Abstract
GFP-like proteins from lancelets (lanFPs) is a new and least studied group that already generated several outstanding biomarkers (mNeonGreen is the brightest FP to date) and has some unique features. Here, we report the study of four homologous lanFPs with GYG and GYA chromophores. Until recently, it was accepted that the third chromophore-forming residue in GFP-like proteins should be glycine and efforts to replace it were in vain. Now, we have the first structure of a fluorescent protein with a successfully matured chromophore that has alanine as the third chromophore-forming residue. Consideration of the protein structures revealed two alternative routes of posttranslational transformation, resulting in either chromophore maturation or hydrolysis of GYG/GYA tripeptide. Both transformations are catalyzed by the same set of catalytic residues, Arg88 and Glu35-Wat-Glu211 cluster, whereas the residues in positions 62 and 102 shift the equilibrium between chromophore maturation and hydrolysis.
Keywords: Fluorescent protein, Lancelet, Branchiostoma floridae, G67A mutation, GFP-like proteins, X-ray crystal structure
Graphical Abstract
INTRODUCTION
The discovery of green fluorescent protein (GFP) has revolutionized the field of molecular biology and biochemistry. Its ability to be expressed in the cells coupled with the unique autocatalytic formation of the chromophore enabled GFP use in a large number of imaging experiments in vivo in real time. Since their first biomarker application in 19941, many new GFP-like proteins with different fluorescence colors have been added to the arsenal of non-invasive biomarkers. To date, fluorescent biomarkers found numerous applications ranging from marking gene activity and protein labeling to tracking whole cells in tissues and GFP-based sensor applications2–3. Development of new equipment and methods of imaging on the one hand and discovery of FPs with new unusual properties on the other (photoactivable FPs, photoswitchable FPs, timers, etc.3) advanced the design of fluorescent biomarkers.
Even though the GFP-like proteins are often derived from completely unrelated species and sometimes have low sequence homology, they exhibit very little variation in the tertiary structure4. The general fold of GFP is an 11-stranded β-barrel with an internal α-helix wound around its principal axis. The chromophore is located in the center of the α-helix, shielded by the tightly packed strands of β-barrel. The encapsulation is doubly beneficial for the fluorescence quantum yield as it protects the chromophore from quenching by water and molecular oxygen and provides a barrier to non-radiative conformational relaxation5. The chromophore matures autocatalytically without the help of external cofactors or enzymes; in original avGFP, it formed from the internal Ser65-Tyr66-Gly67 tripeptide. Chromophore maturation is a three-step process, comprising cyclization, dehydration, and oxidation6–8. Cyclization and dehydration take minutes to complete, whereas the rate-limiting oxidation typically takes hours9–10 and is, in fact, a multistep transformation, including the formation of enolate and peroxy intermediates11–12.
For a long time, it was believed that functional chromophore could be formed from various tripeptides, where the first amino acid residue could be any residue, the second should be an aromatic residue, and the third must be Gly11. Variation of the first two residues, even though the second is limited to four aromatic amino acids, resulted in a vast amount of GFP-like FPs with an emission wavelength range from 430 to 670 nm. Although the second residue of the chromophore-forming triade has to be aromatic to make it fluorescent, chromophore maturation still occurs for non-fluorescent variants with Ser, Leu, and Gly in its place4, 12–14. With several distinct types of chromophore chemistry and structure (e.g., avGFP, Kaede, DsRed, etc.), the third chromophore-forming residue remained unchanged. The attempts to replace Gly67 with the next smallest residue Ala, either destroyed the fluorescence, yielded unstable FPs15, or prevented chromophore maturation16. A thorough direct structural study of GFP variants with non-canonical chromophore tripeptides undertaken by Barondeau et al.11 concluded that substitution Gly67Ala impairs chromophore formation due to steric rather than conformational restrictions imposed by the side chain of Ala67 that has a significant van der Waals collision with the Thr63 carbonyl oxygen12. Thus, for a long time, Gly67 was deemed the only residue that allows a central α-helix with a kinked conformation required for maturation of the chromophore. In this arrangement, the amide nitrogen of Gly67 closely approaches the carbonyl carbon of the residue 65 enabling a nucleophilic attack. It brings Gly67 amide (N) and Thr65 carbonyl into van der Waals contact and does so with the correct orbital orientation, aligning the lone pair of the Gly67 amide N with the C=O π*-orbital. Thus, the FP architecture acts as an enzyme, actively positioning substrate atoms in the reactive conformation, hence reducing the entropic barrier to cyclization5.
Recently, several authors reported a new class of natural GFP-like proteins with non-canonical Gly-Tyr-Gly chromophore found in the genome of the lancelets Branchiostoma lanceolatum and Branchiostoma floridae17–20; this group of FPs had been called lanFPs. While FPs from chordates (including lanFPs) and cnidarians (FPs from corals, anemones, and jellyfish) relate only distantly and share ~20% sequence identity, they have the same fold and the sequence of post-translational events resulting in a fully functional chromophore. However, lanFPs are full of surprises. For example, green, yellow, and red lanFPs with the same Gly-Tyr-Gly chromophore forming tripeptide have drastically different photophysical properties due to the differences in the immediate environment of the chromophore. Although they have a conventional GFP-like chromophore core of conjugated imidazolidone and p-hydroxybenzylidene rings, the maxima of fluorescence bands for laGFP, lanYFP, and laRFP are 502/511, 513/524, and 521/592 nm, respectively. LaGFP has narrow excitation and emission peaks, lanYFP has an unusually high quantum yield (0.95) and extinction coefficient (125,000 M−1cm−1), while laRFP chromophore forms an additional covalent bond with the protein matrix between its Cβ2 atom and hydroxyl oxygen of Tyr6220. Naturally occurring bfloGFPa1 (QY 1.0, EC 120,000 M−1cm−1)21 and monomeric mNeonGreen (QY 0.8, EC 116,000 M−1cm−1)22, genetically engineered from lanYFP are nearly three times brighter than mEGFP, the brightness of which was considered a paramount. LanFPs, thus, present a promising template for the generation of new bright FPs and expansion of the chromophore chemistry.
Some recently reported FPs from Branchiostoma floridae have Ala in the third chromophore-forming position instead of “mandatory” Gly and demonstrate a typical behavior of FPs with a fully functional chromophore23. Here we obtained and analyzed the crystal structures, of the four, recently characterized lanFPs: lanFP6G, lanFP6A, lanFP10G, and lanFP10A and carried out mutagenesis experiments to validate the structural factors enabling the stable GFP-like FP with Ala as the third chromophore-forming residue.
