Abstract
Plant bacterial diseases are routinely managed with scheduled treatments based on heavy metal compounds or on antibiotics; to reduce the negative environmental impact due to the use of such chemical compounds, as pollution or selection of antibiotic resistant pathogens, the integrated control management is required. In the frame of a sustainable agriculture the use of bacterial antagonists, biological agents, plant defence response elicitors or resistant host plant genotypes are the most effective approaches. In this work, cold atmospheric pressure plasma (CAP) was applied to sterile distilled water, inducing the production of a hydrogen peroxide, nitrite and nitrate, and a pH reduction. In particular, an atmospheric pressure dielectric barrier discharge (DBD) has been used to produce plasma activated water (PAW), that was firstly assayed in in vitro experiments and then in planta through application at the root apparatus of tomato plants, against Xanthomonas vesicatoria (Xv), the etiological agent of bacterial leaf spot. Moreover, the transcription abundance of five genes related to the plant defense was investigated in response to PAW treatment.
PAW did not show direct antimicrobial activity against Xv in in vitro experiments, but it enhanced the tomato plants defenses. It was effective in reducing the disease severity by giving relative protections of ca. 61, 51 and 38% when applied 1 h, 24 h and 6 days before the experimental inoculation, respectively. In addition, the experiments highlighted the pal gene involvement in response to the PAW treatments and against the pathogen; its transcription levels resulted significantly high from 1 to 48 h until their decrease 192 h after PAW application.
Introduction
Bacterial leaf spot of tomato caused by Xanthomonas vesicatoria (Xv), is widely spread in all the areas where tomato is cultivated [1,2]. The lack of effective pesticides or resistant tomato cultivars for the management of this bacterial disease, stimulates the efforts to develop sustainable strategies in the framework of the integrated control management strategies [3,4]. The control programmes for bacterial diseases of tomato plants are mainly based on prophylaxis trough diagnostic analysis carried out on seeds, to detect the pathogen presence (latent infections) [5]. Moreover, a certain degree of seed sanitation is achieved through the application of fermentation methods during the seed extraction from tomatoes and/or through physical disinfection procedures (i.e. heat treatments) directly applied to the seeds batches [6,7]. In glasshouse and open field, preventive treatments applied to the plants are based on copper compounds, antibiotics (where it is allowed) and antagonistic bacteria, that are able to reduce the pathogen population and avoid its penetration inside the plant. However, microbial resistance to copper/antibiotics poses a threat to the continued successful use of copper/antibiotics sprays for disease control [8–10]. Therefore, alternative sustainable control methods, as the use of nanoparticles or natural compounds directly active against Xv were studied and employed to control the infections [11–15]. Moreover, since the end of the nineties, the efficacy and the mechanism of action of resistance inducer molecules, acting against the pathogens through responses mediated by the host, were investigated as an alternative to the use of copper/antibiotics compounds [16]. Among all known resistance inducers, those based on the active principle acibenzolar-S-methyl (ASM), a benzo-thiadiazole (BTH) derivative, were found effective against bacterial plant diseases by strengthening the physical barriers, the pre-infectional defenses (e.g. lignins and callose production), and by producing several compounds, directly active against the pathogens (post-infectional defenses). ASM, in fact, mimics the role of salycilic acid (SA), the natural plant activator of systemic acquired resistance (SAR), which culminates in the expression of pathogenesis related proteins (PRs), in particular the PR1a is also kept as SAR marker [17]; through synergistic cross-talking pathways, ASM is also able to elicit the productions of PRs and compounds, dependant to the jasmonic acid/ethylene (JA/ET) pathway, which brings to induced systemic resistance (ISR) [18].
Those two principal defense pathways, following an oxidative burst through the production of reactive oxygen and nitrogen species (RONS), representing the earliest responses of plants to pathogen inoculation and leading to the hypersensitive response (HR) [19,20], are usually activated by the plants depending on pathogen lifestyle: in particular, host plants basically activate SA- or JA/ET-pathways against biotrophic and necrotrophic pathogens, respectively. Moreover, both defenses, dependent on SA- and JA/ET-pathways, appear to provide resistance to pathogens with mixed lifestyles (hemibiotrophs) [21,22]; however, the resistance to some pathogens appears to be through signaling pathways that involve neither of these regulators [18].
Cold atmospheric pressure plasma (CAP) can be generated by directly applying electrical energy to gas (e.g. air or noble gases) and consists of heavy particles (positive and negative ions, atoms, free radicals and excited or non-excited molecules), electrons and UV-rays [23]. CAP can be sustained in ambient air by means of a diversity of plasma source, such as corona discharge, micro hollow cathode discharge, atmospheric pressure plasma jet, gliding arc discharge, one atmospheric uniform glow discharge, plasma needle and dielectric barrier discharge (DBD) [24,25]. The biological effects of CAP treatment has been investigated in human medicine and more recently in agriculture [23,26–28]. During the last decade, different studies highlighted the direct effectiveness of cold plasma treatments in increasing the seed germination and the growth rate of various plant species, and in strengthening plant defences against bacterial pathogens [29–33]. CAP treatment of sterile deionized water (SDW) was shown to produce plasma activated water (PAW) characterized by increased acidity and conductivity, and an increased concentration of nitrates, nitrites and RONS, in particular hydrogen peroxide [28,34,35]. The antimicrobial activity of PAW against bacterial and fungal human pathogens was highlighted in in vitro experiments [28]. In addition, the positive effect of PAW on tomato seeds germination time and rate has been demonstrated [36].
This study was aimed to assess the in vitro antimicrobial activity of PAW against Xv, and its ability to reduce the bacterial leaf spot disease severity by eliciting induced resistance in in planta experiments. Furthermore, transcriptomic analyses were performed in tomato plants treated with PAW to define the transcription kinetics of selected genes regulating pre- and post-infectional plant defense responses as phenylalanine ammonia-lyase (pal), ethylene-response factor (erf1), pathogenesis related protein 1 (pr1a; SAR marker), endochitinase (pr4; ISR marker) and thaumatin-like protein (pr5) genes [18,19,22,37–39].
Materials and methods
PAW production
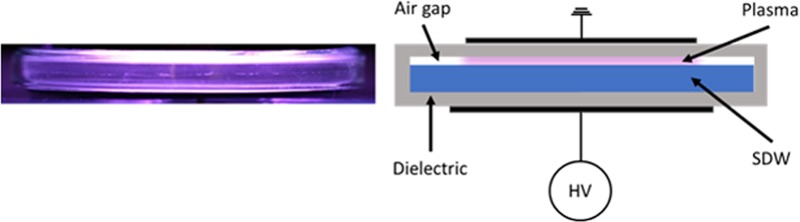
PAW was produced following the methodology described by Laurita et al. [28]. Briefly, CAP was generated by a DBD reactor consisting of a polystyrene cylindrical case (thickness 2 mm, diameter 94 mm) acting both as a liquid container and dielectric barrier (Fig 1). Aliquots of 80 ml of SDW were treated in a closed environment without recirculation; the volume of the reactor was filled with ambient air, which was used as plasma gas. Two circular aluminum foils acted as electrodes: the liquid-side electrode (diameter 89 mm) was connected to a nanosecond pulsed high voltage generator, while the gas-side electrode (diameter 80 mm) was grounded. The plasma source was driven by the HV generator producing pulses with a slow rate of few kV/ns and 50 mJ of energy per pulse (FID GmbH–FPG 20-1NMK).
Fig 1. Water activation through plasma treatment.
Picture (left) and schematic (right) of cold atmospheric pressure plasma (CAP) treatment of sterile distilled water (SDW) by means of nanosecond pulsed DBD for PAW production.
PAW was obtained operating for 10 min the source at a peak voltage of 20 kV, with a pulse repetition frequency of 1 kHz and an air gap of 1 mm and it was applied both in in vitro and in planta experiments between 1 h and 3 h after CAP treatment.
Detection of hydrogen peroxides (H2O2), nitrates (NO3-) and nitrites (NO2-) in PAW.
The pH and concentrations of H2O2, NO3- and NO2- induced by plasma treatment were measured using the Amplex Red Hydrogen Peroxide Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) and Nitrate/Nitrite Colorimetric Assay (Roche, Basel, Switzerland) according to the manufacturer’s protocol. PAW was diluted 100-fold in PBS to avoid any influence of pH on the measurement. A microplate reader (Rayto, Shenzhen, P.R. China) was used to measure solutions absorbances. Measurements were repeated three times.
