Depleting creatine levels in thermogenic adipocytes by inhibiting creatine biosynthesis reduces thermogenesis and causes obesity. However, whether creatine import from the circulation affects adipocyte thermogenesis is unknown. Here we show that deletion of the cell surface creatine transporter (CrT) selectively in fat (AdCrTKO) substantially reduces adipocyte creatine and phosphocreatine levels, and reduces whole body energy expenditure in mice. AdCrTKO mice are cold intolerant and become more obese than wild type animals when fed a high-fat diet. Loss of adipocyte creatine transport blunts diet- and β3-adrenergic-induced thermogenesis, whereas creatine supplementation during high-fat feeding increases whole body energy expenditure in response to β3-adrenergic agonism. In humans, CRT expression in purified subcutaneous adipocytes correlates with lower body mass index and increased insulin sensitivity. Our data indicate that adipocyte creatine abundance depends on creatine sequestration from the circulation. Given that it affects whole body energy expenditure, enhancing creatine uptake into adipocytes may offer an opportunity to combat obesity and obesity-associated metabolic dysfunction.
The escalation in prevalence of obesity worldwide has led to a surge in type 2 diabetes, cardiovascular disease and many cancers1,2. Excess fat storage occurs when caloric intake persistently exceeds caloric expenditure3. The capacity for thermogenic adipose tissues (brown and beige) to dissipate chemical energy offers great potential to combat obesity. Some individuals who are predisposed to obesity exhibit decreased adipose thermogenic capacity4, suggesting that this impairment is relevant in the context of human weight gain. Several pathways in adipose and non-adipose tissues have emerged in recent years to play critical roles in adaptive thermogenesis5–11.
Creatine supports thermogenic respiration by stimulating mitochondrial ATP turnover, in vitro12,13. Importantly, pharmacological reduction of endogenous creatine levels blunts adrenergic-stimulated thermogenesis and core body temperature in vivo, and reduces oxidative metabolism in beige fat and brown adipose tissue (BAT)13–15. Selective inactivation of creatine synthesis in fat (Adipo-Gatm KO mice) causes a reduction in creatine levels in BAT, suppresses whole body energy expenditure, and results in obesity. Critically, creatine supplementation of Adipo-Gatm KO animals prolongs the ability to sustain adrenergic thermogenesis, suggesting that creatine transport can be limiting under certain physiological conditions in vivo, and that adipocyte creatine levels are at least partly regulated by sequestration from the circulation16.
Fat-selective deletion of the creatine transporter depletes adipocyte creatine abundance
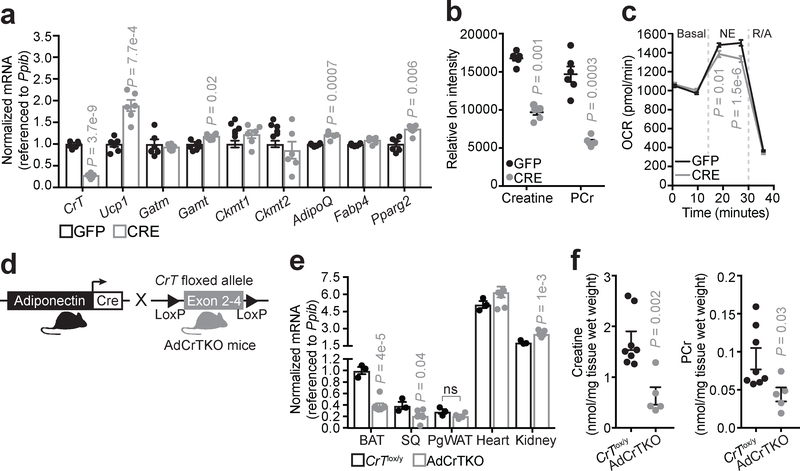
Creatine can be taken up into cells by a creatine transporter, CrT (also known as Slc6a8)17, and the rate of sequestration into BAT from the circulation is substantial as it can match that of skeletal muscle18. To begin to examine the contribution of creatine transport to adipocyte creatine abundance, we purified the BAT stromal vascular fraction from mice containing a floxed allele of CrT19, which we refer to as CrTlox/y mice. CrTlox/y primary brown adipocytes were differentiated in vitro and infected with adenovirus encoding Cre recombinase or green fluorescent protein (Gfp). CrT transcript abundance was significantly reduced (~60%) 6 days following expression of CRE compared to expression of GFP (Fig. 1a). There was no defect in adipocyte differentiation of the cells based on mRNA expression levels of AdipoQ, Fabp4 and Pparg2 (Fig. 1a). Similarly, mitochondrial creatine kinases (Ckmt1 and Ckmt2) and Gatm transcript levels were not changed, but Gamt levels were marginally elevated. We also observed an increase in Ucp1 mRNA abundance upon CrT deletion (Fig. 1a). This potentially compensatory relationship between Ucp1 and genes of creatine metabolism is consistently observed in various murine models13,16,20,21, and is suggestive of parallel thermogenic pathways.
Fig. 1 |. Inactivation of creatine transport depletes creatine abundance in adipocytes.
a, RT-qPCR of primary brown adipocytes (genotype: CrTlox/y) following adenoviral-mediated infection with GFP or Cre recombinase (GFP, n = 6; CRE, n = 6). b, LC-MS analysis of creatine and phosphocreatine (PCr) levels after GFP or CRE adenoviral infection of CrTlox/y primary brown adipocytes (GFP, n = 6; CRE, n = 6). c, Oxygen consumption rate (OCR) of GFP- or CRE-infected primary brown adipocytes (genotype: CrTlox/y) (GFP, n = 17; CRE, n = 19). Norepinephrine (NE) was added acutely at a final concentration of 100 nM. Rotenone and Antimycin A (R/A) were used each at a final concentration of 3 μM to inhibit mitochondrial respiration. d, Cartoon of breeding strategy to generate adipose-specific CrT knockout mice (AdCrTKO). e, RT-qPCR of CrT mRNA from various tissues of CrTlox/y and AdCrTKO animals. Brown adipose tissue (BAT, CrTlox/y, n = 3; AdCrTKO, n = 7), subcutaneous adipose tissue (SQ, CrTlox/y, n = 3; AdCrTKO, n = 6), perigonadal white adipose tissue (PgWAT, CrTlox/y, n = 3; AdCrTKO, n = 4), Heart (CrTlox/y, n = 3; AdCrTKO, n = 7) and Kidney (CrTlox/y, n = 3; AdCrTKO, n = 7). f, NMR analysis of BAT creatine and PCr levels (CrTlox/y, n = 8; AdCrTKO, n = 5). Data are presented as mean ± s.e.m. of biologically independent samples. Two-tailed Student’s t-test (a-c, e, f).
Using liquid chromatography coupled to mass spectrometry (LC-MS), we detected a ~50% reduction in steady state creatine and phosphocreatine (PCr) levels in CRE-infected CrTlox/y primary brown adipocytes compared to GFP-infected cells (Fig. 1b). In contrast, while CRE-infection of Gatmlox/lox brown adipocytes reduced Gatm transcript abundance substantially by 60% (Supplementary Figure 1a), these adipocytes exhibited no change in creatine or PCr levels, in vitro (Supplementary Figure 1b). Thus, while the creatine pool in BAT is partly regulated by GATM-dependent creatine synthesis in vivo16, creatine abundance is primarily regulated by cellular transport by CRT in cultured brown adipocytes in vitro. Consistent with a thermogenic role for creatine, CRE-infected CrTlox/y primary brown adipocytes exhibited decreased norepinephrine-dependent thermogenic respiration, compared to GFP-infected cells (Fig. 1c).
We next explored the role of creatine transport-linked metabolism in vivo. We reduced adipocyte creatine abundance through fat-selective inactivation of CrT by crossing Adiponectin-Cre transgenic animals22 with CrTlox/y mice (Fig. 1d). We hereafter refer to this model as adipose-specific CrT knockout (AdCrTKO) mice. The selective deletion of CrT in adipocytes was evaluated by RT-qPCR analysis of BAT, subcutaneous adipose tissue (SQ), perigonadal white adipose tissue (PgWAT), kidney, and skeletal muscle. Significant depletion of CrT mRNA abundance was detected in bulk BAT and SQ of AdCrtKO animals relative to CrTlox/y controls (BAT: CrTlox/y 1.00 ± 0.06, AdCrTKO 0.39 ± 0.04; SQ: CrTlox/y 0.39 ± 0.06, AdCrTKO 0.21 ± 0.04), but not in PgWAT, kidney or skeletal muscle (Fig. 1e). Moreover, these data demonstrate that, of the adipose depots analyzed, BAT displays the highest CrT expression levels (~3-fold higher than SQ and PgWAT).
We next determined whether CrT deletion perturbs creatine abundance in vivo. Indeed, creatine and PCr levels were significantly reduced in the BAT of AdCrTKO mice compared to CrTlox/y controls, as demonstrated by nuclear magnetic resonance (Creatine: ~60% reduced; PCr: ~40% reduced) (Fig. 1f). Serum creatine levels were unaltered between AdCrTKO and CrTlox/y mice (Supplementary Figure 1c), indicating that loss of adipocyte creatine does not alter systemic abundance of this metabolite. Our previous work with Adipo-Gatm KO animals showed that creatine was depleted by 30–40% in BAT16. Therefore, in terms of relative contributions of the different pathways for creatine accumulation, adipocyte creatine abundance is regulated by both intracellular synthesis and cellular transport in vivo.