MATERIALS AND METHODS
We obtained lanFP6G, lanFP6A, lanFP10G, and lanFP10A proteins, first reported by Baumann et al.18, following the procedures previously described by Roldán-Salgado et al.23 Mutagenesis experiments were carried out using the overlap PCR approach, and the modified genes were cloned as NdeI/XhoI inserts into the homemade pJOQ vector. The UV/Vis and fluorescence spectra of the proteins were recorded in PBS on nanophotometer NP80 (Implen) and the luminescence spectrometer LS55 (Perkin Elmer), respectively. The total concentration of the protein was determined from the intensity of 280 nm absorbance band, and the fraction of matured chromophore was estimated from the intensity of the respective chromophore absorbance bands: 385 and 496 (lanFP6A/G) and 460 nm (lanFP10A/G).
Crystallization
For crystallization, the proteins were transferred to a PBS buffer containing 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM K2HPO4 pH 7.4 and were concentrated to 15 – 31 mg/ml. An initial search for crystallization conditions was carried out using a Mosquito Robotic Crystallization System (TTP LabTech Ltd). In its standard setup robot mixed 0.4 μl of protein with 0.4 μl of well solution and the drops were further incubated against 140 μl of the same reservoir solution at 20 °C for two days. All proteins were tested against Classic, Index, SaltRX, PEG/Ion, PEGRx, and Grid Screen Salt crystallization screens (Hampton Research). To preserve the fluorescent state of the proteins, we used only neutral pH hits for further optimization. Large-scale crystallization was set up by the hanging drop vapor diffusion method at room temperature (20°C). Typically, 2 μl of the protein was mixed with the same amount of well solution and incubated against 0.5 ml of the same well solution for two weeks. The crystallization conditions are given in Table 1S.
Diffraction data collection and processing
X-ray diffraction data were collected at the Advanced Photon Source on SER-CAT 22-BM beamline (Argonne National Laboratory, Argonne, IL). Diffraction intensities were registered on a MAR 225 CCD detector (Rayonix). Before data collection, the crystals were incubated in a cryoprotecting solution consisting of 20% glycerol and 80% of well solution for 10–15 seconds and were flash-frozen in 100 K nitrogen stream. The cryogenic temperature was maintained by a CryoJetXL cooling device (Oxford Cryosystems). Diffraction images were indexed, integrated and scaled with the HKL2000 software24. For data processing statistics see Table 2S.
Structure solution and refinement
The structures of all proteins were solved by molecular replacement method with MOLREP25 using a single monomer of the green fluorescent protein from the lancelet Branchiostoma lanceolatum laGFP (PDB ID: 4HVF)20, excluding its chromophore as a search model. Structure refinement was performed with REFMAC26, COOT27, and PHENIX.REFINE28. Manual structure rebuilding and the addition of ordered solvent molecules were done using COOT. Structure validation was performed with COOT and PROCHECK29, and the refinement statistics are given in Table 3S. We deposited the coordinates and structure factors in the Protein Data Bank under accession codes 6M9Z, 6M9Y, 6MAS, and 6M9X, for lanFP6G, lanFP6A, lanFP10G, and lanFP10A, respectively.
RESULTS
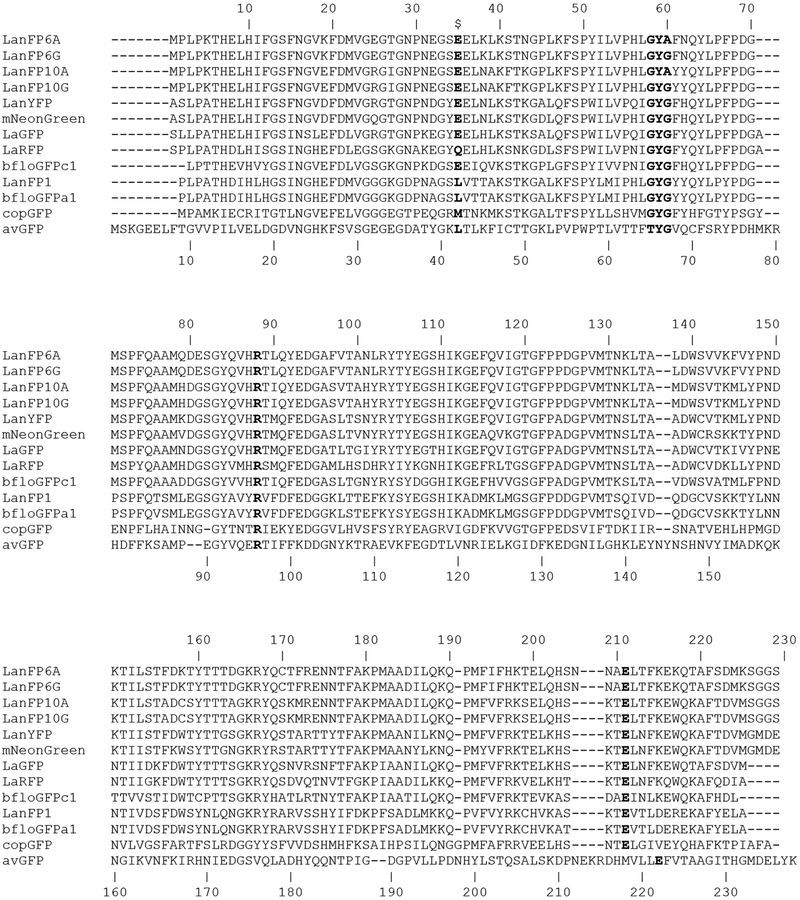
We were fortunate to crystallize a group of four lanFPs, which form two pairs sharing 83.5% of the sequence identity (Fig. 1). The members of each pair, lanFP6G/A, and lanFP10G/A are entirely the same, except for Gly/Ala in position 60 (the third chromophore-forming residue, position 67 in avGFP numbering). The structures of lanFP6G, lanFP6A, lanFP10G, and lanFP10A have been solved at 1.20, 1.35, 1.30, and 1.80Å resolution, respectively (Tables 2S and 3S). The electron density is well defined for all of the structures, and the models are of good quality, as indicated by the refinement statistics (Table 3S). Despite a low sequence identity with avGFP (~20%), lanFPs have the same fold: an 11-stranded β-barrel with an α-helix intertwined around the central axis of the barrel and the chromophore located in the center of α-helix. Like in avGFP, the central α-helix is severely distorted which is a prerequisite for the autocatalytic formation of the chromophore.
