Abstract
Cyclic dinucleotides (CDNs) play central roles in bacterial homeostasis and virulence as nucleotide second messengers. Bacterial CDNs also elicit immune responses during infection when they are detected by pattern recognition receptors in animal cells. Here, we performed a systematic biochemical screen for bacterial signaling nucleotides and discovered a broad family of cGAS / DncV-like nucleotidyltransferases (CD-NTases) that use both purine and pyrimidine nucleotides to synthesize an exceptionally diverse range of CDNs. A series of crystal structures establish CD-NTases as a structurally conserved family and reveal key contacts in the active-site lid that direct purine or pyrimidine selection. CD-NTase products are not restricted to CDNs and also include an unexpected class of cyclic trinucleotide compounds. Biochemical and cellular analysis of novel signaling nucleotides demonstrate that these molecules activate distinct host receptors and thus may modulate the interaction of both pathogens and commensal microbiota with their animal and plant hosts.
Second messenger molecules allow cells to amplify signals, and rapidly control downstream responses. This concept is illustrated in human cells where mislocalized double-stranded DNA stimulates the cytosolic enzyme cyclic GMP–AMP synthase (cGAS) to synthesize the cyclic dinucleotide (CDN) 2′–5′ / 3′–5′ cyclic GMP–AMP (2′3′ cGAMP)1,2. 2′3′ cGAMP diffuses throughout the cell, activates the receptor Stimulator of Interferon Genes (STING), and induces type I interferon and NF-κB responses to elicit protective anti-viral immunity1. Most recently, synthetic CDN analogues have emerged as promising lead compounds for immune modulation and cancer immunotherapy2,3.
CDNs were first identified in bacteria4 and established the foundation for later recognition of the importance of CDN signaling in mammalian cells5. Nearly all bacterial phyla encode CDN signaling pathways, yet enigmatically, all known natural CDN signals are constructed only from purine nucleotides6. CDNs control diverse responses in bacterial cells. For example, cyclic di-GMP coordinates the transition between planktonic and sessile growth, cyclic di-AMP controls osmoregulation, cell wall homeostasis, and DNA-damage responses, and 3′–5′ / 3′–5′ cGAMP (3′3′ cGAMP) modulates chemotaxis, virulence, and exoelectrogenesis7. The human receptor STING also senses these bacterial CDNs as pathogen (or microbe) associated molecular patterns (PAMPs), revealing a direct, functional connection between bacterial and human nucleotide signaling8. However, our understanding of the true scope of immune responses to bacterial signaling nucleotide-products is limited to cyclic dipurine molecules. Here we describe a systematic approach to understanding the diversity of products synthesized by a family of microbial synthases related to the Vibrio cholerae enzyme dinucleotide cyclase in Vibrio (DncV) and its metazoan homolog cGAS9–11.
Discovery of a pyrimidine-containing CDN
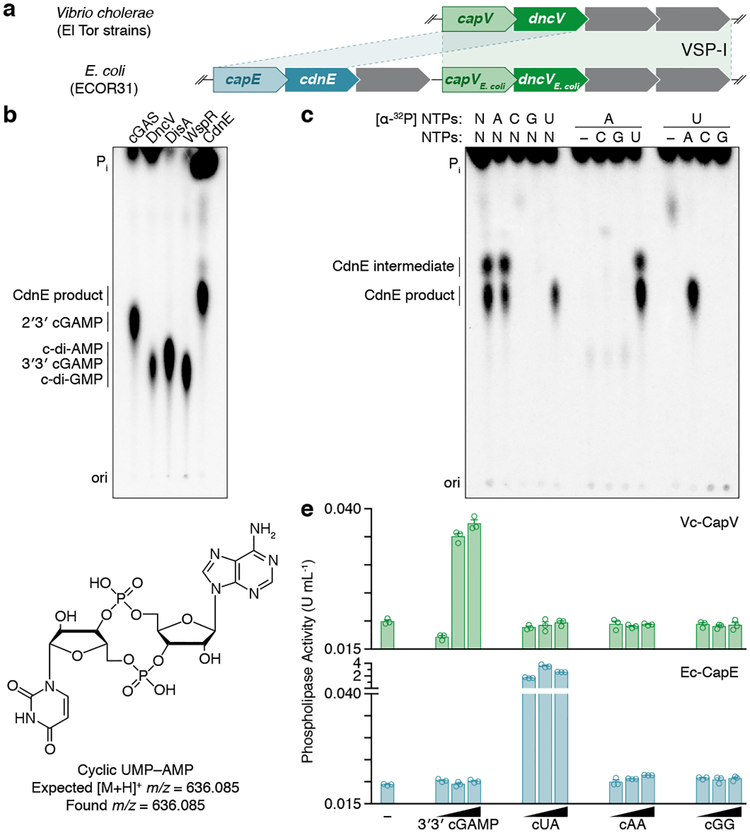
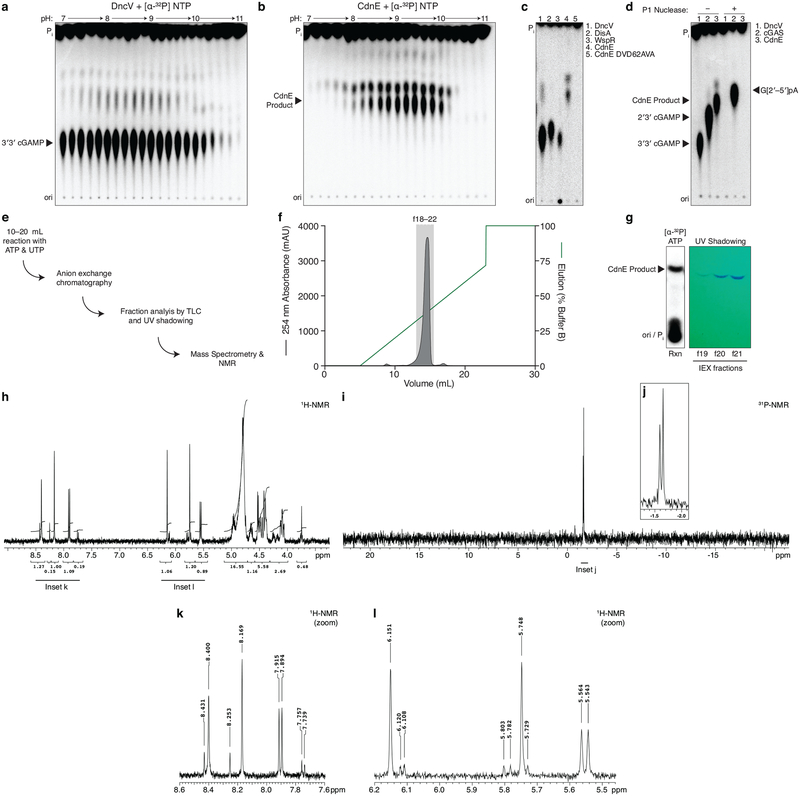
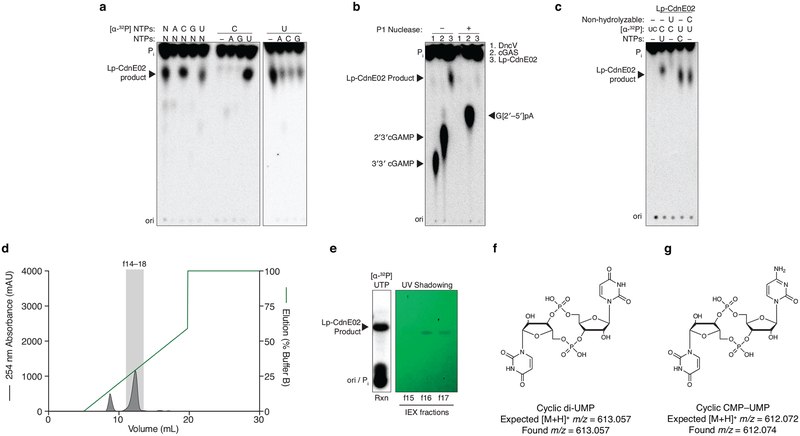
The enzyme DncV synthesizes 3′3′ cGAMP and controls a signaling network on the Vibrio seventh pandemic island-I (VSP-I), a horizontally acquired genetic element present in all current V. cholerae pandemic isolates11–13. While investigating homologs of dncV outside the Vibrionales, we identified an unexpected partial operon in E. coli where dncV is replaced with a gene of unknown function (WP_001593458, here renamed cdnE). The operon architecture implies that cdnE may be an alternative 3′3′ cGAMP synthase (Fig. 1a). We tested this hypothesis by incubating purified CdnE protein with α−32P radiolabeled ATP, CTP, GTP, and UTP and visualized the reaction products using thin-layer chromatography (TLC). CdnE synthesized a product distinct from currently known CDNs (Fig. 1b and Extended Data Fig. 1a and b). Surprisingly, biochemical deconvolution using pairwise assessment of necessary NTPs revealed that ATP and UTP were necessary and sufficient for product formation (Fig. 1c). We analyzed purified product with nuclease digestion, mass spectrometry and NMR (Fig. 1d and Extended Data Fig. 1d–l), and confirmed that the product of CdnE is cyclic UMP–AMP (cUMP–AMP), a hybrid purine–pyrimidine CDN.
Figure 1 |. Bacteria synthesize cyclic UMP–AMP.
a, An E. coli genomic island homologous to the Vibrio seventh pandemic island-I (VSP-I) encodes a 3′3′ cGAMP synthase (dncV) and phospholipase receptor (capV). The ECOR31 island encodes a second capV-like gene (BHF03_01995 encoding WP_001593459, renamed capE) adjacent to a gene of unknown function (BHF03_01990 encoding WP_001593458, renamed cdnE).
b, PEI-cellulose TLC of enzyme reactions. Purified CdnE was incubated with α−32P radiolabeled ATP, CTP, GTP, and UTP. Reactions were terminated by treating with alkaline phosphatase to remove exposed triphosphates. Standards are 2′3′ cGAMP (cGAS), 3′3′ cGAMP (DncV), c-di-AMP (DisA), and c-di-GMP (WspR). Data are representative of 3 independent experiments.
c, Biochemical deconvolution of the CdnE reactions as in (b) after incubating with α−32P labeled and unlabeled NTPs. Data are representative of 3 independent experiments.
d, The CdnE product, confirmed by mass spectrometry (MS) and NMR (see Extended Data Fig. 1e–l).
e, Activation of CapV and CapE by CDNs, tested with no nucleotide added (−) or at 0.1, 1, and 10-fold molar ratios of nucleotide to phospholipase. Enzyme activity is reported in Phospholipase A1 units mL−1. Data are mean ± standard error of the mean (SEM) for n=3 technical replicates and are representative of 3 independent experiments.
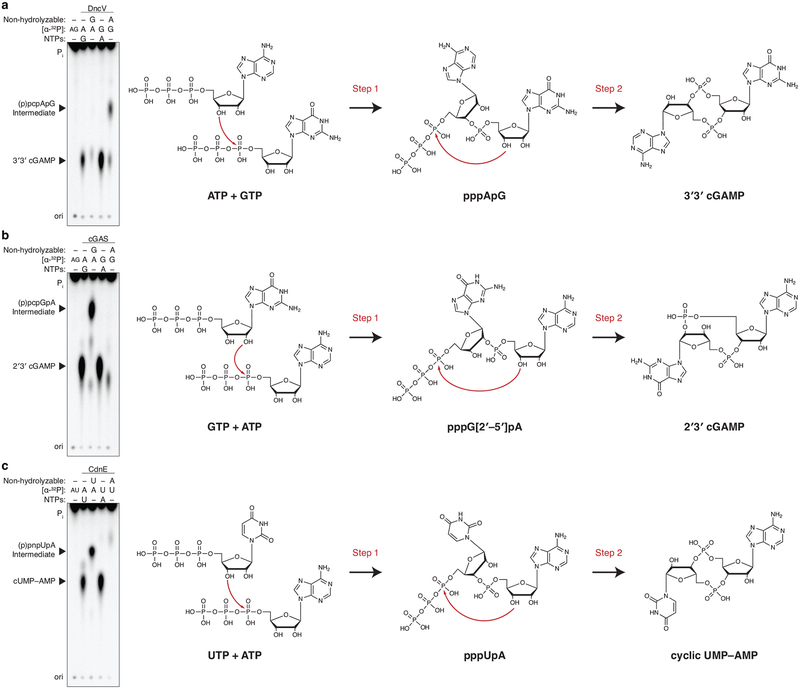
DncV is a structural homolog of cGAS, and each enzyme uses a single active site to sequentially form two separate phosphodiester bonds and release a CDN product10. In spite of no overall sequence homology, careful inspection of the CdnE sequence revealed potential cGAS / DncV-like active site residues (GSYX10DVD), which were essential for catalysis (Extended Data Fig. 1c). Reactions with nonhydrolyzable nucleotides confirmed that CdnE catalyzes synthesis of cUMP–AMP using a sequential path through a pppU[3′–5′]pA intermediate (Extended Data Fig. 2), and revealed that CdnE is likely a divergent enzyme ancestrally related to cGAS and DncV. We therefore renamed the gene cGAS / DncV-like nucleotidyltransferase in E. coli (cdnE).
