Abstract
BACKGROUND:
The purpose of the study was to determine the prevalence and characterize the resistance profiles of Escherichia coli isolated from various clinical specimens by various phenotypic and genotypic methods.
MATERIALS AND METHODS:
A total of 196 consecutive, nonduplicate strains of clinically significant E. coli isolated from various clinical specimens were included in the study. Identification and antimicrobial susceptibility testing was performed by using Vitek-2 system (Biomerieux, France). Phenotypic detection of extended spectrum beta-lactamase (ESBLs), Amp-C-β lactamase (Amp C), and carbapenemase production was done by various combination of disc diffusion methods, minimum inhibitory concentration determination by E-test, followed by polymerase-chain-reaction for the detection of β-lactamase-encoding genes.
RESULTS:
Overall prevalence of ESBLs, Amp C, and carbapenemase production was found to be 88.3%, 42.2%, and 65.1% by the phenotypic detection methods. Our study also revealed high resistance rates against other antibiotics such as cefepime (89%), cefotaxime (95.4%), ceftazidime (85.4%), ceftriaxone (91.8%), cefpodoxime (92.7%), aztreonam (56.3%), piperacillin/tazobactam (89.2%), and ticarcillin/clavulanic acid (76.3%). The most prevalent ESBL gene was blaTEM (67.30%), and least prevalent ESBL gene was blaVEB (2.61%). In case of Amp C, blaFOX gene (21.9%) was predominant. Among the genes encoding for carbapenemases, the most common gene was blaNDM (61.7%) followed by blaVIM (30.8%), blaKPC (10.6%), blaOXA-48 (5.3%), and blaIMP (2.1%).
CONCLUSION:
Our findings suggest a high rate of ESBLs, Amp C, and carbapenemase production among the E. coli isolates. A combination of both phenotypic and genotypic methods would be ideal for better characterization of resistance patterns among the E. coli isolates.
Keywords: Amp-C-β lactamase, carbapenemases, Escherichia coli, extended spectrum beta-lactamases
Introduction
Escherichia coli is one of the most common pathogens among the family Enterobacteriaceae causing various infections worldwide. The increasing spread of resistance to extended generation cephalosporin group of drugs is on the rise in both community and hospital settings. Resistance to cephalosporins is due to the production of extended-spectrum beta-lactamases (ESBLs). However, Amp-C-β lactamase (Amp C) cephalosporinases may also be attributed by the acquisition of a plasmid-mediated Amp C enzyme or due to overexpression of the chromosomally encoded Amp C enzyme in E. coli.[1] Plasmid-mediated Amp C genes are derived from the chromosomal Amp C genes of several members of the family Enterobacteriaceae, including Enterobacter cloacae, Citrobacter freundii, Morganella morganii, and Hafnia alvei. The Amp C is a major clinical concern because these organisms overexpressing it are usually resistant to all the β-lactam drugs, except for cefpirome, cefepime, and the carbapenems.[2] There is also an increasing occurrence of carbapenem resistance in E. coli due to the production of a diverse group of carbapenemases. These enzymes are capable of hydrolyzing almost all β-lactams, including carbapenems. The genes encoding carbapenemases are usually encoded on mobile genetic elements like the plasmids.[3]
The β-lactamase-producing Enterobacteriaceae are mostly cross-resistant to other commonly used class of antibiotics such as aminoglycosides, trimethoprim-sulfamethoxazole, and fluoroquinolones, thus resulting in very few therapeutic options in a clinical setting. These multidrug-resistant organisms have higher mortality rates and medical costs than those due to non-β-lactamase-producing Enterobacteriaceae. Thus, the surveillance of these β-lactamase-producing Enterobacteriaceae is important for clinical care.[4] In the present study, an attempt was made to estimate the prevalence of ESBL, Amp C, and carbapenemases produced by E. coli.
Materials and Methods
Collection of bacterial strains
The study was conducted over a period of 2 years (2013–2015) at the Microbiology laboratory of the Jai Prakash Narayan Apex (JPNA) Trauma Centre of AIIMS hospital, New Delhi. The bacterial strains included in the study consisted of consecutive, nonduplicate, clinically significant E. coli isolates obtained at the Microbiology laboratories of the JPNA Trauma Centre and AIIMS hospital. This study (project code: I-800, grant number: 5/3/3/26/2011-ECD-I) was approved by the Institute Ethics Committee, AIIMS, New Delhi.