Figure 1.
Alignment of the amino acid sequences of twelve lanFPs reported to date and avGFP. Residues in the nearest environment to the chromophore in the lanFP6G/A and lanFP10G/A discussed in the paper are shown in bold.
LanFP6G and lanFP6A
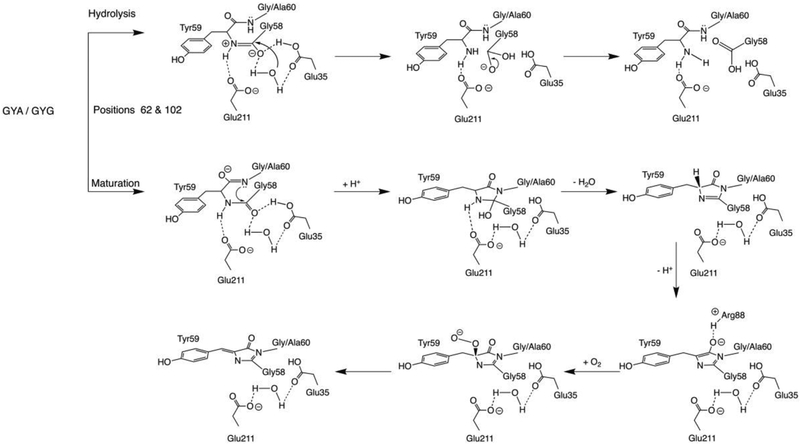
In lanFP6G, post-translational chemistry follows two alternative routes—formation of the classic green chromophore and hydrolysis of the peptide bond between the first and the second chromophore-forming residues (Gly58-Tyr59). Once formed, the chromophore of lanFP6G remains stable, and the protein solution stored at +4° C retains its color and fluorescence for over a year. Formation of lanFP6G chromophore most likely follows the classic route for GFP6, 12. It starts with an attack of the lone pair of the main chain nitrogen of Gly60 (N60) at the carbonyl carbon of Gly58, resulting in the formation of a new C-N bond, subsequent dehydration of the newly formed heterocycle, and oxidation of Cα-Cβ bond of Tyr59.
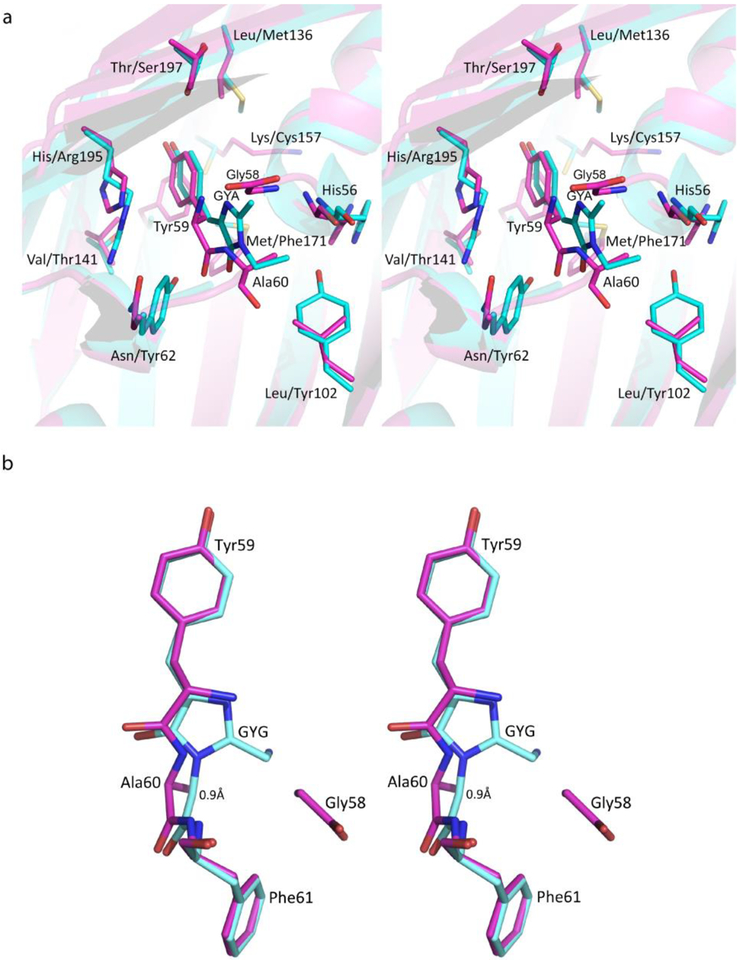
LanFP6A differs from lanFP6G by single amino acid in position 60, Ala instead of Gly. This single-point variation results in a 100% hydrolysis of the Gly-Tyr-Ala tripeptide and a non-fluorescent protein (Fig. 2 and Table 1). Superposition of lanFP6A and lanFP6G structures showed a shift of Ala60 Cα atom by ~ 0.9 Å impairing chromophore formation (Fig. 3). The major cause of the shift is a stereochemical conflict between the side chain of Ala60 and the carbonyl oxygen of His56. Even for hydrolyzed Gly-Tyr-Ala triad, for which hydrolysis has partially resolved the conflict, the distance between the Cβ atom of Ala60 and carbonyl oxygen of His56 remained 3.4 Å. Note that in both lanFP6G and lanFP6A in hydrolyzed GYG/GYA tripeptide Tyr59 adopts two different conformations as the main chain of Gly58 flips 180°. One of them coincides with the position of the matured chromophore in lanFP6G, whereas the other has a distinctly different positioning of Tyr59 arising from hula twist of its side chain.