Bacterial strains
The Xv strain IPV-BO 2684, isolated from symptomatic tomato plants in Italy and stored in the Phytobacteriology collection of the University of Bologna, was grown on the nutritive medium GYCA (glucose, yeast, calcium carbonate agar) [40] for 48 h at 27°C.
In vitro experiments
The in vitro antimicrobial activity of PAW was tested with the diffusion and broth dilution methods recommended by the Clinical and Laboratory Standards Institute [41] and slightly modified, by using LB medium instead of Mueller-Hinton substrate.
Diffusion method
LB agar plates were contaminated with 200 μL of bacterial water suspension calibrated at the spectrophotometer (OD = 0.01600nm; approx. 107 CFU/mL). A drop (60 μL) of each treatment was deposited onto an antibiogram disc (diameter 0.9 cm), previously kept in the middle of each contaminated Petri dish; the plates were then incubated for 24 h at 27°C. The antibacterial effect, shown as an inhibition halo was then measured (mm) by subtracting the antibiogram disc diameter from the halo diameter. SDW and acibenzolar-S-methyl (ASM, 75 ppm) were used as negative controls, while streptomycin sulphate (10,000 ppm) and H2O2 (30%) as positive controls. The assays was repeated three times and the data were elaborated with ANOVA test (p 0.05) using SPSS version 15.0 for Windows.
Broth dilution method
Sterile 50 mL Falcon tubes containing 15 mL LB broth were contaminated with 150 μL of bacterial aqueous suspension calibrated at the spectrophotometer (OD = 0.1600nm; approx. 108 CFU/mL). Each Falcon tube was then added with 300 μL of each treatment and incubated at 27°C for 24 h at 80 rpm in a rotative incubator. The bacterial population was evaluated at 24 h by collecting 1 mL contaminated LB broth. Each sample was 10-fold diluted and, 10 μL from each dilution were dropped onto GYCA medium; the plates were incubated at 27°C for 48–72 h and the bacterial populations were evaluated by counting the colonies. SDW and acibenzolar-S-methyl (ASM, 75 ppm) were used as negative control, while streptomycin sulphate (10,000 ppm) and H2O2 (30%) as positive controls. The assay was repeated three times and the data were elaborated with ANOVA test (p 0.05) using SPSS version 15.0 for Windows.
In planta experiments
Efficacy of PAW against Xv in tomato plants
Under greenhouse conditions three experiments were carried out on two different tomato plant cultivars, cv. Moneymaker and VF-10 [7], disposed in randomized blocks (3 plants x 4 blocks/treatment). The tomato plants were uprooted at 3rd-4th leaf stage and the root apparatus was soaked for 10 min into 500 mL of ASM, (75 ppm; positive control), PAW and SDW (negative control); then, the plants were put back into the pots. The treatments were carried out from 1.5 hours to 7 days before the experimental inoculation with the pathogen (BPI) as shown in Table 1.
Table 1. In planta experiments: Types and time of treatment application carried out at root apparatus of tomato plants (cv. Moneymaker and VF-10) under greenhouse conditions.
| No. Experiment/ Treatments | Application timing (days/hours BPIa) |
|---|---|
| No. 1 and 2 cv. Moneymaker and VF 10 | |
| Sterile distilled water (SDW) | 6 d |
| Plasma activated water (PAW) | 6 d |
| acibenzolar-S-methyl (ASM) | 7 d |
| No. 3 cv. VF-10 | |
| SDW | 1.5 h |
| PAW-1 | 2 d |
| PAW-2 | 1 d |
| PAW-3 | 1.5 h |
| ASM | 7 d |
a BPI: Before pathogen inoculation
The inoculation was carried out by spraying a water suspension (OD600nm = 0.01; ca. 107 CFU/mL), containing Xv strain IPV-BO 2684, on the leaf surfaces; tomato plants were then sealed in polyethylene (PE) bags for two days to favour the water congestion and to allow the pathogen penetration. The disease assessments were carried out by counting the leaf spots (on 5 leaves/plant) 21 days after the experimental inoculation, for a more clear and precise evaluation of the disease severity. The controlled conditions, hold until the disease assessment, were 16 h of day light and 8 h of darkness, 30°C and 24°C during day and night respectively; moreover, the relative humidity (RH) was maintained up to 70–75% according with the pathogen requirements. Data were elaborated with ANOVA test (Duncan's, p 0.05) and the relative protection (RP) of each treatment was calculated [(No. leafspots in SDW control plants—No. leafspots in treated plants)/ No. leafspots in SDW control plants].
Selected symptomatic leaf samples were used for the isolation and identification of the pathogen. The leaf surface was sterilized with 2% sodium hypochlorite. Necrotic lesions were aseptically collected and crushed into a mortar with 2 mL of sterile distilled water; 30 μL of the 10−1 and 10−2 diluted extract were dropped onto GYCA medium and incubated up to 48–72 h. Xv-like colonies were subcultured and identified with molecular assays [5,42].
Evaluation of defense-related gene transcription
Treatments and sample collection
Under greenhouse conditions, tomato plants cv. Moneymaker (2 plants x 4 blocks/treatment) at 3rd-4th leaf stage were uprooted and their root apparatus was drenched into the following treatments: SDW (negative control), PAW, ASM (75 ppm, positive control), jasmonic acid (JA, Sigma cod. J2500, 100 mM, positive control). Two further tomato plant batches, PAW and untreated ones, were inoculated with Xv by spraying a water suspension of the bacterial pathogen (ca. 107 CFU/mL) on the leaf surfaces (PAW+Xv-I and Xv-I, respectively labelled); the inoculated tomato plants were then sealed in polyethylene (PE) bags for two days (humid chamber). Furthermore, untreated and non-inoculated (NT) tomato plants were used as negative control and calibrator for data normalisation.
The three youngest tomato leaves were collected from each replicate/treatment at six time points: 1 h (T0), 7 h (T1), 24 h (T2), 48 h (T3), 120 h (T4) and 192 h (T5). The tissues were flash frozen in liquid nitrogen and stored at -80°C until RNA extraction. The greenhouse conditions were set as 16 h light at 30°C and 8 h dark at 24°C, and the RH was maintained at 70–75% until the end of the experiment.
RNA extraction
Approximately 100 mg of tomato leaves stored at -80°C were ground in liquid nitrogen and homogenized in 1.2 mL of extraction buffer [guanidine iso-thyocianate, 4 M; CH3COO-Na+ (pH 5), 0.2 M; NaEDTA (pH 8), 0.025 M; PVP 40, 2.5%] [43]. The extract was transferred into 2 mL Eppendorf tube and centrifuged at room temperature at 17,500 g for 5 min; 1 mL of the supernatant was added with 100 μL of Na-lauroyl sarcosine 30%, and mixed by inversion. The sample was then incubated at 70°C for 30 min. The total volume of the extract was placed into QIA shredder spin columns, then the extraction continued with Qiagen RNeasy Plant Minikit (cat. N° 74904) in combination with RNase-Free DNase Set (Qiagen; cat. No. 79254) to digest DNA following the instructions of the manufacturer. The total RNA, eluted in 50 μL of nuclease free distilled water, was stored at -20°C. Quantity and quality (ratio A260nm/A280nm) of 3 μL of RNA extracts were evaluated using Tecan Infinite 200 Pro NanoQuant instrument and i-control software (Tecan Group Ltda., Switzerland). The quality was also assayed by loading 5 μL of RNA extracts into a 2% agarose gel; the electrophoresis was performed at 40 V for 100 min in TAE buffer. The gel was stained in ethidium bromide solution (0.03%) for 20 min and distained in distilled water for 5 min; visualisation was then carried out under UV light (312 nm). All RNA samples were diluted at 100 ng/μL before the molecular assays.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Specific primers for RT-qPCR were designed on the sequences of reference and target genes by using Primer Express 2.0 software (Applied Biosystems, Foster City, CA). The primers used for RT-qPCR and the accession numbers of the gene sequences are listed in Table 2. The RT-qPCR assay was performed using ABI7000 v 1.2.3 sequence detection system. Each reaction was carried out with one-step method, in a final volume of 25 μL containing 1 U Murine Leukemia Virus (M-MLV; Promega, cod. 1705), 12.5 μL GoTaq qPCR Master Mix (Promega, No. A6001, SYBR Green/ROX chemistry), 0.25 μL CXR reference dye, primers at 400 nM (β-actin, erf1, pr1a, pr4 and pr5 and 100 nM (pal) (Invitrogen, cod. A9267) and 1 μL of RNA template (100 ng/μL). The primers specificity was evaluated on RNA extracts by dissociation curves analysis, in order to exclude non-specific amplifications or primer dimers presence. The thermal profile was set up as follows: 30 min at 48°C, 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, and 60°C for 1 min. Dissociation curves were performed at 95°C for 15 s, 60°C for 20 s and 95°C for 15 s. Data were analyzed using SDS1.2 software (Applied Biosystems). The efficiency of each primer pair was determined using RNA 10-fold dilution series (calibration curves) to determine the fold changes. Exponential amplification was plotted on a logarithmic scale, and dye fluorescence as a function of cycle number (Rn) was set to 0.32 for each RT-qPCR plate to obtain the cycle threshold (Ct) values.