In Vibrio, DncV controls the activity of cGAMP activated phospholipase in Vibrio (CapV), a patatin-like lipase that is a direct 3′3′ cGAMP receptor encoded in the dncV operon14. cdnE is also preceded by a gene encoding a patatin-like phospholipase (here renamed cUMP–AMP activated phospholipase in E. coli, capE, Fig. 1a) and we hypothesized that this lipase might be activated by cUMP–AMP. CapV and CapE were only activated by the nucleotide synthesized from their adjacently encoded nucleotidyltransferase (Fig. 1e). Importantly, the identification of CapE as a direct cUMP–AMP receptor in E. coli confirms that CdnE produces cUMP–AMP to control downstream signaling. The exquisite specificity of CapE insulates this circuit from 3′3′ cGAMP and other parallel CDN signals, potentially explaining the evolutionary advantage of cUMP–AMP and increased CDN diversity.
Mechanism of pyrimidine discrimination
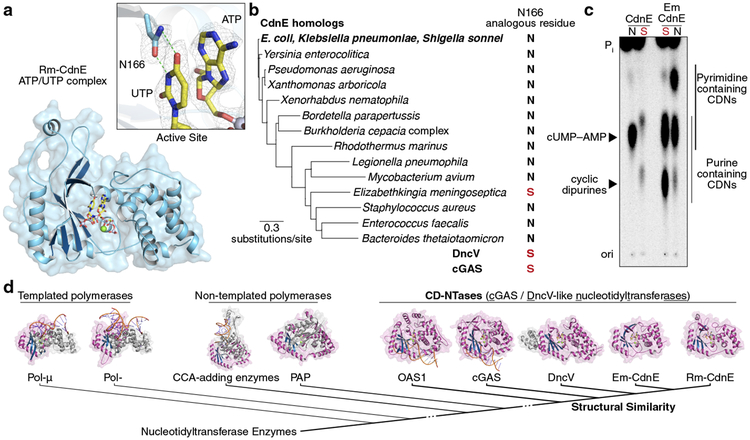
We determined a series of X-ray crystal structures of a CdnE homolog from the thermophilic bacterium Rhodothermus marinus (Rm-CdnE, Fig. 2, Extended Data Fig. 3a, and Supplementary Table 1). CdnE adopts a Pol-β-like nucleotidyltransferase fold highly similar to cGAS and the core of DncV, confirming a shared structural and evolutionary relationship (Fig. 2d). CdnE is more distantly related to other nucleotidyltransferases including non-templated CCA-adding enzymes, poly(A) polymerases, and templated polymerases such as DNA Polymerase β and μ. The human innate immune enzymes cGAS and Oligo Adenylate Synthase 1 (OAS1) are activated through a conformational change induced by binding a double-stranded nucleic acid15. CdnE, like DncV10, is structurally more similar to the “activated” conformation of these two enzymes, consistent with biochemistry demonstrating CdnE is constitutively active and does not require a cognate stimulus in vitro.
Figure 2 |. Conserved active site residues dictate CD-NTase specificity.
a, Crystal structure of Rm-CdnE in complex with nonhydrolyzable ATP and UTP analogs and zoom-in inset of key N166–uridine contacts controlling pyrimidine specificity. Green dotted lines indicate hydrogen bonding and 2Fo−Fc electron density map is contoured at 1 σ.
b, Phylogram of CdnE sequence homologs and their N166 analogous residue determined by sequence alignment (Extended Data Fig. 4a). Red “S’s” highlight cGAS / DncV-like serine residues.
c, CdnE homolog and mutant reactions analyzed by TLC as in Fig. 1b. “N” vs red “S” indicate asparagine or cGAS / DncV-like serine at the N166 analogous position in the tested allele. Side-chains are numbered according to Rm-CdnE sequence. Data are representative of 3 independent experiments. For detailed deconvolution and purine vs pyrimidine migration pattern analysis see Extended Data Fig. 3 and 4.
d, Structure-based comparisons define that Rm-CdnE and Em-CdnE are cGAS / DncV-like nucleotidyltransferase (CD-NTases) with a similar architecture to DncV (4TY0), cGAS (6CTA), and OAS1 (4RWO). CD-NTases are more distantly related to Pol-β-like NTases: Pol-μ (4YD1), Pol-β (4KLQ), CCA-adding enzyme (4X4T), and Poly(A) Polymerase gamma (PAP, 4LT6). The NTase core domain for each enzyme is clustered according to Z-score and colored in magenta/blue.
The structure of Rm-CdnE in complex with nonhydrolyzable ATP and UTP reveals an asparagine side chain (N166) that forms hydrogen bonds with the uracil base and positions the ATP α-P for attack by the 3′ hydroxyl of UTP (Fig. 2a). N166 is located in the same position as a serine residue in the first acceptor nucleotide pocket of both DncV and cGAS (Extended Data Fig. 3b and d), and we hypothesized that this asparagine substitution might be sufficient to dictate CdnE product specificity. Whereas wild-type CdnE robustly synthesized cUMP–AMP, CdnEN166S incorporated almost no UTP and instead predominantly synthesized c-di-AMP (Fig. 2c and Extended Data Fig. 3c). We surveyed CdnE homologs and determined that N166 is nearly universally conserved (Fig. 2b and Extended Data Fig. 4a). An exception is CdnE from the emerging nosocomial pathogen Elizabethkingia meningoseptica (Em-CdnE, Fig. 2b)16, which encodes a serine at the analogous position to N166. Unlike the other CdnE homolog, Em-CdnE robustly synthesized cyclic dipurine products (Fig. 2c and Extended Data Fig. 4b–h). Crystal structures of Em-CdnE bound to its nucleotide substrates demonstrated natural N to S reprogramming in the active-site lid, and re-introduction of the ancestral asparagine at this position reverted Em-CdnE back to preferential hybrid purine–pyrimidine product formation (Supplementary Table 1, Fig. 2c and Extended Data Fig. 4f–i). These data reveal a remarkably low barrier for altering specificity of CdnE and demonstrate that organisms like E. meningoseptica harbor mutations at N166 that reprogram purine and pyrimidine product specificity.
High-resolution structures of cGAS, OAS1, DncV, and two CdnE homologs allowed for the rational definition of shared structural and functional homology. All of these enzymes share three features: (1) a common DNA polymerase β-like nucleotidyltransferase superfamily protein-fold in spite of large sequence divergence, (2) template-independent synthesis of a diffusible molecule through caging of the active site, using a protein scaffold not conserved with more distantly related templated polymerases, and (3) an active site architecture that allows diversification of products and phosphodiester linkage through amino acid substitutions within the active-site lid. We have designated this family of enzymes as CD-NTases (cGAS / DncV-like Nucleotidyltransferases), a structurally and evolutionarily distinct subset of the DNA polymerase β-like nucleotidyltransferase superfamily (Fig. 2d). CD-NTases use distinct enzymatic chemistry and are not structurally related to dimeric GGDEF family c-di-GMP synthases or DAC/DisA family c-di-AMP synthases17,18, and therefore represent a third family of CDN synthase.
CD-NTases and cross-kingdom signaling
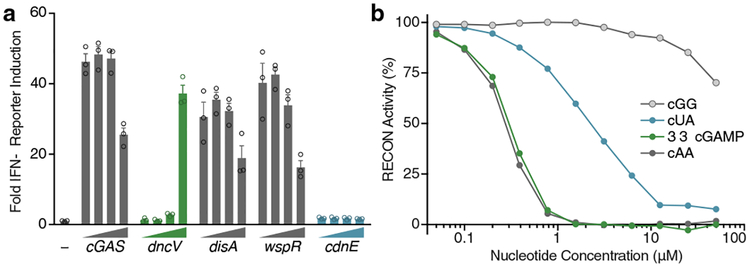
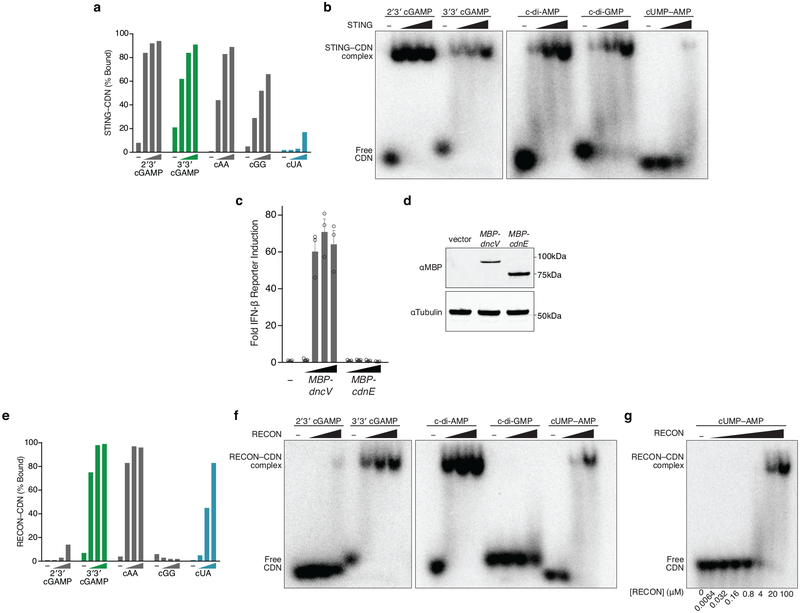
Many bacteria that encode CD-NTases thrive in close proximity to eukaryotic hosts, including humans, plants, and fungi (Fig. 2b). CdnE homologs are found in the intracellular pathogen Shigella sonnei and commensal genera such as Bacteroides (Fig. 2b). Mammals have evolved a sophisticated surveillance system for detecting and initiating immune responses to bacterial products, including CDNs that are secreted or released during bacteriolysis19. Mouse STING detects bacterial c-di-AMP, c-di-GMP, and 3′3′ cGAMP in addition to endogenously produced 2′3′ cGAMP1. We determined if cUMP–AMP was also recognized by STING or other receptors of the innate immune system. STING bound to all four cyclic dipurine molecules with high affinity and activated type I interferon signaling in cells. However, STING was unable to recognize cUMP–AMP in vitro at concentrations known to be sufficient for cyclic dipurine agonists and cUMP–AMP failed to activate STING-dependent type I interferon signaling in cells (Fig. 3a and Extended Data Fig. 5a–d). These data are consistent with previous experiments using chemically synthesized nucleotides20 and were not due to differences in CD-NTase expression levels (Extended Data Fig. 5c and d). In contrast, the recently described mammalian CDN sensor reductase controlling NF-κB (RECON)21 was capable of recognizing cUMP–AMP, and cUMP–AMP inhibited RECON function, albeit with a reduced potency compared to the previously reported inhibition by c-di-AMP and 3′3′ cGAMP (Fig. 3b and Extended Data Fig. 5e–g). RECON bound to cUMP–AMP with a similar low micromolar Kd to that of STING for c-di-GMP (Extended Data Fig. 5g)8, thereby identifying the first host receptor for a naturally occurring purine–pyrimidine hybrid CDN. Whereas the specificity of STING for CDNs is dependent on the presence of two purine bases, RECON requires only the minimal presence of an adenine base in a 3′3′ CDN. These results highlight how discovery of natural bacterial signaling molecules can refine our understanding of host receptor specificity, and demonstrate that the host response may be tuned via multiple receptors that compete for CDNs using distinct rules of engagement.
