Summary
Epigenetic regulation is emerging as a critical mechanism for pancreatic ductal adenocarcinoma (PDA) development. Histone methylation is an important regulatory mechanism, altering chromatin structure and promoter accessibility and causing aberrant gene expression. NSD1 and SETD2 genes encoding two histone H3K36 methyltransferases, are mutated or altered in 8–10% of PDA cases. However, whether there is altered protein expression of NSD1 or SETD2 in PDA and its precursors, and whether they have diagnostic or prognostic utility is unknown.
Tissue microarrays composed of a total of 190 and 192 duplicated cases of PDA (n=74 and 75), metastatic PDA (n=17 and 18), pancreatic intraepithelial neoplasia (PanIN; n=19 and 24), intraductal papillary mucinous neoplasm (IPMN; n=36), mucinous cystic neoplasm (MCN; n=12) and benign pancreatic tissues (n=27 and 32) were analysed for expression of NSD1 and SETD2 by immunohistochemistry. We assessed intensity and percentage of positive cells. NSD1 expression was significantly increased in metastatic PDA compared to benign ducts, primary PDA, and all other lesions combined (p=0.03, 0.02, and 0.03 respectively). Additionally, significantly decreased SETD2 protein expression was found in metastatic PDA and PanIN lesions compared to benign ducts (p=0.04 and 0.007, respectively). High NSD1 expression was associated with clinical stage III/IV disease (p=0.026), tumour grade 2 (p=0.022), use of neoadjuvant therapy (p=0.037), and overall higher clinical stage (p=0.022). There is no significant difference in overall and progression-free survival between NSD1/SETD2 high and low PDA.
Expression of NSD1 and SETD2 is specifically altered in metastatic PDA and some of the PDA precursor lesions, supporting their important role in PDA development and metastasis. In addition, increased NSD1 expression is significantly associated with higher clinical stage and neoadjuvant therapy, suggesting that NSD1 may be a useful prognostic marker.
Keywords: Pancreatic cancer, epigenetics, NSD1, SETD2, histone methylation
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDA) has an abysmal prognosis with a 5-year survival rate of 9%, and is projected to be the second-leading cause of cancer death by 2030.1 Recent whole genome and whole exome sequencing studies in PDA have revealed mutations in genes encoding epigenetic regulators. Epigenetic regulation refers to processes that change gene expression without modification of the underlying DNA sequence through mechanisms including DNA methylation, chromatin remodelling, alteration by non-coding RNAs such as miRNAs, and histone modifications. 2 Examples of potentially important epigenetic alterations in PDA include mutations to the histone demethylase KDM6A, found in over 10% of cases in one whole-genome sequencing study, and the chromatin remodelling gene ARID1A that indicated poor prognosis in a whole-exome sequencing study.2–5
Among the mechanisms of epigenetic regulation, histone methylation and its regulation by histone methyltransferases and demethylases has emerged as a particularly interesting subject in recent studies.2 Two key histone lysine methyltransferases are NSD1 and SETD2. Mutations in NSD1, which functions as a mono- and dimethyltransferase targeting histone H3 at K36 (H3K36), have been implicated in the pathogenesis of acute myeloid leukaemia, HPV-negative head and neck squamous cell carcinoma, lung squamous cell carcinoma, urothelial carcinoma and colorectal carcinoma, and have been associated with increased sensitivity to cisplatin chemotherapy in head and neck squamous cell carcinoma cell lines.6–11 In addition, mutations or altered expression of SETD2, a trimethylase at H3K36, may have a major role in the pathogenesis of some leukaemias and lymphomas as well as solid organ cancers including malignant mesothelioma and thymic carcinoma. Its inactivation or loss of expression has been linked to poor prognosis in chronic lymphocytic leukaemia and gastric and lung adenocarcinoma.12–20 In addition, SETD2 inactivation has been linked with resistance to chemotherapy in some leukaemias.21 Furthermore, alterations in both NSD1 and SETD2 have been recognised as potentially important in pathogenesis of clear cell renal cell carcinoma and gliomas.22–25
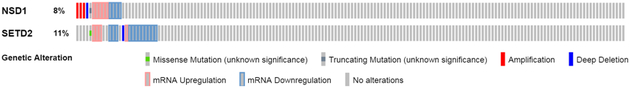
In pancreatic cancer, decreased histone methylation has been shown to be a poor prognostic factor, and one study of commonly mutated genes and loci in pancreatic cancer showed that all tumours had at least one mutation to an epigenetic regulator of histone modification.5 NSD1 is mutated or altered in 8.3% of PDA cases and SETD2 is altered or mutated in 10.7% of PDA cases according to The Cancer Genome Atlas (TCGA) PanCancer Atlas data (Fig. 1).26,27 However, the protein expressions of NSD1 or SETD2 in PDA and its precursor lesions and whether they have prognostic or diagnostic relevance are not known. Hence, in this study we aim to determine whether there is altered protein expression of NSD1 or SETD2 in PDA and its precursor lesions, as well as whether protein expression of either enzyme has diagnostic or prognostic utility.