Phenotypic testing for extended spectrum beta-lactamase production
A total of 196 E. coli isolates were screened for ESBL production by recording the zone diameters of ceftazidime (CZD) (30 μg), cefotaxime (CTX) (30 μg), ceftriaxone (30 μg), cefpodoxime (10 μg), and aztreonam (30 μg) by disc diffusion testing on Muller Hinton Agar using CLSI recommended conditions.[5] ESBL screening also included the following criteria for Escherichia spp., a ceftriaxone, CZD, cefepime, or aztreonam minimum inhibitory concentration (MIC) of ≥2 μg/ml. The isolates showing positive results by screening were tested for further confirmatory and genotypic tests. Phenotypic confirmatory test for ESBL production was performed using CTX (30 μg), CZD (30 μg) discs with and without clavulanate (10 μg) combination disc test method, and E- test for ESBLs according to CLSI guidelines.[5] In E-test, MICs of CTX and CZD with and without clavulanic acid was tested as per the manufacturer's instructions.
Phenotypic testing for Amp-C β-lactamase
For the detection of Amp C cefoxitin, disc diffusion method was used according to the CLSI guidelines.[5] Isolates with zone diameters <14 mm were selected for confirmation of Amp C production. Alternatively, a cefoxitin MIC of ≥32 μg/ml was also used for screening. The isolates which were positive by screening test were further confirmed by three dimensional extract test (TDET), Amp C disk test, boronic acid disk test method, and disk approximation methods.[6,7,8]
Phenotypic testing for carbapenemase and metallo β-lactamases production
Screening for carbapenemase production was done using imipenem (10 μg), meropenem (10 μg), and ertapenem disc (10 μg) according to CLSI recommendations. Resistance to either imipenem, meropenem, or ertapenem disc was considered for further characterization by confirmatory tests and genotypic tests. The modified Hodge test was used for confirmation of carbapenemase production based on CLSI guidelines.[5] For the detection of metallo β-lactamases (MBL), E-test strips (BioMérieux, France) using the imipenem-imipenem/ethylenediaminetetraacetic acid (EDTA) combination, combined disk test (CDT) using CZD-EDTA, imipenem-EDTA discs, meropenem-EDTA discs,[9] double-disk synergy test (DDST) using 2-mercaptopropionic acid in combination with CZD, imipenem, and cefepime was used.[10] For the detection of Klebsiella pneumoniae Carbapenemases (KPC) production, Ertapenem was used as the screening agent for KPC production and the E. coli isolates which were positive by screening test were also subjected to boronic acid disk test.[9,11]
Polymerase-chain-reaction-based identification of β-lactamase genes
The presence of ESBL genes (blaTEM, blaSHV, blaCTX-M, blaPER, and blaVEB), Amp C genes (blaMOX, blaCIT, blaDHA, blaACC, blaEBC, and blaFOX), and carbapenemase-encoding genes (blaIMP, blaVIM, blaOXA, blaKPC, and blaNDM) were detected by polymerase-chain-reaction (PCR) using the primers and cycling conditions from previous published study by our group.[12] The negative control used was PCR mixtures with the addition of water, and positive controls were K. pneumoniae subsp. pneumoniae (ATCC 700603) (ESBL producer, SHV positive), K. pneumonia (ATCC BAA-1144) (low-level Amp C producer), Enterobacter cloacae subsp. cloacae (ATCC BAA-1143) (high-level Amp C producer), K. pneumoniae (ATCC BAA-1705) (KPC positive), and Enterobacter cloacae (ATCC BAA-2468) (NDM1 positive).
Sensitivity, specificity, and statistical analysis
The results for all parameters were calculated as a percentage as applicable. Analysis was carried out using SPSS version 21, (IBM, Armonk, NY, United States of America). Chi-square test was performed to determine the significant difference between variables. P < 0.05 was considered as statistically significant.