Figure 3 |. Immune detection of a pyrimidine containing CDN.
a, In-cell STING reporter assay. Induction of an Interferon-β (IFN-β) reporter in HEK293T cells transfected with a concentration gradient of plasmid overexpressing enzymes as indicated. cGAS synthesizes 2′3′ cGAMP, DncV synthesizes 3′3′ cGAMP, DisA synthesizes c-di-AMP, WspR synthesizes c-di-GMP, and CdnE synthesizes cUMP–AMP. Data are fold induction over vector only, shown as (−), are mean ± SEM for n=3 technical replicates, and are representative of 3 independent experiments.
b, Nucleotide inhibition of RECON enzymatic activity, as measured by oxidation of NADPH co-substrate. X-axis is a log scale and data are representative of 3 independent experiments.
CD-NTases synthesize diverse nucleotide products
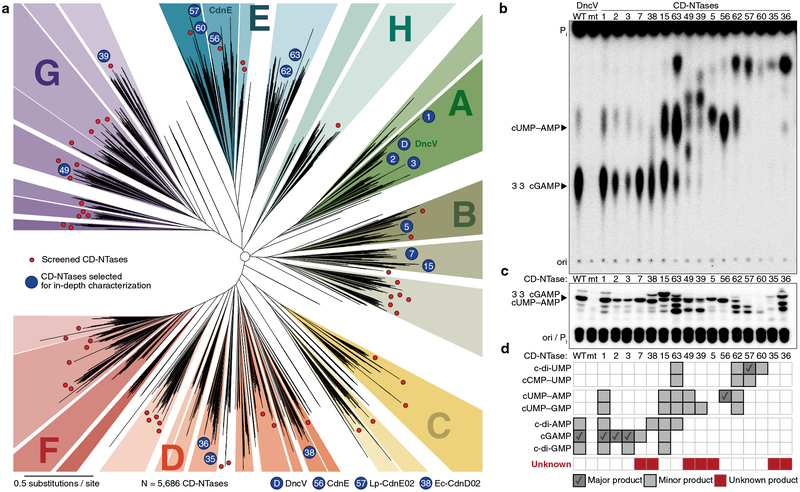
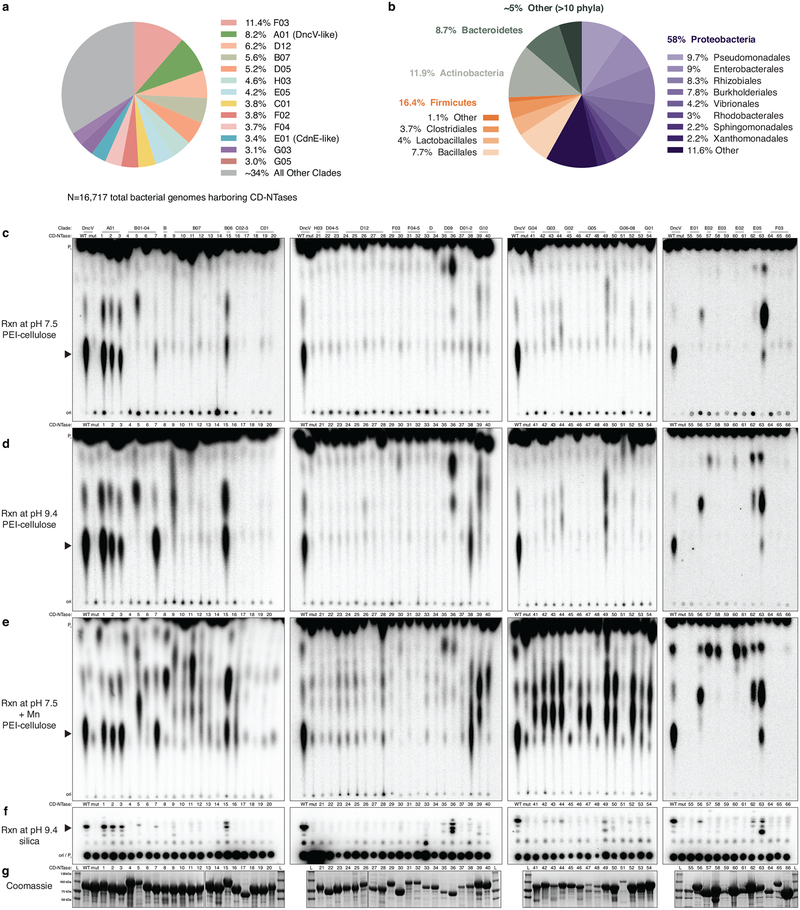
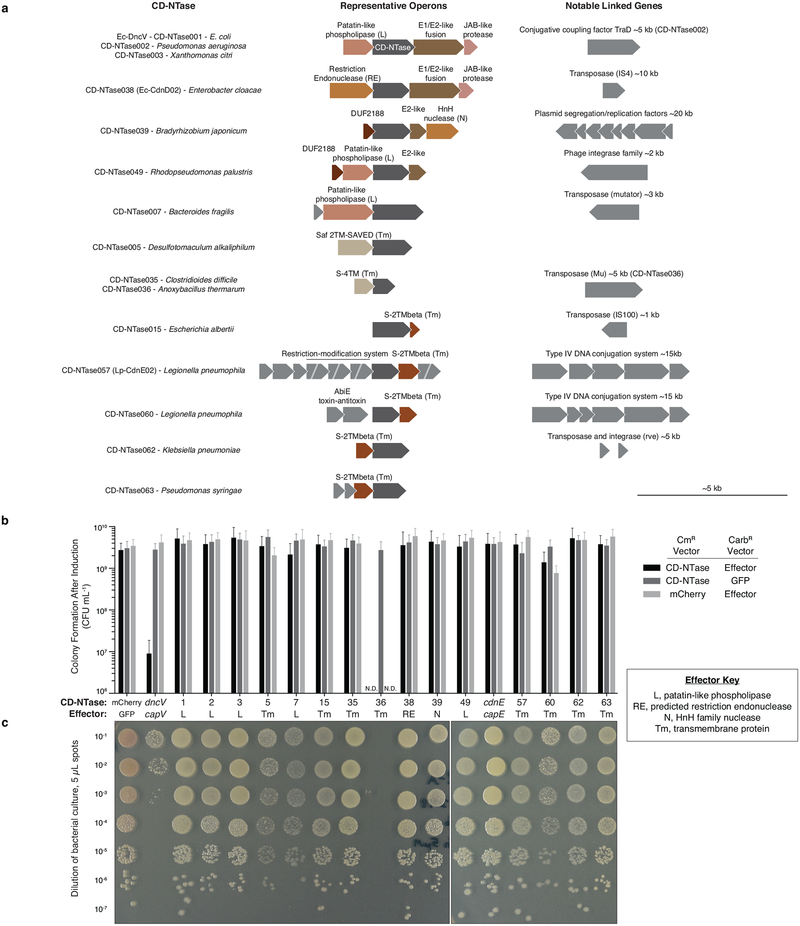
DncV and CdnE evolved from a common ancestor but exhibit dramatic divergence in primary amino acid sequence. We hypothesized that these enzymes comprise only a small fraction of existing bacterial CD-NTase diversity, and that kingdom-wide analysis of the protein family would allow systematic identification of bacterial signaling nucleotides as well as agonists/antagonists of the innate immune system. We therefore coupled bioinformatic analysis with a large-scale, forward biochemical screen to directly uncover additional nucleotide products. Previously, Burroughs et al. used a hidden Markov model derived from cGAS and DncV and conserved operon structures to identify potentially related bacterial proteins22. Building upon this previous analysis, we identified >5,600 unique bacterial enzymes predicted to share common CD-NTase structural features (Fig. 4a, Extended Data Fig. 6a, and Supplementary Table 2). CD-NTases were identified in >10% of bacterial genomes available in the NCBI database, within taxa that span nearly every bacterial phylum (Extended Data Fig. 6b). Bacteria harboring CD-NTase genes include human commensal organisms (e.g., Clostridiales, and Fusobacteria), human pathogens (e.g., Listeria, Shigella, and Salmonella species), extremophiles, and agriculturally significant bacteria (e.g., rhizobia commensals and plant pathogens such as Xanthomonas). Although CD-NTases are found in many different organisms, they are typically not encoded in the core genome and are found in specific strains from each species. Sequence alignments revealed that CD-NTases cluster into roughly eight clades that we designated A–H starting with A for the DncV-harboring clade, E for the CdnE containing clade, and continued to the letter H. We further divided highly-related sequences into clusters, which often grouped bacterial species that occupy a similar niche, such as plant rhizobia in cluster G10 (Fig. 4a and Supplementary Table 2).
Figure 4 |. CD-NTases synthesize diverse nucleotide products and form a family of enzymes abundant in many bacterial phyla.
a, Bioinformatic identification and alignment of ~5,600 predicted CD-NTases found in nearly every bacterial phylum, shown as an unrooted tree. Sequence-related enzymes that are ~10% identical are grouped by lettered clade and similarly colored, enzymes ~25% identical are grouped by cluster in a similar color shade. Circles represent CD-NTase001–066 that were selected as type CD-NTases for a biochemical screen (See Extended Data Fig 6 for additional details). Blue circles denote CD-NTases selected for in-depth characterization and are labeled with CD-NTase numbers from the biochemical screen (see Fig. 4b, CdnE is “56” and DncV is “D”). For additional information, see Supplementary Discussion, Supplementary Table 2, and source data for this figure provided as Supplementary Data.
b and c, PEI-cellulose and silica TLC analysis of 16 CD-NTases selected for in-depth characterization. Activity was analyzed with α−32P radiolabeled NTPs as in Fig. 1b. Wild-type (WT) and catalytically inactive mutant (mt) DncV reactions are included as controls. Screened CD-NTases were numbered CD-NTase001–066. CD-NTase056 is CdnE, CD-NTase057 was renamed Lp-CdnE02, and CD-NTase038 was renamed Ec-CdnD02. These appear as “56”, “57”, and “38” in Fig. 4a, respectively. Data are representative of 3 independent experiments.
d, Identification of CD-NTase products by combining TLC and MS data. CD-NTases that synthesize a major product that could not be matched with a predicted CDNs are denoted as “unknown.”
We purified 66 CD-NTase proteins and tested each for nucleotide product synthesis (Fig. 4a and Extended Data Fig. 6c–g). These proteins were selected as type enzymes from each cluster based on the relevance of the organism from which they were isolated (pathogens, commensals, and bacteria predicted to interact with eukaryotes) and the frequency at which each sequence has been re-isolated from multiple organisms. Recombinant proteins were screened using a broad range of reaction conditions to identify robust activity. Despite encoding an intact active site, no activity was observed from any representative of some CD-NTase clusters. These enzymes may function similarly to human cGAS and OAS1 where a cognate ligand (e.g., dsDNA and dsRNA) is required to stimulate enzyme activity, or it is possible that these clusters may utilize building blocks other than ribonucleotide triphosphates for product synthesis.
The 16 most active CD-NTases were selected for in-depth analysis (Fig. 4b and c). Our previous results established that cyclic dipurine and cyclic purine–pyrimidine hybrid molecules migrate at the bottom and middle of PEI-cellulose TLC plates, respectively. In the collection of active CD-NTase representatives, several enzymes produced products that migrated at the top of the plate, even more rapidly than cUMP–AMP. Further biochemical analysis of CD-NTase057, (renamed Lp-CdnE02) from Legionella pneumophila (strain 12_4117) demonstrated that this class of PEI-cellulose TLC species corresponded to cyclic dipyrimidines, and Lp-CdnE02 synthesized predominantly c-di-UMP (Fig. 4b–d, and Extended Data Fig. 7). Lp-CdnE02 also harbors an asparagine residue analogous to N166 of Rm-CdnE, a feature found in nearly all CD-NTases in clade E but not found in other clades. Mass spectrometry of each CD-NTase reaction coupled with NTP substrate dependency profile and TLC data allowed us to identify the products produced by different CD-NTases and estimate their abundance (Fig. 4d). The 16 active, representative enzymes produced 7 purine, pyrimidine, and purine–pyrimidine hybrid CDN combinations, demonstrating that CD-NTase enzymes synthesize an extraordinarily diverse array of bacterial nucleotide signals (Fig. 4d).
