Abstract
Fluorescence in situ hybridization (FISH) reveals the abundance and positioning of nucleic acid sequences in fixed samples. Despite recent advances in multiplexed amplification of FISH signals, it remains challenging to achieve high levels of simultaneous amplification and sequential detection with high sampling efficiency and simple workflows. Here, we introduce signal amplification by exchange reaction (SABER), which endows oligo-based FISH probes with long, single-stranded DNA concatemers that aggregate a multitude of short complementary fluorescent imager strands. We show that SABER amplifies RNA and DNA FISH signals (5 to 450-fold) in fixed cells and tissues, apply 17 orthogonal amplifiers against chromosomal targets simultaneously, and detect mRNAs with high efficiency. We further apply 10-plexSABER-FISH to identify in vivo introduced enhancers with cell type-specific activity in the mouse retina. SABER represents a simple and versatile molecular toolkit for rapid and cost-effective multiplexed imaging of nucleic acid targets.
INTRODUCTION
Fluorescence in situ hybridization (FISH) allows researchers to interrogate the subcellular distribution of RNA and DNA molecules in fixed cells and tissues through application of complementary probes.1 FISH assays are used for diverse applications such as diagnosing chromosomal abnormalities,2 interrogating three-dimensional genome organization,3 and analyzing gene expression.4,5 FISH is compatible with simultaneous detection of multiple nucleic acid targets and, when combined with sequential imaging methods, the number of detectable targets can be greater than the number of spectrally resolvable fluorophores.3,6,7 Recent approaches that utilize serial rounds of imaging, label removal, and re-labeling of distinct targets, enable researchers to image potentially unlimited numbers of targets. For example, techniques such as DNA-Exchange,8–10 which uses cyclic rounds of hybridization and displacement of fluor-labeled oligos (‘imagers’) bound to probes, can be used to visualize a large number of targets (e.g., up to 84 distinct chromosomal regions in cultured cells11,12 and 33 RNA transcripts in tissues13). For higher levels of multiplexing, multi-round combinatorial labeling allows an exponential number of low abundance targets to be visualized in a linear number of serial imaging rounds, provided the targets are optically resolvable.14–17
Beyond multiplexing, several approaches have been developed to amplify the intensity of quantitative FISH signals. Amplification is particularly relevant in the context of thick tissues, where high levels of autofluorescence, light scattering, and optical aberration can make signal detection challenging. In addition, amplification of signal can shorten imaging times (increased throughput), further reduce requirements on expensive microscopy setups, and potentially reduce cost by lowering the number of probes required. Previous amplification strategies include the targeted deposition of detectable reactive molecules around the site of probe hybridization,18 the targeted assembly of ‘branched’ structures composed of DNA19,20 or locked nucleic acid (LNA) molecules,21 the programmed in situ growth of concatemers by enzymatic rolling circle amplification (RCA)22 or hybridization chain reaction (HCR),23–26 and the assembly of topologically catenated DNA structures using serial rounds of chemical ligation (clampFISH).27
Amplification methods that utilize simultaneous orthogonal amplification, such as HCR and RCA, enable efficient multiplexed visualization of targets in tissue. HCR employs triggered self-assembly of pairs of self-folding hairpin oligos into long concatemeric chains to achieve simultaneous enzyme-free amplification in situ. However, to date only 5 orthogonal HCR concatemers have been demonstrated, potentially due to the complexity of designing multiple complex non-interacting kinetic pathways of triggered autonomous hairpin assembly to operate simultaneously. RCA-based approaches have been used to simultaneously amplify 8 target mRNAs in mouse lung tissue,28 to detect up to 28 mRNA targets simultaneously in cleared 150 μm mouse tissue s, and up to 1,020 spatially separated targets in thin (single cell layer) s.29 While RCA-based methods enable highly multiplexed simultaneous amplification, detection efficiencies for RNA transcripts remain comparable to single-cell RNA sequencing (6–40%) in the best case (STARmap) and are also low for other RCA-based methods such as FISSEQ (0.01–0.2%), and padlock probe-based designs (5–32%),28,29 perhaps due to the complexity of controlling parallel enzymatic reactions in situ.
Here, we sought to develop an amplification method with: (1) programmable high levels of amplification, (2) high orthogonality to allow simultaneous amplification, and (3) high targeting efficiency, including in thick tissues. In addition to these performance characteristics, we further desired the method to be accessible, utilizing: (4) a simple and robust workflow compatible with commonly available imaging platforms and (5) cost-effective and readily available reagents.
We developed an amplification method to meet these performance and accessibility criteria by using a programmable ssDNA synthesis method that we recently developed, the primer exchange reaction (PER)30. We previously demonstrated the growth of long ssDNA concatemers composed of a ‘three-letter code’ of A, T, and C nucleotides from a short (9 nt) DNA primer sequence with PER.30 We found the kinetics of the synthesis reaction are controllable via a number of parameters, providing a simple means of generating concatemers of desired length through in vitro synthesis. Here, we find that these concatemers permit fluorescent signal amplification, as their polymeric structure provides a hybridization scaffold for localizing many fluorescent imager oligos, reminiscent of the sequences found in branched signal amplification approaches.19–21 PER can also be used to synthesize a large number of orthogonal concatemer sequences, and we are able to readily implement multiplexed imaging strategies with cyclic serial readout of the concatemers through hybridization and displacement of imagers (DNA-Exchange).8,9,10 We further establish that these concatemers, designed to have little secondary structure, effectively penetrate thick tissue.
The molecular toolkit we introduce, termed signal amplification by exchange reaction (SABER), harnesses the programmability, orthogonality, and simplicity features of PER to enhance the functionality of oligo-based FISH probes, such as single-molecule RNA FISH probe pools5 and highly complex ‘Oligopaint’ probe sets.31 Briefly, DNA and RNA FISH probes are first chemically synthesized with primer sequences on their 3’ ends, which are extended into PER concatemers in vitro. The ~1–3 hour PER reaction uses a set of widely available and inexpensive reagents similar to PCR. Extended probe sequences are hybridized to targets in situ, and then detected by secondary hybridization of fluorescent imagers, with the options of including intermediate branching concatemers for additional signal amplification or applying serial imaging with DNA-Exchange8,9,10 (Exchange-SABER). In comparison to methods that generate concatemers in situ, this approach allows bulk probe production, quality control, and user-defined adjustment of probe concentration.
We experimentally demonstrate that in different scenarios, SABER can programmably amplify a signal up to 450-fold, can be deployed against 17 different targets simultaneously, and can provide high sampling efficiency of target transcripts for puncta detection and cell type identification in tissue. In a 10-plex FISH assay, we apply SABER to interrogate the activity and specificity of candidate enhancer elements introduced in vivo via the detection of reporter RNAs and retinal cell type markers, and show that reporter RNAs and the plasmids from which they are expressed can be co-detected in a combined RNA/DNA FISH experiment. The method builds directly off standard in situ hybridization protocols, and we efficiently image signal using common widefield (cell) and confocal (tissue) imaging setups. Thus, SABER is an amplification method that enables individually programmable signal amplification for a collection of orthogonal concatemers. SABER concatemers can be deployed in situ simultaneously and with high targeting efficiency even in thick tissues. Furthermore, the simple and robust workflow, compatibility with common microscopes, and cost-effective reagents make the method readily adoptable.
RESULTS
Design of orthogonal sequences for SABER
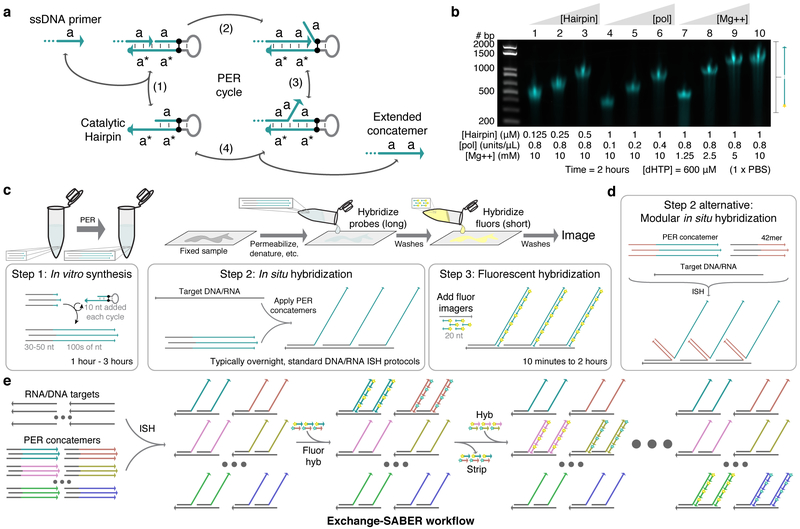
We recently developed the primer exchange reaction (PER) method for autonomously synthesizing arbitrary single-stranded DNA sequences from short DNA primers.30 One embodiment of the reaction uses a catalytic hairpin paired with a strand displacing polymerase and competitive branch migration32 to repeatedly add the same sequence domain onto single-stranded primers (Fig. 1a and Supplementary Fig. 1a). Representative reactant and concatemer sequences are depicted in Supplementary Fig. 1b and Supplementary Fig. 1c, respectively. We found that the length of PER concatemers could be tuned by varying the concentration of polymerase, hairpin, magnesium, or nucleotides, as well as extension time (Fig. 1b and Supplementary Fig. 1d). These concatemers lack G bases to minimize secondary structure and permit G-C pairs to be used as a polymerase terminator sequence within the hairpin in the absence of dGTP in the reaction. We reasoned that the PER-based concatemerization could provide a flexible means to endow FISH probes with repetitive extensions for depositing fluorescent signal. The workflow for this method, which we call signal amplification by exchange reaction (SABER) is depicted in Fig. 1c. PER concatemers are extended onto chemically synthesized probes bearing a 9-mer primer. Following hybridization of extended probes, concatemers are detected by secondary hybridization with imager oligos (Supplementary Fig. 1e). A modular variant of SABER employs 42mer ‘bridge’ sequences to bind target-hybridizing probes to complementary oligos onto which concatemers have been extended, and is deployed in a single hybridization incubation (Fig. 1d and Supplementary Fig. 1f). With either detection scheme, imager oligos can be stripped from their cognate concatemers to reset the signal,8–10 enabling subsequent use of that fluorescence color on a distinct target. With this Exchange-SABER approach, a multitude of PER-concatemerized probe sets can be hybridized to their targets simultaneously and read out in sequential rounds of imaging (Fig. 1e).
