SUMMARY
Glutamate has been implicated in a wide range of brain pathologies and is thought to be metabolized via the astrocyte-specific enzyme glutamine synthetase (GS). We show here that oligodendrocytes, the myelinating glia of the central nervous system, also express high levels of GS in caudal regions like the midbrain and the spinal cord. Selective removal of oligodendrocyte GS in mice led to reduced brain glutamate and glutamine levels and impaired glutamatergic synaptic transmission without disrupting myelination. Furthermore, animals lacking oligodendrocyte GS displayed deficits in cocaine-induced locomotor sensitization, a behavior that is dependent on glutamatergic signaling in the midbrain. Thus, oligodendrocytes support glutamatergic transmission through the actions of GS and may represent a therapeutic target for pathological conditions related to brain glutamate dysregulation.
Graphical Abstract

In Brief
Xin et al. show that mature oligodendrocytes, the myelinating cells of the brain, express the glutamine-synthesizing enzyme glutamine synthetase (GS). Oligodendrocytespecific GS deletion does not impair myelination but disrupts neuronal glutamatergic transmission, thus demonstrating a myelin-independent role for oligodendrocytes in supporting glutamate signaling in the brain.
INTRODUCTION
Glutamate is the major excitatory neurotransmitter in the brain. Following synaptic release, glutamate uptake and degradation are tightly regulated to achieve temporal and spatial signaling specificity and prevent cellular excitotoxicity (Kim et al., 2011; Sattler and Rothstein, 2006; Sheldon and Robinson, 2007). Currently, astrocytes are considered the sole glial cell type that contributes to glutamate uptake and degradation in the CNS (Jayakumar and Norenberg, 2016; Liang et al., 2006; Ortinski et al., 2010; Papageorgiou et al., 2018; Schousboe et al., 2013; Schousboe, 2019; Sun et al., 2017; Tani et al., 2014; Trabelsi et al., 2017; Yuan et al., 2017), as they express high levels of glutamate transporters and glutamine synthetase (GS), an enzyme that converts glutamate into glutamine. In keeping with this view, GS is frequently used as an astrocyte-specific marker (Armbruster et al., 2016; Habbas et al., 2015; Okuda et al., 2014; Papageorgiou et al., 2018; Theofilas et al., 2017; Tong et al., 2014). However, GS expression has also been reported in oligodendrocytes (Bernstein et al., 2014; Takasaki et al., 2010), glial cells known for producing myelin and ensheathing axons in the CNS (Pan and Chan, 2017). Although these results remain controversial (Anlauf and Derouiche, 2013; Jayakumar and Norenberg, 2016; Sun et al., 2017), it is of great physiological and clinical importance to identify all potential cellular and molecular components involved in the life cycle of glutamate. GS deletion from the brain results in neonatal death (He et al., 2010), and mutations in the GS gene produce severe neuropathology in humans (Häberle et al., 2012; Spodenkiewicz et al., 2016). Furthermore, glutamate dysregulation has been implicated in numerous pathological states, including epilepsy, stroke, and substance use disorders, as well as several neurodegenerative diseases (Jayakumar and Norenberg, 2016; Kalivas and Duffy, 1998; Reissner et al., 2015; Sheldon and Robinson, 2007; Spencer and Kalivas, 2017; van der Hel et al., 2005; Yuan et al., 2017). For these reasons, we sought to unequivocally determine whether GS is expressed by oligodendrocytes and, if so, whether oligodendrocyte GS plays a role in maintaining glutamatergic synaptic transmission.
RESULTS
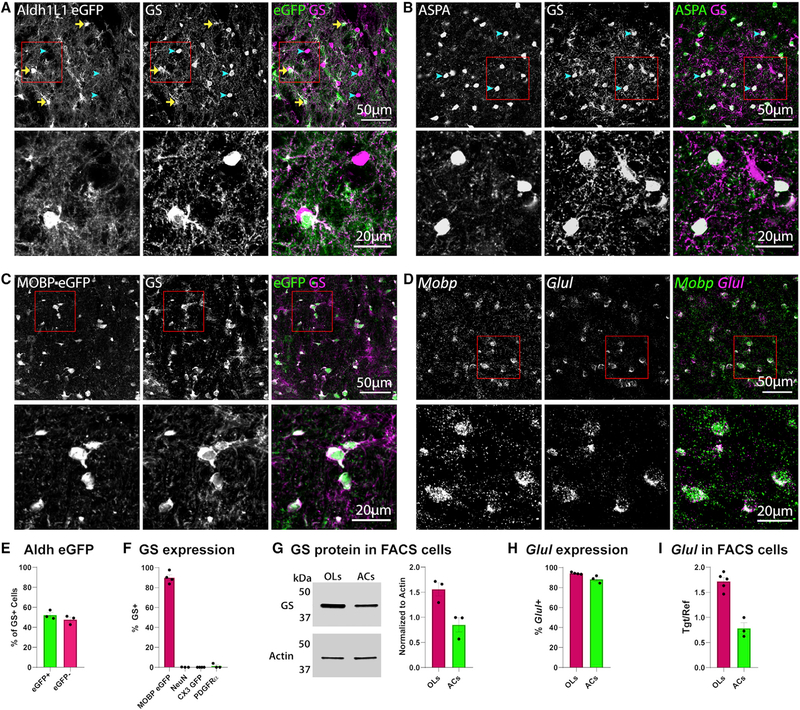
To assist in defining the cellular expression of GS, we immuno-stained brain sections from young adult (P60–75) Aldh1L1-EGFP mice, in which all astrocytes express EGFP (Zhang et al., 2014). Consistent with previous reports of astrocyte GS expression, there were numerous EGFP+ GS+ cells (Figure 1A, yellow arrows). However, we also observed an abundant population of GS+ EGFP− cells (Figure 1A, blue arrowheads; Figure 1E) in subcortical regions of the brain. Immunohistochemistry with aspartoacylase (ASPA), which is selectively expressed by mature oligodendrocytes (Larson et al., 2018), indicated that these GS+ EGFP− cells were oligodendrocytes (Figure 1B).Immunostaining in MOBP-EGFP mice, in which mature oligodendrocytes express EGFP (Larson et al., 2018), also revealed widespread colocalization between EGFP+ oligodendrocytes and GS (Figures 1C and 1F). GS immunoreactivity was absent from cells expressing NeuN (neurons), CX3CR1 GFP (microglia), or PDGFRα(oligodendrocyte progenitors) (Figure 1F; Figures S1A–S1C). As an additional means of comparing astrocyte and oligodendrocyte levels of GS protein, we used fluorescence-activated cell sorting (FACS) to isolate oligodendrocytes or astrocytes from the midbrain of MOBP-EGFP and Aldh1L1-EGFP mice (Figure S1G), respectively, and quantified GS protein by western blot. Again, we detected GS expression in both astrocytes and oligodendrocytes (Figure 1G).
Figure 1. Oligodendrocytes Express GS mRNA and Protein.
(A) Confocal image taken in the ventral midbrain of a P60 Aldh1L1-EGFP mouse, immunolabeled with anti-GS. Yellow arrows indicate EGFP+ GS+ astrocytes; blue arrowheads indicate EGFP−GS+ cells.
(B) Immunostaining for the mature oligodendrocyte marker aspartoacylase (ASPA) and GS in the ventral midbrain of a P60 wild-type mouse; blue arrowheads indicate GS+ oligodendrocytes.
(C) Immunostaining for GS in the ventral midbrain of a P60 MOBP-EGFP mouse.
(D) In situ hybridization for Mobp and Glul in the ventral midbrain of a P63 wild-type mouse. In (A)–(D), the bottom panel is a zoom of the area within the red box in the top image.
(E) Quantification of the percentage of GS+ cells that are EGFP+ or EGFP−; n = 3 mice.
(F) Quantification of % oligodendrocytes (MOBP EGFP+), neurons (NeuN+), microglia (CX3CR1 GFP+), or oligodendrocyte progenitors (PDGFRa+) expressing GS; n = 3–4 mice per cell type. For example images, see Figures S1A–S1C.
(G) Western blot for GS in FACS-isolated oligodendrocytes and astrocytes. The cell number was the same in each sample. For quantification, GS signal was normalized to loading control Actin; n = 3 mice per cell type.
(H) Quantification of % oligodendrocytes or astrocytes expressing Glul mRNA; n = 3–4 mice per cell type. For example images, see Figure S1E.
(I) qPCR for Glul in FACS-isolated oligodendrocytes and astrocytes. For quantification, Glul was normalized to sample Gapdh; n = 3–5 mice per cell type.
(E–I) Error bars indicate SEM.
To corroborate these findings at the mRNA level, we performed in situ hybridization for Glul, the gene that encodes GS. Consistent with immunostaining analysis, Glul probe colocalized with the astrocyte-specific transcript Slc1a2 and the oligodendrocyte-specific transcript Mobp in wild-type mice (Figures 1D and 1H; Figure S1E), as well as Egfp transcript in MOBP-EGFP mice (Figure S1d). Given previous reports of GS mRNA expression in microglia and oligodendrocyte progenitors (Nakajima et al., 2017; Palmieri et al., 2017; Zhang et al., 2014), we also probed for Aif1 (Iba1, expressed in microglia) and Pdgfra (expressed in oligodendrocyte progenitors); neither transcript colocalized with Glul (Figure S1F). Consistent with the western blot data, qPCR analysis of Glul mRNA from FACS-isolated oligodendrocytes or astrocytes (Figures S1G and S1H) detected abundant Glul expression in both cell types (Figure 1I). Collectively, these results indicate that oligodendrocytes express both GS mRNA and protein.