CD-NTases are encoded in conserved operons on mobile genetic elements
A unifying characteristic of almost all CD-NTase-encoding genes is their location within similar operons in predicted mobile genetic elements (Extended Data Fig. 8a). Often genes encoding identical CD-NTase proteins are found in specific strains of unrelated bacterial species, reflecting that these genes are members of the “mobilome” (for detail on identifying CD-NTases within an organism of interest, see Supplementary Discussion). The horizontal acquisition of CD-NTases suggests that they are likely to provide a selective advantage, may not alter species-specific nucleotide signaling networks, and instead alter bacterial physiology via receptors adjacently encoded, similar to capV-dncV and capE-cdnE. Burroughs et al. noted that genes adjacent to CD-NTases are effector-like and are generally involved in biological conflict, including phospholipases, nucleases, and pore-forming agents22. Coexpression of dncV and capV is toxic to E. coli14 and we tested if coexpression of each CD-NTase with its adjacently-encoded, putative receptor was also toxic to E. coli. Expression of dncV-capV was unique in inhibiting colony formation and other CD-NTase-predicted receptor pairs, including the cdnE-capE pair, did not impair bacterial growth (Extended Data Fig. 8b and c). However, it is unclear if CD-NTases are constitutively active in vivo or exhibit regulated enzymatic activity like the metazoan second messenger synthase cGAS. These findings demonstrate that phenotypes observed with Vibrio dncV-capV may not be indicative of general CD-NTase function, and that CD-NTase-containing islands may perform functions such as mediating bacteriophage resistance, modulating bacterial-host interactions, functioning as addiction modules, or regulating bacteriolysis for dissemination of mobile genetic elements.
Bacteria CD-NTase products include cyclic trinucleotide signals
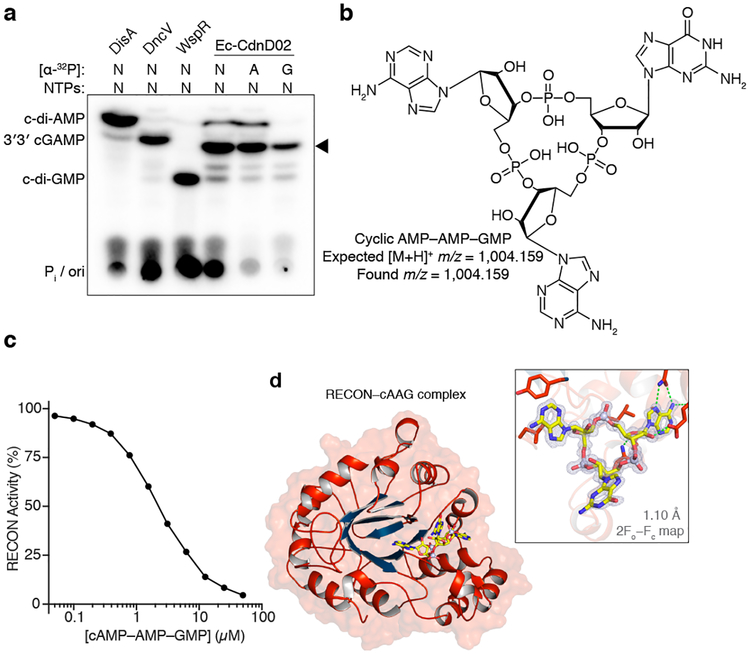
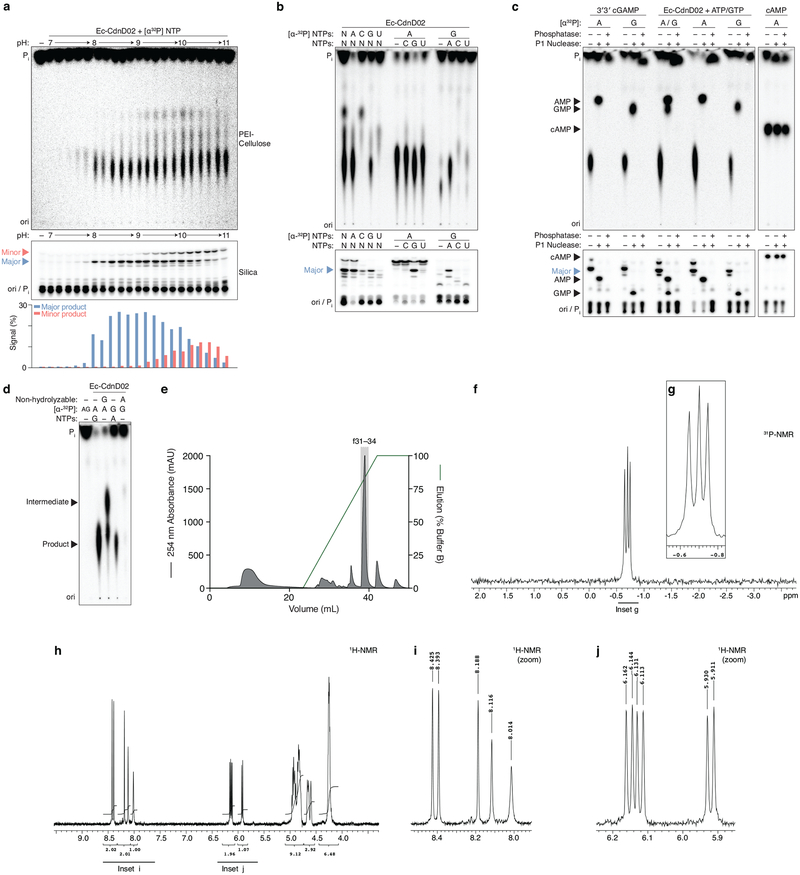
Surprisingly, we were unable to identify expected CDNs by mass spectrometry in some reactions despite visualizing robust product formation by PEI-cellulose TLC. Using orthogonal TLC conditions, these unknown products exhibited distinct migration patterns that suggested existence of unique non-CDN species (Fig. 4c and 4d). We focused on an orphan product of CD-NTase038 (renamed Ec-CdnD02) from Enterobacter cloacae (strain UCI 50) for identification. The Ec-CdnD02 product initially appeared to be a cyclic dipurine by PEI-cellulose TLC, but the major Ec-CdnD02 product displayed a unique migration pattern when analyzed by silica TLC (Fig. 5a and Extended Data Fig. 9a). ATP and GTP were necessary and sufficient for product formation, however, roughly two thirds of the total α32-P was incorporated from ATP and the remaining third from GTP. Consistent with this pattern, re-evaluation of the mass spectrometry data and subsequent biochemical and NMR validation revealed that cyclic AMP–AMP–GMP (cAAG), a cyclic trinucleotide, is the major product of Ec-CdnD02 (Fig. 5b and Extended Data Fig. 9b–j).
Figure 5 |. Bacterial synthesis and host recognition of a cyclic trinucleotide.
a, Silica TLC analysis of the Ec-CdnD02 (CD-NTase038) product and control reactions as in Fig. 1b. The major product of Ec-CdnD02 is indicated with a triangle and incorporates ~2× greater α−32P from ATP than α−32P from GTP. Data are representative of 3 independent experiments.
b, The major product of Ec-CdnD02, cyclic AMP–AMP–GMP (cAAG), confirmed by MS and NMR, see Extended Data Fig. 9 for additional characterization.
c, cAAG inhibition of RECON enzymatic activity, as measured by oxidation of NADPH co-substrate. X-axis is a log scale and data are representative of 3 independent experiments.
d, Co-crystal structure of the host receptor RECON in complex with cAAG, and inset highlighting the cAAG 2Fo−Fc electron density contoured at 1.3 σ. Green dotted lines indicate hydrogen bonding, some RECON–cAAG contacts omitted for clarity, see Extended Data Fig. 10.
Similar to cUMP–AMP, the bacterial cyclic trinucleotide cAAG escaped STING recognition but was detected by RECON, confirming our new definition of STING and RECON ligand specificity (Fig. 5c, Extended Data Fig. 10a and f). We next determined a co-crystal structure of RECON in complex with the Ec-CdnD02 cyclic trinucleotide product (Fig. 5d, Extended Data Fig. 10, and Supplementary Table 1). The structure further confirms that the bacterial cyclic trinucleotide is cAAG and contains exclusively 3′–5′ phosphodiester linkages. The two adenine bases are coordinated in the same adenine and nicotinamide pockets observed in the previous structure of RECON bound to bacterial c-di-AMP21, but unexpectedly RECON E28 makes additional contacts with the third guanine base of the cAAG species as part of an extended base platform not required for CDN recognition. E28 is highly conserved, potentially indicating that RECON may have evolved to allow recognition of additional bacterial or host cyclic trinucleotide species. This unexpected class of nucleotide product reveals that the active site of CD-NTase enzymes can be adapted to synthesize larger cyclic oligonucleotide products, and that host immune receptors are capable of recognizing bacterial cyclic trinucleotide species. Recently, cyclic oligoadenylate synthesized by Cas10 was demonstrated to be a key signaling molecule in type III CRISPR immunity23,24. Although CD-NTases have no homology with Cas10, these parallel findings indicate that larger cyclic oligonucleotide products may be more common in bacterial signaling and host recognition than previously expected.
CD-NTases in health and disease
Our data demonstrate that bacterial CD-NTases are widespread and synthesize diverse CDNs that include pyrimidine nucleotides and additional cyclic trinucleotide compounds. CD-NTases join the GGDEF and DAC/DisA domains, responsible for c-di-GMP and c-di-AMP synthesis17,18, as a third major family of enzymes that control downstream signaling using CDN signals. Distinguishing features of CD-NTases are their location on mobile genetic elements, extreme sequence diversity, and reaction mechanism reliant on a monomeric enzyme active site. Recent evidence demonstrates divergent GGDEF family enzymes produce 3′3′ cGAMP in addition to c-di-GMP25,26, suggesting that the selective pressures driving CD-NTase diversity may also be in effect for GGDEF and DAC/DisA-like synthases.
Understanding the functional role of CD-NTase genes in the biology of bacteria and host-microbe interactions is a major challenge for future studies. Mammalian receptors recognize diverse CD-NTase products, and CD-NTase genes may provide a selective advantage for some bacterium-eukaryote interactions. Our data show that a single mutation in a CD-NTase enables incorporation of pyrimidines and indicate that bacteria may evade or enhance STING signaling by modulating enzyme specificity. The possibility that diverse CDNs and related nucleotide signals produced by prokaryotic CD-NTases act as agonists and inhibitors of innate immunity and other host metabolic pathways provides an important new reservoir of compounds with biotechnology and therapeutic applications.
Methods
Bacterial strains and growth conditions
E. coli was cultivated at 37 °C, shaking, in LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl w/v), and stored in LB plus 30% glycerol at −80 °C unless otherwise indicated. When appropriate, carbenicillin (100 μg mL−1), ampicillin (100 μg mL−1), and chloramphenicol (20–34 μg mL−1) were used. BL21 E. coli (strain CodonPlus (DE3)-RIL transformed with pRARE2, Agilent) was used for all protein expression and DH10β E. coli (strain Top10, Invitrogen) was used for cloning and plasmid propagation. For repression of protein expression from pET vectors, BL21 E. coli was cultivated in MDG medium (0.5% glucose, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 0.25% aspartic acid, and trace metals) with ampicillin and chloramphenicol. For optimum protein expression from pET vectors, BL21 E. coli was cultivated in M9ZB medium (0.5% glycerol, 1% Cas-Amino Acids, 47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, 85.6 mM NaCl, 2 mM MgSO4, and trace metals) with ampicillin and chloramphenicol27.
Cloning and plasmid construction
Cloning and plasmid construction were performed as previously described28. Briefly, for vectors constructed in this study, genes were either amplified from genomic DNA or synthesized as gBLOCKs (Integrated DNA Technologies) with ≥18 base pairs of homology flanking the insert sequence and ligated into restriction endonuclease linearized vector by Gibson assembly. Reactions were transformed into electrocompetent DH10β and selected with appropriate antibiotic plates. Sanger sequencing confirmed each vector was free of mutations within the multiple cloning site. N-terminal 6×His-MBP tag and 6×His-SUMO2 tag fusions were constructed using custom pET16MBP29 or pETSUMO230 vectors, respectively. cGAS standards used Homo sapiens CGAS, DisA standards used Bacillus thuringiensis disA, and WspR standards used Pseudomonas aeruginosa wspR with a D70E constitutively activating mutation31. CD-NTases and their effector coding sequences were codon optimized for bacterial expression (Integrated DNA Technologies) with the exception of genes derived from E. coli strain ECOR3132 (ATCC 35350) and V. cholerae strain C6706. Synthases were overexpressed in mammalian cells from pcDNA4 plasmids as previously described33. For expression of MBP N-terminally tagged dncV and cdnE in mammalian cells, MBP and the fused CD-NTase were codon optimized for expression in human cells. For coexpression with their putative effector genes, N-terminal MBP-tagged CD-NTases were cloned into pBAD3334 modified with a ribosomal binding site and oriT for conjugation. For cloned CD-NTase details see Supplementary Table 2 and for cloned CD-NTase effector details see Supplementary Table 3.