Results
In our study, the clinical isolates were obtained from the following samples which included blood 26/196 (13.2%), sterile body fluids 40/196 (20.4%), endotracheal aspirate 19/193 (9.6%), pus 66/196 (33.6%), and urine 45/196 (22.9%). The distribution of E. coli strains according to the type of specimen is shown in Figure 1. The E. coli strains included in our study revealed high resistance rates against other antibiotics such as cefepime (89%), CTX (95.4%), CZD (85.4%), ceftriaxone (91.8%), cefpodoxime (92.7%), aztreonam (56.3%), piperacillin /tazobactam (89.2%), ticarcillin/clavulanic acid (76.3%), meropenem (63.2%), and doripenem (61.1%) as depicted in Figure 2.
Figure 1.
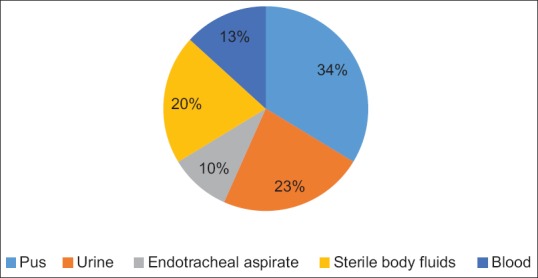
Distribution of Escherichia coli isolates according to the type of specimen
Figure 2.
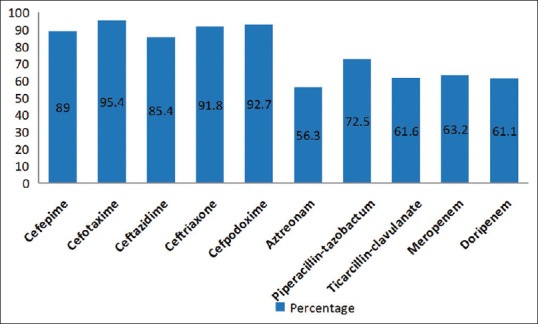
Antibiotic resistance pattern of Escherichia coli isolates
Prevalence of extended spectrum beta-lactamases and Amp C β-lactamases in Escherichia coli isolates
Among the various screening tests evaluated for ESBL production, maximum were detected by the CZD disc diffusion test – 173/196 (88.3%) followed by CTX disc – 165/196 (84.1%), cefpodoxime disc – 163/196 (83.2%), ceftriaxone disc – 162/196 (82.7%), and aztreonam disc – 125/196 (63.8%) diffusion test. In E-test used for screening, CZD E-test detected – 117/196 (76.5%), CTX E-test – 126/196 (63.4%), and ceftriaxone E-test detected – 106/196 (54.1%). Thus, CZD had the highest sensitivity for ESBL screening. Among the 173 E. coli isolates which were positive for ESBL screen by CZD disk diffusion test, 103 (59.53%) were positive by the confirmatory CZD/CZD-clavulanic acid combination disk test. For isolates which were positive for ESBL screen by CTX disk diffusion test, 102 (55.73%) isolates were positive by CTX/CTX-clavulanic acid combination disk test. While in the E-test combination test, 30/117 (25.64%) and 42/126 (33.33%) were detected by CZD/CZD-clavulanic acid and CTX/CTX-clavulanic acid E-test, respectively. Among the 173 isolates, 73 (42.2%) were positive for Amp C production by cefoxitin disk diffusion test and 54/173 (31.2%) were positive by cefoxitin MIC E-test. The isolates which were positive by screening test were tested further by confirmatory tests such as TDET (5.2%), Amp C disk test (8.7%), boronic acid disk test (5.2%), and disk approximation method (7.5%).
Prevalence of carbapenemases and metallo β-lactamase in Escherichia coli isolates
Among the E. coli isolates, 103/196 were suspected to have carbapenem resistance by the screening test. The prevalence of carbapenem resistance was found to be 65% (67/103) by ertapenem disc diffusion test followed by 32.3% (33/103) and 40.7% (42/103) by imipenem disc and meropenem disc diffusion test, respectively. Thus, ertapenem had the highest sensitivity for screening carbapenemases. By confirmatory tests, carbapenem resistance was seen in 24/42 (57.14%) by meropenem MHT and in 24/67 (35.82%) by ertapenem MHT. The highest detection of metallo β-lactamase production was seen in imipenem-imipenem EDTA E-test (88/103 (85.43%). CDT performed using CZD-EDTA, imipenem-EDTA discs, meropenem-EDTA disc detected 35/103 (33.9%), 28/103 (27.1%), and 50/103 (48.5%) isolates, respectively. DDST using 2-mercaptopropionic acid in combination with CZD, imipenem, and cefepime for the detection of MBL production detected 82/103 (79.6%), 28/103 (27.1%), and 76/103 (73.7%), respectively, to be MBL producers. There was significant difference seen between DDST and CDT performed using CZD (P < 0.0001). Around 60% (6/10) were found to be KPC-possessing E. coli isolates by boronic acid disk tests performed using meropenem.