AdCrTKO mice have impaired energy expenditure
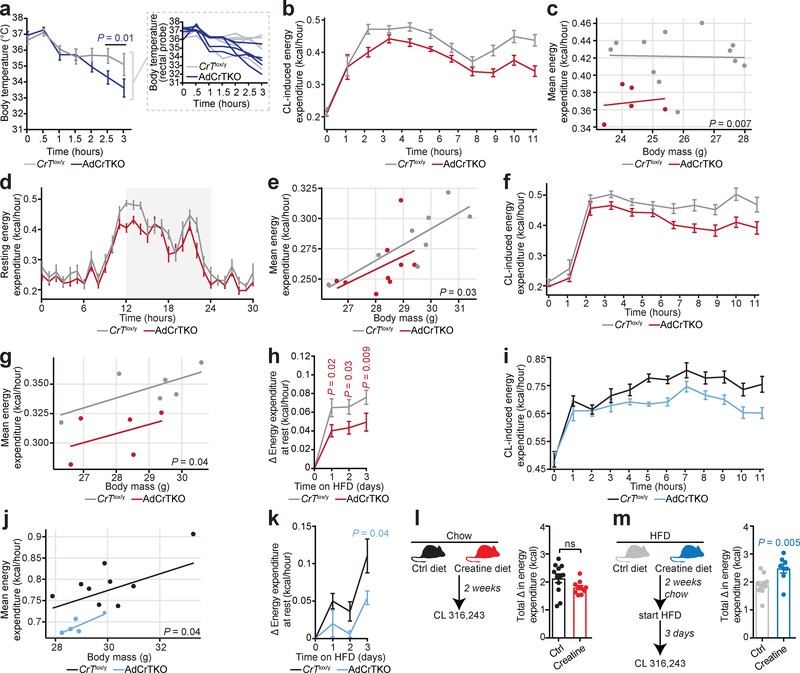
Mice with adipocyte-specific loss of creatine synthesis are unable to maintain thermal homeostasis in response to acute cold (4°C) exposure16. Thus, we explored body temperature defense in AdCrTKO animals. Upon acute challenge to 4°C, the body temperature of AdCrTKO mice dropped at a significantly faster rate than CrTlox/y control mice (CrTlox/y 35.1°C ± 0.69°C, AdCrTKO 33.6°C ± 0.57°C after 3 hours) (Fig. 2a). These data indicate that loss of adipocyte creatine transport may impair thermogenic capacity in vivo. Thermogenic and non-thermogenic factors contribute to the maintenance of body temperature upon exposure to decreased environmental temperature. Thus, we next studied whole body energy expenditure by examining resting and adrenergic energy expenditure in animals relieved from thermal stress at thermoneutral (30°C) housing23. Energy expenditure at rest during chow feeding was not different between AdCrTKO animals and littermate age-matched controls (Supplementary Figure 2a), nor was there any difference between genotypes when accounting for any differences in body mass by ANCOVA (Supplementary Figure 2b). There was no difference in ambulatory movement between genotypes (Supplementary Figure 2c). To examine the direct contribution of adipose tissue to whole-body metabolic rate, we used the relatively adipose-selective β3-adrenergic receptor agonist CL 316,24324, which we refer to as CL. Because adrenergic responsiveness can be masked at subthermoneutral temperatures (when obligatory thermogenesis is not sufficient to maintain thermal homeostasis) we examined the change in energy expenditure of conscious free-moving mice housed at 30°C following intraperitoneal (i.p.) administration of CL. Strikingly, the activation of CL-induced energy expenditure was significantly blunted (~10%) in AdCrTKO animals compared to CrTfl/y controls (Fig. 2b), confirming that loss of creatine transport impairs adrenergic thermogenesis in fat. Next, we applied regression analysis to investigate the effect of inactivation of CrT on the relationship between CL-induced thermogenesis and body mass. The average energy expenditure per unit change in body mass was proportionately and significantly lower in AdCrTKO animals compared to their age-matched littermate controls (Fig. 2c). Thus, ablation of creatine transport into fat reduces whole body metabolic rate in response to adrenergic stimulation.
Fig. 2 |. AdCrTKO mice have impaired energy expenditure.
a, Body temperature of CrTlox/y and AdCrTKO mice (n = 5 mice per genotype, individual mouse temperatures are shown in inset). b, Energy expenditure at 30°C in response to CL 316,243 (CL) at 1 mg kg−1 (CrTlox/y, n = 12; AdCrTKO, n = 5). c, Regression plot of data from b (CrTlox/y, n = 12; AdCrTKO, n = 5). d, Resting energy expenditure in mice acutely (4 days) fed a high fat diet at 30°C (CrTlox/y, n = 8; AdCrTKO, n = 9). e, Regression plot of data from d (CrTlox/y, n = 8; AdCrTKO, n = 9). f, CL-dependent energy expenditure following acute high fat feeding at 30°C (CrTlox/y, n = 6; AdCrTKO, n = 5). g, Regression plot of data from f (CrTlox/y, n = 6; AdCrTKO, n = 5). h, Change in resting metabolic rate at 30°C in response to high fat feeding (CrTlox/y, n = 7; AdCrTKO, n = 9). i, CL-dependent energy expenditure following acute high fat feeding at 22°C (CrTlox/y, n = 9; AdCrTKO, n = 5). j, Regression plot of data from i (CrTlox/y, n = 9; AdCrTKO, n = 5). k, Change in resting metabolic rate at 22°C in response to high fat feeding (CrTlox/y, n = 11; AdCrTKO, n = 4). l, Experimental design for dietary chow creatine supplementation. Sustained energy expenditure (calculated as area under the curve) over 11 hours following i.p. administration of CL at 1 mg kg−1 (control chow diet, n = 12; creatine-supplemented chow diet, n = 10). m, Experimental design for dietary high fat creatine supplementation. Sustained energy expenditure (calculated as area under the curve) over 11 hours following i.p. administration of CL at 1 mg kg−1 (control high fat diet, n = 10; creatine-supplemented high fat diet, n = 8). Data are presented as mean ± s.e.m. of biologically independent samples. Multiple two-tailed Student’s t-tests (a, h, k-m); ANCOVA (c, e, g, j).
Several genes involved in creatine metabolism are regulated at the mRNA level by acute caloric excess16. Based on this, we examined CRT protein abundance in purified brown adipocytes isolated from wild-type (C57BL6/N) mice following acute high fat feeding for 3 days using liquid chromatography tandem mass spectrometry (LC-MS/MS) high resolution quantitative proteomics. Strikingly, CRT protein levels were significantly elevated (20%) following acute high fat feeding (Supplementary Figure 2d), consistent with the idea that creatine transport may be important in triggering an increase in energy expenditure in response to caloric excess, known as diet-induced thermogenesis16,25–27. Thus, we began to explore the interaction of adipocyte creatine transport with diet-induced thermogenesis by examining resting metabolic rate23 following acute exposure (3 days) to high fat feeding. In contrast to chow feeding, acute high fat feeding revealed a significant difference in resting metabolic rate at rest between AdCrTKO animals and littermate age-matched controls (Fig. 2d) and this difference was maintained when accounting for any differences in body mass by ANCOVA (Fig. 2e). There was no difference in ambulatory movement between genotypes (Supplementary Figure 2e). Moreover, adrenergic stimulation of adipocyte thermogenesis was reduced (~10%) in AdCrTKO animals compared to CrTfl/y controls (Fig. 2f), where the average energy expenditure per unit body mass was proportionately and significantly lower in AdCrTKO animals compared to their age-matched littermate controls (Fig. 2g). Next, we monitored the resting metabolic rate of mice at 30°C to determine the change in energy expenditure mediated by high fat feeding itself. Strikingly, AdCrTKO mice had a blunted (~40%) capacity to activate resting metabolic rate in response to high fat feeding, compared to CrTlox/y littermate controls (Fig. 2h and Supplementary Figure 2f). Therefore, loss of adipocyte creatine transport impairs diet-induced thermogenesis.
We next explored the effect of CrT ablation on metabolic rate at standard temperature (22°C) housing. Energy expenditure of chow-fed AdCrTKO mice was similar to CrTlox/y littermates at rest (Supplementary Figure 2g) and when accounting for the variation in body mass of individual mice by ANCOVA (Supplementary Figure 2h). Moreover, the reduced energy expenditure upon CL administration that we detected at 30°C with chow diet feeding was not present in chow-fed animals at 22°C (Supplementary Figure 2i and Supplementary Figure 2j). We hypothesized that the cold stress of 22°C housing might mask adrenergic-mediated creatine-dependent thermogenesis, such that an additional (temperature-independent) thermogenic stimulus might be required to reveal the defect in whole body energy expenditure of AdCrTKO animals. Consistent with this idea, AdCrTKO animals began to exhibit a trend towards reduced energy expenditure at rest, compared to control mice, when fed a high fat diet acutely (Supplementary Figure 2k and Supplementary Figure 2l). However, this trend was not statistically significant. Strikingly, the activation of CL-induced energy expenditure was significantly blunted (~15%) in acute high fat diet-fed AdCrTKO animals compared to CrTlox/y controls (Fig. 2i). Regression analysis demonstrated that the average energy expenditure per unit change in body mass was proportionately lower in AdCrTKO animals compared to their age-matched littermate controls (Fig. 2j). Moreover, AdCrTKO mice exhibited reduced diet-induced thermogenesis, compared to CrTlox/y animals at 22°C (Fig. 2k and Supplementary Figure 2m) Therefore, at 30°C housing, decreased adrenergic thermogenesis is consistently observed in animals with depletion of adipocyte creatine stores, whether chow- or high fat-fed. At 22°C housing, the induction of diet-induced thermogenesis with acute high fat feeding is required to reveal the impairment of adrenergic thermogenesis following depletion of BAT creatine.
Creatine supplementation increases energy expenditure during high fat feeding
Since we hypothesized that creatine transport controls diet-induced thermogenesis, we explored the effect of dietary creatine supplementation on whole body energy expenditure in wild-type mice. Following 2 weeks of consuming creatine-supplemented diet (chow or chow followed by acute high fat), BAT creatine abundance was increased significantly compared to animals receiving no supplementation, as measured by LC-MS (Supplementary Figure 2n). CL-induced activation of whole body energy expenditure was not altered by creatine supplementation under chow fed conditions (Fig. 2l). In contrast, it was significantly increased in creatine-supplemented animals following acute transition to HFD, compared to controls (Fig. 2m). Notably, creatine-supplementation with acute high fat feeding resulted in significantly higher BAT creatine levels compared to creatine-supplementation with chow diet (Supplementary Figure 2n). Together, these data demonstrate that adipocyte creatine transport is critical for supporting diet-induced thermogenesis in vivo. This is caused by either 1) an interaction between adipocyte creatine with certain components of high fat diet, 2) increased sequestration of creatine into BAT upon high fat feeding, or 3) a combination of the two.