Recombinant protein purification
Proteins were purified as previously described30. Briefly, chemically competent BL21 E. coli was transformed with a protein expression plasmid, recovered on MDG plates overnight, cultivated as a 30 mL starter culture in MDG liquid medium overnight at 37 °C with 230 RPM shaking, and used to seed an M9ZB culture at ~1:1000. 25 mL or 2× 1 L M9ZB cultures were cultivated for ~5 h at 37 °C with 230 RPM shaking until OD was 2–3.5 at which time cells were chilled on ice for 20 min, IPTG was added at 0.5 mM and cultures were shifted to 16 °C with shaking 230 RPM overnight. Harvested E. coli was washed in 1× PBS, stored as a flash-frozen pellet at −80 °C or immediately disrupted by sonication in lysis buffer (20 mM HEPES-KOH pH 7.5, 400 mM NaCl, 30 mM imidazole, 10% glycerol, 1 mM DTT). Lysates were clarified by centrifugation, filtered through glass wool, and proteins were purified by affinity chromatography using Ni-NTA (Qiagen) resin and a gravity column. Resin was washed (lysis buffer supplemented to 1 M NaCl), eluted (lysis buffer supplemented to 300 mM imidizole), and eluate was dialyzed overnight at 4 °C (20 mM HEPES-KOH pH 7.5, 300 mM NaCl, 1 mM DTT). For SUMO2 fusion proteins, dialysis was supplemented with ~250 μg of human SENP2 protease (D364–L589, M497A)35. Small-scale preparations of proteins were flash-frozen at this stage and stored at −80 °C in storage buffer (10% glycerol, 20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM TCEP). Where appropriate, proteins were filter-concentrated using centrifugation and a 10 kDa or 30 kDa cut-off column (Millipore Sigma).
For large-scale protein preps (cGAS, DncV, DisA, CdnE, Rm-CdnE, Em-CdnE, STING, RECON, Lp-CdnE02, Ec-CdnD02, CyaA), size exclusion chromatography followed by concentration was performed in storage buffer without glycerol. Initial CD-NTase proteins were purified and screened as N-terminal MBP fusions to increase expression and stability. Proteins were either freshly thawed from −80 °C stocks and immediately used or maintained at −20 °C in a storage buffer with 50% total glycerol. Glycerol stocks of CD-NTases stored at −20 °C retain >90% activity for >6 months and were used for biochemical assays.
Biochemistry and nucleotide synthesis assays
Recombinant CD-NTase reactions combined: 4 μL of 5× reaction buffer (250 mM CAPSO pH 9.4, 175 mM KCl, 25 mM Mg(OAc)2, 5 mM DTT), 2 μL of 10× NTPs, 1 μL [ɑ−32P] NTPs (~1 μCi), 1 μL of candidate enzyme in storage buffer (~20 μM), and a remaining volume of nuclease free water for a total reaction volume of 20 μL. The final reactions (50 mM CAPSO pH 9.4, 50 mM KCl, 5 mM Mg(OAc)2, 1 mM DTT, ≤5% glycerol, 25–250 μM individual NTPs, trace amounts of [ɑ−32P] NTP, 1 μM enzyme) were started with addition of enzyme. Where indicated, pH was altered by replacing CAPSO buffer with appropriate buffer from a StockOptions pH Buffer Kit (Hampton Research). When appropriate, Mg2+ was replaced with an equimolar concentration of Mn2+ (MnCl2). cGAS reactions were carried out with Tris at pH 7.5 and supplemented with 1 μM ISD45 dsDNA36. Reactions were carried out with 25 μM of each indicated NTP for Figures 1b, 2c, and 4b–c and for Extended Data Figures 1d, 2a–c, 4c, 6c–f, 7a–c, 8d. Reactions in all other figures were carried out with 250 μM NTP. The NTP/[ɑ−32P] NTPs in Fig. 1b are cGAS (ATP, GTP/[ɑ−32P] GTP), DncV (ATP, GTP/[ɑ−32P] ATP), DisA (ATP /[ɑ−32P] ATP), WspR (GTP /[ɑ−32P] GTP), and CdnE (NTP /[ɑ−32P] NTP). Nuclease P1 treated reactions in Extended Data Figures 1d and 7c are cGAS (ATP, GTP/[ɑ−32P] ATP), DncV (ATP, GTP/[ɑ−32P] GTP), CdnE (ATP, UTP/[ɑ−32P] ATP), and Lp-CdnE02 (CTP, UTP/[ɑ−32P] UTP). Where indicated, nonhydrolyzable nucleotides [Ap(c)pp, Gp(c)pp, Cp(c)pp, or Up(n)pp (Jena Bioscience)] were used at 25 μM.
Reactions were incubated for 2 h at 37 °C prior to analysis unless otherwise stated. Reactions were stopped by addition of 5 U of alkaline phosphatase (New England Biolabs) which removed triphosphates on remaining NTPs and converted the remaining nucleotide ɑ−32P to 32Pi allowing visualization of cyclized species. After a ≥20 min incubation, 0.5 μL (PEI-cellulose) or 1 μL (silica) of the reaction was spotted 1.5 cm from the bottom of the TLC plate, spaced 0.8 cm apart. When migration of Pi appeared inconsistent (e.g. due to variability in pH) samples were diluted 10-fold in 100 mM sodium acetate pH 5.2. 20 cm × 20 cm F-coated PEI-cellulose TLCs (Millipore) were developed in 1.5 M KH2PO4 (pH 3.8) until the buffer front reached ~1 cm from the top; 20 cm × 10 cm F-coated silica HP-TLC plates (Millipore) were developed in 11:7:2 1-propanol: NH4OH: H2O in a chemical fume hood for 1 h. Plates were dried and exposed to a phosphorscreen prior to detection by Typhoon Trio Variable Mode Imager system (GE Healthcare).
Nucleotide synthesis and purification
Cyclic dinucleotides and oligonucleotides were produced in large scale using previously described methods37 with the following changes. Small-scale nucleotide synthesis assays were scaled up to 10–40 mL reactions with final conditions of 50 mM CAPSO pH 9.4, 12.5–50 mM K/NaCl, 5–20 mM Mg(OAc)2, 1 mM DTT, ≤5% glycerol, 250 μM individual NTPs, and 1 μM enzyme. A 20 μL aliquot of the larger reaction was removed and [ɑ−32P] NTPs were added to monitor reaction progress. Reactions were incubated for 24 h at which time 5 U mL−1 of alkaline phosphatase (New England Biolabs) was added and the reaction was further incubated for 2–24 hours. Reactions were heat inactivated at 65 °C for 30 min, diluted to a final salt concentration of 12.5 mM, and purified by anion exchange chromatography and FPLC (either 1 mL Q-sepharose column or Mono Q 4.6/100 PE, GE Healthcare). The column was washed with water and 1 mL fractions were collected during a gradient elution with 2 M ammonium acetate. Fractions harboring the appropriate product were identified by A260 and silica TLC, visualizing the nucleotide products by UV-shadowing, imaging using a handheld camera, and comparing migration to paired, radiolabeled reactions detected by phosphorimaging. Selected fractions were concentrated by evaporation and re-suspended in 30 μL of nuclease free water for MS. For NMR, nucleotides were further purified using size chromatography (Superdex 30 Increase 10/300 GL, GE Healthcare). The column was equilibrated with H2O running buffer and 1 mL fractions were collected, identified by A260, pooled, and evaporated. Concentrations of purified nucleotides were estimated from A260 using the estimated extinction coefficients based on RNA oligonucleotides: cUMP–AMP ε=22,800 L mole−1 cm−1, cAAG ε=37,000 L mole−1 cm−1.
Mass spectrometry
ESI-LC/MS analysis was performed using an Agilent 6530 QTOF mass spectrometer coupled to a 1290 infinity binary LC system operating the electrospray source in positive ionization mode. All samples were chromatographed on an Agilent ZORBAX Bonus-RP C18 column (4.6 × 150 mm; 3.5 μm particle size) at 50 °C column temperature. The solvent system consisted of 10 mM ammonium acetate (A) and methanol (B). The HPLC gradient with a flow rate of 1 ml min−1 starts at 5% B, holds for 2 min and then increases over 12 min to 100% B. Identification of CDNs and cAAG was performed by targeted mass analysis for exact masses and formulae for all possible CDNs and cAAG using Profinder software (version B.06.00 build 6.0.606.0, Agilent).
NMR
All NMR experiments were conducted on a Varian 400-MR spectrometer (9.4 T, 400 MHz). Samples were prepared by re-suspending evaporated nucleotide samples in 500 μL D2O supplemented with 5 mM TMSP (3-(trimethylsilyl)propionic-2,2,3,3-d4) at 27 °C. Data were processed and figures were generated using VnmrJ software (version 2.2C). 1H and 31P chemical shifts are reported in parts per million (ppm). J coupling constants are reported in units of frequency (Hertz) with multiplicities listed as s (singlet), d (doublet), and m (multiplet). These data appear in the figure legends of each NMR spectra.
Phospholipase assay
Patatin-like lipases were assayed as previously described38. Briefly, CapV and CapE were produced recombinantly and catalytic activity was measured using the EnzChek Phospholipase A1 Assay Kit (Invitrogen) according to the manufacturer’s instructions. Phospholipases (250 nM) were incubated with 2.5, 0.25, or 0.025 μM CDN. c-di-AMP (Invivogen), 3′3′ cGAMP (Invivogen), and c-di-GMP (Biolog) were purchased as chemical standards, cUMP–AMP was purified as described above. Assays were monitored fluorometrically (Ex = 460 nm / Em = 515 nm) for 60 min at ~90 s intervals at room temperature using a Biotek Synergy plate reader. Slope of each reaction in the linear range was used to calculate activity (Linear regression/straight line analysis, Prism 7.0c). A PLA1 standard curve from 20–0.02 U was used to interpolate phospholipase activity. Emission was monitored at a gain of 100 and/or 50 in order to extend the linear range of the assay.
Crystallization and structure determination
CdnE homologs were crystallized in apo form or in complex with nucleotide substrates at 18 °C using hanging drop vapor diffusion. Purified Rm-CdnE and Em-CdnE were diluted on ice to 7–10 mg ml−1 and used immediately to set trays. Alternatively, co-complex crystals were grown by first incubating Rm-CdnE and Em-CdnE in the presence of ~10 mM total combined nucleotide concentration and 10.5 mM MgCl2 on ice for 30 min. RECON–cAAG co-complex crystals were grown by pre-incubating ~10 mg ml−1 RECON (K68A K70A) with 1 mM of purified Ec-CdnD02 product. Following incubation, 2 μl hanging drops were set at a ratio of 1:1 or 1.2:0.8 (protein:reservoir) over 350 μl of reservoir in Easy-Xtal 15-Well trays (Qiagen). Optimized crystallization conditions were as follows: Apo Rm-CdnE 100 mM Tris-HCl pH 7.5, 10–20% ethanol; Rm-CdnE–Apcpp–Upnpp 0.24 M sodium malonate, 24% PEG-3350; Apo Em-CdnE 21 mM sodium citrate pH 7.0, 100 mM HEPES-KOH pH 7.5, 16% PEG-5000 MME; Em-CdnE–GTP–Apcpp 100 mM tri-sodium citrate pH 6.4, 10% PEG-3350; Em-CdnE–pppApA 100 mM tri-sodium citrate pH 7.0, 8% PEG-3350; RECON–cAAG 0.1 M NaOAc, 1.0 M LiCl, 30% PEG-6000. Crystals grew in 3–30 days, and all crystals were harvested using reservoir solution supplemented with 10–25% ethylene glycol using a nylon loop except Apo Rm-CdnE crystals were harvested using NVH oil. X-ray diffraction data were collected at the Advanced Light Source (beamlines 8.2.1 and 5.0.1) and the Advanced Photon Source (beamlines 24-ID-C and 24-ID-E).
