Abstract
A wide variety of organic dyes form, under certain conditions, clusters know as J- and H-aggregates. Cyanine dyes are such a class of molecules where the spatial proximity of several dyes leads to overlapping electron orbitals and thus to the creation of a new energy landscape compared to that of the individual units. In this work we create artificial H-aggregates of exactly two cyanine 3 dyes by covalently linking them to a DNA molecule with controlled sub-nanometer distances. The absorption spectra of these coupled systems exhibit a blue-shifted peak, whose intensity varies depending on the distance between the dyes and the rigidity of the DNA template. Simulated vibrational resolved spectra, based on molecular orbital theory, excellently reproduce the experimentally observed features. Circular dichroism spectroscopy additionally reveals distinct signals, which indicates a chiral arrangement of the dye molecules. Molecular dynamic simulations of a Cy3-Cy3 construct including a 14-base pair DNA sequence verified chiral stacking of the dye molecules.
Introduction
The manipulation of light on the nanometer scale is of fundamental importance for designing efficient light harvesting and photonic devices.1–4 Fluorescent emitters such as quantum dots, fluorescence dyes, and NV centers are highly sensitive to their environment. In particular, organic fluorescent molecules are known to aggregate and stack on top of each other under certain conditions, resulting in changes of their absorption and emission properties.5–11 Many applications demand the precise tuning of the dye’s spectroscopic properties, such as dipole orientation and coupling strength, for which control over their interactions is highly desired.4,12,13 Aggregation of fluorescent molecules has been vastly exploited by tuning experimental parameters such as solvent polarity, ionic concentration and temperature.14–16 Nonetheless, these methods are rather non-specific and do not yield fine control over the resulting aggregate, making it difficult to fully characterize the resulting supramolecular dye structure.8,17 Natural light harvesting complexes, on the other hand, offer an example of full control over spectral coupling. In nature, a protein scaffold organizes the relative position and orientation of chromophores, so that their coupling strength and optical properties can be tuned into the desired frequency window.1,18
In a similar way DNA base pair recognition offers unprecedented spatial control over the assembly of nanometer-scale objects, including optically active components, such as dyes,19 quantum dots20 and metallic nanoparticles.21,22 Simple double-stranded DNA constructs have already been employed in a variety of photonics applications, which require a precise spatial organization of dyes, such as the creation of artificial aggregates of dyes. For example, the aggregation of cyanine dyes can occur via their intercalation in the minor groove of a DNA double strand.23 With the goal of improving the control over the positioning of dyes, DNA bases were substituted with the molecule of interest, for example methyl red,24 biphenyl and bipyridyl.25 In a different conjugation scheme, porphyrin aggregates have been created on a DNA strand through covalent linkage of porhyrin rings to thymine bases in a defined sequence.26,27 Owing to the wide variety of commercially available dyes that are covalently linked to DNA bases, covalent coupling today is the method of choice for imaging and energy transfer studies.
In this work we exploit this excellent addressability of DNA strands and in particular thymine bases to induce cyanine dye dimerization in a controlled manner and create new molecular excitonic states. With the ultimate goal of mimicking multi-dye coupling similar to that found in light harvesting complexes, we are here interested in studying interactions between pairs of Cyanine 3 (Cy3) dyes brought into close proximity in synthetic DNA single- and double-strands. Our DNA – dye hybrid structures are characterized by absorption spectroscopy and circular dichroism (CD). Due to the a priori design of the system, the interaction between the dyes can further be modeled by Time-Dependent Density Functional Theory (TD-DFT) calculations, allowing the comparison of spectral properties with simulated absorption spectra. Finally, the dynamics of the dye – DNA complex is investigated through molecular dynamics (MD) simulations.
