Abstract
Helicobacter pylori infection and a high salt diet are each risk factors for gastric cancer. In this study, we tested the hypothesis that environmental salt concentration influences the composition of the H. pylori exoproteome. H. pylori was cultured in media containing varying concentrations of sodium chloride, and aliquots were fractionated and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We identified proteins that were selectively released into the extracellular space, and we identified selectively released proteins that were differentially abundant in culture supernatants, depending on the environmental salt concentration. We also used RNA-seq analysis to identify genes that were differentially expressed in response to environmental salt concentration. The salt-responsive proteins identified by proteomic analysis and salt-responsive genes identified by RNA-seq analysis were mostly non-concordant, but the secreted toxin VacA was salt-responsive in both analyses. Western blot analysis confirmed that VacA levels in the culture supernatant were increased in response to high salt conditions, and quantitative RT-qPCR experiments confirmed that vacA transcription was upregulated in response to high salt conditions. These results indicate that environmental salt concentration influences the composition of the H. pylori exoproteome, which could contribute to the increased risk of gastric cancer associated with a high salt diet.
Keywords: Helicobacter pylori, salt, sodium chloride, VacA, toxin, protein secretion, exoproteome, gastric cancer, peptic ulcer disease, proteomics, transcription
Introduction
Helicobacter pylori persistently colonizes the stomach in about 50% of the world’s population [1, 2]. In most individuals there are no adverse effects. Conversely, the presence of H. pylori is associated with an increased risk of peptic ulcer disease and gastric cancer [2, 3]. An important goal is to identify H. pylori-infected individuals who are at the highest risk of developing gastric cancer, so that these individuals can be targeted for therapeutic intervention.
H. pylori strains from unrelated individuals exhibit a high level of genetic diversity, and this variation among strains is an important factor that influences the risk of gastric cancer [4–6]. For example, H. pylori strains vary in the production of a pore-forming toxin known as VacA. The VacA proteins secreted by some strains cause vacuolation (and many other alterations) in gastric epithelial cells, whereas VacA proteins secreted by other strains lack vacuolating activity [6–9]. H. pylori strains producing highly active forms of VacA (designated type s1/i1/m1) are associated with a higher risk of gastric cancer, compared to strains producing less active forms of the protein (designated s2/i2/m2) [6, 10]. H. pylori strains also vary in the presence or absence of a chromosomal region known as the cag pathogenicity island [11–16] and in the production of outer membrane proteins (BabA, SabA and HopQ) mediating adherence to host cells [6, 17–19] H. pylori strains containing the cag PAI, producing active forms of VacA, and producing BabA, SabA and type I HopQ are associated with a higher risk of gastric cancer and peptic ulcer disease, compared to strains lacking the cag PAI, producing inactive forms of VacA, and lacking these adhesins [6, 20].
The risk of gastric cancer is influenced not only by strain-specific bacterial constituents, but also by genetic variation among human hosts, environmental factors, and dietary factors [21]. Epidemiologic studies in many regions of the world have detected an increased risk of gastric cancer associated with a high salt diet [21, 22]. One study reported that a high salt diet increased the risk of H. pylori-induced gastric cancer in a Mongolian gerbil model [23], and effects of a high salt diet on gastric pathology in mouse models of H. pylori infection also have been reported [24]. There are multiple possible mechanisms by which a high salt diet might enhance gastric cancer risk. For example, high salt conditions in the stomach may have direct effects on H. pylori that influence the interactions between the bacteria and the gastric mucosa. In support of this hypothesis, several studies detected alterations in H. pylori gene expression or proteomic composition in response to high salt conditions [25–29]. H. pylori growth in media containing high salt concentrations augments CagA production and alters the expression of genes encoding outer membrane proteins [27, 28, 30, 31].
Within the stomach, H. pylori localizes in the gastric mucus layer and attaches to gastric epithelial cells, but rarely invades human cells. H. pylori-induced alterations in the gastric mucosa have been attributed, at least in part, to the actions of secreted H. pylori proteins. CagA is translocated from adherent bacteria into human cells through a type IV secretion system [12–16]. VacA is secreted through a type V secretion system and released into the extracellular space as a soluble protein [7, 32, 33]. Many other H. pylori proteins are also released into the extracellular space [34–37], either by specific secretion pathways or non-specifically as a consequence of bacterial autolysis.
Previous studies identified H. pylori proteins released into the extracellular space using proteomic methods, typically involving either 2D gel electrophoresis and or other protein separation methods, followed by mass spectrometric analysis [34–37]. Most of the previous studies endeavored to distinguish between H. pylori proteins selectively released into the extracellular space and proteins non-specifically released as a consequence of autolysis. In the current study, we tested the hypothesis that environmental sodium chloride concentration influences the composition of the H. pylori exoproteome. Specifically, we cultured H. pylori in media containing varying concentrations of sodium chloride, and then analyzed the composition of the exoproteome under each of the culture conditions tested. In parallel, we used RNA-seq methodology to analyze effects of environmental sodium chloride concentration on H. pylori gene transcription and identified salt-responsive genes encoding proteins that are released into the extracellular space. We report that high salt conditions alter the abundance of numerous H. pylori proteins released into the extracellular space. Increased levels of VacA in the extracellular space in response to high salt conditions is relevant to the increased risk of gastric cancer observed in persons who consume a high salt diet.
Methods
Bacterial strains and culture conditions
H. pylori strain 26695 was passaged on Trypticase soy agar plates containing 5% sheep blood at 37° C in room air supplemented with 5% CO2. Liquid cultures were grown in a modified form of sulfite-free Brucella broth [38] containing only low molecular mass (< 3 kDa) components (Brucella broth filtrate), supplemented with 1X cholesterol (Gibco), as described previously [37]. The use of this medium facilitates subsequent mass spectrometric analysis with minimal interference from high molecular mass proteins present in unfractionated Brucella broth. The Brucella broth used for routine H. pylori culture contains 0.5% added sodium chloride. In the current study, we also used modified forms of Brucella broth filtrate containing higher levels of added sodium chloride (1.0% or 1.25% NaCl).
Bacteria were harvested from plates and inoculated into liquid medium (10 ml volume, initial OD600 = 0.02) with the compositions described above (0.5% added sodium chloride). The seed cultures were grown with shaking for about 18 h, and then were inoculated into fresh Brucella broth filtrate [containing varying amounts of NaCl (0.5%, 1.0%, or 1.25%)] at an initial OD600 of 0.02. A previous study showed that the composition of the exoproteome is growth phase-dependent [37], so we collected aliquots at two different time points (24 h and 36 h post-inoculation, corresponding to late log and stationary phase). The culture aliquots were centrifuged at 4500 x g at 4° C, yielding broth culture supernatants and bacterial pellets that were processed as described below.
Processing of broth culture supernatants
Culture supernatants were processed as described previously [37]. Specifically, the supernatants were passed through a 0.22 μm filter to remove any residual bacteria. Filtered supernatants were concentrated using a 10 kDa cut-off centrifugal filter ultrafiltration unit. The concentration of any residual low molecular mass Brucella broth components was reduced by buffer exchange with Tris-buffered saline. The resulting preparations were centrifuged at 100,000 x g for 2 h at 4° C to remove outer membrane vesicles and other insoluble components, and the samples was further concentrated using centrifugal filter ultrafiltration units (10 kDa cut-off). Protein concentrations were determined by bicinchoninic acid (BCA) assay (Pierce).
Bacterial fractionation
Bacteria were fractionated into preparations enriched in membrane components or soluble components (cytoplasmic and periplasmic proteins), as described previously [28, 37]. Specifically, the bacterial pellets were washed twice in TNKCM buffer [50 mM Tris (pH 7.4), 100 mM NaCl, 27 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2] and re-suspended in suspension lysis buffer [50 mM Tris (pH 7.4) containing 1 mM MgCl2 and EDTA-free protease inhibitor cocktail (Roche)]. The re-suspended pellets were sonicated (5 pulses, 20 seconds on/40 seconds off, 20% amplitude), and lysates were centrifuged at 4500 x g at 4° C. The resulting supernatant was collected and subjected to ultracentrifugation at 100,000 x g at 4° C for 2 h. The resulting insoluble fraction (pellet) is enriched in membrane proteins, and the soluble fraction (supernatant) is enriched in cytoplasmic and periplasmic proteins [37, 39]. Both the supernatants and solubilized protein fractions were concentrated by ultrafiltration with a 10 kDa cutoff membrane, and protein concentrations were quantified by BCA assay (Pierce).