Data were processed with XDS and AIMLESS39 using the SSRL autoxds script (A. Gonzalez, Stanford SSRL). Experimental phase information for Rm-CdnE was determined using data collected from crystals grown with selenomethionine-substituted protein as previously described10. 4 sites were identified with HySS in PHENIX40, and an initial map was calculated using SOLVE/RESOLVE41. Model building was completed in Coot42 prior to refinement in PHENIX. Following model completion, the Apo Rm-CdnE structure was used for molecular replacement to determine the nucleotide bound structures. Rm-CdnE models were not sufficient to phase Em-CdnE data, but a minimal core Rm-CdnE active-site model was able to successfully determine the substructure and assist experimental phasing with data collected from a native crystal using sulfur single-wavelength anomalous dispersion at a minimal accessible wavelength (~7,235 eV). 16 heavy sites were identified in HySS that correspond to 12 sulfur, and 4 phosphate sites in the Em-CdnE–pppApA structure, and Em-CdnE model building was completed as for Rm-CdnE. RECON–cAAG data were phased by molecular replacement using the previously determined RECON–c-di-AMP structure (PDB 5UXF21), and model building was manually completed in Coot. X-ray data for refinement were extended according to I/σ resolution cut-off of ~1.5 and CC* correlation and Rpim parameters. Final structures were refined to stereochemistry statistics for Ramachandran plot (favored/allowed), rotamer outliers, and MolProbity score as follows: Rm-CdnE Apo, 98.6%/1.4%, 0.4% and 0.98; Rm-CdnE–Apcpp–UpNpp, 98.9%/1.1%, 0.4% and 1.25; Em-CdnE Apo 97.8%/2.2%, 0.8% and 1.08, Em-CdnE–GTP–Apcpp, 97.8%/2.2%, 0.8%, and 1.05, Em-CdnE–pppApA, 98.1%/1.9%, 0.8%, and 1.29. See Supplementary Table 1 and the Data Availability section for deposited PDB codes.
Structural comparisons
Structure-based comparisons used to define the conserved CD-NTase architecture in Fig. 2d were calculated using the Dali server and enzymes were clustered according to Z-score43 of the core NTase domain. Structures used include: DncV (4TY010), cGAS (6CTA30), OAS1 (4RWO44), Poly(A) Polymerase gamma (PAP, 4LT645), CCA-adding enzyme (4X4T46), Pol-β (4KLQ47), and Pol-μ (4YD148).
In the text and in figures, side-chains are numbered according to Rm-CdnE sequence. The analogous residue to N166 from Rm-CdnE in E. coli CdnE is N174, in Em-CdnE is S169, in DncV is S259, and in human cGAS is S378. RECON crystal structures with cAAG, c-di-AMP (5UXF21), and NAD (3LN3) were superimposed using PyMol (version 1.7.4.4), and all structure figures were prepared using PyMol.
Gel shift assays
In vitro binding assays were performed as previously described33. Briefly, recombinant Mus musculus STING or RECON, at 4, 20, or 100 μM was incubated with radiolabeled nucleotide (≤1 μM final concentration) in gel shift buffer for final conditions of 50 mM Tris pH 7.5, 60 mM KCl, 5 mM Mg(OAc)2, and 1 mM DTT. Experiments were prepared by combining 1 μL of α−32P labeled nucleotide, 2 μL 5× gel shift buffer, 5 μL of nuclease free water, and started by addition of 2 μL of recombinant protein in storage buffer. α−32P labeled nucleotides were produced with 25 μM of each NTP and ~1 μCi of each [α−32P] NTP in the following conditions: cGAS (ATP, GTP/[ɑ−32P] GTP), DncV (ATP, GTP/[ɑ−32P] ATP), DisA (ATP /[ɑ−32P] ATP), WspR (GTP /[ɑ−32P] GTP), CdnE (ATP, UTP /[ɑ−32P] UTP), and Ec-CdnD02 (ATP, GTP/[ɑ−32P] GTP).
After 30 min of equilibration, bound and free nucleotide were separated by 6% native PAGE in 0.5× TBE buffer, gels were dried, and exposed to a phosphorscreen prior to detection by Typhoon Trio Variable Mode Imager system (GE Healthcare). Gel shifts were quantified with ImageQuant 5.2 and the percent bound nucleotide was calculated as a proportion of total bound and free nucleotide for each lane, after subtraction of background signal.
Cellular assays for interferon-β induction
In-cell assays were performed as previously described33. Briefly, HEK293T cells were transfected using Lipofectamine2000 in 96-well format with: a control plasmid constitutively expressing Renilla luciferase (2 ng pRL-TK), a reporter plasmid expressing interferon-β inducible firefly luciferase (20 ng), a plasmid expressing Mus musculus STING (5 ng), and a 5-fold dilution series of pcDNA4-based plasmids expressing a nucleotidyltransferase (1.2, 6, 30, 150 ng). 2′3′ cGAMP was produced with mouse cGAS, 3′3′ cGAMP was produced with V. cholerae DncV, cyclic di-AMP (cAA) was produced with Bacillus subtilis DisA, cyclic di-GMP (cGG) was produced with P. aeruginosa WspR. Luciferase production was quantified after 24 h and firefly luciferase was normalized to Renilla, which was then normalized to empty nucleotidyltransferase vector used at 150 ng.
Cell lines
HEK293T cells were used to measure a reporter plasmid expressing interferon-β inducible firefly luciferase. Cell lines were originally provided by the ATCC, no methods were used for authentication, and cell lines were not tested for mycoplasma.
Western blot analysis
CD-NTase in-cell expression levels were verified by Western blot of lysed cells. Confluent HEK293T cells were seeded 24 h prior to transfection at a dilution of 1:4 in a 6-well dish. Cells were transfected with 2 μg of plasmid using Lipofectamine2000. At 24 h post transfection cells were harvested by washing cells from the dish using Hanks Buffered Saline Solution, pelleted at low speed, and flash frozen. Pelleted cells were lysed by re-suspending the pellet in 400 μL 1x LDS buffer (ThermoFisher Scientific) + 5% β-mercaptoethanol, boiling for 5 min, and vigorously vortexing. Samples were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with primary antibodies 1:5,000 Rabbit anti-MBP (Millipore Cat# AB3596, RRID:AB_91531) and 1:10,000 Mouse anti-Tubulin (Millipore Cat# MABT205, RRID:AB_11204167), followed by secondary antibodies at 1:10,000 IRDye 680RD Goat anti-Rabbit IgG (LI-COR Biosciences Cat# 925–68071, RRID:AB_2721181) and IRDye 800CW Goat anti-Mouse IgG (LI-COR Biosciences Cat# 925–32210, RRID:AB_2687825). Stained membrane was imaged using a LI-COR Odyssey CLx imager.
RECON enzyme assay
Activity assays were performed as previously described21. Briefly, a 2-fold dilution series of nucleotide from 50–0.05 μM was incubated in 1× PBS with 200 μM NADPH (co-substrate) and 25 μM 9,10-Phenanthrenequinone (substrate). The reactions were started with the addition of RECON to a final concentration of 0.5 μM and absorbance at 340 nm was monitored at 20 s intervals for 20 min. The slope of each reaction in the linear range (20–250 s) was used to calculate activity (Linear regression/straight line analysis, Prism 7.0c). Values were normalized to reactions with no CDN/nucleotide signaling molecule added, which defined 100% activity. Nucleotides were produced or purchased as described above and cAAG was purified as described above.
Bioinformatics and tree construction
To bioinformatically map CD-NTase-like enzymes in bacteria, we extended a previous analysis by Burroughs et al. that combined iterative BLAST analysis, secondary structure predictions, and hidden Markov models to collect DncV-like proteins and their genomic context22. We identified homologs of each of these 1300 identified proteins by BLAST analysis of the NCBI non-redundant protein database, then combined these datasets to identify >5600 CD-NTase-like genes. The dataset was then manually curated (Geneious Software) according to shared CD-NTase structural and active-site features. Bacterial genomes and sequences were aligned using MAFFT FFT-NS-2 algorithm v7.38849, a BLOSUM62 scoring matrix, an open gap penalty of 2, and an offset value of 0.123. Proteins with large truncations or lacking the essential DNA polymerase β-like nucleotidyltransferase residues [ie. GS…(D/E)X(D/E)…(D/E)] were removed. The tree was generated from the MAFFT alignment using a Jukes-Cantor genetic distance model, Neighbor-Joining method, no outgroup, and resampled by Bootstrap for 100 replicates sorted by topologies. The unrooted tree is used to represent global CD-NTase diversity and does accurately reflect the specific evolutionary relationship between the major CD-NTase A–H clades. The aligned sequences along with pairwise identity comparisons were extracted and used to define clades and clusters. A cluster was defined as >10 CD-NTases that share >24.5 % identity to the sequence preceding each in the alignment. For clarity, 14 poorly aligned CD-NTases were excluded from the tree and are indicated in Supplementary Table 2. The full dataset organized by order from the alignment and containing pairwise comparison of protein identity to each preceding gene is available as Supplementary Table 2 and as source data for Fig. 4a. For detail on identifying CD-NTases within an organism of interest or organisms encoding a CD-NTase of interest, see Supplementary Discussion.
Each sequence was identified from the nonredundant database of protein sequences and, at times, represents identical proteins translated from genes found in multiple bacteria. For this reason, additional metadata was extracted for each sequence from the NCBI Identical Protein Groups (IPG) database. The number of “Protein Accessions” in IPG was used as a surrogate quantification of the number of isolated bacterial genomes that harbor each CD-NTase. At the time of access (02/03/2018) 5,686 nonredundant CD-NTases sequences were identified representing a total of 16,717 genomes. At that time, 130,135 bacterial genomes had been deposited in the NCBI Genome database, leading to the crude approximation of 12.8% of genomes harboring CD-NTase genes. As some of these genomes may harbor more than one CD-NTase and the IPG database can overestimate number of genomes encoding a given protein we have estimated that >10% of bacterial genomes sequenced encode CD-NTases. Taxonomic analysis was performed using metadata associated with each nonredundant CD-NTase record in NCBI and when multiple bacteria were represented by one identical sequence the highest common taxonomical group was used. IPG and Taxonomic data are also found in Supplementary Table 2.
Type CD-NTase enzymes were manually selected from clusters based on the relevance of the organism from which they were isolated (i.e. human or plant pathogen/commensal organism), their predicted aptness for in vitro expression (thermophilic organisms or isolates from E. coli), the similarity of their operon to the DncV/CdnE operons, and the number of identical protein sequences represented by each unique sequence. CD-NTase001 was selected as and additional control and is encoded by dncVE. coli in ECOR3150.
CD-NTase screen
Each type CD-NTase gene was codon optimized for E. coli, synthesized (IDT), cloned as an N-terminal 6×His-MBP-fusion, and the protein was purified from a 25 mL culture. E. coli growth, protein induction, and bacterial disruption were performed as described above. Lysates were clarified by centrifugation and Ni-NTA affinity purification was performed as described above with gravity columns replaced by spin columns at 100 × g. Buffer exchange of eluted proteins was performed by concentrating the eluate using a 0.5 mL 10 kDa cut-off spin column (Ambion) followed by dilution with storage buffer and re-concentration 3× (final imidazole concentration ~0.3 mM). Proteins were analyzed for nucleotide synthesis fresh and flash-frozen for storage at −80 °C. For biochemical screen, ATP/CTP/GTP/UTP were used at 25 μM each and incubated overnight with the reaction conditions indicated using methods described above. 1 μL of screened protein (~1 μg) was added to the reaction and the same volume assessed by SDS-PAGE followed by coomassie staining, shown in Extended Data Fig. 6g.
Fig. 4d was manually constructed based on known TLC migration patterns which guided CDN identification in each sample. The quantity of ions detected by MS relative to other CD-NTases was used to determine if products were a major or minor constituent. On PEI-Cellulose, cyclic dipurines migrate similarly, cyclic purine–pyrimidine hybrids migrate similarly, and cyclic dipyrimidines migrate similarly; on Silica c-di-AMP migrates uniquely, cyclic UMP–AMP and cGAMP migrate similarly, c-di-GMP and c-di-UMP migrate similarly, and cGMP–UMP and cCMP–UMP migrate similarly, these cannot be distinguished.