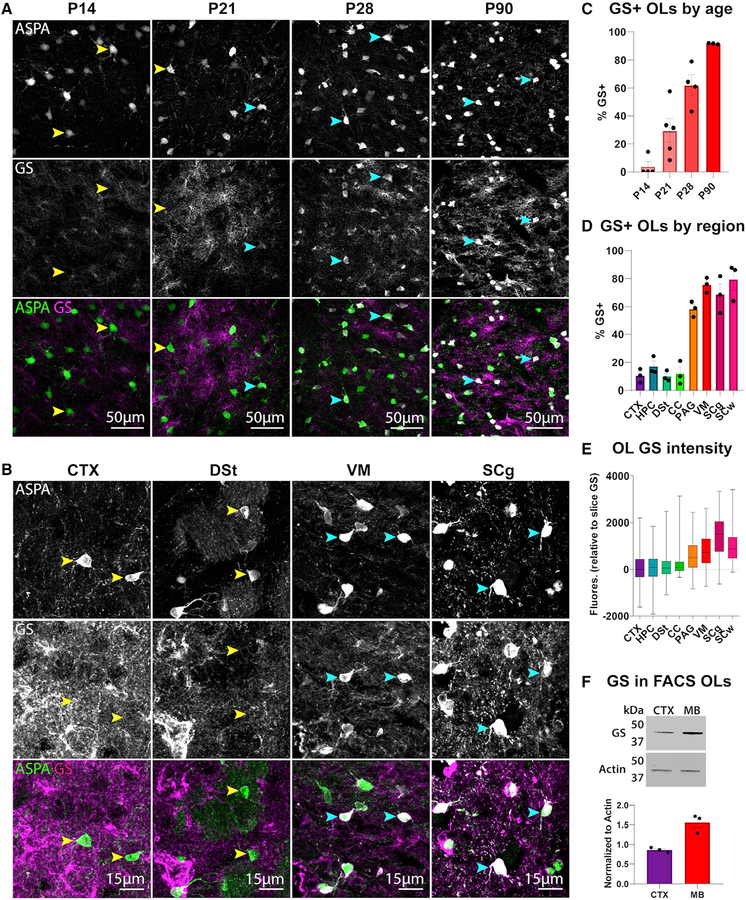
Given the widespread reports of GS expression being exclusive to astrocytes, we hypothesized that GS expression in oligodendrocytes may be temporally restricted. Indeed, analysis of GS expression in wild-type animals at several developmental time points revealed that oligodendrocyte GS expression only emerged at P21 and continued to increase in subsequent weeks (Figures 2A and 2C). In addition, GS intensity was much higher in oligodendrocytes in caudal regions, such as the ventral midbrain and spinal cord, than in rostral regions like the cortex and hippo-campus (Figures 2B, 2D, and 2E; Figure S2A). This pattern held true for both the percentage of GS-expressing oligodendrocytes (Figure 2D; F = 43.02, p < 0.0001) and the fluorescence intensity of GS in oligodendrocytes relative to overall GS intensity in the section (Figure 2E; Kruskal-Wallis test, p < 0.0001). Western blot of GS in oligodendrocytes isolated from the cortex or midbrain also revealed a significant difference in regional GS expression (Figure 2F; t = 5.051, p = 0.0072). A similar pattern emerged when we quantified Glul mRNA (Figures S2B and S2D). This regional heterogeneity in oligodendrocyte GS expression was partially maintained when we cultured oligodendrocyte progenitors from the midbrain or cortex and differentiated them into oligodendrocytes in vitro (Figures S2C and S2E), suggesting some degree of cell-intrinsic GS regulation. The late onset and regional heterogeneity of oligodendrocyte GS expression may in part explain why GS has thus far been predominantly associated with astrocytes.
Figure 2. Oligodendrocyte GS Expression Appears at 3 Weeks Postnatally and Is Regionally Heterogeneous.
(A) Immunostaining of ASPA and GS in the ventral midbrain of wild-type mice at 14, 21, 28, and 90 days postnatally.
(B) Immunostaining of ASPA and GS in the cortex (CTX), dorsal striatum (DSt), ventral midbrain (VM), and spinal cord gray matter (SCg) of P60 wild-type mice. In (A) and (B), yellow arrowheads indicate GS− oligodendrocytes, and blue arrowheads indicate GS+ oligodendrocytes.
(C) Percentage of ASPA+ oligodendrocytes expressing GS per animal at different ages. Percentages are calculated from two sections per animal and averaged; n = 3–4 mice per age.
(D) Quantification of % oligodendrocytes expressing GS in each region; n = 3 mice. One-way ANOVA, F(7,16) = 43.02, p < 0.0001.
(E) Box and whisker plot of GS fluorescence intensity within ASPA+ oligodendrocytes relative to slice GS fluorescence; values represent GS fluorescence within individual oligodendrocytes minus mean slice GS fluorescence. Whiskers represent minimum and maximum values; n = 500–950 cells per region sampled from 3 animals. Kruskal-Wallis statistic = 1789, p < 0.0001.
(F) Western blot for GS from FACS-isolated oligodendrocytes in the cortex or midbrain (MB); n = 3 mice per region. Cell number was the same in each sample. For quantification, GS signal was normalized to loading control Actin. Unpaired t test, t = 5.051, df = 4, p = 0.0072.
(C, D, and F) Error bars indicate SEM.
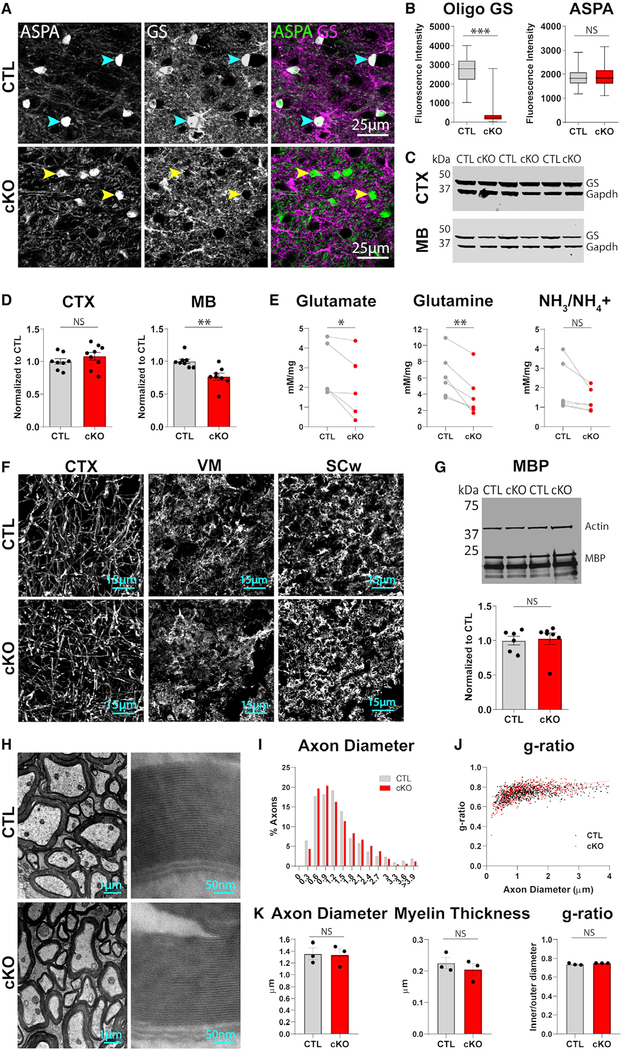
To determine the functional significance of oligodendrocyte GS expression, we generated oligodendrocyte-specific GS knockout mice by crossing MOG-iCre mice (Hövelmeyer et al., 2005) with floxed GS mice (He et al., 2010). Robust GS immunoreactivity was evident in oligodendrocytes of littermate controls (MOG-iCre−/−;GSfl/fl; control [CTL]), but not in MOG-iCre+/−; GSfl/fl (oligodendrocyte GS knockout [cKO]) mice (Figures 3A and 3B; Figure S3A; GS intensity Kolmogorov-Smirnov test [K-S] D = 0.9871, p < 0.0001); in contrast, astrocyte GS immuno-reactivity remained high in cKO mice (Figure S3B). Consistent with the immunohistochemical pattern of GS expression, knockout of oligodendrocyte GS significantly reduced tissue levels of GS in the midbrain, but not in the cortex (Figures 3C and 3D; midbrain [MB] GS, t = 3.609, p = 0.0028; cortex [CTX] GS t = 1.073, p = 0.3004). To examine the effect of this reduction on tissue glutamate homeostasis, we measured GS substrates in midbrain tissue using colorimetric assays. We detected reductions in glutamate and glutamine in cKO animals (Figure 3E; glutamate paired t = 2.852, p = 0.0463; glutamine paired t = 5.037, p = 0.004), but no differences in tissue levels of ammonia and/or ammonium (Figure 3E; paired t = 2.075, p = 0.0927), glutathione (Figure S3C; paired t = 1.684, p = 0.1908), or NADH (Figure S3C; paired t = 0.8253, p = 0.4697), which also incorporate glutamate in their synthetic pathways. Thus, removal of oligodendrocyte GS selectively impacts tissue levels of glutamate and glutamine.
