Introductory
Multiple Sclerosis (MS) is characterised by an immune system attack targeting myelin, which is produced by oligodendrocytes (OLs). We performed single-cell transcriptomic analysis of OL lineage cells from the spinal cord of mice induced with experimental autoimmune encephalomyelitis (EAE), which mimics several aspects of MS. We found unique OLs and OL precursor cells (OPCs) in EAE and uncovered several genes specifically alternatively spliced in these cells. Surprisingly, EAE-specific OL-lineage populations expressed genes involved in antigen processing and presentation via major histocompatibility complex class I and II (MHC-I and -II), and in immunoprotection, suggesting alternative functions of these cells in a disease context. Importantly, we found that disease-specific oligodendroglia are also present in human MS brains and that a substantial number of genes known to be susceptibility genes for MS, so far mainly associated with immune cells, are expressed in the OL lineage cells. Finally, we demonstrate that OPCs can phagocytose and that MHC-II expressing OPCs can activate memory and effector CD4+ T cells. Our results suggest that OLs and OPCs are not passive targets but instead active immunomodulators in MS. The disease-specific OL lineage cells, for which we identify several biomarkers, may represent novel direct targets for immunomodulatory therapeutic approaches in MS.
The adaptive immune system is currently thought to be the most likely aetiological component for MS, although microglia have been suggested to also have a role1,2. We have shown that OLs, whose myelin is thought to be a passive target of the immune system in MS, are heterogeneous in mouse3,4. To investigate whether specific OL populations are targeted in MS, we isolated single cells from the spinal cord of control (treated with Complete Freund's Adjuvant, CFA) or EAE mice (Fig. 1a). Cells from EAE mice were collected at the peak of the disease (score=3, indicating total hindlimb paralysis, Fig. 1b) and we performed Smart-seq25 single cell RNA-seq (Fig. 1a). OL-lineage cells were isolated by fluorescence activated cell sorting (FACS) GFP+ cells from EAE-induced Pdgfra-H2B-GFP transgenic mice6 (enriching for OPCs and “young” OLs, that had differentiated recently), and Pdgfra-Cre-LoxP-GFP7, (containing mainly “old” OL-lineage cells labelled with GFP since early development, but also “young” OLs and OPCs; Fig. 1a and Supplementary Fig. 1a).
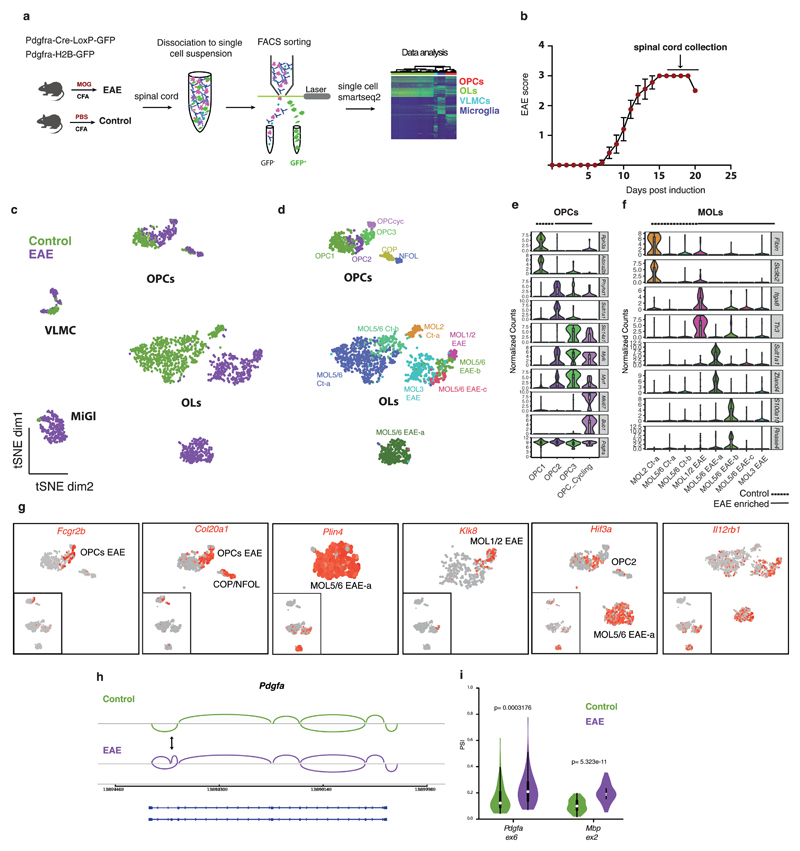
Figure 1. Single cell RNA-sequencing of oligodendrocyte (OL) lineage cells in response to experimental autoimmune encephalomyelitis (EAE) uncovers new disease-specific populations and disease markers.
a, Schematic overview of the methodology used to perform single-cell RNA-seq of the OL lineage cells. b, Clinical score of the diseased animals used in the study (n=12 mice; data represented as mean ± s.e.m.; only animals that reached score 3 and one that reached score 2.5 were used in this study). c, t-SNE plots of all cells sequenced showing the segregation of cells derived from Complete Freund's Adjuvant (CFA) controls and EAE (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 794 for controls and 971 for EAE). d, t-SNE plots of all cells sequenced representing different cell populations within OL lineage cells. Mature oligodendrocyte (MOL) identities were defined according to marker genes identified in ref. 4 (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 745 for controls and 707 for EAE). e-f, Violin plots depicting the expression of specific markers for OL precursor cells (OPC) (e) and for MOL clusters (f) (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 116 for OPC controls and 132 for OPC EAE and 626 for MOL controls and 575 for MOL EAE). Violin plots are centered around the median with interquartile ranges and shape represents cell distribution. g, t-SNE plots of disease-specific markers for OL lineage cells (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 745 for controls and 707 for EAE). h, Schematic representation of the exon 6 inclusion in the Pdgfa gene in response to EAE. i, Violin plot representing the PSI (proportion of spliced isoform) in controls (MOL2 Ct-a depicted in green) and EAE (MOL1/2 EAE depicted in purple) of the alternative spliced exons in Pdgfa and Mbp genes. PSI=0 means totally excluded and PSI=1 totally included (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 56 for MOL2 Ct-a and 49 for MOL1/2 EAE; Pdgfa ex6: p=0.0003176 and Mbp ex2: p=5.323e-11 by two-sided Wilcoxon rank sum test with continuity correction; Violin plots are centered around the median with interquartile ranges and shape represents cell distribution. VLMCs - vascular and leptomeningeal cells; MiGl - Microglia-like cells.
EAE mice spinal cord cells segregated from CFA control mice, with a subset of clusters uniquely found in EAE (Fig. 1c, Supplementary Fig. 1b,e). Clustering analysis (GeneFocus pipeline, see Methods and Supplementary Fig. 2) revealed thirteen OL lineage clusters, including four clusters of OPCs, one differentiation-committed oligodendrocyte precursor (COP), one newly formed OL (NFOL) and eight mature OL (MOLs; Fig. 1d and Supplementary Fig. 1b). We also identified vascular and leptomeningeal cells (VLMCs)3,4, consistent with the labelling of this population with Pdgfra, and microglia-like cells (which, while they could also include macrophages, we refer to as microglia; Fig. 1c and Supplementary Fig. 1b), which could reflect transient Pdgfra expression in these cells, or phagocytosis of myelin-associated GFP.
EAE-enriched OPC populations comprised OPCcyc (cycling), OPC2 and OPC3; distinct from the OPC1 population found in healthy mice (Supplementary Fig. 1e) that shares transcriptional profile similar to previously identified postnatal and adult mouse OPCs3,4 (Fig. 1e and Supplementary Fig. 3d-f). Dimensionality reduction using non-negative matrix factorization (NNMF) yielded a rank of 8 components (see Methods). Removal of components 7 and 8 that correlated to S and G2 phase cell-cycle genes revealed that OPCcyc can be further deconvoluted, suggesting that the OPCcyc population describes a mixture of cell state transitions from OPC1, OPC2 and OPC3 (Supplementary Fig. 3). Comparison of OPC populations showed specific expression of genes such as Rph3a, Adora2b in OPC1, Phyhd1, Sult1a1 in OPC2 and Slc14a1 in OPC3 (Fig. 1e and Supplementary Table 1). OPC2 and OPC3 clusters displayed unique expression of Fcgr2b, Mylk, Lgals1 (Fig. 1e, g, Supplementary Fig. 4a and Supplementary Table 1), when compared to OPC1, and increased levels of Myrf (Fig. 1e), a transcription factor necessary for myelination8, and decreased levels of Hes1 and Hes5 (Supplementary Fig. 4a), that keep OPCs in a progenitor state9. The COPs and NFOLs expressing Col20a1 (Fig. 1g) and Syt4 (Supplementary Fig. 4a) were mostly from EAE mice. Thus, OPCs undergo proliferation and differentiation in EAE10,11, but also transition to previously unidentified transcriptional states.
MOLs presented several markers identified in Marques et al.4, and could be subcategorized accordingly (Fig. 1d, f and Supplementary Fig. 1c). EAE-associated MOL lineage populations expressed unique genes, absent or very low expressed in healthy controls. Klk8, Itga8, Tlr3, Trim34a were enriched in MOL1/2-EAE; Plin4, Sult1a1 and Zfand4 in MOL5/6-EAE-a; and S100a10, RNAse4 and Tnfrsf1a, a MS susceptibility gene,12 in MOL5/6-EAE-b (Fig. 1f, g, Supplementary Fig. 4a and Supplementary Table 1). We found disease-specific markers in OL lineage cells such as Igtp, Nlrc5, and Serpina3n (Fig. 2b and Supplementary Fig. 4a). Il12rb1 was present in all MOL populations found in EAE mice (Fig. 1g). All OL lineage clusters comprised cells from both transgenic mouse lines, with the exception of MOL5/6-EAE-a, from Pdgfra-H2B-GFP mice, which might thus be composed of “young” OLs (Supplementary Fig. 1e).