Fig. 1.
Frequencies of mutations and altered mRNA expressions of NSD1 and SETD2 in pancreatic ductal adenocarcinoma (TCGA database, n=149).
MATERIALS AND METHODS
Case selection and clinicopathological variables
The Institutional Review Board at the University of Michigan approved the study (protocol number: HUM00098128). Patients with pancreas resections for pancreatitis, cystic neoplasms, or PDA from 2002 to 2015 at the University of Michigan Health System were included in the study. All pathology slides were reviewed and diagnosis confirmed by a gastrointestinal pathologist (JS). The electronic medical record was examined for clinical and demographic patient information. Date of surgery and date of last patient contact were recorded from the electronic medical record. Deaths were confirmed from the Social Security Death Index. Clinical staging was analysed using the American Joint Committee on Cancer 8th edition staging system.28 For patients who received neoadjuvant treatment, clinical stage was analysed based on pre-treatment tumour size, while pathological parameters of tumour size, grade, lymph node status, and peripancreatic, duodenal and common bile duct extension were analysed based on the post-treatment surgical specimen.
Tissue microarrays
All haemotoxylin and eosin (H&E) slides were reviewed and diagnoses confirmed by a gastrointestinal pathologist (JS) and corresponding areas were carefully selected and marked. Duplicated 1 mm diameter adjacent tissue cores from the same lesion in a total of 311 patient tissue samples were selectively punched/extracted and transferred to recipient tissue array blocks. Five tissue microarrays (TMAs) were set up according to a standard protocol as previously described.29 H&E staining was performed on each TMA block using standard protocol, and unstained slides were prepared for immunohistochemical (IHC) staining.
Immunohistochemical analysis and scoring
IHC staining of the TMAs was completed using the standard protocol and the following antibody titrations: SETD2 (KMT3A; Ab69836; 1:50; Abcam, UK) and NSD1 (ABE1009; 1:200; Millipore Sigma, USA). Overall a total of 190 and 192 (SETD2 and NSD1, respectively) duplicated tissue cores were present, including primary PDA (n=74 and 75), PDA metastases (n=17 and 18), pancreatic intraepithelial neoplasia (PanIN; n=19 and 24), intraductal papillary mucinous neoplasm (IPMN; n=36), mucinous cystic neoplasm (MCN; n=12) and benign pancreatic ducts (n=32 and 27). The intensity of the staining was recorded as: 0, negative; 1, weak; 2, moderate; 3, strong. A moderate to strong nuclear staining pattern is expected in normal cells which can serve as internal positive controls. A minor cytoplasmic component is also expected for SETD2. The percentage of positive cells was also assessed by a semiquantitative estimate of all lesional or benign ductal epithelial cells in each tissue core. The final IHC staining score was calculated as the staining intensity multiplied by the percentage of positive cells. The cases were divided into NSD1 high versus low and SETD2 high versus low groups with the median as the cut-off since there was no previous study to the best of our knowledge. Scoring was completed by a gastrointestinal pathologist (ME) blinded to clinicopathological data and patient outcomes, with confirmation of the scoring by a second gastrointestinal pathologist (JS) on a subset of cases.
Statistical analysis
ANOVA models were used to compare expression of SETD2 or NSD1 in PDA and its precursor lesions.
To evaluate correlation of clinicopathological characteristics with NSD1 or SETD2 expression in patients with PDA, ANOVA models were used for categorical clinicopathological variables and simple linear regression models were used for continuous clinicopathological variables. Kaplan–Meier methods were used to estimate overall survival and progression-free survival and log-rank tests were used to compare the survival functions between NSD1 or SETD2 groups (both NSD1 and SETD2 were dichotomised into low and high score groups using corresponding medians). Significance was determined if p<0.05. All analyses were conducted using SAS (version 9.4, SAS Institute, USA).