Analysis of samples by mass spectrometry
Samples prepared as described above were run 2 cm into a 10% BisTris NuPAGE gel. Gels were stained with Coomassie blue and an in-gel trypsin digest was performed. Single dimensional LC-MS/MS was performed using ThermoFisher LTQ equipped with a nano-electrospray source and attached to a Nanoacuity (Waters) HPLC unit with an autosampler [37]. Peptides were resolved via reversed phase separation (90 min total cycle time). Peptide MS/MS spectra were acquired data-dependently with one full scan MS followed by 5 MS/MS scans. The peptide MS/MS spectral data were queried using SEQUEST (full tryptic specificity) and searched against the H. pylori 26695 protein database, to which both common contaminants and reversed versions of H. pylori protein sequences had been added. Peptide identifications were filtered and collated to proteins using Scaffold 4 (Proteome Systems). Protein identifications required a minimum of 2 unique peptides per protein, and were filtered to a 5% false discovery rate (both peptide and protein).
Analysis of mass spectrometry data
H. pylori was cultured under each of the experimental conditions on three separate days, and the resulting samples were analyzed by mass spectrometry. Spectral counts (peptides assigned to a given protein) detected in LC-MS/MS analyses of the three independent samples from each condition were summed and normalized. To identify proteins that were selectively released into the extracellular space, proteomic data for the broth culture supernatant fraction were compared to proteomic data for the cytoplasmic/periplasmic fraction, using previously described methods [37]. Specifically, for each protein, we compared the relative abundance of assigned spectra in the supernatant to the relative abundance of assigned spectra in the cytoplasm/periplasm fraction by calculating an enrichment value (Esup = % abundancesup/% abundanceCP/PP)[37]. To facilitate further analysis, the levels of enrichment were expressed as log2 values (log2 Esup). Proteins enriched in the supernatant compared to the cytoplasm/periplasm fraction are assigned positive log2 Esup values, while those enriched in the cytoplasm compared to the supernatant are assigned negative log2 Esup values.
As an additional analytic approach for identifying selectively released proteins, the significance of differences in numbers of assigned spectra in supernatants compared to cytoplasm/periplasm fractions was calculated using a Fisher’s exact test (FET) with Benjamini-Hochberg (BH) multiple test correction. Selectively released proteins were defined as proteins with log2 Esup value ≥ 1 (a 2-fold or greater abundance in the supernatant vs the cytoplasm/periplasm) and exhibiting statistically significant differences based on Fisher’s Exact test with Benjamini-Hochberg correction (false discovery rate = 0.1). This approach successfully discriminated between proteins previously reported to be selectively released into the extracellular space [37] and ribosomal proteins (data not shown).
To assess the effect of salt concentration on the relative abundance of selectively released proteins detected in culture supernatant, assigned spectral counts for individual proteins detected in LC-MS/MS analyses of supernatants from all cultures containing 0.5% salt (both 24 h and 36 h time points) were summed, and a similar approach was used for spectral counts from all cultures containing 1.0% salt or 1.25% salt. The numbers of spectral counts in samples from different conditions were then compared. Proteins exhibiting a ≥2-fold difference in the number of normalized spectral counts when comparing cultures containing high salt concentrations (1.0% or 1.25%) with cultures containing routine salt concentrations (0.5% NaCl) were considered to be salt-responsive. This analysis was restricted to selectively released proteins (Supplemental Table S1) for which a minimum of 10 spectral counts were detected in either high salt or low salt conditions.
Western blotting
H. pylori was inoculated into Brucella broth filtrate supplemented with cholesterol and containing 0.25%, 0.5%, 1.0% or 1.25% NaCl, at a starting OD600 of ~0.02, as described above. After 24 h and 36 h, aliquots of the cultures were removed and centrifuged at 4500 x g for 15 minutes. The supernatant fraction was concentrated using a 10 kDa cut-off centrifugal filter ultrafiltration unit, and protein concentrations were determined using a BCA protein assay (Pierce). In addition to analyses of cultures grown in Brucella broth filtrate, H. pylori were also inoculated into unfractionated Brucella broth supplemented with cholesterol and containing either 0.5% or 1.25% NaCl at OD600 ~0.30 and incubated for 1 h with shaking. This latter condition mimicked the H. pylori growth conditions used for RNA analyses (see below). Supernatant samples were processed as described above.
Western blot analyses were performed using samples standardized by protein concentration (40 ug of total protein per sample). Proteins were separated on a gradient (4–20%) acrylamide gel and transferred to a nitrocellulose membrane. Membranes were immunoblotted using rabbit polyclonal antisera to VacA [40] followed by an anti-rabbit secondary antibody conjugated to horseradish peroxidase (Promega). Enhanced chemiluminescent reaction-generated signals were detected using x-ray film.
RNA isolation
H. pylori was grown in unfractionated Brucella broth supplemented with 1X cholesterol (Gibco) to an OD600 of ~0.5, and was subcultured into Brucella broth containing either 0.5% NaCl or 1.25% NaCl at a starting OD600 of ~0.3. Following growth for 1 h, the cultures were centrifuged, bacterial pellets were resuspended in RNAlater (Ambion) for 40 min, and the pellets were stored at −80° C. RNA was prepared using TRIzol reagent (Ambion), using the manufacturer’s instructions. All RNA samples were digested with RQ1 RNase-free DNase (Promega) to remove contaminating DNA, before being subjected to a clean-up step using Qiagen RNeasy columns.
Preparation of RNA-seq library and analysis
The preparation of RNA-seq libraries and subsequent analysis was performed as described previously [29]. Briefly, RNA quality was determined using 2100 Bioanalyzer (Agilent), and 200 ng of DNase-treated total RNA (RNA integrity number greater than 9) was used to generate rRNA-depleted/mRNA-enriched libraries using TruSeq Ribo-Zero bacterial RNA kits (Illumina). Library quality was assessed using the 2100 Bioanalyzer (Agilent), and libraries were analyzed using KAPA library quantification Kits (KAPA Biosystems). Pooled libraries were subjected to 75-bp paired-end sequencing according to the manufacturer’s protocol (Illumina HiSeq 3000). Bcl2fastq2 conversion software (Illumina) was used to generate demultiplexed Fastq files. Six independent RNA-seq libraries prepared from H. pylori cultures grown in media containing either 0.5% or 1.25% NaCl (three cultures of each) were sequenced. The numbers of sequence reads for each sample ranged from 2.28 x 107 to 5.12 x 107.
RNA-seq data were trimmed to remove all bases below a quality of Q3, and adapter sequences were removed using FastQ quality control software (FaQCs) [41]. Kallisto pseudocounting [42] was applied to all annotated genes in the H. pylori 26695 reference genome [43] (GenBank accession number GCA_000008525.1). The expected counts fields from Kallisto outputs were used for all analysis steps. Transcripts associated with a total of 1,562 H. pylori genes were identified by RNA-seq. The EdgeR [44] package for R was used to analyze count files. Data from individual samples were normalized within EdgeR and analyzed using the generalized linear model (GLM).
The RNA-seq data were analyzed by calculating for each gene a transcript abundance ratio (number of sequence reads from bacteria grown in high salt conditions divided by the number of sequence reads from bacteria grown in routine conditions). The mean ± SD of all the calculated transcript abundance ratios for the 1562 identified genes was 1.01 ± 0.3. Specific genes were classified as upregulated or downregulated in response to high salt conditions (1.25% NaCl) if the calculated transcript ratios were ≥2 standard deviations above or below the mean transcript abundance ratios (i.e. ≥1.61 or ≤0.62), with a false discovery rate (FDR) of <0.05.
