nimwegen E, and Hand-schin C (2016). The Genomic Context and Corecruitment of SP1 Affect ERRα Coactivation by PGC-1α in Muscle Cells. Mol. Endocrinol 30, 809–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, and Goldberg AL (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears DD, Hsiao G, Hsiao A, Yu JG, Courtney CH, Ofrecio JM, Chapman J, and Subramaniam S (2009). Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc. Natl. Acad. Sci. USA 106, 18745–18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, and Kaestner KH (2012). Transcriptional networks in liver and intestinal development. Cold Spring Harb. Perspect. Biol 4, a008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HQ, Zhu JS, and Baron AD (1999). Dose-response relationship of insulin to glucose fluxes in the awake and unrestrained mouse. Metabolism 48, 965–970. [DOI] [PubMed] [Google Scholar]
- Steele AM, Shields BM, Wensley KJ, Colclough K, Ellard S, and Hattersley AT (2014). Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 311, 279–286. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45 (D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, and Kahn CR (2006). Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol 7, 85–96. [DOI] [PubMed] [Google Scholar]
- Thiebaud D, Jacot E, DeFronzo RA, Maeder E, Jequier E, and Felber JP (1982). The effect of graded doses of insulin on total glucose uptake, glucose oxidation, and glucose storage in man. Diabetes 31, 957–963. [DOI] [PubMed] [Google Scholar]
- Tokarz VL, MacDonald PE, and Klip A (2018). The cell biology of systemic insulin function. J. Cell Biol 217, 2273–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Prajapati S, Park KJ, Kelly NJ, Yamamoto Y, and Gaynor RB (2006). IKK alpha regulates estrogen-induced cell cycle progression by modulating E2F1 expression. J. Biol. Chem 281, 6699–6706. [DOI] [PubMed] [Google Scholar]
- Villegas VE, and Zaphiropoulos PG (2015). Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci 16, 3251–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, and Chen T (2012). Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin. Drug Metab. Toxicol 8, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Besser D, Luca E, and Stoffel M (2003). Insulin regulates the activity of forkhead transcription factor Hnf-3beta/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc. Natl. Acad. Sci. USA 100, 11624–11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, and Smyth GK (2010). ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, and Qu LH (2010). Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52, 1431–1442. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, Nakakuki M, Takahashi A, Suzuki H, Sone H, et al. (2004). SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J. Biol. Chem 279, 12027–12035. [DOI] [PubMed] [Google Scholar]
- Yang L, Li P, Yang W, Ruan X, Kiesewetter K, Zhu J, and Cao H (2016). Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab 24, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, and Kahn CR (2004). Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc. Natl. Acad. Sci. USA 101, 16525–16530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip L, and Fathman CG (2014). Type 1 diabetes in mice and men: gene expression profiling to investigate disease pathogenesis. Immunol. Res 58, 340–350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data described here is deposited in Gene Expression Omnibus (GEO) under accession number GEO: GSE117741. Expression data from muscle and liver microarrays of HFD C57BL/6 mice are under accession number GEO: GSE123394. The metabolic phenotype of these mice has been described (Almind and Kahn, 2004). Data-sets from STZ-treatment in mouse muscle GEO: GSE1659 (Kivela¨ et al., 2006) and liver GEO: GSE39752 (Franko et al., 2014) were downloaded from GEO.