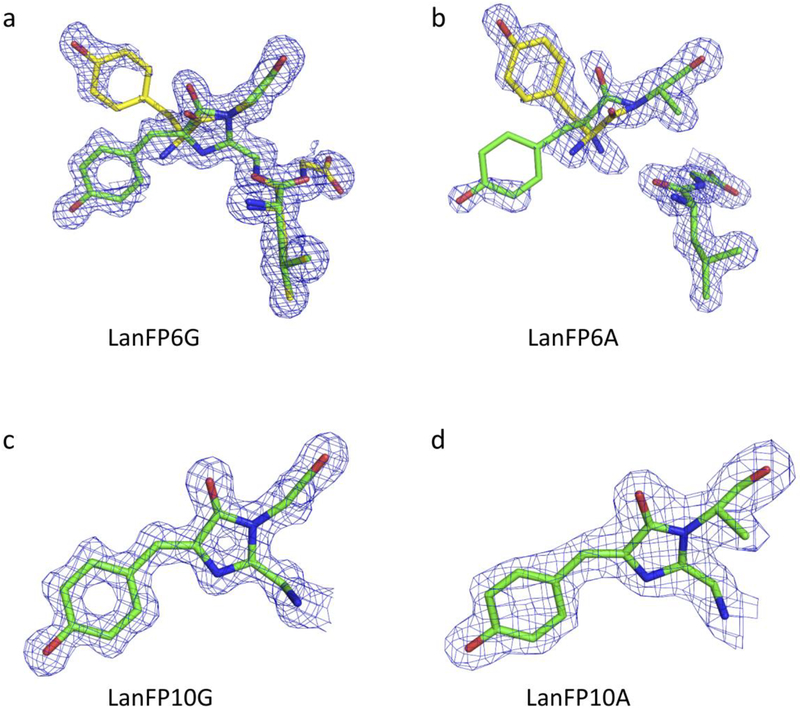
Figure 2.
2Fo-Fc electron density for the chromophores of lanFP6G, lanFP6A, lanFP10G, and lanFP10A at 1σ-level: (a) showing the presence of two species in lanFP6G corresponding to hydrolyzed GYG tripeptide and matured GYG chromophore; (b) showing the presence of two conformations of hydrolyzed GYG tripeptide in lanFP6A; and (c) and (d) showing the presence of a single species of mature chromophore in lanFP10G and lanFP10A.
Table 1.
Photophysical properties of lanFP6A/G, lanFP10A/G, and their mutants.
| Variant | λabs (nm) | λex (nm) | λem (nm) |
|---|---|---|---|
| lanFP6A | Non-fluorescent protein | ||
| lanFP6A/L102Y | Non-fluorescent protein | ||
| lanFP6A/N62Y | 385, 494 | 499 | 519 |
| lanFP6A/N62Y/L102Y | 385, 494 | 494 | 515 |
| lanFP6A/E35A/E211A | Non-fluorescent protein | ||
| lanFP6A/E35Q/E211Q | 390, 490 | 497 | 515 |
| lanFP6G | 385, 496 | 501 | 519 |
| lanFP6G/E35A | 380, 493 | 510 | 533 |
| lanFP6G/E211A | 385, 434 | 474 | 519 |
| lanFP6G/E35A/E211A | Non-fluorescent protein | ||
| lanFP6G/E35Q/E211Q | 390, 490 | 506 | 519 |
| lanFP10G | 462 | 492 | 502 |
| lanFP10A | 462 | 492 | 502 |
| lanFP10A/Y62N | 320 (shoulder) | 361 | 441 |
| lanFP10A/Y102L | 472 | 499 | 510 |
| lanFP10A/Y62N/Y102L | 450 | 489 | 498 |
| lanFP10A/E35A | 469 | 495 | 510 |
| lanFP10A/E211A | Non-fluorescent protein | ||
Figure 3.
Immediate chromophore environment and geometry. (a) Key residues and the immediate chromophore environment of lanFP6A (magenta; His56, Asn62, and Leu102) and lanFP10A (cyan; His56, Tyr62, and Tyr102) chromophore. (b) The geometry of the chromophore: 0.9 Å shift of the main chain of Ala60 in lanFP6A (magenta) relative to that of Gly60 in lanFP6G (cyan) arising from Ala60 steric conflict with His56.
LanFP10G and lanFP10A
Two other lancelet FPs, lanFP10G and lanFP10A are both fluorescent and have a fully matured Gly-Tyr-Gly or Gly-Tyr-Ala chromophore, respectively (See Fig. 4, and Table 1). They are completely identical aside from the third chromophore-forming residue. Unlike lanFP6A, lanFP10A contains Tyr102 (Leu102 in lanFP6A). The bulky side chain of Tyr102 pushes His56 away from Ala60 by ~1 Å resolving the stereochemical conflict between Additionally, the bulky side chain of Tyr62 (Asn62 in lanFP6A) pushes chromophore-forming Tyr59 along the α-helix axis towards its N-terminal end closing the distance between Ala60(N) and Gly58(C=O) and favoring tripeptide cyclization. Thus, successful maturation of the chromophore with the third chromophore-forming residue Ala is possible but requires sufficient space to resolve stereochemical conflicts in its immediate environment. In lanFP10G, Tyr62 shifts cyclization-hydrolysis equilibrium towards chromophore formation and suppresses hydrolysis.
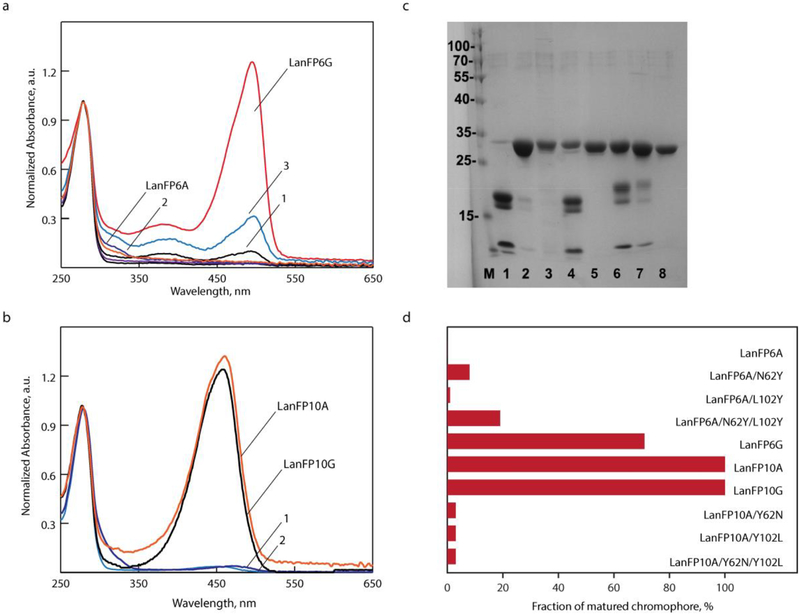
Figure 4.