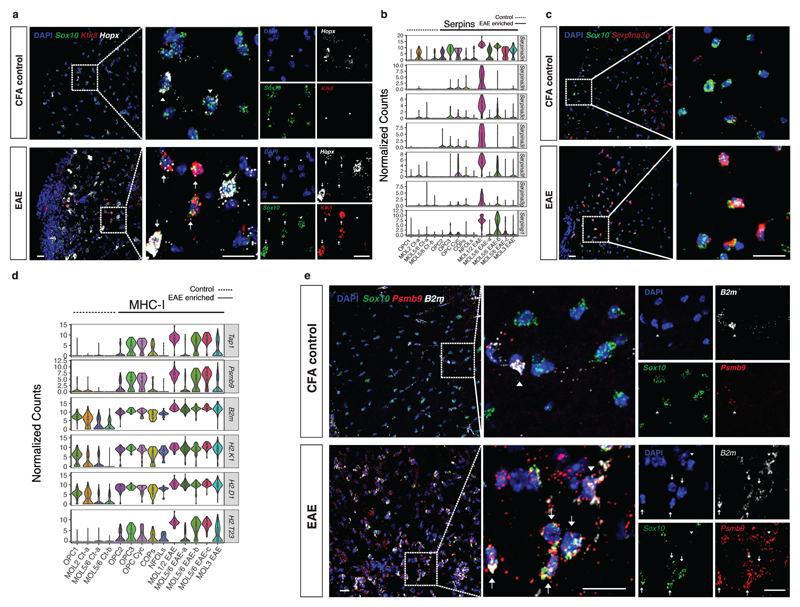
Figure 2. Expression of immunoprotective and adaptive immunity genes in response to EAE.
a, RNAscope ISH representing a spinal cord from CFA control and EAE mice marked with probes for Sox10, Klk8 and Hopx, markers of MOL1/2-EAE. Dashed boxes shown at higher magnification highlight regions with higher densities of MOL1/2 cells. Arrows depict triple positive cells in EAE and arrowheads depict Hopx positive OL lineage cells negative for Klk8. Representative images, n=3 biologically independent mouse spinal cord samples. Scale bars - 20μm b, Violin plots representing Serpina3 family of genes in OL lineage cell populations derived from CFA controls and EAE mice. (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 745 for controls and 707 for EAE). Violin plots are centered around the median with interquartile ranges and shape represents cell distribution. c, RNAscope ISH representing a spinal cord from CFA control and EAE mice marked with probes for Sox10 and Serpina3n. Representative images, n=3 biologically independent mouse spinal cord samples. Scale bars - 20μm. d, Violin plots representing MHC class I related genes in OL lineage cell populations derived from CFA controls and EAE mice. (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 745 for controls and 707 for EAE). Violin plots are centered around the median with interquartile ranges and shape represents cell distribution. e, RNAscope ISH representing a spinal cord from CFA control and EAE mice marked with probes for Sox10, and MHC-I related genes, Psmb9 and B2m. Arrows depict triple positive cells, arrowheads depict cells double positive for Psmb9/B2m and negative for Sox10 that do not belong to the OL lineage. Representative images, n=3 biologically independent mouse spinal cord samples, scale bars - 20μm.
To identify gene-transcription trends, we decomposed the dataset into components using NNMF (Supplementary Fig. 4b) and found 19 robust gene modules, including two that were uniquely associated with EAE: module 1, present in a subset of EAE cells comprising genes related to interferon response pathways and MHC-I and -II genes, and module 13 restricted to the MOL5/6-EAE-a population, which contained genes such as Plin4, Hif3a, and Fam107a (Supplementary Fig. 4b). Similar results were obtained when performing GO/Reactome analysis (Supplementary Fig. 4c and 5a, and Supplementary Table 3). We also uncovered 360 genes that were alternatively spliced in EAE OL-lineage cells, with either exon exclusion or inclusion (Supplementary Table 2). Gene Ontology (GO)/reactome analysis showed that genes with alternatively spliced exons in EAE OPCs were related to regulation of transcriptional elongation from RNA Pol II (exon exclusion), membrane trafficking (exon inclusion), and RNA-splicing components (exon exclusion; Supplementary Fig. 5b and Supplementary Table 3). We also found evidence of alternative splicing in EAE in several genes involved in myelination and MS, including the myelin genes Mbp (as previously reported13), Mobp, Pdgfa and the Ifih1 gene, in which MS-associated polymorphisms have been found14 (Fig. 1.h, i Supplementary Fig. 5c and Supplementary Table 2).
We performed RNAscope in situ hybridization (ISH) and immunohistochemistry (IHC) in EAE spinal cords, against several markers of the identified clusters, such as Klk8, which specifically marks MOL1/2-EAE (Fig. 1g) and Hopx, which marks MOL24. While control spinal cords exhibited sparse Klk8 expression we observed a strong induction of Klk8 in a subset of Hopx+/Sox10+ MOL2 in EAE mice (Fig. 2a). We also confirmed the presence of PLIN4 protein (enriched in MOL5/6 EAE-a) by performing IHC on spinal cords and found elevated GFP/PLIN4 double positive cells (from the Pdgfra-H2B-GFP mouse) and Sox10/PLIN4 (Supplementary Fig. 6a) in EAE mice. Thus, Klk8 and Plin4 are new markers of subpopulations of EAE-responsive MOLs.
All oligodendroglia in EAE displayed elevated Serpina3n expression (Fig. 2b), encoding a serine protease inhibitor that has been shown to reduce EAE severity and induce neuroprotection15. We confirmed the increase in Serpina3n in EAE by RNAscope ISH, especially notable within Sox10+ cells (Fig. 2c). The expression of this molecule reached such high levels in EAE, that we could no longer detect single dots. Interestingly, we found expression of additional genes of the Serpina3 family, including Serpina3h, Serpina3c, Serpina3i, Serpina3f and Serpina3g specific to MOL1/2-EAE (Fig. 2b), which could suggest that this population might be protected against direct damage by T- or NK-cells. This population also expresses the gene Serping1 (Fig. 2b) encoding for an inhibitor of complement activation, another possible mechanism of immunoprotection.
MHC-I genes such as H2-K1, H2-D1, H2-T23, B2m, and genes necessary for antigen processing and binding to MHC-I molecules, such as Psmb9, Tap1 and Tap2 were significantly increased upon EAE induction in OL lineage cells (Fig. 2d and Supplementary Table 1). Thus, EAE OLs might be direct targets of cytotoxic T cells, which is consistent with their expression of myelin epitopes. Surprisingly, however, we found that OPCs also exhibited a robust increase in the expression of these molecules (Fig. 2d), suggesting that OPCs might also be targeted during the disease, despite not expressing myelin proteins. To validate the expression of MHC-I molecules in oligodendroglia cells, we have performed RNAscope ISH and observed an upregulation of B2m molecules in Sox10+ cells in EAE spinal cords as well as the presence of Psmb9 molecules in Sox10+ cells in EAE that were absent in Sox10+ cells in control spinal cords (Fig. 2e).
MHC-II genes are thought to be restricted to microglia/macrophages in MS, and not present in OLs in active lesions16. Strikingly, we found the expression of all key genes required for a MHC-II mediated response, including H2-aa, H2-ab1, H2-eb1, Cd74 and Ctss, in specific subsets of OPCs and OLs (Fig. 3a). To confirm the presence of these RNA transcripts in OL lineage cells, we performed RNAscope ISH in the EAE and control spinal cords using probes targeting Cd74 and H2-Eb1 (for MHC-II), in combination with Aif1 (for microglia), Sox10 (for OL lineage cells), Klk6 (for MOL2) and Ptprz1 (for OPC) (Fig. 3b-d, Supplementary Fig. 6b, and Supplementary Videos V1-V6). While we observed microglia processes enwrapping OLs (Supplementary video V7), a small proportion of OLs/OPCs could be confidently assigned with double positive Cd74/Sox10 and few or no molecules for Aif1 (Fig. 3d and Supplementary Videos V1-V6). We also combined RNAscope ISH with immunohistochemistry and found triple positive OPCs for GFP (from EAE Pdgfra-H2B-GFP spinal cord), Ptprz1 and Cd74 (Fig. 3c and Supplementary video V2). We also performed IHC for MHC-II and the OL markers OLIG2 and SOX10, and identified double positive cells for OLIG2/MHC-II and SOX10/MHC-II oligodendroglia in the spinal cord of EAE mice (Fig. 3e and Supplementary Fig. 6c). We estimated that in the lesion areas of the white matter SOX10/MHC-II double oligodendroglia constitute about 3,4% (+/-1,76 SD) of the SOX10+ cells. We performed IBA-1 staining to distinguish between SOX10/MHC-II oligodendroglia and MHC-II expressing macrophages/microglia and we could distinguish the two cell types. Nevertheless, IBA-1 could still be observed in SOX10/MHC-II oligodendroglia although with a lower expression, which could be due to induction of some Aif1 molecules (corresponds to IBA-1 protein) in oligodendroglia in EAE, by for instance interferon-gamma (IFNγ)17. Control CFA-treated spinal cord OPCs/OLs did not express MHC-II molecules neither at the RNA (RNAscope ISH) nor protein level (IHC; Fig. 3b and data not shown).