Material and methods
The constructs used in this work consist of a synthetic DNA strand where one or two Cy3 molecules are linked to tymine bases through NHS coupling and the corresponding, unmodified, complementary sequence. The DNA strands were purchased from IBA GmbH (Gottingen, DE). The following sequences were used:
Monomer 5’-ATC GTA TC Tcy3 GTG TCT ATG CTA -3’
Dimer 1 base distance 5’-ATCGTATC Tcy3G Tcy3GTCTATGCTA-3’
Dimer 0 base distance 5’-ATCGTATCTG Tcy3 Tcy3TGTCTATGCTA-3’
Tcy3 indicates the modified thymine bases.
To hybridize the modified strand with their complement a mixture of both strands with a 1 to 1 ratio was mixed with buffer containing 1xTE (10 mM Tris, 1 mM EDTA) and 150 mM NaCl at pH 8.0. The solution was heated to 65°C for 5 min and slowly cooled to room temperature over 2h.
The absorption measurements were performed using a 10 mm optical path length quartz cuvette (Hellma-analytics) and a V-650 Spectrometer (Jasco) with 0.5 nm resolution and 1 s/point integration time. The CD signal was acquired with Chirascan-Plus Circular Dichroism Spectrometer (Applied Photo-physics Ltd) 1 nm resolution and 1 s/point integration time and the samples (DNA constructs concentration 10 μM in 1xTE and 150 mM NaCl buffer) placed in a 3 mm optical path length quartz cuvette (Hellma-analytics). Absorption and CD measurements were all performed at room temperature.
We also test a construct with a different Cy3-DNA linker. In this case one Cy3 molecule is located on each of two complementary DNA strands, the dye effectively acts as a substitute for the sugar-base complex at a specific location on the oligonucleotide. This substitution is accomplished by conjugating the 3' and 5' ends of two shorter strands to the two free hydroxy groups of the dye (SI Scheme 1). Such modified oligos and the relative complements were purchased from IDT (Integrated DNA Technologies Inc). The following sequences were used:
Dimer N=0
5'- ATT CAG ATT TTT TTT TTT TTT TTT TTT T/iCy3/TT TTT AGT TGA A -3’
5'- TTC AAC TAA AAA /iCy3/AAA AAA AAA AAA AAA AAA AAA TCT GAA T -3'
Dimer N=6
5'- ATT CAG ATT TTT TTT TT TTT TT/iCy3/TTTTTTTT TTT AGT TGA A -3'
5'- TTC AAC TAA AAA /iCy3/AAA AAA AAA AAA AAA AAA AAA TCT GAA T -3'
Monomer:
5'- ATT CAG ATT TTT TTT TT TTT TTTTTTTTTT TTT AGT TGA A -3'
5'- TTC AAC TAA AAA /iCy3/AAA AAA AAA AAA AAA AAA AAA TCT GAA T -3'
The doubly labeled Cy3 strands were hybridized by mixing equimolar amounts of the two complementary strands at a final concentration of 2 μM in 1xTE, followed by heating to 70° C and linear cooling to 25° C over 45 minutes in a thermocycler (Bio-Rad Laboratories, Inc.).
Absorbance measurements were performed using a Cary 300 Bio UV-Visible Spectrophotometer (Agilent Technologies) with 1 nm resolution and 0.1 s/point integration time. The DNA duplex samples were diluted to a final concentration of 0.7 μM in 1xTE in a low volume cuvette (100 μl) (10 mm path length) (Sigma-Aldrich). CD spectra were recorded on an Applied Photo-physics Chirascan circular dichroism spectropolarimeter using a 1 mm path length quartz cuvette. CD measurements were performed at room temperature over a range of 450-600 nm using a response time of 0.5 s, 1 nm pitch and 0.5 nm bandwidth. The recorded spectra represent a smoothed average of three scans, zero-corrected at 600 nm and normalized (Molar ellipticity θ is quoted in 105 deg cm2 dmol−1). The absorbance of the buffer was subtracted from the recorded spectra. The sample was diluted to a final concentration of 5 μM in 1xTE.