Characterization of lanFP6G, lanFP6A, and lanFP10A mutants testing positions 62 and 102. (a) Absorbance spectra of lanFP6G and lanFP6A (1) lanFP6A/N62Y; (2) lanFP6A/L102Y; and (3) lanFP6A/N62Y/L102Y) and (b) lanFP10A mutants at pH 7.5 ((1) lanFP10A/Y102L and (2) lanFP10A/Y62N/Y102L). Spectra are normalized to the absorbance at 280 nm for each protein. (c) Denaturating SDS-PAGE gel: (1) lanFP6A, (2) lanFP6A/N62Y, (3) lanFP6A/L102Y, (4) lanFP6A/N62Y/L102Y, (5) lanFP10A, (6) lanFP10A/Y62N, (7) lanFP10A/Y102L, and (8) lanFP10A/Y62N/Y102L. (d) Fraction of matured chromophore in lanFP6A and lanFP10A probing positions 62 and 102.
Key residues enabling maturation of GYA chromophore
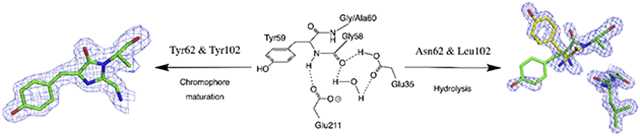
LanFP6A and lanFP10A differ from each other by 37 amino acid residues; 18 of them are very similar, 6 are weakly similar, and 13 are completely different. 6 of 37 residues (61, 62, 102, 136, 155, and 195) are located within 4 Å from the chromophore. Residues 61, 62, and 102 are positioned near the imidazolidone ring, and 136, 155, and 195 are close to the p-hydroxybenzylidene moiety. As both lanFP6A and lanFP10A have an identical arrangement of the catalytic residues, the difference in the outcome of their posttranslational chemistry is presumably determined by the residues adjacent to the imidazolidone ring: Phe61, Asn62, and Leu102 in lanFP6A and Tyr61, Tyr62, and Tyr102 in lanFP10A. The X-ray structure of lanFP10A shows that bulky tyrosines 62 and 102 rearrange the residues near the chromophore and make additional space to accommodate the side chain of Ala60. In lanFP6A, Asn62, and Leu102 are too small to push away His56 and could neither make enough space for the side chain of Ala60 nor position the main chain atoms of GYA triade favorably for the tripeptide cyclization.
Since Phe61 and Tyr61 are turned away from the chromophore-forming tripeptide and could not affect its posttranslational transformations, we concentrated our mutagenesis efforts on the residues 102 and 62 (Fig. 4 and Table 1). In lanFP6A, which is a non-fluorescent protein with a fully hydrolyzed chromophore-forming triad, a single point mutation Leu102Tyr yielded a non-fluorescent variant with suppressed hydrolysis. A single point mutation Asn62Tyr caused a partial restoration of the fluorescence with the same fluorescence bands as for homologous lanFP6G. Combination of Asn62Tyr and Leu102Tyr introduced in lanFP6A further increased the fraction of mature chromophore, confirming the critical role of the two residues (Fig. 4). Introduction of Asn62 and/or Leu102 into lanFP10A resulted in three variants with the drastically reduced amount of mature chromophore and substantial hydrolysis of GYA tripeptide. Surprisingly, a single point mutation in lanFP10A, Tyr62Asn caused partial hydrolysis and yielded in a chromophore with unexpected violet fluorescence (λex/λem of 361/441 nm) and absorbance band that appears as 320 nm shoulder of 280 nm peak (Fig. 4).
Catalytic residues affecting chromophore maturation and hydrolysis
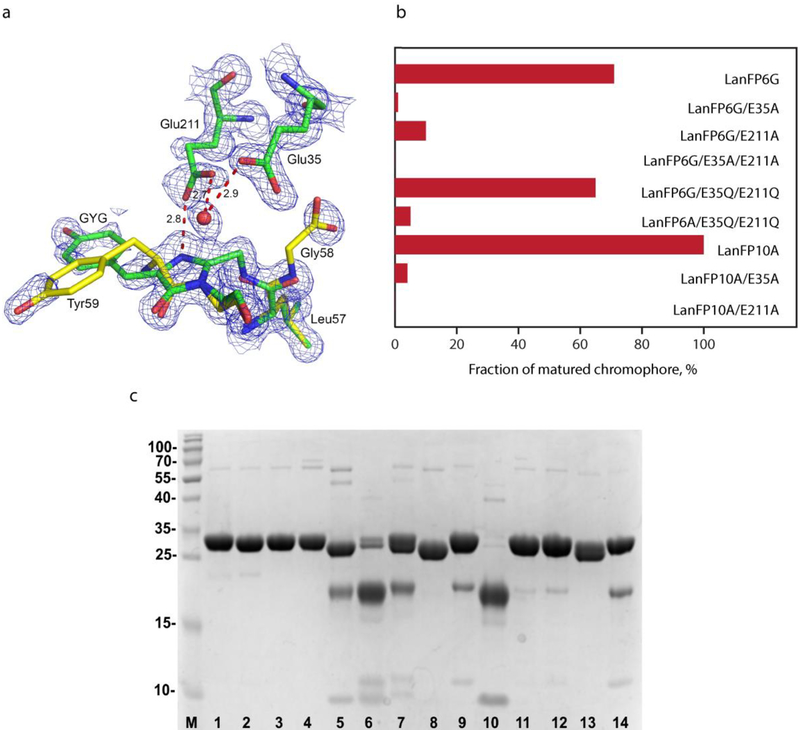
While considering the factors providing for hydrolysis of the chromophore-forming triad in lanFP6G/A, we have noticed the spatial arrangement of the nearby Glu35, Glu211, and the water molecule (Fig. 5). We used mutagenesis (Table 1) to verify the roles of Glu35 (Leu42 in avGFP) and Glu211 (Glu222 in avGFP) in hydrolysis taking place in lanFP6G/A. In lanFP6G, for which posttranslational chemistry results in both chromophore formation and hydrolysis, replacement of either Glu35Ala or Glu211Ala drastically reduced chromophore maturation but did not block hydrolysis; replacement of both Glu with Ala resulted in intact tripeptide. For lanFP10A, substitution of either one or both glutamates with Ala halted maturation and hydrolysis. Replacement of both glutamates with glutamines in lanFP6G/E35Q/E211Q and lanFP6A/E35Q/E211Q resulted in chromophore maturation and partial suppression of hydrolysis. In lanFP6G, glutamines have been nearly as effective as glutamates in promoting chromophore maturation, whereas lanFP6A variant yielded ~5% of GYA chromophore.