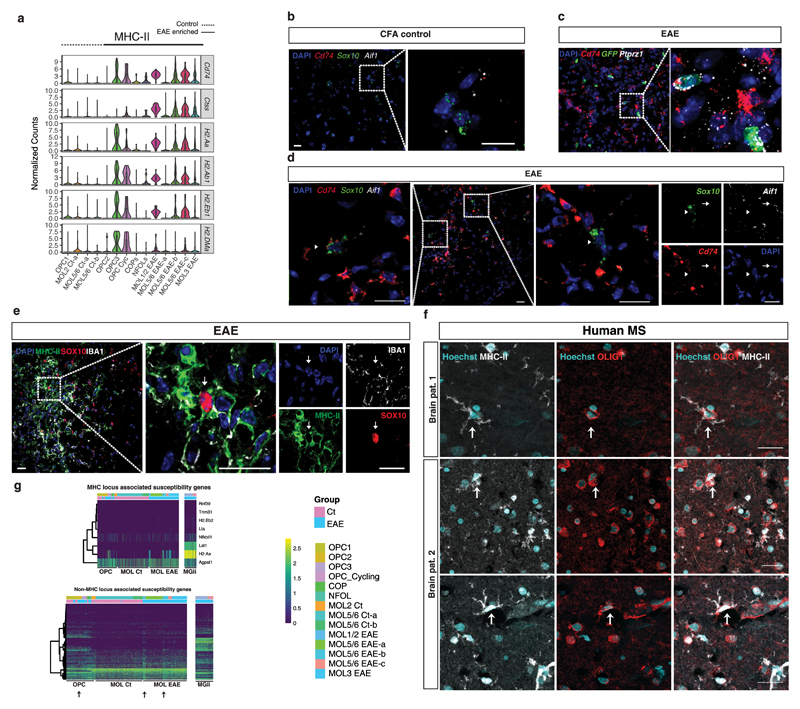
Figure 3. Expression of MHC class II and MS susceptibility genes in the OL lineage cells in response to EAE and in MS.
a Violin plots representing MHC class II related genes in OL lineage cell populations derived from CFA controls and EAE mice. (n=4 biologically independent mouse spinal cord samples per condition; total number of cells is 745 for controls and 707 for EAE). Violin plots are centered around the median with interquartile ranges and shape represents cell distribution. b-d, RNAscope ISH representing spinal cords from CFA control and EAE mice marked with probes for MHC-II (Cd74), OL lineage cells (Sox10 and Ptprz1/Pdgfra-H2B-GFP specific for OPCs) and microglia (Aif1). Dashed boxes shown at higher magnification represent both OL lineage cells expressing MHC-II (arrowhead), and microglia cells expressing MHC-II (arrow). Representative images, n=3 biologically independent mouse spinal cord samples per condition, scale bars - 20μm. e, Immunohistochemistry showing OL lineage cells (positive for Sox10) co-expressing MHC-II protein, in a lesion in the spinal cord of EAE mice. Representative images, n=3 biologically independent mouse spinal cord samples per condition, scale bars - 20μm. f, Immunohistochemistry in human samples from two MS patients representing OLIG1 positive OLs expressing MHC-II (arrows). Representative images, n=2 biologically independent human brain samples. Scale bars - 25μm. g, Expression-based heat maps for the MS susceptibility genes (from ref. 1) in microglia and OL lineage cells. MHC locus and non-MHC locus related genes are represented in both microglia and all OL lineage cells populations analyzed in this study.
We found several interferon responsive genes in MHC-II expressing OL/OPC populations, such as Ifih1, Iigp1, Trim34, Irf7, Irgm1, Irgm2, Igtp and Zbp1 (Supplementary Fig. 7a). It has been shown that IFNγ can trigger endoplasmic reticulum stress in OLs18. Indeed, we found many genes involved in protein processing in endoplasmic reticulum differentially expressed in EAE (Supplementary Fig. 7b). Transcription factors such as Nlrc5, a transactivator of the MHC-I19, was expressed upon disease in all OL lineage cells, and Ciita, induced by the IFNγ pathway and activator of MHC-II genes, was expressed in a subset of these cells (Supplementary Fig. 4a). Thus, OL/OPC populations in the spinal cord appear to have the capacity to activate MHC genes upon EAE induction.
To determine if these populations also occur in human MS, we performed IHC for MHC-II and the OL markers OLIG2 and OLIG1 in postmortem brain tissue from two MS patients. As in mouse model EAE, we observed cells in which immune cell-derived MHC-II processes appear to touch/enwrap OLIG2+ nuclei (Supplementary Fig. 6d arrowheads and Supplementary video V8) but also OLIG2 and OLIG1 cells positive for MHC-II without any immune cell in their neighborhood (Fig. 3f and Supplementary Fig. 6d arrows, and Supplementary video V9), indicating that indeed human OLs can express adaptive immunity proteins in the context of MS.
Several MS susceptibility variants including MHC and autosomal non-MHC locus and the associated putative susceptibility genes were recently described1. Comparison of these loci/genes with RNA-seq from bulk brain led to the inference that peripheral immune system cells and brain resident microglia were the most likely cell types to contribute to MS susceptibility1. To examine if MS susceptibility genes were also expressed in OL lineage cells in EAE, we analyzed the expression of the mouse homologs for genes associated with the 200 non-MHC, 32 MHC loci, and 19 X-chromosomal loci in our single-cell transcriptomics dataset (Fig. 3g and Supplementary Fig. 6e and Supplementary Table S4). Interestingly, the MHC-II gene H2.Aa (human HLA-DQA1) was enriched not only in microglia, but also in EAE-derived OPCs and MOLs (Fig. 3g). Strikingly, several OL lineage cells expressed non-MHC locus associated susceptibility genes (Fig. 3g and Supplementary Table S4). This was particular strong for OPCs (importantly both from control and EAE mice) and MOL1/2-EAE populations (Fig. 3g). OPCs expressed 61% of these genes, which was in the same range as microglia (67%). Moreover, while we found many genes are equally expressed in all the populations, there were also genes enriched in OL lineage cells (such as Bcas1 and Sox8), MOLs (Prr5l), OPCs (such as Kcnh8, Pkia, Pitpnm2) and microglia (such as Plek; Supplementary Table S4). Interestingly, a subset of genes starts to be expressed and is upregulated in response to the disease in MOL populations, in particular MOL1/2-EAE.
To investigate the mechanism triggering MHC-II expression in OL lineage cells and the functional implications in the development of the disease, we performed co-cultures of GFP+ OPCs (from Sox10Cre-GFP mice brains) and CD45+ immune cells isolated from the spinal cord of EAE mice (Fig. 4a). After 72 hours of co-culture, 4.4% (+/-2.42 SD) of the total GFP+ OPCs in culture were positive for MHC-II (Fig. 4b). We also performed co-cultures of OPCs with CD45+ immune cells from CFA mice (most likely comprising of microglia/macrophages and few invading leukocytes from the periphery) and did not find any GFP/MHC-II double positive OPCs in these conditions (Supplementary Fig. 8a). This indicates that factors secreted by EAE-specific immune cells are inducing MHC-II expression in OPCs. Indeed, while MHC-II expression has been reported to be excluded from OLs in MS16, expression of these molecules in rat OPCs and OLs has been observed, upon dexamethasone and IFNγ treatment20. As CD4 lymphocytes such as Th1 and memory T cells are interferon-producing cells, we hypothesized that the observed effect in the co-cultures was mediated by lymphocyte derived IFNγ. As such, we treated OPCs with IFNγ (100 ng/ml), dexamethasone (1μM) and the combination of both for 3 days (Fig. 4c). We observed the induction of expression of MHC-II molecules, in OPCs and OLs, only in IFNγ treated cells, alone or in combination with dexamethasone, both at the RNA level (RNAscope ISH) and protein level (shown by ICC; Fig. 4d and Supplementary Fig. 8b-c). We did not find any positive cell for Aif1 RNA molecules in our MACS OPC primary cultures, nor IBA-1 positive cells, indicating that microglia were absent and excluding the possibility that MHC-II RNA and protein were derived from microglia processes. qPCR analysis further confirmed these results. While genes such as Plin4, were not affected by IFNγ treatment but only to dexamethasone, we observed a dramatic induction of interferon responsive genes (Ifit2, Ifih1) MHC-II genes (Cd74) and MHC-II and I transactivators (Ciita, Nlrc5) (Fig. 4e). Thus, our results indicate that MHC-II genes induction in EAE OL lineage cells is possibly mediated by IFNγ.