Table 2. Primer sequences used for qRT-PCR.
| Gene (Accession No.) |
Primer | Sequence (5’ - 3’) | Dissociation curve (°C) | Reference |
|---|---|---|---|---|
|
β-actin (KY008745) |
Forward | AGCTCCTCCATTGAAAAGAACTATG | 73.0 | This study |
| Reverse | GGTAATAACTTGTCCATCAGGCAA | |||
|
pr1a (KY609511) |
Forward | TGTTGGTGGAAAAATGTGTGGA | 74.5 | This study |
| Reverse | GAGTTGCGCCAGACTACTTGAGT | |||
|
pr4 (KY609512) |
Forward | TATGAACGTTAGGGCAACGTATCA | 73.0 | This study |
| Reverse | CAGTTTATGTTTTGCGGATTGTACA | |||
|
pr5 (KY609513) |
Forward | CCAGTTTAGCAACCTAGATTTCTGG | 74.0 | This study |
| Reverse | TTAAATCCATCGACTAAAGAAATGTCC | |||
|
pal (KY614301) |
Forward | TCAGCACTTTGGACATGGTTAGTC | 75.0 | This study |
| Reverse | AGAACTTCAATTCCTTGCAAATCC | |||
|
erf1 (KY614303) |
Forward | AACTCAATGGCTAGGGCTTGTTT | 75.0 | This study |
| Reverse | TTTGCTATTTTCTGTCCACTTCAAAG |
All gene transcription levels were reported as Mean Normalised Transcription relative to β-actin, used as the reference gene [44,45]. Gene transcription was determined by the 2−ΔΔCT method adjusted by amplification efficiency for each trancripts [46]. The mean data obtained were analysed by ANOVA (p 0.05) followed by Student-Newman-Keuls (SNK) multiple range test.
Results and discussion
The plasma treatment of SDW induced the production of NO2-, NO3- and H2O2. As reported by Laurita et al. [28], NO2- completely disappeared few minutes after the plasma treatment, due to its reaction with H2O2 in acidic liquids. One hour after treatment, H2O2 and NO3-concentrations (approx. 20 mg/L and 120 mg/L, respectively) in PAW resulted stable for at least 2 hours at room temperature (Table 3); so the delay time between the production and the PAW treatment of plants was mantained between 1 h and 3 h in order to have stable concentrations of H2O2 and NO3-.
Table 3. pH, hydrogen peroxide and nitrate nitrite concentrations in SDW exposed for 10 min to nanosecond pulsed DBD, 1 and 3 h after treatment.
| pH | Hydrogen peroxide [mg/l] | Nitrates [mg/l] | |
|---|---|---|---|
| Untreated solution | 5,5±0,1 | 0 | 0 |
| 1 h delay time | 2,78±0,47 | 22±0,24 | 121,4±25,9 |
| 3 h delay time | 2,78±0,47 | 21,1±0,47 | 126±2,3 |
Antimicrobial activity
PAW did not show antimicrobial activity against Xv in the in vitro experiments using the diffusion method; the results were comparable to those of SDW and ASM (negative controls). Meanwhile, the positive controls (streptomycin and H2O2) inhibited the pathogen growth causing a mean inhibition haloes of 3.0 cm and 3.8 cm, respectively (S1 Fig).
The lack of PAW inhibition activity against Xv was confirmed in the experiments carried out using the broth dilution method. 24 h after contamination, the bacterial population in the Falcon tubes added with PAW rose up to approx. 109 CFU/mL, as occurred in the negative controls (SDW and ASM); on the contrary, streptomycin sulphate and H2O2 completely killed the strain IPV-BO 2684 (S2 Fig). In a recent work [28], in which the efficacy of PAW against Candida albicans and Staphylococcus aureus was studied using the time-kill method [47], it was pointed out that the loss of PAW antimicrobial activity was related to the post discharge chemistry: 16 minutes after the treatment, the concentration of H2O2 resulted 100 μM (ca. 0.0003%), whereas NO2- was not detectable by the instruments, and PAW did not show any efficacy against the two pathogens. The lack of antimicrobial activity was related to the absence of peroxynitrous acid (ONOOH), NO2- and to the low concentration of H2O2. In the PAW used for the in vitro experiments of the present study, ONOOH and NO2- were not detected, and H2O2 concentration resulted low as well [28], since the experiments were carried out more than 1 h after plasma treatment: this partially explains the lack of PAW efficacy against Xv. In addition, the assays used in the in vitro experiments of this study were based on diffusion and broth dilution methods, as suggested by Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS) [41]; these methods differed from those proposed and used in Laurita et al. (2015) [28] and in the other studies in which PAW was employed [48,49]. In those studies, it was taken into account not just the time elapsed between the PAW production and its use in the in vitro experiments, but also the effect of exposure time of the bacterial suspension to higher volumes of PAW: Laurita et al. [28] added 10 mL of PAW to 100 μL of pathogen suspension and evaluated the decrease of pathogen concentration at different time points after the treatment (time-kill method).
The presence of H2O2 and NO3-, and the low pH suggested the possible use of PAW in controlling plant pathogens, as activator of a defence mechanisms mediated by the plant host. The reactive oxygen species (ROS), such as ∙O2, ∙OH and H2O2, are known to be one of the upstream responses of the plant during a pathogen attack [19,50–54].
The in planta experiments conducted on tomato plants were therefore aimed to evaluate the indirect effect of PAW (applied at the roots) against Xv (experimentally inoculated at the leaves), through the host-mediated induced resistance.
Induced resistance in planta
In all the experiments carried out in planta no phytotoxicity or reduction in the vegetative growth was observed in PAW treated non-inoculated plants (used as control).
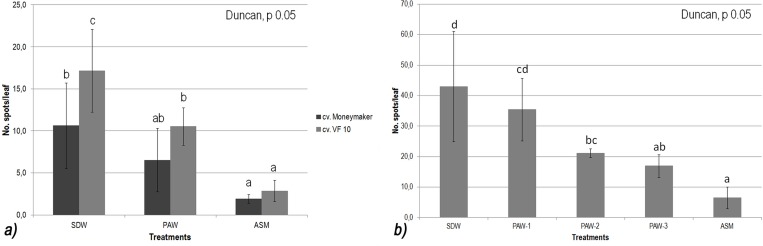
In the first experiment, tomato plants cv. Moneymaker treated with PAW 6 days BPI, showed a medium level of disease severity (ca. 6 spots/leaf) compared to the plants treated with SDW (negative control; ca. 11 spots/leaf) and those treated with ASM (2 spots/leaf) (Fig 2); the RP of PAW was approx. 38%.
Fig 2. In planta experiments on two different tomato plant cultivars.
a) Results of the in planta efficacy of plasma activated water applied 6 days before pathogen inoculation at root apparatus (PAW-R), compared to the negative (SDW-R) and positive controls (ASM-R), against bacterial leaf spot caused by Xanthomonas vesicatoria (strain IPV-BO 2684) on tomato plants cv. Moneymaker (dark grey histogram) and cv. VF 10 (grey histogram) grown under greenhouse conditions. b) Results of the in planta efficacy of plasma activated water applied at root apparatus at different times before pathogen inoculation (BPI) (PAW-1: 2 days BPI, PAW-2: 1 day BPI, PAW-3: 1.5 hours BPI), compared to that of the negative (SDW) and positive controls (ASM), against bacterial leaf spot caused by Xanthomonas vesicatoria (strain IPV-BO 2684) on tomato plants cv. VF-10 grown under greenhouse conditions. Different letters denote significant differences according to the Duncan's test (p 0.05).