AdCrTKO mice become more obese than control littermates upon high fat feeding
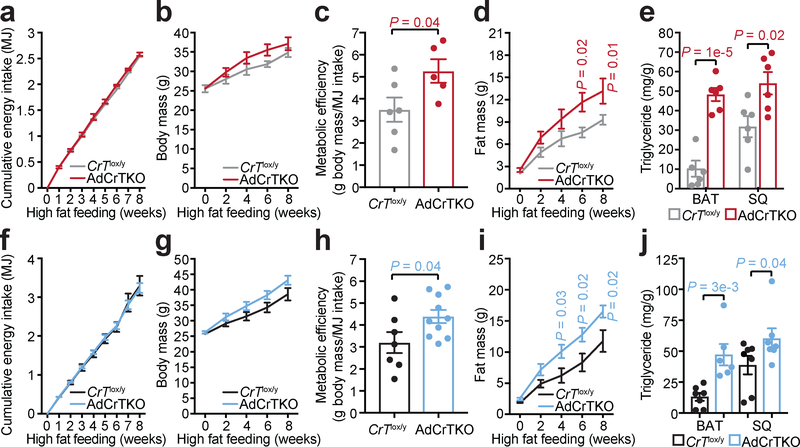
The decreased energy expenditure of AdCrTKO animals, particularly under high fat feeding conditions, prompted us to explore their propensity for diet-induced obesity. AdCrTKO mice and age-matched CrTlox/y littermate controls were challenged with ad libitum feeding of high fat diet over 8 weeks at two environmental temperatures (30°C and 22°C). At 30°C housing, cumulative food intake was indistinguishable between AdCrTKO mice and CrTlox/y animals (Fig. 3a), while the body mass of AdCrTKO mice increased over controls (Fig. 3b). Metabolic efficiency is a hallmark of obesity28, representing the fraction of assimilated energy that is stored somatically. AdCrTKO mice exhibited higher metabolic efficiency than CrTlox/y control mice (CrTlox/y 3.51 ± 0.55, AdCrTKO 5.26 ± 0.54) (Fig. 3c), strongly suggesting that fat-specific deletion of CrT may increase the propensity toward diet-induced obesity. Congruent with this hypothesis, AdCrtKO animals accreted a significantly greater quantity of fat mass compared to CrTlox/y animals (Fig. 3d), while lean mass was indistinguishable between genotypes (Supplementary Figure 3a). Most striking was the triglyceride (TG) abundance in individual fat depots. Consistent with CrT deficiency causing adipocyte thermogenic impairment, BAT TG content from AdCrTKO animals was significantly greater (~5-fold) than BAT TG content from CrTlox/y mice, and SQ TG content of AdCrTKO animals nearly doubled that of CrTlox/y mice (Fig. 3e). The obese phenotype exhibited by AdCrTKO animals was not observed when these animals were fed a chow diet (Supplementary Figure 3b and Supplementary Figure 3c). Similarly, there was no difference in lean mass between genotypes (Supplementary Figure 3d).
Fig. 3 |. AdCrTKO mice become obese on a high fat diet.
a, Cumulative energy intake in megajoules (MJ), b, body mass, c, metabolic efficiency and d, fat mass over 8 weeks of high fat feeding at 30°C (CrTlox/y, n = 6; AdCrTKO, n = 5). e, Triglyceride (TG) abundance in BAT and SQ following 12 weeks of high fat feeding at 30°C (BAT and SQ: CrTlox/y, n = 6; AdCrTKO, n = 6). f, Cumulative energy intake (MJ), g, body mass, h, metabolic efficiency and i, fat mass over 8 weeks of high fat feeding at 22°C (CrTlox/y, n = 7; AdCrTKO, n = 10). j, TG abundance in BAT and SQ following 8 weeks of high fat feeding at 22°C (BAT: CrTlox/y, n = 7; AdCrTKO, n = 6; SQ: CrTlox/y, n = 7; AdCrTKO, n = 7). Data are presented as mean ± s.e.m. of biologically independent samples. Two-tailed Student’s t-tests (c, h); multiple two-tailed Student’s t-tests (a, b, d-g, i, j).
At 22°C housing, cumulative food intake was identical between AdCrTKO mice and CrTlox/y animals (Fig. 3f), while the body mass of AdCrTKO mice trended higher than controls (Fig. 3g). AdCrTKO mice exhibited significantly higher metabolic efficiency than CrTlox/y control mice (CrTlox/y 3.2 ± 0.48, AdCrTKO 4.39 ± 0.30), further consistent with the hypothesis that deficiency in creatine transport into fat causes obesity (Fig. 3h). Indeed, the fat mass of AdCrtKO animals expanded greater than fat mass from CrTlox/y animals following high fat feeding (Fig. 3i), while lean mass was similar between genotypes (Supplementary Figure 3e). BAT TG levels from AdCrTKO animals was significantly greater (~3-fold) than BAT TG content from CrTlox/y mice, and SQ TG content of AdCrTKO animals nearly doubled that of CrTlox/y mice (Fig. 3j). Obesity did not occur in AdCrTKO mice fed chow diet (Supplementary Figure 3f and Supplementary Figure 3g), nor was there a change in lean mass (Supplementary Figure 3h). Thus, the consistent findings of obesity at the whole body level and TG accumulation in specific depots (BAT and SQ) at 30°C and 22°C, along with prior work16, strongly indicate that adipocyte creatine depletion causes obesity due to increased metabolic efficiency and impaired diet-induced thermogenesis.
AdCrTKO mice exhibit increases in kininogen expression in BAT and SQ
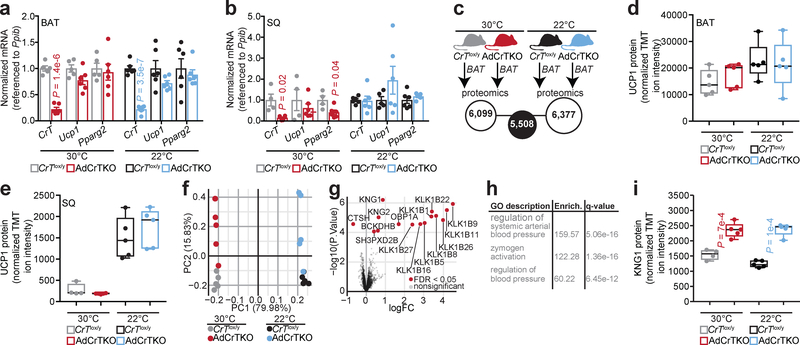
Inactivation of creatine transport into adipose tissue did not alter Ucp1 mRNA levels in BAT or SQ between AdCrTKO and CrTlox/y animals, regardless of housing temperature (Fig. 4a and Fig. 4b). Quantitative proteomics was next used to determine the global BAT expression profile of AdCrTKO animals compared to CrTlox/y littermate controls, housed at 30°C or 22°C. We were primarily interested in defining a common signature resulting from CrT inactivation, irrespective of housing temperature (Fig. 4c). Consistent with the mRNA levels, we did not detect any difference in BAT or SQ UCP1 protein expression between AdCrTKO and CrTlox/y animals regardless of housing temperature (Fig. 4d and Fig. 4e), nor was there any significant difference in mitochondrial proteins (Supplementary Data 3), suggesting that the thermogenic impairment exhibited by AdCrTKO animals is not a result of reduced UCP1-dependent thermogenesis or general mitochondrial abundance. We identified 6,099 and 6,377 unique proteins from BAT of mice housed at 30°C or 22°C, respectively (Fig. 4c, Supplementary Data 1 and Supplementary Data 2). Intersection of these two data sets resulted in identification of 5,508 proteins (Fig. 4c and Supplementary Data 3). We next performed unbiased clustering of the samples using proteins exhibiting the highest variance across all conditions and an empirical Bayes method to find proteins differentially expressed between AdCrTKO and CrTlox/y BAT consistently at 30°C and 22°C (Fig. 4f). This analysis yielded a set of 15 differentially expressed proteins at a false discover rate (FDR) of 0.05 (Fig. 4g and Supplemental Figure 3i). Gene ontology analysis on the differentially expressed proteins indicated an enrichment of proteins with known functions in the control of blood pressure (Fig. 4h). Specifically, kininogen 1 and 2 (KNG1 and KNG2) along with several kallikreins (involved in proteolytic processing of kininogens) were elevated in AdCrTKO compared to CrTlox/y BAT (Fig. 4g and Supplemental Figure 3i). Consistent with KNG1 expression in BAT, SQ from AdCrTKO animals also exhibited significantly elevated KNG1 protein expression at both 30°C and 22°C (Fig. 4i).