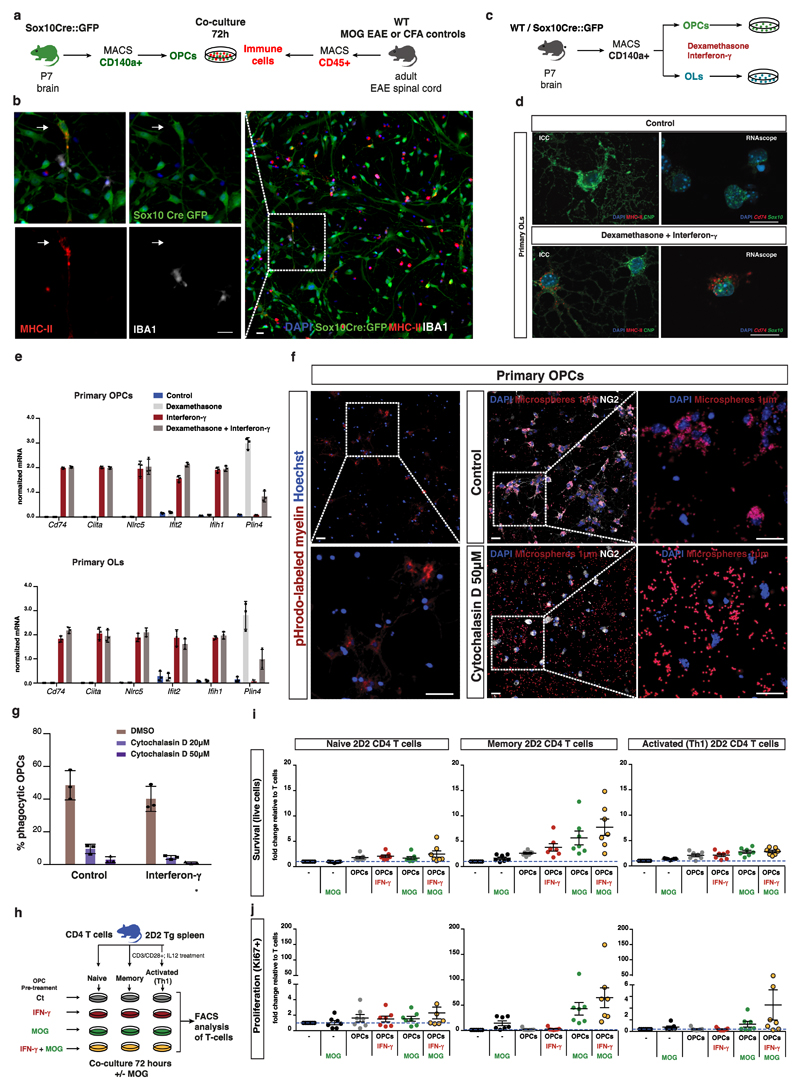
Figure 4. OPCs express MHC-II in response to interferon-γ, exhibit phagocytic capacity and regulate T cell survival and proliferation.
a, Schematic representation of co-culture between OPCs isolated from Sox10-Cre-GFP mice and CD45+ immune cells isolated from EAE mice or CFA controls. b, Immunohistochemistry showing OL lineage cells (positive for Sox10) co-expressing MHC-II protein, but not the microglia marker IBA1, upon co-culture with CD45+ immune cells from EAE mice. Representative images, n=3 independent experiments, scale bars - 20μm. c, Schematic representation of treatment of OPCs and OLs with interferon-γ (100 ng/ml) and dexamethasone (1μM). d, OLs cultured with dexamethasone and interferon-γ for 72h express MHC-II as represented by Cd74/Sox10 and MHC-II/CNP double staining in RNAscope ISH and immunocytochemistry, respectively. Representative images, n=3 independent experiments. Scale bars - 20μm. e, qRT-PCR analysis for MHC and interferon responsive genes on primary OPCs and OLs treated with interferon-γ and dexamethasone, n=3 independent experiments per condition; data represented as mean ± s.d. f, Uptake of pHrodo-labeled myelin and 1μm diameter fluorescent microspheres by primary NG2+ OPCs after 6 and 24 hours, respectively, and upon treatment with 50μM of cytochalasin D. Representative images, n=3 independent experiments. Scale bars - 20μm. g, Quantification of the percentage of OPCs uptaking microspheres in the presence or absence of IFN–γ (100 ng/ml) and upon treatment with 20 and 50μM of cytochalasin D. n=3 independent experiments per condition; data represented as mean ± s.d. h, Schematic representation of co-culture between primary OPCs, treated with interferon-γ and/or MOG 35-55 peptide, and three different types of CD4 lymphocytes (naïve, memory and activated Th1) from 2D2 transgenic mice, where a high proportion of CD4 T-lymphocytes express a T-cell receptor specific for MOG35-55 peptide. i-j, Graph plots obtained from FACS analysis of naïve, memory and activated Th1 2D2 CD4 T cells after 72 hours of co-culture with OPCs. General survival (dead cell exclusion marker) and proliferation (CD4+ Vβ11+ Ki67+) were assessed. Numbers of cells in the live gate as well as in the Ki67+ gate were estimated and reflect survival and proliferation since the same cell numbers were seeded onto different wells. Plots represent the averages of the assessed values for the different conditions divided by the values of the control (T cells only) for fold change differences. n=7 independent experiments; data represented as mean ± s.e.m.
To uncover the role of immunocompetent OPCs in the disease, we first investigated if OPCs were capable of phagocytosis, by adding 1μm diameter fluorescent microspheres to OPC cultures for 24h. OPCs could indeed uptake several microspheres, with at least 48% of them exhibiting this capability (Fig. 4f). Moreover, treatment with the phagocytosis inhibitor cytochalasin D at two different concentrations for 24 hours, lead to a remarkable reduction in microsphere uptake (Fig. 4f, g). IFNγ treatment did not alter OPC phagocytic capacity (Fig. 4g). To further investigate whether OPCs can phagocytise myelin, we incubated OPCs for 6 hours with pHrodo-labeled myelin (a pH-sensitive fluorogenic dye that fluoresce red in phagosomes21) and found that OPCs were able to uptake myelin as observed by the red fluorescence staining of live OPCs (Fig. 4f). Altogether, these data demonstrate that OPCs exhibit phagocytic activity and strongly suggests that OPCs are capable of taking up myelin debris in a disease context.
Next, we addressed the impact of MHC-II expressing OPCs on the survival, proliferation and cytokine production of CD4+ T cells derived from 2D2 mice, where T cells express the T cell receptor for the MOG35-55 peptide22. OPCs (controls or pre-treated with IFNγ and/or MOG35-55 peptide) were co-cultured for 72 hours with either naïve, memory or effector T cells (naïve cells that had been pre-activated in vitro in the presence of IL-12, acquiring a Th1 phenotype). T cells were subsequently assessed by flow cytometry for survival (cell dead exclusion), proliferation (Ki67+), as well as for expression of IFNγ and TNF (Fig. 4h). Co-culturing with non-stimulated OPCs lead to an increase in numbers of surviving naïve, activated and memory T cells. Survival was further enhanced in memory T cells when OPCs were pre-treated in IFNγ and MOG peptide suggesting an MHC-II mediated effect (Fig. 4i, Supplementary Figs. 8e and 9). Regarding proliferation, naïve CD4+ T cells seemed unaffected by OPC co-culture (Fig. 4j and Supplementary Figs. 8e and 9), as expected, since these cells have a high threshold of activation provided in secondary lymphoid tissue. In contrast, we found a higher number of Ki67+ memory CD4+ T cells when co-cultured with OPCs in the presence of MOG peptide. Similar effects were observed for Th1 CD4+ T cells which proliferated more in the presence of OPCs and MOG peptide. These results also suggest that the proliferation effect is via OPC-MOG peptide presentation to memory and Th1 T cells. Of notice, the presence of MOG peptide, not necessarily together with IFNγ, was enough to induce proliferation. This indicates that production of IFNγ by memory and Th1 cells may provide a positive regulatory feedback loop. We thus investigated if the increase in survival/proliferation was accompanied by cytokine production. Indeed, memory CD4+ T cells displayed more cells producing both TNF and IFNγ when co-cultured with OPCs in the presence of MOG peptide (Supplementary Fig. 8d). In contrast, the presence of OPCs per se triggered an increase in the number of Th1 CD4+ T cells producing IFNγ and TNF (Supplementary Fig. 8d).
OLs are widely seen as a passive target of a dysregulated immune system, but there is a growing body of evidence of immunomodulatory factors expressed in OLs such as cytokines/chemokines and their receptors23–27. Our results indicate that both OPCs and MOLs undergo a transcriptional overhaul during chronic inflammatory demyelination. The selective expression of immunoprotective, innate and adaptive immunity genes in OPCs and OLs in EAE suggests potential mechanisms of protection and an immune function in the context of disease. Since several of these genes are susceptibility genes for MS, the OL lineage might have a more central role in the origin and progression of MS than previously thought.
OL cell death has been recently shown to lead to an adaptive auto-immune response28, which could imply an initial dysregulation in cells of the OL lineage as a triggering event in MS and support the “inside-out” hypothesis for the aetiology of the disease28. Whether the new identified OL lineage cell states play a role on the aetiology of MS, by acting as antigen presenting cells and triggering immunologic attack or reinforcing the disease-initiating event from the periphery, or both, will require further investigation. In either case, our results indicate that OL lineage cells may not simply be passive targets of the immune system in MS, but rather central players which may be targeted in therapeutic approaches for MS.
Online Methods
Information regarding methods used in this paper can be found below and also in the Life Sciences Reporting Summary.
Animals
Mouse lines used in this study included C57BL/6NJ wild type (WT) mice, Pdgfra-Cre-LoxP-GFP7, Pdgfra-H2B-GFP knock-in mice6, Sox10Cre-LoxP-GFP29 and C57Bl/6 2D2 transgenic mice22. Pdgfra-Cre-LoxP-GFP mice are a strain of mice obtained originally by crossing mice with Cre recombinase under the control of a Pdgfra genomic DNA fragment (with a C57BL/6NJ genetic background; The Jackson Laboratories, CA, USA) with reporter mice RCE:loxP-GFP (with CD1 background; Gord Fishell, NYU Neuroscience Institute) to label the complete OL lineage. Sox10-Cre-LoxP-GFP mice are a strain of mice obtained originally by crossing mice with Cre recombinase under the control of the Sox10 promoter (The Jackson Laboratories, CA, USA; with a C57BL/6 genetic background) with reporter mice RCE:loxP-GFP (with CD1 background) to label the complete OL lineage. Pdgfra-H2B-GFP, with a C57BL/6NJ background, presents an H2B-eGFP fusion gene expressed under the promoter of the OPC marker, Pdgfra. Mice homozygous for the knock-in targeted mutation have an embryonic lethal phenotype, with half of the embryos failing to survive past embryonic day 12.5 and the remainder failing to survive beyond embryonic day 15.5 (https://www.jax.org/strain/007669). Only heterozygote mice were used, in which Pdgfra is expressed mainly in OPCs but also in some extent in the early stages of OL differentiation, due to GFP half-life. Animals were used in adult stage, between 10-12 weeks old and both genders were included. The following light/dark cycle was used: dawn 6.00-7.00; daylight 07.00-18.00; dusk 18-00-19.00; night 19.00-06.00. A maximum of 5 adult mice per IVC-cage of type II Allentown. Breedings were done with 1 male and up to 2 females. All experimental procedures performed followed the guidelines and recommendations of local animal protection legislation and were approved by the local committee for ethical experiments on laboratory animals in Sweden (Stockholms Norra Djurförsöksetiska nämnd).