The results of the second in planta experiment on tomato cv. VF-10 (Fig 2), confirmed that PAW treatment applied 6 days BPI was able to significantly reduce the number of bacterial leaf spots (11 spots/leaf), with respect of SDW (17 spots/leaf), meanwhile, ASM treated plants showed again the lowest statistical number of spots per leaf (ca. 3 spots/leaf). The RP provided by PAW application was approx. 38%. as occurred in the first experiment on tomato cv. Moneymaker. It was clear that the tomato plants cv. Moneymaker resulted less susceptible to Xv than those of cv. VF-10; even though, the RP among the treatments were comparable, thus confirming the efficacy of PAW in reducing the disease severity.
The third experiment, carried out on cv. VF-10 plants, was characterized by a consistently higher disease pressure with respect of the previous two experiments. The disease severity on plants treated with PAW 2 days BPI (PAW-1) resulted lower (ca. 35 spots/leaf) than that observed on SDW treated plants (ca. 43 spots/leaf), but statistically higher with respect of plants treated with PAW applied 1 day (PAW-2) and 1.5 hours (PAW-3) BPI (ca. 21 and 17 spots/leaf respectively). RPs provided by PAW-1, PAW-2 and PAW-3 were ca. 18%, 51% and 61% respectively; ASM treated tomato plants showed the statistical lowest disease severity (ca. 6 spots/leaf) among all treated plants (Fig 2), giving the highest levels of RPs which ranged approx. 82% to 85% in the three experiments as expected [12,16,17]. These results confirmed the efficacy of PAW as possible resistance inducer against bacterial diseases of tomato plants.
The time elapsed between PAW application and pathogen inoculation played an important role in the plant defences induction. In all in planta experiments, it was observed a reduction of disease severity in the treatments with PAW; in particular, the highest RP was provided by PAW applied 1.5 h or 1 day BPI. The results suggested that the host mediated response was triggered by the PAW treatment and the induced disease reduction was related to the time elapsed between the treatment application and the pathogen inoculation. The ROS and/or RNS, induced by CAP treatment in SDW, might be linked to signalling pathways in tomato that establish a correlation between the disease reduction and the activation of defence mechanisms, and that were investigated with the transcriptomic analysis on genes related to the main induced resistance pathways [19].
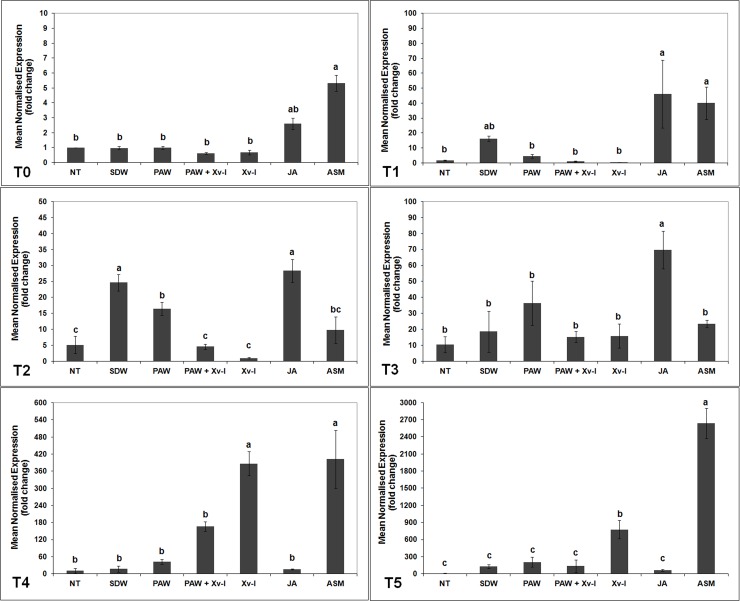
Gene transcription
Five genes (pal, erf1, pr1a, pr4 and pr5), regulating pre- and post-infectional plant defence responses [18,19,22,37–39] were assayed by RT-qPCR on cv. Moneymaker, to evaluate their transcription abundance in leaves after the root application of different treatments and/or inoculation with the pathogen. The β-actin resulted a reliable reference gene, because its abundance in PAW treated plants, and in the negative (NT and SDW) and positive (ASM and JA) control plants were equivalent. The use of the negative control treatment (SDW), whose type of application was similar to that of the other treatments (PAW, ASM and JA), allowed to measure the intensity of the plant defence response: the application method, in fact, could have brought side stress effects dependant to the wounds produced during the treatment application [39].
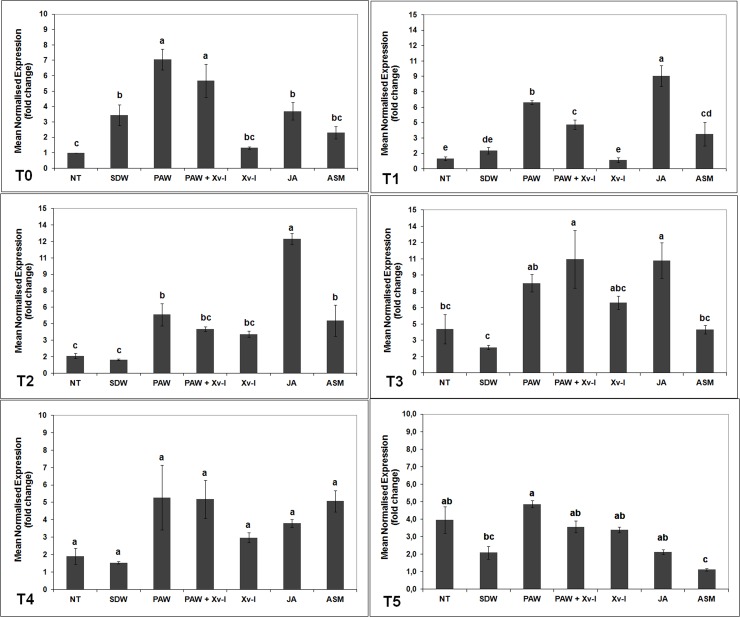
pal gene
One hour after treatment (T0), the number of pal gene transcripts significantly increased in tomato plants treated with PAW and in those treated with PAW and inoculated with the pathogen (PAW+Xv-I) by ca. 7.0- and 7.5-fold, respectively, when compared to untreated and non-inoculated plants (NT, ca. 1-fold), to those pre-treated with SDW (3.4-fold) and to those inoculated with the pathogen (Xv-I, 1.3-fold). The plants treated with JA (positive control) showed a statistical increase of pal gene transcripts, compared to NT, SDW and Xv-I ones; nonetheless, such increase resulted statistically lower than that observed in plants treated with PAW (Fig 3). After 7 h (T1), the pal gene transcripts increase evaluated in the treatments PAW and PAW+Xv-I resulted lower (ca. 6.5- and 4.4-fold, respectively) compared to those observed at T0, since it resulted significantly higher than that found in NT, SDW and Xv-I (ca. 1-, 1.8- and 0.9-fold, respectively). In the treatment ASM (positive control), a higher transcript abundance was present compared to that of negative controls (NT and SDW) and of Xv-I, but lower than that resulted in PAW, PAW+Xv-I and JA; in particular, the plants treated with JA showed the highest increase of transcription activity (ca. 9.0-fold, Fig 3).
Fig 3. pal transcription kinetics in tomato leaves.
The graph shows the pal induction at each time point (T0: 1 h; T1: 7 h; T2: 24 h; T3: 48 h; T4: 120 h; T5: 192 h) after treatment/inoculation and the standard error (±SE). Different letters denote significant differences according to the SNK test (p 0.05). NT: non-treated/non-inoculated; SDW: treated plants at root apparatus with sterile distilled water; PAW: treated plants at root apparatus with plasma activated water; PAW+Xv-I: treated plants at root apparatus with plasma activated water and inoculated with Xanthomonas vesicatoria sprayed at leaves; Xv-I: inoculated with Xanthomonas vesicatoria sprayed at leaves; JA: treated plants at root apparatus with jasmonic acid; ASM: treated plants at root apparatus with acibenzolar-S-methyl.