Computational methods
We generate simulated vibrational resolved absorption spectra using Time-Dependent Density Functional Theory (TD-DFT) and a Polarizable Continuum Model (PCM). All ab initio calculations were carried out with the program package Gaussian09.28 The functional B3LYP and the basis set 6-31G(d) were used throughout. To account for solvent effects the Polarizable Continuum Model29 was used. For the optimization of the dimer the dispersion correction D3 from Grimme30 was added. The simulation of the vibrationally resolved spectra was done using the implementation of Santoro et al.31 in Gaussian09.28 The Franck-Condon method and a spectral broadening of 300 cm-1 was used.
The molecular dynamics (MD) simulations were carried out with the program Gromacs 5.1.32,33 The following DNA sequence was used: 5’-TAT CTG Tcy3Tcy3T GTC TA-3’. A starting structure for the double stranded DNA was generated with model.it.34 The amber99bsc035,36 force field was used for the DNA and GAFF37 for the Cy3 molecule. The parameter for Cy3 were taken from Graen et al.,38 the ones for the modified thymine and the linker were generated using ANTECHAMBER relying on HF/6-31G(d) optimizations. The simulations were performed in TIP3P water,39 sodium ions were added to neutralize the system. A rectangular box with side lengths of 55x55x43 Å was used. After energy minimization the system was first equilibrated for 200 ps while restraining the DNA, then again for 1 ns without any constraints, before running production simulations.
Results and discussion
Dyes located at distances below ~10 nm can exchange energy via non-radiative, incoherent dipole-dipole interaction known as Förster Resonance Energy Transfer (FRET).40 When the molecules are brought into even closer proximity, to distances comparable to their physical size, they can instead have coherent energy exchange.41 This type of interaction gives rise to new energy states compared to those observed in individual dyes. Consequently, this coupling creates a new energy landscape and therefore leads to new optical properties, which can be quantified via absorption spectroscopy.5,7,23,42,43
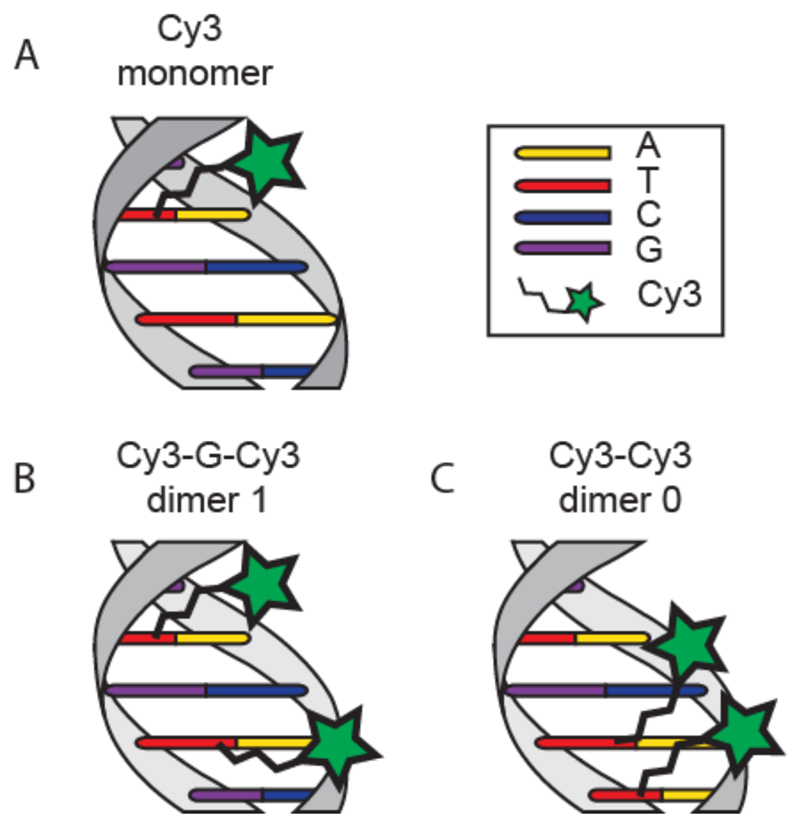
Our system of study is a short DNA duplex consisting of a single-stranded DNA (ssDNA) oligonucleotide carrying either one or two Cy3 molecules, which are covalently attached to selected thymine bases through NHS coupling (Scheme S1.a) and an unmodified complementary DNA sequence. We arranged the dyes in two different configurations: Dimer 1 (Cy3-G-Cy3) contains two Cy3 dyes coupled to a thymine with one unmodified guanine in between while Dimer 0 (Cy3-Cy3) has no separating base (Figure 1b and 1c). A single labeled strand serves as a control sample (Figure 1a).