Figure 3. Mice Lacking Oligodendrocyte GS Have Reduced Tissue Levels of GS Substrates but Normal Patterns of Myelination.
(A) Immunostaining for ASPA and GS in the ventral midbrain of control (CTL) and conditional knockout (cKO) mice. Blue arrowheads indicate GS+ oligodendrocytes, and yellow arrowheads indicate GS− oligodendrocytes.
(B) Box and whisker plots of GS and ASPA fluorescence within ASPA+ oligodendrocytes; whiskers represent minimum and maximum values. GS K-S D = 0.9871, p < 0.0001; n = 150−190 cells, 2 animals per group. ASPA K-S D = 0.09755, p = 0.438; n = 150 190 cells, 2 animals per group.
(C) Western blot of GS and loading control Gapdh in the microdissected cortex or midbrain of control and cKO animals.
(D) Quantification of GS protein relative to sample Gapdh. Cortex unpaired t = 1.073, df = 15, p = 0.3004; n = 8−9 animals per group. Midbrain unpaired t = 3.609, df = 14, p = 0.0028; n = 8 animals per group.
(E) Quantification of GS metabolites in microdissected midbrains via colorimetric assays. One control and one cKO animal were processed in parallel per day; control and cKO values from the same day were paired for analysis. Glutamate paired t = 2.852, df = 4, p = 0.0463, n = 5 animals per group. Glutamine paired t = 5.037, df = 5, p = 0.004, n = 6 animals per group. NH3/NH4+ paired t = 2.075, df = 5, p = 0.0927, n = 6 animals per group.
(F) Immunostaining for MBP in the cortex, ventral midbrain (VM), and spinal cord white matter (SCw) of control and cKO animals.
(G) Western blot for MBP and loading control Actin in microdissected midbrain of control and cKO animals. Quantification of MBP protein relative to sample Gapdh, unpaired t = 0.2463, df = 11, p = 0.8100; n = 6[C0]7 animals per group.
(H) TEM images of myelinated axons in spinal cord of control and cKO animals.
(I) Histogram of axon diameter (feret) in control (673 axons) and cKO (671 axons) animals.
(J) Scatterplot of axon diameter versus g-ratio (inner feret diameter/outer feret diameter). Linear regression control R2 = 0.1713, p < 0.0001; cKO R2 = 0.2448, p < 0.0001.
(K) Axon diameter by animal, unpaired t = 0.1311, df = 4, p = 0.902; myelin thickness by animal, unpaired t = 0.7996, df = 4, p = 0.4688; g-ratio by animal, Welch-corrected t = 1.738, df = 2.029, p = 0.2225; n = 3 animals per group.
(D, G, and K) Error bars indicate SEM.
To determine whether oligodendrocyte GS deletion impairs myelination, we measured myelin protein levels by western blot and immunostained for MBP in control and cKO animals. We detected no differences in myelin protein levels (Figure 3G; Figure S3E; MBP t = 0.2463, p = 0.8100; CNP t = 0.8652, p = 0.4054) or patterns of myelination (Figure 3F; Figure S3D) between the two groups. To evaluate myelin integrity at the ultrastructural level, we performed EM analysis of myelinated axons in the spinal cord (Figure 3H; Figure S3F), a white matter tract that exhibited high oligodendrocyte GS expression in wild-type animals (Figures 2D and 2E; Figure S2A). We observed no differences in axon diameter or area (Figures 3I and 3K; Figure S3G; axon diameter t = 0.1311, p = 0.902; axon area t = 0.3318, p = 0.7567), myelin thickness (Figure 3K; t = 0.7996, p = 0.4688), g-ratio (Figures 3J and 3K; Welch-corrected t = 1.738, p = 0.2225), or the relationship between g-ratio and axon diameter (Figure 3J; Figure S3G; slope of best-fit lines, t = 1.011, p = 0.3691), indicating that oligodendrocyte GS expression is not required for myelination.
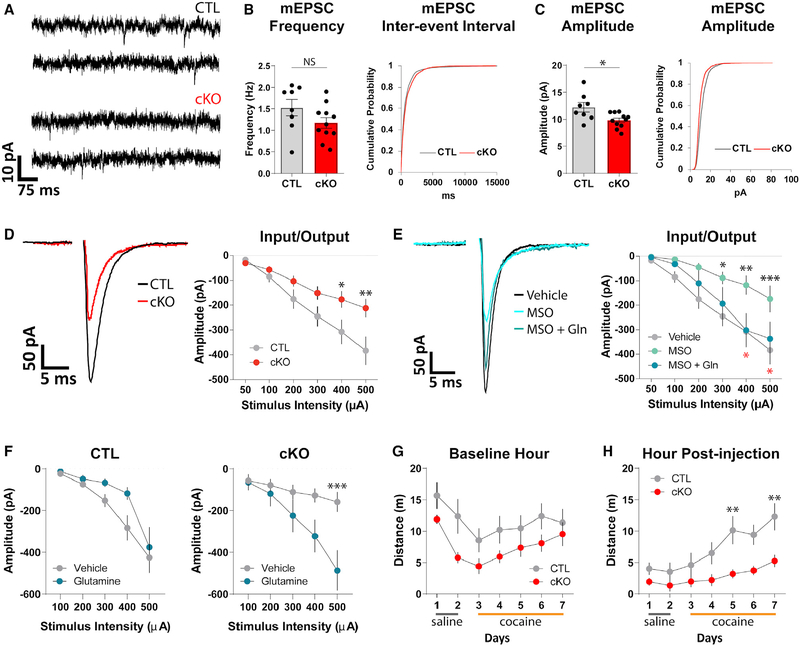
To determine whether oligodendrocyte GS deletion disrupts neuronal glutamate transmission, we performed slice whole cell patch clamp recordings from dopamine neurons in the ventral midbrain, where we had observed high GS expression in wild-type (Figure 2B) and control (Figure 3A) animals. Oligodendrocyte GS deletion did not affect the spontaneous firing rate (t = 1.129, p = 0.264), membrane resistance (t = 0.1098, p = 0.9133), and cell capacitance (t = 0.3577, p = 0.7229) of dopamine neurons (Figures S4A–S4E), indicating no gross differences in neuronal health. However, the amplitude of miniature excitatory post-synaptic currents (mEPSCs) was significantly reduced in cKO animals (Figures 4A and 4C; Figure S4F; t = 2.682, p = 0.0158), without significant changes in mEPSC frequency (Figure 4B; Figure S4F; t = 1.605, p = 0.1269), rise time (Figure S4G; t = 0.09417, p = 0.9261), or decay tau (Figure S4H; t = 0.8004, p = 0.4345). Furthermore, when we electrically evoked synaptic glutamate release, we observed a significant reduction in the amplitude of evoked EPSCs (eEPSCs) measured from cKO animals across a range of stimulating intensities (Figure 4D; F = 4.705, p = 0.0359). To clarify whether this deficit is due to a requirement for ongoing GS activity or, perhaps, a manifestation of earlier deficits that occurred during development, we acutely inhibited GS in control slices using the pharmacological GS inhibitor L-methionine sulfoximine (MSO). Pre-incubation with MSO (5 mM) led to a significant reduction in eEPSC amplitude, recapitulating the deficit we detected in cKOs (Figure 4E; F = 4.351, p = 0.0193). This reduction could be prevented by pre-incubation with both MSO and exogenous glutamine (Gln; 500 mM) (Figure 4E). Although we cannot distinguish the relative contribution of oligodendrocyte GS and astrocyte GS activity in these pharmacological experiments, the fact that acutely inhibiting GS produces a similar deficit as seen in our cKO animals suggests that ongoing production of glutamine by oligodendrocyte GS is required for proper glutamatergic transmission in the midbrain. In line with this interpretation, pre-incubation with glutamine significantly increased the eEPSC amplitude in cKO slices but not in control slices (Figure 4F; control slices F = 3.18, p = 0.0905; cKO slices F = 4.964, p = 0.0406), suggesting that a decrease in glutamine production underlies the deficits observed in cKO animals.
Figure 4. Oligodendrocyte GS Deletion Disrupts Synaptic Glutamate Transmission in the Midbrain and Impairs Cocaine-Induced Locomotor Sensitization.
(A) Example traces of mEPSCs.
(B) Quantification of mEPSC frequency and cumulative probability plot of inter-event intervals of all mEPSC events. mEPSC frequency unpaired t = 1.605, df = 17, p = 0.1269; n = 8−11 cells, 4 animals per group.
(C) Quantification and cumulative probability plot of amplitudes of all mEPSC events. mEPSC amplitude unpaired t = 2.682, df = 17, p = 0.0158; n = 8−11 cells, 4 animals per group.
(D) Example traces and quantification of evoked EPSCs. Repeated-measures ANOVA (stimulus intensity 3 genotype), main effect of genotype F(1,41) = 4.705, p = 0.0359; main effect of stimulus intensity F(5,205) = 54.02, p < 0.0001; interaction F(5,205) = 5.636, p < 0.0001; n = 20−23 cells, 8−9 animals per group.
*posthoc p < 0.05, **posthoc p < 0.01.