Fig. 4 |. Increased kininogen expression from BAT and SQ of AdCrTKO mice.
a, RT-qPCR from BAT of mice housed at 30°C (CrTlox/y, n = 5; AdCrTKO, n = 6), or 22°C (CrTlox/y, n = 6; AdCrTKO, n = 6). b, RT-qPCR from SQ of mice housed at 30°C (CrTlox/y, n = 4; AdCrTKO, n = 6), or 22°C (CrTlox/y, n = 6; AdCrTKO, n = 6). c, Cartoon of BAT proteomics experiment. d, Relative UCP1 protein abundance in BAT from mice housed at 30°C (CrTlox/y, n = 5 [light gray]; AdCrTKO, n = 5 [red]) or 22°C (CrTlox/y, n = 5 [black]; AdCrTKO, n = 5 [blue]). e, Relative UCP1 protein abundance in SQ from mice housed at 30°C (CrTlox/y, n = 4 [light gray]; AdCrTKO, n = 5 [red]) or 22°C (CrTlox/y, n = 5 [black]; AdCrTKO, n = 5 [blue]). f, Principal Component Analysis of proteomics results using the 50 proteins with highest variance across all samples (n = 5 mice per group). g, Volcano plot showing expression fold changes (log base 2) between AdCrTKO and CrTlox/y controls from both 30°C and 22°C housing, compared to their associated p values. Red dots represent proteins with significant changes at 0.05 FDR (CrTlox/y, n = 10; AdCrTKO, n = 10). h, Gene ontology (GO) enrichment of differentially abundant proteins (CrTlox/y, n = 10; AdCrTKO, n = 10). i, Relative KNG1 protein abundance in SQ from mice housed at 30°C (CrTlox/y, n = 4 [light gray]; AdCrTKO, n = 5 [red]) or 22°C (CrTlox/y, n = 5 [black]; AdCrTKO, n = 5 [blue]). Box plots were generated in Graphpad Prism using default parameters. Boxes stretch from the 25th to the 75th percentile; black horizonal line within each box represents the median. Data are presented as mean ± s.e.m. of biologically independent samples. Two-tailed Student’s t-tests (a-d, i); moderated t-tests using the limma (v3.32.10) package in R51,52 as justified in53, with P values being adjusted for multiple testing using the Benjamini-Hochberg approach (g, h).
AdCrTKO mice incur adaptive increases in the cold-inducible HK isoform of Kng1
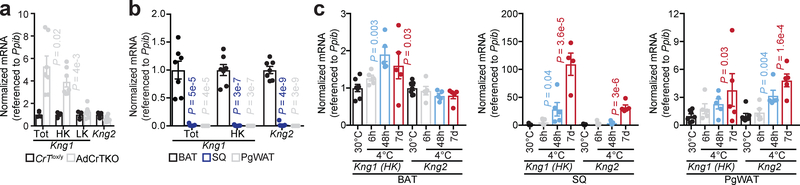
Adipose tissue plays an important role in maintaining vascular homeostasis29, and BAT vasculature is critical for optimal thermogenic function. Obesity can impair BAT-mediated thermogenesis partly through capillary rarefaction, and obesity has been posited to be associated with impaired arterial distensibility30,31. Conversely, increasing BAT perfusion can increase glucose uptake into BAT, which may (indirectly) support thermogenic respiration32,33. However, Kng1 mRNA has been identified in several reports to be a BAT-enriched secreted protein, and to be elevated in SQ fat upon cold exposure34,35, suggesting that KNG1 may have a thermogenic function. Thus, kininogen upregulation in AdCrTKO BAT and SQ, suggested that this was an adaptive response to counter the impaired thermogenesis caused by creatine depletion. Alternative splicing leads to two Kng1 isoforms, yielding a high molecular weight kininogen (HK) and low molecular weight kininogen (LK). Using RT-qPCR, we compared the mRNA abundance of Kng1 and Kng2 between AdCrTKO and CrTlox/y BAT. We detected a significant increase in Kng1 mRNA abundance in AdCrTKO compared to CrTlox/y BAT when using primers that detect all Kng1 isoforms or primers specific to the HK isoform (Fig. 5a). In contrast, the LK isoform of Kng1, as well as Kng2 were not differentially expressed at the mRNA level (Fig. 5a). These data suggest that in AdCrTKO BAT, KNG1 and KNG2 protein up-regulation (Fig. 4g) are at least partly regulated at the transcriptional and post-transcriptional level, respectively, and that the HK KNG1 isoform is the primary KNG1 isoform elevated in response to creatine depletion in adipose tissue.
Fig. 5 |. AdCrTKO mice incur adaptive increases in the cold-inducible HK isoform of Kng1.
a, RT-qPCR of kininogen isoforms in BAT (CrTlox/y, n = 3; AdCrTKO, n = 7). b, RT-qPCR of kininogen isoforms from mice housed at 30°C (n = 7). c, RT-qPCR of kininogen isoforms from mice housed at 30°C (n = 7), or exposed to 4°C for 6 hours (n = 5), 48 hours (n = 5), and 7 days (n = 5). Data are presented as mean ± s.e.m. of biologically independent samples. Two-tailed Student’s t-tests (a); one-way ANOVA (b, c).
Next, we examined the expression of Kng1 and Kng2 mRNA between different fat depots, as well as their regulation following acute and chronic exposure to cold (4°C), compared to 30°C housing. Kng1 and Kng2 expression was highly enriched in BAT, compared to SQ and pgWAT (Fig. 5b). Furthermore, abundance of the HK isoform of Kng1 was significantly increased in BAT, SQ and pgWAT following 4°C exposure (Fig. 5c). Kng2 levels were significantly elevated in the SQ and pgWAT (but not BAT) upon exposure to environmental cold (Fig. 5c). Together, our and others findings suggest a potential role for kininogens (particularly KNG1) in adaptive thermogenesis. If true, elevated KNG1 and KNG2 abundance at the protein level in AdCrTKO BAT is consistent with an adaptive response resulting from impaired creatine-dependent thermogenesis.
CRT levels in human adipocytes correlates with lower BMI and increased insulin sensitivity
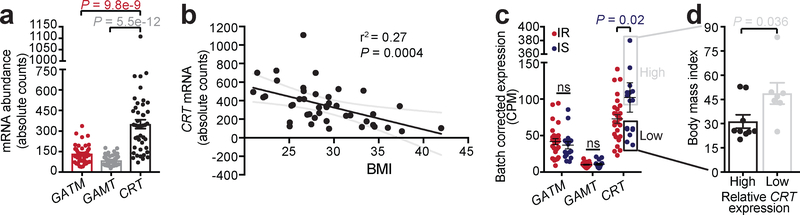
Creatine metabolism is highly selective for BAT compared to white fat in humans8,36. A recent analysis of ~2,850 18F-FDG PET/CT scans from 1,644 human subjects demonstrated that BAT creatine energetics may be a significant predictor of total activated human BAT37. Moreover, in a recent study of human BAT-mediated postprandial thermogenesis, 52 genes significantly correlated with UCP1 expression, with two of them being the mitochondrial creatine kinases (CKMT1A and CKMT2)38. We investigated the expression of genes that regulate the abundance of creatine in purified human subcutaneous adipocytes of 43 patients covering a wide BMI range, undergoing elective plastic surgery. A diagnosis of diabetes was a criterion for exclusion, as the focus here was on proximal changes associated with obesity and insulin resistance, rather than downstream changes resulting from hyperglycemia (and drug treatments). We compared the mRNA expression of genes that regulate creatine abundance (CRT, GATM, and GAMT) by RNA sequencing. CRT was the most abundant, followed by GATM, and then GAMT (Fig. 6a). Strikingly, CRT abundance was inversely and significantly correlated with body mass index (BMI) of patients (Fig. 6b), suggesting a possible causal relationship between decreased creatine transport and human obesity. Furthermore, when patients were stratified into insulin-sensitive (IS) and insulin-resistant (IR) groups, CRT expression was significantly lower in IR patients, compared to IS patients, whereas GATM and GAMT expression was not different between these groups (Fig. 6c). Finally, the IS group could be sub-stratified into those with relatively low or high CRT expression. The sub-group with low CRT expression had a significantly greater BMI, compared to the sub-group with high CRT expression (Fig. 6c and Fig. 6d). Thus, the strong inverse correlation of CRT transcript levels with BMI and insulin resistance suggests that creatine transport-mediated energetics plays an important role in human adipocyte metabolism. Importantly, the IS sub-group data (Fig. 6d) indicate that CRT expression can be uncoupled from insulin-resistance, but not BMI, suggesting that creatine energetics is primarily associated with obesity, and that insulin resistance may arise consequently, second to obesity, in the population with low CRT expression.
Fig. 6 |. CRT expression in human adipocytes is negatively correlated with obesity and insulin resistance.
a, mRNA abundance of GATM, GAMT, and CRT by RNA-sequencing (n = 43). b, Pearson correlation of CRT mRNA expression with body mass index (BMI) (n = 43). Black line, linear regression; gray curved lines, 95% confidence intervals. c, mRNA abundance of GATM, GAMT, and CRT in patients stratified as insulin resistant (IR) and insulin sensitive (IS) (IS, n = 16; IR, n = 27). d, BMI of IS patients with relatively high (n = 9) or low (n = 7) CRT mRNA abundance. Data are presented as mean ± s.e.m. of biologically independent samples. One-way ANOVA (a); pearson correlation (b); multiple two-tailed Student’s t-tests (c); two-tailed Student’s t-tests (d).
Conclusions
Several properties of creatine transport suggest that it functions as a mediator for diet-induced thermogenesis: (i) It is required to support full thermogenic activation upon pharmacological adrenergic stimulation, (ii) its loss substantially reduces activation of whole body energy expenditure in response to caloric excess, (iii) selective modulation of energy expenditure by creatine occurs primarily during high fat feeding, and (iv) loss of creatine transport increases metabolic efficiency, without effecting food intake, leading to obesity. It is important to note that creatine-dependent thermogenesis does not require Ucp1 inactivation to reveal its physiological relevance, and that UCP1 protein levels are modestly elevated in BAT of AdCrTKO animals compared to CrTlox/y littermates.
While we detected a statistically significant decrease (80%) in BAT CrT mRNA levels at both 30°C and 22°C, CrT abundance was only significantly reduced (85%) in SQ from animals housed at 30°C (Fig. 4b). Interestingly, conditions where CL-dependent energy expenditure is lower in AdCrTKO mice compared to controls (Fig. 2b, Fig. 2f, Fig. 2i and Supplemental Figure 2i) correspond well with conditions where CrT abundance is significantly reduced in both BAT and SQ (Fig. 4a and Fig. 4b). These data support the idea that BAT and SQ both contribute to creatine-dependent thermogenesis.