Figure 1.
Schematic representation of experimental dye-DNA constructs with relative nomenclature and dye positions. A) Cy3 monomer, only one thymine base is modified with the dye. B) Cy3 “dimer 1” with one base (guanine) separation between dyes. C) Cy3 “dimer 0” consisting of two cy3 molecules linked to adjacent bases.
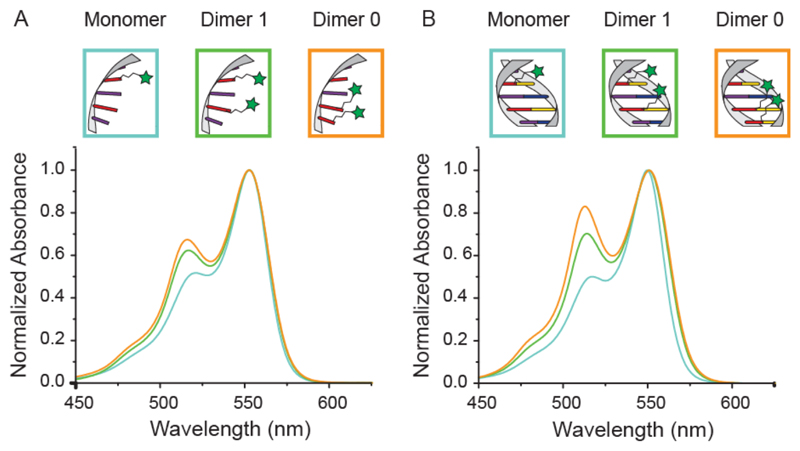
To investigate the interactions between the dyes in their different configurations we performed absorption spectroscopy measurements on both the ssDNA and the dsDNA constructs. The results are summarized in Figure 2. For the ssDNA constructs (Figure 2a) the Cy3 monomer displays the expected absorption maximum at 550 nm with two small shoulders at 520 nm and at 480 nm (Supporting Information Figure S1). Constructs containing two Cy3 molecules display a new, blue-shifted peak around 512 nm (hypsochromic peak), which increases in intensity as the dyes get closer to each other. Additionally, the shoulder in the region around 480 nm becomes more pronounced in the 2-dye constructs. Similar trends are observed in the dsDNA constructs (Figure 2b) but with an even more pronounced intensity increment of the blue-shifted hypsochromic peak. The spectral change is so pronounced that it is visible with bare eyes (see Supporting Information Figure S2). As the only structural difference compared to the ssDNA constructs is the presence of the hybridized, unmodified complement, we conclude that the higher rigidity of the duplex (persistence length PdsDNA ~ 50 nm) compared to ssDNA (PssDNA ~ 1 nm) leads to a decreased orientation fluctuation of the two dyes and therefore to a modified spectrum. 44 Further, the average distance between two bases in a single-stranded configuration is on the order of 6 Å while it is 3.4 Å in dsDNA45 also resulting in closer proximity of the dyes in the duplex configuration. The fact that the relative distance between dyes plays a crucial role in the strength of their interaction becomes obvious when comparing the increased intensity of the hypsochromic peak of the construct with 0 bases in between the dyes with that containing a spacer guanine. We attribute the formation of the hypsochromic peak to Cy3 – Cy3 interaction and not to interaction of the dyes with adjacent DNA bases as the single-labeled control sample does not display any additional spectral peaks.
Figure 2.
Absorbtion spectra of the dye-DNA constructs. A) Normalized absorption of the ssDNA and B) dsDNA constructs. The light blue curves correspond to the cy3 monomer, while the green and orange curves correspond to the Cy3 dimer with 1 base and 0 base distance respectively.