(E) Example traces and quantification of evoked EPSCs. Repeated-measures ANOVA (stimulus intensity 3 treatment), main effect of treatment F(2,41) = 4.351, p = 0.0193; main effect of stimulus intensity F(5,205) = 46.11, p < 0.0001; interaction F(10,205) = 2.309, p = 0.0136; n = 11−20 cells, 4−8 animals per group. *posthoc Veh versus MSO p < 0.05, **posthoc Veh versus MSO p < 0.01, ***posthoc Veh versus MSO p < 0.001. *posthoc MSO versus MSO+Gln p < 0.05.
(F) Quantification of evoked EPSC amplitude with vehicle and glutamine pre-incubation. Control repeated-measures ANOVA (stimulus intensity x treatment), main effect of treatment F(1,19) = 3.18, p = 0.0905; main effect of stimulus intensity F(4,76) = 26.18, p < 0.0001; interaction F(4,76) = 1.083, p = 0.371; n = 9−12 cells,4−6 animals per group. cKO repeated-measure ANOVA (stimulus intensity x treatment), main effect of treatment F(1,16) = 4.964, p = 0.0406; main effect of stimulus intensity F(4,64) = 18.94, p < 0.0001; interaction F(4,64) = 7.247, p < 0.0001; n = 8−10 cells, 3−4 animals per group.
(G) Daily distance traveled during baseline hour. Repeated-measures ANOVA (day 3 genotype), main effect of day F(6,96) = 13.06, p < 0.0001; main effect of genotype F(1,16) = 2.784, p = 0.1147; interaction F(6,96) = 1.289, p = 0.2697; n = 8−10 animals per group.
(H) Daily distance traveled following drug injection. Repeated-measures ANOVA (day 3 genotype), main effect of day F(6,96) = 9.992, p < 0.0001; main effect of genotype F(1,16) = 7.775, p = 0.0132; interaction F(6,96) = 2.109, p = 0.0591; n = 8−10 animals per group. **posthoc p < 0.01.
(B–H) Error bars indicate SEM.
Changes in glutamate signaling in the ventral midbrain are associated with the acquisition of certain psychoactive drug-related behaviors (Mahler et al., 2013; Mao et al., 2011; Ungless et al., 2001; Wang et al., 2012). In particular, cocaine-induced locomotor sensitization (i.e., a progressive escalation of locomotor activity in response to repeated cocaine exposure) requires elevations in midbrain glutamate (Kalivas and Duffy, 1998). Therefore, we investigated whether the deficits in midbrain glutamatergic transmission we detected in cKO animals would affect their behavioral response to repeated injections of cocaine. Animals were placed in open field boxes outfitted with infrared beams to measure locomotor activity. Following 1 h of baseline activity, animals received an intraperitoneal injection of saline (days 1 and 2, for habituation; Figure S4I) or 10 mg/kg cocaine (days 3–7). By day 7, both control and cKO animals displayed a large cocaine-induced increase in locomotor activity (Figure S4I). Quantification of distance traveled in the hour prior to drug injection revealed no significant differences in baseline activity between groups (Figure 4G; F = 2.784, p = 0.1147). However, cKO animals moved significantly less than control animals in the hour following cocaine injection (Figure 4H; F = 7.775, p = 0.0132). Thus, oligodendrocyte GS deletion significantly impacts performance in a midbrain-dependent behavior.
DISCUSSION
In the present study, we report an unexpected myelin-independent role for oligodendrocytes in supporting glutamatergic transmission. This aspect of oligodendrocyte function is mediated by expression of the glutamate-metabolizing enzyme GS. Oligodendrocyte GS expression appears late in development and is regionally heterogeneous, with higher expression in more caudal regions of the CNS. Selective deletion of oligodendrocyte GS disrupted midbrain glutamatergic synaptic transmission and cocaine-induced locomotor sensitization, a behavior that requires glutamate signaling in the midbrain. These results represent a significant departure from the canonical view of glutamate metabolism in the CNS (Jayakumar and Norenberg, 2016; Rose et al., 2013), which posits that glutamate and glutamine are exchanged solely between astrocytes and neurons.
Our data also revealed an unexpected dimension of regional oligodendrocyte heterogeneity. The differences in oligodendrocyte GS expression across the CNS may reflect different regional needs for glutamate processing but may also be due to differences in cell-intrinsic metabolic demands. In addition to metabolizing glutamate and synthesizing glutamine, GS also metabolizes ammonia, which can be a toxic byproduct of amino acid biosynthesis (Rose et al., 2013). Although the present study focused on the impact of oligodendrocyte GS deletion on neuronal glutamate transmission, GS may play other cell-autonomous roles in oligodendrocytes related to ammonia processing. Further investigation of mechanisms regulating GS expression in oligodendrocytes, as well as potential roles of GS in cell-autonomous oligodendrocyte processes, may yield important insights into regional oligodendrocyte specialization and function.
Although GS is widely considered to be astrocyte-specific (Armbruster et al., 2016; Habbas et al., 2015; Okuda et al., 2014; Papageorgiou et al., 2018; Theofilas et al., 2017; Tong et al., 2014), there have been reports of GS expression, particularly at the mRNA level, in oligodendrocyte progenitors and microglia (Nakajima et al., 2017; Palmieri et al., 2017; Zhang et al., 2014). Within the ventral midbrain of naive young adult mice, we did not detect expression of GS mRNA or protein in these cell types. However, it is possible that they express GS in other brain regions or upregulate GS expression in aging or pathological contexts (Nakajima et al., 2017; Palmieri et al., 2017), especially given the sensitivity of oligodendrocyte progenitors and microglia to injury and disease (Hickman et al., 2018; Hughes et al., 2013).
The differential effects of glutamine pre-incubation on control and cKO slices suggest that the deficit in glutamatergic signaling we observed in cKO animals is at least partially due to decreased glutamine availability. However, it is unclear whether glutamine pre-incubation was able to increase EPSC amplitude in cKO slices by directly replenishing the presynaptic glutamine pool used for glutamate synthesis. It is also possible that glutamine indirectly enhances transmission efficacy by acting as an energy substrate for axons (Amaral, 2013). In either case, these data indicate an ongoing requirement for oligodendrocyte GS activity in supporting glutamate transmission, at least in a brain region where GS is robustly expressed by oligodendrocytes.
How might oligodendrocytes exchange GS substrates with neurons? Some studies have indicated that oligodendrocytes express transporters that are permissible to glutamate and glutamine, such as EAAC1 (DeSilva et al., 2009), SNAT3 (Marques et al., 2018), and ASCT2 (Scopelliti et al., 2018), which would allow for a direct exchange of GS substrates with axons. Furthermore, we previously reported extensive gap junction coupling between midbrain oligodendrocytes and astrocytes (Xin et al., 2019). As molecules smaller than 1 kDa can theoretically pass freely through gap junctions (Berg et al., 2002), these structural connections may enable exchange of GS substrates between astrocytes and oligodendrocytes and thus neurons by way of astrocyte transporters. Intriguingly, the temporal expression pattern of the astrocyte connexin 30 (CX30) closely parallels that of oligodendrocyte GS, with expression onset around 2 weeks postnatally (Kunzelmann et al., 1999; Nagy et al., 1999). Spatially, there is also correspondence between CX30 expression and oligodendrocyte GS expression; both are enriched in hindbrain regions and low in white matter tracts (Griemsmann et al., 2015; Nagy et al., 1999). Whether glutamate and glutamine exchange occur directly via oligodendrocyte transporters or via astrocyte intermediaries will require additional functional studies, ideally combined with cell-type specific deletion of connexins or transporters. Elucidating the downstream mechanisms that enable oligodendrocyte-neuron interactions in the context of glutamate and glutamine shuttling will be important for furthering our understanding of glutamate processing in the brain.
Finally, these results point to oligodendrocytes as potential players, and perhaps even therapeutic targets, in the numerous CNS pathologies involving glutamate dysregulation (Jayakumar and Norenberg, 2016; Sheldon and Robinson, 2007). Indeed, there is evidence that impaired oligodendrocyte function and/or myelination is accompanied by changes in brain glutamate levels (Gao et al., 2018; Maheras et al., 2018; Nantes et al., 2017; Takanashi et al., 2014; Werner et al., 2001). Moving forward, studies on the role of glutamate in various pathological states should consider the possibility of dual roles for oligodendrocytes as victim and perpetrator of glutamate-related CNS dysfunction.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Antonello Bonci (antonello.bonci@nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Unless indicated, both male and female mice were used, and the number of males and females in each analysis group was balanced. No significant effect of sex was observed in data analyses. Ages of mice used in each experiment are described in the Method Details section below. Mice were housed in normal light dark (lights on 7am, lights off 7pm) cycle and had ad libitum access to food and water. Mice were group housed 2–5 to a cage and had no prior history of drug administration, surgery, or behavioral testing. All experiments were performed in strict accordance with protocols approved by the Animal Care and Use Committee at NIDA. Wild-type mice (C57BL/6J; JAX 000664) were purchased from Jackson Laboratories. Aldh1L1-eGFP mice (OFC789Gsat/ 011015-UCD) were purchased from MMRRC and maintained as heterozygotes by crossing with FVB mice. MOBP-eGFP mice (IN1Gsat) were originally obtained from MMRRC and maintained as homozygotes. CX3CR1-eGFP mice were originally obtained from Jackson Labs (JAX 005582) and maintained as homozygotes. MOG-iCre mice were generated by Dr. Ari Waisman (Hövelmeyer et al., 2005). GSfl/fl mice were generated by Dr. Wouter Lamers (He et al., 2010). For experiments using MOG-iCre x GSfl/fl mice, GSfl/fl mice were bred with MOG-iCre+/− GSfl/fl mice to obtain a mix of MOG-iCre− GSfl/fl (“control”) and MOG-iCre+ GSfl/fl (“cKO”) pups.