Figure 1: SABER-FISH design and workflow.
a, Primer exchange reaction (PER) cycle.30 A primer with domain a on its 3’ end binds (reversibly) to a catalytic hairpin (step 1) and gets extended with a new a domain by a strand displacing polymerase (step 2). Competitive branch migration (step 3)32 displaces the extended primer, which can then dissociate (step 4). Primers undergo the cycle repeatedly to form long repetitive concatemeric sequences. b, Length programmability of PER concatemers. Hairpin concentration, polymerase concentration, magnesium concentration, and incubation time can be adjusted to control PER kinetics and therefore concatemer length. Time and dHTP (dATP, dTTP, and dCTP) concentration were fixed at 2 hours and 600 μM, respectively. See also Supplementary Fig. 1d. c, SABER workflow. First, large batches of probe sequences with primers on their 3’ ends can be concatemerized using PER. These extensions are then hybridized to DNA and RNA targets in fixed samples following standard in situ hybridization (ISH) protocols. This is followed by a short step that hybridizes fluor- conjugated oligos (imagers) to complementary concatemers. d, An alternative method uses 42mer ‘bridge’ sequences to couple probes to concatemer sequences. This method allows the same 42mer barcodes with PER concatemers to be re-used for different targets. e, Exchange-SABER workflow. Optionally, fluorescent signal can be rapidly stripped from concatemers without disrupting probe binding, allowing exchange of imagers8,9,10 and reuse of the same fluorescence channels through multiple rounds of imaging.
Sequence orthogonality in all aspects of the design was considered to ensure robust and specific targeting of fluorescent signal with multiplexed SABER. We used the OligoMiner pipeline33 to computationally design orthogonal ‘oligopaint’ probe sequences with homology to targets of interest. The pipeline vets sequences for orthogonality against the relevant target genome, and single-strandedness and melting temperature constraints further filter sequences. FISH probes are hybridized under conditions close to their melting temperature to increase specificity of binding (Supplementary Fig. 1g). A similar design process, but with sequences drawn from blocks of orthogonal sequences34, was used to generate 84 orthogonal 42mer bridge sequences. To successfully deploy a large number of orthogonal concatemers simultaneously using SABER, we also needed to design many orthogonal PER concatemer sequences. We used NUPACK35–37 to model on- and off-target interactions for sets of 50 probes, fifty 42mer bridges, and 50 computationally designed PER sequences in their respective incubation conditions (2×SSCT with 50% formamide at 42°C for ISH and 1×PBS at 37°C for fluorescent hybridization, see Supplementary Fig. 1h). Dimerization probabilities for these sets of sequences were also modeled (Supplementary Fig. 1i).
SABER effectively amplifies fluorescent signals
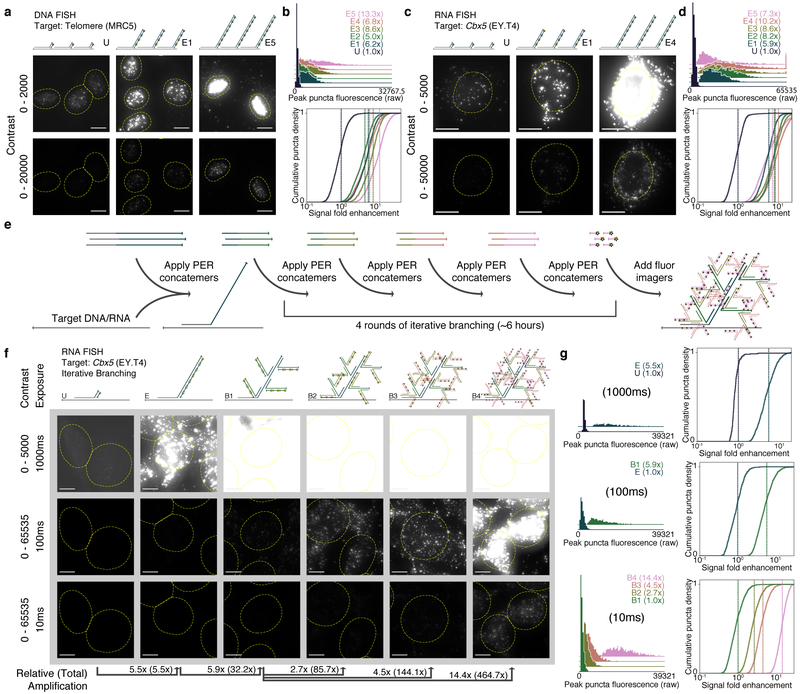
We applied SABER to DNA and RNA targets with known distribution in cell culture samples. First, a DNA oligo with homology to the human telomere sequence was extended to 5 different lengths (conditions E1–E5) using varied concentrations of hairpin (Supplementary Fig. 2a). The fluorescence resulting from hybridization with probes of each length, and a probe having an unextended sequence with a single binding site for imagers (condition U), was visualized using fluorescence microscopy (Fig. 2a and Supplementary Fig. 2b). A custom CellProfiler38 pipeline was used to identify and quantify puncta within cell nuclei (Supplementary Fig. 2c). Distributions of peak puncta fluorescence values for all conditions were measured (Fig. 2b, top), and fluorescence fold enhancement was estimated by subtracting background and dividing by the mean of the unextended condition (Fig. 2b, bottom). We estimated 6.2×, 5.0×, 8.6×, 6.8×, and 13.3× fold enhancement for conditions E1 through E5 relative to the unextended (i.e. unamplified) probe. See Supplementary Fig. 2d for additional analyses.
Figure 2: SABER effectively amplifies fluorescent signals.
a, Microscopy images for unextended (U) and two different lengths of PER-concatemerized probes (E1, E5) targeting the human telomere sequence. See Supplementary Fig. 2b for images of E2, E3, and E4. b, Distributions of peak fluorescence intensity values for CellProfiler38- detected puncta (top, fold enhancements in legend). Normalized background-subtracted cumulative distribution functions showing fold enhancement over the unextended condition (U), with vertical lines depicting means (fold enhancement, bottom). See Supplementary Fig. 2d for additional analyses. n(puncta): n(U)=1,895; n(E1)=1,846; n(E2)=1,876; n(E3)=2,011; n(E4)=2,190; n(E5)=2,006. c, Images for unextended (U) and two extension lengths (E1, E4) of a 122 probe pool targeting the mouse Cbx5 mRNA transcript. See Supplementary Fig. 2f for additional images. d, Distributions analogous to part (B) from CellProfiler38-identified puncta within cell bodies. Mean lines shown on distributions. See Supplementary Fig. 2g for additional analyses. n(puncta): n(U)=1,720; n(E1)=1,588; n(E2)=1,649; n(E3)=2,099; n(E4)=2,884; n(E5)=3,279. e, Schematic showing how multiple rounds of PER concatemer binding (branching) can be used to further increase signal. f, Representative images for samples with unextended probes (U), extended (E), and up to four rounds of branching (B1 through B4) are shown. g, Relative fluorescence was compared as previously but with no background subtraction to estimate amplification fold enhancement. In total, amplification fold enhancement over unextended probes was estimated to be 465×. Mean lines shown on distributions. See Supplementary Fig. 3e for additional analyses. n(puncta, exposure time): n(U, 1000ms)=262; n(E, 1000ms)=928; n(E, 100ms)=717; n(B1, 100ms)=1,821; n(B1, 10ms)=1,818; n(B2, 10ms)=1,796; n(B3, 10ms)=2,059; n(B4, 10ms)=1,344. Scale bars: 10 μM.
Next, the process was repeated for a set of 122 probes designed to target the mouse Cbx5 mRNA transcript. Here, for technical utility, we employed a large probe set to ensure unamplified signal could be robustly visualized and quantified. The probes were pooled together, extended to five lengths (Supplementary Fig. 2e), and visualized (Fig. 2c and Supplementary Fig. 2f). Puncta within cell bodies were segmented for analysis (Fig. 2d and Supplementary Fig. 2g). The first four extension lengths showed increasing levels of amplification (5.9×, 8.2×, 8.6×, and 10.2× fold enhancement for conditions E1 through E4), but the longest extension (condition E5) showed a dropoff (7.3×), indicating the importance of extension length programmability available through modulation of the parameters described above. In general, our results indicate that extension lengths between about ~250 and 750 nt provide robust, though not substantially different, levels of amplification.
Multiple rounds of PER concatemer hybridization can further increase fluorescent signal level by creating branched concatemeric structures19–21 (Supplementary Fig. 3a). A secondary round of hybridization binds PER concatemers with 30 nt homology to the primary probe concatemer. With a similar pipeline, branching amplification was visualized and quantified for several branch concatemer lengths targeting Cbx5 mRNA transcripts (Supplementary Fig. 3b). We observed amplification levels of up to 35.5× fold enhancement (Supplementary Fig. 3c and additional analyses in Supplementary Fig. 3d). Multiple rounds of branching can result in even higher levels of amplification. We implemented one to four rounds of branching (conditions B1 to B4, Fig. 2e) on top of probe concatemers targeting the Cbx5 mRNA transcript (Fig. 2f). After feature segmentation, max pixel values within identified puncta were quantified only under exposure times where puncta were reliably identified (Fig. 2g and Supplementary Fig. 3e). In total, signal fold enhancement was estimated to be 32.2×, 85.7×, 144.1×, and 464.7× for one, two, three, and four levels of branching, respectively.
SABER enables robust transcript detection in tissue
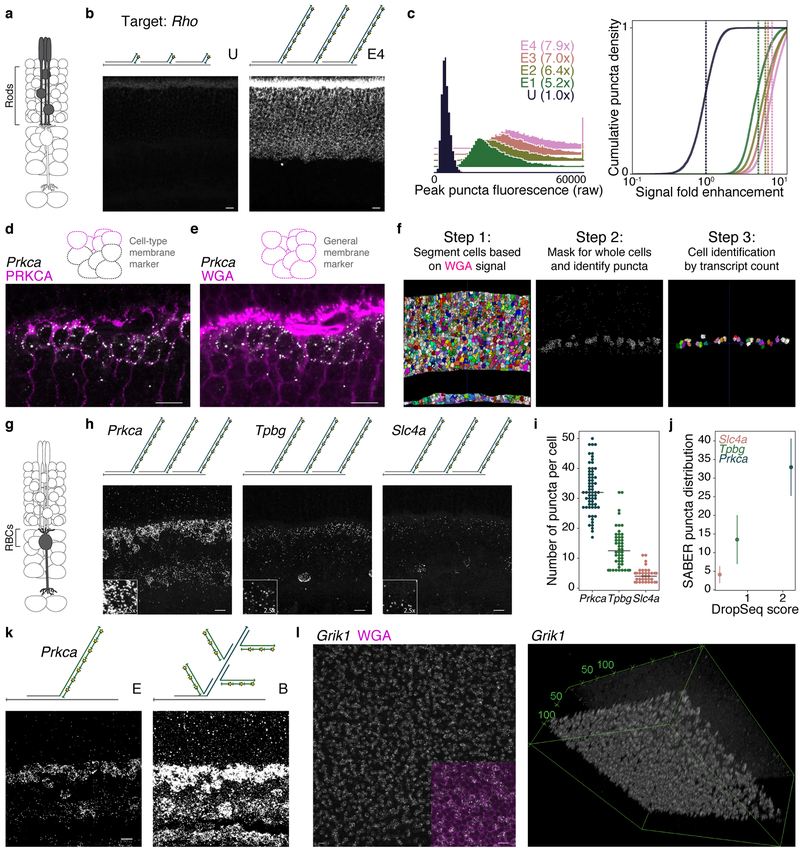
We next asked whether SABER could be used to amplify RNA FISH signal in tissue sections. The mouse retina has been extensively characterized by single cell transcriptomics (scRNA-seq)39,40, providing a useful point of comparison to assess the target specificity and quantifiability of FISH using SABER probes. We first compared unextended probes to PER-extended probes by targeting Rhodopsin (Rho), expressed exclusively in rod photoreceptors (Fig. 3a). Here, an exceptionally abundant mRNA was selected to permit visualization of unamplified signal in tissue sections. Fluorescent signal was localized to the photoreceptor layer (Fig. 3b), and we observed 5.2×, 6.4×, 7.0×, and 7.9× fold enhancement for increasing extension lengths (conditions E1 through E4) versus unextended probes (condition U) (Fig. 3c and SNR analysis in Supplementary Fig. 4a). FA-fixed cryosections cut to 35–40 μm thickness were used for these and subsequent experiments.