RESULTS
Expression of NSD1 and SETD2 in benign and neoplastic pancreatic epithelium
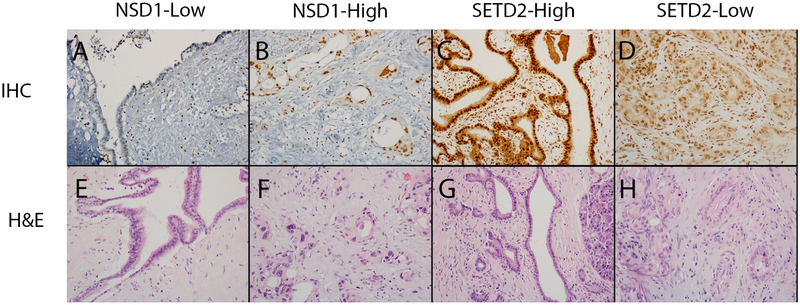
H&E and IHC of NSD1 and SETD2 were performed on five TMAs containing benign pancreas, PDA (both primary lesions and metastases, which were analysed separately) and its precursor lesions (IPMN/MCN and PanINs) using standard protocol. Representative examples of benign pancreas and PDA are shown in Fig. 2. All cases except three PDA cases showed at least some nuclear staining of NSD1 (expression score range 0–270). Interestingly, NSD1 expression in the metastatic PDA (118.3) was significantly higher than all other lesions combined (88.9), primary PDA (87.2), or benign ducts (84.1) (p=0.03, 0.02, and 0.03, respectively, Table 1). However, the overall difference, which refers to whether there is a difference between any pairs of groups, was not statistically significant (p=0.227). This is not surprising since there is no significant difference in NSD1 expression between most other groups. Cystic neoplasms (IPMN/MCN, 95.4) and PanIN (86.7) had a mean NSD1 expression score comparable to primary PDA (87.2) (Table 1).
Fig. 2.
Representative images of immunohistochemical analyses (A–D) and H&E (E–H) of NSD1 and SETD2 proteins in benign pancreas (A, C, E, G) and pancreatic ductal adenocarcinoma (B, D, F, H) tissues.
Table 1.
Expression of NSD1 in PDA and its precursor lesions
| Diagnosis | n | Expression score, mean ± SD |
p value (0.227 overall difference) |
|---|---|---|---|
| Normal | 27 | 84.1 ± 43.9 | 0.03 (vs metastatic PDA) |
| PanIN | 24 | 86.7 ± 64.2 | |
| IPMN/MCN | 48 | 95.4 ± 61.7 | |
| PDA (primary tumour) | 75 | 87.2 ± 48.3 | 0.02 (vs metastatic PDA) |
| PDA (metastasis) | 18 | 118.3 ± 61.8 | 0.03 (vs others) |
IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystic neoplasm; PanIN, pancreatic intraepithelial neoplasia; PDA, pancreatic ductal adenocarcinoma; SD, standard deviation; vs, versus.
While all cases showed at least some nuclear SETD2 expression (expression score range 40–300), analysis of SETD2 expression showed significantly decreased SETD2 protein expression in metastatic PDA (144.7) and PanIN (134.2) lesions compared to benign ducts (176.6) (p=0.04 and 0.007 respectively, Table 2). There was also a trend toward decreased SETD2 protein expression in cystic neoplasms (157.1) and primary PDA (157.8) compared to normal pancreas, but they were not statistically significant (p=0.166 and 0.129 respectively, Table 2). The decrease in SETD2 protein overall also did not reach statistical significance (p=0.138).
Table 2.
Expression of SETD2 in PDA and its precursor lesions
| Diagnosis | n | Expression score, mean ± SD |
p value (0.138 overall difference) |
|---|---|---|---|
| Normal | 32 | 176.6 ± 58.8 | |
| PanIN | 19 | 134.2 ± 47.4 | 0.007 (vs normal) |
| IPMN/MCN | 48 | 157.1 ± 64.1 | |
| PDA (primary tumour) | 74 | 157.8 ± 53.8 | |
| PDA (metastasis) | 17 | 144.7 ± 44.3 | 0.04 (vs normal) |
IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystic neoplasm; PanIN, pancreatic intraepithelial neoplasia; PDA, pancreatic ductal adenocarcinoma; SD, standard deviation; vs, versus.
