Summary
Reprogramming cell fate during the first stages of embryogenesis requires that transcriptional activators gain access to the genome and remodel the zygotic transcriptome. Nonetheless, it is not clear if the continued activity of these pioneering factors is required throughout zygotic genome activation or if they are only required early to establish cis-regulatory regions. To address this question, we developed an optogenetic strategy to rapidly and reversibly inactivate the master regulator of genome activation in Drosophila, Zelda. Using this strategy, we demonstrated that continued Zelda activity is required throughout genome activation. We showed that Zelda binds DNA in the context of nucleosomes and suggest that this allows Zelda to occupy the genome despite the rapid division cycles in the early embryo. These data identify a powerful strategy to inactivate transcription factor function during development and suggest that reprogramming in the embryo may require specific, continuous pioneering functions to activate the genome.
Graphical Abstract

eTOC summary
Pioneer factors activate the zygotic genome following fertilization. Using optogenetic inactivation, McDaniel et al. demonstrate that continued activity of the Drosophila transcription factor Zelda is required to activate gene expression, and that Zelda binds to nucleosomes. This pioneering activity may facilitate the establishment of nucleosome-depleted regions during rapid replication cycles.
Introduction
The initial stages of embryonic development require that the fertilized germ cells be reprogrammed to the totipotent cells of the early embryo. During this time, the zygotic genome is transcriptionally quiescent and development is driven by maternally supplied mRNAs and proteins (Newport and Kirschner, 1982; Tadros and Lipshitz, 2009). Transcriptional activation of the zygotic genome is a gradual process that consists of an initial minor wave of genome activation followed by a major wave of activation, which occurs hours to days after fertilization (Harrison and Eisen, 2015). This highly conserved maternal-to-zygotic transition (MZT) must be precisely executed as failure to eliminate the maternal products or activate the zygotic genome is lethal to the embryo. The essential reprogramming that occurs during this conserved transition shares features with reprogramming in culture.
The MZT in Drosophila melanogaster occurs over the first few hours of development. At this time, the embryo is rapidly replicating its genome through a series of nuclear divisions within a shared cytoplasm. Because these divisions occur approximately every ten minutes, there is only time for a synthesis (S) phase followed by mitosis (M), with no gap phases (Foe and Alberts, 1983). Zygotic genome activation (ZGA) occurs gradually within the context of these rapid division cycles. Transcription initiates around the eighth nuclear cycle (NC8) with the expression of transcription factors required for embryonic patterning, cellularization, and sex determination (ten Bosch et al., 2006; Pritchard and Schubiger, 1996). There is a major wave of zygotic genome activation at cycle 14 with hundreds of genes increasing in expression (Anderson and Lengyel, 1979; McKnight and Miller Jr., 1976).
The transcription factor Zelda (ZLD; Zinc-finger early Drosophila activator) is essential for activation of the zygotic genome (Liang et al., 2008). zld is maternally deposited as an mRNA and is translationally up-regulated one hour after fertilization at approximately NC8 (Harrison et al., 2010; Liang et al., 2008). At this time, ZLD binds to thousands of cis-regulatory regions, and ZLD-bound regions remain largely unchanged through cycle 14 (Harrison et al., 2011; Nien et al., 2011). Early ZLD binding establishes or maintains regions of accessible chromatin, and this activity potentiates DNA occupancy by additional transcription factors, including those expressed during the minor wave of ZGA (Foo et al., 2014; Schulz et al., 2015; Sun et al., 2015; Xu et al., 2014; Yanez-Cuna et al., 2012). Activators of zygotic transcription in other species (Oct4/Pou5f3 and Nanog in zebrafish, Nfy and Dux in mice, OCT4 and DUX4 in humans) share with ZLD this ability to define regions of accessible chromatin (Hendrickson et al., 2017; Liu et al., 2018; Lu et al., 2016; Soufi et al., 2012). Nonetheless, it is not known whether these activators are required throughout ZGA to drive gene expression or whether their ability to potentiate transcription-factor binding during the early nuclear cycles is sufficient to activate gene expression later during the major wave of transcriptional activation at NC14.
The ability of ZLD to potentiate chromatin accessibility, transcription-factor binding and gene expression are characteristic of pioneer transcription factors, such as Oct4, which are instrumental in activating the zygotic genome in zebrafish and humans as well as reprogramming in culture (Gao et al., 2018; Lee et al., 2013; Leichsenring et al., 2013; Takahashi and Yamanaka, 2006). In culture, Oct4, Sox2 and Klf4 engage naive chromatin, to facilitate additional transcription-factor binding and modulate gene expression (Soufi et al., 2012). The ability to bind DNA even when it is wrapped around histones, in nucleosomes, likely enables Oct4 to bind closed chromatin and initiate reprogramming (Soufi et al., 2015). Despite the known properties shared between ZLD and additional reprogramming factors, it is unclear whether ZLD shares with these other pioneer factors the ability to bind nucleosomal DNA.
Here we combine genome engineering with an optogenetic strategy to rapidly and reversibly inactivate ZLD at precise time points during the MZT. Using this strategy, we demonstrate that ZLD is continuously required to drive gene expression during both the minor and major waves of zygotic genome activation. Because ZLD is excluded from chromatin during mitosis (Dufourt et al., 2018; Staudt et al., 2006), the continued requirement for ZLD throughout the MZT necessitates that ZLD rapidly regain access to the genome during the intervening S phase. We show that ZLD possesses the ability to bind DNA in the context of nucleosomes, a defining feature of pioneer factors, and suggest that this pioneering activity allows ZLD to rapidly rebind chromatin following nuclear division. Based on these data, we propose that the pioneering function of ZLD is required throughout the MZT to facilitate reprogramming of the zygotic genome.
Results
Rapid inactivation of ZLD using optogenetic manipulation
We designed an optogenetic system to enable blue-light-mediated, rapid inactivation of ZLD. We engineered an N-terminal Arabidopsis cryptochrome 2 (CRY2)-tagged version of endogenous ZLD based on our previous tagging strategies (Hamm et al., 2017; Kennedy et al., 2010; Liu et al., 2008). Flies carrying this edited allele were homozygous viable and fertile, demonstrating that the CRY2 tag alone does not interfere with ZLD function in the absence of acute blue-light exposure. While this optogenetic system is normally composed of two parts, the CRY2 light-responsive module and the CIBN dimerization partner (Guglielmi et al., 2015; Kennedy et al., 2010), it was recently demonstrated that an N-terminal CRY2 tag on the transcription factor Bicoid (BCD) results in blue-light-mediated inhibition (Huang et al., 2017). Thus, we tested whether, in the absence of the CRY2 dimerization partner, blue light could similarly result in inactivation of CRY2-tagged ZLD. In contrast to the viability and fertility observed under standard conditions, embryos laid by homozygous CRY2-ZLD female flies failed to gastrulate when laid and raised in blue light (107/112), phenocopying embryos lacking ZLD (Liang et al., 2008).
To more precisely test for blue-light-mediated inactivation, we exposed embryos to blue light during nuclear cycles (NC) 10-14, encompassing both the minor and major waves of ZGA. Embryos expressed His2Av-RFP, which was used to determine precise staging based on nuclear density (Figure 1A) (Lott et al., 2011). Embryos carrying only His2Av-RFP did not respond noticeably to the blue-light exposure and proceeded to gastrulate 60 minutes after entry into NC14 (Figure 1B). By contrast, CRY2-ZLD embryos exposed to blue light from NC10-14 failed to undergo gastrulation and showed nuclear fallout, reminiscent of embryos lacking maternally provided zld (Figure 1B) (Liang et al., 2008). Western blots and immunostaining revealed that ZLD protein levels remained unchanged upon exposure to blue light, and ZLD remained localized to the nucleus (Figures 1C, D). Chromatin immunoprecipitation coupled with quantitative PCR demonstrated that, as predicted, CRY2-ZLD occupies the Sryα promoter, but that blue-light exposure dramatically reduces chromatin occupancy at this locus (Figure 1E). Together these data suggest that blue-light exposure results in conformational changes in the CRY2-tagged protein and that this, not protein degradation, causes the inactivation. Upon blue-light exposure embryos heterozygous for CRY2-tagged zld proceed normally through the MZT (111/113 gastrulate), demonstrating that the untagged protein is able to retain access to the genome and activate gene expression. These results are largely consistent with what was previously observed for BCD (Huang et al., 2017).