Cyanine dye molecules are known to interact with each other via π-orbital stacking thereby forming H-aggregates in aqueous solutions.11,16,17 The appearance of the blue-shifted peak in our experiments is consistent with this picture of H-aggregation of two polarized molecules with parallel dipoles.4,10,42,43 To demonstrate that the dimer formation is a result of the controlled positioning of the dyes mediated by the DNA scaffold and not caused by spontaneous aggregation, we performed a series of control experiments: First, the observed spectra were not significantly affected by varying the concentration of the constructs (Supporting Information Figure S3). Second, in order to exclude the possibility of H-aggregation being mediated by DNA groove intercalation,23 we mixed free dye with unmodified dsDNA at varying ratios and again observed no changes of the spectra compared to those originating from mono-dye-labeled samples (Supporting Information Figure S4).
We also tested a different construct where one single Cy3 dye was located on each of two complementary DNA strands and we again observed the same spectral changes that are typical for H-aggregate dimerization (Supporting Information Figure S5).
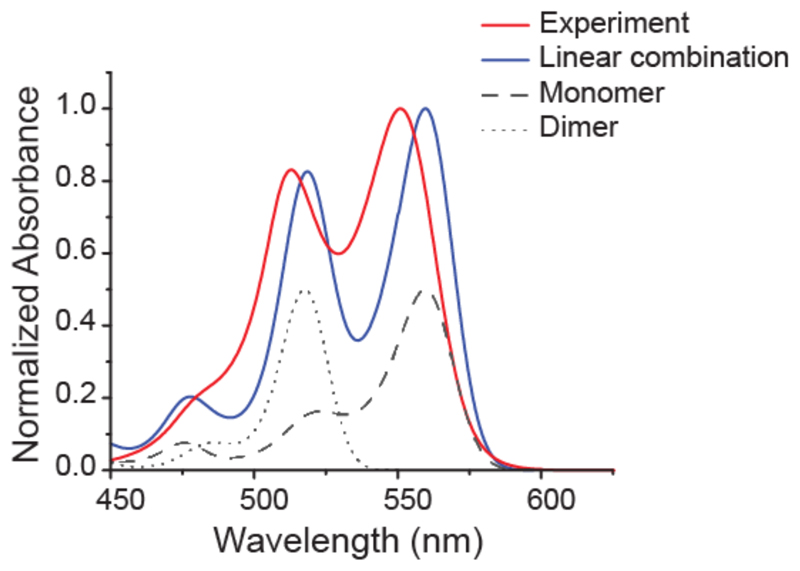
Simulated vibrational resolved absorption spectra were generated using Time-Dependent Density Functional Theory (TD-DFT) and a Polarizable Continuum Model (PCM) was applied. To reduce the computational cost, we neglected the surrounding DNA and chose a model system (Supporting Information Figure S6) consisting only of the π-system, which is responsible for the absorption properties of the dye. Figure 3 shows the simulated absorption spectrum for the strongly dipole-allowed S1 state of the monomer and its excellent agreement with our experiments. The 0-0 band is located at 560 nm with a smaller peak at 523 nm, characteristic for cyanine dyes.46,47 This transition corresponds to an excitation from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO), as shown in Table 1.
Figure 3.
Comparison of weighted linear combination of TD-DFT simulated spectra (solid red line) and experimentally obtained spectra from the construct “dimer 0” on dsDNA (solid blue line). Simulated spectra of monomer and dimer are shown in black (dashed and dotted lines corresponding to monomer and dimer, respectively). The intensities of the experimental spectra were fitted best when assuming one-third of dimer and two-third of monomer contribution.
Table 1.