It is still debated what rodent housing temperature most precisely mimics human thermal conditions39,40. The results presented herein were derived from experiments conducted at 30°C and 22°C, and so the drive for heat production to combat heat loss was either eliminated or mildly present, respectively. Importantly, irrespective of housing temperature, adipocyte creatine transport is critical for mitigating fat mass gain, indicating that it may have important clinical relevance to human obesity.
Our and others prior work, using in vitro and chemical approaches, suggested a critical role for adipose tissue creatine metabolism in enhancing energy expenditure through stimulation of ATP turnover8,12–16,36,41. While the phenotype of the currently available tissue-specific genetically-engineered mouse models (Adipo-Gatm KO and AdCrTKO) substantiate the initial mechanistic interpretations in isolated mitochondria, it is currently unknown whether the thermogenic role for creatine in vivo is more complex than the proposed mechanism of mitochondrial ATP turnover. This is obviously the focus of our current research efforts. Nevertheless, evidence in support of creatine playing a key role in adipocyte thermogenesis has accumulated in recent years from work independent from ours. Global CrT knockout mice, which exhibit similar levels of creatine depletion as Adipo-Gatm KO and AdCrTKO animals, have increased body fat stores compared to controls, despite a decrease in food intake42. Creatine kinase U-type (Ckmt1) and brain-type creatine kinase (Ckb) double knockout animals are cold-sensitive and have impaired capacity to activate thermogenic respiration in response to norepinephrine administration43, providing additional supporting evidence for a role of creatine metabolism in thermoregulation in vivo. Future work in this area will require the generation of new animal models with fat-selective creatine kinase deletion. These models will undoubtedly be useful to further delineate the molecular mechanism of creatine-dependent thermogenesis and its role in obesity and metabolic disease. The current findings, in conjunction with our and others prior work, place adipocyte creatine energetics as a central regulator of whole body energy expenditure and obesity.
Methods
Animals
Mice were housed at 22°C under a 12 hr light/dark cycle with free access to food and water until 8 weeks of age. CrTlox/y animals were obtained from the Jackson Laboratory [B6(SJL)-Slc6a8tm1.1Clar/J), Stock No: 020642]. Adiponectin-Cre mice [B6;FVB-Tg(Adipoq-cre)1Evdr/J, Stock No: 028020], maintained on a C57BL/6J background, were bred to CrTlox/y animals to generate experimental groups. All experiments used age-matched male littermates and were conducted at either 30°C or 22°C. Animals were housed in groups of 3 unless otherwise stated. Animal experiments were performed according to procedures approved by the Animal Resource Centre at McGill University and comply with guidelines set by the Canadian Council of Animal Care. Experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee (IACUC) of the Beth Israel Deaconess Medical Center.
High fat feeding
AdCrTKO mice and age-matched littermate controls (CrTlox/y) were genotyped at 2 weeks of age. At three weeks of age, mice were weaned by genotype into groups of three per cage, where they were housed at 22°C and fed a chow diet. Mice of different genotypes were housed in cages side by side to limit variability in exposure to differential temperatures within the housing facility. At 8 weeks of age, mice (housed three per cage) were either transferred to incubators set at 30°C or maintained in the housing room at 22°C. High fat diet (initiated at 8 weeks of age) was a rodent diet (OpenSource Diets, D12492) with 60% kcal% fat, 20% kcal% carbohydrate, and 20% kcal% protein. C57BL/6NJ mice (Jackson labs; stock No: 005304) were used as wild type mice for examining CRT protein levels of after acute high fat feeding.
Calorimetric measurements
Indirect calorimetry studies were conducted at the Brigham and Women’s Hospital (BWH) Metabolic Core facility. Animals were housed individually in metabolic chambers maintained at 30°C or 22°C under a 12 hr light/dark cycle with free access to food and water. The mice were acclimated to metabolic cages for 24 hours prior to measurements. Whole body metabolic rate was measured using the Oxymax open-circuit indirect calorimeter, Comprehensive Lab Animal Monitoring System (CLAMS, Columbus Instruments), available to the Brigham and Women’s Hospital Metabolic Phenotyping Core.
Diet-Induced Thermogenesis
Animals were housed individually in metabolic chambers maintained at 30°C or 22°C under a 12 hr light/dark cycle with free access to food and water. Resting metabolic rate at each ambient temperature (30°C or 22°C) was calculated from the mean of three lowest energy expenditure (kcal) readings. Change in energy expenditure (kcal) in response to high fat feeding was plotted as the absolute increase in resting metabolic rate from last day of chow feeding.
Metabolic efficiency
Measured as body mass accumulation as a function of cumulative energy intake.
Body Composition
Body composition was examined with Echo MRI (Echo Medical Systems, Houston, Texas) using the 3-in-1 Echo MRI Composition Analyzer.
Adrenergic activation of metabolic rate
Animals were housed individually in metabolic chambers maintained at 30°C or 22°C under a 12 hr light/dark cycle with free access to food and water. Mice were injected with CL 316,243 (dissolved in saline) intraperitoneally at 1 mg kg−1 at 0600 hours. Mice were subsequently placed back in their metabolic cages, and oxygen consumption was monitored.
Metabolite analyses of primary brown adipocytes by mass spectrometry
Creatine and phosphocreatine were profiled with General Metabolics, LLC, and executed at General Metabolics’ labs according to previously published methodology44. Briefly, 2×106 adipocytes were washed three times with fresh pre-warmed (37°C) 75 mM ammonium carbonate, pH 7.4. Metabolites were extracted with 700 μl of pre-heated (75°C) extraction solvent [70% (v/v) ethanol (absolute) in HPLC-grade water]. Extraction solvent was kept on cells for 3 minutes. Samples were rapidly transferred to a dry ice/ethanol bath. Samples were centrifuged 16,000 g × 10 minutes at 4°C. Equal volume of extract supernatant was recovered and stored at −80°C until metabolite analysis. The analysis was performed on a platform consisting of an Agilent Series 1100 LC pump coupled to a Gerstel MPS2 autosampler and an Agilent 6520 Series Quadrupole Time-of-flight mass spectrometer equipped with an electrospray source operated in negative and positive mode. The flow rate was 150 μl/min of mobile phase consisting of isopropanol/water (60:40, v/v) buffered with 5 mM ammonium carbonate at pH 9 for negative mode and methanol/water (60:40, v/v) with 0.1% formic acid at pH 3 for positive mode. For online mass axis correction, 2-propanol (in the mobile phase), taurocholic acid, and hexakis(1H, 1H, 3Htetrafluoropropoxy)phosphazine (HP-0921, Agilent Technologies) for negative mode and creatinine, reserpine, and HP-0921 for positive mode were added to the mobile phase. Mass spectra were recorded in profile mode from m/z 50 to 1000 with a frequency of 1.4 spectra/s for 0.48 min using the highest resolving power (4 GHz HiRes). Source temperature was set to 325°C with 5 l/min drying gas and a nebulizer pressure of 30 psig. Fragmentor, skimmer, and octopole voltages were set to 175, 65, and 750 V, respectively.
Metabolite analyses of BAT by mass spectrometry
Creatine was profiled at the Rosalind and Morris Goodman Cancer Research Centre Metabolomics Core Facility. Briefly, BAT (10mg) was combined with 1.14 ml of methanol/ HPLC-grade water (50:50, v/v), 0.66 ml of ice cold acetonitrile (ACN) (at −20°C or colder), and 4 ceramic beads (2.8 mm diameter). The mixture was homogenized in a bead beater (Qiagen TissueLyser) for 2 minutes at 30Hz. Next, samples were transferred into a 5 ml tube containing 1.8 ml of ice-cold dichloromethane and 0.9 ml of ice-cold HPLC-grade water. The samples were vortexed for 1 minute, incubated on ice for 10 minutes and centrifuged at 4,000 rpm × 10 minutes at 1°C. Water soluble metabolites in the upper polar phase were collected and dried using a chilled speed-vac set at 4°C. Samples were resuspended in 50 μl of HPLC-grade water before LC-MS analysis. For targeted metabolite analysis and semi-quantitative concentration determination of creatine, samples were injected onto an Agilent 6430 Triple Quadrupole (QQQ)-LC-MS/MS. Chromatography was achieved using a 1290 Infinity ultra-performance LC system (Agilent Technologies, Santa Clara, CA, USA) consisting of vacuum degasser, autosampler and a binary pump. Mass spectrometer was equipped with an electrospray ionization (ESI) source and samples were analyzed in positive mode. Multiple reaction monitoring was optimized on authentic metabolite standards. The quantifying and qualifying ion transitions for creatine were 132.1→90.1 and 132.1→44.2 respectively. Source gas temperature and flow were set at 350°C and 10 l/min respectively, nebulizer pressure was set at 40 psi and capillary voltage was set at 3500V. Chromatographic separation of creatine was achieved using an Intrada Amino Acid column 3 μm, 3.0×150mm (Imtakt Corp, JAPAN). The chromatographic gradient started at 100% mobile phase B (0.3% formic acid in ACN) with a 3-minute gradient to 27% mobile phase A (100 mM ammonium formate in 20% ACN / 80% water) followed with a 19.5-minute gradient to 100% A at a flow rate of 0.6 ml/minute. This was followed by a 5.5-minute hold time at 100% mobile phase A and a subsequent re-equilibration time (7 minutes) before next injection. For all LC-MS analyses, 5 μl of sample was injected. The column temperature was maintained at 10°C. Creatine eluted at 7.9 minutes. Relative concentrations were determined from external calibration curves. Data were analyzed using MassHunter Quant (Agilent Technologies). No additional corrections were made for ion suppression; thus, concentrations are relative, not absolute.