Genotypic characterization of extended spectrum beta-lactamase, Amp C, and carbapenemase -producing Escherichia coli isolates
The prevalence of blaTEM, blaSHV, blaCTX-M, blaPER, and blaVEB β-lactamases genes for ESBL was found to be 67.3%, 16.81%, 63.26%, 19.4%, and 2.4%, respectively. The prevalence of Amp C genes blaMOX, blaCIT, blaDHA, blaACC, blaEBC, and blaFOX was found to be 6.86%, 9.16%, 2.5%, 0.5%, 0%, and 21.9%, respectively. The most common carbapenemase encoding gene detected was blaNDM-1 in 58/94 (61.7%) followed by blaVIM (30.8%), blaKPC (10.6%), blaOXA-46 (5.3%), and blaIMP (2.1%).
Discussion
Emergence of ESBLs, Amp C, and carbapenemase -producing Enterobacteriaceae presents a significant diagnostic and therapeutic challenge in the management of infections. The risk factors leading to infections with such multidrug-resistant organisms include increased length of hospital stay, prolonged stay in the intensive care units, arterial or urinary catheterization, and exposure to various broad-spectrum antibiotics. The detection of these resistant isolates on a daily basis in a clinical laboratory is quite challenging.[13]
The prevalence of ESBL in our study was estimated to be 88.3%. The prevalence of ESBLs reported from various other studies from India ranges from 60% to 80%.[14,15] Combined disk diffusion test was more sensitive than the E-test combination test used for the detection of ESBL. The above finding was consistent with other studies.[13,14] According to CLSI for Enterobacteriaceae, MIC ≥2 μg/ml against cefpodoxime, CZD, aztreonam, CTX, and ceftriaxone is regarded as a possible ESBL producer. In our study, the MIC of isolates resistant to third-generation cephalosporin was in the range of 2–64 μg/ml against the CZD. About 68.3% of the isolates had an MIC of 64 μg/ml, and these were clinical isolates from pyogenic infections.
Although there are no CLSI guidelines for the detection of Amp C production, reduced cefoxitin susceptibility was taken as an indicator of Amp C production.[16] In our study, the Amp C production among E. coli was 42.2% which was concordant with other studies from India.[17,18] We observed coexistence of ESBL and Amp C in 19.6% of the E. coli isolates which was consistent with previous reports.[19] In our study, the ESBL and Amp-C producers were highly resistant to other antibiotics such as aztreonam (56.3%), piperacillin/tazobactam (89.2%), ticarcillin/clavulanic acid (76.3%), meropenem (63.2%), and doripenem (61.1%).