Analysis of vacA by real time RT-qPCR
H. pylori total RNA was isolated using Trizol reagent (Gibco). RNA samples were digested with RQ1 RNAse-free DNAse (Promega) and subjected to a clean-up step using RNAeasy columns (Qiagen), as described above. Purified RNA (100 ng) was then used for cDNA synthesis with random hexamer primers using the iScript cDNA synthesis kit (Biorad). Real-time reverse transcription quantitative PCR (RT-qPCR) was completed using 1:20 dilutions of cDNA. Control reactions were performed in the absence of reverse transcriptase. Real time analysis was performed using an ABI real-time PCR machine and SYBR green fluorophore (iTaq universal SYBR mix; Bio-Rad), with transcript abundance determined using the ΔΔCT method. In addition to analyzing expression of vacA and other genes of interest, we analyzed expression of the housekeeping genes gyrB (DNA gyrase subunit B) and atpA (encoding ATP synthase F1 α subunit) and 16S rRNA as controls. Prior to analysis, the transcript levels of target genes in each sample were normalized to the abundance of the 16S rRNA internal control. The normalized transcript signals from high-salt conditions and routine conditions were then compared. The primers used for real-time RT-qPCR analysis were as follows: 5’-ACAACAAACACACCGCAAAA- 3’ and 5’ CCTGAGACCGTTCCTACAGC −3’ for vacA; 5- GGAGTACGGTCGCAAGATTAAA −3’ and 5– CTAGCGGATTCTCTCAATGTCAA −3’ for 16S rRNA; 5’- CGTGGATAACGCTGTAGATGAGAGC - 3’ and 5’ -GGGATTTTTTCCGTGGGGTG −3’ for gyrB; and 5’-CTTCACGCAATTCGCTTCTG-3’ and 5’-AAGCCCTTAGCCCCAGCATA-3’ for atpA.
Results
Analysis of selectively released proteins
To determine the effect of environmental salt concentration on composition of the H. pylori exoproteome, we cultured H. pylori in media containing three different concentrations of sodium chloride (0.5%, 1.0%, or 1.25%), as described in Methods. Culture medium containing 0.5% NaCl is used for routine culture of H. pylori, whereas 1.0% or 1.25% NaCl concentrations correspond to elevated salt concentrations. Aliquots were removed at 24 h and 36 h post-inoculation (corresponding to late-log phase and stationary phase), and the samples were centrifuged to yield bacterial pellets and culture supernatants. The broth culture supernatant fractions were processed to remove outer membrane vesicles and other insoluble components, and the bacterial pellets were processed to yield a soluble cellular fraction enriched in cytoplasmic and periplasmic proteins, as described in Methods. The broth culture supernatant and soluble cellular fractions were then analyzed by LC-MS/MS. H. pylori can potentially release proteins into the extracellular space through specific secretion pathways or non-specifically as a result of autolysis [34–37, 45]. To identify proteins that were selectively released into the extracellular space, we used a previously described approach [37] in which the relative abundance of individual proteins in the supernatant fraction is compared to the relative abundance of the corresponding proteins in the cytoplasmic/periplasmic fraction, as described in Methods.
Twenty-five proteins met the criteria for selective release into the extracellular space (as defined in Methods) in all 6 of the culture conditions tested [i.e. medium containing 0.5%, 1.0%, or 1.25% NaCl, and time points of 24 h or 36 h] (Supplemental Table S1), and 35 proteins met criteria for selective release in at least 5 of the 6 conditions tested (Table 1). These included several proteases [HP0657, HP1012, and HP1019 (serine protease, HtrA)], several cysteine-rich proteins belonging to the Sel-1-like protein family (HP0211, HP0235, and HP1098, designated HcpA, HcpE and HcpC, respectively)[46, 47], HP0104 (2’,3’-cyclic-nucleotide 2’-phosphodiesterase, CpdB), and HP0410 (neuraminyllactose-binding hemagglutinin homolog). Many of the proteins listed in Table 1 [including HP0175 [48–50], HP0211 (HcpA) [51, 52], HP1019 (serine protease, HtrA) [53, 54], HP1118 (γ-glutamyltranspeptidase, GGT) [55–57], HP1173 [58], HP1286 [59, 60], and HP1454] [61] are capable of causing alterations in host cells. The majority of the proteins listed in Table 1 are predicted to contain Sec-dependent signal sequences. Many of the proteins listed in Table 1 were also identified as selectively released proteins in a previous study that analyzed H. pylori protein release when cultured in medium containing 0.5% sodium chloride [37](Table 1, asterisks). Proteins identified as selectively released under all conditions in the current study but not the previous study [37] included HP0175 (cell binding factor 2, also known as peptidyl prolyl cis,trans-isomerase) and HP0389 (superoxide dismutase). These data confirm that specific proteins are selectively released into the extracellular space under routine culture conditions, and indicate that most of these proteins are also selectively released under high salt conditions.
Table 1.
Proteins selectively released into the extracellular space under ≥5 culture conditions
| Gene/Protein | Description | Enrichment of proteins in culture supernatant compared to the soluble bacterial fraction under the indicated culture conditions a,b | ||||
|---|---|---|---|---|---|---|
| 0.5 % NaCl | 1.0% NaCl | 1.25% NaCl | Average Esupc | Predicted cleavage sited | ||
| HP0104*e | 2',3'-cyclic-nucleotide 2'-phosphodiesterase (CpdB) | 5.88 | 6.08 | 6.28 | 6.08 | 18–19 |
| HP0129* | hypothetical protein | 2.64 | 2.71 | 5.54 | 3.63 | 21–22 |
| HP0175 | cell binding factor 2; peptidyl prolyl cis,transisomerase | 1.22 | 1.85 | 2.18 | 1.75 | 26–27 |
| HP0176 | fructose-bisphosphate aldolase (Tsr) | 1.34 | 1.53 | 1.23 | 1.37 | - |
| HP0211* | hypothetical secreted protein (HcpA) | 2.74 | 4.99 | 3.64 | 3.79 | 25–26 |
| HP0224 | peptide methionine sulfoxide reductase (MsrA) | 0.94 | 1.30 | 1.45 | 1.23 | 22–23 |
| HP0231* | hypothetical protein; DsbK (thioloxidoreductase) | 2.04 | 2.51 | 2.81 | 2.45 | 26–27 |
| HP0235* | hypothetical secreted protein (HcpE) | 3.57 | 4.61 | 3.83 | 4.00 | 24–25 |
| HP0298* | dipeptide ABC transporter, periplasmic dipeptide-binding protein (DppA) | 2.74 | 4.38 | 3.87 | 3.66 | 22–23 |
| HP0304* | alginate lyase | 5.10 | 3.86 | 5.12 | 4.69 | 19–20 |
| HP0323* | membrane bound endonuclease (Nuc); NucT | 2.96 | 2.95 | 3.65 | 3.19 | 16–17 |
| HP0377* | thiol:disulfide interchange protein (DsbC), putative | 2.92 | 4.65 | 5.03 | 4.20 | 24–25 |
| HP0389 | superoxide dismutase (SodB) | 1.60 | 1.23 | 1.54 | 1.46 | - |
| HP0410* | putative neuraminyllactose-binding hemagglutinin homolog (HpaA paralog) | 4.49 | 3.97 | 4.36 | 4.27 | 24–25 |
| HP0485* | catalase-like protein | 2.97 | 3.31 | 4.25 | 3.51 | 24–25 |
| HP0630 | modulator of drug activity (Mda66) | 1.51 | 1.72 | 1.67 | 1.63 | - |
| HP0657* | processing protease (YmxG) | 2.34 | 2.68 | 2.63 | 2.55 | 20–21 |
| HP0871* | CDP-diglyceride hydrolase (Cdh) | 3.81 | 4.14 | 5.05 | 4.33 | 21–22 |
| HP0953* | hypothetical protein | 1.98 | 2.93 | 3.22 | 2.71 | 20–21 |
| HP0973* | hypothetical protein | 2.35 | 4.25 | 3.71 | 3.44 | 28–29 |
| HP1012* | protease (PqqE) | 2.46 | 2.15 | 2.54 | 2.38 | 26–27 |
| HP1019* | serine protease (HtrA) | 4.89 | 5.60 | 5.84 | 5.44 | - |
| HP1098* | hypothetical secreted protein (HcpC) | 3.08 | 3.63 | 3.93 | 3.55 | 25–26 |
| HP1118* | gamma-glutamyltranspeptidase (Ggt) | 2.81 | 3.52 | 3.83 | 3.39 | 27–28 |
| HP1164 | thioredoxin reductase (TrxB); flavodoxin: quinone reductase (FqrB) | 1.80 | 1.49 | 1.79 | 1.69 | - |
| HP1173* | hypothetical protein | 3.77 | 5.79 | 6.96 | 5.51 | 26–27 |
| HP1186* | carbonic anhydrase | 1.97 | 4.16 | 3.87 | 3.33 | 18–19 |
| HP1227* | cytochrome c553 | 1.81 | 2.99 | 3.13 | 2.64 | 19–20 |
| HP1285* | 5’-nucleotidase, lipoprotein e(P4) family | 2.90 | 3.48 | 4.15 | 3.51 | - |
| HP1286* | polyisoprenoid-binding protein (YceI) | 1.92 | 2.18 | 2.90 | 2.34 | 17–18 |
| HP1454* | Lpp20 domain containing protein | 2.39 | 3.54 | 5.03 | 3.65 | 19–20 |
| HP1458 | thioredoxin-2 (Trx 2) | 1.69 | 1.40 | 1.79 | 1.63 | - |
| HP1561* | iron(III) ABC transporter, periplasmic iron-binding protein (CeuE1) | 2.52 | 2.89 | 3.09 | 2.83 | 31–32 |
| HP1562* | iron(III) ABC transporter, periplasmic iron-binding protein (CeuE2) | 2.62 | 2.98 | 3.25 | 2.95 | 29–30 |
| HP1564* | putative outer membrane protein | 2.21 | 2.45 | 2.79 | 2.48 | - |
| Averagef | 2.59 | 3.17 | 3.52 | 3.09 | ||
Esup values (log2) were calculated and analyzed as described in Methods. The reported data represent the average of Esup values (log2) from broth cultures grown under the indicated conditions for 24 and 36 h. All proteins listed in the table were selectively released in at least 5 of the 6 conditions tested (see Supplemental Table 1).