Vertical excitation energies Evert, oscillator strengths f and the involved transitions for the lowestlying excited states of monomer and stacked dimer obtained with TD-DFT/B3LYP/6-31G(d) in water (PCM). The calculations are performed for a model system with inserted sulfur atoms instead of C(CH3)2 shown in Supporting Figure S6. For the monomer there is only one excitation of importance with an energy of 2.61 eV. In the case of the dimer only the S4 state contributes significantly to the absorption and is clearly blue-shifted compared to the monomer with an energy of 2.8 eV. The corresponding orbitals can be seen in Supporting Information Figure S7.
| State | Transition | Weight (%) | Evert (eV) | F | |
|---|---|---|---|---|---|
| Monomer | S1 | HOMO -> LUMO | 100 | 2.61 | 1.6014 |
| Dimer | S1 | HOMO-1 -> LUMO HOMO -> LUMO+1 |
11 89 |
2.18 | < 5*10-5 |
| S2 | HOMO-1 -> LUMO+1 HOMO -> LUMO |
40 60 |
2.22 | 0.0424 | |
| S3 | HOMO-1 -> LUMO HOMO -> LUMO+1 |
89 11 |
2.43 | < 5*10-5 | |
| S4 | HOMO-1 -> LUMO+1 HOMO -> LUMO |
60 40 |
2.80 | 2.8945 |
Stacking two of these monomers in a parallel dipole configuration leads to the dimer structure shown in Supporting Information Figure S6. Both molecules are completely flat and the distance between their molecular planes is 3.4 Å, as by design in the “Dimer 0” construct. In this configuration, the HOMO and LUMO of the monomers combine resulting in four different orbitals and hence, four different excited states. Each new state consists of a positive and negative linear combination of the monomer’s HOMO and LUMO energy states. The contributions of the S1 and S3 states combination results in a zero net contribution. The S2 and the S4 states consist of the same transitions, but with inverse weight. The S2 state has a vertical excitation energy of 2.22 eV and is thus red-shifted compared to the monomer. Its oscillator strength is two orders of magnitude smaller (0.0424) than the that of the S4 state and is, as a consequence, the weak component in molecular exciton theory.48,49 In contrast, the S4 state is blue-shifted (2.80 eV) and has a significant oscillator strength (2.8945) that is almost twice the monomer value. It is the strong component in molecular exciton theory and the only relevant transition here. The resulting absorption spectrum is shown in Figure 3. It consists of the 0-0 transition at 518 nm with almost none vibrational progression. Comparing this outcome to the monomer, we observe a blue shift of the absorption signal by 42 nm resulting from dimerization. The hypsochromic peak observed in the experimental spectrum is 38 nm shifted from the monomer absorption, which is very close to the difference displayed by the simulated spectra. The absolute simulated values of the vertical excitation of the monomer and dimer are shifted bathochromatically in respect to the experimental peaks by 0.04 and 0.03 eV, respectively, which constitutes very good agreement for an ab initio method such as TD-DFT.50 This overall agreement between experimental data and theory strengthens our hypothesis of controlled H-aggregation.
The simulations show either the original peak for the monomers at 560 nn or the blue-shifted peak for the dimers at 518 nm. In our dimer samples we observe not only the blue-shifted peak, but also the original peak indicating that not all dyes form the H-aggregate state or that this state is not formed permanently, as also indicated by the MD simulations discussed below (see MD video included as SI). To account for the mixed spectrum of the monomer and dimer species, we created a linear combination of the two simulated spectra. The superimposed modeled spectrum fitted the experimental spectrum best with one-third contribution from the dimer population and two thirds originating from the monomers. Also the appearance of the more pronounced shoulder at 480 nm could this way be attributed to the convolution of the two minor peaks of the monomer and dimer spectra around these wavelengths.