Figure 3: Transcript detection and quantification in retina tissue.
a, Schematic representation of retinal cell type (rods) targeted. b, SABER-FISH detection of Rho transcript with an unextended probe set (condition U) versus PER-extended probes (extended, E4). c, Quantification of SABER-FISH signal intensity for Rho transcript detection with unextended probe (U) and probes of varying concatemer lengths (E1–E4). Mean lines shown on distributions. n(puncta): n(U)=11,159; n(E1)=16,426; n(E2)=19,848; n(E3)=16,217; n(E4)=18,051 d, Combined Prkca transcript detection and PRKCA protein detection by IF demonstrating speci- ficity of transcript detection and localization of transcripts relative to cell boundaries. e, Fluorophore-conjugated WGA outlining of cell bodies in the retina. f, Computational pipeline for assignment of puncta to discrete cells in 3D tissue sections using cell segmentation41 and custom MATLAB pipeline (PD3D) for puncta detection and assignment. g, Schematic representation of retinal cell type (rod bipolar cells, RBCs) targeted. h, SABER-FISH detection of transcripts for three genes with differential expression levels and highly enriched expression in RBCs. i, Swarm plot of SABER puncta per RBC with a median line overlay. j, For the three markers, transcript counts per RBC as detected by SABER (y-axis) are plotted against average number of transcripts detected per RBC in a Drop-seq dataset (‘Drop-seq score’), showing similar relative transcript abundance between methods. 40 Means with standard deviations of SABER puncta per cell are shown. n(cells): n(Slc4a)=45; n(Tpbg)=48; n(Prkca)=63. k, Detection of Prkca transcript with and without branch amplification. l, Whole mount retina detection of Grik1 transcript. Maximum intensity projection of en face view in the inner nuclear layer, inset shows WGA counterstain (magenta, left). 3D volume rendering of a Z-stack (right). Scale bars: 10 μm.
We next tested the performance of SABER in the detection of lower abundance transcripts, choosing rod bipolar cells (RBCs), a single type of bipolar interneuron that has been extensively profiled by scRNA-seq39,40 as a test population. Specificity of FISH was confirmed by co-detection of the Prkca transcript and PRKCA protein, an established RBC marker (Fig. 3d and Supplementary Fig. 4b). Since the ability to quantify detected transcripts per cell is important to assess the performance of SABER, we sought a generalizable and unbiased method for defining cell boundaries. We found that fluorophore-conjugated wheat germ agglutinin (WGA) effectively outlined retinal cells (Fig. 3e), enabling 3D cell segmentation using ACME,41 an open-source software for membrane-based watershed segmentation (Fig. 3f). A Laplacian of Gaussian method was used to localize fluorescent SABER puncta in 3D (Supplementary Fig. 4c) with robust thresholding (Supplementary Fig. 4d). Cells could then be assigned cellular identities based on both marker gene transcript counts and laminar position in the tissue (Supplementary Fig. 4e). While WGA-based segmentation is limited by the inability to resolve neuronal processes, this limitation also applies to dissociated single retinal cells, and it is therefore a relevant method for use in comparisons to scRNA-seq.
We selected three transcripts for quantification that are highly enriched among RBCs40 (Fig. 3g) and that are expressed at low (Slc4a), moderate (Tpbg), or high (Prkca) levels (Supplementary Fig. 4b). After imaging (Fig. 3h) and transcript quantification (Fig. 3i), we found the relative transcript abundance for these genes in RBCs as detected by SABER-FISH closely paralleled the relative abundance observed by Drop-seq (Fig. 3j). Sampling of transcripts by SABER was approximately 15× fold higher than observed for Drop-seq-profiled cells, where cells had been sequenced to an average depth of 8,200 reads per cell for comprehensive classification of bipolar cell types (50× fold deeper sequencing of Drop-seq libraries improves transcript detection probability by up to ~2× fold40). We also observed an 8.8× fold increase in signal intensity using a single round of branching compared to simple extension of the Prkca probe set (Fig. 3k and Supplementary Fig. 4f). Transcript counts for branched probe Prkca detection were similar to the unbranched condition (Supplementary Fig. 4g) and still closely paralleled Drop-seq values (Supplementary Fig. 4h). Branching also permitted robust detection of transcripts with a 12 oligo probe set (Supplementary Fig. 4i). A concern in the application of pre-extended probes to tissue was the ability of long DNA strands to penetrate. We tested SABER-FISH in FA-fixed flat-mounted retinas (~150 μm thickness), modifying the tissue protocol to have longer incubation and wash times, and observed effective labeling of mRNA in bipolar cells of the inner nuclear layer (Fig. 3l and Supplementary Fig. 5a) for a variety of probe lengths (see Supplementary Fig. 5b for quantification and Supplementary Fig. 5c for images).
SABER enables spectrally multiplexed imaging
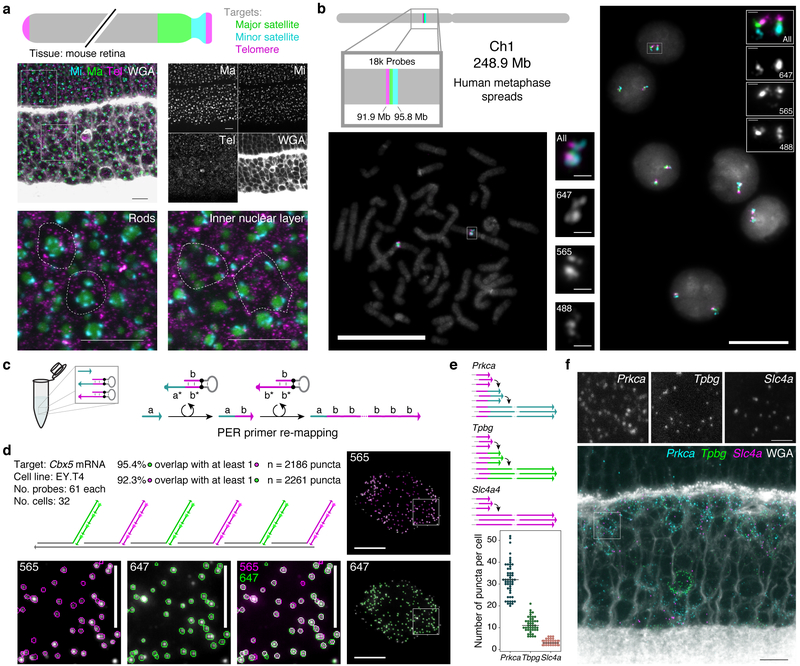
Probes that target distinct transcripts can be visualized simultaneously by appending orthogonal concatemer sequences detectable by imager oligos with distinct fluorophores. Three repetitive regions of the mouse chromosome - major satellite, minor satellite, and telomere - were visualized simultaneously in mouse retinal tissue using this approach (Fig. 4a), permitting observation of the distinctive chromatin architecture of rod photoreceptors.42 Another three-color visualization was performed in human metaphase spreads and interphase cells to target three adjacent positions on Chromosome 1 (Fig. 4b). In total, 18,000 probes targeting a 3.9 Mb region were mapped to three colors, which all co-localized as expected. Intronic and exonic sequences were also separately detected for the Dll1 mRNA transcripts in developing retina (Supplementary Fig. 6a), a distinction that is useful as a method to probe transcription kinetics.
Figure 4: SABER enables spectrally multiplexed imaging.
a, Multiplexed SABER-FISH in mouse retina. Mouse major satellite, minor satellite, and telomere chromosomal regions were detected with orthogonal SABER concatemers. Magnified views show distinct organization of these chromosome regions in rods compared to inner nuclear layer cells. Dashed outlines indicate approximate cell boundaries based on WGA staining. Scale bars: 10 μm. b, Multiplexed SABER- FISH on metaphase spreads. Three adjacent positions on human chromosome 1 were visualized using a bridge strategy (Fig. 1d) in metaphase spreads and interphase cells. Scale bars: 20 μm. c, Primer re-mapping with PER. Primers (e.g. with domain a) can be concatemerized with a different repetitive sequence (e.g. primer domain b) using a stepwise PER hairpin (e.g. that appends b to a)30 and a standard repetitive hairpin. d, Single molecule co-localization. Primer re-mapping was used to map two halves of the Cbx5 probe pool (used in Fig. 2) to distinct concatemer sequences, and two-color co-localization was visualized (see also Supplementary Fig. 6b) and quantified. In total, 92.3% of identified puncta in the 565 channel ovelapped with puncta in 647, and 95.4% of identified puncta in 647 overlapped with puncta in the 565 channel. Scale bars: 10 μm (cells), 5 μm (panels). n(puncta): n(565)=2,261; n(647)=2,186. e, Primer re-mapping and 3-color visualization in retina tissue. The Tpbg and Prkca probe sets from Fig. 3 were re-mapped to two new primers to enable simultaneous visualization and quantification of Prkca, Tpbg, and Slc4a transcripts. Median lines shown on distributions. Representative images are depicted in panel f,. n(cells): n(Slc4a)=38; n(Tpbg)=45; n(Prkca)=52. Scale bars: 2.5 μm, 10 μm (overlay).
We previously showed how PER cascades can be programmed to autonomously undergo differential synthesis pathways by changing the hairpins present in solution.30 This flexibility to program sequences allows us to take existing probe sets and change the sequence of the PER concatemer synthesized onto them. Fig. 4c shows an example of how a primer a can be mixed with two hairpins to produce a concatemer with repeats of sequence b. The first of two hairpins appends the b sequence 3’ from the a sequence, and then a second hairpin repeatedly adds the b sequence to generate a concatemer with a different PER primer sequence than the original one. See Supplemental Protocols for further information about designing these PER primer re-mapping reactions.