Figure 1:
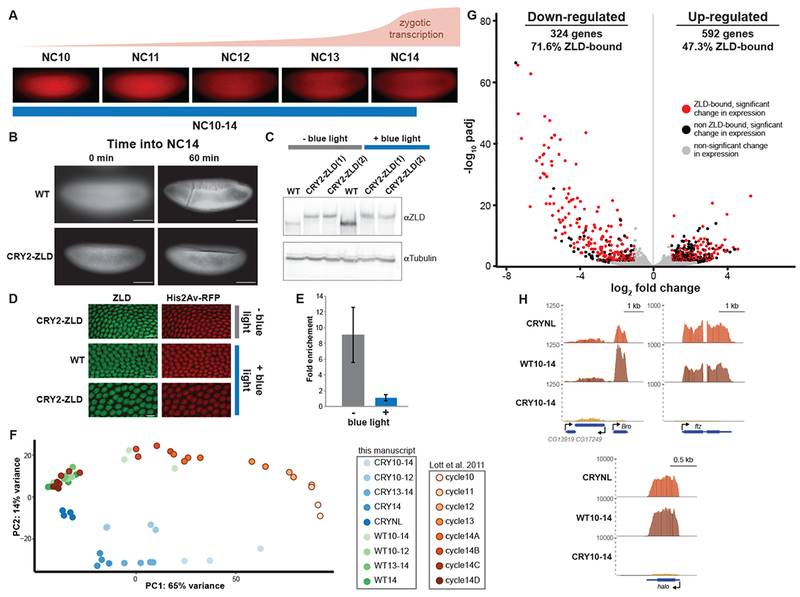
Rapid inactivation of ZLD using optogenetic manipulation. (A) Representative images of His2AvRFP expressing embryos in the identified nuclear cycles (NC) with approximate timing and levels of ZGA shown above. Blue line below indicates timing of blue-light exposure for embryos harvested for RNA-seq analyzed in G and H. (B) His2Av-RFP images of embryos exposed to blue light starting at NC10. Both wild-type (WT) embryos and CRY2-tagged ZLD (CRY2-ZLD) were imaged at the initiation of NC14 (0 min) or near the completion of NC14 (60 min). Scale bar is 100 μm. (C) Immunoblot for ZLD on embryos 2-3 hour after egg laying both with (+) and without (−) blue-light exposure. Two different lines of CRY2-tagged ZLD expressing embryos are included. (D) Immunostaining for ZLD (green) and His2Av-RFP (red) on embryos harvested 2-3 hour after egg laying expressing His2AvRFP alone (WT) or CRY2-ZLD with and without exposure to blue light starting at egg laying. Scale bar is 10 μm. (E) ChIP-qPCR analysis of ZLD binding to the promoter of Sryα in the presence (+) and absence (−) of blue light. Fold enrichment is compared to a region of Act5C not bound by ZLD. Error bars show the standard deviation (n = 2). (F) Distribution of datasets based on principal components for all the single-embryo data generated in this manuscript as well as the staged embryos from Lott et al. 2011. (G) Volcano plot highlighting the hundreds of genes that were mis-regulated in CRY2-tagged ZLD expressing embryos upon blue-light exposure from NC10 through NC14. Red dots indicate genes with proximal ZLD ChIP-seq peaks that change in gene expression upon blue-light exposure. Black dots indicate additional genes that change in expression upon blue-light exposure (adjusted p-value < 0.05, fold change > 2). Gray dots indicate genes that do not significantly change expression upon blue-light exposure. (see also Figure S1 and Table S1) (H) Genome browser tracks of RNA-seq data from wild-type (WT10-14) or CRY2-tagged ZLD (CRY10-14) embryos exposed to blue light for nuclear cycles 10-14 or CRY2-tagged ZLD mbryos without blue-light exposure (CRYNL). Tracks are shown for three previously identified, ZLD-responsive genes, Bro, ftz, and halo.
Having demonstrated that blue-light exposure of embryos expressing CRY2-tagged ZLD phenocopies maternal zld depletion, we used single-embryo RNA-sequencing (RNA-seq) to test whether blue-light treatment specifically inhibits the ability of ZLD to activate transcription. We exposed individual embryos expressing CRY2-tagged ZLD to blue light starting immediately after entry into NC10 as determined by nuclear density and exit from mitosis. We maintained blue-light exposure constantly, except to image the His2Av-RFP for staging purposes, and harvested embryos 30 minutes following entry into NC14 (CRY10-14). To control for the effects of blue light, we extracted RNA from His2Av-RFP embryos exposed to the identical blue-light regime (WT10-14). We also performed RNA-seq on CRY2-ZLD expressing embryos that had not been exposed to blue light to control for any changes in gene expression caused by the CRY2 tag or genetic background (CRYNL). For each condition, we sequenced RNA from at least three embryos, and the high-degree of reproducibility amongst our replicates (Figure S1) allowed us to identify differences in gene expression between the three experimental conditions. We used principal component analysis to compare our RNA-seq data sets to each other and also to RNA-seq from precisely staged single embryos (Lott et al., 2011). This analysis showed that wild-type embryos exposed to blue light had expression profiles similar to embryos during the beginning of NC14 (cycle 14A and 14B), while data from CRY10-14 embryos formed a distinct cluster, indicative of dramatic differences in transcript levels under these conditions (Figure 1F).
We identified genes that were significantly mis-regulated in the blue-light-exposed, CRY2-ZLD-expressing embryos as compared to identically staged, blue-light-exposed, control embryos (WT10-14) (Love et al., 2014). Because embryos expressing CRY2-tagged ZLD had gene expression differences compared to wild-type embryos even in the absence of blue light (Figure 1F), we excluded from our analysis any gene expression changes that were the result of the addition of the CRY2 tag (see STAR Methods for details). 324 genes were down-regulated and 592 genes were up-regulated upon exposure to blue light in embryos expressing CRY2-tagged ZLD (Figure 1G, Table S1). Significantly down-regulated genes showed larger changes in gene expression and these changes had lower p-values than for up-regulated genes (Figure 1G). The down-regulated set was highly enriched for zygotically expressed genes (Figure S1B), and in comparison to up-regulated genes, down-regulated genes were enriched for having a proximal ZLD-binding site (p = 9.41 × 10−13, two-tailed Fisher’s exact test) (Harrison et al., 2011). Furthermore, down-regulated genes included a large number of previously identified ZLD-target genes, including Bro, ftz and halo (Figure 1H) (Liang et al., 2008). Whereas Bro transcripts were significantly decreased in our CRY10-14 samples as compared to wild-type controls, immediately adjacent genes were largely unaffected, demonstrating the specificity of our approach (Figure 1H). In contrast to the down-regulated genes, up-regulated genes were enriched for maternal expression (Figure S1B), similar to genes up-regulated when zld is maternally depleted (Liang et al. 2008) and suggesting their up-regulation was due to indirect effects of inactivating ZLD. The sequencing data showed widespread down-regulation of zygotically expressed ZLD-target genes upon blue-light exposure. Together our data demonstrate that blue light disables N-terminally CRY2-tagged ZLD, establishing this as a powerful strategy for rapidly inactivating transcription factors within a developing organism.