Clinicopathological and demographic data of PDA cases
Demographic and clinicopathological data were recorded for patients with PDA in order to correlate these data with NSD1 and SETD2 expression levels. Parameters for the 67 patients with PDA are summarised in Table 3. The median age was 64 years (age range 39–83 years), and 34 of 67 (50.7%) patients were females. Of the 67 patients whose cases were included in the analysis, 64 had sufficient PDA tissue for the analysis of NSD1 expression, while 65 had sufficient PDA tissue for the analysis of SETD2 expression. Most PDAs occurred in the head of the pancreas (57/67, 86.4%). Jaundice, weight loss, abdominal pain, and diabetes were common presentations. The median body mass index (BMI) was 28.4 (range 15.2–48.1). Smoking and alcohol use was also common. The median serum CA19-9, total bilirubin, and alkaline phosphatase were 156, 4.4, and 305, respectively. The median tumour size was 2.5 cm (range 0.5–10 cm). Most of the patients were clinically staged as IIB (34/67, 50.7%) according to the new 8th edition of the AJCC Cancer Staging Manual, and had peripancreatic tumour extension (56/67, 83.6%). PDA was moderately-differentiated (G2) in 52.4%, poorly-differentiated (G3) in 30.1%, and well-differentiated (G1) in 17.5% of patients. Around half of the patients had perineural and angiolymphatic invasion, and duodenal involvement. About a third of the patients had lymph node metastases and common bile duct tumour involvement. Twelve (17.9%) patients received neoadjuvant chemotherapy and/or radiation prior to resection.
Table 3.
Demographic and clinicopathological characteristics of patients with PDA (n=67)
| Characteristic | n (%) |
|---|---|
| Age, years, median (range) | 64 (39–83) |
| Sex | |
| Female | 34 (50.7) |
| Male | 33 (49.3) |
| Tumour site | |
| Body/tail | 9 (13.6) |
| Head | 57 (86.4) |
| Jaundice | 48 (71.6) |
| Weight loss | 41 (63.1) |
| Abdominal pain | 28 (41.8) |
| Preoperative diabetes | 15 (22.4) |
| BMI, median (range) | 28.4 (15.2–48.1) |
| Smoking (pack-years), median (range) | 10 (0–100) |
| Alcohol use | |
| Low/moderate | 60 (90.9) |
| Heavy | 6 (9.1) |
| Serum CA19-9, median (range) | 156 (0–8691) |
| Total bilirubin, median (range) | 4.4 (0.2–29.7) |
| Alkaline phosphatase, median (range) | 305 (48–3629) |
| Tumour size, cm, median (range) | 2.5 (0.5–10) |
| Clinical stage (AJCC8) | |
| IA | 3 (4.5) |
| IB | 10 (14.9) |
| IIA | 1 (1.5) |
| IIB | 34 (50.7) |
| III | 19 (28.4) |
| IV | 0 (0) |
| Tumour grade | |
| Well (G1) | 11 (17.5) |
| Moderate (G2) | 33 (52.4) |
| Poor (G3) | 19 (30.1) |
| Lymph node metastases present | 19 (28.4) |
| Perineural invasion present | 37 (56.1) |
| Angiolymphatic invasion present | 31 (46.3) |
| Common bile duct extension present | 25 (37.3) |
| Peripancreatic extension present | 56 (83.6) |
| Duodenal extension present | 35 (52.2) |
| Neoadjuvant treatment | 12 (17.9) |
AJCC8, American Joint Committee on Cancer 8th edition cancer staging manual; BMI, body mass index; CEA, carcinoembryonic antigen; PDA, pancreatic ductal adenocarcinoma.
Correlation of clinicopathological characteristics with NSD1/SETD2 expression in PDA patients
High NSD1 expression was significantly associated with stage III/IV disease compared to stage I/IIA disease (p=0.026), tumour grade G2 compared to G3 (p=0.022), and neoadjuvant therapy (p=0.037) (Table 4). There was also a statistically significant association between overall higher pre-treatment clinical stage and high NSD1 expression (p=0.022). PDA with increased NSD1 also tended to have more lymph node metastasis and perineural invasion, but these potential associations were also not statistically significant. Increased NSD1 expression was not significantly associated with other clinicopathological features in our analysis. In addition, low SETD2 protein expression was significantly associated with a lower alkaline phosphatase value (p=0.026) and higher BMI (p=0.045) (Table 5). Decreased SETD2 expression was not significantly associated with other clinicopathological features.