This re-mapping strategy is useful for cases where multiple probe sets have been synthesized with the same PER primer on the oligonucleotide 3’ ends but must be independently detected. By using PER primer re-mapping to impart new, orthogonal, concatemer sequences on some of these probe sets, each can be detected as a distinct color despite sharing an identical 9 nt sequence at the 5’ end of the concatemer. The Cbx5 probe set (used in Fig. 2) was split into two pools, and each pool was re-mapped to a new primer sequence. This enabled a two-color visualization of Cbx5 transcripts (Fig. 4d and Supplementary Fig. 6b), where half of the probes were mapped to the 565 channel and the other half were mapped to 647. We found that 92.3% of puncta identified in the 647 channel overlapped with puncta in the 565 channel, and conversely that 95.4% of puncta identified in the 565 channel overlapped with puncta in the 647 channel. These numbers further indicate that SABER-FISH probes can enable detection of a large fraction of available transcripts at the single-molecule level. We also evaluated primer re-mapping for two of the three RBC-expressed genes evaluated in Fig. 3 (Prkca and Tpbg) to simultaneously detect the three transcript species originally synthesized with identical primers (see Fig. 4e and Supplementary Fig. 6c for analysis and Fig. 4f for images).
Exchange-SABER enables fast exchange for highly multiplexed sequential imaging in cells and tissues
Higher levels of multiplexing can be achieved by iteratively detecting nucleic acid targets using DNA-Exchange8,9,10. We use formamide to rapidly destabilize short fluorescent imager strands without destabilizing the primary probe, permitting re-use of spectral channels. By modeling the melting temperatures of 20mer imagers, 42mer bridge sequences, and FISH probe sequences (Supplementary Fig. 7a), we predicted that 50–60% formamide in 1×PBS would effectively de-stabilize imagers without significantly affecting the underlying probe or 42mer bridge sequence stability.
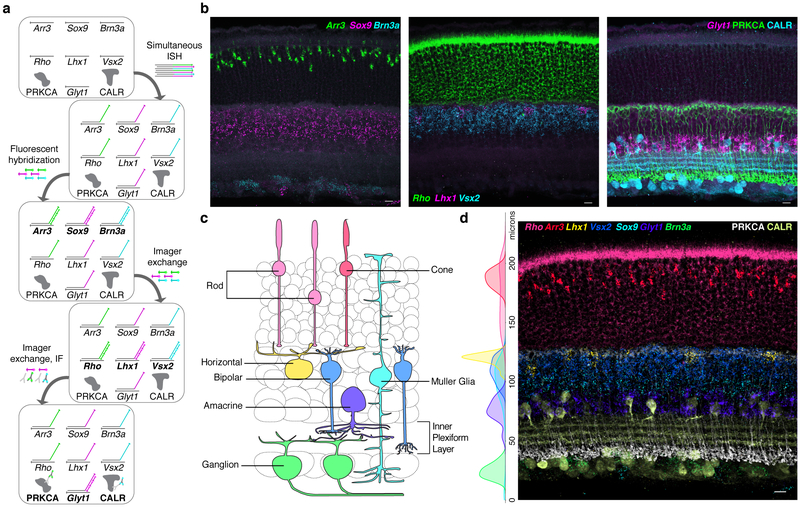
We tested this approach, which we call Exchange-SABER, in retinal tissue. Neural tissues typically display high cell type heterogeneity, requiring multiplexed detection methods for comprehensive identification of cellular populations. We aimed to detect all seven major cell classes in the retina (Cone, Rod, Horizontal, Bipolar, Amacrine, Ganglion, and Müller glia cells) using SABER probes against established markers. A pool of seven primary FISH probes were hybridized simultaneously, and detected in three sequential rounds of secondary fluorescent oligo hybridization (Fig. 5a). Imager oligo exchange occurred effectively in tissues, permitting re-use of spectral channels (Fig. 5b), and we observed the expected laminar separation of the cell classes (Fig. 5c) after overlaying and quantifying the channels (Fig. 5d). Following serial FISH probe detection, protein epitopes were still detectable by immunofluorescence (IF) and tissue integrity appeared well-preserved, with sublaminae of the inner plexiform layer (IPL) clearly discernible.
Figure 5: Exchange-SABER for detection of cell types in retina tissue.
a, Workflow for serial FISH detection of 7 transcripts and IF detection of two protein targets. b, Three FISH detection cycles for identification of 7 retinal cell populations. IF was performed after the 3rd FISH detection. c, Schematic representation of the retinal cell classes detected. d, All nine channels overlaid following puncta thresholding, with additional tissue autofluorescence background subtracted by a Gaussian filter masking channels around detected puncta. Side plot: Marker positive segmented cells plotted by distance from the inner limiting membrane. n(cells)=649. Scale bars: 10 μm.
We additionally confirmed that DNase I and Exonuclease I enzymes could be used to strip both primary SABER probes and imagers in tissue, while preserving mRNA integrity, as assayed by the ability to perform a second round of mRNA detection (Supplementary Fig. 7b). Therefore, high multiplexing using SABER is achievable by both a large selection of concatemer sequences (Exchange-SABER) and the ability to recycle concatemer sequences through primary probe digestion and rehybridization43. While the probe digestion approach is slower than Exchange-SABER as it requires iterative (long) ISH steps, rather than sequential (fast) fluorescent hybridization steps, it allows re-use of the same PER hairpin and imager sequences and is therefore a simple way to reduce the upfront cost of reagents.
We further used Exchange-SABER to directly compare two-color primary concatemer detection to two-color branching-based detection of a non-endogenous target mRNA (Cas9, Supplementary Fig. 8a and Supplementary Fig. 8b) and compared it to co-expressed GFP signal (Supplementary Fig. 8c and Supplementary Fig. 8d) to further confirm the specificity of SABER signals. As expected, all four SABER signals showed strong colocalization (Supplementary Fig. 8e and Supplementary Fig. 8f), further increasing our confidence that branched and standard SABER-FISH signals correspond to true target transcripts.
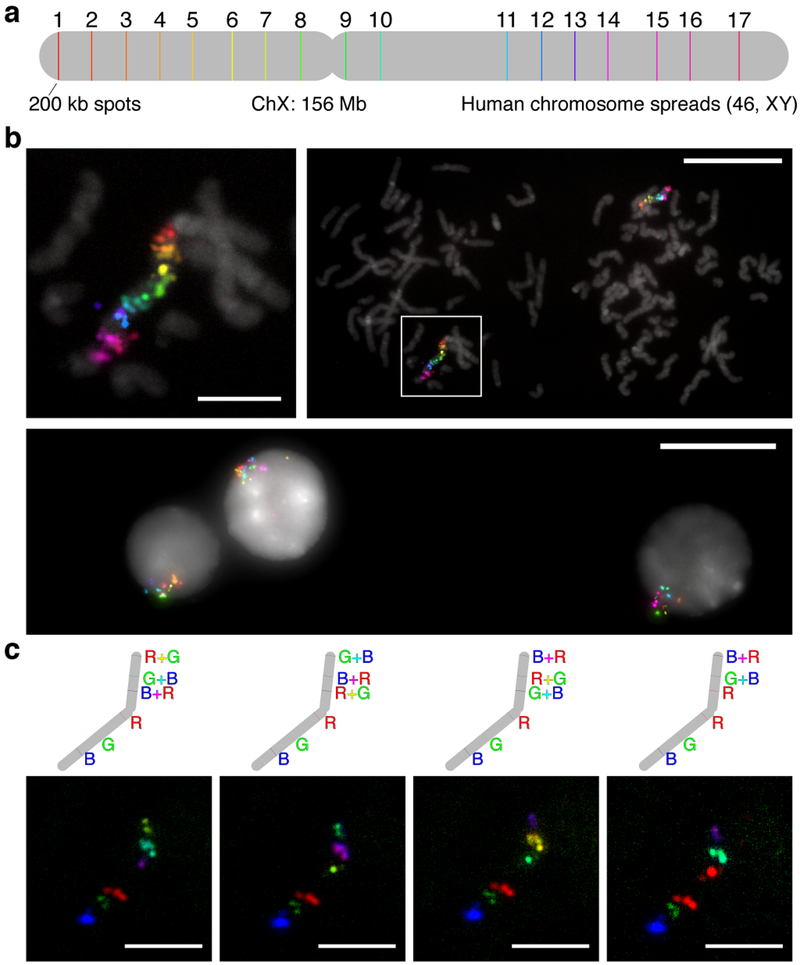
We also evaluated Exchange-SABER in human metaphase spreads and interphase cells, with 17 colors targeting seventeen 200 kb regions spread along the Human X chromosome (Fig. 6a). Metaphase spreads validate the directional coloring of targets, and interphase cells depict X chromosome territories within their nuclei. In total, 17 colors (6 hybridizations) were imaged in 7 hours, including stripping, re-hybridization, field of view finding, and Z-stack imaging times (Fig. 6b and Supplementary Fig. 9a, additional images in Fig. 6c). Such brief incubation times are aided by the improved reaction kinetics conferred by the presence of many imager binding sites within concatemers, as not all binding sites must be saturated to discern signal.
Figure 6: Sequential imaging of chromosomal targets using Exchange-SABER.
a, Schematic of 17 targeted regions along the human X chromosome (width to scale). Each set of probes per spot had different 42mer barcode sequences appended to their 3’ ends (Fig. 1d). Seventeen 42mer bridge sequences concatemerized with 17 different PER primers were co-hybridized. b, Individual color channels on DAPI. 6 hybridizations, targeting 3, 3, 3, 3, 3, and 2 spots, respectively, that took course over a single day were used to image the 17 colors. c, 17-color overlays on DAPI. The representative metaphase spread from part (B) is shown overlaid on DAPI at two length scales (top left, top right). Interphase cells showing the X chromosome territories were also captured (bottom). d, Combinatorial 6-color SABER imaging. As a step toward increasing multiplexing with SABER amplification further, we demonstrated mapping six of the spots on the chromosome to 4 different 6-color combinations. Scale bars: 5 μm (spreads), 20 μm (fields of view).
We next took the same metaphase sample and applied new, combinatorial fluorescent hybridizations. Each hybridization targeted the same set of 6 regions, mapping them to each of three colors in addition to the pairwise combinations of the three colors, achieved by co-hybridizing complementary imager oligos conjugated with different fluorophores. This process was repeated four times (~4 hours total), each with a different 6-color mapping (Fig. 6d). The success of this detection scheme indicates SABER should be compatible with combinatorial imaging strategies for future highly multiplexed and amplified target detection.