Twentyfour hours after treatment/inoculation (T2), the pal gene transcription activity in the treatments PAW and PAW+Xv-I resulted still higher (ca. 5.3- and 4.0-fold) than that in the control treatments NT and SDW (ca. 1.6- and 1.2-fold), while it was comparable to that observed in Xv-I (ca. 3.6-fold). The tomato plants treated with ASM produced pal gene transcriptional activity of ca. 4.8-fold, higher than that evaluated in NT and SDW; plants treated with JA showed the highest transcripts increase up to ca. 12.2-fold (Fig 3). After 48 h (T3), a general increase of the pal transcripts was recorded in all treatments, but it was still possible to evaluate statistical differences among them: in PAW and PAW+Xv-I tomato plants, the number of transcripts significantly increased (ca. 8.3- and 10.5-fold) with respect of those in NT and SDW (ca. 4.0- and 2.3-fold), whose transcription of pal resulted similar to that of plants treated with ASM (ca. 4.0-fold). On the contrary, plants treated with JA showed high number of transcripts (ca. 10.4-fold), comparable to that observed in the treatment PAW+Xv-I (Fig 3, T3). After 120 and 192 h (T4 and T5, respectively), the transcriptional level of pal gene decreased in all treatments: however at T4 the transcription resulted apparently higher in PAW, PAW+Xv-I, Xv-I, JA and ASM (ca. 5.3-, 5.2-, 3.0-, 3.8- and 5.0-fold), in comparison to that of NT and SDW (ca. 1.9- and 1.5-fold, respectively), but the differences were not significant. Finally at T5, the pal gene transcription in PAW decreased down to ca. 4.9-fold, still being higher in comparison to that found in the treatments NT, SDW, PAW+Xv-I, Xv-I, JA and ASM (ca. 4.0-, 2.1-, 3.6-, 3.4-, 2.2- and 1.1-fold) (Fig 3).
The rapid induction of pal gene transcription 1 hour after PAW treatment might be explained by the PAW chemical composition, characterised by low pH and the presence of H2O2, inducing the plant hypersensitive response (HR), and also acting as signal to induce local and systemic resistance [54–60]. The combination of these chemical characteristics of PAW could allow a similar, and in few cases, larger increase of pal gene transcription in comparison with positive controls (ASM and JA).
In Fig 3, it can also be observed the effect of the treatment application method (explantation, root drenching and transplant): the tomato plants treated with SDW (negative control) showed an increase of pal gene transcripts after 1 hour, significantly higher in comparison to NT and Xv-I plants, even though such increase resulted lower of that of PAW treatments.
Moreover, the tomato plants responded against the pathogen (Xv-I) 24 h (T2) after the experimental inoculation, and they increased the pal gene transcription up to ca. 3.6-fold, higher than that evaluated in negative control treatments (NT and SDW). The increase of pal gene transcription was evaluated until 48 h (T3) as almost double in comparison to that observed at 24 h thus confirming that the plants started at a late time to activate these resistance defences. On the other hand, at 48 h the plants treated with PAW in combination with pathogen inoculation (PAW+Xv-I) resulted the highest in the pal gene transcript abundance, comparable to those of JA treatment. This was due to the response of the tomato plants to the infection plus the response to PAW treatment. Interestingly, the RP on plants treated with PAW 48 h before the inoculation with the pathogen resulted ca. 18%. Still, the results highlighted the pal involvement in response to PAW treatments and against the pathogen.
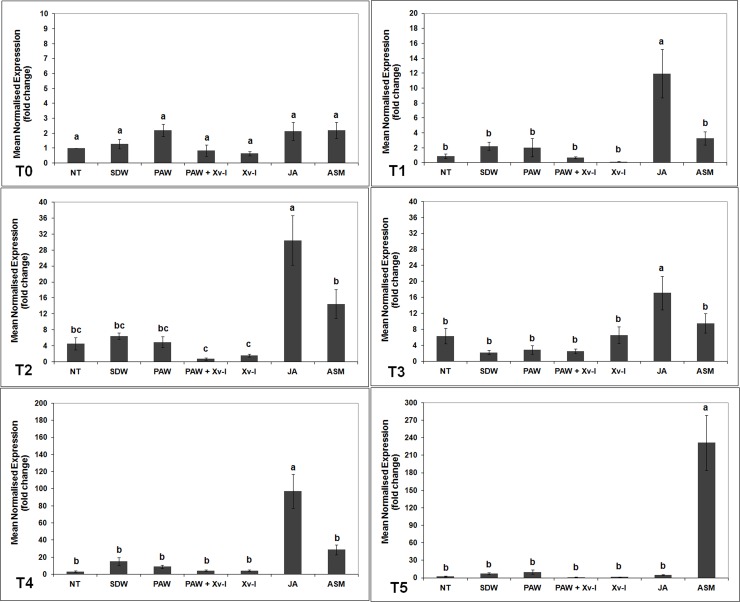
erf1 gene
In the plants belonging to PAW and PAW+Xv-I treatments, it was not shown a significant increase in the abundance of erf1 gene transcriptional activity from T0 to T5 in comparison to the negative controls (NT and SDW). Seven h (T1) after treatment, a low increase of erf1 gene transcription was observed in PAW, PAW+Xv-I and SDW (ca. 5.2-, 4.6- and 5.7-fold, respectively), when compared to that of NT and Xv-I (ca. 3.0- and 2.7-fold, respectively) (Fig 4), very likely due to the stress occurred during the treatment application [39]. Even the inoculation with the pathogen (Xv-I) did not elicit the erf1 gene transcription in none of the time points (from T0 to T5), suggesting that pathogen infection did not involve the ethylene pathway as first response. On the contrary, in the tomato plants treated with JA, the abundance of erf1 transcripts increased since the beginning of the experiment. At 1 h (T0), the transcription resulted approx. 8.0-fold, and after 7 h (T2) it reached 12.0-fold, significantly higher than in the plants of negative control treatments (NT and SDW) and in those inoculated with the strain IPV-BO 2684 (Xv-I) at both time points, thus demonstrating the activation of JA/ethylene pathway which usually brings to ISR [22,61,62]. The transcriptional level of erf1 gene in plants treated with ASM resulted statistically higher than that of negative control treatments at 7 (T1), 24 (T2) and 120 h (T4) (ca. 11.3-, 5.1- and 10.2-fold, respectively) (Fig 4). The delay of erf1 gene transcription in plants treated with ASM, with respect of those treated with JA active since T0, confirmed the presence of inter-signaling pathways between the salicylic acid (SA) and JA/ethylene pathways [22,63].
Fig 4. erf1 transcription kinetics in tomato leaves.
The graph shows the erf1 induction at each time point (T0: 1 h; T1: 7 h; T2: 24 h; T3: 48 h; T4: 120 h; T5: 192 h) after treatment/inoculation and the standard error (±SE). Different letters denote significant differences according to the SNK test (p 0.05). NT: non-treated/non-inoculated; SDW: treated plants at root apparatus with sterile distilled water; PAW: treated plants at root apparatus with plasma activated water; PAW+Xv-I: treated plants at root apparatus with plasma activated water and inoculated with Xanthomonas vesicatoria sprayed at leaves; Xv-I: inoculated with Xanthomonas vesicatoria sprayed at leaves; JA: treated plants at root apparatus with jasmonic acid; ASM: treated plants at root apparatus with acibenzolar-S-methyl.