Table 4.
Correlation of clinicopathological characteristics with NSD1 expression in patients with PDA
| Characteristic | Estimate for increase NSD1 score a |
SE | p value |
|---|---|---|---|
| Tumour site: head vs body/tail | −24.8 | 18.9 | 0.194 |
| Jaundice | 7.04 | 14.3 | 0.624 |
| Weight loss | 14.4 | 13.3 | 0.284 |
| Abdominal pain | −13.8 | 12.8 | 0.286 |
| Preoperative diabetes | −20.1 | 16.0 | 0.215 |
| BMI (kg/m2) | −0.44 | 1.16 | 0.704 |
| Smoking (pack-years) | 0.24 | 0.31 | 0.434 |
| Alcohol use: heavy vs low/moderate | −16.8 | 20.8 | 0.421 |
| Serum CA19-9 | −0.0013 | 0.0044 | 0.761 |
| Total bilirubin | −0.11 | 0.94 | 0.905 |
| Alkaline phosphatase | −0.0022 | 0.0124 | 0.861 |
| Tumour size | −7.8 | 4.4 | 0.080 |
| Clinical stage (AJCC8) | |||
| Overall difference | 0.022 b | ||
| Stage III/IV vs stage I/IIA | 46.0 | 17.3 | 0.026 b |
| Stage IIB vs stage I/IIA | 15.1 | 15.8 | 0.605 |
| Stage III/IV vs stage IIB | 30.9 | 13.9 | 0.076 |
| Tumour grade | |||
| Overall difference | 0.038 b | ||
| G1 vs G2 | −31.4 | 17.0 | 0.070 |
| G1 vs G3 | 1.8 | 18.5 | 0.924 |
| G2 vs G3 | 33.2 | 14.1 | 0.022 b |
| Lymph node metastases present | 20.0 | 13.9 | 0.155 |
| Perineural invasion present | 9.7 | 12.7 | 0.447 |
| Angiolymphatic invasion present | −5.9 | 12.7 | 0.642 |
| Common bile duct extension present | −7.3 | 13.0 | 0.573 |
| Peripancreatic extension present | −25.5 | 17.1 | 0.142 |
| Duodenal extension present | 0.9 | 12.7 | 0.945 |
| Neoadjuvant treatment | 34.6 | 16.2 | 0.037 b |
Estimate of effect for increase in NSD1 score.
p<0.05.
AJCC8, American Joint Committee on Cancer 8th edition cancer staging manual; BMI, body mass index; CEA, carcinoembryonic antigen; PDA, pancreatic ductal adenocarcinoma; SE, standard error.
Table 5.
Correlation of clinicopathological characteristics with SETD2 expression in patients with PDA
| Characteristic | Estimate for decrease SETD2 score a |
SE | p value |
|---|---|---|---|
| Tumour site: head vs body/tail | −31.4 | 18.7 | 0.098 |
| Jaundice | −15.1 | 14.6 | 0.304 |
| Weight loss | 2.3 | 13.6 | 0.867 |
| Abdominal pain | 11.4 | 13.4 | 0.397 |
| Preoperative diabetes | −3.5 | 15.6 | 0.822 |
| BMI (kg/m2) | 2.2 | 1.1 | 0.045 b |
| Smoking (pack-years) | 0.17 | 0.33 | 0.619 |
| Alcohol use: heavy vs low/moderate | −29.2 | 22.5 | 0.200 |
| Serum CA19-9 | 0.0003 | 0.0046 | 0.944 |
| Total bilirubin | −1.83 | 0.95 | 0.058 |
| Alkaline phosphatase | −0.030 | 0.013 | 0.026 b |
| Tumour size | 1.1 | 4.5 | 0.810 |
| Clinical stage (AJCC8) | |||
| Overall difference | 0.637 | ||
| Stage III/IV vs stage I/IIA | 21.1 | 22.4 | 0.350 |
| Stage IIB vs stage I/IIA | 11.2 | 16.9 | 0.511 |
| Stage III/IV vs stage IIB | 9.9 | 18.7 | 0.599 |
| Tumour grade | |||
| Overall difference | 0.600 | ||
| G1 vs G2 | −12.3 | 18.7 | 0.514 |
| G1 vs G3 | −20.8 | 20.5 | 0.315 |
| G2 vs G3 | −8.5 | 15.8 | 0.592 |
| Lymph node metastases present | 2.5 | 14.7 | 0.864 |
| Perineural invasion present | 13.4 | 13.4 | 0.320 |
| Angiolymphatic invasion present | −7.5 | 13.2 | 0.571 |
| Common bile duct extension present | −10.0 | 13.5 | 0.459 |
| Peripancreatic extension present | 8.9 | 17.6 | 0.614 |
| Duodenal extension present | −18.1 | 13.0 | 0.169 |
| Neoadjuvant treatment | 17.5 | 16.9 | 0.303 |
Estimate of effect for decrease in SETD2 score.