Raw data with complete list of selectively released proteins (1 or more condition) are shown in Supplementary Table 1.
Average Esup values (log2) for each protein, based on analysis of 6 culture conditions.
Predicted signal sequence cleavage site, based on use of SignalP 5.0 [71]. Numbers represent positions of amino acids at the site of predicted cleavage. Dash (−) indicates that no signal sequence is predicted.
Asterisks (*) represent proteins also selectively released in Snider et al. [37]
Average Esup value for all of the listed proteins.
Culturing H. pylori in medium containing increased salt concentrations could potentially lead to multiple bacterial alterations that affect the composition of the exoproteome. For example, high salt conditions might alter the release of proteins from the bacterial surface into the culture supernatant, might alter expression of genes encoding released proteins, or might promote bacterial autolysis. As an initial approach for distinguishing among these possibilities, we assessed whether high salt conditions changed the selectivity of release of the proteins listed in Table 1 (i.e., resulting in higher or lower log2 Esup values than observed under routine culture conditions). As shown in Table 1, culturing H. pylori in high salt conditions was associated with an increased selectivity of release of the listed proteins (mean log2 Esup values of 2.59 for cultures containing 0.5% salt, 3.17 for cultures containing 1.0% salt, and 3.52 for cultures containing 1.25% salt). In contrast, culturing H. pylori in high salt conditions did not result in any change in the selectivity of release of ribosomal proteins (data not shown). These results indicate that H. pylori growth in medium containing elevated salt concentrations does not promote autolysis.
89 proteins met the criteria for selective release in fewer than 5 of the conditions tested (Supplemental Table 1). Examples included numerous outer membrane proteins (OMPs) and VacA (vacuolating toxin) (Supplemental Table 1). Selective release of OMPs was mainly detected at the 24 h time point (Supplemental Table 1). Collectively, these data indicate that numerous proteins are selectively released into the extracellular space under both routine conditions and high salt conditions (Table 1).
Selectively released proteins exhibiting altered relative abundance in culture supernatant in response to high salt conditions
We next sought to identify the subset of selectively released proteins that differed in relative abundance when comparing supernatant fractions from cultures grown in medium containing 0.5% sodium chloride with supernatant fractions from cultures grown in medium containing higher levels of sodium chloride. As described in Methods, we calculated ratios of spectral counts (high salt vs normal salt) for the selectively released proteins listed in Supplementary Table 1. Numerous OMPs and VacA (as well as multiple other proteins) were selectively released into the supernatant and were more abundant in one or both of the high salt conditions (1.0% and/or 1.25% NaCl) compared to routine salt conditions (Table 2). A smaller number of proteins were more abundant in supernatants from cultures grown in the normal salt condition (0.5% salt) compared to supernatants from cultures grown in the higher salt conditions (i.e. decreased abundance in high salt conditions) (Table 2). These experiments demonstrate that the proportional abundance of specific proteins in the extracellular space is altered during H. pylori growth under high salt conditions compared to growth under routine conditions.
Table 2.
Selectively released proteins that exhibit altered abundance in supernatant in response to high salt conditions
| Protein | Description | Fold changea | ||
|---|---|---|---|---|
| 1.0% vs. 0.5% | 1.25% vs. 0.5% | Predicted cleavage siteb | ||
| Downregulated | ||||
| HP0089 | Pfs; Mtn; MqnB | (0.75) | 0.44 | - |
| HP0966 | P-loop NTPase and CrfC domain containing protein | (0.73) | 0.24 | - |
| HP1526 | exodeoxyribonuclease (LexA) | (0.86) | 0.41 | - |
| Upregulated | ||||
| HP0097 | hypothetical protein | 2.05 | 2.55 | 15–16 |
| HP0127 | outer membrane protein (Omp4, HorB) | 5.62 | 19.46 | 20–21 |
| HP0130 | hypothetical protein | (1.92) | 2.35 | 17–18 |
| HP0227 | outer membrane protein (Omp5, HopM) | 21.78 | 23.57 | 18–19 |
| HP0229 | outer membrane protein (Omp6, HopA) | 9.40 | 14.53 | 16–17 |
| HP0252 | outer membrane protein (Omp7, HopF) | 10.24 | 27.37 | 23–24 |
| HP0317 | outer membrane protein (Omp9, HopU) | 3.10 | 6.29 | 20–21 |
| HP0472 | outer membrane protein (Omp11, HorE) | (1.62) | 2.19 | 18–19 |
| HP0671 | outer membrane protein (Omp14, HorF) | 7.93 | 27.37 | 23–24 |
| HP0686 | iron(III) dicitrate transport protein (FecA1) | 12.55 | 27.37 | 17–18 |
| HP0709 | S-adenosylmethionine hydrolase | 3.48 | 3.09 | 21–22 |
| HP0710 | putative outer membrane protein | 6.53 | 16.45 | 16–17 |
| HP0797 | flagellar sheath adhesin (HpaA) | 6.14 | 13.20 | 27–28 |
| HP0863 | lipoprotein, putative; plasminogen-binding protein (PgbB) | 5.62 | 19.46 | 17–18 |
| HP0884 | hypothetical protein | 2.15 | 2.95 | - |
| HP0887 | vacuolating cytotoxin (VacA) | 8.77 | 10.58 | 33–34 |
| HP0896 | outer membrane protein (Omp19, BabB) | (1.25) | 5.52 | 19–20 |
| HP0912 | outer membrane protein (Omp20, HopC, AlpA) | 5.85 | 11.74 | 21–22 |
| HP0913 | outer membrane protein (Omp21, HopB, AlpB) | 2.52 | 6.52 | 41–42 |
| HP1083 | putative outer membrane protein | 4.19 | 7.42 | 26–27 |
| HP1117 | hypothetical secreted protein (HcpX) | (1.43) | 3.17 | 23–24 |
| HP1126 | colicin tolerance-like protein (TolB) | 2.10 | 2.70 | 16–17 |
| HP1177 | outer membrane protein (Omp27, HopQ) | 2.96 | 7.24 | 21–22 |
| HP1243 | outer membrane protein (Omp28, BabA) | 17.16 | 74.74 | 20–21 |
| HP1376 | (3R)-hydroxymyristoyl-(acyl carrier protein) dehydratase (FabZ) | (1.14) | 2.26 | - |
| HP1395 | outer membrane protein (Omp30, HorL) | (1.74) | 4.61 | 21–22 |
| HP1456 | membrane-associated lipoprotein (Lpp20) | 2.16 | 3.63 | 21–22 |
| HP1461 | cytochrome c551 peroxidase | 7.38 | 2.92 | 18–19 |
| HP1469 | outer membrane protein (Omp31, HorJ) | 2.43 | 3.90 | 17–18 |
| HP1501 | outer membrane protein (Omp32, HorK) | 4.19 | 19.13 | - |
| HP1512 | iron-regulated outer membrane protein (FrpB) | 3.51 | 10.25 | 21–22 |
To identify selectively released proteins that differed in proportional abundance in the culture supernatant in response to salt concentration, we analyzed proteomic data for the proteins listed in Suppl. Table S1. The fold change in response to high salt conditions was calculated as a ratio (total spectral counts from 24 h and 36 h time points in 1.0% or 1.25% high salt conditions compared to total spectral counts in the 0.5% condition). The analysis was restricted to proteins with ≥10 total assigned spectral counts in either the 0.5% NaCl condition or high salt conditions. Proteins with fold change values >2 or <2 were considered salt-responsive. The proteins listed met these criteria in at least one of the two high salt conditions. Values in parentheses did not have fold values > 2 for that condition or did not meet the criteria for ≥10 total assigned spectral counts.