Experimental Autoimmune Encephalomyelitis (EAE)
For the induction of chronic EAE, animals were injected subcutaneously with an emulsion of MOG35-55 in complete Freud’s adjuvant (CFA; EK-2110 kit from Hooke Laboratories) followed by the administration of pertussis toxin in PBS (0,2μg per animal), for two consecutive days (all according to manufacturer’s instructions). Spinal cords and cerebellum were collected at the peak/chronic stage of the disease with clinical score=3 representing limp tail and complete paralysis of hind legs. Animals that did not reach this clinical score were not analyzed in this study. Additional animals were injected subcutaneously with a CFA emulsion (CK-2110 kit from Hooke Laboratories) and analyzed as controls.
Tissue dissociation for single-cell RNA sequencing, FACS analysis and sequencing data processing
Cells were isolated from the spinal cord of P90 Pdgfra-H2B-GFP and Pdgfra-Cre-LoxP-GFP mice. Tissue was then dissociated into a single cell suspension, as previously described in Marques et al 2016, with some modifications. Mice were perfused with oxygenated cutting solution (87 mM, NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 75 mM sucrose, 20 mM glucose, 1 mMCaCl2, and 2 mM MgSO4) and the brain was quickly dissected and dissociated using the Adult brain dissociation kit from Miltenyi (130-107-677) following the manufacturer’s instructions (red blood cells removal step was not included). After debris removal, cell suspension was filtered with 30μm filter (Partec) and processed by FACS. Spinal cord single GFP+ cells were selected in a BD Influx sorter and collected into a 384 plate for SmartSeq2, according to procedures described in5. SmartSeq2 raw data was processed according to procedures described in5. Smart-seq2 paired-end reads were trimmed with cutadapt 1.8.030 and aligned with STAR 2.5.1b31 to mm10 genome assembly, only uniquely mapped reads were used for downstream analyses.
Primary OPC cell culture
Mice brains from P7 pups were removed and dissociated in single-cell suspensions using the Neural Tissue Dissociation Kit (P; Miltenyi Biotec, 130-092-628) according to the manufacturer’s protocol. OPCs were obtained with MACS with CD140a microbeads following the manufacturer’s protocol (CD140a Microbead kit, Miltenyi Biotec 130-101-547). Alternatively to MACS, GFP+ OPCs derived from Sox10-GFP mice were collected with FACS following brain dissociation. Cells were seeded in poly-L-lysine (O/N; Sigma P4707) plus fibronectin (1h; Sigma F1141) coated dishes and grown on proliferation media comprising DMEM/Gmax (ThermoFisher Scientific 10565018), N2 media (ThermoFisher Scientific 17502048), Pen/Strep (ThermoFisher Scientific 15140122), NeuroBrew (Miltenyi 130-093-566), bFGF 20ng/ml (Peprotech 100-18B) and PDGF-AA 10ng/ml (Peprotech, 100-13A). For OPC differentiation, cells were left for 2 days in medium without bFGF and PDGF-AA. Cells were treated either with Dexamethasone (1μM), Interferon-gamma (100ng/mL; R&D, 485-MI-100) or the combination of the two for 72 hours. For the phagocytosis experiments, 1μL per ml of media of pHrodo-labeled myelin and 0.5μL per ml of media of Fluoresbrite® Polychromatic Red Microspheres 1μm (Polysciences, Inc, 18660-5) were added to OPCs for 6h and 24h, respectively. Hoechst (ThermoFisher Scientific, 62249) was added at a dilution 1:1000 to OPCs incubated with pHrodo-labeled myelin for live-cell fluorescent staining of the DNA and nuclei. Phagocytosis inhibition experiments were performed by adding two different concentrations of cytochalasin D (Sigma, C8273) 20μM and 50μM in the presence or absence of Interferon-gamma (100ng/mL) for 24 hours. Cells were thoroughly washed before fixation.
pHrodo-labeled myelin preparation: myelin sheaths were obtained by dissection and mechanical homogenization of PBS perfused mouse brains. Pure myelin was then isolated by several steps of density separation by ultra-centrifugation in sucrose solutions32,33. After several cleaning steps we incubated the purified myelin with pHrodo red (Thermo Fisher, P36600) for 30 minutes following the manual instructions, followed by extensive washing in PBS/Hepes.
OPC/CD45+ EAE co-culture experiments
CFA and EAE mice were perfused in PBS and the spinal cords were collected. Immune cells (lymphocytes, monocytes and macrophages) were then isolated using the adult Brain Dissociation Kit (Miltenyi Biotec, 130-107-677) according to the manufacturer’s protocol and collected with MACS with CD45+ microbeads (CD45 Microbead kit, Miltenyi Biotec 130-052-301) according to the manufacturer’s protocol. CD45+ cells (150-200,000) were added on top of cultured OPCs (300,000 cells) and incubated for 72hours.
OPC/T cell co-culture experiments
Naïve and memory T cells were isolated from C57Bl/6 2D2 transgenic mice that express a TCR specific for the MOG 35-55 peptide22. Splenocytes were isolated by tissue disruption through a 100μm mesh and erythrocytes were lysed with ACK buffer (Invitrogen). Splenocytes were depleted from all non-T cells using a naive CD4+ T cell Isolation Kit (130-104-453; Miltenyi), and the flow through was further sorted into CD44 low (naïve) and CD44 high (memory) cells using anti-CD44 beads from the same kit. Cells were frozen in FCS containing 10% DMSO for subsequent experiments. Activated T cells were generated from naïve cells stimulated in vitro with plate-bound anti-CD3 (BD; 555273; 2μg/ml) and soluble anti-CD28 (BD; 553294; 1μg/ml) in the presence of 10ng/ml IL-12 (R&D Systems, 419-ML) for 3 days, followed by 3 days expansion in 10ng/ml IL-2 (R&D Systems, 402-ML) and cytokine starvation for 24h prior to co-culture (at the exception of one n which was not deprived from cytokines).
OPCs (6 n’s obtained with MACS and one n with FACS of GFP+ cells from Sox10-GFP mice) were seeded at a density of 20,000 cells per well in a 96-well plate and pre-treated with interferon-gamma (100ng/mL) and/or MOG 35-55 peptide (Anaspec, AS-60130-5) at 40μg/ml for 72hours. Cells were washed twice in PBS. Naïve, memory and activated T cells were both seeded alone (with or without MOG 35-55 peptide at 40μg/ml) and on top of control or stimulated OPCs at 2x105 cells/well in 96 well plates with MOG 35-55 peptide at 40μg/ml and incubated at 37°C. Media used for T cells as well as for T cell:OPC co-culture was RPMI (Sigma, R8758), 10% FBS (v/v; Sigma F7524), Penicillin-Streptomycin (Sigma, P4458, 100 U/ml). After 72h, cells were stimulated for 5h at 37°C in media containing 50ng/ml PMA (Sigma, P1585), 1μg/ml Ionomycin (Sigma, I0634) and 1μg/ml GolgiPlug (BD, 555029), to boost cytokine secretion, followed by staining for CD4 PE-Dazzle (BioLegend, 100566), Ki67 V450 (BD, 561281), anti-TCR Vbeta11 BV510 (BD, 743677), TNF PE-Cy7 (eBioscience, 25-7423-82), IFNg APC (BD, 554413) as well as dead cell exclusion dye near-IR (Invitrogen, L10119). Surface staining at 4°C for 30 minutes was followed by fixation and permeabilization using an intracellular staining kit (eBioscience, 88-8824-00), followed by intracellular staining. For the relative assessment of cell survival and proliferation, samples were resuspended in equal volume and acquired by fixed time with a Gallios Flow Cytometer (Beckman Coulter) and analyzed using a Kaluza Flow Cytometry Analysis Software (Beckman Coulter). Numbers of cells in the live gate as well as in the Ki67-positive gate reflect survival and proliferation since the same cell numbers were seeded onto different wells. All conditions were run in three biological replicates for each of the seven experiments. Averages of the assessed values for the different conditions were divided by the values of the control (T cells only) for fold differences.