Coexpression of CD-NTases and effectors in E. coli
CD-NTases chosen for in-depth analysis were cloned into an arabinose-inducible, chloramphenicol resistant pBAD33 plasmid. Putative CD-NTase effector genes were selected based on proximity, if they were classified as involved in biological conflict22, and based on analogous operon architecture to known effector phospholipases. Effector genes were codon optimized for E. coli and cloned into pETSUMO230, a carbenicillin resistant vector that is IPTG inducible in BL21-DE3 E. coli (ThermoFisher Scientific). Three pairs of vectors were assessed for each CD-NTase-effector pair: (1) cogant CD-NTase + effector, (2) CD-NTase + GFP, (3) mCherry + effector. Fluorescent proteins were used as negative controls. Vectors were co-transformed into electrocompetent BL21-DE3 E. coli, selected with both relevant antibiotics, and maintained under non-inducing conditions (0.2% glucose). Overnight bacterial cultures were serially diluted into LB and 5 μL was spot plated on selective medium containing 5 μM IPTG and 0.2% arabinose. Colony formation was quantified and images were taken at ~24 h.
Extended Data
Extended Data Figure 1 |. Detailed characterization of CdnE, a cUMP–AMP synthase.
a and b, Titration of reaction buffer pH in steps of 0.2 pH units. Recombinant DncV or CdnE was incubated with α−32P radiolabeled ATP, CTP, GTP, and UTP at varying pH and the reactions were treated with alkaline phosphatase and visualized by PEI-cellulose TLC. CdnE activity is optimal at pH ~9.4, and this reaction condition was used in further experiments. Data are representative of 2 independent experiments.
c, PEI-cellulose TLC of products after incubation of indicated enzyme, wild-type CdnE, or active site mutant CdnE with α−32P radiolabeled ATP, CTP, GTP, and UTP as in Fig. 1b. Mutations that ablate the CdnE Mg2+-coordinating, active-site residues eliminate all detectable activity. Data are representative of 3 independent experiments.
d, Nuclease P1 sensitivity of CDN products. The endonuclease P1 specifically cleaves 3′–5′, canonical phosphodiester bonds. DncV and CdnE products are completely digested in the presence of P1 and alkaline phosphatase, whereas only one bond of 2′3′ cGAMP is susceptible to digestion, producing the linear G[2′–5′]pA product. Data are representative of 3 independent experiments.
e, Workflow of nucleotide production for MS and NMR analysis.
f, Anion exchange chromatography of a CdnE reaction with ATP and UTP, eluted with a gradient of Buffer B (2 M ammonium acetate) by FPLC. Individual fractions were concentrated prior to pooling for further analysis.
g, Anion exchange chromatography (IEX) fractions from f were separated by silica TLC, visualized by UV shadowing, and compared to a radiolabeled reaction to confirm the appropriate A254 peak. Fractions were pooled and concentrated prior to MS and NMR analysis.
h, k, and l, 3′3′ cyclic uridine monophosphate–adenosine monophosphate proton NMR spectra and associated zoomed in datasets. 1H NMR (400 MHz): δΗ 8.40 (s, 1H), 8.17 (s, 1H), 7.90 (d, J = 8.2 Hz, 1H), 6.15 (s, 1H), 5.75 (s, 1H), 5.55 (d, J = 8.2 Hz, 1H), 5.00–4.90 (m, 2H), 4.80 (d, J = 4.5 Hz, 1H) 4.70–4.61 (m, 1H), 4.55–4.38 (m, 4H), 4.17–4.02 (m, 2H)
i and j, 3′3′ cyclic uridine monophosphate–adenosine monophosphate phosphate NMR spectra and associated zoomed in dataset. 31P{1H} NMR (162 MHz): δP −1.59 (s, 1P), −1.65 (s, 1P).
Extended Data Figure 2 |. DncV, cGAS, and CdnE reaction order.
a–c, Incubation of CD-NTase enzymes with nonhydrolyzable nucleotides traps reaction intermediates and identifies reaction order. Left, PEI-cellulose TLC analysis of reactions as in Fig. 1b where individual NTPs have been replaced with nonhydrolyzable nucleotide analogs. Exposed gamma phosphates are labeled in parenthesis to indicate these are removed by phosphatase treatment prior to analysis. Data are representative of 3 independent experiments. Right, previously determined reaction mechanisms (DncV and cGAS)10,51 and proposed reaction mechanism for CdnE.
Extended Data Figure 3 |. Detailed analysis of Rm-CdnE.
a, A thermophilic homolog of CdnE from Rhodothermus marinus (Rm-CdnE) also synthesizes cUMP–AMP. Recombinant proteins were incubated with α−32P radiolabeled NTPs as indicated at either 37 °C (CdnE) or 70 °C (Rm-CdnE) and the reactions were visualized by PEI-cellulose TLC as in Fig. 1b. Data are representative of 2 independent experiments.
b, Active site of Rm-CdnE superimposed with structures of cGAS (6CTA30) and DncV (4TY010).
c, The analogous position to N166 was mutated in CdnE to a serine and that protein, CdnEN166S, was characterized in depth. Reactions analyzed as in Fig. 1b, demonstrate that CdnEN166S loses pyrimidine-specificity. Data are representative of 2 independent experiments.
d, Structure-based sequence alignment of CD-NTases, annotated with secondary structure features of Rm-CdnE and human cGAS (6CTA30). Mg2+ coordinating active site residues are highlighted in red, and the analogous residues to Rm-CdnE N166 are highlighted in orange.
Extended Data Figure 4 |. Detailed structural analysis of Em-CdnE.
a, Sequence alignment of CdnE homologs in Fig. 2c, annotated with Rm-CdnE secondary structure features. Mg2+ coordinating active site residues are highlighted in red, and the analogous residues to Rm-CdnE N166 are highlighted in orange. WP_050915017, Yersinia enterocolitica; WP_096075289, Pseudomonas aeruginosa; WP_104644370, Xanthomonas arboricola; WP_010848498, Xenorhabdus nematophila; WP_015040391, Bordetella parapertussis; WP_006482377, Burkholderia cepacia complex; WP_014072508, Rhodothermus marinus (Rm-CndE); WP_042646516, Legionella pneumophila; WP_062886322, Mycobacterium avium; WP_016200549, Elizabethkingia meningoseptica (Em-CdnE); WP_031901603, Staphylococcus aureus; WP_050492554, Enterococcus faecalis; WP_062695386, Bacteroides thetaiotaomicron.
b, Biochemical deconvolution of Em-CdnE, which has a natural serine substitution at the N166 analogous site. Recombinant protein was incubated with NTPs as indicated and reactions were visualized as in Fig. 1b. Data are representative of 3 independent experiments.
c, Reactions of Em-CdnE incubated with α−32P radiolabeled NTPs and nonhydrolyzable nucleotide analogs as indicated, and visualized as in Fig. 1b. Data are representative of 3 independent experiments.
d, Anion exchange chromatography of an Em-CdnE reaction with ATP and GTP, eluted with a gradient of Buffer B (2 M ammonium acetate) by FPLC. Individual fractions were concentrated prior to pooling for further analysis.
e, Anion exchange chromatography (IEX) fractions from d were separated by silica TLC, visualized by UV shadowing, and compared to a radiolabeled reaction to confirm the appropriate peak. Fractions were pooled and concentrated prior to MS analysis. MS confirmed synthesis of products with masses corresponding to c-di-AMP, cGAMP, and c-di-GMP.
f, Crystal structure of Em-CdnE in complex with GTP and nonhydrolyzable ATP capturing the “1st state” structure prior to NTP hydrolysis. Mg2+ ions are omitted for clarity.
g, Zoom-in cut-away of the active site of f, confirming position of a serine at the analogous site to Rm-CdnE N166. Mg2+ ions are shown in green. Nucleotide and metal 2Fo−Fc electron density is contoured at 1 σ.
h, Zoom-in cut-away of the active site of Em-CdnE–pppApA structure, capturing the “2nd state” after the first reaction has occurred to form a linear intermediate, but prior to CDN formation. Mg2+ ions are shown in green. Nucleotide and metal 2Fo−Fc electron density is contoured at 1 σ.
i, Biochemical deconvolution of mutant Em-CdnE reverted to the ancestral asparagine at the N166 analogous site. This mutant loses preference for producing cyclic dipurine molecules and instead produces more pyrimidine-containing CDN products. Reactions were visualized as in Fig. 1b. Data are representative of 2 independent experiments.
Extended Data Figure 5 |. cUMP–AMP-recognition helps define innate immune receptor specificity.
a and e, Quantification of nucleotide interactions with the host receptors STING or RECON, measured with radiolabeled nucleotide bound to a concentration gradient of host protein, separated in a native PAGE gel shift (0, 4, 20, 100 µM protein). Data are quantification of gels in b and f and representative of n=2 independent experiments.
b- and f, Native PAGE gel shift analysis of STING or RECON complex formation with indicated radiolabeled CDNs. Proteins are titrated at 0 (–), 4, 20, and 100 µM. For quantification see a and e. STING readily binds all cyclic dipurine species but does not form a high-affinity complex with cUMP–AMP. RECON readily binds all 3′3′ CDN species that contain at least one adenine base, including cUMP–AMP. Data are representative of 2 independent experiments.
c, In-cell STING reporter assay. Induction of an IFN-β reporter in HEK293T cells transfected with a concentration gradient of plasmid overexpressing enzymes as indicated. DncV and CdnE were expressed with N-terminal MBP tags and IFN-β reporter induction was compared as fold over empty vector, shown as (−). Data are mean ± SEM for n=3 technical replicates, and are representative of 2 independent experiments.
d, Western blot of MBP-tagged DncV and CdnE expressed from plasmids analyzed in c to validate in vivo expression. Data are representative of 2 independent experiments. Gel source data are available as Supplementary Figure 1.
g, Gel shift analysis as in f, with protein titration to measure the relative affinity of the RECON–cUMP–AMP interaction. Protein concentrations listed below. Data are representative of 2 independent experiments.
Extended Data Figure 6 |. A biochemical screen of CD-NTases from diverse bacterial genera.
a-, Chart of the number of bacterial genomes (N = a total 16,717) that harbor CD-NTases from clusters in Fig. 4a. See also Supplementary Table 2.
b-, Taxa of genome sequenced bacteria isolated with unique CD-NTase genes, bold face type indicate phyla, Proteobacteria and Firmicutes are further divided by order and visualized by shades of color.
c-f-, Type CD-NTases interrogated for product synthesis. Purified proteins were incubated with α−32P radiolabeled NTPs under different reaction conditions (indicated pH and divalent cation) and reaction products were visualized by either PEI-cellulose or Silica TLC as in Fig. 1b and Fig. 4c.
g, CD-NTase expression level and purity. Coomassie stained SDS-PAGE analysis of purified CD-NTase enzymes used in each reaction. Data are representative of 2 independent experiments.
Extended Data Figure 7 |. Detailed biochemical analysis of Lp-CdnE02.
a-, Biochemical deconvolution of Lp-CdnE02 (CD-NTase057) as in Fig. 1c, demonstrates specific synthesis of cyclic dipyrimidine products. Data are representative of 3 independent experiments.
b, Nuclease sensitivity of the Lp-CdnE02 product, as described in Extended Data Fig. 1d. Data are representative of 3 independent experiments.
c, Incubation of Lp-CdnE02 with nonhydrolyzable nucleotides, as described in Extended Data Fig. 2. Nonhydrolyzable UTP completely blocks the reaction, indicating the first step requires attack of the α-P from UTP. However, the product formed when nonhydrolyzable CTP is present cannot be distinguished from c-di-UMP in this assay, and it is unclear if the reaction proceeds through a pppCpU reaction intermediate. Data are representative of 3 independent experiments.
d, Anion exchange chromatography of a Lp-CdnE02 reaction with UTP and CTP, eluted with a gradient of Buffer B (2 M ammonium acetate) by FPLC. Individual fractions were concentrated prior to pooling for further analysis.
e, Anion exchange chromatography (IEX) fractions from d were separated by silica TLC, visualized by UV shadowing, and compared to a radiolabeled reaction to confirm the appropriate peak. Fractions were pooled and concentrated prior to MS analysis.
f, MS confirmed synthesis of c-di-UMP as the major product of Lp-CdnE02.
g, MS confirmed synthesis of cCMP–UMP as a minor product of Lp-CdnE02.