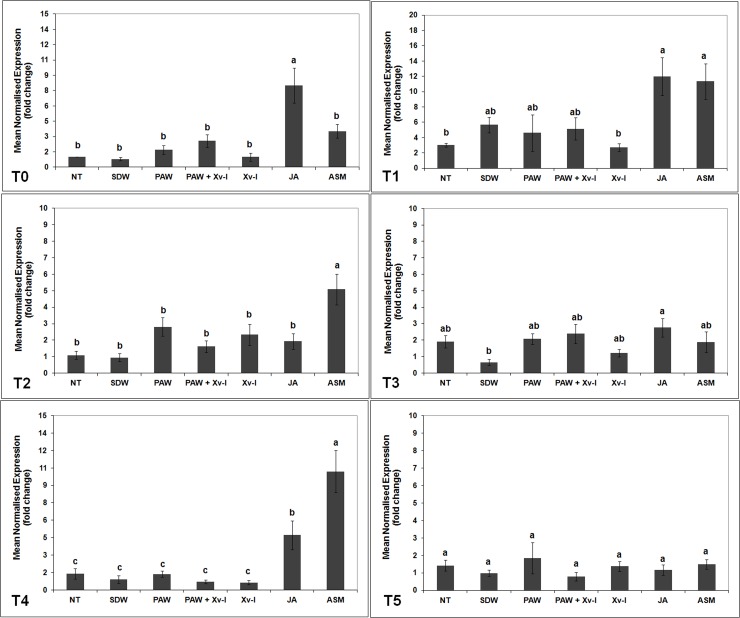
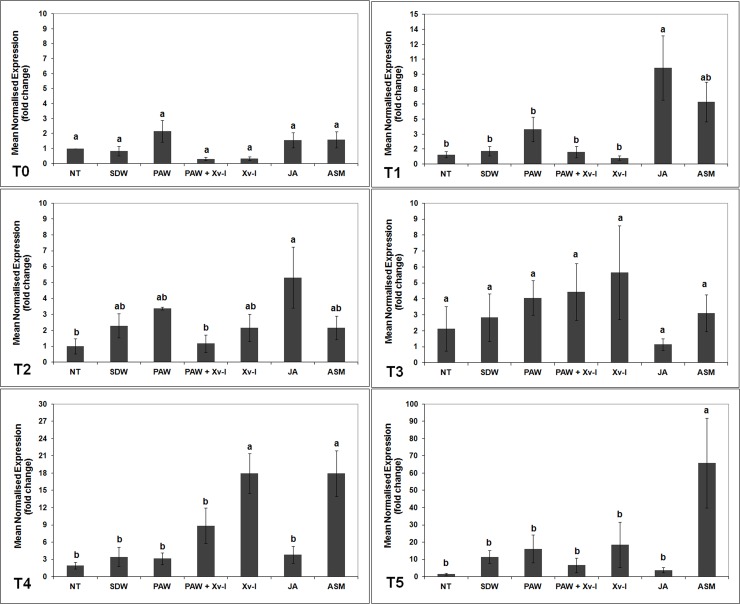
pr1a, pr4, and pr5 genes
The transcription kinetics of pr1a, pr4 and pr5 genes, encoding post-infection pathogenesis-related proteins (PRs), did not show significant increases in the tomato plants treated with PAW and PAW+Xv-I, when compared to those of NT and SDW treatments, at all time points evaluated (Figs 5–7).
Fig 5. pr 1a transcription kinetics in tomato leaves.
The graph shows the pr 1a induction at each time point (T0: 1 h; T1: 7 h; T2: 24 h; T3: 48 h; T4: 120 h; T5: 192 h) after treatment/inoculation and the standard error (±SE). Different letters denote significant differences according to the SNK test (p 0.05). NT: non-treated/non-inoculated; SDW: treated plants at root apparatus with sterile distilled water; PAW: treated plants at root apparatus with plasma activated water; PAW+Xv-I: treated plants at root apparatus with plasma activated water and inoculated with Xanthomonas vesicatoria sprayed at leaves; Xv-I: inoculated with Xanthomonas vesicatoria sprayed at leaves; JA: treated plants at root apparatus with jasmonic acid; ASM: treated plants at root apparatus with acibenzolar-S-methyl.
Fig 7. pr 5 transcription kinetics in tomato leaves.
The graph shows the pr 5 induction at each time point (T0: 1 h; T1: 7 h; T2: 24 h; T3: 48 h; T4: 120 h; T5: 192 h) after treatment/inoculation and the standard error (±SE). Different letters denote significant differences according to the SNK test (p 0.05). NT: non-treated/non-inoculated; SDW: treated plants at root apparatus with sterile distilled water; PAW: treated plants at root apparatus with plasma activated water; PAW+Xv-I: treated plants at root apparatus with plasma activated water and inoculated with Xanthomonas vesicatoria sprayed at leaves; Xv-I: inoculated with Xanthomonas vesicatoria sprayed at leaves; JA: treated plants at root apparatus with jasmonic acid; ASM: treated plants at root apparatus with acibenzolar-S-methyl.
The plants inoculated with the pathogen highlighted a statistical increase of pr1a gene transcript quantity after 120 h (T4) (ca. 387.0-fold) in comparison to that registered in NT (ca. 11.0-fold) and SDW treatments (ca. 17.0-fold) (Fig 5). As concerns the plants treated with JA, pr1a transcripts abundance significantly increased from T0 (1 h) to T3 (48 h); in this last time point, the transcriptional activity rose at the highest level with respect of all time points (ca. 69.8-fold). In the further time points (T4 and T5) such transcription level decreased and it was comparable to that of NT and SDW (Fig 5). The ASM treatment induced a significant increase of pr1a gene, which resulted higher than that observed in NT and SDW, in particular, transcript number reached ca. 5.3- and 40.0-fold at 1 h and 7 h, respectively; on the other hand, at 120 h (T4) the pr1a gene transcription level was ca. 402.0-fold, comparable to Xv-I plants (ca. 387.0-fold). At 192 h (T5), the number of transcripts in ASM plants was the highest reaching ca. 2600.0-fold (Fig 5), while that of Xv-I plants resulted ca. 777.0-fold. The increase of pr1a in response to pathogen infection suggests the activation of SA pathway, which in known to be related to biotrophic pathogens [22;64].
On the other hand, significant increase of pr4 gene transcript quantity in Xv-I plants was also found after 120 h (T4) (ca. 18.0-fold), this in comparison to that recorded in NT (ca. 2.0-fold) and SDW treatments (ca. 3.5-fold) (Fig 6); this demonstrates that pr4 gene is also involved in plant resistance, through the activation of JA/ethylene pathway (basically elicited by necrotrophic pathogens), against Xv, thus confirming its hemibiotrophic lifestyle [22,64–66]. In the JA treatment, an early rise of the pr4 gene transcripts started at 7 h (ca. 9.6-fold), and resulted the highest until 24 h (ca. 5.3-fold); this transcription decreased to the baseline until the last time point (192 h, Fig 6). In plants treated with ASM, the pr4 gene transcript level rose to high levels as well, at 120 h and 192 h it reached ca. 18.0- and 65.8-fold, respectively.
Fig 6. pr 4 transcription kinetics in tomato leaves.
The graph shows the pr 4 induction at each time point (T0: 1 h; T1: 7 h; T2: 24 h; T3: 48 h; T4: 120 h; T5: 192 h) after treatment/inoculation and the standard error (±SE). Different letters denote significant differences according to the SNK test (p 0.05). NT: non-treated/non-inoculated; SDW: treated plants at root apparatus with sterile distilled water; PAW: treated plants at root apparatus with plasma activated water; PAW+Xv-I: treated plants at root apparatus with plasma activated water and inoculated with Xanthomonas vesicatoria sprayed at leaves; Xv-I: inoculated with Xanthomonas vesicatoria sprayed at leaves; JA: treated plants at root apparatus with jasmonic acid; ASM: treated plants at root apparatus with acibenzolar-S-methyl.
As concerns Xv inoculated tomato plants (Xv-I), they did not show rise of pr5 gene transcription at any time point; in fact, it was similar to that observed in negative controls (Fig 7), thus indicating that pr5 gene might be not involved in plant resistance against Xv within 192 h after inoculation with the pathogen. On the contrary, in the JA treated plants, pr5 gene transcription levels started at 7 h (ca. 11.9-fold) until the highest level at 120 h (ca. 97.2-fold) (Fig 7). Furthermore, ASM induced the highest level of pr5 gene transcription at 192 h (T5) resulted ca. 230.0-fold after treatment application.
In tomato plants inoculated with the pathogen (Xv-I), though the induction of the pr1a and pr4 gene transcript was evaluated, that of pr5 and erf genes, usually involved in both SA and JA/ethylene pathways [22,65], respectively, did not resulted.
Conclusions
PAW did not show antimicrobial activity against Xv in the in vitro experiments using the diffusion and dilution methods.