RNA extraction, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Spinal cord and cerebellum were collected from WT control and EAE mice. OPCs and OLs from in vitro cultures were collected in 700μl Qiazol. RNA was extracted with the miRNeasy microkit (Qiagen, 217084) according to manufacturer’s protocols. Contaminating DNA was degraded by treatment of the samples with RNase-free DNase (Qiagen, 79254) in column. 0.35-1µg of RNA from each sample was reversed transcribed for 1h with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813) including RNase inhibitor (Applied Biosystems, N8080119). A reverse transcriptase negative (RT-) control was included for each sample. Both the cDNA and the RT- were diluted 1:5 in RNase/DNAse free water for qRT-PCR.
qRT-PCR reactions were run on a StepOnePlus™ System (Applied Biosystems) in duplicate and with RT- reactions to control for genomic DNA. Fast SYBR® Green Master Mix (Applied Biosystems, 4385616) was used according to the manufacturer’s instructions, each PCR reaction had a final volume of 10μl and 1–2.5μl of diluted cDNA or RT-. The running conditions were 20 seconds at 95°C, followed by 40 cycles of 3 seconds of 95°C and 30 seconds of 60°C, then 15 seconds at 95°C, 1 minute at 60°C and 15 seconds at 95°C. A melting curve was obtained for each PCR product after each run, to control for primer dimers and gene-specific peaks. Tbp and Gapdh were run as housekeeping genes. Relative standard curves were generated for each gene to determine relative expression (CT values are converted to arbitrary quantities of initial template per sample). Expression levels were then obtained by dividing the quantity by the value of the geometric mean of the housekeeping genes. PCR primers sequences (design according 34) used are the following: Pdgfa short (F: CGTCAAGTGCCAGCCTTC and R: GCACACTCCAGGTGTTCCTC), Pdgfa long (F: TGAAAGAGGTCCAGGTGAGG and R: CCTTTTCCTTTTCCGCTTTT), Mbp exon2 (F: GCTTCTTTAGCGGTGACAGG and R: CCTTGTACATGTGGCACAGC), Mbp exon1 (F: TGGCCACAGCAAGTACCAT and R: AGTCAAGGATGCCCGTGT), Tbp (F: GGGGAGCTGTGATGTGAAGT and R: CCAGGAAATAATTCTGGCTCA), Gapdh (F: GAGAAACCTGCCAAGTA and R: AGACAACCTGGTCCTCA), Cd74 (F: CTGGATGAAGCAGTGGCTCT and R: CCCAGGCCAGAAGATAGGTC), Ciita (F: CTGGCACAGGTCTCTCCAGT and R: TACTGAGGCTGCTTGAAGGG), Nlrc5 (F: CCGTGGTACTCACATTTGCC and R: CCTTCGAGATCTCTGGGACA), Ifit2 (F: AAGGCAGAGGAAGAGGTTGC and GTCGCAGATTGCTCTCCAGT), Ifih1 (F: ATGTCTTGGACACTTGCTTCG and R: CTGACTCATTCCCGCTGTTT) and Plin4 (F: ACACAGTGGCCACAGGACTT and R: GGTCACCGTGTCCTTAGTGC).
Tissue processing for Immunofluorescence and RNAscope ISH
For immunohistochemistry and RNAscope ISH mice were perfused with PBS followed by 4% PFA. Spinal cords from EAE and control mice were dissected and post-fixed with 4% PFA for 1h, at 4ºC. The tissues were embedded into OCT (Tissue-Tek), frozen in dry ice and sectioned coronally (20 μm thickness). Sections were stored at –80°C.
For immunocytochemistry and RNAscope ISH in cultured cells, cells were fixed in 4% formaldehyde for 10 minutes and washed in PBS.
Immunohistochemistry and immunocytochemistry (mouse and human samples)
For immunocytochemistry cells were incubated overnight at 4ºC with the following primary antibodies: CNP (Abcam ab6319, Mouse 1:200), MHC-II (anti-I-A/I-E; BD Bioscience rat 1:600), GFP (Abcam ab13970, Chicken 1:1000), NG2 (Millipore AB5320, Rabbit 1:200), IBA-1 (Wako 019-19741, rabbit 1:400) in PBS/0.5%Triton/10% normal donkey serum (Sigma, D9663). Cells were washed with PBS and then incubated for 2 hours with Alexa Fluor-conjugated antibodies (Invitrogen).
Spinal cord sections were incubated overnight at 4ºC in the following primary antibodies: OLIG2 (R&D, Goat 1:200), SOX10 (R&D, Goat 1:100), PLIN4 (Sigma, Rabbit 1:200), IBA-1 (Wako 019-19741, rabbit 1:400) and MHC-II (anti-I-A/I-E; BD Bioscience rat 1:600) diluted in PBS/0.5% Triton/10% normal donkey serum. After washing the section with PBS, secondary Alexa Fluor-conjugated antibodies (Invitrogen) diluted in PBS/0.5% Triton/10% normal donkey serum were incubated for 2h at room temperature. Slides were mounted with mounting medium containing DAPI (Vector, H-1200) and kept at 4ºC until further microscopic analysis. Images from sections were taken of the spinal cord using a Zeiss LSM700 Confocal, and processed in ImageJ.
Human brain 4μm paraffin sections, from regions with high microglia activity (thus active lesions) were dewaxed in a descending EtOH-row. After antigen retrieval and endogenous peroxidase quenching, they were incubated over night at 4 degrees with the following primary antibodies: OLIG1 (Abcam, 68105, 1:100), OLIG2 (Atlas, HPA003254, 1:200) and MHC-II (Dako, M0775 1:100) diluted in TBS/0.3% Triton/20% normal horse serum. After washing with TBS/0.001% Triton, they were incubated for 1 hour at room temperature with Goat F(ab) Anti-Rabbit IgG H&L (HRP) and Goat F(ab) Anti-Mouse IgG H&L (HRP; ab7171 and ab6823 abcam, 1:500). After washing in TBS/0.001% Triton, the fluorescence was visualized with the tyramide kits from Perkin Elmer (NEL745B001KT and NEL744B001KT). The human tissue used in this study comprised one female and one male, with ages of 38 and 50 years old both with secondary progressive multiple Sclerosis. No patient had specific disease treatment. Post mortem MS tissue was provided via a UK prospective donor scheme with full ethical approval from the UK Multiple Sclerosis Tissue Bank (MREC/02/02/39).
RNAscope ISH
RNAscope ISH was performed on cultured cells and spinal cord sections from controls and EAE mice with probes for mouse Cd74, H2-Eb1, Aif1, Sox10, Klk6, Hopx, Ptpzr1, Serpina3n, B2m, Psmb9 and Klk8 all purchased from ACD. RNAscope ISH protocol for sections was performed following manufacturer’s instructions with minor modifications (ACD, RNAscope® Multiplex Fluorescent Detection Reagents v2 Cat. No.323110). Briefly, sections were placed on a hot plate (100°C) with 1x target retrieval (Pretreatment Reagents Cat. No. 322381 and 322000) for 5 min followed by 2 steps of washes of 2 min and 1 wash with 100% ethanol for 2min. Protease IV was applied on top of the sections and incubated for 20min at RT followed by 2 washes of 2 min each. Probes were diluted 1:50 in the C1 probe, hybridized for 2h at 40°C and washed twice in wash buffer (RNAscope® Wash Buffer Reagents Cat. No. 310091). Amplification steps were performed by incubating with v2Amp1 (30min), v2Amp2 (30min) and v2Amp3 (15min) at 40°C with washes of 2x2min in between steps (RNAscope® Multiplex Fluorescent Detection Reagents v2 Cat. No. 323110). Sections were incubated v2HRP-C1 for 15min at 40°C and washed twice in wash buffer for 2min. TSA conjugated fluorophores were diluted in 1:1500 in TSA buffer (RNAscope® Multiplex TSA Buffer Cat. No. 322809) and incubated for 30min at 40°C followed by 2 washes of 2min and HRP blocker incubation for 30min at 40°C. The last steps were performed subsequently for v2HRP-C2 and v2-HRP-C3. To combine the RNAscope ISH with immunofluorescence for GFP, sections were further blocked in block with 5% NDS in 0.3% PBS-Tx100, 1h, RT and incubated with chicken anti-GFP (abcam, ab13970 1:200,) overnight at 4°C. Sections were further incubated with goat anti-chicken (Alexa Fluor 488 1:500) for 2h at RT and with DAPI (1:5000) for 2 min.
Confocal Microscopy and cell counting
Images from sections of both Immunofluorescence and RNAscope ISH were acquired for the spinal cord using a Zeiss LSM700 Confocal, and processed in Fiji/ImageJ. Images from the human tissue were acquired using a Leica SP8 Confocal, Images were processed in Fiji/ImageJ. Estimation of the percentage of the double MHC-II/SOX10 positive cells in the spinal cord of EAE mice was performed by counting the double positive cells present in the lesion areas of the white matter divided by the total SOX10+ cells. The final percentage is represented as an average of 3 EAE mice. For the co-cultures the number of GFP/MHC-II double positive cells were divided by the total number of GFP+ cells. The final percentage is represented as an average of 3 independent experiments. For estimation of the percentage of OPCs that phagocyte microspheres, only OPCs with more than 10 microspheres surrounding the nucleus were considered as phagocytic OPCs. While this might have led to an underestimation of phagocytic OPCs, this way we have avoided counting OPCs with adherent and extracellular microspheres as phagocytic OPCs. Videos were obtained from z-stacks after processing with Imaris image analysis software.
Clustering analysis using GeneFocus pipeline
Quality Control
Cells were clustered using a custom-made approach, which we developed and refer as GeneFocus. We generated an iterative Level 1 and Level 2 clustering pipeline, taking advantage of diffusion mapping and spatial autocorrelation metrics to define relevant genes and cell clusters. First cells were analysed and quality control (QC) filtering was applied using the scater 1.6.0 in R. We applied cut-offs for the expression level 200000 total FPKM and number of genes expressed 2500, resulting in 2304 and 1777 cells pre- and –post QC respectively. Median FPKM values were 395721, and median gene counts were 3694 genes.