Application of SABER for quantitative in situ reporter assay
We next investigated whether the ability of SABER to provide multiplexed, amplified detection of both RNA and DNA sequences in tissues could be applied to reporter assays involving the introduction of exogenous DNA elements. An ideal reporter assay would permit simultaneous quantification of the expression of reporter molecules, the number of introduced DNA constructs encoding the reporters, and the expression of endogenous markers. We applied SABER to the detection of reporter RNAs transcribed from plasmids bearing isolated cis-regulatory modules (CRMs). First, reporter sequences were cloned downstream of a minimal promoter and validated independently for the ability to report CRM activity, using upstream insertion of a validated bipolar cell enhancer44 (Supplementary Fig. 10a), followed by electroporation in vivo into the retina (Supplementary Fig. 10b).
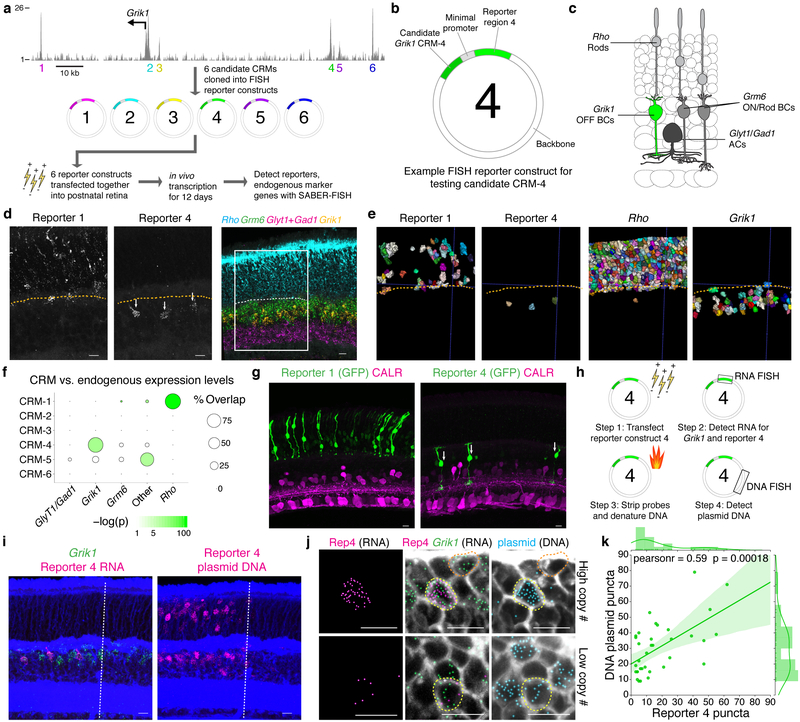
We applied this reporter set to evaluate the behaviors of previously uncharacterized CRMs using a 10-plex SABER-FISH experiment. We selected for investigation candidate CRMs in the vicinity of the gene Grik1, which encodes a kainite-family glutamate receptor subunit with strong and enriched expression in most OFF bipolar cells (Types 2, 3a, 3b, 4),40 as few genetic tools exist to specifically label this population in vivo. Candidate CRMs were identified by inspection of retina chromatin accessibility to DNase I (data from the ENCODE project45) in a genomic interval proximal to the transcription start site (Fig. 7a). Six candidate DNA sequences (CRMs 1–6) were inserted independently upstream of distinct reporters, and the reporter set was introduced as a pool into the retina (Fig. 7a and Fig. 7b). We used SABER to detect all six reporters as well as four markers of cell types accessible by postnatal retina electroporation (Grm6: ON bipolar cells; Glyt1 and Gad1: amacrine cells; Rho: rods)46 (Fig. 7c). Images (Fig. 7d and Supplementary Fig. 10c) and segmentation results (Fig. 7e) indicated that one reporter, corresponding to CRM-4, was selectively expressed in Grik1+ cells (Supplementary Fig. 10d). A second reporter (CRM-1) showed instead abundant and specific expression in Rho+ cells (rods) (Fig. 7d and Fig. 7e) . We estimated the specificity of CRM activity in different cell types by comparing reporter expression to endogenous cell-type marker expression. We first calculated cell type abundances, determining that 9% of electroporated cells expressed endogenous Grik1 transcripts. A hypergeometric test was used to evaluate the probability of observing the empirically determined positive patterns for each CRM, yielding highly significant p-values (p value of 1.27×10−125 for CRM-1/Rho and p value of 1.03×10−15 for CRM-4/Grik1, Fig. 7f). Specificity of CRM activity was also confirmed using a GFP fluorescence assay (Fig. 7g).
Figure 7: SABER-FISH enables detection of in vivo RNA reporters for enhancer activity analysis.
a, Mouse retina DNase I hypersensitive regions (EN- CODE45) in the vicinity of the Grik1 start site and workflow for the reporter screen experiment. Vertical axis represents absolute read density. b, Representation of key components of the reporter plasmid. c, Schematic representation of neuronal cell types electroporated in the postnatal retina. Grik1 expression distinguishes OFF bipolar cells from ON bipolar cells (Grm6+). d, Representative images of two expressed reporters (single channel) and four endogenously expressed genes. Dashed line indicates the approximate position of the outer plexiform layer. Box indicates the area of magnification shown for the single channel reporter expression images. e, Relevant reporter and endogenous gene-expressing cells displayed following cell segmentation. f, Quantification of the percent of reporter positive cells that are positive for each marker probed. ‘Other’ refers to expression in cells not positive for any marker tested, and may include Müller Glia and Type 1 bipolar cells. Dot size corresponds to the percent of CRM-positive cells that are positive for each endogenous marker. Dot color reflects the p-value for a hypergeometric test plotted on a logarithmic color scale. n(cells): n(CRM-1)=440; n(CRM-4)=18; n(CRM-5)=25. g, Expression of GFP driven by CRM-1 and CRM-4 following retina electroporation. Rods (left) are identifiable by position in the outer nuclear layer, and OFF bipolars (right, arrows) are identifiable by bipolar morphology and lamination in the upper layers of the inner plexiform layer, labeled by Calretinin (CALR). h, Experimental design for reporter RNA and DNA sequential detection. i, Representative image of an electroporated retina with detection of Reporter 4 RNA, Grik1 RNA, and plasmid DNA. Dashed line indicates the approximate location of the electroporation patch boundary. j, Magnified images showing detected Reporter 4 RNA puncta in cells with Grik1 expression and plasmid DNA (yellow outline) but not in Grik1+ cells lacking plasmid DNA (orange outline). k, Quantification of detected Reporter 4 RNA puncta plotted against detected plasmid DNA puncta in Grik1+ cells. Pearson correlation coefficient (pearsonr) and p value shown. Shaded region indicates 95% confidence interval. n(cells)=35. Scale bars: 10 μm.
We investigated whether SABER could be used to co-detect reporter-encoding plasmids and reporter transcripts. Commonly used methods for dense introduction of exogenous DNAs in vivo (e.g. electroporation, AAVs, cationic lipids) can result in a broad distribution of DNA copy number per cell, with cell type-specific biases in transfection rates. This variability can impede assessment of CRM activity (reporter transcripts per reporter DNA). Measurements of enhancer specificity must account for cell type abundance of transfected cells which may also be highly variable. We applied co-detection of the reporter RNA and the plasmid DNA backbone to determine transfected cell type distributions and plasmid load (Fig. 7h). The CRM-4 reporter was singly electroporated into the retina, followed by detection and quantification of reporter RNA, Grik1 transcript, and a 2.8 kb region of the plasmid backbone in the same electroporated cell populations (Fig. 7i). We examined the relationship between number of plasmids and number of reporter transcripts in Grik1+ cells (Fig. 7j) and observed a significant correlation (Pearson correlation coefficient of 0.59, p value of 0.00018, Fig. 7k) that is not observed when comparing plasmid copy number to endogenous Grik1 expression (Supplementary Fig. 10e, analysis in Supplementary Fig. 10f). Variation in transcript number per plasmid may represent selective silencing of plasmids in particular cells, or differences in CRM-4 activity between distinct Grik1-expressing OFF bipolar cell types.
DISCUSSION
The Primer Exchange Reaction (PER) method is a versatile tool for creating user-defined assembly of short sequences using a catalytic hairpin structure. Here we apply the telomerase-like mode of PER to achieve enhanced FISH signals. PER is used to synthesize concatemers of user-defined length on smFISH and Oligopaint-style tiling probes in vitro. These concatemers provide a scaffold for concentrating fluorescent signal via secondary hybridization, a method we name ‘SABER.’ Through the application of SABER to a large array of different sample and target types we demonstrated the strength and flexibility of this approach: (1) Programmable signal amplification levels between 5× and 450×. (2) Multiplexing with at least 17 orthogonal concatemers applied simultaneously. (3) High detection efficiency in tissue. (4) Applicability to diverse targets (RNA, DNA, endogenous and exogenously introduced). Additionally, SABER can also be applied for detecting protein targets via DNA-conjugated antibodies (upcoming work, see preprint47).
SABER was also practical to apply in terms of workflow and cost. SABER works with minimally pre-treated tissues using standard hybridization protocols, and we effectively labeled thick tissue sections and whole retinas without the application of tissue clearing or gels. Probe sets ordered unpurified and unmodified can be amplified in bulk using PER in vitro with a single enzymatic step, allowing enough material for dozens of experiments to be prepared in one step, avoiding the need for long amplification steps in situ, and reducing cost. The combined cost of all oligos and enzymes is currently estimated to be less than $5 per target per experiment (120 μL ISH solution) and could be further reduced with additional optimization and bulk synthesis (see Supplementary Note 10).
The analytic pipeline demonstrated here is similarly straightforward to use. 3D cell segmentation combined with SABER permitted quantification of transcripts on a single cell level, and relative transcript abundance closely correlated with Drop-seq measurements. SABER is therefore well suited to accompany scRNA-seq in transcriptomic studies. SABER, which is compatible with IF, can be used to link scRNA-seq-defined populations to positions in tissue such that morphological stains, or labels that permit post-hoc identification of recorded cells, can be integrated with cell type identification.
Detection of complex pools of reporters or barcodes is another area where effective multiplexed FISH technologies can be applied. In cell culture, FISH-based barcode detection has been employed for the analysis of lineage,48 and barcode reading is an element of STARmap probe design.29 Here we demonstrated the utility of SABER for assaying the activity of isolated candidate enhancer sequences introduced in vivo. Simultaneous detection of reporter expression and cell type markers with single cell resolution was required for assessing cell-type specificity of CRM activity. Using SABER we were able to detect reporters across a broad range of expression levels and assay DNA plasmid copy number in the same cells, providing a tool to quantify enhancer strength and specificity. As an effective and simple method to robustly detect RNA and DNA sequences in cells and tissue, SABER enables the characterization of abundances, identities, and localizations of complex sets of endogenous and introduced nucleic acids.