p<0.05.
AJCC8, American Joint Committee on Cancer 8th edition cancer staging manual; BMI, body mass index; CEA, carcinoembryonic antigen; PDA, pancreatic ductal adenocarcinoma; SE, standard error.
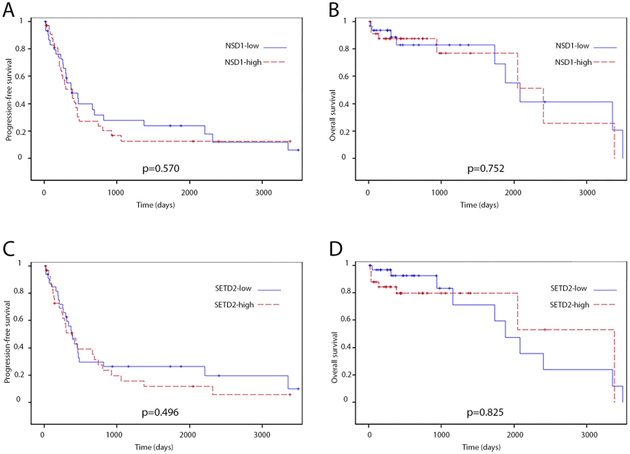
Influence of NSD1/SETD2 expression on survival outcomes in PDA patients
Expression of NSD1/SETD2 was not associated with a difference in progression-free survival or overall survival in PDA patients (Fig. 3). The difficulty in reaching statistical significance was possibly due to the variability of the protein levels in neoplastic lesions and the difficulty of establishing statistical significance in a cohort where only 8–10% of patients would be expected to have mutations or significantly altered mRNA expression based on prior data.26,27
Fig. 3.
Kaplan–Meier progression-free (A, C) and overall (B, D) survival curves comparing pancreatic ductal adenocarcinoma patients with high or low NSD1 (A, B), and high or low SETD2 (C, D) expression.
DISCUSSION
Epigenetic regulation genes, including histone modification genes, are frequently mutated in PDA. Of these genes, NSD1 and SETD2 have been found by genome sequencing to be frequently mutated in PDA.26,27 However, protein expressions of NSD1/SETD2 in PDA and its precursor lesions, including PanIN, IPMN, and MCN, are not known. Furthermore, whether expression of NSD1/SETD2 is correlated with clinicopathological characteristics and survival outcomes of PDA patients is also not known.
In the present study, we showed a significantly higher protein expression of NSD1 in metastatic PDA compared to normal pancreas, primary PDA, or any other tissues in the TMA. This finding is consistent with the mRNA expression and gene mutation data from the TCGA PanCancer Atlas where the majority of NSD1 mutations were amplifications and NSD1 mRNA level was frequently upregulated (Fig. 1).26,27 Furthermore, NSD1 protein was also mildly increased in precursor lesions (PanIN, IPMN, and MCN), suggesting that NSD1 may play an important role in PDA tumourigenesis and progression. The lack of statistical significance is most likely due to the relatively low sample number, low frequency of NSD1 alteration in PDA, and the large range of scores. Compared to PDA, other solid organ cancers with frequent NSD1 alteration, including head and neck squamous cell carcinomas, clear cell renal cell carcinomas, and gliomas, more often showed inactivation and decreased expression of NSD1.6,8,10,22,24 While NSD1 inactivating mutations have been associated with a good prognosis in laryngeal squamous cell carcinoma, our study did not show prognostic significance for the expression of NSD1 in PDA, again probably related to the relatively low sample number and variability of NSD1 protein expression.30 Similarly, the TCGA provisional data survival analysis based on mutation and mRNA data also failed to show prognostic significance for NSD1 alteration in PDA.26,27
Our results showed that protein expression of SETD2 was significantly decreased in metastatic PDA and PanIN lesions compared to normal pancreas. This finding is consistent with the results from the TCGA PanCancer Atlas showing a predominance of mRNA downregulation and inactivating mutations in PDA patients (Fig. 1).26,27 Interestingly, SETD2 expression seems to be the lowest in PanIN lesions, which may suggest the early role of SETD2 in PDA development. SETD2 has also shown inactivation and/or decreased expression in other malignancies such as clear cell renal cell carcinomas, gastric cancers and acute leukaemias.20–22,31 Low expression of SETD2 in gastric cancer and loss of function mutations to the gene in lung adenocarcinoma have been associated with poor survival, but the TCGA provisional data for pancreatic cancer does not identify a significant effect of altered SETD2 on prognosis.19,20,26,27 Similarly, in our IHC analysis of SETD2 protein expression, we did not find any prognostic significance for SETD2 protein expression in PDA.