Figure 5.
LanFP6G and lanFP10A mutants aimed at positions 35 and 211. (a) LanFP6G chromophore, hydrolyzed tripeptide, and Glu35-Wat-Glu211 cluster. (b) Fraction of matured chromophore for lanFP6A/G and lanFP10A variants. (c) SDS-PAGE gel: (1) lanFP10A, (2) lanFP10A/E35A, (3) lanFP10A/E211A, (4) lanFP10A/E35A/E211A, (5) lanFP6G, (6) lanFP6G/E35A, (7) lanFP6G/E211A, (8) lanFP6G/E35A/E211A, (9) lanFP6G/E35Q/E211Q, (10) lanFP6A, (11) lanFP6A/E35A, (12) lanFP6A/E211A, (13) lanFP6A/E35A/E211A, and (14) lanFP6A/E35Q/E211Q.
DISCUSSION
The primary difference between the two pairs of lanFPs that we considered is the presence of hydrolysis, partial or complete, in lanFP6A/G and its absence in lanFP10A/G. After analyzing X-ray crystal structures that we obtained and carrying out site-specific mutagenesis, we concluded that the amino acid residues in positions 35, 211, 62, and 102 play the key role in the posttranslational chemistry of four examined lanFPs. While the first two residues are important for formation of the chromophore and hydrolysis, two other shift the equilibrium between them.
The extensive studies of cnidarian FPs demonstrated that their posttranslational chemistry is catalyzed by water and two highly conservative residues Arg96 and Glu2227, 11–12, 15. In four examined lanFPs, the set of catalytic residues comprises Arg88 and Glu211-Wat-Glu35 cluster. In lanFP6G and lanFP10A variants, replacement of either of glutamates with Ala tumbled chromophore maturation, indicating essential role of both positions. Alanines in both positions 35 and 211 produced intact chromophore-forming tripeptide. Finally, replacement of both glutamates with glutamines decreased hydrolysis and resulted in chromophore formation with conventional maturation rates, and in case of lanFP6G/E35Q/E211Q, showed yields nearly as good as parental protein (Fig. 5). Recently, we reported laRFP with a Gln35-Wat-Gln211 cluster near the chromophore to be the first wild-type FP without catalytic glutamate that has a conventional maturation rate for green-emitting chromophore intermediate20. The mutually hydrogen-bonded Gln211 and Gln35 form a direct hydrogen bond with the cyclic chromophore unit, whereas Asp142 was identified as a base required for maturation of the chromophore. In lanFP6G/E35Q/E211Q, the only nearby base is Glu173, H-bonded with the chromophore carbonyl through Arg88. Note that other lanFPs do not have a basic group equivalent to Glu173 in lanFP6G/A and lanFP10G/A or Asp142 in laRFP (Fig.1). Thus, in four examined lanFPs, both Glu35 and Glu211 are highly desirable for efficient formation of the chromophore but could be replaced with glutamines without substantial loss in the chromophore maturation efficacy. A pair of glutamates then plays two roles: it aligns carbonyl carbon of Gly58 and amino-group of Gly/Ala60 favorably for tripeptide cyclization and acts as a base. Glutamines, occupying the same position, provide only for tripeptide alignment favorable for cyclization, whereas Glu173 plays the role of a base.
For cnidarian FP, Barondeau et al.11 observed hydrolysis of GAG and GSG chromophore-forming tripeptide between the first and the second chromophore-forming residues for variants with non-aromatic central residue; aromatic amino acids in this position yielded a mature chromophore. The authors pointed out that the same structural features that favor peptide cyclization might also favor peptide hydrolysis. They suggested that the side chain steric interactions for the first chromophore-forming residue determined its propensity to hydrolysis and noted that the side chain larger than Gly inhibits either the conformations necessary for hydrolysis or the approach of water molecules to the carbonyl carbon. In lanFP6G/A, the first chromophore-forming residue is also glycine, but peptide backbone fragmentation occurs for chromophore-forming tripeptide that has an aromatic amino acid in the middle. Recreating an intact GYG/GYA tripeptide in lanFP6A/G, we found that Glu211 forms a direct H-bond with Tyr59 amide nitrogen and carbonyl oxygen of Gly58, whereas Glu35 forms both direct and water-mediated H-bonds with the Gly58 carbonyl oxygen and a water-mediated H-bond with the amide nitrogen of the residue 60. Replacement of either of glutamates with Ala did not stop hydrolysis; replacement of both glutamates with glutamines decreased it but did not stop it either (Fig. 5), and only alanine at both positions preserved the intact tripeptide and showed no signs of hydrolysis. From the structure of laRFP20, we know that the presence of two glutamines at positions 35 and 211 preserves water molecule H-bonded with both of them. By analogy with laRFP, we could expect that in lanFP6G/E35Q/E211Q and lanFP6A/E35Q/E211Q, such water molecule is present as well. When the tripeptide geometry is only moderately favorable or unfavorable for cyclization, the presence of this water molecule results in partial (lanFP6G) or complete (lanFP6A) hydrolysis of the tripeptide. The appearance of hydrolysis in lanFP10A/Y62N and lanFP10A/Y102L corroborates this suggestion as they have chromophore-forming tripeptide in the environment unfavorable for cyclization.