In cases where targets are spatially separated, there is further opportunity to combine SABER amplification with multi-round combinatorial barcoding. Such barcoding methods allow an exponential number of unique, non-overlapping targets to be visualized in a linear number of rounds.14–17 These multiplexing strategies could benefit from signal amplification, and recent studies have been able to combine signal amplification with combinatorial barcoding (e.g. seqFISH combined with HCR43 and RCA paired with a sequencing-by-ligation readout29). For hybridization-based, high-targeting efficiency spatial genomic and transcriptomic methods, SABER may offer the unique potential for rapid one-step deployment of a large set of orthogonal amplifiers for subsequent combinatorial readout.
ONLINE METHODS
Expanded methods details can be found in Supplemental Experimental Procedures. All procedures were performed at room temperature, except where otherwise specified. Step-by-step protocols for probe design, oligo ordering, PER concatemerization, RNA FISH in cells, and RNA FISH in tissues can be found in the Supplemental Protocols file.
Cell culture
MRC-5 (human, ATCC CCL-171) and HEK293T cells (human, ATCC CRL-1573) were grown in Dulbecco’s modified Eagle medium (Gibco #10564) supplemented with 10% (vol/vol) serum (Gibco #10437 for MRC-5 and Peak Serum PS-FB2 for HEK293Ts), 50 U / mL penicillin, and 50 μg / mL streptomycin (Gibco #15070). EY.T4 embryonic fibroblasts (mouse)49 were grown in Dulbecco’s modified Eagle medium supplemented with 15% (vol/vol) serum, 50 U / mL penicillin, and 50 μg / mL streptomycin. All cells were cultured at 37°C in the presence of 5% CO2.
Tissue
All animal experiments were conducted in compliance with the protocol IS00001679, approved by the Institutional Animal Care and Use Committees (IACUC) at Harvard University. Experiments were performed on tissue collected from wild-type male and female CD1 IGS mice (Charles River). Tissue were collected at postnatal day P17 for all experiments with the exception of electroporated retinas used for reporter assays (P13) and retinas in Supplementary Fig. 5 (P25).
Sequence design
Oligopaint FISH probe sets31 were designed with the OligoMiner pipeline33 and accessed for the mm10 and hg38 whole-genome probe sets hosted at https://yin.hms.harvard.u/oligoMiner/list.html. For RNA FISH probes, the genomic locations of the exons and/or introns of the target gene were acquired from the UCSC Genome Browser50 and used in combination with the ‘intersectBed’ utility of BedTools51 to isolate probe oligos targeting the RNA features of interest. The 42mer bridge sequences were drawn from blocks of orthogonal barcode sequences34 catenated together. 42mer bridge sequences and PER sequences (designed to have high single-strandedness and low cross-talk as evaluated with NUPACK35–37) were vetted using a similar screening process as for probe sequences (with Bowtie,52 Jellyfish,53 and BLAST54). Oligo probes targeting the mouse major satellite, minor satellite, and telomere repeats were adapted from ref.55 Strands were ordered unpurified or HPLC-purified from IDT or Twist. See Supplemental Experimental Procedures for more detailed information and a list of sequences, as well as Supplemental Protocols for step-by-step instructions for designing and ordering probes and PER sequences.
PER concatemerization
Typically, 100 μL reactions were prepared with final concentrations of:1×PBS, 10mM MgSO4, 400 – 1000 units / mL of Bst LF polymerase (NEB M0275L or McLab BPL-300), 300 – 600 μM each of dATP/dCTP/dTTP (NEB M0275L), 100 nM of Clean.G hairpin, 50 nM - 1μM hairpin(s), and water to 90 μL (see Supplementary Fig. 1a for a visual representation). We previously reported this type of preliminary incubation with the Clean.G hairpin as a strategy to filter out a small amount of contaminant nucleotides that can halt the extension (see Fig. S6 from ref30). After incubation for 15 minutes at 37°C, 10 μL of 10 μM primer was added, and the reaction was incubated another 1–3 hours followed by 20 minutes at 80°C to heat inactivate the polymerase. PER extension solutions were directly diluted into ISH solutions or, in the case of the 17-color chromosome walk (Fig. 6) and tissue experiments, purified and concentrated using a MinElute (Qiagen #28004) kit with distilled water elution to reduce volume and salt concentration from the reaction condition. Lengths of concatemers were evaluated by diluting 1 μL of in vitro reaction with 19 μL water. Samples were then run on 1% E-Gel EX agarose gels (Thermo Fisher G402001) for 10 minutes alongside a 1kB Plus dsDNA Ladder (Invitrogen) to estimate length, and imaged with the Sybr Gold channel on a Typhoon FLA 9000 scanner. See Supplementary Experimental Procedures for complete PER sequence, extension, and purification conditions for each experiment and Supplemental Protocols for a step-by-step PER protocol.
FISH in fixed cell chambers
8-well chambers (Ibidi #80827) were seeded with MRC-5 cells and grown in a tissue culture incubator (37°C with 5% CO2). Samples were rinsed in 1×PBS, fixed in a 1×PBS + 4% (wt/vol) paraformaldehyde solution (10 minutes), and rinsed with 1×PBS. Chambers were then optionally stored at 4°C for up to two weeks before continuing to the in situ hybridization (ISH) step.
3D DNA ISH closely followed previous protocols.21,31,33,56 After fixation, samples were rinsed in 1×PBS (1 minute), permeabilized in 1×PBS with 0.5% (vol/vol) Triton X-100 (10 minutes), washed in 1×PBS + 0.1% (vol/vol) Tween-20 (1×PBSTw) (2 minutes), incubated in 0.1N HCl (5 minutes), and washed in 2×SSC + 0.1% (vol/vol) Tween-20 (2×SSCT) (1×1 minute, 1×2 minutes). Samples were incubated in 2×SSCT + 50% (vol/vol) formamide (5 minutes), transferred to fresh 2×SSCT + 50% (vol/vol) formamide at 60°C (at least 1 hour), and then wells were loaded with 125 μL ISH solution comprising 2×SSCT, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, 400 ng / μL RNase A (EN0531, Thermo Fisher), and each PER extension at ~67 nM final concentration (1:15 dilution from 1μM PER). After denaturation at 80°C (3 minutes), samples were incubated overnight at 44°C on a flat-block thermocycler (Mastercycler Nexus, Eppendorf).
RNA ISH was performed similarly to 3D DNA FISH, but with a shortened protocol. After fixation, samples were rinsed in 1×PBS (1 minute), permeabilized in 1×PBS with 0.5% (vol/vol) Triton X-100 (10 minutes), washed in 1×PBSTw (1 minute). Samples were then transferred to 2×SSCT (1 minute) before wells were loaded with 125 μL of ISH solution comprising 2×SSCT, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, and PER extension at ~67 nM final concentration. After denaturation at 60°C (3 minutes), chambers were incubated overnight 42°C on a flat-block thermocycler.
After (DNA or RNA) ISH, 200 μL pre-warmed 2×SSCT (at 60°C) was added, and the hybridization solution was aspirated. Samples were then washed in pre-warmed 2×SSCT at 60°C (4×5 minutes) and in 2×SSCT at room temperautre (2×2 minutes). Samples going directly to fluorescent hybridization were then transferred to 1×PBS and washed (1 minute and then fresh solution). Samples were optionally held at 4°C (1–2 overnights) before continuation with branching or fluorescent hybridization protocols.
Samples with one round of branching were washed in 2×SSCT at room temperature (2 minutes) before wells were loaded with branch hybridization solutions comprising 2×SSCT, 30% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, and PER extension at ~67 nM final concentration (1:15 dilution from 1μM PER) and hybridizing at 37°C for 1.5 hours. Samples were then washed as before in 2×SSCT (4×5 minutes at 60°C with pre-warmed buffer and 2×2 minutes at room temperature). For iterative branching experiments, the branching and washing steps were performed repeatedly with slightly less stringent conditions: 30°C for 1 hour for hybridization, 55°C for the heated washes, and only one of the two 2×SSCT washes at room temperature. After branching, samples were transferred to 1×PBS and washed (1 minute and then fresh solution). Samples were then typically held at 4°C overnight before continuation with fluorescent hybridization. See Supplemental Protocols for a note about choosing the right branch hybridization condition for a given set of sequences.
For fluorescent hybridization, samples were rinsed in 1×PBS, and then wells were loaded with 125 μL fluorescent hybridization solution comprising 1×PBS and 1 μM fluorescent imager strands. Samples were incubated at 37°C (1 hour), washed in pre-warmed 1×PBS at 37°C (5 minutes and 2×2 minutes), rinsed with 1×PBS at room temperature, and transferred to SlowFade Gold + DAPI (Thermo Fisher S36939) for diffraction-limited imaging. See Supplemental Experimental Procedures for additional details and Supplemental Protocols for step-by-step instructions for performing RNA FISH in cells.
Metaphase DNA FISH
Human metaphase chromosome spreads on slides (XX 46N or XY 46N, Applied Genetics Laboratories) were denatured in 2×SSCT + 70% (vol/vol) formamide at 70°C (90 seconds) and then transferred to ice-cold 70% (vol/vol) ethanol (5 minutes), to 90% (vol/vol) ethanol (5 minutes), and to 100% ethanol (5 minutes). Slides were air-dried before 25 μL of ISH solution comprising 2×SSCT, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, 400 ng / μL RNase A (EN0531, Thermo Fisher), probe pools with bridges at ~500 nM, and PER extensions at 96 or 192 nM final concentration was added. The hybridization solution was sealed underneath a coverslip with rubber cement, and the slide was placed into a humidified chamber inside an air incubator at 45°C overnight. After hybridization, samples were washed in 2×SSCT at 60°C (15 minutes) and in 2×SSCT at room temperature (2×5 minutes).
For spectral imaging (Fig. 4b), slides were transferred to 1×PBS and then dried before 25 μL fluorescent hybridization solution comprising 1×PBS with 1 μM fluorescent imager strands was added. After covering the hybridization solution with a coverslip, slides were put into a humidified chamber and incubated in an air incubator at 37°C (1 hour). Slides were then washed three times (1×15 minutes and 2×5 minutes) at 37°C in pre-warmed 1×PBS and dried. 12 μL of SlowFade Gold + DAPI (Thermo Fisher S36939) was added and sealed underneath a coverslip with nail polish before imaging.