The other interesting finding of our study is that increased protein level of NSD1 is significantly associated with higher clinical stage and neoadjuvant therapy which, especially in our institution, usually suggests borderline resectable or unresectable PDA. There is also a trend where patients with higher NSD1 present with more lymph node metastasis and perineural invasion. These findings are intriguing because they suggest that NSD1 may be used as a prognostic marker where higher expression implicates higher stage and more lymph node metastasis. However, the small sample sizes of these subgroups (patients with stage III/IV or neoadjuvant treated PDA) hampered the significance of this finding. Larger cohort studies are needed to further confirm this observation. The significance of the associations between SETD2 and BMI and alkaline phosphatase is not clear and also needs more study.
Epigenetic regulators are attractive therapeutic candidates for PDA, including the histone methyltransferase EZH2 which is often upregulated in PDA and other cancers. Several selective EZH2 inhibitors are now in development.2,32 In addition, recent studies show potential therapeutic implications of NSD1 and SETD2 mutations or alterations in other neoplasms. In a study in cell lines of head and neck squamous cell carcinoma, loss of function mutations in NSD1 increase sensitivity to cisplatin.7 However, no studies have examined the therapeutic implications in malignancies such as PDA where we show a potential increase in NSD1 expression. In contrast to NSD1 inactivation, SETD2 inactivation has been associated with worsened response to therapy. SETD2 loss decreases the sensitivity of leukaemia cell lines to DNA-damaging agents and increases resistance to the DNA-damaging chemotherapeutic agent cytarabine in a mouse model of MLL-rearranged leukaemia.12 Our study suggests that SETD2 expression may likewise be decreased in PDA. DNA-damaging therapies such as gemcitabine are a cornerstone to treatment of PDA, so further research into the role of SETD2 in the chemotherapy sensitivity of PDA will be essential.
Limitations to our study include the relatively small cohort for survival analysis; given that only 8–10% of patients would be expected to have NSD1 and SETD2 alterations. In addition, the retrospective nature of our study limits the ability to validate NSD1 or SETD2 as biomarkers. Furthermore, while we felt it was important to include PDA patients with neoadjuvant therapy so that our results would be generalisable to this population, it is not known if or how neoadjuvant therapy may affect the expression of NSD1 or SETD2. Our study is also limited by the use of TMA in our IHC analysis. While TMA eliminates the variation of staining between different batches of IHC experiments, the small tissue core size may lead to sampling bias and may not be representative of the wider lesional or normal tissue. Furthermore, IHC analysis has its own set of limitations such as using different cut-offs, relatively subjective scoring techniques, and antibody specificity. Finally, because no studies have specifically analysed the relation between SETD2 and NSD1 mutation status and IHC expression, additional studies delineating this relationship could help inform the possible utility of IHC analysis for these potential biomarkers.
Despite these limitations, our study adds to the knowledge of NSD1 and SETD2 as frequently mutated epigenetic regulators in PDA, as it is the first to examine changes of protein expression in these two markers in PDA and its precursor lesions, and their correlation with clinicopathological characteristics and patient outcome. We showed potential roles for these markers in prognostic implications, and as therapeutic options are tailored to the epigenetic profile of PDA in the future, our study may help to guide treatment options as well. Future objectives for investigating the role of NSD1 and SETD2 in PDA and its precursors include correlation of mutations with IHC staining, and investigating the biological impact of NSD1/SETD2 mutations on tumourigenesis, metastasis, and treatment resistance.