Continued ZLD activity is necessary for the major wave of zygotic genome activation
During the MZT, ZLD is required for the expression of hundreds of genes. ZLD is instrumental in defining chromatin accessibility at cis-regulatory regions and recruiting additional transcription factors, including BCD, Dorsal (DL), and Twist (TWI) (Foo et al., 2014; Schulz et al., 2015; Sun et al., 2015; Xu et al., 2014; Yanez-Cuna et al., 2012). These pioneering characteristics are shared between ZLD and other master regulators of cell fate (Iwafuchi-Doi and Zaret, 2016). Nonetheless, it is unclear whether the activity of pioneering factors is required continually to maintain gene expression or only transiently to license sites for the binding of additional factors that can then maintain gene expression (Figure 2A). Our ability to rapidly inactivate ZLD upon blue-light exposure poised us to directly test if continued pioneering activity is required to drive gene expression.
Figure 2:
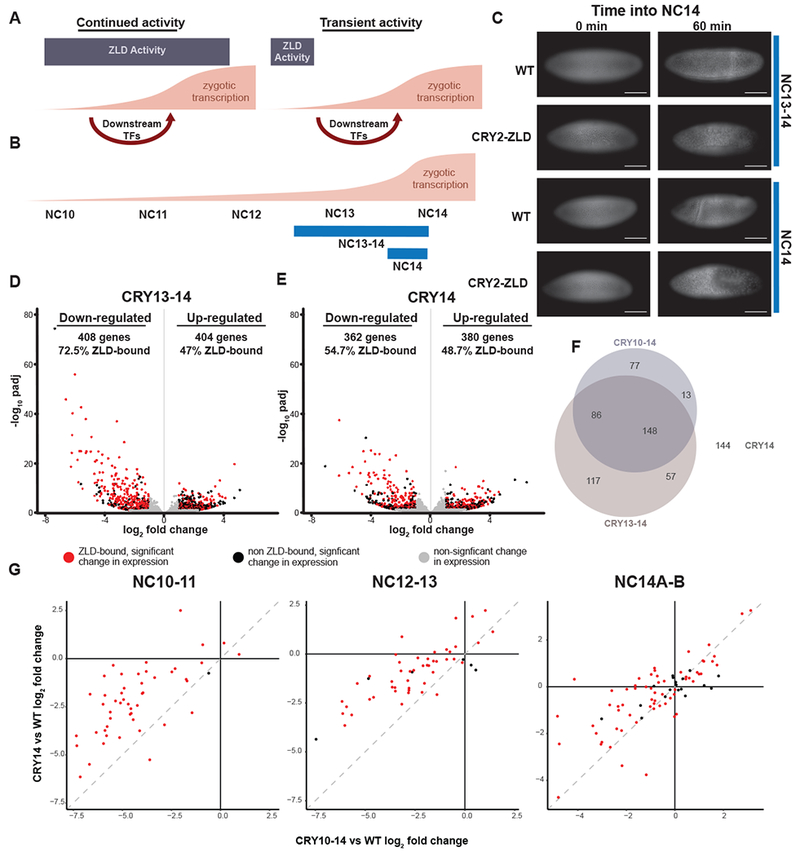
Continued ZLD activity is required to drive the major wave of ZGA. (A) Models depicting the possible requirements for ZLD during ZGA. On the left, ZLD is required through ZGA to potentiate transcription-factor binding and gene expression. On the right, ZLD is only required early, during the minor wave of ZGA. Downstream transcription factors then drive the major wave of ZGA. (B) The approximate timing and levels of ZGA are shown with blue lines below indicating the timing of blue-light exposure for embryos harvested for RNA-seq analyzed in D and E. (C) His2Av-RFP images of embryos exposed to blue light starting at NC13 or NC14 as indicated. Both wild-type (WT) embryos and CRY2-tagged ZLD (CRY2-ZLD) were imaged at the initiation of NC14 (0 min) or near the completion of NC14 (60 min). Scale bar is 100 μm. (D-E) Volcano plots highlighting the hundreds of genes that were mis-regulated in CRY2-tagged ZLD expressing embryos upon blue-light exposure from NC13 through NC14 (see also Table S2) (D) or from the beginning of NC14 (see also Table S2) (E). Red dots indicate genes with proximal ZLD ChIP-seq peaks that change in gene expression upon blue-light exposure. Black dots indicate additional genes that change in expression upon blue-light exposure (adjusted p-value < 0.05, fold change > 2). Gray dots indicate genes that do not significantly change expression upon blue-light exposure. (F) Venn diagram of the overlap amongst down-regulated genes in CRY2-ZLD embryos exposed to blue light from NC10-14, from NC13-14, and NC14. (G) Log2 fold change of gene expression over wild type in CRY2-ZLD embryos exposed to blue light only during NC14 was plotted against log2 fold change for CRY2-ZLD embryos exposed to blue light throughout ZGA (NC10-14). Zygotically expressed genes were analyzed based upon the timing at which gene expression initiated (indicated above each plot: NC10-11, NC12-13, NC14A-14B). Dotted gray lines mark a one-to-one correlation between the changes in gene expression in both blue-light regimes.
To test the necessity of ZLD during the major wave of ZGA at NC13-14, we exposed CRY2-tagged ZLD embryos to blue light starting either at the entry into NC13 or NC14 (Figure 2B). Even these abbreviated blue-light exposures caused gastrulation defects specific to the CRY2-tagged ZLD expressing embryos (Figure 2C), suggesting an effect on ZLD-mediated gene expression. To determine the extent to which ZLD is required for genome activation during the major wave of ZGA, we exposed embryos to blue light starting at either NC13 (CRY13-14) or the beginning of NC14 (CRY14) and harvested them for RNA-seq 30 minutes into NC14. We again controlled for effects of blue-light exposure by harvesting His2AV-RFP embryos that had undergone an identical blue-light regime (WT13-14 and WT14). Principal component analysis demonstrated that the blue-light-exposed His2Av-RFP embryos showed expression patterns similar to wild-type embryos at stages 14C and 14D (Figure 1F). The fact that this abbreviated blue-light exposure results in gene expression profiles mirroring stages 14C and 14D rather than cycles 14A and 14B, as seen in embryos exposed to blue light for a longer period of time, suggests that prolonged blue-light exposure results in a minor developmental delay. Thus to control for this developmental delay, we always used paired His2AvRFP-expressing embryos exposed to identical blue-light conditions as controls to determine the effects on gene expression profiles of CRY2-induced ZLD inactivation.
We identified hundreds of genes mis-regulated in embryos in which ZLD was inactivated either during NC13-14 or NC14 (Figure 2D, E, and Table S2). Similar to the gene expression profiles in embryos in which ZLD was inactivated throughout ZGA, the hundreds of down-regulated genes were significantly enriched for genes with proximal ZLD-binding sites and there was a large degree of overlap in these down-regulated genes amongst the three different conditions (Figure 2F). Given the remarkably similar changes in gene expression caused by abbreviated and prolonged ZLD inhibition, we wanted to ensure that we were specifically inactivating ZLD during blue-light exposure and not more generally perturbing ZLD-target gene expression. We specifically analyzed the change in gene expression of sets of genes that initiate expression at different times during ZGA (Li et al., 2014; Lott et al., 2011). Importantly, as we would predict, genes that initiate expression during NC10-11 were more down-regulated in embryos that were exposed to blue light starting at NC10 as compared to NC14 (Figure 2G). By contrast, genes that initiate expression during NC14 had a similar magnitude change in expression on blue-light exposure in both conditions (Figure 2G). Thus, our system provides a time-resolved method of inactivating ZLD in a blue-light-dependent manner. Together, our analyses demonstrate that prolonged ZLD activity is required during the major wave of genome activation and that early ZLD activity at cis-regulatory elements is not sufficient to activate gene expression later in embryonic development. Thus, ZLD is required continually through the MZT to maintain proper gene expression.