For the metaphase walk (Fig. 6), slides were transferred to 1×PBS after the post-FISH washes and then dried. A flow chamber (~50 μL volume) was constructed using a coverslip attached to the slide using double-sided tape to allow fluid exchange, and samples were re-hydrated in 1×PBS. Each hybridization comprising 1×PBS with 10% (wt/vol) dextran sulfate and 1 μM fluorescent imager strands was incubated at room temperature for 15 minutes. (For combinatorial hybridizations with PER concatemers mapped to two colors, each fluorescent imager strand was included at 500 nM, retaining the 1 μM overall concentration). After washing with 200 μL (~4×flow through the chamber), SlowFade Gold + DAPI (Thermo Fisher S36939) was added for diffraction-limited imaging. Between each fluorescent hybridization, previous imager strands were stripped with formamide: SlowFade was washed out with 200 μL of 1×PBS, followed by stripping with a total of 1.6 mL of 1×PBSTw containing 60% (vol/vol) formamide flowed through over the course of 15 minutes. After stripping, slides were washed with 200 μL 1×PBS, allowed to sit for 2 minutes, and washed twice more with a total of 400 μL 1×PBS before adding the next fluorescent hybridization solution. The slide was stored overnight at 4°C after the first six hybridizations before subsequent stripping and hybridization steps.
Cas9 colocalization experiments
The Cas9 colocalization experiments employed the fixation, RNA FISH, fluorescent exchange, branch hybridization, and fluorescent hybridization methods described above. See Supplemental Experimental Procedures for full details.
Retinal histology
Neural retinas were dissected in 1×PBS, fixed for 25 minutes at room temperature in 4% formaldehyde, 1×PBS solution, and frozen in O.C.T. solution (50%O.C.T., 15% Sucrose, 0.5×PBS). Cryosections cut to 35 or 40 μm were adhered to Poly-D-Lysine-coated (Sigma P6407) 8-well Ibidi chamber slides and dried. Prior to hybridization, tissues were washed in 1×PBS with Tween-20 (Sigma P9416) at 0.1% (vol/vol) (1×PBSTw), then pretreatment consisting of a mild proteinase K exposure (1.5 μg / mL, 15 minutes) followed by post-fixation and acetic anhydride treatment as described previously.40 Sections were incubated at 43°C in a hyb oven in wash hyb (40% formamide, 2×SCC pH 7, 1% Tween-20) for 30 minutes preceding addition of pre-warmed probe/hyb solution. Probe concentrations were determined by nanodrop and probes were added to a final mass of 1μg each per well (120 μL volume) in hyb solution (40% formamide, 2×SSC pH 7, 1% Tween-20, and 10% dextran sulfate (Sigma, D8906)). Following overnight incubation (18–24 hours), slides were washed 2×30 minute in 40% formamide wash hyb, 2×45 minutes in 25% formamide wash hyb, and 2×15 minutes in 2×SSCTw (0.1% tween). For fluorescent detection slides were washed three times in 1×PBSTw at room temperature, and then transferred to 37°C for hybridization and subsequent wash steps. Detection oligos were diluted to a concentration of 1 μM in a 1×PBS solution with 0.2% Tween-20 and 10% dextran sulfate. This solution was incubated with the sample for 2 hours, and then washed 4×7 minutes in 1×PBSTw. Imaging was performed in 80% glycerol mounting media (80% glycerol, 1×PBS, 20mM Tris pH 8, and 2.5 mg / mL of propyl gallate). For serial detections, fluorescent oligos were stripped with a solution of 50% formamide in 1×PBS at room temperature (3×5 minutes washes), and washed 3×2 minutes in 1×PBSTw.
Whole mount stainings were conducted with a similar protocol but with extended hybridization and wash times. Whole mount retinas were held in place within chamber slides by application of a nylon mesh (SEFAR-NITEX 03–64/45) cut to size, layed over the retina, and glued to the chamber with Gorilla Glue. For detection of DNA, retinas were treated as described for RNA FISH with the addition of a 5 minute treatment in a 1N HCl, 0.5M NaCl solution, followed by a 15 minute incubation at 80°C in a solution of 50% formamide and 2×SSC on a preheated metal block prior to the primary probe hyb. For RNA and DNA co-detection, DNA denaturation and hybridization was conducted following RNA detection. RNAse A (Thermo Fisher EN0531) was added to the primary probe hybridization solution for DNA FISH at a concentration of 200 ng / μl. We subsequently performed optimization experiments with tissue RNA FISH indicate that most pretreatment steps were unnecessary and that several steps in the protocol can be performed more rapidly. Proteinase K, post-fixation, and acetic anhydride steps were eliminated and sections were only washed 3×5 minutes in 1×PBSTw prior to addition of wash hyb. See Supplemental Experimental Procedures for additional details and Supplemental Protocols for step-by-step instructions for performing retinal RNA FISH.
Reporter Assay
Reporter sequences were derived from full or partial sequences of genes commonly expressed heterologously in mammals: dCas9 (template: Addgene #60954), Lacz, Cre, and Luciferase. Reporters were designed to be within a size range of 1–1.6kb and targetable by 22–24 primary probes. A reporter plasmid with a minimal TATA promoter (Stagia3)44 was used, replacing the GFPiAP ORF of this plasmid with the described reporter sequences. Reporters were electroporated into mouse pups via subretinal injection at P1 as described46 at a concentration of 500 ng / μl for each construct. Retina DNase I hypersensitivity data (GEO:GSM1014198) available through the ENCODE consortium45 was accessed via the UCSC genome browser.50 See Supplemental Experimental Procedures for additional details.
Microscopy
Imaging of iterative branching samples was conducted on an inverted Zeiss Axio Observer Z1 using a 100x Plan-Apochromat Oil N.A. 1.40 objective and LED light source. Remaining cell and metaphase samples imaged on a Nikon Eclipse Ti-E microscope by using a CFI PlanApo 100x Oil (N.A. 1.45) objective and LED light source. All tissue images were acquired on a Zeiss Axio Observer Z1 inverted microscope equipped with an LSM780 single point scanning confocal attachment (405, 488, 561, 594, and 633 laser lines). See Supplementary Experimental Procedures for additional information about the microscope setups.
Image processing
Maximum projections taken on the Nikon Eclipse Ti-E microscope were processed using the Nikon Elements software, and by Zeiss ZEN software for non-confocal images taken on the Zeiss Axio Observer 1. Images were then processed with Fiji/ImageJ.57,58 Multicolor overlays of cells and metaphase spreads were generated using Python scripts written to mimic the ‘screen’ behavior of Photoshop, which also allowed automatic cropping, contrasting, and DAPI alignment. Most images presented in the main and supplemental figures utilized max projections of Z-stacks, with the exception of the metaphase spread image in Fig. 4b, the interphase image in Fig. 4b, and the metaphase spreads in Fig. 6, for which single in-focus z slices for each hybridization were utilized to create overlays. For cell images with nuclei outlined, nuclear outlines were first automatically generated using CellProfiler38,59 analysis pipelines (see below), and then these outlines were automatically outlined and then re-styled in Adobe Illustrator. Scale bars were added either in Adobe InDesign or Adobe Illustrator based on expected pixel size scaling. For retina images, maximum intensity projections were generated in ZEN 2.3 lite. Multicolor overlays were generated using the screen setting in Adobe Photoshop, and brightness and contrast were adjusted for display purposes using Adobe Photoshop or Fiji/ImageJ.58 For whole mount volume visualizations the ImageJ plugins 3D Viewer and Volume Viewer were employed. For quantification of intensities and puncta detection in retina tissue, MATLAB and the Image Processing Toolbox were used (MATLAB and Image Processing Toolbox Release R2018a, The MathWorks, Inc., Natick, Massachusetts, United States). Open Microscopy Environment’s Bio-Formats library60 was used for image file manipulation, including import of image stacks to the MATLAB environment.
In all cases except where otherwise noted in figure legends, images were only contrasted to improve signal visibility by changing the min (black) and max (white) values. Tissue autofluorescence was subtracted for Fig. 4d using puncta detection (see below) and background masking for each marker. High-resolution images of detected puncta (as shown in Fig. 7j) were generated by resizing punta centroid images using bicubic interpolation and dilation based on empirically estimated puncta size. See Supplementary Experimental Procedures for more detailed information.
QUANTIFICATION AND STATISTICAL ANALYSIS
Puncta quantification in cells
Maximum intensity projections in Z were created using Nikon Elements software from raw mutlichannel Z-stacks. These max projections were then inputted into CellProfiler 3.0,38,59 in which an automated image analysis pipeline was constructed to identify nuclei, cell bodies, and FISH foci and to calculate background-subtracted maximum pixel intensity of each segmented focus. For intensity quantification experiments, the same pipeline was used for all conditions being compared. For cases where the number of puncta per cell or nucleus was calculated, a parent-child relationship was established between the FISH foci and the respective cellular or subcellular feature. For fold enhancement calculations in Fig. 2b, Fig. 2d, and Supplementary Fig. 3c, background was calculated as the mean of the image pixels masked for the detected puncta. Background-subtracted peak intensity distributions of puncta were then divided by the average of the corresponding distribution for the unextended condition. Cumulative density distributions depicted in Fig. 2g did not subtract background for fold enhancement calculations because the sample was very crowded. In Fig. 4d, co-localization of puncta was assumed if any pixels within the detected area of a punctum in one channel overlapped with any pixels corresponding to a punctum detected in the other channel. See Supplemental Experimental Procedures for a table containing cell counts, puncta counts, and data for amplification, SNR, puncta counts per body, and puncta area distributions.
Drop-seq data processing
Bipolar cell Drop-seq data40 was processed according to the markdown accompanying the manuscript using class file class.R (provided at https://github.com/broadinstitute/BipolarCell2016. All 10,888 cells identified present in cluster 1 (corresponding to Rod bipolar cells) following Louvain clustering and cluster merging were used for plotting average number of transcripts per cell. This analysis discards cells considered of poor quality with fewer than 500 detected genes per cell.
Retina image analysis
Serially detected retina images were aligned based on the WGA stain using intensity-based image registration algorithms in the MATLAB Image Processing Toolbox. Cells were segmented by applying the open-source membrane-based segmentation software, ACME,41 to WGA images. For serial imaging, cell segmentation was performed on the WGA channel from a single session and applied to all registered channels. After automated segmentation, results were verified and visualized using ITK-SNAP.61
Puncta were detected in 3D Z-stack images using a Laplacian of Gaussian method,62 similar to the analagous 2-D pipeline implemented in.5 See Supplementary Fig. 4c–d and Data and Code Availability sections for details. Intensity quantifications in tissue were done by performing puncta detection, taking the maximum pixel intensity of each puncta, and subtracting the average background intensity for each image. See Supplemental Experimental Procedures for additional information on image analysis as well as puncta and numbers.
A universal threshold was applied to call cells positive for each marker based on the distribution of puncta per cell (see Supplementary Fig. 4e for details). Thresholds were 15, 5, and 2 for Prkca, Tpbg, and Slc4a, respectively. For quantification of reporter RNA versus plasmid DNA, a threshold of 2 puncta per cell was used for CRM-4 reporter RNA, 7 puncta per cell for endogenous Grik1, and 3 puncta per cell for plasmid DNA.