Nuclear magnetic resonance (NMR) spectroscopy
Brown fat tissue samples (~20 mg) were homogenized and extracted with a mixture of ice-cold solvents water/methanol/chloroform (H2O/MeOH/CHCl3 1:1:1, v:v:v). The water/methanol phase was separated and dried in a speedvac. The extract was reconstituted in a 550 μl mixture containing 500 μl of phosphate buffer pH 7.4 and 50 μl of 1mM of TSP-d4 in D2O, vortexed for 20 seconds and transferred to 5 mm NMR tubes. The NMR spectra were acquired on a Bruker 600 MHz Avance III HD spectrometer equipped with a BBI room temperature probehead and SampleJet auto sampler (Bruker Biospin, Rheinstetten, Germany). 1H NMR spectra were recorded using 1D noesy pulse sequence with presaturation (noesygppr1d), collecting 256 scans with calibrated 90-degree pulse (~11 μs), 4.55 second acquisition time, and 4 second relaxation delay. Metabolites were identified and quantified using Chenomx NMR suite 8.2 software, by fitting the spectral lines of library compounds into the recorded NMR spectrum of tissue extract. The quantification was based on peak area of TSP-d4 signal. The metabolite concentrations are exported as μM in NMR sample and normalized to wet tissue mass (nmol/mg of tissue).
Body temperature
A mouse rectal probe (World Precision Instruments) was used to examine body temperature. Mice were group-housed (3 per cage) at 30°C for 5 days. Body temperature was measured in individually housed mice at 4°C following acclimation at 30°C for 5 days.
Triglyceride quantitation
Triglyceride (TG) content in BAT and SQ tissues was measured using the High Sensitivity TG Assay Kit (Sigma Aldrich, Cat. No. MAK264). Five standard solutions were prepared at 40, 80, 120, 160 and 200 pmol/well in 50 μl of TG buffer. 10 mg of BAT and SQ tissues were weighed and homogenized in 100μl of cold TG buffer. To ensure that the lipid layer was not separated from the aqueous phase, samples were boiled for 30–45 seconds. Insoluble materials were removed by centrifuging the samples at 12,000g for 5 minutes. Supernatant, including the entire fat layer, was transferred into clean tubes. Each sample was then diluted 5000× with cold TG buffer and transferred to a 96-well plate at a final volume of 50μl. 2 μl of lipase was added to each sample which were subsequently incubated at 37ºC for 20 minutes. Following the hydrolysis of TG’s to glycerol and fatty acids, 50 μl of the master reaction mix was added to the wells and the samples were incubated at 37ºC for 30 minutes. Fluorescence intensity was measured at λex = 535/λem = 587 nm. To control for NADH and NADPH background signal, every sample was also analyzed in the absence of the TG enzyme mix. The reading from this control was subtracted from the sample reading.
Stromal vascular fraction isolation of mouse brown preadipocytes
Interscapular BAT stromal vascular fraction (SVF) was obtained from 3-week-old animals (following genotyping). Interscapular BAT was dissected, washed in PBS, minced, and digested for 45 minutes at 37°C in isolation buffer (1.5 mg ml-1 collagenase B (Sigma), 123 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 5 mM glucose, 100 mM HEPES, and 4 % BSA). The tissue suspension was filtered through a 100 μm cell strainer and centrifuged at 600 g for 5 min to pellet the SVF, which was subsequently resuspended in adipocyte culture medium (DMEM/F12 (Wisent), 10 % FBS and 0.1 % PenStrep), filtered through a 40 μm cell strainer, centrifuged at 600 g for 5 min, resuspended and plated in adipocyte culture medium onto polystyrene cell culture dish (Corning). Cells were maintained at 37°C in 10% CO2. Cells from 5 animals were plated on a single 10 cm dish, passaged twice to ten 10 cm dishes. Cells from each 10 cm dish were frozen and stored in liquid nitrogen in freezing media (10% DMSO, 90% FBS) at 80% confluency.
Adipocyte differentiation
Preadipocytes were grown to post-confluency and differentiation was induced with adipogenic cocktail (1μM rosiglitazone, 0.5mM IBMX, 5μM dexamethasone, 0.114μg ml-1 insulin, 1nM T3, and 125μM Indomethacin). Cells were re-fed every 48 hours with maintenance cocktail (1μM rosiglitazone and 0.5μg ml-1 insulin). Cells were fully differentiated by day 6.
Adenoviral cloning, construction, and transduction
Cre and Gfp were amplified with PCR, gel extracted and cloned into the pENTR™Directional TOPO entry vector (Invitrogen) according to the manufacturer’s instructions. Entry vector clones were shuttled into the pAD/CMV/V5-DEST Gateway® vector, and digested with PacI (NEB). 293A cells were plated at 0.5×106 cells per well of a 6-well dish and transfected 12 hours later with 3μg of PacI-linearized adenoviral plasmid. Crude adenovirus was generated according to the manufacturer’s instructions (Invitrogen), and then amplified by infecting 293A cells (500μl crude virus per 15cm dish of 293A cells). Virus was purified using the Fast Trap Adenovirus Purification and Concentration Kit (EMD Millipore), according to the manufacturer’s instructions. Three 15 cm dishes were used for each preparation. Primary adipocytes were transduced with purified adenovirus on day 3.5 of differentiation.
Deletion of CrT in vitro
Preadipocytes from the BAT of CrTlox/y animals was isolated. On day 3.5 following the induction of differentiation, adipocytes were infected with GFP- or CRE-expressing adenovirus (1:500 dilution). After 12 hours of infection, virus was removed and the cells were refed with maintenance cocktail every 48 hours thereafter. All experiments using this system were conducted from adipocytes at day 10 of differentiation, which was determined empirically to allow for sufficient time for CrT transcript levels to decrease significantly.
Cellular respirometry
Primary brown preadipocytes were plated at 15,000 cells per well of a XF24 V7 cell culture microplate and differentiation was induced 12 hours later. On day 10 post-differentiation, adipocytes were washed once and maintained for ~45min in unbuffered DMEM, supplemented with 1mM pyruvate. The protocol for the XFe24 was set to analyze cellular respirometry using a mix:wait:measure ratio of 4:2:2 minutes. Norepinephrine (100 nM) was used to stimulate thermogenic respiration. A mix of Rotenone and Antimycin A (3 μM each) was used to inhibit mitochondrial respiration.
Purification of mature mouse brown adipocytes
Interscapular BAT was digested in isolation buffer (HBSS (Sigma), 2mg/ml Collagenase B (Worthington), 1mg/ml soybean trypsin inhibitor (Worthington) and 4% BSA with continuous shaking at 37°C for 45 minutes. The tissue suspension was filtered through a 100 μm cell strainer, and spun at 30g for 5 minutes at room temperature. The infranatant was removed and adipocytes were washed with 20 ml ice-cold PBS, and centrifuged again. Infranatant was removed and cells were snap frozen in liquid nitrogen until downstream analysis.
Collection of human adipose tissue samples
Subcutaneous adipose tissue was collected under IRB 2011P000079 (approved by the Beth Israel Deaconess Medical Center Committee on Clinical Investigations) from subjects recruited from plastic surgeon operating room schedule at Beth Israel Deaconess Medical Center in consecutive fashion as scheduling permitted to process the sample. Inclusion criteria: healthy male and female subjects, ages 18–64 receiving abdominal surgery. Exclusion criteria were diagnosis of diabetes, any subjects taking insulin-sensitizing medications like thiazolidinediones or metformin, chromatin modifying enzymes such as valproic acid, and drugs known to induce insulin resistance such as mTOR inhibitors (e.g. sirolimus, tacrolimus) or systemic steroid medications. Fasting serum was collected and tested for insulin, glucose, free fatty acids, and lipid panel in CLIA-approved lab. Body Mass Index (BMI) measures were derived from electronic medical record and confirmed by self-report and measures of insulin resistance, the homeostasis model assessment-estimated insulin resistance index (HOMA-IR) and revised quantitative insulin sensitivity check index (QUICKI) were calculated45,46. Female subjects in 1st and 4th quartiles for either HOMA-IR or QUICKI and matched for age and BMI were processed for RNAseq. Human participants who donated adipose tissue provided informed consent.
Purification of mature human adipocytes
Whole tissue subcutaneous adipose specimens were freshly collected from the operating room. Skin was removed and adipose tissue was cut into 1–2 inch pieces and rinsed thoroughly with 37ºC PBS to remove blood. Cleaned adipose tissue pieces were quickly minced by placing through an electric grinder with 3/16” hole plate and 400ml of sample was placed in 2L wide-mouthed erlenmeyer culture flask with 100ml of freshly prepared blendzyme (Roche Liberase TM Research Grade #05401127001 in PBS, at a ratio of 6.25mg/50ml) and shaken in a 37ºC shaking incubator at 120 RPM for 15–20 minutes to digest until sample appeared uniform. Digestion was stopped with 100ml of freshly made KRB (0.12X HBSS, 15mM HEPES), supplemented with 2% Bovine Serum Albumin. Digested tissue was filtered through a 300 μM sieve and washed with KRB/Albumin and flowthrough until only connective tissue remained. Sample was centrifuged at 233 g for 5 minutes at room temperature, clear lipid later removed, and floated adipocyte supernatant was collected, aliquoted and flash frozen in liquid nitrogen.
RNA isolation from mature human adipocytes
Total RNA from ~400 μl of thawed floated adipocytes was isolated in Trizol reagent (Invitrogen) following the manufacturer’s instructions. For RNA-seq library construction, mRNA was purified from 100 ng of total RNA using the Ribo-Zero rRNA removal kit (Epicentre) to deplete ribosomal RNA and converted into double stranded cDNA using NEBNext mRNA Second Strand Synthesis Module (E6111L). cDNA was subsequently tagmented and amplified for 12 cycles by using Nextera XT DNA Library Preparation Kit (Illumina FC-131). Sequencing libraries were analyzed Qubit and Agilent Bioanalyzer, pooled at a final loading concentration of 1.8pM, and sequenced on a NextSeq500. Sequencing reads were demultiplexed using bcl2fastq and aligned to the mm10 mouse genome using HISAT247. PCR duplicates and low-quality reads were removed by Picard (https://broadinstitute.github.io/picard). Filtered reads were assigned to the annotated transcriptome and quantified using featureCounts48.