ZLD activity during the major wave of ZGA can activate gene expression
Inhibition of ZLD activity during the major wave of ZGA showed the continued requirement for ZLD activity in driving widespread gene activation. Therefore, we wanted to test whether ZLD activity during only the major wave of ZGA would be able to activate gene expression. To test this, we exposed CRY2-tagged ZLD embryos to blue light to inactivate ZLD during the minor wave of ZGA, NC10-12, and then removed the blue light as the embryos entered NC13 (Figure 3A). As expected, given the essential role of early activated genes for development, these embryos failed to gastrulate (Figure 3B). To determine the effect on gene expression, we harvested embryos for RNA-seq. While we identified hundreds of mis-regulated genes, the fold change and significance of this mis-expression was much less than was identified when ZLD was inactivated during NC10-14 (Figure 3C, D and Table S3). When all the genes that were mis-expressed in any of the four blue-light conditions were compared, it was clear that inactivation throughout the ZGA (NC10-14) had the most pronounced effect on gene expression as compared to inactivation of ZLD during only a subset of NC10-14 (Figure 3E). These changes in expression were mirrored when ZLD was inhibited during the major wave of ZGA (NC13-14 and NC14) (Figure 3E). By contrast, if ZLD activity was only inactivated during the minor wave of ZGA (NC10-12), gene expression of many ZLD-target genes was down-regulated to a lesser extent than upon exposure throughout ZGA (NC10-14) (Figure 3D, E). This rescue in gene expression, suggests that, when blue-light exposure was removed at the beginning of cycle 13, ZLD activity was restored and could drive target-gene expression during the major wave of ZGA.
Figure 3:
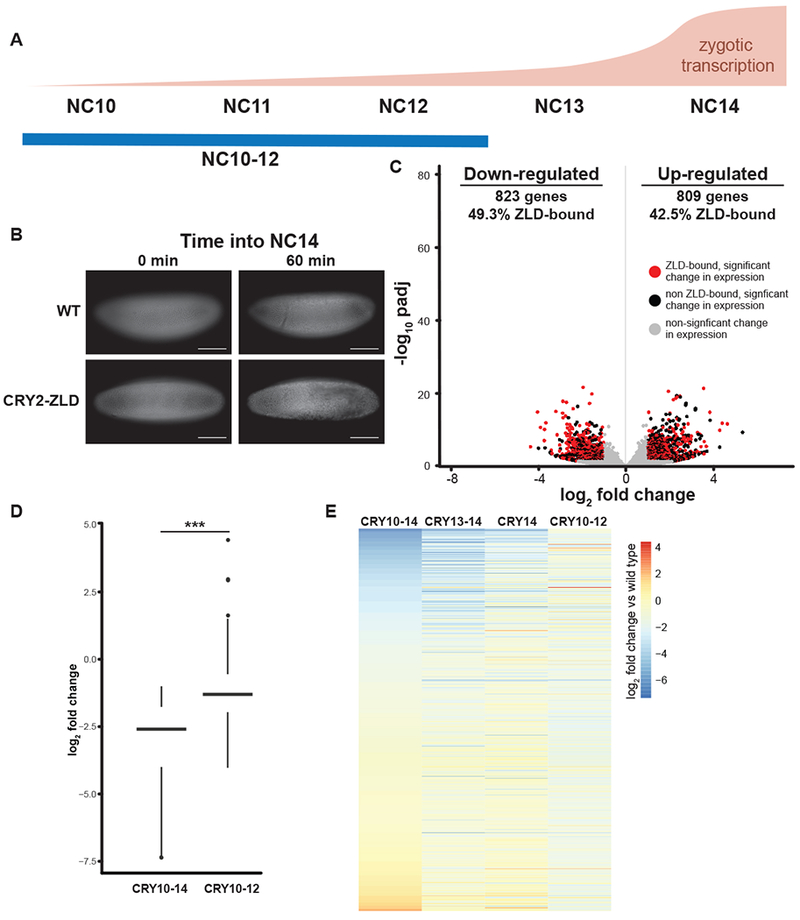
ZLD activity restricted to nuclear cycles 13-14 is able to drive gene expression. (A) The approximate timing and levels of ZGA are shown with blue lines below indicating the timing of blue-light exposure for embryos harvested for RNA-seq analyzed in C. (B) His2Av-RFP images of embryos exposed to blue light starting from NC10 though NC12. Both wild-type (WT) embryos and CRY2-tagged ZLD (CRY2-ZLD) were imaged at the initiation of NC14 (0 min) or near the completion of NC14 (60 min). Scale bar is 100 μm. (C) Volcano plot highlighting the hundreds of genes that were mis-regulated in CRY2-tagged ZLD expressing embryos upon blue-light exposure from NC10 through NC12. Red dots indicate genes with proximal ZLD ChlP-seq peaks that change in gene expression upon blue-light exposure. Black dots indicate additional genes that change in expression upon blue-light exposure (adjusted p-value < 0.05, fold change > 2). Gray dots indicate genes that do not significantly change expression upon blue-light exposure. (see also Table S3) (D) Box plot of the log2 fold change in gene expression upon blue-light exposure either from NC10-14 (CRY10-14) or NC10-12 (CRY10-12) for the set of ZLD-bound genes that are down-regulated in CRY2-tagged ZLD expressing embryos. ***, p = 7.64 × 10−33 (two-sided Wilcoxian rank sum test) (E) Heat map of all the genes that significantly changed in gene expression upon blue-light exposure in any of the four conditions used in this study (indicated above). Genes are ordered based on the levels of change in embryos exposed to blue light from NC10-14.
ZLD binds sequence specifically to nucleosomal DNA
Prior work demonstrated that ZLD shared with classically defined pioneer factors the capacity to establish or maintain chromatin accessibility and, in so doing, define cis-regulatory regions (Foo et al., 2014; Schulz et al., 2015; Sun et al., 2015; Xu et al., 2014; Yanez-Cuna et al., 2012). Furthermore, ZLD occupies thousands of loci throughout the four mitotic cycles that occur during ZGA (NC10-14) (Harrison et al., 2011). Yet, unlike these classically defined pioneer factors, ZLD does not remain bound to the mitotic chromosomes (Dufourt et al., 2018; Staudt et al., 2006). Our optogenetic approach demonstrated that continued ZLD activity is necessary for the embryo to progress through the MZT. During this time in development the embryonic genome undergoes rapid rounds of replication and division. Therefore, the relatively stable occupancy of ZLD-binding sites as assayed by ChIP-seq indicates that ZLD binding is consistently reestablished following each mitotic division (Harrison et al., 2011). Thus, the capacity of ZLD to rapidly and reproducibly rebind to the genome following mitosis is required for embryonic development.
Pioneer factors are defined by binding to DNA even in the context of nucleosomes, a property that could allow ZLD to rapidly access the genome after mitosis (Iwafuchi-Doi and Zaret, 2016). To test whether ZLD binds nucleosomal DNA, we identified a region of the bottleneck (bnk) locus that is bound by ZLD throughout the MZT and at which a nucleosome is established in the absence of ZLD (Figure 4A), suggesting ZLD is instrumental in promoting accessibility at this region (Sun et al., 2015). We generated nucleosomes using 160 bp of DNA that centered on this ZLD-binding site, which contains four ZLD-binding motifs. In electromobility shift assays (EMSAs), full-length recombinant ZLD bound to naked bnk DNA and to the same DNA incorporated into nucleosomes (Figure 4B, S2, and S3). Recombinant protein containing only the C-terminal DNA-binding domain also bound to DNA and nucleosomes, but with decreased affinity as compared to the full-length protein, suggesting additional domains might be required for optimal DNA and nucleosome affinity (Figure 4C, S2, and S3). Increasing the concentration of the DNA-binding domain in these reactions resulted in multiple shifted species, suggesting multiple ZLD-binding motifs can be occupied as protein concentration increases (Figure S2C). This interaction was confirmed with independently generated nucleosomes and is not unique for the bnk sequence as ZLD similarly bound to nucleosomes containing two non-canonical ZLD-binding motifs (CAGGCAG) (Figure S4). Thus, nucleosome binding by ZLD was reproducible between laboratories and on multiple, ZLD-motif containing substrates. Immunoblots confirmed that these shifted species contained histone H3, demonstrating that ZLD-binding alone does not evict histones from the nucleosome (Figures 4D, E and S3B).