Predicted signal sequence cleavage site, based on use of SignalP 5.0 [71]. Numbers represent positions of amino acids at the site of predicted cleavage. Dash (−) indicates that no signal sequence is predicted.
Release of VacA toxin in response to high salt conditions
As described above, the relative abundance of VacA in culture supernatant was higher in cultures containing elevated sodium chloride concentrations than in supernatant from cultures containing 0.5% salt. Western blot analysis of samples from 3 independent cultures using a polyclonal antibody to the 88 kDa secreted VacA protein confirmed that VacA levels in the culture supernatant were increased in cultures containing high salt concentrations, compared to cultures containing 0.5% salt (Figure 1A).
Figure 1.

VacA protein and transcript levels are increased in response to high salt concentration. H. pylori was cultured in Brucella broth filtrate supplemented with cholesterol and containing the indicated concentrations of NaCl (0.25%, 0.5%, 1.0% and 1.25%). (A) Western blot analysis of VacA in culture supernatant samples from 24 and 36 h time points (standardized at 40 μg protein per sample). (B) Cultures were inoculated at an OD600 of about 0.3, and samples from a 1 h time point (100 μg protein per sample) were analyzed by Western blotting. The results are representative of analyses of three independent sets of cultures. (C) H. pylori was cultured for 1 h in Brucella broth-cholesterol containing 0.5% NaCl or 1.25% NaCl. Transcript abundance was analyzed by RT-qPCR. Normalized transcripts signals from bacteria grown in 1.25% NaCl were compared to those from bacteria grown in 0.5% NaCl. RNA was isolated from four independent experiments. The mean and standard error of the mean are shown. A significant difference (P < 0.05, 1-way ANOVA) was observed when comparing the effect of high salt on vacA transcript levels, compared to effects on control genes (gyrB and atpA).
We also analyzed whether VacA levels in the culture supernatant were increased after exposure of the bacteria to high salt conditions for a relatively short time period (1 h). H. pylori was grown overnight and subcultured at OD600 ~0.3 into either the standard medium (0.5% NaCl) or medium containing 1.25% NaCl. As shown in Figure 1B, higher levels of VacA were detected in H. pylori supernatants from cultures containing 1.25% NaCl (Figure 1B) than in supernatants from cultures containing 0.5% NaCl. Therefore, the Western blotting data corroborate the results obtained by analysis of mass spectrometry data, and provide further evidence indicating that the levels of VacA in the extracellular space are increased in response to high salt conditions.
Under routine culture conditions, the 88 kDa VacA toxin and a small VacA peptide are released into the extracellular space, and the carboxy-terminal VacA β-barrel domain remains associated with the bacteria [8, 62]. To determine whether H. pylori exposure to high salt conditions altered this pattern of release, we analyzed proteomic data for samples containing elevated salt concentrations and mapped the location of detected VacA peptides relative to the full-length VacA amino acid sequence (Figure 2A). Nearly all of the VacA peptides detected in the supernatant mapped to the secreted 88 kDa toxin. VacA peptides corresponding to the predicted C-terminal β-barrel domain of VacA were not detected in the culture supernatant, similar to what has been observed previously when analyzing VacA secretion in medium containing 0.5% salt [37]. For comparison, we analyzed the distribution of peptides assigned to two salt-responsive OMPs [(HP0710 (HomA), HP1177 (HopQ), Table 2)]. We detected peptides in the supernatant that were mapped throughout the entire length of the HopQ and HomA outer membrane proteins, including the predicted C-terminal β-barrel domains (Figure 2B and C) [63].
Figure 2.
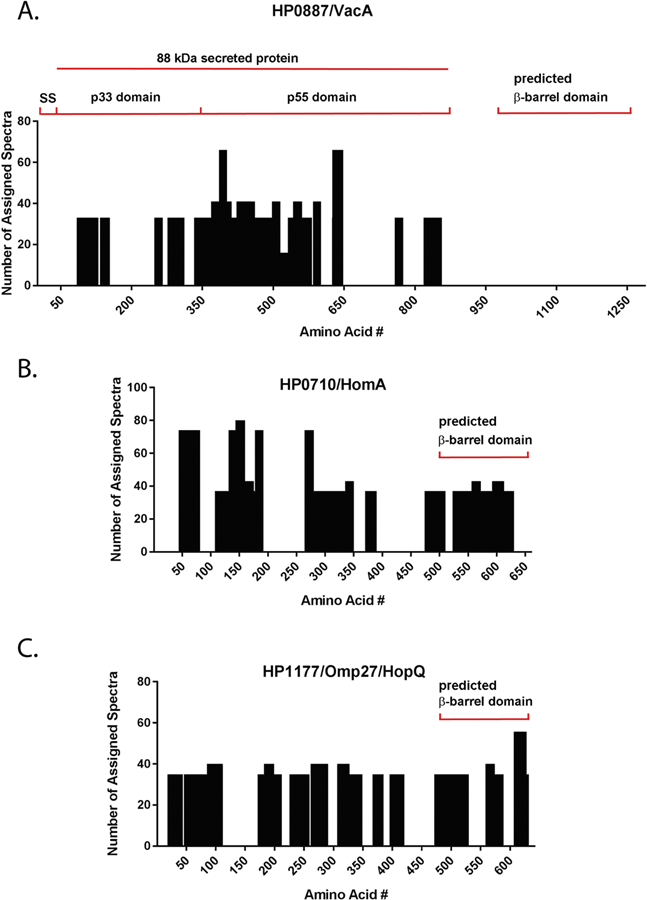
Analysis of VacA and Omp peptide distribution. H. pylori was grown in high salt conditions (1.25% NaCl for 24 h), and broth culture supernatants were processed as described in Methods. Spectral counts for peptides assigned exclusively to VacA, HomA, or HopQ were analyzed. The x -axis corresponds to the amino acid position for each protein and the y-axis indicates the number of assigned spectra. Regions of VacA corresponding to a signal sequence (SS), p33 domain, p55 domain, and C-terminal β-barrel domain are shown. The C-terminal portions of HomA and HopQ are predicted to have a β-barrel architecture, based on use of the program BOCTOPUS, a transmembrane β-barrel topology prediction tool (http://boctopus.bioinfo.se/)[63].
Effect of high-salt conditions on vacA transcription
To further investigate the mechanisms by which high salt conditions led to increased levels of VacA in the extracellular space, we performed real-time reverse transcription (RT)-PCR analysis. We compared vacA transcription in H. pylori cultured for one hour in medium containing 0.5% NaCl compared to medium containing 1.25% NaCl, as described in Methods. RT-qPCR analysis confirmed that vacA transcript levels were significantly upregulated in a high salt environment, compared to transcription of control genes ( gyrB and atpA) (p<0.05, 1-way ANOVA) (Figure 1C).