Primary cell culture
Pregnant CD1 mice were ordered from Jackson labs and housed in normal light dark cycle with ad libitum access to food and water. P5–P7 pups were used to make primary oligodendrocyte progenitor cell cultures.
METHOD DETAILS
Immunohistochemistry
Mice aged postnatal day 14 (“P14”), 21 (“P21”), 28–29 (“P28”), 58–69 (“adult”), or 87–90 (“P90”) were deeply anesthetized with Euthasol (sodium pentobarbital 150 mg/kg and sodium phenytoin 19.2mg/kg, Virbac) and perfused transcardially with 4% paraformaldehyde in 1X PBS. Brain tissue was isolated and postfixed in this solution for 4 hr at 4°C, then stored in 1X PBS with 0.1% NaAz. Coronal brain sections (50 μm) were prepared on a vibratome in chilled 1X PBS. For quantification of GS intensity in different brain regions, brains were sucrose protected (30% in PBS) prior to flash freezing and sectioned on a cryostat (20 μm). Free-floating sections (both vibratome and cryostat sectioned) were permeabilized/blocked with 0.2% Triton X-100 and 5% normal donkey serum in 1X PBS for 1hr at room temperature. Sections were incubated with primary antibodies prepared in 0.2% Triton X-100 and 5% normal donkey serum in 1X PBS at 4°C overnight. Sections were incubated with secondary antibodies in 5% normal donkey serum in 1X PBS for 2hr at room temperature. Primary antibodies and concentrations used are listed in the Key Resources Table. Primary antibodies have been validated for use in immunohistochemistry in mouse tissue, in published literature, and on the manufacturer’s websites. Secondary antibodies used included the following: Alexa Fluor 488−, 594−, or 647−conjugated secondary antibodies to rabbit, mouse, goat, chicken, rat or guinea pig (1:500; all raised in donkey; Jackson ImmunoResearch). No sex differences were observed (e.g., GS fluorescence in midbrain, K-S D = 0.0553, p = 0.8944).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-GS | Millipore Sigma | Cat# MAB302; RRID AB_2110656 |
| Rabbit anti-PDGFRα | Cell Signaling | Cat# 3174; RRID AB_2162345 |
| Rabbit anti-ASPA | GeneTex | Cat# GTX113389; RRID AB_2036283 |
| Chicken anti-MBP | Aves | Cat# MBP; RRID AB_2313550 |
| Mouse anti-NeuN | Millipore Sigma | Cat# MAB377; RRID AB_2298767 |
| Rabbit anti-GS | Abcam | Cat# Ab73593; RRID AB_2247588 |
| Mouse anti-CNP | Millipore Sigma | Cat# MAB326; RRID AB_2082608 |
| Mouse anti-MBP | Biolegend | Cat# 808401; RRID AB_2564741 |
| Mouse anti-Gapdh | Santa Cruz | Cat# Sc-32233; RRID AB_627679 |
| Rabbit anti-Actin | Sigma | Cat# A5060; RRID AB_2766001 |
| Donkey anti-Rabbit | Li-Cor | Cat# 926–32213; RRID AB_621848 |
| Donkey anti-Mouse | Li-Cor | Cat# 926–68072; RRID AB_10953628 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-Methionine sulfoximine (‘MSO’) | Sigma | Cat# M5379 |
| L-glutamine | Sigma | Cat# G3126 |
| Critical Commercial Assays | ||
| Glutamate Colorimetric Assay Kit | Biovision | Cat# K629 |
| Glutamine Colorimetric Assay Kit | Biovision | Cat# K446 |
| Ammonia Colorimetric Assay Kit | Biovision | Cat# K370 |
| Glutamate Dehydrogenase Activity Assay Kit | Abcam | Cat# Ab102527 |
| QuantiChrom Glutathione (GSH) Assay Kit | Bioassays | Cat# DIGT-250 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Aldh1L1-eGFP | MMRRC | OFC789Gsat/ 011015-UCD |
| Mouse: MOBP-eGFP | MMRRC | IN1Gsat |
| Mouse: CX3CR1-eGFP | Jackson Laboratory | JAX 005582 |
| Mouse: MOG-iCre | Dr. Ari Waisman | Hövelmeyer et al., 2005 |
| Mouse: GSfl/fl | Dr. Wouter Lamers | Heetal., 2010 |
| Oligonucleotides | ||
| See Table S1 | n/a | n/a |
| Software and Algorithms | ||
| FIJI | NIH | https://fiji.sc/; RRID SCR_002285 |
| Microsoft Office 365 Excel | Microsoft | RRID SCR_016137 |
| Prism 8 | Graphpad | RRID SCR 002798 |
Image acquisition and analysis
For all experiments, confocal fluorescence images were acquired with an Olympus FV1000 microscope. For quantification of GS+ cell density and GS intensity, stacks of confocal images were acquired using a 20X objective and a 1.5 μm z-interval and analysis was performed on maximum projections of confocal stacks within ImageJ software. For quantification of GS intensity in slice, regions of interest (ROIs) were manually delineated in ImageJ based on MOBP-eGFP or ASPA immunoreactivity using the freehand tool, and mean intensity for individual ROIs was extracted; similar patterns were observed when using integrated density in the place of mean intensity. For all imaging analyses, at least 2 images from separate brain sections were analyzed per mouse to obtain a summed value for that mouse.
Tissue dissociation and flow cytometry
MOBP-eGFP or Aldh1L1-eGFP mice, aged 2–3mo, were deeply anesthetized with Euthasol and perfused transcardially with sodium-substituted artificial cerebral spinal fluid (NMDG ACSF) containing, in mM, 92 NMDG, 20 HEPES, 25 glucose, 30 NaHCO3, 1.2 NaH2PO4, 2.5 KCl, 5 sodium ascorbate, 3 sodium pyruvate, 2 thiourea, 10 MgSO4, and 0.5 CaCl2 (pH7.35, ~300 mOsm, saturated with 95%O2/5% CO2). Brains were rapidly extracted; cortex and midbrain were microdissected on ice. Microdissected regions were minced and transferred to microtubes containing ice-cold Hank’s Buffered Salt Solution (no Ca2+/Mg2+). Tissue was dissociated into a single cell suspension using the Miltenyi Neural Tissue Dissociation Kit (version ‘P’). Briefly, tissue was enzymatically digested for 30min at 37°C and manually triturated using fire-polished glass Pasteur pipets with progressively smaller diameter openings (final pipet ~0.4mm diameter) (Rubio et al., 2016). Cell suspensions were pelleted and resuspended in PBS for sorting via flow cytometry. Samples were sorted using a BD Biosciences FACS Aria I cell sorter into either 200mL of extraction buffer from the Arcturus PicoPure RNA isolation kit for qPCR, or 50mL PBS for western blot. Cell suspensions from wild-type tissue were used to establish eGFP-negative gates. Oligodendrocytes and astrocytes were identified based on forward scatter (FSC), side scatter (SSC), and FITC channel intensity (see Figure S1G). Samples from individual animals were not pooled. Using tissue microdissected from individual animals, we collected an average of 53052 ± 15841 cortical oligodendrocytes, 52318 ± 18446 midbrain oligodendrocytes, 48461 ± 20432 cortical astrocytes, and 67385 ± 4511 midbrain astrocytes (per animal). For each sample, 70000 eGFP-cells were also collected for validation of cell-type specific qPCR primers and probes.
RNA isolation
RNA from sorted cells was isolated using the PicoPure RNA isolation kit (Arcturus Bioscience). Cells were sorted into Eppendorf tubes containing 200 μL PicoPure RNA extraction buffer and, following the sort, samples were incubated at 42°C for 30 min, and stored in RNase-free tubes at −80°C until further processing. Column filtration, washing, and elution of RNA from the columns was performed according to manufacturer’s instructions in section IV.C of the PicoPure RNA Isolation Kit User Guide.
cDNA synthesis and RT-PCR analysis of gene expression
Single strand cDNAs were synthesized with Superscript III first strand cDNA synthesis kit (Invitrogen, Life Technologies), according to the manufacturer’s protocol. Duplex RT-PCR assays were performed on technical duplicates using a FAM-labeled probe for each target gene, and a VIC-labeled probe for the endogenous control gene (Gapdh), along with TaqMan Advanced Fast PCR Master Mix (Cat# 4444963; Life Technologies). To avoid amplification of genomic DNA contamination, primers and probes that amplify across target gene exon-exon junctions were selected when possible. RT-PCR reactions were run in a 7500 Fast TaqMan instrument using the program: 95°C hold for 20 s, followed by 40 cycles of 95°C denaturation for 3 s, and 60°C annealing and extension for 30 s. Calculations of relative expression from Ct data were carried out according to User Bulletin #2 for ABI Prism 7900 Sequence Detection System. For each target gene, the average Ct value for the endogenous control (Gapdh) was subtracted from the average Ct value for the target gene, to obtain ΔCt. The relative expression was then plotted as 2-ΔCt. All TaqMan assays and custom primers/probes that were used are detailed in the Key Resources Table. No sex differences were observed (e.g., Midbrain Glul in oligodendrocytes, t = 0.5093, p = 0.6457).