Spatial Gene-filtering
Cells were normalized (see supplemental code in Github) and feature selection was performed using a support vector model from the e1071 R package (https://CRAN.R-project.org/package=e1071). The resulting expression matrix was then inputted into a custom pipeline performing iterative gene-filtering in the following manner: we converted the expression matrix into a transition matrix using destiny 2.6.1 in R35 with the input being the first 30 principal components of the expression matrix. Subsequently, a distance matrix was created from the transition matrix, after which the MoransI autocorrelation metric was computed for each gene using the spdep 0.6-15 R package36. Genes were filtered according to the mean MoransI computed for all the genes in the expression matrix, and this mean MoransI was set as a general threshold for subsequent gene filtering. After this initial filtering, a new diffusion map was computed and we iteratively repeated this process until all genes within the gene set remained above the determined threshold of spatial correlation set during the first round of gene filtering. A similar approach was performed using the transition matrix as a distance metric. A cut-off value was calculated using a support vector model that predicts a mean distance as a function of the population size in which the gene is expressed and the mean distance observed. We iterated the distance matrix until we obtained a gene set equal or smaller than the covered gene set obtained from the MoransI metric. A joint diffusion map was then created.
Level 1 Clustering
Level 1 clustering was established by estimating the ideal number of clusters by silhouette width using the principal components of the transition matrix, from which the number was estimated on the basis of an elbow plot. We estimated clusters using the factoextra 1.0.5 package (https://CRAN.R-project.org/package=factoextra) and hierarchical clustering using Wards metric. We determined 5 initial clusters representing OPCs and early OLs, MOLs, VLMCs and pericytes, and microglial cells.
Level 2 Clustering
To determine more specific clustering and relevant genes we performed the iterative gene filtering on each of the subclusters. The threshold for the MoransI metric was established to be the mean MoransI of the gene set resulting from the second gene filtering round. This resulted in a final gene set for each sub-cluster varying between 800-1000 spatially correlated genes per Level 1 sub-cluster. Clusters were estimated by silhouette width and the final clustering was evaluated using the resulting heatmap of the filtered genes.
For validation of the GeneFocus pipeline, we extracted the expression information from GEO from37 and were able to identify the disease-associated microglia cluster previously described37 (Supplementary Fig. 2). To achieve out final clustering result, we also removed cells that were clustered in clusters with less than 3 cells, resulting in a total number of 1765 cells. These cells seemed to be doublets based on mixed expression profiles.
Gene Modules
To generate the gene-modules, we performed non-negative matrix factorization, where ranks were established using a measure of mutual information with the elbow method. We established that 30 ranks were the optimal rank to decompose the dataset in. To only select robust components from the decomposed matrix, we performed PCA on the components and established an optimal k number of clusters based on wards metric. All component values falling within a cluster were summed after which the new concatenated components were filtered based on a threshold of a Pearson correlation of 0.5 with any gene in the dataset. This resulted in 15 components that revealed the major gene trends underlying the data.
Differential expression and pathway visualization
Differential expression analysis was performed using the MAST R package v1.4.138. Pathway visualization was performed using the clusterProfiler package v3.6.039.
Comparison between OPCs in the current dataset and OPCs during development4
The R package MetaNeighbor was used, found at (https://github.com/maggiecrow/MetaNeighbor).
Gene ontology and pathway analysis
Most significantly differentially expressed genes were selected for each of the GO analyses. For each cell type, unique top differentially upregulated genes in EAE and unique top differentially upregulated genes in controls were selected. For comparisons between cell types, uniquely differentially expressed genes for each cell category were selected. In order to make gene sets comparable, top 100 genes were selected. GO and pathway analysis was performed with Cytoscape 3.5.140 plug-in ClueGO v2.3.341 with settings, GO Biological process (23.02.2017) and REACTOME pathways (01.03.2017), showing only pathways with P-val <= 0.05, minimum 5 genes per cluster, and default settings.
In accordance with the NNMF analysis, OPCs and OLs from EAE mice shared an enrichment for genes involved in the positive regulation of adaptive immune response, positive regulation of T cell mediated cytotoxicity, response to interferon and antigen processing and presentation of endogenous peptides via MHC-I (Supplementary Fig. 4b). GO/Reactome analysis for the enriched genes in individual clusters indicated that OPC2-EAE was enriched in positive regulation of T cell mediated cytotoxicity, antigen presentation, among other processes (Supplementary Fig. 5). MOL1/2-EAE exhibited enrichment in similar processes, but also additional processes related with glycosylation (Supplementary Fig. 5), which has been implicated in the activation of adaptive immune activation42. In contrast to these populations, “young” (Pdgfra-H2B-GFP) MOL5/6-EAE-a exhibited less pronounced upregulation in immune related GO-terms (Supplementary Fig. 5). Instead, this Plin4+ MOL cluster is enriched in genes related to lipid modification, gliogenesis, and intrinsic apoptotic signaling pathway (with anti-apoptotic genes Bcl2l1, Cdkn1a and pro-apoptotic genes Bnip3, Ppp1r13b; Supplementary Table 1), among other categories (Supplementary Fig. 5).
Alternative splicing analysis
Alternatively spliced cassette exons were identified with BRIE 0.1.343 in the cell types clusters from Smart-seq2 data. Cassette exons annotations were extracted from Gencode.vM12 from protein-coding genes according to the parameters in 43. For each of the cells classified as MOL1/2, MOL5/6 and OPC, the fraction of exon inclusion was calculated (PSI), with default parameters. Then, for each cell type, cell to cell pair-wise comparisons were performed, comparing EAE-assigned with control-assigned cells. Significantly alternative spliced candidates were selected with the following parameters, Bayes factor > 10, delta PSI between the two cells > 0.2 and a minimum number of significant comparisons for each cell type (MOL12 >= 20, OPC >= 20 and MOL56 >= 100). In order to get more specific alternatively spliced exons, the candidate events were selected only when they were uniquely spliced in one direction for each cell type, for instance events that are only skipped in EAE and events only included in EAE. Violin plots of selected events were plotted using all the PSI values for that specific alternative exon in EAE and in control, Wilcoxon rank sum test with continuity correction was used for significance test. Visualization of the alternative spliced events junctions reads was done with the merged bam files and the sashimi plot option in IGV44.
MS susceptibility genes
Genes assigned to human MS susceptibility SNPs were transform to Mus musculus annotation using BioMart 45. Human chrX associated SNPs from ref 2 were assigned to hg19 GENCODEv19 gene annotations with Bedtools v2.25.046, with parameters windowbed -60000. Recovered genes were transform to Mus musculus gene symbols using BioMart.
Ethics approval and consent to participate
The collection of all animal samples was performed according to the guidelines and recommendations of local animal protection legislation and were approved by the local committee for ethical experiments on laboratory animals in Sweden (Stockholms Norra Djurförsöksetiska nämnd). Human post mortem MS tissue was provided via a UK prospective donor scheme with full ethical approval from the UK Multiple Sclerosis Tissue Bank (MREC/02/02/39). All participants gave prospective pre-mortem written consent for their brains to be banked and used for research.
Code availability
Code used for single cell RNA-Seq analysis is available at https://github.com/Castelo-Branco-lab/GeneFocus.
Data availability
A web resource for browsing differential gene expression data for the single cell data can be accessed at https://ki.se/en/mbb/oligointernode. Raw data is deposited in GEO, accession number GSE113973. Code used for single cell RNA-Seq analysis is available at https://github.com/Castelo-Branco-lab/GeneFocus.
Supplementary Material
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots) and Klk6 (green dots). A double positive cell is further highlighted in the video and represents an MOL2 cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), nuclear GFP (from Pdgfra-H2B-GFP knock-in mice labeling OPCs) and Ptprz1 (white dots). A triple positive cell is further highlighted in the video and represents an OPC expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). At least 2 double positive cells are further highlighted in the video and represent an OL lineage cells expressing MHC-II and few Aif1 molecules.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for H2.eb1 (red dots) and Sox10 (green dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A microglia-derived process touching an OL lineage cell is further highlighted in the video.
IHC performed in human brain tissue from one MS patient marked with antibodies for MHC-II (white) and OLIG1 (green). A MHC-II positive cell that resides between two OLIG1 positive cells is further highlighted.
IHC performed in human brain tissue from one MS patient marked with antibodies for MHC-II (white) and OLIG1 (green). A double positive cell representing an OL lineage cell expressing MHC-II is further highlighted in the video.
One Sentence Summary.
Single-cell RNA-seq of a mouse model of multiple sclerosis uncovers new oligodendrocyte populations, putative disease markers and suggests new mechanisms underlying the pathogenesis of disease.
Acknowledgements
We would like to thank Alessandra Nanni, Ahmad Moshref, Johnny Söderlund for laboratory management and support. We thank Single Cell Genomics Facility, WABI Long Term Bioinformatic Support (Leif Wigge) at SciLifeLab, the FACS facilities at CMB (Belinda Panagel), Science for Life Laboratory, the National Genomics Infrastructure (NGI) and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. We want to acknowledge Antonio Gigliotti Rothfuchs for advice and reagents, Marek Bartosovic for assistance, Rasmus Berglund and Marie N’diaye for providing the tools to perform the phagocytosis experiments. The bioinformatics computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX, Uppsala University. Post mortem MS tissue used for IHC was provided via a UK prospective donor scheme with full ethical approval from the UK Multiple Sclerosis Tissue Bank (MREC/02/02/39). D.P.V. would like to acknowledge the University of Sydney HPC service at The University of Sydney for providing HPC resources that have contributed to the research reported in this paper. This work was supported in part by a University of Sydney HPC Grand Challenge Award. D.P.V. was supported in part by a Boehringer Ingelheim Travel Grant. C.f.-C. is funded by a Wellcome Trust Investigator award. AW is funded by UK Multiple Sclerosis Society. SJ is funded by European Union, Horizon 2020, Marie-Skłodowska Curie Actions, grant number EC reference number 789492. A.M.F. by the European Committee for Treatment and Research of Multiple Sclerosis (ECTRIMS). EA is funded by European Union, Horizon 2020, Marie-Skłodowska Curie Actions, grant SOLO, number 794689. Work in G.C.-B.’s research group was supported by Swedish Research Council (grant 2015-03558), European Union (Horizon 2020 Research and Innovation Programme/ European Research Council Consolidator Grant EPIScOPE, grant agreement number 681893), Swedish Brain Foundation (FO2017-0075), Ming Wai Lau Centre for Reparative Medicine, Petrus och Augusta Hedlunds Foundation ((grants M-2014-0041 and M-2016-0428) and Karolinska Institutet.