Figure 4:
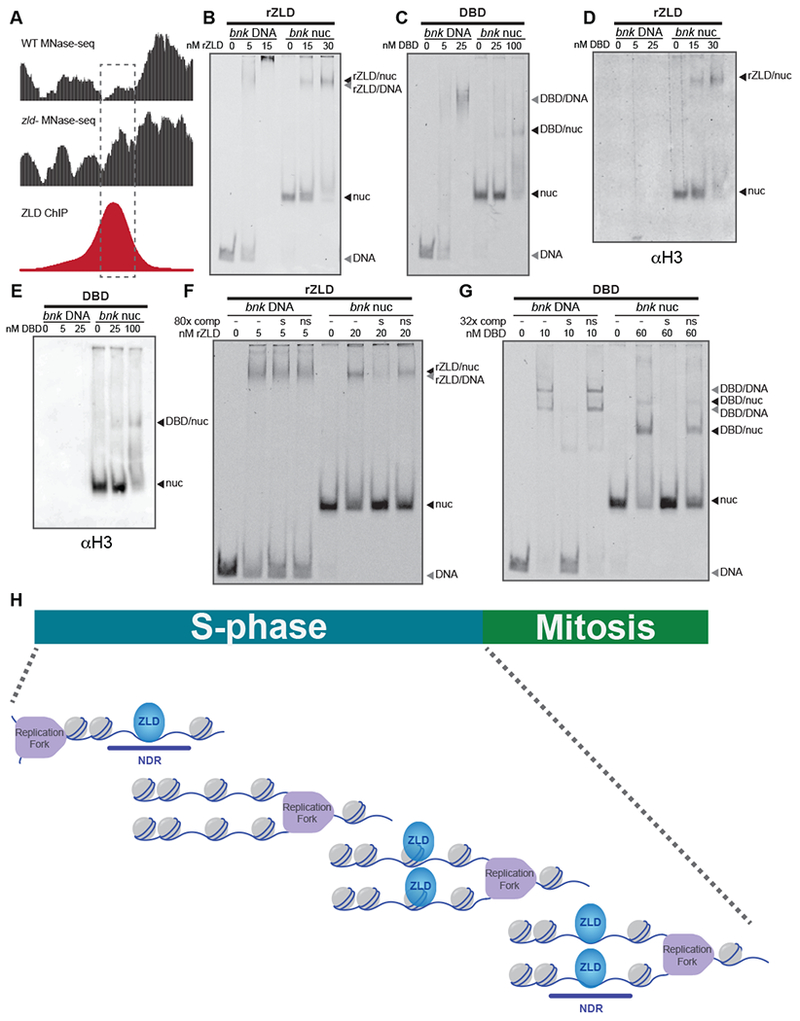
ZLD binds nucleosomal DNA with sequence specificity. (see also Figures S2, S3, and S4) (A) Genome browser tracks for a region of the bnk locus used to reconstitute nucleosomes. MNase-seq from wild-type embryos (WT) or embryos depleted for maternal zld (zld−) (Sun et al., 2015) and ZLD ChIP-seq (Harrison et al., 2011) are shown. Dashed rectangle highlights the region used to reconstitute nucleosomes. (B) Representative EMSA showing the affinity of increasing amounts of recombinant full-length ZLD (rZLD) to Cy5-labeled bnk DNA or bnk nucleosomes. (C) EMSA with increasing amounts of recombinant ZLD DNA-binding domain (DBD) to Cy5-labeled bnk DNA or bnk nucleosomes. (D-E) Protein from the EMSAs shown in (B) or (C) transferred to PVDF and blotted for histone H3. (F) EMSA demonstrating the specificity of binding of full-length ZLD (rZLD) to bnk DNA and bnk nucleosomes. Reactions included either no double-stranded oligonucleotide competitor (−), unlabeled, specific, double-stranded oligonucleotide competitor containing a canonical ZLD-binding motif (s), or unlabeled, non-specific, double-stranded oligonucleotide competitor containing a mutated ZLD-binding motif (ns). (G) EMSAs demonstrating the specificity of binding of the ZLD DNA-binding domain (DBD) to bnk DNA and bnk nucleosomes. Reactions were incubated with competitor as in (F). In all reactions, labelled probes were at a concentration of 2.5 nM. (H) Model of ZLD binding to nucleosomes following the replication fork to mediate rapid re-establishment of accessible, nucleosome-depleted regions (NDR) during the short (5-14 minute) S-phases of the early embryonic nuclear cycles (NC10-13). Because ZLD is largely absent from the mitotic chromosomes, rapid DNA binding by ZLD during S-phase is required to maintain chromatin occupancy.
To determine the specificity of these interactions, we performed EMSAs in the presence of unlabeled specific or non-specific DNA sequences (Figure S2B). These competition experiments showed that excess unlabeled probe containing a canonical ZLD-binding motif, but not non-specific probe, could displace complexes of recombinant ZLD and nucleosomes (Figures 4F and S4E). Thus, ZLD binding to nucleosomes relies, at least in part, on the underlying DNA sequence. By contrast, neither specific nor non-specific competitors could compete for binding of full-length ZLD to DNA (Figure 4F and S4E). Under similar conditions, excess of specific competitor could compete with both naked DNA and nucleosomes for binding to the DNA-binding domain alone, suggesting that regions outside of the DNA-binding domain facilitate non-specific DNA binding in vitro (Figure 4G and S4E). We had previously shown that a mutation in the second zinc finger of the DNA-binding domain (ZnF4) could disrupt DNA binding (Hamm et al., 2015, Figure S2C). Full-length ZnF4 mutant protein still bound to bnk DNA and nucleosomes (Figures S3 and S4). However, competition experiments demonstrated that this nucleosomal binding could be competed by both specific and non-specific competitors (Figure S3 and S4). Together, these experiments suggest that both sequence-driven binding, mediated by the DNA-binding domain, and non-specific interactions, facilitated by additional regions of the protein, contribute to the binding of full-length ZLD to nucleosomes. Thus, ZLD binds to nucleosomes with sequence specificity in vitro, and this pioneering characteristic may facilitate the ability of ZLD to rapidly bind chromatin following mitosis.
Discussion
Using an optogenetic strategy to rapidly and reversibly inactivate transcription-factor function, we demonstrated that ZLD, the master regulator of zygotic genome activation, is required throughout the MZT to drive gene expression. Inactivation of ZLD during the major wave of ZGA resulted in embryonic lethality and a failure to robustly activate hundreds of target genes. During this time in development, the early embryonic nuclear cycles consist of alternating replication (S) and division phases (M) without any gap phases (Figure 4G), and ZLD is largely absent from the condensed mitotic chromosomes (Dufourt et al., 2018; Staudt et al., 2006). Thus, to drive gene expression throughout these cycles, ZLD must be able to quickly and reproducibly access its binding motifs during S phase. Recent data suggests that DNA is incorporated into nucleosomes immediately following the replication fork and that subsequent activity is required to reestablish nucleosome positioning (Blythe and Wieschaus, 2016; Ramachandran and Henikoff, 2016). We propose that the ability to bind nucleosomal DNA enables ZLD to rapidly access its DNA-binding sites during S phase and, together with other factors, drive gene expression. Therefore, the unique ability of pioneering transcription factors to recognize their binding sites within the context of nucleosomes could provide them the capacity to define cis-regulatory regions in rapidly cycling cell populations like those of the early embryo or embryonic stem cells.