Among the various tests performed for the detection of MBLs, CDT performed with meropenem and EDTA showed the highest sensitivity 58.8% (95% confidence interval [CI] - 48.2%–68.6%) but low specificity rate 66.6% (95% CI - 35.4%–87.9%), whereas CDT performed with imipenem and EDTA exhibited a better specificity 77.7% (45.2%–93.6%), and among the DDST, CZD with 2MPA showed the highest sensitivity 87.2% (95% CI - 79%–92.5%) but low specificity of 33.3% (95% CI - 12%–64.5%). Previously published studies had found that the above methods to be a better predictor of MBL production with high sensitivity and specificity rates;[20,21] however, in our study, we have found that the above methods were not a true predictor of MBL production because of lower sensitivity and specificity rates. In case of boronic acid disk test for the detection of KPCs, meropenem was found to be the most sensitive substrate, which was similar to previously published studies.[11,22]
The prevalence of carbapenemase-producing E. coli in our hospital setting was found to be as high as 65%. This was similar to the prevalence rates obtained in similar studies from other parts of India.[23,24] Phenotypic tests showing reduced carbapenem susceptibility are used for the identification of carbapenemase-producing organisms. The carbapenems have varied MICs for different carbapenemase producers. In our study, ertapenem was found to be an ideal candidate for screening the carbapenemase producers. This is consistent with other previously published studies.[9,25] Ertapenem has also been proposed to be the most suitable carbapenem for the identification of KPC producers which harbors low-level resistance to carbapenems.[26] Most of the carbapenem-resistant E. coli isolates were also resistant to other antimicrobial agents that were routinely used in the laboratory as observed in other studies.[27,28] Our study also revealed high resistance rates against other antibiotics such as cefepime (89%), CTX (95.4%), CZD (85.4%), ceftriaxone (91.8%), cefpodoxime (92.7%), aztreonam (56.3%), piperacillin/tazobactam (89.2%), and ticarcillin/clavulanic acid (76.3%). The occurrence of coresistance to other antibiotic class of drugs may be due to plasmids or insertion sequences which transmits these resistance determinants seen in CRE isolates.[27] In our study, the most common genes encoding ESBL, Amp C, and carbapenemase genes in E. coli was blaTEM, 67.3% blaFOX (21.9%) and blaNDM-1 (61.7%). The above finding was coherent to other studies from India.[29]
Conclusion
The present study highlights the high burden of ESBL, Amp C, and carbapenemase-producing multidrug-resistant E. coli isolates from a tertiary care center in India. This situation is alarming as these multidrug-resistant organisms are refractory to the commonly used antibiotics in clinical setting. A strong antimicrobial stewardship program is the need of the hour along with emphasis on strict hospital infection control practices to prevent the dissemination of these multidrug-resistant organisms.
Financial support and sponsorship
We acknowledge the financial support of Indian Council of Medical Research (ICMR) (project code: I-800, grant number: 5/3/3/26/2011-ECD-I) for funding the study.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Baudry PJ, Nichol K, DeCorby M, Mataseje L, Mulvey MR, Hoban DJ, et al. Comparison of antimicrobial resistance profiles among extended-spectrum-beta-lactamase-producing and acquired AmpC beta-lactamase-producing Escherichia coli isolates from Canadian intensive care units. Antimicrob Agents Chemother. 2008;52:1846–9. doi: 10.1128/AAC.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodford N, Reddy S, Fagan EJ, Hill RL, Hopkins KL, Kaufmann ME, et al. Wide geographic spread of diverse acquired AmpC beta-lactamases among Escherichia coli and Klebsiella spp. In the UK and Ireland. J Antimicrob Chemother. 2007;59:102–5. doi: 10.1093/jac/dkl456. [DOI] [PubMed] [Google Scholar]
- 3.Hrabák J, Študentová V, Jakubů V, Adámková V, Dvořáková L, Balejova M, et al. Prevalence study on carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates in Czech hospitals – Results from Czech part of European survey on carbapenemase – Producing Enterobacteriaceae (EuSCAPE) Epidemiol Mikrobiol Imunol. 2015;64:87–91. [PubMed] [Google Scholar]
- 4.Sheng WH, Badal RE, Hsueh PR. SMART Program. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for monitoring antimicrobial resistance trends (SMART) Antimicrob Agents Chemother. 2013;57:2981–8. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M100S27 | Performance Standards for Antimicrobial Susceptibility Testing. 2017. [Last accessed on 2017 Dec 19]. Available from: https://www.clsi.org/standards/products/microbiology/documents/m100/