Footnotes
Accession codes and data availability: A web resource for browsing differential gene expression data for the single cell data can be accessed at https://ki.se/en/mbb/oligointernode. Raw data is deposited in GEO, accession number GSE113973. Code used for single cell RNA-Seq analysis is available at https://github.com/Castelo-Branco-lab/GeneFocus.
Author Contributions: A.M.F., D.V.B and G.C.-B. conceived the project, designed the study and interpreted results. A.M.F., S.M. and A.O.G-C. performed EAE model and A.M.F and S.M. collected single cells to generate single-cell sequencing data. D.V.B and E.A. performed computational analyses. A.M.F and M.M. designed, performed and analyzed most in vitro OPC experiments and S. performed the phagocytosis experiments together with A.M.F. A.O.G-C. and A.M.F. designed, performed and analyzed all experiments involving co-cultures with immune cells. S.J., A.W. and C.f.-C. provided the human postmortem MS brain tissue and performed the human IHC analysis. D.P.V. provided support for computational analysis. E.M.F. provided RNAscope ISH expertise and performed all videos. A.M.F, D.V.B and G.C.-B. wrote the manuscript with feedback from all authors.
Competing interests:
The G.C-B research group has received funding from F. Hoffmann – La Roche, Ltd. for another research project in the area.
Additional information: Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
References
- 1.Patsopoulos N, et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. 2017 doi: 10.1101/143933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skene NG, Grant SG. Identification of Vulnerable Cell Types in Major Brain Disorders Using Single Cell Transcriptomes and Expression Weighted Cell Type Enrichment. Front Neurosci. 2016;10:16. doi: 10.3389/fnins.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marques S, et al. Transcriptional Convergence of Oligodendrocyte Lineage Progenitors during Development. Dev Cell. 2018;46:504–517 e507. doi: 10.1016/j.devcel.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 6.Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- 7.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emery B, et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogata T, et al. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. Neuroscience. 2011;176:132–141. doi: 10.1016/j.neuroscience.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Tripathi RB, Rivers LE, Young KM, Jamen F, Richardson WD. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. J Neurosci. 2010;30:16383–16390. doi: 10.1523/JNEUROSCI.3411-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zawadzka M, et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6:578–590. doi: 10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capello E, Voskuhl RR, McFarland HF, Raine CS. Multiple sclerosis: re-expression of a developmental gene in chronic lesions correlates with remyelination. Annals of neurology. 1997;41:797–805. doi: 10.1002/ana.410410616. [DOI] [PubMed] [Google Scholar]
- 14.Enevold C, et al. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. Journal of neuroimmunology. 2009;212:125–131. doi: 10.1016/j.jneuroim.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Haile Y, et al. Granzyme B-inhibitor serpina3n induces neuroprotection in vitro and in vivo. Journal of neuroinflammation. 2015;12:157. doi: 10.1186/s12974-015-0376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SC, Raine CS. Multiple sclerosis: oligodendrocytes in active lesions do not express class II major histocompatibility complex molecules. Journal of neuroimmunology. 1989;25:261–266. doi: 10.1016/0165-5728(89)90145-8. [DOI] [PubMed] [Google Scholar]
- 17.Sibinga NE, Feinberg MW, Yang H, Werner F, Jain MK. Macrophage-restricted and interferon gamma-inducible expression of the allograft inflammatory factor-1 gene requires Pu.1. J Biol Chem. 2002;277:16202–16210. doi: 10.1074/jbc.M200935200. [DOI] [PubMed] [Google Scholar]
- 18.Lin W, Harding HP, Ron D, Popko B. Endoplasmic reticulum stress modulates the response of myelinating oligodendrocytes to the immune cytokine interferon-gamma. J Cell Biol. 2005;169:603–612. doi: 10.1083/jcb.200502086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nature reviews. Immunology. 2012;12:813–820. doi: 10.1038/nri3339. [DOI] [PubMed] [Google Scholar]
- 20.Bergsteindottir K, Brennan A, Jessen KR, Mirsky R. In the presence of dexamethasone, gamma interferon induces rat oligodendrocytes to express major histocompatibility complex class II molecules. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9054–9058. doi: 10.1073/pnas.89.19.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosius Lutz A, et al. Schwann cells use TAM receptor-mediated phagocytosis in addition to autophagy to clear myelin in a mouse model of nerve injury. Proc Natl Acad Sci U S A. 2017;114:E8072–E8080. doi: 10.1073/pnas.1710566114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettelli E, et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeis T, Enz L, Schaeren-Wiemers N. The immunomodulatory oligodendrocyte. Brain research. 2016;1641:139–148. doi: 10.1016/j.brainres.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 24.Peferoen L, Kipp M, van der Valk P, van Noort JM, Amor S. Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology. 2014;141:302–313. doi: 10.1111/imm.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeis T, Schaeren-Wiemers N. Lame ducks or fierce creatures? The role of oligodendrocytes in multiple sclerosis. J Mol Neurosci. 2008;35:91–100. doi: 10.1007/s12031-008-9042-1. [DOI] [PubMed] [Google Scholar]
- 26.Moyon S, et al. Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:4–20. doi: 10.1523/JNEUROSCI.0849-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh JL, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17:121–130. doi: 10.1038/nn.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traka M, Podojil JR, McCarthy DP, Miller SD, Popko B. Oligodendrocyte death results in immune-mediated CNS demyelination. Nat Neurosci. 2016;19:65–74. doi: 10.1038/nn.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka T, et al. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 31.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norton WT, Poduslo SE. Myelination in rat brain: method of myelin isolation. J Neurochem. 1973;21:749–757. doi: 10.1111/j.1471-4159.1973.tb07519.x. [DOI] [PubMed] [Google Scholar]
- 33.Larocca JN, Norton WT. Isolation of myelin. Curr Protoc Cell Biol. 2007;Chapter 3:Unit3 25. doi: 10.1002/0471143030.cb0325s33. [DOI] [PubMed] [Google Scholar]
- 34.Zeisel A, Yitzhaky A, Bossel Ben-Moshe N, Domany E. An accessible database for mouse and human whole transcriptome qPCR primers. Bioinformatics. 2013;29:1355–1356. doi: 10.1093/bioinformatics/btt145. [DOI] [PubMed] [Google Scholar]
- 35.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 36.Bivand R, Hauke J, Kossowski T. Computing the Jacobian in Gaussian Spatial Autoregressive Models: An Illustrated Comparison of Available Methods. Geographical Analysis. 2013;45:150–179. doi: 10.1111/gean.12008. [DOI] [Google Scholar]
- 37.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell. 2017;169:1276–1290 e1217. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Killcoyne S, Carter GW, Smith J, Boyle J. Protein Networks and Pathway Analysis Methods in Molecular Biology. Humana Press; 2009. pp. 219–239. [DOI] [PubMed] [Google Scholar]
- 41.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfert MA, Boons GJ. Adaptive immune activation: glycosylation does matter. Nature chemical biology. 2013;9:776–784. doi: 10.1038/nchembio.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Sanguinetti G. BRIE: transcriptome-wide splicing quantification in single cells. Genome Biol. 2017;18 doi: 10.1186/s13059-017-1248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2014;47:11.12.11–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots) and Klk6 (green dots). A double positive cell is further highlighted in the video and represents an MOL2 cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), nuclear GFP (from Pdgfra-H2B-GFP knock-in mice labeling OPCs) and Ptprz1 (white dots). A triple positive cell is further highlighted in the video and represents an OPC expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). At least 2 double positive cells are further highlighted in the video and represent an OL lineage cells expressing MHC-II and few Aif1 molecules.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for H2.eb1 (red dots) and Sox10 (green dots). A double positive cell is further highlighted in the video and represents an OL lineage cell expressing MHC-II.
RNAscope ISH representing a mouse spinal cord from EAE mice marked with probes for Cd74 (red dots), Sox10 (green dots) and Aif1 (white dots). A microglia-derived process touching an OL lineage cell is further highlighted in the video.
IHC performed in human brain tissue from one MS patient marked with antibodies for MHC-II (white) and OLIG1 (green). A MHC-II positive cell that resides between two OLIG1 positive cells is further highlighted.
IHC performed in human brain tissue from one MS patient marked with antibodies for MHC-II (white) and OLIG1 (green). A double positive cell representing an OL lineage cell expressing MHC-II is further highlighted in the video.
Data Availability Statement
A web resource for browsing differential gene expression data for the single cell data can be accessed at https://ki.se/en/mbb/oligointernode. Raw data is deposited in GEO, accession number GSE113973. Code used for single cell RNA-Seq analysis is available at https://github.com/Castelo-Branco-lab/GeneFocus.