Pioneer factors are distinctive in that they can bind to nucleosomal DNA, establish or maintain accessible chromatin and facilitate transcription factor binding to cis-regulatory regions. For the defining pioneer factor FoxA1, it is evident that these three properties are functionally related with nucleosome binding driving chromatin accessibility and subsequent transcription-factor binding (Cirillo et al., 2002; Iwafuchi-Doi et al., 2016). Nonetheless, for additional factors with pioneering functions it remains unclear if these features are necessarily related. Here we have shown that ZLD shares with canonical pioneer factors the ability to bind DNA in the context of nucleosomes, and prior work demonstrated that ZLD mediates chromatin accessibility and enables chromatin binding by multiple additional transcription factors, including BCD, DL, and TWI (Foo et al., 2014; Schulz et al., 2015; Xu et al., 2014; Yanez-Cuna et al., 2012). Recent data have revealed additional mechanistic insight into pioneering activity, showing that ZLD forms nuclear hubs or microenvironments (Dufourt et al., 2018; Mir et al., 2018). These ZLD-mediated hubs are thought to concentrate BCD, and likely other transcription factors, driving chromatin occupancy (Mir et al., 2017). Thus, the ability of ZLD to bind nucleosomes may facilitate hub formation rather than directly affecting nucleosome stability. Understanding the relationship between pioneering activity, chromatin accessibility and hub formation will be critical to determining how pioneer factors drive genomic reprogramming in the early embryo.
Our demonstration of blue-light-induced inactivation of CRY2-tagged ZLD, combined with the prior demonstration that a similar strategy could be used to inhibit BCD (Huang et al., 2017), suggests that CRY2-mediated inactivation may be a generalizable strategy for precisely restricting transcription-factor activity within a developmental context. Using this strategy, we demonstrated that the ability of the master regulator ZLD to reprogram the transcriptome of the early embryo requires continued activity and that defining cis-regulatory regions early in development is not sufficient to prime the genome for later activation. These data suggest that other early embryonic reprogramming factors, such as Pou5f3 and Oct4, may similarly be required throughout early embryonic development to reprogram the embryo. Future studies using CRY2-tagged factors in other systems will determine whether reprogramming factors in general are required not only to initiate widespread transcriptional changes, but also to continue to drive transcription to allow for developmental reprogramming.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for fly stocks or other reagents should be directed to and will be fulfilled by Lead Contact Melissa Harrison (mharrison3@.wisc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly stocks
All stocks were grown on molasses food at 25°C. Fly strains used in this study: His2AvRFP (II) (Bloomington Drosophila Stock Center (BDSC) #23651) and CRY2-ZLD (this study); His2AvRFP (II).
METHOD DETAILS
Cas9-mediated generation of CRY2-ZLD embryos
Cas9-mediated genome engineering as described in (Hamm et al., 2017) was used to generate the N-terminal CRY2-tagged ZLD. The sequence encoding GFP in the double-stranded DNA (dsDNA) donor plasmid described in Hamm et al. 2017 was replaced with sequence encoding CRY2 (Addgene #26866) with Gibson assembly. In addition to the CRY2 tag, the dsDNA donor contained 1-kb homology arms flanking the N-terminal region of the ZLD open-reading frame and a 3xP3-DsRed cassette flanked by the long-terminal repeats of the piggyback transposase for selection and subsequent removal. The purified plasmid was injected into w1118; PBac{y[+mDint2]=vas-Cas9}VK00027 (BDSC#51324) embryos by BestGene Inc. Lines with successful integration were identified by DsRed expression. The DsRed cassette was then cleanly excised using piggyBac transposase. The entire locus was sequenced to ensure the CRY2 tag was incorporated without errors.
Imaging live embryos
Homozygous His2AvRFP or CRY2-ZLD; His2AvRFP embryos were dechorionated in 50% bleach for 3 minutes and exposed to light using a 470 +/− 20 nm filter in front of an epifluorescence light source on a Nikon ECLIPSE Ti-E microscope for the times indicated. Embryos were switched to a 572 +/− 35 nm excitation filter for 30 msec every 30 seconds and imaged with a 632 +/− 60 nm emission filter light to allow the use of His2Av-RFP to determine nuclear density. Images were acquired immediately following entry into NC 14 and 60 minutes into NC 14. Nuclear cycles were determined by nuclear density (calculated based on the number of nuclei/2500 μm2) after the preceding mitosis.
Immunostaining embryos
0-3 hr His2AvRFP and CRY2-ZLD; His2AvRFP embryos were dechorionated in 50% bleach for 3 minutes and fixed for 25 minutes in 4% formaldehyde-saturated heptanes in 1.3XPBS, 67 mM EGTA pH 8.0. Fixed embryos were washed with methanol and 100% ethanol. They were rehydrated in 1XPBS + 0.1% Triton X-100 (PT), washed and then blocked in PT +5% normal goat serum and 0.1% BSA. Embryos were incubated overnight with anti-ZLD antibody (1:500) at 4°C (Harrison et al., 2010), washed, and then incubated for 2 hrs with goat anti-rabbit IgG DyLight 488 conjugated secondary antibody (Thermo Scientific #35552). Embryos were imaged at 40X using a Nikon A1R-2I+ confocal microscope.
Chromatin immunoprecipitations and qPCR
Chromatin immunoprecipitations were performed as described previously (Blythe and Wieschaus, 2015). Briefly, 1000 stage 5 embryos were collected, dechorionated in 50% bleach for 3 minutes, fixed for 15 minutes in 4% formaldehyde and then lysed in 1 ml of RIPA buffer (50 mM Tris-HCl pH 8.0, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, and 150 mM NaCl). The fixed chromatin was then sonicated for 20 seconds 11 times at 20% output and full duty cycle (Branson Sonifier 250). Chromatin was incubated with 4-6 μg of anti-ZLD antibody overnight at 4°C, and then bound to 50 μl of Protein A magnetic beads (Invitrogen). The purified chromatin was then washed, eluted, and treated with 90 μg of RNAseA (37°C, for 30 minutes) and 100 μg of Proteinase K (65°C, overnight). The DNA was purified using phenol/chloroform extraction and concentrated by ethanol precipitation. Each sample was resuspended in 25 μl of water and analyzed via qPCR for enrichment (Promega). Fold enrichment of binding to the Sryα promoter was calculated relative to a control region of Act5C.
Immunoblots
Proteins were transferred to 0.45 μm Immobilon-P PVDF membrane (Millipore) in transfer buffer (25 mM Tris, 200 mM Glycine, 20% methanol) for 75 min at 500mA at 4°C. The membranes were blocked with 5%−10% non-fat dry milk in TBST for 20 min at room temperature and then incubated with anti-histone H3 antibody (1mg/ml, 1:5000 dilution, Abcam #ab1791), anti-ZLD (1:750, Harrison et al. 2010), or anti-Tubulin (DM1A) (1:5000 Sigma #T6199), overnight at 4°C. The secondary incubation was performed with goat anti-rabbit IgG-HRP conjugate (1:10,000 dilution, Bio-Rad #1706515) or anti-mouse IgG-HRP conjugate (1:10000 dilution, Bio-Rad #172-1011) for 1 hr at room temperature. Blots were treated with SuperSignal West Pico PLUS chemiluminescent substrate (Thermo-Scientific) and visualized using the Azure Biosystems c600.