Nevertheless, in the in planta experiments against bacterial leaf spot of tomato, the disease severity resulted lower when PAW was applied 1 and 24 h before the pathogen inoculation, providing RP of 61% and 51%, respectively. When tomato plants, belonging to different cultivars, were treated using PAW six days before the inoculation, the RPs were reduced to ca. 38%. In addition, the transcriptomic experiments highlighted the pal involvement in response to PAW treatments and against the pathogen; pal gene transcription levels resulted significantly high between 1 and 48 h until their decrease, after 192 h from PAW application.
These results indicate that pal gene might be a key factor in response to PAW pre-treatments, nonetheless, further investigations are needed to explain the lack of PRs transcript increase. In particular the studies should deeply investigate as concerns the phenylpropanoid pathway, which leads the production of phytoalexins and phenolic substances, or the enhancement of different pathways and to the transcription of defence related genes or transcription factors not investigated in the present study.
Supporting information
The histogram shows the inhibition haloes of Xanthomonas vesicatoria growth resulted from in vitro assays by using diffusion method.
(TIF)
The histogram shows the inhibition of Xanthomonas vesicatoria population in in vitro assays by using broth dilution method.
(TIF)
Acknowledgments
The Authors would like to acknowledge Eng. Petr Lukes (Institute of Plasma Physics) for the support on the measurement of reactive species in liquid.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Jones J, Bouzar H, Stall R, Almira E, Roberts P, Bowen B, et al. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. International Journal of Systematic Bacteriology. 2000; 50: p. 1211–1219. [DOI] [PubMed] [Google Scholar]
- 2.Jones J, Lacy G, Bouzar H, Stall R, Schaad N. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic Applied Microbiology. 2004; 27: p. 755–762. 10.1078/0723202042369884 [DOI] [PubMed] [Google Scholar]
- 3.Louws F, Wilson M, Campbell H, Cuppels D, Jones J, Shoemaker P, et al. Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Disease. 2001; 85: p. 481–488. 10.1094/PDIS.2001.85.5.481 [DOI] [PubMed] [Google Scholar]
- 4.Byrne JM, Dianese AC, Ji P, Campbell HL, Cuppels DA, Louws FJ, et al. Biological control of bacterial spot of tomato under field conditions at several locations in North America. Biological Control. 2005; 32(3): p. 408–418. [Google Scholar]
- 5.EPPO. PM 7/110 (1) Xanthomonas spp. (Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. Bulletin OEPP/EPPO. 2013; 43: p. 7–20. [Google Scholar]
- 6.Chambers S, Merriman P. Perennation and control of Pseudomonas tomato in Victoria. Austr. J. Agric. Res. 1975; 26: p. 657–663. [Google Scholar]
- 7.Giovanardi D, Biondi E, Ignjatov M, Jevtic R, Stefani E. Impact of bacterial spot outbreaks on the phytosanitary quality of tomato and pepper seeds. Plant Pathology. 2018; 67(5): p. 1168–1176, ISSN: 1365-3059, 10.1111/ppa.12839 [DOI] [Google Scholar]
- 8.Cooksey DA. Copper uptake and resistance in bacteria. Molecular Microbiology. 1993; 7(1): p. 1–5. [DOI] [PubMed] [Google Scholar]
- 9.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, & Aarts HJ. Acquired antibiotic resistance genes: an overview. Frontiers in microbiology. 2011; 2 (203): p. 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Hendawy H, Osman M, Sorour N. Biological control of bacterial spot of tomato caused by Xanthomonas campestris pv. vesicatoria by Rahnella aquatilis. Microbiological Research. 2005; 160: p. 343–352. 10.1016/j.micres.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 11.Paret ML, Vallad GE, Averett DR, Jones JB, and Olson SM. Photocatalysis: effect of light-activated nanoscale formulations of TiO2 on Xanthomonas perforans and control of bacterial spot of tomato. Phytopathology. 2013; 103: p 228–236. 10.1094/PHYTO-08-12-0183-R [DOI] [PubMed] [Google Scholar]
- 12.Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Molecular Plant Pathology. 2015; 16(9): p. 907–920. 10.1111/mpp.12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzaglia A, Fortunati E, Kenny JM, Torre L, Balestra GM. Nanomaterials in Plant Protection. Nanotechnology in Agriculture and Food Science. 2017; 7: p. 115–133. [Google Scholar]
- 14.Cortesi R, Quattrucci A, Esposito E, Mazzaglia A, and Balestra GM. Natural antimicrobials in spray-dried microparticles based on cellulose derivatives as potential eco-compatible agrochemicals. Journal of Plant Disease Protection. 2017; 124: p. 269–278. [Google Scholar]
- 15.Fortunati E, Mazzaglia A, Balestra GM. Sustainable control strategies for plant protection and food packaging sectors by natural substances and novel nanotechnological approaches. Journal of The Science of Food and Agriculture. 2019; 99(3): p. 986–1000. 10.1002/jsfa.9341 [DOI] [PubMed] [Google Scholar]
- 16.Obradovic A, Jones J, Momol M, Olson S, Jackson L, Balogh B, et al. Integration of biological control agents and systemic acquired resistance inducers against bacterial spot on tomato. Plant Diseases. 2005; 89: p. 712–716. [DOI] [PubMed] [Google Scholar]
- 17.Oostendorp M, Kunz W, Dietrich B, Staub T. Induced disease resistance in plants by chemicals. European Journal of Plant Pathology. 2001; 107: p. 19–28. [Google Scholar]
- 18.van Loon LC, Rep M, Pieterse CMJ. Significance of Inducible Defense-related Proteins in Infected Plants. Annual review in Phytopathology. 2006; 44: p. 7.1–7.28. [DOI] [PubMed] [Google Scholar]
- 19.Lamb C. and Dixon RA. The Oxidative Burst In Plant Disease Resistance Annual Reviews Plant Physiology Plant Molecular Biology. 1997; 48: p. 251–275. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC. and Hwang BK. Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annum. Planta. 2005; 221: p. 790–800. 10.1007/s00425-005-1488-6 [DOI] [PubMed] [Google Scholar]
- 21.Thaler JS, Owen B, Higgins WJ. The Role of the Jasmonate Response in Plant Susceptibility to Diverse Pathogens with a Range of Lifestyles. Plant Physiology. 2004; 35: p. 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Loon LC, Geraats BP, and Linthorst HJ. Ethylene as a modulator of disease resistance in plants. Trends in plant science. 2006; 11(4): p. 184–191. 10.1016/j.tplants.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 23.Heinlin J, Morfill G, Landthaler M, Stolz W, Isbary G, Zimmermann J, et al. Plasma medicine: possible applications in dermatology. Journal Deutschen Dermatologischen Gesellschaft. 2010; 8: p. 968–976. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Xu G, Wang T. Non-thermal plasma approaches in CO2 utilization. Fuel Process. 1999; 58: p. 119–134. [Google Scholar]
- 25.Nehra V, Kumar A, Dwivedi H. Atmospheric non-thermal plasma sources. International Journal of Engineering. 2008; 2(1): p. 53–68. [Google Scholar]
- 26.Moreau M, Orange N, Feuilloley M. Non-thermal plasma technologies: New tools for bio-decontamination. Biotechnology Advances. 2008; 26: p. 610–617. 10.1016/j.biotechadv.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 27.Kostov K, Rocha V, Koga-Ito C, B M, Algatti M, Honda R, et al. Bacterial sterilization by a dielectric barrier discharge (DBD) in air. Surface Coatings Technology. 2010; 204(18–19): p. 2954–2959. [Google Scholar]
- 28.Laurita R, Gherardi M, Colombo V, Lukes P. Chemical analysis of reactive species and antimicrobial activity of water treated by nanosecond pulsed DBD air plasma. Plasma-Liquid Interactions. 2015; 3(2): p. 53–61. [Google Scholar]
- 29.Carvalho R, Carvalho A, Silva M, Demarquette N, Assis O. Use of thin films obtained by plasma polymerization for grain protection and germination enhancement. Quimica Nova. 2005; 28 (6): p. 1006–1009. [Google Scholar]
- 30.Moreau M, Feuilloley M, Orange N, Brisset J. Lethal effect of the gliding arc discharges on Erwinia spp. Journal of Applied Microbiology. 2005; 98: p. 1039–1046. 10.1111/j.1365-2672.2004.02535.x [DOI] [PubMed] [Google Scholar]