Differential expression of H. pylori genes in response to high salt
To investigate the effects of high salt concentrations on H. pylori gene transcription, we used RNA-seq analysis to compare the transcriptomes of bacteria cultured for 1 h in medium containing 0.5% or 1.25% NaCl. Based on the criteria described in the Methods, 173 genes were differentially expressed in response to varying salt concentration (127 downregulated and 46 upregulated in response to high salt conditions) (Supplemental Table 2). In many cases, we detected differential expression of multiple genes that are predicted to be transcribed within the same operons. Based on the predicted H. pylori operon structure reported by Sharma et al. [64], we identified 27 operons that were downregulated and 5 operons that were upregulated in response to high salt conditions (Figure 3). Sixteen of the 124 selectively released proteins shown in Supplemental Table 1 were salt-responsive, based on RNA-seq analyses (Table 3).
Figure 3.
Operons regulated in response to salt. Multiple salt-responsive genes identified using RNA-seq were mapped to the same operons. Fifteen genes upregulated in response to high salt conditions were mapped to 5 operons (green arrows), and 68 downregulated genes were mapped to 27 operons (red arrows). Black arrows indicate genes for which transcription was unchanged. Gene numbers and protein functions are listed.
Table 3.
Selectively released proteins encoded by genes that were salt responsive in RNA-seq experiments
| Gene/Protein | Description | Fold changea (1.25% vs 0.5%) |
|---|---|---|
| HP0003 | 3-deoxy-d-manno-octulosonic acid 8-phosphate synthetase (KdsA) | 0.61 |
| HP0089 | Pfs; Mtn; MqnB | 0.50 |
| HP0204 | hypothetical protein | 0.54 |
| HP0618 | adenylate kinase (Adk) | 0.46 |
| HP0630 | modulator of drug activity (Mda66) | 0.57 |
| HP0865 | deoxyuridine 5’-triphosphate nucleotidohydrolase (Dut) | 0.60 |
| HP0871 | CDP-diglyceride hydrolase (Cdh) | 2.77 |
| HP0887 | vacuolating cytotoxin (VacA) | 1.81 |
| HP1098 | hypothetical secreted protein (HcpC) | 0.35 |
| HP1099 | 2-keto-3-deoxy-6-phosphogluconate aldolase (Eda) | 0.58 |
| HP1117 | hypothetical secreted protein (HcpX) | 0.54 |
| HP1118 | gamma-glutamyltranspeptidase (Ggt) | 0.49 |
| HP1186 | carbonic anhydrase | 1.85 |
| HP1299 | methionine amino peptidase (Map) | 0.54 |
| HP1458 | thioredoxin-2 (Trx2) | 0.43 |
| HP1469 | outer membrane protein (Omp31, HorJ) | 0.59 |
H. pylori was grown in Brucella broth containing either 0.5% or 1.25% NaCl. RNA-seq was performed as described in the methods. The fold change in transcript levels in response to high salt conditions was then calculated as a ratio (number of RNA-seq reads from cultures containing 1.25% NaCl compared to reads from cultures containing 0.5% NaCl).
We hypothesized that many of the genes encoding salt-responsive proteins identified in the proteomic analysis might be differentially transcribed in response to variations in salt concentration. To test this hypothesis, we compared the list of salt-responsive proteins identified in the proteomic analysis (Table 2) with the list of salt-responsive genes identified in the RNA-seq analysis. Overall, there was relatively little concordance in the set of salt-responsive proteins or genes identified by these two approaches. Transcription of HP0089 was downregulated in response to high salt concentrations, and the corresponding protein exhibited decreased abundance in culture supernatants in response to high salt conditions. Upregulated transcription of vacA (HP0887) in response to high salt concentrations was consistent with elevated levels of these proteins detected in culture supernatant under high salt conditions. Upregulated transcription of vacA (HP0887) in response to high salt concentrations was also consistent with the results of RT-qPCR experiments (Figure 1).
DISCUSSION
Consumption of a high salt diet is an important risk factor for development of gastric adenocarcinoma [21, 22, 24, 65, 66]. In response to high salt conditions in vitro, H. pylori undergoes alterations in gene transcription and altered production of multiple proteins [25, 27, 30, 31]. These changes potentially influence gastric cancer risk. In this manuscript, we tested the hypothesis that growth of H. pylori in a high salt environment alters the composition of the exoproteome. We also used RNA-seq methods to analyze the effects of high salt concentrations on the H. pylori transcriptome and compared the transcriptomic results with proteomic results.
Previous studies identified H. pylori proteins that are selectively released into the culture supernatant under routine culture conditions [34–37], and the current experiments showed that the most of these proteins are also selectively released under high salt conditions. Many of these proteins are predicted to have signal sequences that facilitate translocation of the proteins across the inner membrane [37], but the mechanism underlying selective release across the outer membrane is unknown.
One of the key findings in the current study was the detection of higher levels of the VacA toxin in culture supernatants of cultures containing high salt concentrations, compared to supernatants from cultures grown in conventional culture medium. RNA-seq and RT-qPCR experiments showed that vacA transcription was increased in response to high salt concentrations. Therefore, we presume that the increased levels of VacA in culture supernatant in response to high salt conditions are attributable, at least in part, to increased vacA transcription. Notably, the magnitude of change in vacA transcription detected by RNA-seq or quantitative RT-qPCR experiments was substantially less than the magnitude of change in VacA levels detected in proteomic analyses or Western blotting experiments. A previous proteomic analysis showed that levels of VacA protein detected in the membrane fraction of H. pylori grown in high salt conditions were reduced, in comparison to VacA levels in corresponding fractions from bacteria grown in low-salt conditions [28]. Therefore, it seems likely that high salt conditions not only upregulate vacA transcription, but also may have other effects such as promoting VacA release from the outer membrane into the extracellular space.
Several previous studies analyzed effects of high salt concentrations on VacA transcription or production. One study detected increased vacA transcription after exposure of H. pylori to high salt conditions for 15 to 60 minutes, using an RNAse protection assay [67], and another study detected increased vacA transcription in response to high salt conditions for 1 h, using RT-qPCR methods [68]. In contrast, other studies did not detect alterations in vacA transcription in bacteria exposed to high salt conditions for longer time periods [30, 31]. The current study confirms that vacA transcription increases in response to high salt conditions for 1 h.
Consistent with results of previous studies [37], we detected multiple OMPs in the culture supernatant. We detected increased abundance of outer membrane proteins in the culture supernatants of cultures containing high salt concentrations, compared to supernatants of cultures grown in conventional culture medium. Hop outer membrane proteins have multiple features in common with autotransporter proteins [69], and therefore, we hypothesized that portions of Hop outer membrane proteins corresponding to passenger domains might be translocated across the outer membrane and released into the extracellular space. Instead, we detected all portions of the outer membrane proteins selected for analysis (including the predicted C-terminal beta-barrel domains) in the culture supernatant. Although the methodology for the current study included the use of ultracentrifugation to remove outer membrane vesicles from culture supernatant, we speculate that the outer membrane proteins detected in the culture supernatant might be components of small vesicles or membrane fragments that were not completely removed by ultracentrifugation. A recent study reported that H. pylori outer membrane vesicles exhibit varying properties at different stages of the growth cycle [70], which provides a possible explanation for the detection of outer membrane proteins in the culture supernatant mainly at the 24 hour time point compared to later time points.
RNA-seq studies showed that growth of H. pylori in high salt conditions leads to altered transcription of numerous H. pylori genes, including genes encoding proteins that are selectively released into the culture supernatant. In agreement with the results of a previous RNA-seq analysis [31], the magnitude of salt-induced changes in transcription observed in the current study were relatively small. Differences in the set of salt-responsive genes identified in the current study compared to a previous study [31] are presumably attributable to differences in the time points selected for analysis, as well as the use of a different H. pylori strain in the previous study. The mechanisms by which high salt conditions alter H. pylori gene transcription are not yet understood.
We hypothesized that differences in the relative abundance of proteins in culture supernatants of cultures grown in high salt conditions compared to routine conditions might result from effects of high salt conditions on H. pylori gene transcription. However, there was relatively little overlap in the list of salt-responsive genes identified by RNA-seq compared to the list of salt-responsive proteins identified in proteomic studies. The limited concordance may be partly attributable to differences in the time points at which the proteomic and transcriptomic analyses were performed. In addition, it seems likely that the effect of high salt conditions on composition of the exoproteome is not dependent solely on transcriptional changes, but also may reflect the capacity of high salt conditions to stimulate release of proteins from the outer membrane into the extracellular space.