Gene expression analysis (RT-qPCR)
Total RNA was extracted from frozen tissue using TRIzol (Invitrogen), purified with RNeasy Mini spin columns (QIAGEN) and reverse transcribed using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). The resultant cDNA was analyzed by RT–qPCR. Briefly, 20 ng cDNA and 150 nmol of each primer were mixed with GoTaq qPCR Master Mix (Promega). Reactions were performed in a 384-well format using a CFX384 Real-time PCR system (Bio-rad). Normalized mRNA expression was calculated using the ΔΔCt method, using Ppib mRNA as the reference gene. List of primer sequences can be found in Supplementary Table 1.
BAT lysis, protein digest, and peptide TMT-labeling
BAT was mechanically lysed in ice-cold lysis buffer (50 mM TRIS pH 8.5, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF and EDTA free protease inhibitor cocktail in 2% SDS. The lysate was centrifuged at 10,000g for 10 minutes. Protein content was measured using a BCA assay (Thermo Scientific, Rockford, IL); disulfide bonds were reduced with 5 mM TCEP and cysteine residues alkylated with iodoacetamide (14 mM). Protein lysates were purified by methanol-chloroform precipitation and digested overnight with LysC (Wako, Japan) in a ½00 enzyme/protein ratio in 2 M urea and 25 mM Tris-HCl, pH 8.5. The digest was acidified with 10 % formic acid (FA) to a pH of ~ 2–3 and subjected to C18 solid-phase extraction (50 mg SPE) (Sep-Pak, Waters, Milford, MA). Isobaric labeling of the digested peptides was accomplished with 10-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific, Rockford, IL). Reagents, 5 mg, were dissolved in 252 μl acetonitrile (ACN) and ¼ of the solution were added to 100 μg of peptides dissolved in 100 μl of 200 mM HEPES, pH 8.5. After 1 hour (room temperature), the reaction was quenched by adding 3 μl of 5% hydroxylamine. Labeled peptides were combined and acidified prior to C18 SPE on Sep-Pak cartridges (50 mg).
Basic pH reversed-phase separation (bpHrp)
TMT labeled peptides were solubilized in 500 μl solution containing 5% ACN/10 mM ammonium bicarbonate, pH 8.0 and separated by an Agilent 300 Extend C18 column (5 μm particles, 2.6 mm ID and 220 mm in length). An Agilent 1100 binary pump coupled with a photodiode array (PDA) detector (Thermo Scientific) was used to separate the peptides. A 45-minute linear gradient from 18% to 45% acetonitrile in 10 mM ammonium bicarbonate pH 8.0 (flow rate of 0.8 ml/min) separated the peptide mixtures into a total of 96 fractions (36 seconds). A total of 96 Fractions were consolidated into 24 samples in a checkerboard fashion, acidified with 20 μl of 20% FA and vacuum dried to completion. Each sample was re-dissolved in 12 μl 5% FA/ 5% ACN, and desalted prior to LC-MS/MS analysis.
Liquid chromatography separation and tandem mass spectrometry (LC-MS/MS)
Data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with a Proxeon EASY-nLC 1200 LC pump (Thermo Fisher Scientific). Peptides were separated on a 75 μm inner diameter microcapillary column packed with 35 cm of Accucore C18 resin (2.6 μm, 100 Å, ThermoFisher Scientific). Peptides were separated using a 3-hour gradient of 6–27% acetonitrile in 0.125% FA with a flow rate of 400 nl/min. The data were acquired using a mass range of m/z 350–1350, resolution 120,000, AGC target 1 × 106, maximum injection time 100 ms, dynamic exclusion of 120 seconds for the peptide measurements in the Orbitrap. Data dependent MS2 spectra were acquired in the ion trap with a normalized collision energy (NCE) set at 35%, AGC target set to 1.8 × 104 and a maximum injection time of 120 ms. MS3 scans were acquired in the Orbitrap with a HCD collision energy set to 55%, AGC target set to 1.5 × 105, maximum injection time of 150 ms, resolution at 15,000 and with a maximum synchronous precursor selection (SPS) precursors set to 10.
Data processing and spectra assignment
A compendium of in-house developed software was used to convert mass spectrometric data (Raw file) to the mzXML format, as well as to correct monoisotopic m/z measurements and erroneous assignments of peptide charge state49. All experiments used the Mouse UniProt database (downloaded 10 April 2017) where reversed protein sequences and known contaminants such as human keratins were appended. SEQUEST searches were performed using a 20 ppm precursor ion tolerance, while requiring each peptide’s amino/carboxy (N/C) terminus to have trypsin protease specificity and allowing up to two missed cleavages. 10-plex TMT tags on peptide N termini and lysine residues (+229.162932 Da) and carbamidomethylation of cysteine residues (+57.02146 Da) were set as static modifications while methionine oxidation (+15.99492 Da) was set as variable modification. A MS2 spectra assignment false discovery rate (FDR) of less than 1% was achieved by applying the target-decoy database search strategy49. Filtering was performed using an in-house linear discrimination analysis method to create one combined filter parameter from the following peptide ion and MS2 spectra metrics: Sequest parameters XCorr and ΔCn, peptide ion mass accuracy and charge state, in-solution charge of peptide, peptide length and mis-cleavages. Linear discrimination scores were used to assign probabilities to each MS2 spectrum for being assigned correctly and these probabilities were further used to filter the dataset with an MS2 spectra assignment FDR of smaller than a 1% at the protein level50.
Determination of TMT reporter ion intensities and quantitative data analysis
For quantification, a 0.03 m/z window centered on the theoretical m/z value of each the 10 reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were further de-normalized based on their ion accumulation time for each MS2 or MS3 spectrum and adjusted based on the overlap of isotopic envelopes of all reporter ions (as determined by the manufacturer). The total signal intensity across all peptides quantified was summed for each TMT channel, and all intensity values were adjusted to account for potentially uneven TMT labeling and/or sample handling variance.
Proteomic data processing and quality control
Two proteomic assays were performed to determine expression changes between CrTlox/y and AdCrTKO animals at 30°C (n=10) and 22°C (n=10) conditions. Expression measurements were successfully obtained across all samples in both assays for 5508 unique proteins. We did not consider proteins that could not be measured in all conditions. Expression values were log2-transformed and their distribution visualized as a mean of quality control. We then performed a principal component analysis (PCA) of expression values for the 50 proteins showing the highest variance across samples using the prcomp() function in R (v3.4.2.). It is important to note, because 30°C and 22°C BAT were processed independently in our proteomics work-flow, most differences were probably due to batch effects, and not temperature. Regardless, while the samples segregated by assay on the first principal component, a clear difference was observed between the CrTlox/y and AdCrTKO genotypes on the second principal component. Similar results were obtained when using 100 or 200 highest variance peptides. The PCA plot was produced in R using ggplot2 (v3.0.0) and ggfortify (v0.4.5.) package.
Differential protein expression
We determined proteins exhibiting differential expression between CrTlox/y and AdCrTKO BAT at 30°C (n=10) and 22°C (n=10) by performing moderated t-tests using the limma (v3.32.10) package in R51,52 as justified in53. P values were adjusted for multiple testing using the Benjamini-Hochberg approach. In R code, this translates to:
fit <- eBayes(lmFit(expression, model.matrix(~temperature + genotype))) results = topTable(fit, coef=“genotypeKO”, number = Inf, adjust.method = “BH”, lfc = 0).
This led to 15 proteins differentially expressed at 0.05 FDR. The volcano plot was produced with the ggplot2 (v3.0.0) package in R.
Gene ontology enrichment
To find gene ontology terms that are enriched in the set of differentially expressed peptides (15) with respect to the complete set of tested peptides (5,508), we used the GOrilla tool54 with the “two unranked lists of genes” option and a p-value threshold of 10e-3 (Mus musculus organism, database update “Aug 18, 2018”).
Statistical analyses
Results are presented as mean ±s.e.m. Unpaired two-tailed Student’s t-test for pairwise comparison, one-way ANOVA for multiple comparisons, F-test for linear regression, and ANCOVA for in vivo metabolic analyses were used to calculate P values to determine statistical differences. Significance was considered as p < 0.05. Statistical analysis and plotting for metabolic studies was performed in the R programming language with CalR, a custom package for analysis of indirect calorimetry using analysis of covariance with a graphical user interface. Mice were randomly assigned to treatment groups for in vivo studies. n values represent biological replicates for cultured cell experiments or individual animals for in vivo experiments. Specific details for n value are noted in each figure legend.
Data availability
All proteomic data generated or analyzed during this study are included in this published article (and its supplementary information files). Additional data that support the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Supplementary Data 1 | Proteomics inventory from AdCrTKO and CrTlox/y controls at 30°C (CrTlox/y, n = 5; AdCrTKO, n = 5).
Supplementary Data 2| Proteomics inventory from AdCrTKO and CrTlox/y controls at 22°C (CrTlox/y, n = 5; AdCrTKO, n = 5).
Supplementary Data 3 | Cross-referenced proteomics inventory from AdCrTKO and CrTlox/y controls at 30°C and 22°C (CrTlox/y, n = 10; AdCrTKO, n = 10).
Acknowledgements
This work was supported by the Canadian Institutes of Health Research (CIHR; PJT-159529), Goodman Cancer Research Centre and McGill University New Investigator Program, and NIH/NIDDK K99 Pathway to Independence award (to L.K.). We acknowledge funding from a Canderel Fellowship (to J.F.R.). We acknowledge technical assistance from the McGill/GCRC Metabolomics core facility. The GCRC Metabolomics Core Facility is funded by the Dr. John R and Clara M. Fraser Memorial Trust, the Terry Fox Foundation, the Québec Breast Cancer Foundation, and McGill University. We acknowledge funding from NIH R01HL 85744 and U24DK100469 Mayo Clinic Metabolomics Resource Core (to P.D.), AHA 13POST14540015 and NIH/NIDDK P30 DK057521 (to L.T.), NIH/NIDDK P30 DK057521, NIH/NIDDK R01 DK102173, and R01 ES017690 (to E.D.R) and NIH DK31405 and JPB Foundation (to B.M.S.).