- 31.Moreau M, Feuilloley M, Veron W, Meylheuc T, Chevalier S, Brisset J, et al. Gliding Arc Discharge in the potato pathogen Erwinia carotovora subsp. atroseptica: Mechanism of lethal action and effect on membrane-associated molecules. Applied and Environmental Microbiology. 2007; 73(18): p. 5904–5910. 10.1128/AEM.00662-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, He X, Li L, Li J, Shao H, Xu Q, et al. Effect of cold plasma treatment on seed germination and growth of wheat. Plasma Science and Technology,. 2014; 16(1): p. 54–58. [Google Scholar]
- 33.Jiang J, Lu Y, Li J, Li L, He X, Shao H, et al. Effect of seed treatment by cold plasma on the resistance of tomato to Ralstonia solanacearum (bacterial wilt). Plos One. 2014; 9(5): p. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park D, Davis K, Gilani S, Alonzo C, Dobrynin D, Friedman G, et al. Reactive nitrogen species produced in water by non-equilibrium plasma increase plant growth rate and nutritional yield. Current Applied Physics. 2013; 13(1): p. 19–29. [Google Scholar]
- 35.Lukes P, Dolezalova E, Sisrova I, Clupek M. Aqueous-phase chemistry and bactericidaleffects from an air discharge plasma incontact with water: evidence for theformation of peroxynitrite through apseudo-second-order post-dischargereaction of H2O2 and HNO2. Plasma Sources Science and Technology. 2014; 23(1): p. 1–15. [Google Scholar]
- 36.Sivachandiran L, Khacef A. Enhanced seed germination and plant growth byatmospheric pressure cold air plasma: combine deffect of seed and water treatment. RSC Advances. 2017; 7(4): p. 1822–1832. [Google Scholar]
- 37.Lund ST, Stall RE, and Klee HJ. Ethylene regulates the susceptible response to pathogen infection in tomato. The Plant Cell. 1998; 10(3): p. 371–382. 10.1105/tpc.10.3.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler A, Schwindling S, Conrath U. Benzothiadiazole-Induced Priming for Potentiated Responses to Pathogen Infection, Wounding, and Infiltration of Water into Leaves Requires the NPR1/NIM1 Gene in Arabidopsis Plant Physiology. 2002; 128: p. 1046–1056. 10.1104/pp.010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tournier B, Sanchez-Ballesta M, Jones B, Pesquet E, Regad F, Latche A, et al. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Letters. 2003; 550: p. 149–154. [DOI] [PubMed] [Google Scholar]
- 40.Dye DW. The inadequacy of the usual determinative tests for the identification of Xanthomonas spp. New Zealand Journal of Science. 1962; 5(4): p. 393–416. [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. M100-S17: Performance Standards for Antimicrobial Susceptibility Testing; 16th informational supplement Wayne, PA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 42.Koenraadt H, van Betteray B, Germain R, Hiddink G, Jones J, Oosterhof J, et al. Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Horticulturae. 2009; 808: p. 99–102. [Google Scholar]
- 43.Ratti C, Budge G, Ward L, Clover G, Rubies-Autonell C, Henry C. Detection and relative quantitation of Soil-borne cereal mosaic virus (SBCMV) and Polymyxa graminis in winter wheat using real-time PCR (TaqMan). Journal Virology Methods. 2004; 122: p. 95–103. [DOI] [PubMed] [Google Scholar]
- 44.Danve C. Defence gene expression in the tomato-Verticillium pathosystem [Thesis of Master of Science]. Faculty of Graduate Studies, University of Guelph; 2010.
- 45.Murshed R, Lopez-Lauri F, Sallanon H. Effect of water stress on antioxidant systems and oxidative parameters in fruits of tomato (Solanum lycopersicon L. cv. Micro-tom). Physiology and Molecular Biology of Plants. 2013; 19(3): p. 363–378. 10.1007/s12298-013-0173-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl M. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research. 2001; 29(9): p. 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carson CF, Mee BJ, Riley TV. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrobial Agents and Chemotherapy. 2002; 46(6): p. 1914–1920. 10.1128/AAC.46.6.1914-1920.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Julák J, Scholtz V, Kotúčová S, Janoušková O. The persistent microbicidal effect in water exposed to the corona discharge. Physica Medica. 2012; 28: p. 230–239. 10.1016/j.ejmp.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 49.Traylor M, Pavlovich M, Karim S, Hait P, Sakiyama Y, Clark D, et al. Long-term antibacterial efficacy of air plasma-activated water. Journal of Physics D: Applied Physics. 2011; 44(47): p. 1–4. [Google Scholar]
- 50.Arasimowicz M, and Floryszak-Wieczorek J. Nitric oxide as a bioactive signaling molecule in plant stress responses. Plant Science. 2007; 172(5): p. 876–887. [Google Scholar]
- 51.Peng M, Kuć J. Peroxidase-generated hydrogen peroxide as a source of antifungal activity in vitro and on tobacco leaf disks. Phytopathology. 1992; 82: p. 696–699. [Google Scholar]
- 52.Doke N. Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal wall components of Phytophthora infestans and specific inhibition of the reaction by suppressors of hypersensitivity. Physiological Plant Pathology. 1983; 23(3): p. 359–367. [Google Scholar]
- 53.Grant J, Yun B, Loake G. Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant Journal. 2000; 24(5): p. 569–582. [DOI] [PubMed] [Google Scholar]
- 54.Miller G, Schlauch K, Tam R, Cortes D, Torres M, Shulaev V, et al. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science Signaling. 2009; 2(84): p. 45–49. [DOI] [PubMed] [Google Scholar]
- 55.Alvarez M, Pennell R, Meijer P, Ishikawa A, Dixon R, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998; 92: p. 773–784. [DOI] [PubMed] [Google Scholar]
- 56.Dat J, Vandenabeele S, Vranova E, van Montagu M, Inze D, van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cellular and Molecular Life Sciences. 2000; 57(5): p. 779–795. 10.1007/s000180050041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuzniak E, Urbanek H. The involvement of hydrogen peroxide in plant responses to stresses. Acta Physiologiae Plantarum. 2000; 22(2): p. 195–203. [Google Scholar]
- 58.Miller G, Shulaev V, Mittler R. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum. 2008; 133(3): p. 481–489. 10.1111/j.1399-3054.2008.01090.x [DOI] [PubMed] [Google Scholar]
- 59.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti V, Vandepoele K, et al. ROS signaling: the new wave?. Trends in Plant Science. 2011; 16: p. 300–309. 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 60.Suzuki N, Miller G, Salazar C, Mondal A, Shulaev E, Cortes F, et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell. 2013; 25: p. 3553–3569. 10.1105/tpc.113.114595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lorenzo O, Piqueras R, Sánchez-Serrano J, Solano R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell. 2003; 15(1): p. 165–178. 10.1105/tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorenzo O, Solano R. Molecular players regulating the jasmonate signalling network. Current Opinion in Plant Biology. 2005; 8(5): p. 532–540. 10.1016/j.pbi.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 63.Cellini L, Fiorentini L, Buriani G, Yu J, Donati I, Cornish D, et al. Elicitors of the salicylic acid pathway reduce bacterial canker of kiwifruit. Annals of Applied Biology. 2014; 165(3): p. 441–413. [Google Scholar]
- 64.Michelotti V, Lamontanara A, Buriani G, Orrù L, Cellini A, Spinelli F, et al. Comparative transcriptome analysis of the interaction between Actinidia chinensis var. chinensis and Pseudomonas syringae pv. actinidiae in absence and presence of acibenzolar-S-methyl. BMC genomics. 2018; 19(1), 585 10.1186/s12864-018-4967-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necro- trophic pathogens. Annual Review Phytopathology. 2005; 43: p. 205–222. [DOI] [PubMed] [Google Scholar]
- 66.Bari R, Jones JDG. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009; 69: p. 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The histogram shows the inhibition haloes of Xanthomonas vesicatoria growth resulted from in vitro assays by using diffusion method.
(TIF)
The histogram shows the inhibition of Xanthomonas vesicatoria population in in vitro assays by using broth dilution method.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.