Reporter specificity analysis
The plasmid DNA FISH images were used to estimate that 52% of electroporated cells were rods, 9% were positive for Grik1 endogenous RNA, 18% were ON bipolar cells, 12% Müller Glia, and 9% amacrine cells. These numbers were estimated directly from the data based on plasmid DNA detection, Grik1 RNA expression, and the known cell body localization of each cell type. Reporter specificity was evaluated using a hypergeometric test63 to define the probability of observing the empirical positive cell patterns for each CRM. For a given CRM-driven reporter (CRM 1–6) and endogenous gene (Rho, Grm6, Grik1, Glyt1/Gad1, Other), we can consider CG+ , the number of CRM reporter-positive cells that are positive for the endogenous gene:
Where,
CRM+ = Set of cells positive for CRM-driven reporter RNA
GENE+ = Set of cells positive for endogenous gene
CG+ = |CRM+ ∩ GENE+| = Total # cells positive for both the CRM and gene
N = Total # cells that CRM-reporter plasmid DNA
n = Total # GENE+ in the plasmid-receiving population
C = |CRM+| = Total # cells observed positive for CRM reporter RNA
We took N =1,500 as an estimate of the total cell population assayed for each CRM based on the number of plasmid-positive cells observed in the DNA FISH experiment. In any one 240 μm×240 μm electroporated retinal region, we estimate that approximately 300 cells plasmid DNA based on the automated cell segmentation. Cells were analyzed across 5 similar retinal regions, yielding a total population size of approximately 1,500 cells. With this estimate, n can be inferred based on the proportion of each endogenous marker within the electroporated population. Both C and CG+ were directly measured.
Statistics and reproducibility
All retina histology experiments were conducted at least twice on separate occasions with similar results. For figure displays, quantification was performed for at least three retinal sections from one animal from a single experiment with the exception of the distance plot in Fig. 5d which is specific to the image shown. The exact number of biological replicates for all experiments is listed in Supplemental Experimental Procedures. Cell experiments were performed a number of times with similar results before the final data were quantified for a single experiment. Further internal controls, including quantitative comparison to sequencing data, co-localization of signal from independent probe sets, negative controls with probes or other elements missing, and signals matching expected morphologies, further increase our confidence in the consistency and reproducibility of the technique applied in multiple contexts.
Plotting and visualization
Most plots and some image overlays were generated in Python, using the Matplotlib,64 Seaborn,65 NumPy,66 Pandas,67 PIL, and Biopython68 libraries. Data was imported either in CSV format or read in from CellProfiler38,59 output files. The plot in Fig 7f was generated using the ggballoonplot function of ggpubr,69 a package for ggplot270 in R.71 All box-plots were generated using the Seaborn65 software default settings.
CODE AVAILABILITY
The complete set of CellProfiler38,59 pipelines used as well as example input images for each are available at https://github.com/brianbeliveau/SABER. PD3D, a package of MATLAB functions for detecting SABER puncta (or other fluorescent puncta) in 3D and assigning puncta to cells in a watershed segmentation is available at https://github.com/ewest11/PD3D. Any remaining scripts for image processing and plotting will be made available upon request.
DATA AVAILABILITY
All raw and processed data will be made available upon request.
Supplementary Material
Acknowledgements:
The authors thank B. Fields, S. Kennedy, J. A. Abed, T. Wu, T. Ferrante, N. Liu, F. Dannenberg, M. Cicconet, P. M. Llopis and The Microscopy Resources on the North Quad (MicRoN) at Harvard Medical School for fruitful discussions and technical support. We also thank the ENCODE consortium and Dr. Stamatoyannopoulos (UW) for retina DHS data. This work was supported by the National Institutes of Health (under grants 1R01EB018659–01 to P.Y., 1UG3HL145600 to P.Y., 1R01GM124401 to P.Y., 1-U01-MH106011–01 to P.Y., 5K99EY028215–02 to S.W.L., and a T32 training grant GM096911 supporting E.R.W.), the Office of Naval Research (under grants N00014-16-1-2410 to P.Y. and N00014-18-1-2549 to P.Y.), the National Science Foundation (under grant CCF-1317291 to P.Y. and a Graduate Research Fellowship to J.Y.K.), the Howard Hughes Medical Institute (C.L.C.), the Damon Runyon Cancer Research Foundation (under a fellowship to B.J.B.), the Uehara Memorial Foundation (under a fellowship to H.M.S.), the Human Frontier Science Program (under fellowship LT000048/2016-L to S.K.S.), EMBO (under a fellowship ALTF 1278–2015 to S.K.S.), and the Wyss Institute’s Molecular Robotics Initiative (MRI) (P.Y., J.Y.K., B.J.B.).
Footnotes
Competing Financial Interests: A provisional US patent has been filed based on this work. P.Y. is co-founder of Ultivue Inc. and NuProbe Global.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts, Peng Yin (py@hms.harvard.edu), Constance L. Cepko (cepko@genetics.med.harvard.edu), and Brian J. Beliveau (beliveau@uw.edu). Step-by-step protocols, links to software, and additional resources will continue to be posted online at http://saber.fish or http://saber-fish.net/.
REFERENCES
- 1.Pardue ML & Gall JG Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proceedings of the National Academy of Sciences of the United States of America 64, 600–4 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riegel M Human molecular cytogenetics: From cells to nucleotides 37, 194–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolzer A et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biology 3, 0826–0842 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Femino AM, Fay FS, Fogarty K & Singer RH Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nature methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schröck E et al. Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Lubeck E & Cai L Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature methods 9, 743–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature Methods 11, 313 EP – (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schueder F et al. Universal Super-Resolution Multiplexing by DNA Exchange. Angewandte Chemie - International Edition 56, 4052–4055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y et al. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Letters 17, 6131–6139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bintu B et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Codeluppi S et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nature Methods 15, 932–935 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M & Cai L Single-cell in situ RNA profiling by sequential hybridization. Nature methods 11, 360–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque MJ & Raj A Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nature Methods 10, 246–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S et al. Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerstens HM, Poddighe PJ & Hanselaar AG A novel in situ hybridization signal amplification method based on the deposition of biotinylated tyramine. The journal of histochemistry and cytochemistry 43, 347–352 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Player AN, Shen SP, Kenny D, Antao VP & Kolberg JA Single-copy gene detection using branched DNA (bDNA) in situ hybridization. Journal of Histochemistry and Cytochemistry 49, 603–611 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Wang F et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. Journal of Molecular Diagnostics 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beliveau BJ et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nature Communications 6, 7147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lizardi PM et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification (1998). [DOI] [PubMed]
- 23.Dirks RM & Pierce NA Triggered amplification by hybridization chain reaction. Proceedings of the National Academy of Sciences 101, 15275–15278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi HMT et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nature Biotechnology 28, 1208 EP – (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi HM, Beck VA & Pierce NA Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah S et al. Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development 92, 2862–2867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouhanifard SH et al. ClampFISH detects individual nucleic acid molecules using click chemistry–based amplification. Nature Biotechnology 37, 84 EP – (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagendran M, Riordan DP, Harbury PB & Desai TJ Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife 7, e30510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishi JY, Schaus TE, Gopalkrishnan N, Xuan F & Yin P Programmable autonomous synthesis of single-stranded DNA. Nature Chemistry (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beliveau BJ et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proceedings of the National Academy of Sciences 109, 21301–21306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CS, Davis RW & Davidson N A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. Journal of Molecular Biology 48, 1–22 (1970). [DOI] [PubMed] [Google Scholar]
- 33.Beliveau BJ et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proceedings of the National Academy of Sciences (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Q, Schlabach MR, Hannon GJ & Elledge SJ Design of 240,000 orthogonal 25mer DNA barcode probes. Proceedings of the National Academy of Sciences 106, 2289–2294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dirks RM & Pierce NA A partition function algorithm for nucleic acid secondary structure including pseudoknots. Journal of Computational Chemistry 24, 1664–1677 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Dirks RM & Pierce NA An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. Journal of Computational Chemistry 25, 1295–1304 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Dirks RM, Bois JS, Schaeffer JM, Winfree E & Pierce NA Thermodynamic Analysis of Interacting Nucleic Acid Strands. SIAM Review 49, 65–88 (2007). [Google Scholar]
- 38.Carpenter AE et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shekhar K et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosaliganti KR, Noche RR, Xiong F, Swinburne IA & Megason SG ACME: automated cell morphology extractor for comprehensive reconstruction of cell membranes. PLoS computational biology 8, e1002780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solovei I et al. Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell 137, 356–368 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Shah S, Lubeck E, Zhou W & Cai L In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92, 342–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emerson MM & Cepko CL Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Developmental Biology 360, 241 – 255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium TEP et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57 EP – (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuda T & Cepko CL Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proceedings of the National Academy of Sciences 101, 16–22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saka SK et al. Highly multiplexed in situ protein imaging with signal amplification by Immuno-SABER. Preprint at https://www.biorxiv.org/content/10.1101/507566v1 (2018).
- 48.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107 EP – (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yildirim E, Sadreyev RI, Pinter SF & Lee JT X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nature Structural & Molecular Biology 19, 56–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent WJ et al. The Human Genome Browser at UCSC. Genome Research 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan AR & Hall IM BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marçais G & Kingsford C A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. Journal of Molecular Biology 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 55.Casanova M et al. Heterochromatin Reorganization during Early Mouse Development Requires a Single-Stranded Noncoding Transcript. Cell Reports 4, 1156–1167 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Beliveau BJ, Apostolopoulos N & Wu C Visualizing genomes with Oligopaint FISH probes. Current Protocols in Molecular Biology 2014, 14.23.1–14.23.20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ : 25 years of image analysis HISTORICAL commentary NIH Image to ImageJ : 25 years of image analysis. Nature Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McQuin C et al. CellProfiler 3.0: Next-generation image processing for biology. PLOS Biology 16, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linkert M et al. Metadata matters: access to image data in the real world. The Journal of Cell Biology 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yushkevich PA et al. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 31, 1116–1128 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Marr D & Hildreth E Theory of Edge Detection. Proceedings of the Royal Society of London Series B 207, 187–217 (1980). [DOI] [PubMed] [Google Scholar]
- 63.Plaisier S, Taschereau R, Wong J & Graeber T Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Research 38, e169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunter JD Matplotlib: A 2D graphics environment. Computing In Science & Engineering 9, 90–95 (2007). [Google Scholar]
- 65.Waskom M et al. mwaskom/seaborn: v0.8.1 (September 2017) (2017).
- 66.Oliphant TE A guide to NumPy (2006).
- 67.McKinney W Data Structures for Statistical Computing in Python In van der Walt S & Millman J (eds.) Proceedings of the 9th Python in Science Conference, 51 – 56 (2010). [Google Scholar]
- 68.Cock PJA et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kassambara A ggpubr: ‘ggplot2’ Based Publication Ready Plots (2018). R package version 0.1.7.
- 70.Wickham H ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; New York, 2016). [Google Scholar]
- 71.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed data will be made available upon request.