In situ hybridization using RNAscope
Aldh1L1-eGFP and MOBP-eGFP mice aged P60-P65 were deeply anesthetized with an intraperitoneal injection of Euthasol. Animals irresponsive to toe pinches were decapitated, and brains were rapidly extracted and flash frozen in isopentane. Brains were directly collected onto SuperFrost microscope slides at 14μm using a cryostat. Sections were dehydrated with ethanol and incubated with RNAscope probes according to the manufacturer’s instructions for the RNAscope® Multiplex Fluorescent Assay. Sections were imaged as a Z stack on the confocal microscope described above at a 1 mm z-interval. No sex differences were observed in the RNAscope dataset (e.g., % Glul+ oligodendrocytes, t = 0.4029, p = 0.726).
Cell culture
For cell culture experiments, CD1 P5–P7 pups were used, with 3–5 animals per experimental group. Pups were not sexed prior to sacrifice. Pups were rapidly decapitated and brain tissue was immediately placed in ice-cold HBSS with no Ca2+ and Mg2+(ThermoFisher Scientific, #14175103). Cortices were resected from the rest of the brain, and motor and frontal cortices were dissected with additional care taken to avoid including any calossal white matter. Midbrain tissue was dissected by another investigator at the same time. Afterward, midbrain and cortical tissue was then processed in parallel using Neural Tissue Dissociation kit according to the manufacturer’s instructions (#130-092-628). OPC purification was performed using anti-O4 isolation beads (#130-094-543), MS columns (#130-042-201), and a MiniMACs Separator (130-042-102), according to the manufacturer’s instructions (all Miltenyi Biotec). Cells were spotted at the density 12–15 thousand cells/well on Poly-D-lysine (Milipore-Sigma, # P6407, 10 μg/ml) coated glass coverslips (Warner Instruments, #64–0702) in 50–80 μL of OPC culture medium and allowed to adhere for 20 minutes in a tissue culture incubator maintained at 37°C and 5% CO2. Afterward, the wells were gently flooded with the culture medium up to the total volume of 250–300 μl. OPC culture medium consisted of the following: DMEM:F12 (ThermoFisher Scientific, #11330032), N2B (Stem Cell Technologies, 1:1000 stock, #7156), SM1 (Stem Cell Technologies, 1:50, #5711), Penicillin/Streptomycin (ThermoFisher Scientific, 1:200 stock), N-Acetyl-Cysteine (Sigma-Milipore, #A8199, 5 μg/ml), forskolin (Calbiochem, #344270, 5uM), CNTF (Peprotech, #450–50, 10ng/ml), PDGF-AA (Peprotech, #100–13A, 20ng/ml), NT-3 (#450–03B, 1ng/ml). To induce OPC differentiation, the day after cell plating OPC culture medium was replaced with the medium no longer containing PDGF-AA and NT3, but supplemented with T3 (Milipore-Sigma, #T6397, 4ng/ml). Cells were then cultured for 4–6 additional days to ensure uniform differentiation. Cells were fixed with 4% PFA for 12–15 minutes, washed 3×5min with PBS and processed for staining. Data were collected from three separate cohorts.
Western blotting
For tissue western blots, control and cKO mice aged P90-P105 were deeply anesthetized with an intraperitoneal injection of Euthasol. Animals irresponsive to toe pinches were decapitated, and midbrains were dissected out on ice, then flash frozen in isopentane. Tissues were homogenized in RIPA lysis buffer (Cell Signaling Technology) and protein concentration for each sample was quantified with a Bio-Rad Protein Assay. A total of 30 mg of proteins per sample was loaded for each run.
For FACS-isolated cells, samples were diluted with PBS down to the same number of cells across groups. Laemmli buffer with 2-mercaptoethanol were added to samples (1 part Laemmli to 3 parts sample volume). Mixtures were boiled on a preheated hot plate for 5 minutes at 95°C. 20 μL per sample were loaded for each run.
Membranes were blocked overnight with Li-Cor Blocking Buffer at 4C, then incubated with primary antibodies diluted in the same buffer for 2 hours at RT, washed, then incubated with secondary antibodies for 1.5 hours at RT. Antibodies used are listed in the Key Resources Table. Membranes were scanned using a LI-COR Odyssey Image System. Band intensities were measured using ImageJ software; proteins of interest were normalized to reference genes (Gapdh or Actin). Sex of samples was not tracked during processing.
Tissue measurement of metabolites
One control and one cKO mouse, aged P50–55 and sex matched, were sacrificed, and brain tissues processed, in parallel on each day of an experiment. Animals were deeply anesthetized with an intraperitoneal injection of Euthasol. Animals irresponsive to toe pinches were perfused with ice-cold PBS. Brains were rapidly extracted and midbrains were dissected out on ice, then flash frozen in pre-chilled isopentane. Tissue samples were then prepared according to kit manufacturers’ instructions. The following kits were used for detection of metabolites: Biovision Glutamate Colorimetric Assay Kit (cat. K629), Biovision Glutamine Colorimetric Assay Kit (cat. K556), Biovision Ammonia Colorimetric Assay Kit (cat. K370), Abcam Glutamate Dehydrogenase Activity Assay Kit (for NADH, cat. Ab102527), Bioassays QuantiChrom Glutathione (GSH) Assay Kit (cat. DIGT-250). Kit reactions produced stable colorimetric signals that were measured as absorbance in a specific wavelength by a plate reader (BioTek Snyergy H1 Hybrid Reader). Plates included vehicle wells with no sample for background subtraction. For each tissue sample, two technical replicates were included per well and averaged. No sex differences were observed (e.g., Glutamine t = 1.106, p = 0.3839).
Slice electrophysiology
Control and cKO mice aged P52-P65 were deeply anesthetized with an intraperitoneal injection of Euthasol. Animals irresponsive to toe pinches were decapitated, and brains were rapidly extracted and sectioned using a vibratome (Leica VT-1200). The cutting ACSF contained (in mM) 92 NMDG, 20 HEPES, 25 glucose, 30 NaHCO3, 1.2 NaH2PO4, 2.5 KCl, 5 sodium ascorbate, 3 sodium pyruvate, 2 thiourea, 10 MgSO4, and 0.5 CaCl2 (pH7.35, ~300mOsm). 200 μm horizontal slices containing ventral midbrain were collected and recovered for 10min in NMDG ACSF at 32°C, after which slices were transferred to a holding ACSF solution identical to the cutting solution, except it contained 1mM MgCl2, 2mM CaCl2, and 92mM NaCl instead of NMDG; slices were left to recover for 1h at RT prior to beginning of recording. All ACSF solutions were saturated with 95% O2 and 5% CO2.
For recording, a single slice was transferred to a heated chamber (34–35°C) and perfused with normal ACSF (2.5ml min−1) using a peristaltic pump (WPI). The ACSF used to perfuse slices during recording contained (in mM): 125 NaCl, 2.5 KCl, 1.25 sodium phosphate, 1 MgCl2, 2.4 CaCl2, 26 sodium bicarbonate, and 11 glucose. Cells were identified on an upright microscope equipped for differential interference contrast (DIC) microscopy (Olympus BX51WI). VTA dopamine neurons were identified based on location (medial to the medial nucleus of the optic tract), morphology, and the presence of low frequency (0.5–5Hz) tonic firing (Wanat et al., 2008). Whole-cell patch-clamp recordings were made using a MultiClamp 700B amplifier (1kHz low-pass Bessel filter and 10kHz digitization) with pClamp 10.3 software (Molecular Devices). Voltage-clamp recordings were made using borosilicate glass pipets (King Precision Glass Inc., KG-33 ID1.00 OD 1.50) with resistance 1.5–2.5ΩU, filled with internal solution containing (in mM): 117 cesium methanesulfonate, 20 HEPES, 0.4 EGTA, 2.8 NaCl, 5 TEA-Cl, 2.5 Mg-ATP, and 0.25 Na-GTP, pH7.2–7.3 and 280–285mOsm. Current-clamp recordings were made with internal solution containing (in mM): 125 potassium gluconate, 0.05 EGTA, 10 HEPES, 6.27 KCl, 5 Na2-phosphocreatine, 4 Mg-ATP, and 0.3 Na-GTP, pH7.3 and 285mOsm. Input resistance was continually monitored throughout recording; cells in which input resistance rose above 25MU were excluded from analysis. Membrane potentials were not corrected for junction potentials.
Spontaneous firing rate was assessed in a cell attached configuration. For evoked EPSC experiments, electrical stimuli were delivered using a bipolar stimulating electrode (FHC, MX21AES) placed anterior to the recorded cell, voltage clamped at −60mV, in the presence of picrotoxin (100 μM) and APV (50 μM). For MSO and glutamine recordings, slices were pre-incubated for1–2hrs at RT either in holding ACSF described above (‘vehicle’), in holding ACSF with 5mM L-Methionine sulfoximine (‘MSO’; Sigma M5379), in holding ACSF with 5mM MSO and 500 mM L-glutamine (‘MSO+gln’; Sigma G3126), or in holding ACSF with 500 μM L-glutamine (‘Glutamine’). Analysis was performed offline using Clampfit (v 10.6). mEPSCs were pharmacologically isolated by having tetrodotoxin (1 μM), picrotoxin (100 μM), and APV (50 μM) present throughout the experiment. 200–250 events per cell were analyzed, using a threshold of 2X the baseline noise. Analysis of mEPSCs was performed offline using the MiniAnalysis program (v 6.0). Experimenter was naive to animal genotype during acquisition and analysis of data. No sex differences were observed (e.g., Spontaneous firing frequency, t = 1.25, p = 0.2178).