- 6.Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated ampC beta-lactamases in Enterobacteriaceae lacking chromosomal ampC beta-lactamases. J Clin Microbiol. 2005;43:3110–3. doi: 10.1128/JCM.43.7.3110-3113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahid M, Malik A, Agrawal M, Singhal S. Phenotypic detection of extended-spectrum and AmpC beta-lactamases by a new spot-inoculation method and modified three-dimensional extract test: Comparison with the conventional three-dimensional extract test. J Antimicrob Chemother. 2004;54:684–7. doi: 10.1093/jac/dkh389. [DOI] [PubMed] [Google Scholar]
- 8.Thomson KS. Extended-spectrum-beta-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48:1019–25. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asthana S, Mathur P, Tak V. Detection of carbapenemase production in gram-negative bacteria. J Lab Physicians. 2014;6:69–75. doi: 10.4103/0974-2727.141497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623–9. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsakris A, Kristo I, Poulou A, Themeli-Digalaki K, Ikonomidis A, Petropoulou D, et al. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J Clin Microbiol. 2009;47:362–7. doi: 10.1128/JCM.01922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khurana S, Mathur P, Kapil A, Valsan C, Behera B. Molecular epidemiology of beta-lactamase producing nosocomial gram-negative pathogens from North and South Indian hospitals. J Med Microbiol. 2017;66:999–1004. doi: 10.1099/jmm.0.000513. [DOI] [PubMed] [Google Scholar]
- 13.Sageerabanoo S, Malini A, Mangaiyarkarasi T, Hemalatha G. Phenotypic detection of extended spectrum β-lactamase and amp-C β-lactamase producing clinical isolates in a tertiary care hospital: A preliminary study. J Nat Sci Biol Med. 2015;6:383–7. doi: 10.4103/0976-9668.160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaur J, Mahajan G, Chand K, Sheevani, Chopra S. Enhancing phenotypic detection of ESBL in AmpC co-producers by using Cefepime and Tazobactam. J Clin Diagn Res. 2016;10:DC05–8. doi: 10.7860/JCDR/2016/15264.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bora A, Ahmed GU, Hazarika NK. Phenotypic detection of extended spectrum β-lactamase and AmpC β-lactamase in urinary isolates of Escherichia coli at a tertiary care referral hospital in Northeast India. J Coll Med Sci Nepal. 2013;8:22–9. [Google Scholar]
- 16.Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791–6. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arora S, Bal M. AmpC beta-lactamase producing bacterial isolates from Kolkata hospital. Indian J Med Res. 2005;122:224–33. [PubMed] [Google Scholar]
- 18.Manoharan A, Sugumar M, Kumar A, Jose H, Mathai D, Khilnani GC, et al. Phenotypic & molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian medical centers. Indian J Med Res. 2012;135:359–64. [PMC free article] [PubMed] [Google Scholar]
- 19.Singhal S, Mathur T, Khan S, Upadhyay DJ, Chugh S, Gaind R, et al. Evaluation of methods for AmpC beta-lactamase in gram negative clinical isolates from tertiary care hospitals. Indian J Med Microbiol. 2005;23:120–4. doi: 10.4103/0255-0857.16053. [DOI] [PubMed] [Google Scholar]
- 20.Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H, et al. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 2008;61:548–53. doi: 10.1093/jac/dkm535. [DOI] [PubMed] [Google Scholar]
- 21.Franklin C, Liolios L, Peleg AY. Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006;44:3139–44. doi: 10.1128/JCM.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsakris A, Themeli-Digalaki K, Poulou A, Vrioni G, Voulgari E, Koumaki V, et al. Comparative evaluation of combined-disk tests using different boronic acid compounds for detection of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceaec clinical isolates. J Clin Microbiol. 2011;49:2804–9. doi: 10.1128/JCM.00666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair PK. Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbiol Infect Dis. 2013;3:207–10. [Google Scholar]
- 24.Exner M, Bhattacharya S, Christiansen B, Gebel J, Goroncy-Bermes P, Hartemann P, et al. Antibiotic resistance: What is so special about multidrug-resistant gram-negative bacteria? GMS Hyg Infect Control. 2017;12:Doc05. doi: 10.3205/dgkh000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V, et al. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18:432–8. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 26.Birgy A, Bidet P, Genel N, Doit C, Decré D, Arlet G, et al. Phenotypic screening of carbapenemases and associated β-lactamases in carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2012;50:1295–302. doi: 10.1128/JCM.06131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27–34. doi: 10.2147/IDR.S127177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.AlTamimi M, AlSalamah A, AlKhulaifi M, AlAjlan H. Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci. 2017;24:155–61. doi: 10.1016/j.sjbs.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhary U, Agarwal S, Raghuraman K. Identification of extended spectrum beta lactamases, AmpC and carbapenemase production among isolates of Escherichia coli in North Indian tertiary care centre. Avicenna J Med. 2018;8:46–50. doi: 10.4103/ajm.AJM_156_17. [DOI] [PMC free article] [PubMed] [Google Scholar]