In the examined lanFPs, residue 62 is one of the two residues that determine the course of posttranslational events. The structures of lanFP10A/G revealed that Tyr62 brings Gly58 carbonyl carbon and Gly/Ala60 amide nitrogen closer together in a cyclization-favorable alignment. If a bulky residue occupies position 62, the equilibrium of posttranslational events is shifted towards the formation of the chromophore (lanFP10G). If residue 62 is small, the tripeptide is left to statistical probability and steric conflicts with the nearest environment to determine the outcome of posttranslational events. If carbonyl carbon of Gly58 is attacked by amide nitrogen of the residue 60, the chromophore is formed. If the nearby water molecule attacks carbonyl carbon of Gly58, the chromophore-forming tripeptide undergoes hydrolysis. In case of GYG tripeptide and no bulky residue in position 62, the lack of steric conflict with the immediate chromophore environment shifts the equilibrium towards maturation of the chromophore (~70%), but still gets a sizable amount of the tripeptide hydrolyzed (lanFP6G). In the case of GYA chromophore with an unresolved steric conflict between Ala60 and His56, posttranslational modification outcome shifts entirely towards hydrolysis (lanFP6A). Our observations are in a good agreement with the fact that all lanFPs with successfully matured chromophore reported to date (i.e., in all these lanFPs, the equilibrium is shifted towards chromophore formation), have position 62 occupied by a bulky residue (Fig. 1): Tyr (laGFP, laRFP, copGFP, LanFP1, and bfloGFPa1) or His (lanYFP, mNeonGreen, and bfloGFPc1).
For a long time, it was generally accepted that the chromophore of GFP-like proteins could be formed from the tripeptide in which the first residue could be any, the second residue should be an aromatic, and the third residue must be Gly. Barondeau et al. demonstrated that in avGFP, a modeled Ala side chain for residue 67 had a significant van der Waals collision with the carbonyl oxygen of Thr63 and pointed out that the requirement for the third chromophore-forming Gly comes from steric rather than conformational or chemical limitations11. At present, we crystallized and examined the first GFP-like FP with Ala as the third chromophore-forming residue. We found that the residue essential for maturation of GYA chromophore is Tyr102. In lanFP10A, the presence of Tyr102 resolves the stereochemical conflict between carbonyl group of His56 and methyl group of Ala60, enabling satisfactory accommodation of Ala side chain and chromophore maturation. In combination with a bulky Tyr62, it shifts the hydrolysis-cyclization equilibrium towards chromophore maturation. In the absence of bulky residue 102 in the homologous lanFP6A, the steric conflict between Ala60 and His56 shifts equilibrium towards complete hydrolysis of GYA tripeptide, whereas lanFP10A/Y102L, lacking bulky 102 residue, demonstrated a drastic drop in the chromophore maturation.
Scheme 1 summarizes two possible outcomes of posttranslational modifications in lanFP6A/G and lanFP10A/G. Our findings agree with the conclusions of Barondeau et al. and additionally demonstrate that in GFP-like proteins the third chromophore-forming residue could be different from Gly, provided there is enough space for its side chain. The choice of appropriate amino acids is limited to the residues with a small side chain by the tight nature of the chromophore environment required to preserve the architecture of β-barrel essential for maturation of the chromophore. In all lanFPs reported to date, even with a GYG chromophore, a bulky residue occupies the position 102: Tyr (lanYFP, mNeonGreen, and laGFP, bfloGFPc1), His (laRFP), or Phe (bfloGFPa1, copGFP, and lanFP1)20–22, 30–31, making lanFPs a suitable platform for variation of the third chromophore-forming residues.
Scheme 1.
Posttranslational modifications of GYG/GYA tripeptide in lanFP6A/G and lanFP10A/G with two alternative outcomes: hydrolysis of tripeptide and maturation of the chromophore.
While verifying the role of Tyr62, we obtained a violet-emitting variant lanFP10A/Y62N (λabs 320 nm shoulder to 280 nm peak, λex/λem 360/440 nm, Fig. 1S). The lack of absorbance in the visible range indicate the impaired formation of the imidazolidone ring of the chromophore, yet the presence of violet fluorescence points at the conjugation system extended beyond Tyr59. Unfortunately, poor resolution of 320 nm band that overlaps with 280 nm peak does not permit an adequate quantification of the matured chromophore. SDS-PAGE gel indicates that a substantial part of lanFP10A/Y62N undergoes hydrolysis of the chromophore-forming tripeptide. At present, we do not know the exact structure of the chromophore accountable for the violet fluorescence, but its properties indicate that with further optimization it could serve as an interesting template for generation violet-emitting FPs.
CONCLUSIONS
Here, we systematically considered four lanFPs: lanFP6G, lanFP6A, lanFP10G, and lanFP10A. We discovered an active role of Glu35-Wat-Glu211 cluster in the posttranslational transformations, resulting in either maturation of the chromophore or hydrolysis of the chromophore-forming tripeptide. We also found that the equilibrium between the chromophore maturation and hydrolysis is shifted towards chromophore maturation when a bulky residue occupies position 62, Tyr or His (in some lanFPs). The absence of bulky residue 62 results in lanFPs with the chromophore-forming tripeptide prone to hydrolysis. We demonstrated that maturation of GFP-like FP with Ala as the third chromophore-forming residue requires resolution of the stereochemical conflict between Ala side chain and the immediate chromophore environment. In our particular case, this was achieved by Tyr102 that resolved the stereochemical conflict between Ala60 and the main chain carbonyl oxygen of the nearby His56, making space to accommodate the side chain of Ala60.
Supplementary Material
Highlights.
GFP-like fluorescent proteins from lancelet Branchiostoma floridae (lanFPs) have a mature chromophore with alanine as the third chromophore-forming residue (GYA).
In lanFPs, there are two alternative routes of posttranslational transformation, resulting in either chromophore maturation or hydrolysis of GYG/GYA tripeptide.
Both transformations are catalyzed by the same set of residues, Arg88 and Glu211-Wat-Glu35 cluster present in most lanFPs.
The residues in positions 62 and 102 rule the chromophore maturation/hydrolysis equilibrium.
ACKNOWLEDGEMENTS
Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, and Office of Basic Energy Sciences under Contract No.W-31-109-Eng-38. This project was also funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH contract HHSN261200800001E, and the Intramural Research Program of the NIH, Frederick National Laboratory, Center for Cancer Research as well as with the grant from Russian Foundation for Basic Research (grant 19-04-00107). Oligonucleotide synthesis provided by Eugenio López-Bustos and Santiago Becerra-Ramírez, as well as DNA sequencing provided by Jorge A. Yáñez and Ana Yanci Alarcón are highly appreciated. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPORTING INFORMATION
Supporting information for this paper is available free of charge on the JMB Publications website.