Cocaine-induced locomotor sensitization
Control and cKO mice aged P150-P165 were placed in a locomotor chamber outfitted with infrared beams to detect locomotor activity (Accuscan, Columbus, OH, USA). Animals were monitored for two hours each day, under IR light. After the first hour (‘Baseline’), mice received intraperitoneal injections of saline (days 1 and 2) or cocaine (10mg/kg, days 3–7), then were placed back in the chamber for an additional hour (‘Post-injection’). Experimenter was blinded to animal genotype. Locomotor data were collected from three separate cohorts of animals. Prior to the experiment, animals were naive to any behavioral or drug manipulation. Total horizontal distance traveled was used to assess locomotor sensitization to cocaine. No sex differences were observed (e.g., Baseline hour, F(1,16) = 0.7191, p = 0.4089).
Electron Microscopy
Control and cKO mice aged P73-P78 were perfused with 60mL (5.5mL/min) 4% paraformaldehyde (freshly prepared from EM grade prill form paraformaldehyde), 2% glutaraldehyde, 3mM MgCl2, in 0.1M sodium cacodylate buffer, pH7.2. After perfusion, spinal cords were dissected out and placed in fresh fixative overnight at 4°C. After buffer rinse, samples were postfixed in reduced 2% osmium tetroxide, 1.6% potassium ferrocyanide in buffer (2 hr) on ice in the dark. Following a dH2O rinse, samples were stained with 2% aqueous uranyl acetate (0.22μm filtered, 1 hr, dark), dehydrated in a graded series of ethanol, propylene oxide and embedded in Eponate 12 (Ted Pella) resin. Samples were polymerized at 60°C overnight. Thin sections, 60–90nm, were cut with a diamond knife on the Reichert-Jung Ultracut E ultramicrotome and picked up with copper slot (1 × 2 mm) grids. Grids were stained with 2% uranyl acetate and observed with a Philips/FEI BioTwin CM120 TEM at 80kV. Images were captured with an AMT CCD (1K × 1K) camera.
For analysis, axons were traced in Fiji ImageJ (Schindelin et al., 2012) using the freehand tool. A minimum of 200 axons per animal were traced. Inner and outer axon diameters represent feret diameters calculated by ImageJ. G-ratio was defined as inner feret diameter divided by outer feret diameter. Myelin thickness was defined as the outer feret diameter minus the inner feret diameter, divided by two. No sex differences were observed (e.g., g-ratio t = 1.19, p = 0.300).
QUANTIFICATION AND STATISTICAL ANALYSIS
All graphed values are shown as mean ± SEM. Statistical details of experiments (statistical tests used, statistical values, exact n, what n represents) can be found in the figure legends, and abbreviated statistics are also included in the results section. The number of animals included in each experiment was based on standards established in the literature and not predetermined by statistical methods. Data were entered in Microsoft Excel; statistics were performed using GraphPad Prism 8. Statistical significance was defined as having a p value of less than 0.05. Tests for normality and equal variances were used to determine the appropriate statistical test to employ. For datasets that did not pass the normality test in Prism, non-parametric tests (Kolmogorov-Smirnov test for two groups, Kruskal-Wallis test for more than two groups) were used. All reported t tests were two-tailed, with Welch’s correction when group variances were significantly different. For colorimetric experiments, paired t tests were used; for all other experiments, unpaired t tests were used. For experiments with more than two groups, one-way ANOVAs were used. For experiments with more than one variable, two-way ANOVAs were used. For experiments with repeated-measurements from the same animals, two-way repeated-measures ANOVAs were used. Welch-corrected ANOVA tests were used when group variances were significantly different. Linear regression was used to test dependence of g-ratio on axon diameter.
Supplementary Material
Highlights.
Oligodendrocytes (OLs) express glutamine synthetase (GS)
OL GS expression is low in rostral regions and high in caudal regions of the brain
OLs do not require GS for survival or myelination
Loss of OL GS disrupts neuronal glutamate signaling and glutamate-dependent behavior
ACKNOWLEDGMENTS
The authors would like to thank Raffaello Cimbro at the Johns Hopkins Bay-view Immunomics Core (supported by NIAMS grant P30 AR-070254) for assistance with flow cytometry; Dr. LaToya Roker and the Johns Hopkins School of Medicine Microscope Facility for assistance with EM sample processing and imaging; Dr. Francois Vautier, Joni McKenzie, and the NIDA IRP Transgenic Breeding staff for assistance with animal husbandry; Drs. Lindsay De Biase, Richard Roth, and Ayesha Sengupta for helpful discussions and feedback during manuscript writing; and Dr. Gail Seabold for proofreading. This work was supported by the NIDA Intramural Research Program of the NIH and an NSF fellowship (grant 1232825) to W.X.; a fellowship from the National Multiple Sclerosis Society to Y.A.M.; and a grant from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation to D.E.B.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j. celrep.2019.04.094.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amaral AI (2013). Effects of hypoglycaemia on neuronal metabolism in the adult brain: role of alternative substrates to glucose. J. Inherit. Metab. Dis 36, 621–634. [DOI] [PubMed] [Google Scholar]
- Anlauf E, and Derouiche A (2013). Glutamine synthetase as an astrocytic marker: its cell type and vesicle localization. Front. Endocrinol 4, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster M, Hanson E, and Dulla CG (2016). Glutamate Clearance Is Locally Modulated by Presynaptic Neuronal Activity in the Cerebral Cortex. J. Neurosci 36, 10404–10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, and Stryer L (2002). Gap Junctions Allow Ions and Small Molecules to Flow between Communicating Cells In Biochemistry, Fifth Edition (W.H. Freeman; ). [Google Scholar]
- Bernstein H-G, Bannier J, Meyer-Lotz G, Steiner J, Keilhoff G, Dobrowolny H, Walter M, and Bogerts B (2014). Distribution of immunoreactive glutamine synthetase in the adult human and mouse brain. Qualitative and quantitative observations with special emphasis on extra-astroglial protein localization. J. Chem. Neuroanat 61–62, 33–50. [DOI] [PubMed] [Google Scholar]
- DeSilva TM, Kabakov AY, Goldhoff PE, Volpe JJ, and Rosenberg PA (2009). Regulation of glutamate transport in developing rat oligodendrocytes.J. Neurosci 29, 7898–7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yin X, Edden RAE, Evans AC, Xu J, Cao G, Li H, Li M, Zhao B, Wang J, and Wang G (2018). Altered Hippocampal GABA and Glutamate Levels and Uncoupling from Functional Connectivity in Multiple Sclerosis. Hippocampus 28, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griemsmann S, Höft SP, Bedner P, Zhang J, von Staden E, Beinhauer A, Degen J, Dublin P, Cope DW, Richter N, et al. (2015). Characterization of Panglial Gap Junction Networks in the Thalamus, Neocortex, and Hippo-campus Reveals a Unique Population of Glial Cells. Cereb. Cortex 25, 3420–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbas S, Santello M, Becker D, Stubbe H, Zappia G, Liaudet N, Klaus FR, Kollias G, Fontana A, Pryce CR, et al. (2015). Neuroinflamma-tory TNFa Impairs Memory via Astrocyte Signaling. Cell 163, 1730–1741. [DOI] [PubMed] [Google Scholar]
- Häberle J, Shahbeck N, Ibrahim K, Schmitt B, Scheer I, O’Gorman R, Chaudhry FA, and Ben-Omran T (2012). Glutamine supplementation in a child with inherited GS deficiency improves the clinical status and partially corrects the peripheral and central amino acid imbalance. Orphanet J. Rare Dis 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Hakvoort TBM, Vermeulen JLM, Labruyère WT, De Waart DR, Van Der Hel WS, Ruijter JM, Uylings HBM, and Lamers WH (2010). Glutamine synthetase deficiency in murine astrocytes results in neonatal death. Glia 58, 741–754. [DOI] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, and El Khoury J (2018). Microglia in neurodegeneration. Nat. Neurosci 21, 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hövelmeyer N, Hao Z, Kranidioti K, Kassiotis G, Buch T, Frommer F, von Hoch L, Kramer D, Minichiello L, Kollias G, et al. (2005). Apoptosis of oligodendrocytes via Fas and TNF-R1 is a key event in the induction of experimental autoimmune encephalomyelitis. J. Immunol 175, 5875–5884. [DOI] [PubMed] [Google Scholar]
- Hughes EG, Kang SH, Fukaya M, and Bergles DE (2013). Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci 16, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakumar AR, and Norenberg MD (2016). Glutamine Synthetase: Role in Neurological Disorders. Adv. Neurobiol 13, 327–350. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, and Duffy P (1998). Repeated cocaine administration alters extracellular glutamate in the ventral tegmental area. J. Neurochem 70, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee SG, Kegelman TP, Su ZZ, Das SK, Dash R, Dasgupta S, Barral PM, Hedvat M, Diaz P, et al. (2011). Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: opportunities for developing novel therapeutics. J. Cell. Physiol 226, 2484–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann P, Schröder W, Traub O, Steinhäuser C, Dermietzel R, and Willecke K (1999). Late onset and increasing expression of the gap junction protein connexin30 in adult murine brain and long-term cultured astrocytes. Glia 25, 111–119. [DOI] [PubMed] [Google Scholar]