Acknowledgments
Conflicts of interest and sources of funding: Research reported in this manuscript was supported in part by National Cancer Institute of the National Institutes of Health under award number K08CA234222 (JS) and an AP project grant from the Department of Pathology at the University of Michigan (JS). The authors state that there are no conflicts of interest to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Silverman BR, Shi J. Alterations of epigenetic regulators in pancreatic cancer and their clinical implications. Int J Mol Sci 2016; 17: E2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015; 6: 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann KM, Ward JM, Yew CC, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA 2012; 109: 5934–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papillon-Cavanagh S, Lu C, Gayden T, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 2017; 49: 180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui N, Huang JK, Bojorquez-Gomez A, et al. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther 2018; 17: 1585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan K, Shin JH, Tay JK, et al. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep 2017; 7: 17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi M, Vasekar M, Grivas P, et al. Relationship of smoking status to genomic profile, chemotherapy response and clinical outcome in patients with advanced urothelial carcinoma. Oncotarget 2016; 7: 52442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jo YS, Kim MS, Yoo NJ, Lee SH. NSD1 encoding a histone methyltransferase exhibits frameshift mutations in colorectal cancers. Pathology 2016; 48: 284–6. [DOI] [PubMed] [Google Scholar]
- 11.Jaju RJ, Fidler C, Haas OA, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001; 98: 1264–7. [DOI] [PubMed] [Google Scholar]
- 12.Skucha A, Ebner J, Schmollerl J, et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun 2018; 9: 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hylebos M, Van Camp G, Vandeweyer G, et al. Large-scale copy number analysis reveals variations in genes not previously associated with malignant pleural mesothelioma. Oncotarget 2017; 8: 113673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yabe M, Miranda RN, Medeiros LJ. Hepatosplenic T-cell Lymphoma: a review of clinicopathologic features, pathogenesis, and prognostic factors. Hum Pathol 2018; 74: 5–16. [DOI] [PubMed] [Google Scholar]
- 15.Saito M, Fujiwara Y, Asao T, et al. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis 2017; 38: 1084–91. [DOI] [PubMed] [Google Scholar]
- 16.Moffitt AB, Ondrejka SL, McKinney M, et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J Exp Med 2017; 214: 1371–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph NM, Chen YY, Nasr A, et al. Genomic profiling of malignant peritoneal mesothelioma reveals recurrent alterations in epigenetic regulatory genes BAP1, SETD2, and DDX3X. Mod Pathol 2017; 30: 246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker H, Rose-Zerilli MJ, Larrayoz M, et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia 2016; 30: 2179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol 2017; 28: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Raghoonundun C, Chen W, et al. SETD2 indicates favourable prognosis in gastric cancer and suppresses cancer cell proliferation, migration, and invasion. Biochem Biophys Res Commun 2018; 498: 579–85. [DOI] [PubMed] [Google Scholar]
- 21.Mar BG, Chu SH, Kahn JD, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 2017; 130: 2631–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su X, Zhang J, Mouawad R, et al. NSD1 Inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res 2017; 77: 4835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge S, Li B, Li Y, et al. Genomic alterations in advanced gastric cancer endoscopic biopsy samples using targeted next-generation sequencing. Am J Cancer Res 2017; 7: 1540–53. [PMC free article] [PubMed] [Google Scholar]
- 24.Berdasco M, Ropero S, Setien F, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA 2009; 106: 21830–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomura M, Mukasa A, Nagae G, et al. Distinct molecular profile of diffuse cerebellar gliomas. Acta Neuropathol 2017; 134: 941–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. New York: Springer, 2010. [Google Scholar]
- 29.Nguyen N, Bellile E, Thomas D, et al. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck 2016; 38: 1074–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peri S, Izumchenko E, Schubert AD, et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun 2017; 8: 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licht JD. SETD2: a complex role in blood malignancy. Blood 2017; 130: 2576–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomberk GA, Iovanna J, Urrutia R. The promise of epigenomic therapeutics in pancreatic cancer. Epigenomics 2016; 8: 831–42. [DOI] [PMC free article] [PubMed] [Google Scholar]