In summary, these data indicate that H. pylori exposure to high environmental salt concentrations results in alterations of the exoproteome. When taken together with several previous papers showing effects of salt concentrations on H. pylori cellular proteins and H. pylori gene transcription [25, 28, 30, 31], the data indicate that many properties of the bacteria are altered in response to changes in environmental salt concentration. We propose that these salt-induced changes contribute to the increased risk of gastric cancer observed in H. pylori-infected individuals who consume a high salt diet.
Supplementary Material
Significance.
Helicobacter pylori-induced alterations in the gastric mucosa have been attributed, at least in part, to the actions of secreted H. pylori proteins. In this study, we show that H. pylori growth in high salt concentrations leads to increased levels of a secreted VacA toxin. Salt-induced alterations in the composition of the H. pylori exoproteome is relevant to the increased risk of gastric cancer associated with consumption of a high salt diet.
Highlights.
We define effects of salt concentration on the Helicobacter pylori exoproteome.
We define effects of salt concentration on the H. pylori transcriptome.
Levels of VacA toxin increase in response to high salt concentrations.
vacA transcription is upregulated in response high salt concentrations.
The findings help to explain how a high salt diet influences stomach cancer risk.
Acknowledgments:
The work described in this paper was supported by the National Institutes of Health (CA116087, AI039657, AI118932) and the Department of Veterans Affairs (1I01BX004447). Proteomics experiments were supported by the Vanderbilt Digestive Diseases Research Center (P30 DK058404), and the Vanderbilt Cancer Center (P30 CA068485).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Atherton JC, Blaser MJ, Coadaptation of Helicobacter pylori and humans: ancient history, modern implications, J Clin Invest 119(9) (2009) 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cover TL, Blaser MJ, Helicobacter pylori in health and disease, Gastroenterology 136(6) (2009) 1863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Suerbaum S, Michetti P, Helicobacter pylori infection, N Engl J Med 347(15) (2002) 1175–1186. [DOI] [PubMed] [Google Scholar]
- [4].Blaser MJ, Berg DE, Helicobacter pylori genetic diversity and risk of human disease, J Clin Invest 107(7) (2001) 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Suerbaum S, Josenhans C, Helicobacter pylori evolution and phenotypic diversification in a changing host, Nat Rev Microbiol 5(6) (2007) 441–452. [DOI] [PubMed] [Google Scholar]
- [6].Cover TL, Helicobacter pylori Diversity and Gastric Cancer Risk, MBio 7(1) (2016) e01869–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cover TL, Blaser MJ, Purification and characterization of the vacuolating toxin from Helicobacter pylori, J Biol Chem 267(15) (1992) 10570–5. [PubMed] [Google Scholar]
- [8].Telford JL, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z, et al. , Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease, J Exp Med 179(5) (1994) 1653–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cover TL, Blanke SR, Helicobacter pylori VacA, a paradigm for toxin multifunctionality, Nat Rev Microbiol 3(4) (2005) 320–32. [DOI] [PubMed] [Google Scholar]
- [10].Atherton JC, Cao P, Peek RM Jr., Tummuru MK, Blaser MJ, Cover TL, Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration, J Biol Chem 270(30) (1995) 17771–7. [DOI] [PubMed] [Google Scholar]
- [11].Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B, A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island, PLoS Genet 6(8) (2010) e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hatakeyama M, Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis, Cell Host Microbe 15(3) (2014) 306–316. [DOI] [PubMed] [Google Scholar]
- [13].Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R, Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion, Science 287(5457) (2000) 1497–1500. [DOI] [PubMed] [Google Scholar]
- [14].Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL, Molecular and Structural Analysis of the Helicobacter pylori cag Type IV Secretion System Core Complex, MBio 7(1) (2016) e02001–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tegtmeyer N, Neddermann M, Asche CI, Backert S, Subversion of host kinases: a key network in cellular signaling hijacked by Helicobacter pylori CagA, Mol Microbiol 105(3) (2017) 358–372. [DOI] [PubMed] [Google Scholar]
- [16].Backert S, Tegtmeyer N, Fischer W, Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system, Future Microbiol 10(6) (2015) 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cao P, Cover TL, Two different families of hopQ alleles in Helicobacter pylori, J Clin Microbiol 40(12) (2002) 4504–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadstrom T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarstrom L, Boren T, Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation, Science 297(5581) (2002) 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T, Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging, Science 279(5349) (1998) 373–377. [DOI] [PubMed] [Google Scholar]
- [20].Berthenet E, Yahara K, Thorell K, Pascoe B, Meric G, Mikhail JM, Engstrand L, Enroth H, Burette A, Megraud F, Varon C, Atherton JC, Smith S, Wilkinson TS, Hitchings MD, Falush D, Sheppard SK, A GWAS on Helicobacter pylori strains points to genetic variants associated with gastric cancer risk, BMC Biol 16(1) (2018) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cover TL, Peek RM Jr., Diet, microbial virulence, and Helicobacter pylori-induced gastric cancer, Gut Microbes 4(6) (2013) 482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tsugane S, Salt, salted food intake, and risk of gastric cancer: epidemiologic evidence, Cancer Sci 96(1) (2005) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM Jr., Algood HM, Cover TL, High Dietary Salt Intake Exacerbates Helicobacter pylori-Induced Gastric Carcinogenesis, Infect Immun 81(6) (2013) 2258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fox JG, Dangler CA, Taylor NS, King A, Koh TJ, Wang TC, High-salt diet induces gastric epithelial hyperplasia and parietal cell loss, and enhances Helicobacter pylori colonization in C57BL/6 mice, Cancer Res 59(19) (1999) 4823–4828. [PubMed] [Google Scholar]
- [25].Gancz H, Jones KR, Merrell DS, Sodium chloride affects Helicobacter pylori growth and gene expression, J Bacteriol 190(11) (2008) 4100–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Loh JT, Torres VJ, Cover TL, Regulation of Helicobacter pylori cagA expression in response to salt, Cancer Res 67(10) (2007) 4709–4715. [DOI] [PubMed] [Google Scholar]
- [27].Loh JT, Friedman DB, Piazuelo MB, Bravo LE, Wilson KT, Peek RM Jr., Correa P, Cover TL, Analysis of Helicobacter pylori cagA promoter elements required for salt-induced upregulation of CagA expression, Infect Immun 80(9) (2012) 3094–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Voss BJ, Loh JT, Hill S, Rose KL, McDonald WH, Cover TL, Alteration of the Helicobacter pylori membrane proteome in response to changes in environmental salt concentration, Proteomics Clin Appl 9(11–12) (2015) 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Loh JT, Lin AS, Beckett AC, McClain MS, Cover TL, Role of a stem-loop structure in Helicobacter pylori cagA transcript stability, Infect Immun (2018). [DOI] [PMC free article] [PubMed]
- [30].Loh JT, Torres VJ, Cover TL, Regulation of Helicobacter pylori cagA expression in response to salt, Cancer Res 67(10) (2007) 4709–15. [DOI] [PubMed] [Google Scholar]
- [31].Loh JT, Beckett AC, Scholz MB, Cover TL, High-Salt Conditions Alter Transcription of Helicobacter pylori Genes Encoding Outer Membrane Proteins, Infect Immun 86(3) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fischer W, Buhrdorf R, Gerland E, Haas R, Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori, Infect Immun 69(11) (2001) 6769–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cover TL, Blanke SR, Helicobacter pylori VacA, a paradigm for toxin multifunctionality, Nat Rev Microbiol 3(4) (2005) 320–332. [DOI] [PubMed] [Google Scholar]