Blue-light exposure and single-embryo RNA-seq
Single His2AvRFP or CRY2-ZLD; His2AvRFP embryos were dechorionated in 50% bleach for 3’, mounted in halocarbon (700) oil (Sigma) on a coverslip, and exposed to light using a 470 +/− 20 nm filter in front of an epifluorescence light source on a Nikon ECLIPSE Ti-E microscope for the times indicated. Embryos were switched to a 572 +/− 35 nm excitation filter for 30 msec every 30 seconds and imaged with a 632 +/− 60 nm emission filter light to allow the use of His2Av-RFP for precise developmental staging. Embryos were individually exposed to blue light and collected sequentially to ensure that each embryo was precisely timed and received the same amount of blue-light exposure. The only exceptions were the CRYNL embryos, which did not receive any blue light exposure and were collected 30 minutes into NC14. The nuclear cycle was identified following mitosis based on nuclear density (calculated by the number of nuclei/2500 μm2) and confirmed by timing of entry into nuclear cycle 14. Thirty minutes into nuclear cycle 14, embryos were picked into Trizol (Invitrogen #15596026) with 200 μg/ml glycogen (Invitrogen #10814010) and pierced with a 27G needle to release the RNA for 5 minutes. RNA was extracted and RNA-seq libraries were prepared using the TruSeq RNA sample prep kit v2 (Ilumina). 100-bp reads were obtained using an Illumina HiSeq2500 sequencer, generating between 16,434,144 - 49,784,627 reads per sample.
Protein expression and purification
MBP-ZLD1117-1487 (ZLDDBD) was purified from E. coli as described previously (Hamm et al., 2015), with the exclusion of the final dialysis step. Briefly, protein was bound to amylose resin (New England Biolabs) and eluted with 20 mM maltose. Full-length wild-type and mutant ZLD protein were prepared using baculovirus infection and affinity purification as described previously (Harrison et al., 2010). Baculovirus stocks were amplified for four days in Sf9 cells grown in Grace’s media (Himedia) supplemented with 10% fetal bovine serum. Hi5 cells were grown in ESF 921 media (Expression Systems) and were infected with 1 ml of freshly amplified virus. After three days, the cells were collected on ice, centrifuged, washed with PBS containing 5 mM MgCl2, and centrifuged once more. The cells were resuspended in hypotonic buffer (15 mM HEPES, 15 mM KCl, 2 mM MgCl2, 0.02% Tween, 10% glycerol, 2 mM βME, 1 mM EGTA, 0.4 mM PMSF, Roche complete protease inhibitor cocktail), flash frozen in liquid nitrogen, and kept at −80°C. Cell suspensions were thawed and dounced. KCl was then added to bring the concentration to 300 mM, and the lysate was cleared by centrifugation (10,000 rpm for 10 min at 4°C, 16,000rpm for 10 min at 4°C). 20 μM PMSF and 7 50 μL pre-washed anti-flag M2 affinity beads (Sigma) were added to the extract and incubated at 4°C for at least 2 hours. The beads were washed and applied to a chromatography column (BioRad) for additional washing. Protein was eluted twice (total volume: 3ml) in buffer containing 150 mM KCl and 200 μg /mL Flag peptide following a 30 min incubation with end-over-end mixing at 4°C. The fractions were pooled and further concentrated via injection onto a Mono Q PC 1.6/5 anion exchange chromatography column (GE Healthcare Life Sciences) connected to an ÄKTAmicro system (GE Healthcare Life Sciences) equilibrated in 150 mM KCl buffer. Using gradient elution, ZLD was eluted ~400 mM KCl. Protein concentration was determined using SYPRO-Red (Invitrogen) stained SDS-PAGE gels using BSA protein standards as a reference. Gels were visualized with a Typhoon FLA9000 using the SYPRO-Red fluorescence setting (excitation at 532nm).
Nucleosome reconstitution
The 160 bp bnk DNA fragment (dm3_chr3R:27018900-27019059) was amplified from Drosophila genomic DNA using one Cy5-labeled primer and one unlabeled primer (see bnk and primer sequences below). The DNA was ethanol precipitated and purified with AxyPrep Mag PCR Clean-up (Axygen) using a 1.8x ratio of beads to sample. The nucleosomes were reconstituted using a standard salt-urea gradient. Underlined sequences are ZLD-binding motifs.
bnk:TCCTTTTTTACTTTCATAGCTTAGGTAGTGATCTCAGGTAGTTTCCCGGAATTAAGTTAGGCTGGGTATCGCCTATCGGGAGCGGCTACCTGAACTTTTGGCACCAGCTGTCGGGGGTGAAAACTGGACCAGGTAGTCTTTAGAAGTGCACCTATATAAG
Primers:
KS111+ Cy5-TCCTTTTTTACTTTCATAGCTT
KS112 CTTATATAGGTGCACTTCTAAA
Electromobility shift assays
Cy5-labelled probe (DNA or nucleosome) at 2.5 nM (50 fmol/reaction) was incubated with recombinant ZLD protein in buffer containing: 5 ng poly[d-(IC)], 12.5 mM HEPES, 0.5 mM EDTA, 0.5 mM EGTA, 5% glycerol, 93.75 μM ZnCl2, 0.375 mM DTT, 75 μM PMSF, 0.075 mg/ml BSA, 5 mM MgCl2, 0.00625% NP-40, 50 mM KCl at room temperature for 60 min. These conditions are similar to conditions previously used to test nucleosome binding (Cirillo et al., 2002; Hsu et al., 2015; Sekiya et al., 2009), allowing comparison between different proteins. For competition assays, a 32 to 320-fold molar excess of unlabeled, double-stranded scute probe (sc WT or sc MUT) was added to the binding reaction (see sequences below, wild-type or mutated ZLD-binding motifs are underlined). Reactions were run in a 4% non-denaturing polyacrylamide gel in 0.5X Tris-borate-EDTA. Gels were visualized with a Typhoon FLA9000 using Cy5 fluorescence setting (excitation at 635 nm).
scute wild-type (specific) GAGAGAGACTACCTGTGGCTCACT
scute mutant (nonspecific) GAGAGAGAGTAGTTCTGGCTCACT
The EMSAs in Figure S4 were performed similarly, but used 40 fmol Cy5-labelled probe (NRCAM DNA or NCRAM nucleosome) in buffer containing: 10 mM Tris-HCl (pH7.5), 1 mM MgCl2, 10 μM ZnCl2, 1 mM DTT, 50 mM KCl, 3 mg/mL BSA, and 5% glycerol at room temperature for 30 min. Competition experiments were performed by incubating the Cy5-labeled probes with ZLD protein for 30 min followed by an additional incubation of 30 min with unlabeled competitor DNA. Underlined sequence is ZLD-binding motif.
NRCAM:
GATCCATTACTTCTGAAACAGATGACTCCCAGCAGCTGCTGCCTGTGGCCCACAGGGCTTCCTGCCCTGCATGACAGCTGCACATCACATCCTGTGGTCATACTACTTCAGCCGCTTCTACGGCCAGATACAAAAGTGGGTGGGGAACATAGGCAAGGGAT
QUANTIFICATION AND STATISTICAL ANAYLSIS
RNA-seg read alignment and processing
To remove adapter and low-quality sequences, raw RNA-seq reads were trimmed using Trimmomatic v0.36 (Bolger et al., 2014). Reads < 70 bp after trimming were discarded. Trimmed reads were aligned to the BDGP D. melanogaster genome release 6 (dm6) using hisat2 v2.1.0 with the following non-default parameters: −k 2 (Kim et al., 2015). Reads aligning to multiple locations were discarded.