- Larson VA, Mironova Y, Vanderpool KG, Waisman A, Rash JE, Agar-wal A, and Bergles DE (2018). Oligodendrocytes control potassium accumulation in white matter and seizure susceptibility. eLife 7, e34829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S-L, Carlson GC, and Coulter DA (2006). Dynamic regulation of synaptic GABA release by the glutamate-glutamine cycle in hippocampal area CA1. J. Neurosci 26, 8537–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheras KJ, Peppi M, Ghoddoussi F, Galloway MP, Perrine SA, and Gow A (2018). Absence of Claudin 11 in CNS Myelin Perturbs Behavior and Neurotransmitter Levels in Mice. Sci. Rep 8, 3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler SV, Smith RJ, and Aston-Jones G (2013). Interactions between VTA orexin and glutamate in cue-induced reinstatement of cocaine seeking in rats. Psychopharmacology 226, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Gallagher K, and McGehee DS (2011). Nicotine potentiation of excitatory inputs to ventral tegmental area dopamine neurons. J. Neurosci 31, 6710–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, van Bruggen D, Vanichkina DP, Floriddia EM, Munguba H, Väremo L, Giacomello S, Falcão AM, Meijer M, Björklund ÅK , et al. (2018). Transcriptional Convergence of Oligodendrocyte Lineage Progenitors during Development. Dev. Cell 46, 504–517.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JI, Patel D, Ochalski PAY, and Stelmack GL (1999). Connexin30 in rodent, cat and human brain: selective expression in gray matter astrocytes, co-localization with connexin43 at gap junctions and late developmental appearance. Neuroscience 88, 447–468. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Kanamatsu T, Koshimoto M, and Kohsaka S (2017). Micro-glia derived from the axotomized adult rat facial nucleus uptake glutamate and metabolize it to glutamine in vitro. Neurochem. Int 102, 1–12. [DOI] [PubMed] [Google Scholar]
- Nantes JC, Proulx S, Zhong J, Holmes SA, Narayanan S, Brown RA, Hoge RD, and Koski L (2017). GABA and glutamate levels correlate with MTR and clinical disability: Insights from multiple sclerosis. Neuroimage 157, 705–715. [DOI] [PubMed] [Google Scholar]
- Okuda H, Tatsumi K, Morita S, Shibukawa Y, Korekane H, Horii-Hayashi N, Wada Y, Taniguchi N, and Wanaka A (2014). Chondroitin sulfate proteoglycan tenascin-R regulates glutamate uptake by adult brain astrocytes. J. Biol. Chem 289, 2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, Dong J, Mungenast A, Yue C, Takano H, Watson DJ, Haydon PG, and Coulter DA (2010). Selective induction of astrocytic gliosis generates deficits in neuronal inhibition. Nat. Neurosci 13, 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri EM, Menga A, Lebrun A, Hooper DC, Butterfield DA, Maz-zone M, and Castegna A (2017). Blockade of Glutamine Synthetase Enhances Inflammatory Response in Microglial Cells. Antioxid. Redox Signal 26, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, and Chan JR (2017). Regulation and dysregulation of axon infrastructure by myelinating glia. J. Cell Biol 216, 3903–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou IE, Valous NA, Lahrmann B, Janova H, Klaft Z-J, Koch A, Schneider UC, Vajkoczy P, Heppner FL, Grabe N, et al. (2018). Astrocytic glutamine synthetase is expressed in the neuronal somatic layers and down-regulated proportionally to neuronal loss in the human epileptic hippo-campus. Glia 66, 920–933. [DOI] [PubMed] [Google Scholar]
- Reissner KJ, Gipson CD, Tran PK, Knackstedt LA, Scofield MD, and Kalivas PW (2015). Glutamate transporter GLT-1 mediates N-acetylcysteine inhibition of cocaine reinstatement. Addict. Biol 20, 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose CF, Verkhratsky A, and Parpura V (2013). Astrocyte glutamine synthetase: pivotal in health and disease. Biochem. Soc. Trans 41, 1518–1524. [DOI] [PubMed] [Google Scholar]
- Rubio FJ, Li X, Liu QR, Cimbro R, and Hope BT (2016). Fluorescence Activated Cell Sorting (FACS) and Gene Expression Analysis of Fos-expressing Neurons from Fresh and Frozen Rat Brain Tissue. J. Vis. Exp Published online August 27, 2016 10.3791/54358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler R, and Rothstein JD (2006). Regulation and dysregulation of gluta-mate transporters In Neurotransmitter Transporters, Sitte HH and Freissmuth M, eds. (Springer; ), pp. 277–303. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe A (2019). Metabolic signaling in the brain and the role of astrocytes in control of glutamate and GABA neurotransmission. Neurosci. Lett 689, 11–13. [DOI] [PubMed] [Google Scholar]
- Schousboe A, Bak LK, and Waagepetersen HS (2013). Astrocytic Control of Biosynthesis and Turnover of the Neurotransmitters Glutamate and GABA. Front. Endocrinol 4, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopelliti AJ, Font J, Vandenberg RJ, Boudker O, and Ryan RM (2018). Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nat. Commun 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon AL, and Robinson MB (2007). The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem. Int 51, 333–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, and Kalivas PW (2017). Glutamate Transport: A New Bench to Bedside Mechanism for Treating Drug Abuse. Int. J. Neuropsychopharmacol 20, 797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spodenkiewicz M, Diez-Fernandez C, Rüfenacht V, Gemperle-Britschgi C, and Häberle J (2016). Minireview on Glutamine Synthetase Deficiency, an Ultra-Rare Inborn Error of Amino Acid Biosynthesis. Biology (Basel) 5, E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cornwell A, Li J, Peng S, Osorio MJ, Aalling N, Wang S, Benraiss A, Lou N, Goldman SA, and Nedergaard M (2017). SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J. Neurosci 37, 4493–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi J, Nitta N, Iwasaki N, Saito S, Tanaka R, Barkovich AJ, and Aoki I (2014). Neurochemistry in shiverer mouse depicted on MR spectroscopy. J. Magn. Reson. Imaging 39, 1550–1557. [DOI] [PubMed] [Google Scholar]
- Takasaki C, Yamasaki M, Uchigashima M, Konno K, Yanagawa Y, and Watanabe M (2010). Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur. J. Neurosci 32, 1326–1336. [DOI] [PubMed] [Google Scholar]
- Tani H, Dulla CG, Farzampour Z, Taylor-Weiner A, Huguenard JR, and Reimer RJ (2014). A local glutamate-glutamine cycle sustains synaptic excitatory transmitter release. Neuron 81, 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilas P, Steinhäuser C, Theis M, and Derouiche A (2017). Morphological study of a connexin 43-GFP reporter mouse highlights glial heterogeneity, amacrine cells, and olfactory ensheathing cells. J. Neurosci. Res 95, 2182–2194. [DOI] [PubMed] [Google Scholar]
- Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, Anderson MA, Mody I, Olsen ML, Sofroniew MV, and Khakh BS (2014). Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci 17, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi Y, Amri M, Becq H, Molinari F, and Aniksztejn L (2017). The conversion of glutamate by glutamine synthase in neocortical astrocytes from juvenile rat is important to limit glutamate spillover and peri/extrasynaptic activation of NMDA receptors. Glia 65, 401–415. [DOI] [PubMed] [Google Scholar]
- Ungless MA, Whistler JL, Malenka RC, and Bonci A (2001). Single cocaine exposure in vivo induces long-term potentiation in dopamine neurons. Nature 411, 583–587. [DOI] [PubMed] [Google Scholar]
- van der Hel WS, Notenboom RGE, Bos IWM, van Rijen PC, van Veelen CWM, and de Graan PNE (2005). Reduced glutamine synthetase in hippocampal areas with neuron loss in temporal lobe epilepsy. Neurology 64, 326–333. [DOI] [PubMed] [Google Scholar]
- Wanat MJ, Hopf FW, Stuber GD, Phillips PE, and Bonci A (2008). Corticotropin-releasing factor increases mouse ventral tegmental area dopa-mine neuron firing through a protein kinase C-dependent enhancement of IH. J. Physiol 586, 2157–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, You Z-B, and Wise RA (2012). Heroin self-administration experience establishes control of ventral tegmental glutamate release by stress and environmental stimuli. Neuropsychopharmacology 37, 2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner P, Pitt D, and Raine CS (2001). Multiple sclerosis: altered gluta-mate homeostasis in lesions correlates with oligodendrocyte and axonal damage. Ann. Neurol 50, 169–180. [DOI] [PubMed] [Google Scholar]
- Xin W, Schuebel KE, Jair K, Cimbro R, Biase LMD, Goldman D, and Bonci A (2019). Ventral midbrain astrocytes display unique physiological features and sensitivity to dopamine D2 receptor signaling. Neuropsychopharmacology 44, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Zhang Z-W, and Li Z-L (2017). Cell Death-Autophagy Loop and Glutamate-Glutamine Cycle in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci 10, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.