Extended Data Figure 8 |. CD-NTases are encoded in conserved, poorly understood operons on mobile genetic elements.
a-, Operon structure for CD-NTases selected for in-depth characterization showing the conserved protein domains in CD-NTase adjacent genes (see Fig. 4a–c). Conserved operons were first identified by Burroughs et al. and operons are vertically organized by similarity to one another22. Where found, linked genes demonstrating CD-NTases are encoded on mobile genetic elements are indicated.
b, CD-NTases and their adjacently encoded “effector” proteins were coexpressed in E. coli and bacterial colony formation was quantified by spot dilution analysis. CD-NTases were inducibly expressed from a chloramphenicol resistant (CmR) vector and effectors were inducibly expressed from a carbenicillin resistant (CarbR) vector. Bacteria harboring cognate CD-NTase / effector plasmids or control plasmids were plated on medium containing both inducers and incubated for 24 h at 37 °C. Data were not determined (N.D.) for CD-NTase036 because the effector was toxic to E. coli under non-inducing conditions. Data are the mean ± SEM of 3 independent experiments.
c, Spot dilution analysis of bacteria harboring the cognate CD-NTase-effector pair indicated, as quantified in b. The CD-NTase036-effector pair was not analyzed in this assay. Colony morphology indicates a potential interaction for some combinations, however, it is unclear how specific or significant this may be. Data are representative of 3 independent experiments.
Extended Data Figure 9 |. Detailed biochemical analysis of Ec-CdnD02.
a-, Titration of reaction buffer pH in steps of 0.2 pH units. Recombinant Ec-CdnD02 was incubated with α−32P radiolabeled NTPs at varying pH and the reactions were analyzed and visualized by PEI-cellulose or silica TLC as in Fig. 1b and Fig. 4c. Silica TLC identified two products, denoted the major (blue triangle) and minor (red triangle) product. Quantification of TLC spots is shown below. Data are representative of 2 independent experiments.
b-, Biochemical deconvolution of Ec-CdnD02, recombinant protein was incubated with NTPs as indicated and analyzed by TLC as in a. Data are representative of 3 independent experiments.
c-, Nuclease digestion of the Ec-CdnD02 product. Conventional nuclease digestion includes addition of a phosphatase, in this experiment reactions were first treated with Antarctic phosphatase to remove remaining NTPs then heat inactivated. Next, reactions were either untreated, treated with nuclease P1 (specific for 3′–5′ phosphodiester bonds) only, or treated with nuclease P1 and phosphatase to remove exposed phosphate groups. 3′3′ cGAMP (DncV) and Ec-CdnD02 product are digested into AMP and GMP constituents, which are phosphatase sensitive. cAMP (CyaA) is insensitive to P1 digestion and cyclic monophosphates are phosphatase resistant. These data demonstrate that the Ec-CdnD02 product does not contain a cyclic monophosphate. Data are representative of 3 independent experiments.
d, Incubation of Ec-CdnD02 with nonhydrolyzable nucleotides, as described in Extended Data Fig. 2. Nonhydrolyzable ATP completely blocks the reaction, indicating the first step requires attack of the α-P from ATP. However, when nonhydrolyzable GTP is present the possible intermediates [pp(c)pGpA, pp(c)pGpApA, or pppApA] cannot be distinguished in this assay and it is unclear how the reaction proceeds. Silica TLC is not suited for analyzing nonhydrolyzable nucleotides because they do not migrate beyond the origin. Data are representative of 3 independent experiments.
e, Anion exchange chromatography of a Ec-CdnD02 reaction with ATP and GTP, eluted with a gradient of Buffer B (2 M ammonium acetate) by FPLC. Individual fractions were concentrated prior to pooling for further analysis.
f and g, 3′3′3′ tricyclic adenosine monophosphate–adenosine monophosphate–guanosine monophosphate (cAAG) NMR spectra and associated zoomed in dataset. 31P{1H} NMR (162 MHz): δP −0.65 (s, 1P), −0.70 (s, 1P), −0.75 (s, 1P).
h-j, 3′3′3′ tricyclic adenosine monophosphate–adenosine monophosphate–guanosine monophosphate (cAAG) proton NMR spectra and associated zoomed in datasets. 1H NMR (400 MHz): δΗ 8.43 (s, 1H), 8.39 (s, 1H), 8.19 (s, 1H), 8.12 (s, 1H), 8.01 (s, 1H), 6.15 (d, J = 7.0 Hz, 1H), 6.12 (d, J = 7.0 Hz, 1H), 5.92 (d, J = 7.5 Hz, 1H), 5.00–4.78 (m, 6H), 4.69–4.58 (m, 3H), 4.3–4.2 (m, 6H).
Extended Data Figure 10 |. Structural analysis of cAAG inhibition of RECON.
a, cAAG interactions with STING or RECON, radiolabeled nucleotide incubated with a concentration gradient of each protein, separated in a native PAGE gel shift (0, 4, 20, 100 µM protein). Data are representative of 2 independent experiments.
b, Co-crystal structure of the RECON–cAAG complex shown as cartoon (left) and surface (right).
c, Overlay and orientation of RECON ligands cAAG, c-di-AMP (5UXF), co-substrate NAD (3LN3) demonstrate three individual binding pockets.
d, Schematic representation of residues from RECON that interact with cAAG. Green dotted lines indicate hydrogen bonding, grey dotted lines indicate hydrophobic interactions.
e, Zoom-in cutaways of individual RECON binding pockets as in d.
f, Mammalian innate-immune sensors recognize CD-NTase products with overlapping specificities. 2′3′ cGAMP and c-di-GMP are detected by STING; 3′3′ cGAMP and c-di-AMP are detected by both STING and RECON; and cUMP–AMP and cAAG are detected by RECON.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Kacie McCarty and Victor Cabrera for technical assistance; Stephen Wilson for helpful advice; Michelle Reniere for critical reading of the manuscript; Tera Levin for input on CD-NTase tree construction; Thomas Wyche and Matthew Henke from the HMS ICCB Longwood Screening Facility and the staff at the HMS East Quad NMR Facility for their advice and technical support; Apurva Govande and Wen Zhou for generously providing purified proteins; Chris Miller for assistance with X-ray crystallography data collection; and the members of the Mekalanos and Kranzusch labs for helpful advice and discussions. This work was funded by the Claudia Adams Barr Program for Innovative Cancer Research (P.J.K.), Richard and Susan Smith Family Foundation (P.J.K.), Charles H. Hood Foundation (P.J.K.), a Cancer Research Institute CLIP Grant (P.J.K.), NIH/NIAID R01AI018045 and R01AI026289 (J.J.M.), the Searle Scholars Program (A.S.Y.L.), and a Sloan Research Fellowship (A.S.Y.L.). A.T.W. is supported as a fellow of The Jane Coffin Childs Memorial Fund for Medical Research, C.C.d.O.M. is supported as a Cancer Research Institute/Eugene V. Weissman Fellow, and B.R.M. is supported by the NIH T32 Cancer Immunology training grant (5T32CA207021–02). X-ray data were collected at the Lawrence Berkeley National Lab Advanced Light Source beamlines 8.2.1 and 8.2.2, this work used Northeastern Collaborative Access Team beamlines 24-ID-C and 24-ID-E (P30 GM124165), a Pilatus detector (S10RR029205), an Eiger detector (S10OD021527) and Argonne National Laboratory Advanced Photon Source (DE-AC02–06CH11357).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Competing interests: Harvard Medical School and the Dana-Farber Cancer Institute have patents pending for CD-NTase technologies on which the authors are inventors.
Data availability
All data supporting the findings of this study are available within the article, the associated supplementary and source materials, or deposited in the PDB database. X-ray crystallographic coordinates and structure factor files are available from the PDB: Rm-CdnE Apo (6E0K); Rm-CdnE Upnpp, Apcpp (6E0L); Em-CdnE Apo (6E0M); Em-CdnE GTP, Apcpp (6E0N); Em-CdnE pppA[3′–5′]pA (6E0O); RECON cAAG (6M7K). CD-NTase sequences and CD-NTase encoding bacteria are available as Supplementary Table 2, CD-NTase effector genes sequences are available as Supplementary Table 3, and CD-NTase alignments for tree construction provided as source data for Fig. 4a are available as Supplementary Data. Source gel images are available in Supplementary Figure 1.
References
- 1.Wu J & Chen ZJ Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Corrales L et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Reports 11, 1018–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu J et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Science Translational Medicine 7, 283ra52–283ra52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross P et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Danilchanka O & Mekalanos JJ Cyclic Dinucleotides and the Innate Immune Response. Cell 154, 962–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson JW & Breaker RR The lost language of the RNA World. Sci Signal 10, eaam8812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krasteva PV & Sondermann H Versatile modes of cellular regulation via cyclic dinucleotides. Nat. Chem. Biol 13, 350–359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kranzusch PJ et al. Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity. Cell 158, 1011–1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies BW, Bogard RW, Young TS & Mekalanos JJ Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu D et al. Origins of the current seventh cholera pandemic. Proc. Natl. Acad. Sci. U.S.A. 113, E7730–E7739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dziejman M et al. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proceedings of the National Academy of Sciences 99, 1556–1561 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severin GB et al. Direct activation of a phospholipase by cyclic GMP-AMP in El Tor Vibrio cholerae. Proceedings of the National Academy of Sciences 56, 201801233–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornung V, Hartmann R, Ablasser A & Hopfner K-P OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol 14, 521–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jean SS, Lee WS, Chen FL, Ou TY & Hsueh PR Elizabethkingia meningoseptica: an important emerging pathogen causing healthcare-associated infections. J. Hosp. Infect 86, 244–249 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Jenal U, Reinders A & Lori C Cyclic di-GMP: second messenger extraordinaire. Nat Rev Micro 325, 279 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Corrigan RM & Gründling A Cyclic di-AMP: another second messenger enters the fray. Nat Rev Micro 11, 513–524 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Woodward JJ, Iavarone AT & Portnoy DA c-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science 328, 1703–1705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C et al. Synthesis of All Possible Canonical (3’−5’-Linked) Cyclic Dinucleotides and Evaluation of Riboswitch Interactions and Immune-Stimulatory Effects. J. Am. Chem. Soc 139, 16154–16160 (2017). [DOI] [PubMed] [Google Scholar]
- 21.McFarland AP et al. Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 46, 433–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burroughs AM, Zhang D, Schäffer DE, Iyer LM & Aravind L Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Research 43, 10633–10654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G & Siksnys V A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Niewoehner O et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Hallberg ZF et al. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3’, 3’-cGAMP). Proc. Natl. Acad. Sci. U.S.A. 113, 1790–1795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson JW et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proceedings of the National Academy of Sciences 112, 5389–5394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 27.Studier FW Protein production by auto-induction in high density shaking cultures. Protein Expression and Purification 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Whiteley AT et al. c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Molecular Microbiology 104, 212–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranzusch PJ & Whelan SPJ Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proceedings of the National Academy of Sciences 108, 19743–19748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W et al. Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 174, 300–311.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulasakara H et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3’−5’)-cyclic-GMP in virulence. Proceedings of the National Academy of Sciences 103, 2839–2844 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert S, Dufke S, Sorsa J & Heesemann J A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Molecular Microbiology 51, 837–848 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Kranzusch PJ et al. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Molecular Cell 59, 891–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman LM, Belin D, Carson MJ & Beckwith J Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reverter D & Lima CD Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol 13, 1060–1068 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Stetson DB & Medzhitov R Recognition of Cytosolic DNA Activates an IRF3-Dependent Innate Immune Response. Immunity 24, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Sureka K et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158, 1389–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaspar AH & Machner MP VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 4560–4565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabsch W XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger TC Reciprocal-space solvent flattening. Acta Crystallogr. D Biol. Crystallogr 55, 1863–1871 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Holm L & Laakso LM Dali server update. Nucleic Acids Research 44, W351–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohöfener J et al. The Activation Mechanism of 2’−5’-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity. Structure 23, 851–862 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Nausch LWM, Martin G, Keller W & Doublié S Crystal structure of human poly(A) polymerase gamma reveals a conserved catalytic core for canonical poly(A) polymerases. J Mol Biol 426, 43–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn C-D, Wilusz JE, Zheng Y, Beal PA & Joshua-Tor L On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160, 644–658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freudenthal BD, Beard WA, Shock DD & Wilson SH Observing a DNA Polymerase Choose Right from Wrong. Cell 154, 157–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon AF, Gosavi RA, Kunkel TA, Pedersen LC & Bebenek K Creative template-dependent synthesis by human polymerase mu. Proc. Natl. Acad. Sci. U.S.A. 112, E4530–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato K, Ishii R, Hirano S, Ishitani R & Nureki O Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase. Structure 23, 843–850 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Gao P et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.