Footnotes
Competing interests. The authors declare no competing interests
References
- 1.Lengyel E, Makowski L, DiGiovanni J & Kolonin MG Cancer as a Matter of Fat: The Crosstalk between Adipose Tissue and Tumors. Trends Cancer 4, 374–384, doi: 10.1016/j.trecan.2018.03.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Twig G et al. Body-Mass Index in 2.3 Million Adolescents and Cardiovascular Death in Adulthood. N Engl J Med 374, 2430–2440, doi: 10.1056/NEJMoa1503840 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ravussin E et al. Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med 318, 467–472, doi: 10.1056/NEJM198802253180802 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Jung RT, Shetty PS, James WP, Barrand MA & Callingham BA Reduced thermogenesis in obesity. Nature 279, 322–323 (1979). [DOI] [PubMed] [Google Scholar]
- 5.Hofmann WE, Liu X, Bearden CM, Harper ME & Kozak LP Effects of genetic background on thermoregulation and fatty acid-induced uncoupling of mitochondria in UCP1-deficient mice. J Biol Chem 276, 12460–12465, doi: 10.1074/jbc.M100466200 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Liu X et al. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. J Clin Invest 111, 399–407, doi: 10.1172/JCI15737 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mottillo EP et al. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. J Lipid Res 55, 2276–2286, doi: 10.1194/jlr.M050005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller S et al. Proteomic Analysis of Human Brown Adipose Tissue Reveals Utilization of Coupled and Uncoupled Energy Expenditure Pathways. Sci Rep 6, 30030, doi: 10.1038/srep30030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowland LA, Maurya SK, Bal NC, Kozak L & Periasamy M Sarcolipin and uncoupling protein 1 play distinct roles in diet-induced thermogenesis and do not compensate for one another. Obesity (Silver Spring) 24, 1430–1433, doi: 10.1002/oby.21542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ukropec J, Anunciado RP, Ravussin Y, Hulver MW & Kozak LP UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J Biol Chem 281, 31894–31908, doi: 10.1074/jbc.M606114200 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med 23, 1454–1465, doi: 10.1038/nm.4429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertholet AM et al. Mitochondrial Patch Clamp of Beige Adipocytes Reveals UCP1-Positive and UCP1-Negative Cells Both Exhibiting Futile Creatine Cycling. Cell Metab 25, 811–822 e814, doi: 10.1016/j.cmet.2017.03.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazak L et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655, doi: 10.1016/j.cell.2015.09.035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakatsuki T et al. Thermogenic responses to high-energy phosphate contents and/or hindlimb suspension in rats. Jpn J Physiol 46, 171–175 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Yamashita H et al. Increased growth of brown adipose tissue but its reduced thermogenic activity in creatine-depleted rats fed beta-guanidinopropionic acid. Biochim Biophys Acta 1230, 69–73 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Kazak L et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab 26, 660–671 e663, doi: 10.1016/j.cmet.2017.08.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitch CD, Shields RP, Payne WF & Dacus JM Creatine metabolism in skeletal muscle. 3. Specificity of the creatine entry process. J Biol Chem 243, 2024–2027 (1968). [PubMed] [Google Scholar]
- 18.Berlet HH, Bonsmann I & Birringer H Occurrence of free creatine, phosphocreatine and creatine phosphokinase in adipose tissue. Biochim Biophys Acta 437, 166–174 (1976). [DOI] [PubMed] [Google Scholar]
- 19.Skelton MR et al. Creatine transporter (CrT; Slc6a8) knockout mice as a model of human CrT deficiency. PLoS One 6, e16187, doi: 10.1371/journal.pone.0016187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Choi J, Aja S, Scafidi S & Wolfgang MJ Loss of Adipose Fatty Acid Oxidation Does Not Potentiate Obesity at Thermoneutrality. Cell Rep 14, 1308–1316, doi: 10.1016/j.celrep.2016.01.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazak L et al. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proc Natl Acad Sci U S A 114, 7981–7986, doi: 10.1073/pnas.1705406114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eguchi J et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13, 249–259, doi: 10.1016/j.cmet.2011.02.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speakman JR, Krol E & Johnson MS The functional significance of individual variation in basal metabolic rate. Physiol Biochem Zool 77, 900–915, doi: 10.1086/427059 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Bloom JD et al. Disodium (R,R)-5-[2-[[2-(3-chlorophenyl)-2-hydroxyethyl]-amino] propyl]-1,3-benzodioxole-2,2-dicarboxylate (CL 316,243). A potent beta-adrenergic agonist virtually specific for beta 3 receptors. A promising antidiabetic and antiobesity agent. J Med Chem 35, 3081–3084 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Himms-Hagen J, Hogan S & Zaror-Behrens G Increased brown adipose tissue thermogenesis in obese (ob/ob) mice fed a palatable diet. Am J Physiol 250, E274–281 (1986). [DOI] [PubMed] [Google Scholar]
- 26.Bachman ES et al. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297, 843–845, doi: 10.1126/science.1073160 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Rothwell NJ & Stock MJ A role for brown adipose tissue in diet-induced thermogenesis. Nature 281, 31–35 (1979). [DOI] [PubMed] [Google Scholar]
- 28.Leibel RL & Hirsch J Diminished energy requirements in reduced-obese patients. Metabolism 33, 164–170 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Eringa EC et al. Regulation of vascular function and insulin sensitivity by adipose tissue: focus on perivascular adipose tissue. Microcirculation 14, 389–402, doi: 10.1080/10739680701303584 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Singhal A et al. Influence of leptin on arterial distensibility: a novel link between obesity and cardiovascular disease? Circulation 106, 1919–1924 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Shimizu I et al. Vascular rarefaction mediates whitening of brown fat in obesity. J Clin Invest 124, 2099–2112, doi: 10.1172/JCI71643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernande L et al. Relationship of brown adipose tissue perfusion and function: a study through beta2-adrenoreceptor stimulation. J Appl Physiol (1985) 120, 825–832, doi: 10.1152/japplphysiol.00634.2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hankir MK & Klingenspor M Brown adipocyte glucose metabolism: a heated subject. EMBO Rep 19, doi: 10.15252/embr.201846404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cereijo R et al. CXCL14, a Brown Adipokine that Mediates Brown-Fat-to-Macrophage Communication in Thermogenic Adaptation. Cell Metab, doi: 10.1016/j.cmet.2018.07.015 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Rosell M et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab 306, E945–964, doi: 10.1152/ajpendo.00473.2013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson PA et al. Gene expression in human brown adipose tissue. Int J Mol Med 27, 227–232, doi: 10.3892/ijmm.2010.566 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Gerngross C, Schretter J, Klingenspor M, Schwaiger M & Fromme T Active Brown Fat During (18)F-FDG PET/CT Imaging Defines a Patient Group with Characteristic Traits and an Increased Probability of Brown Fat Redetection. J Nucl Med 58, 1104–1110, doi: 10.2967/jnumed.116.183988 (2017). [DOI] [PubMed] [Google Scholar]
- 38.M UD et al. Postprandial Oxidative Metabolism of Human Brown Fat Indicates Thermogenesis. Cell Metab, doi: 10.1016/j.cmet.2018.05.020 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Fischer AW, Cannon B & Nedergaard J Optimal housing temperatures for mice to mimic the thermal environment of humans: An experimental study. Mol Metab 7, 161–170, doi: 10.1016/j.molmet.2017.10.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speakman JR & Keijer J Not so hot: Optimal housing temperatures for mice to mimic the thermal environment of humans. Mol Metab 2, 5–9, doi: 10.1016/j.molmet.2012.10.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada S et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev 30, 2551–2564, doi: 10.1101/gad.287953.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perna MK et al. Creatine transporter deficiency leads to increased whole body and cellular metabolism. Amino Acids 48, 2057–2065, doi: 10.1007/s00726-016-2291-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streijger F et al. Mice lacking brain-type creatine kinase activity show defective thermoregulation. Physiol Behav 97, 76–86, doi: 10.1016/j.physbeh.2009.02.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuhrer T, Heer D, Begemann B & Zamboni N High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem 83, 7074–7080, doi: 10.1021/ac201267k (2011). [DOI] [PubMed] [Google Scholar]
- 45.Katz A et al. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab 85, 2402–2410, doi: 10.1210/jcem.85.7.6661 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Matthews DR et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360, doi: 10.1038/nmeth.3317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214, doi: 10.1038/nmeth1019 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Huttlin EL et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189, doi: 10.1016/j.cell.2010.12.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smyth GK Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article3, doi: 10.2202/1544-6115.1027 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Kammers K, Cole RN, Tiengwe C & Ruczinski I Detecting Significant Changes in Protein Abundance. EuPA Open Proteom 7, 11–19, doi: 10.1016/j.euprot.2015.02.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48, doi: 10.1186/1471-2105-10-48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 | Proteomics inventory from AdCrTKO and CrTlox/y controls at 30°C (CrTlox/y, n = 5; AdCrTKO, n = 5).
Supplementary Data 2| Proteomics inventory from AdCrTKO and CrTlox/y controls at 22°C (CrTlox/y, n = 5; AdCrTKO, n = 5).
Supplementary Data 3 | Cross-referenced proteomics inventory from AdCrTKO and CrTlox/y controls at 30°C and 22°C (CrTlox/y, n = 10; AdCrTKO, n = 10).
Data Availability Statement
All proteomic data generated or analyzed during this study are included in this published article (and its supplementary information files). Additional data that support the findings of this study are available from the corresponding authors on reasonable request.