REFERENCES
- 1.Chalfie M; Tu Y; Euskirchen G; Ward WW; Prasher DC, Green fluorescent protein as a marker for gene expression. Science 1994, 263 (5148), 802–5. [DOI] [PubMed] [Google Scholar]
- 2.Wiedenmann J; Oswald F; Nienhaus GU, Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges. IUBMB Life 2009, 61 (11), 1029–42. [DOI] [PubMed] [Google Scholar]
- 3.Chudakov DM; Matz MV; Lukyanov S; Lukyanov KA, Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev 2010, 90 (3), 1103–63. [DOI] [PubMed] [Google Scholar]
- 4.Ong WJ; Alvarez S; Leroux IE; Shahid RS; Samma AA; Peshkepija P; Morgan AL; Mulcahy S; Zimmer M, Function and structure of GFP-like proteins in the protein data bank. Mol Biosyst 2011, 7 (4), 984–92. [DOI] [PubMed] [Google Scholar]
- 5.Craggs TD, Green fluorescent protein: structure, folding and chromophore maturation. Chem Soc Rev 2009, 38 (10), 2865–75. [DOI] [PubMed] [Google Scholar]
- 6.Cubitt AB; Heim R; Adams SR; Boyd AE; Gross LA; Tsien RY, Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 1995, 20 (11), 448–55. [DOI] [PubMed] [Google Scholar]
- 7.Barondeau DP; Tainer JA; Getzoff ED, Structural evidence for an enolate intermediate in GFP fluorophore biosynthesis. J Am Chem Soc 2006, 128 (10), 3166–8. [DOI] [PubMed] [Google Scholar]
- 8.Pletneva NV; Pletnev VZ; Lukyanov KA; Gurskaya NG; Goryacheva EA; Martynov VI; Wlodawer A; Dauter Z; Pletnev S, Structural evidence for a dehydrated intermediate in green fluorescent protein chromophore biosynthesis. J Biol Chem 2010, 285 (21), 15978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heim R; Cubitt AB; Tsien RY, Improved green fluorescence. Nature 1995, 373 (6516), 663–4. [DOI] [PubMed] [Google Scholar]
- 10.Reid BG; Flynn GC, Chromophore formation in green fluorescent protein. Biochemistry 1997, 36 (22), 6786–91. [DOI] [PubMed] [Google Scholar]
- 11.Barondeau DP; Kassmann CJ; Tainer JA; Getzoff ED, Understanding GFP posttranslational chemistry: structures of designed variants that achieve backbone fragmentation, hydrolysis, and decarboxylation. J Am Chem Soc 2006, 128 (14), 4685–93. [DOI] [PubMed] [Google Scholar]
- 12.Barondeau DP; Putnam CD; Kassmann CJ; Tainer JA; Getzoff ED, Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc Natl Acad Sci U S A 2003, 100 (21), 12111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barondeau DP; Kassmann CJ; Tainer JA; Getzoff ED, Understanding GFP chromophore biosynthesis: controlling backbone cyclization and modifying post-translational chemistry. Biochemistry 2005, 44 (6), 1960–70. [DOI] [PubMed] [Google Scholar]
- 14.Rosenow MA; Huffman HA; Phail ME; Wachter RM, The crystal structure of the Y66L variant of green fluorescent protein supports a cyclization-oxidation-dehydration mechanism for chromophore maturation. Biochemistry 2004, 43 (15), 4464–72. [DOI] [PubMed] [Google Scholar]
- 15.Sniegowski JA; Lappe JW; Patel HN; Huffman HA; Wachter RM, Base catalysis of chromophore formation in Arg96 and Glu222 variants of green fluorescent protein. J Biol Chem 2005, 280 (28), 26248–55. [DOI] [PubMed] [Google Scholar]
- 16.Wood TI; Barondeau DP; Hitomi C; Kassmann CJ; Tainer JA; Getzoff ED, Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis. Biochemistry 2005, 44 (49), 16211–20. [DOI] [PubMed] [Google Scholar]
- 17.Deheyn DD; Kubokawa K; McCarthy JK; Murakami A; Porrachia M; Rouse GW; Holland ND, Endogenous green fluorescent protein (GFP) in amphioxus. Biol Bull 2007, 213 (2), 95–100. [DOI] [PubMed] [Google Scholar]
- 18.Baumann D; Cook M; Ma L; Mushegian A; Sanders E; Schwartz J; Yu CR, A family of GFP-like proteins with different spectral properties in lancelet Branchiostoma floridae. Biology Direct 2008, 3 (1), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomati EK; Manning G; Deheyn DD, Amphioxus encodes the largest known family of green fluorescent proteins, which have diversified into distinct functional classes. BMC Evol Biol 2009, 9, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pletnev VZ; Pletneva NV; Lukyanov KA; Souslova EA; Fradkov AF; Chudakov DM; Chepurnykh T; Yampolsky IV; Wlodawer A; Dauter Z; Pletnev S, Structure of the red fluorescent protein from a lancelet (Branchiostoma lanceolatum): a novel GYG chromophore covalently bound to a nearby tyrosine. Acta Crystallogr D Biol Crystallogr 2013, 69 (Pt 9), 1850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bomati EK; Haley JE; Noel JP; Deheyn DD, Spectral and structural comparison between bright and dim green fluorescent proteins in Amphioxus. Sci Rep 2014, 4, 5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaner NC; Lambert GG; Chammas A; Ni Y; Cranfill PJ; Baird MA; Sell BR; Allen JR; Day RN; Israelsson M; Davidson MW; Wang J, A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 2013, 10 (5), 407–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roldan-Salgado A; Sanchez-Barreto C; Gaytan P, LanFP10-A, first functional fluorescent protein whose chromophore contains the elusive mutation G67A. Gene 2016, 592 (2), 281–90. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z; Minor W, Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 25.Vagin A; Teplyakov A, Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 1), 22–5. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN; Skubak P; Lebedev AA; Pannu NS; Steiner RA; Nicholls RA; Winn MD; Long F; Vagin AA, REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011, 67 (Pt 4), 355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P; Lohkamp B; Scott WG; Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 2), 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laskowski RA; Macarthur MW; Moss DS; Thornton JM, Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography 1993, 26, 283–291. [Google Scholar]
- 30.Lin MZ; McKeown MR; Ng HL; Aguilera TA; Shaner NC; Campbell RE; Adams SR; Gross LA; Ma W; Alber T; Tsien RY, Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol 2009, 16 (11), 1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilmann PG; Battad J; Petersen J; Wilce MC; Dove S; Devenish RJ; Prescott M; Rossjohn J, The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily. J Mol Biol 2006, 359 (4), 890–900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.