RNA-seq analysis
Reads were assigned to annotated genes using featureCounts v1.5.3 (Liao et al., 2014) using default parameters and the UCSC annotation (r6.20). The resultant table of read counts was imported into R v3.4.0 and differential expression was determined using DESeq2 v1.14.1. (Love et al., 2014). Genes with < 50 reads across all samples were filtered out. Differential expression (adjusted p-value < 0.05 and fold change > 2) was determined using the standard DESeq2 analysis. To identify ZLD-target genes, previously determined ZLD ChIP peaks were assigned to the nearest gene (Harrison et al., 2011). Zygotically and maternally expressed genes, as well as the onset of zygotic gene expression, were previously defined (Li et al., 2014; Lott et al., 2011).
Identification of nonspecific gene expression changes
To identify nonspecific gene expression changes due to the CRY2 tag or blue-light exposure, RNA-seq data were compared to published RNA-seq data from precisely-staged single embryos (Lott et al., 2011). Published data were processed and realigned to the dm6 genome as described above. Principle component analysis (PCA) revealed that wild-type embryos exposed to blue light from nuclear cycles 10-14 clustered with early cycle 14 embryos, while all other wild-type, blue-light-exposed embryos clustered with late cycle 14 embryos, suggesting that extended blue-light exposure caused a developmental delay in the WT10-14 embryos. To identify changes in gene expression caused by blue-light exposure alone, we determined the differential gene expression between wild-type embryos exposed to blue-light from cycles 10-14 (WT10-14) and Lott et al. cycle 14A-B embryos, identifying 1,225 genes (Table S4). To identify changes due to the CRY2 tag, we determined differential gene expression between CRY2-ZLD expressing embryos without blue-light exposure (CRYNL) and Lott et al. cycle 14C-D, identifying 1,591 genes (Table S4). The combined list of 2,440 genes was excluded from subsequent analyses, resulting in a total of 8107 genes analyzed for differential expression. All comparisons for differential gene expression were between wild-type and CRY2-ZLD embryos with identical exposure to blue light.
DATA AND SOFTWARE AVAILABILITY
All RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO http//www.nicbi.nlm.nih.gov/geo) under accession number GSE121157. Raw image files are available through Mendeley Data at doi:10.17632/yd93nw42cp.1
Supplementary Material
Table S1 related to Figure 1. List of genes misregulated in CRY10-14 embryos.
Table S2 related to Figure 2. Lists of genes misregulated in CRY13-14 or CRY14 embryos.
Table S3 related to Figure 3. List of genes misregulated in CRY10-12 embryos.
Table S4 related to STAR Methods. Lists of genes misregulated in CRY-ZLD embryos or His2AvRFP embryos upon blue-light exposure.
Highlights.
An N-terminal CRY2 tag allows rapid and reversible transcription-factor inactivation
Zelda activity is required throughout zygotic genome activation in Drosophila embryos
Zelda is a pioneer factor that binds with sequence specificity to nucleosomal DNA
Nucleosome binding by pioneer factors may enable genome access during S-phase
Acknowledgments
We would like to thank members of the Harrison, Lewis, and Zaret labs for helpful feedback. We acknowledge the assistance of Elle Grevstad and the University of Wisconsin Madison Biochemistry Optical Core along with the University of Wisconsin Madison Biotechnology Sequencing Center. Work in the Harrison lab was supported by grants from the National Institutes of General Medical Sciences (R01GM111694), the American Cancer Society (RSG DDC-130854) and a Vallee Scholar Award. K.Z. was supported by NIH-R01GM36477, and P.L. was supported by the Greater Milwaukee Foundation and the Sidney Kimmel Foundation for Cancer Research. T.G. and K.S. were supported by NIH National Research Service award T32 GM007215.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Anderson KV, and Lengyel JA (1979). Rates of synthesis of major classes of RNA in Drosophila embryos. Dev Biol 70, 217–231. [DOI] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2015). Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2016). Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, and Cline TW (2006). The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967–1977. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, and Zaret KS (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289. [DOI] [PubMed] [Google Scholar]
- Dufourt J, Trullo A, Hunter J, Fernandez C, Lazaro J, Dejean M, Morales L, Nait-Amer S, Schulz KN, Harrison MM, et al. (2018). Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun. 9, 5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, and Alberts BM (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci 61, 31–70. [DOI] [PubMed] [Google Scholar]
- Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien CY, Kirov N, Shvartsman SY, and Rushlow CA (2014). Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol 24, 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. (2018). Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 173, 248–259. e15. [DOI] [PubMed] [Google Scholar]
- Guglielmi G, Barry JD, Huber W, and De Renzis S (2015). An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Dev. Cell 35, 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm DC, Bondra ER, and Harrison MM (2015). Transcriptional Activation Is a Conserved Feature of the Early Embryonic Factor Zelda That Requires a Cluster of Four Zinc Fingers for DNA Binding and a Low-complexity Activation Domain. J. Biol. Chem. 290, 3508–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm DC, Larson ED, Nevil M, Marshall KE, Bondra ER, and Harrison MM (2017). A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster. PLOS Genet. 13, e1007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, and Eisen MB (2015). Transcriptional Activation of the Zygotic Genome in Drosophila. In Current Topics in Developmental Biology, pp. 85–112. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, and Cline TW (2010). Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol 345, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, and Eisen MB (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HT, Chen HM, Yang Z, Wang J, Lee NK, Burger A, Zaret K, Liu T, Levine E, and Mango SE (2015). Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science (80-. ). 348, 1372–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Amourda C, Zhang S, Tolwinski NS, and Saunders TE (2017). Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, and Zaret KS (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, and Zaret KS (2016). The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell 62, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, and Giraldez AJ (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, and Onichtchouk D (2013). Pou5f1 Transcription Factor Controls Zygotic Gene Activation In Vertebrates. Science (80-. ). 341, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, and Eisen MB (2014). Establishment of regions of genomic activity during the maternal to zygotic transition. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wang W, Hu S, Wang X, and Zhang Y (2018). Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res. 28, 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, and Lin C (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, and Eisen MB (2011). Noncanonical Compensation of the Zygotic X Transcription in Early Drosophila melanogaster Development Revealed through Single-Embryo RNA-Seq. PLoS Biol. 9, e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, and Zhang Y (2016). Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 165, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, and Miller OL Jr. (1976). Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8, 305–319. [DOI] [PubMed] [Google Scholar]
- Mir M, Reimer A, Haines JE, Li X-YY, Stadler M, Garcia H, Eisen MB, and Darzacq X (2017). Dense bicoid hubs accentuate binding along the morphogen gradient. Genes Dev. 31, 1784–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Harrison MM, Darzacq X, and Eisen MB (2018). Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 7, e40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, and Kirschner M (1982). A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696. [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, and Rushlow C (2011). Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard DK, and Schubiger G (1996). Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10, 1131–1142. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, and Henikoff S (2016). Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell 165, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, and Harrison MM (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, and Zaret KS (2009). Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 23, 804–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, and Zaret KS (2012). Facilitators and Impediments of the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell 151, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, and Zaret KS (2015). Pioneer Transcription Factors Target Partial DNA Motifs on Nucleosomes to Initiate Reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, and Vorbruggen G (2006). Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell 17, 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nien C-Y, Chen K, Liu H-Y, Johnston J, Zeitlinger J, and Rushlow C (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, and Lipshitz HD (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, and Small S (2014). Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 28, 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, and Stark A (2012). Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res 22, 2018–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 related to Figure 1. List of genes misregulated in CRY10-14 embryos.
Table S2 related to Figure 2. Lists of genes misregulated in CRY13-14 or CRY14 embryos.
Table S3 related to Figure 3. List of genes misregulated in CRY10-12 embryos.
Table S4 related to STAR Methods. Lists of genes misregulated in CRY-ZLD embryos or His2AvRFP embryos upon blue-light exposure.
Data Availability Statement
All RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO http//www.nicbi.nlm.nih.gov/geo) under accession number GSE121157. Raw image files are available through Mendeley Data at doi:10.17632/yd93nw42cp.1