- [34].Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, Meyer TF, Jungblut PR, Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori, Infect Immun 70(7) (2002) 3396–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim N, Weeks DL, Shin JM, Scott DR, Young MK, Sachs G, Proteins released by Helicobacter pylori in vitro, J Bacteriol 184(22) (2002) 6155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Smith TG, Lim JM, Weinberg MV, Wells L, Hoover TR, Direct analysis of the extracellular proteome from two strains of Helicobacter pylori, Proteomics 7(13) (2007) 2240–2245. [DOI] [PubMed] [Google Scholar]
- [37].Snider CA, Voss BJ, McDonald WH, Cover TL, Growth phase-dependent composition of the Helicobacter pylori exoproteome, J Proteomics 130 (2016) 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hawrylik SJ, Wasilko DJ, Haskell SL, Gootz TD, Lee SE, Bisulfite or sulfite inhibits growth of Helicobacter pylori, J Clin Microbiol 32(3) (1994) 790–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Voss BJ, Gaddy JA, McDonald WH, Cover TL, Analysis of Surface-Exposed Outer Membrane Proteins in Helicobacter pylori, J Bacteriol 196(13) (2014) 2455–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schraw W, Li Y, McClain MS, van der Goot FG, Cover TL, Association of Helicobacter pylori vacuolating toxin (VacA) with lipid rafts, J Biol Chem 277(37) (2002) 34642–34650. [DOI] [PubMed] [Google Scholar]
- [41].Lo CC, Chain PS, Rapid evaluation and quality control of next generation sequencing data with FaQCs, BMC Bioinformatics 15 (2014) 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nat Biotechnol 34(5) (2016) 525–527. [DOI] [PubMed] [Google Scholar]
- [43].Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou LX, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weldman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Weidman JM, Fujii C, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC, The complete genome sequence of the gastric pathogen Helicobacter pylori, Nature 388(6642) (1997) 539–547. [DOI] [PubMed] [Google Scholar]
- [44].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26(1) (2010) 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, Dunn BE, Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis, Infect Immun 64(3) (1996) 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cao P, McClain MS, Forsyth MH, Cover TL, Extracellular release of antigenic proteins by Helicobacter pylori, Infect Immun 66(6) (1998) 2984–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ogura M, Perez JC, Mittl PR, Lee HK, Dailide G, Tan S, Ito Y, Secka O, Dailidiene D, Putty K, Berg DE, Kalia A, Helicobacter pylori evolution: lineage-specific adaptations in homologs of eukaryotic Sel1-like genes, PLoS Comput Biol 3(8) (2007) 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Basak C, Pathak SK, Bhattacharyya A, Pathak S, Basu J, Kundu M, The secreted peptidyl prolyl cis,trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4- and apoptosis signal-regulating kinase 1-dependent manner, J Immunol 174(9) (2005) 5672–5680. [DOI] [PubMed] [Google Scholar]
- [49].Halder P, Datta C, Kumar R, Sharma AK, Basu J, Kundu M, The secreted antigen, HP0175, of Helicobacter pylori links the unfolded protein response (UPR) to autophagy in gastric epithelial cells, Cell Microbiol 17(5) (2015) 714–729. [DOI] [PubMed] [Google Scholar]
- [50].Amedei A, Munari F, Bella CD, Niccolai E, Benagiano M, Bencini L, Cianchi F, Farsi M, Emmi G, Zanotti G, de Bernard M, Kundu M, D’Elios MM, Helicobacter pylori secreted peptidyl prolyl cis, trans-isomerase drives Th17 inflammation in gastric adenocarcinoma, Intern Emerg Med 9(3) (2014) 303–309. [DOI] [PubMed] [Google Scholar]
- [51].Deml L, Aigner M, Decker J, Eckhardt A, Schutz C, Mittl PR, Barabas S, Denk S, Knoll G, Lehn N, Schneider-Brachert W, Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent, Infect Immun 73(8) (2005) 4732–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dumrese C, Slomianka L, Ziegler U, Choi SS, Kalia A, Fulurija A, Lu W, Berg DE, Benghezal M, Marshall B, Mittl PR, The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype, FEBS Lett 583(10) (2009) 1637–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schroder P, Sewald N, Backert S, Schneider G, Wessler S, Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion, EMBO Rep 11(10) (2010) 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, Asche CI, Boehm M, Loessner H, Figueiredo C, Naumann M, Palmisano R, Solcia E, Ricci V, Backert S, Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery, Cell Host Microbe 22(4) (2017) 552–560 [DOI] [PubMed] [Google Scholar]
- [55].Shibayama K, Kamachi K, Nagata N, Yagi T, Nada T, Doi Y, Shibata N, Yokoyama K, Yamane K, Kato H, Iinuma Y, Arakawa Y, A novel apoptosis-inducing protein from Helicobacter pylori, Mol Microbiol 47(2) (2003) 443–51. [DOI] [PubMed] [Google Scholar]
- [56].Schmees C, Prinz C, Treptau T, Rad R, Hengst L, Voland P, Bauer S, Brenner L, Schmid RM, Gerhard M, Inhibition of T-cell proliferation by Helicobacter pylori gamma-glutamyl transpeptidase, Gastroenterology 132(5) (2007) 1820–1833. [DOI] [PubMed] [Google Scholar]
- [57].Oertli M, Noben M, Engler DB, Semper RP, Reuter S, Maxeiner J, Gerhard M, Taube C, Muller A, Helicobacter pylori gamma-glutamyl transpeptidase and vacuolating cytotoxin promote gastric persistence and immune tolerance, Proc Natl Acad Sci U S A 110(8) (2013) 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tavares R, Pathak SK, Induction of TNF, CXCL8 and IL-1beta in macrophages by Helicobacter pylori secreted protein HP1173 occurs via MAP-kinases, NF-kappaB and AP-1 signaling pathways, Microb Pathog 125 (2018) 295–305. [DOI] [PubMed] [Google Scholar]
- [59].Li J, Meng FL, He LH, Zhang JZ, Secreted protein HP1286 of Helicobacter pylori strain 26695 induces apoptosis of AGS cells, Biomed Environ Sci 25(6) (2012) 614–619. [DOI] [PubMed] [Google Scholar]
- [60].Tavares R, Pathak SK, Helicobacter pylori Secreted Protein HP1286 Triggers Apoptosis in Macrophages via TNF-Independent and ERK MAPK-Dependent Pathways, Front Cell Infect Microbiol 7 (2017) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Capitani N, Codolo G, Vallese F, Minervini G, Grassi A, Cianchi F, Troilo A, Fischer W, Zanotti G, Baldari CT, de Bernard M, D’Elios MM, The lipoprotein HP1454 of Helicobacter pylori regulates T-cell response by shaping T-cell receptor signalling, Cell Microbiol 21(5) (2019) e13006. [DOI] [PubMed] [Google Scholar]
- [62].Nguyen VQ, Caprioli RM, Cover TL, Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin, Infect Immun 69(1) (2001) 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hayat S, Elofsson A, BOCTOPUS: improved topology prediction of transmembrane beta barrel proteins, Bioinformatics 28(4) (2012) 516–522. [DOI] [PubMed] [Google Scholar]
- [64].Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J, The primary transcriptome of the major human pathogen Helicobacter pylori, Nature 464(7286) (2010) 250–5. [DOI] [PubMed] [Google Scholar]
- [65].Fox JG, Rogers AB, Ihrig M, Taylor NS, Whary MT, Dockray G, Varro A, Wang TC, Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific, Cancer Res 63(5) (2003) 942–950. [PubMed] [Google Scholar]
- [66].Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM Jr., Algood HM, Cover TL, High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis, Infect Immun 81(6) (2013) 2258–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gancz H, Jones KR, Merrell DS, Sodium chloride affects Helicobacter pylori growth and gene expression, J Bacteriol 190(11) (2008) 4100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Amilon KR, Letley DP, Winter JA, Robinson K, Atherton JC, Expression of the Helicobacter pylori virulence factor vacuolating cytotoxin A (vacA) is influenced by a potential stem-loop structure in the 5’ untranslated region of the transcript, Mol Microbiol 98(5) (2015) 831–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Coppens F, Castaldo G, Debraekeleer A, Subedi S, Moonens K, Lo A, Remaut H, Hop-family Helicobacter outer membrane adhesins form a novel class of Type 5-like secretion proteins with an interrupted beta-barrel domain, Mol Microbiol 110(1) (2018) 33–46. [DOI] [PubMed] [Google Scholar]
- [70].Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M, Helicobacter pylori Growth Stage Determines the Size, Protein Composition, and Preferential Cargo Packaging of Outer Membrane Vesicles, Proteomics (2018) e1800209. [DOI] [PubMed]
- [71].Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H, SignalP 5.0 improves signal peptide predictions using deep neural networks, Nat Biotechnol 37(4) (2019) 420–423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