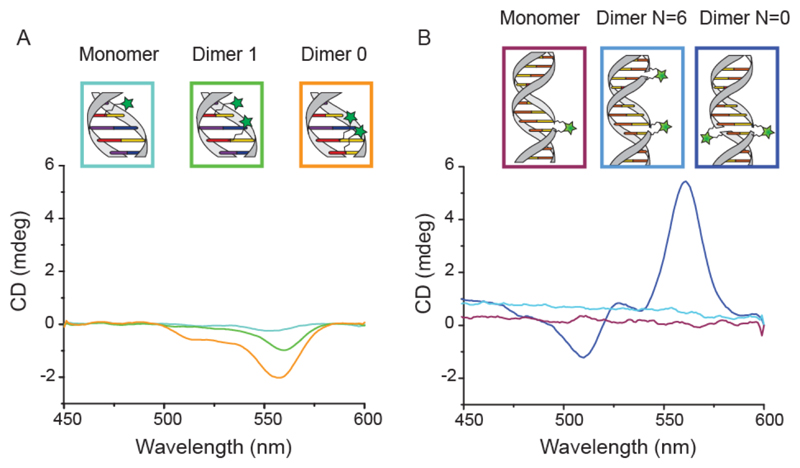
In the simulations, the dyes are stacked one above the other in a parallel dipole configuration, where the energy level of the excited state corresponds to the main absorption peak of the dimer spectrum. To test for chiral arrangements of the dyes we also conducted circular dichroism (CD) measurements. All constructs containing two cyanines in close proximity exhibited a measureable CD signal (see Figure 4). In contrast, all samples with only a single dye did not exhibi chiral signatures, indicating that CD transfer, i.e. coupling between the chiral dsDNA and the achiral individual dyes, is not the origin of the recorded CD.51 In accordance with this observation, the Cy3 dimer with dyes in closest proximity displays the strongest CD signal. The same behavior has been reported for DNA-porphyrin constructs.27 Our MD simulations further confirm that the measured signal is not a result of Cy3 – DNA intercalation but a result of chiral stacking of the dyes. Interestingly, the constructs with the doubly anchored Cy3 exhibit even stronger CD signals and a different spectral shape compared to that of the thymine-modified constructs. Previous studies have shown that doubly labeled Cy3 can intercalate between DNA bases,52 suggesting that dyes can stack on top of each other but also between adjacent base pairs resulting in strong, bisignate CD signals.51,53 Generally, CD spectroscopy cannot be used to gather full structural information and additional modeling would be required to quantitatively understand the recorded spectra.54 Nevertheless, we can infer that all the constructs showing the hypsochromic shift also exhibit chiral arrangements of dyes and that different strategies of linking the same dyes to DNA duplexes lead to structural and spectral variations.
Figure 4.
CD measurements of all dsDNA constructs. A) Spectra of thymine-labeled Cy3-dsDNA constructs, showing an increasing CD signal as the distance between dyes decreases. B) Spectra of doubly labeled Cy3-dsDNA constructs, showing a CD signal for the dimer with 0 bases in between dyes and no signal for both the monomer and the dimer with 6 bases in between dyes.
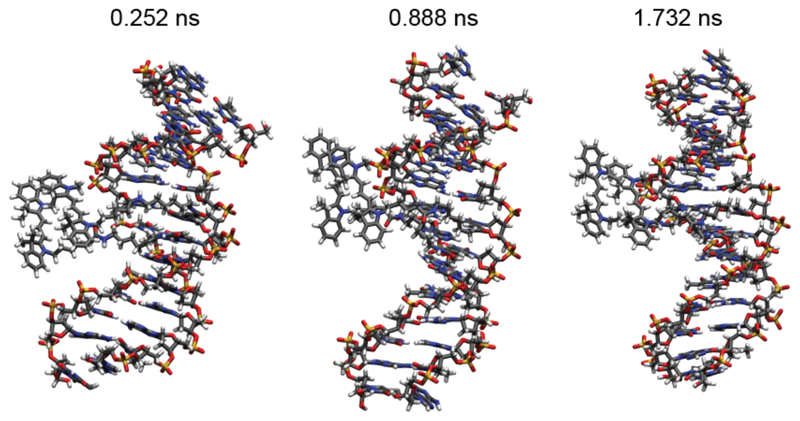
Molecular dynamics simulations of the Cy3-Cy3 construct including a 14-base pair DNA sequence were used to investigate the dynamics of the system. We considered the experimental Cy3 molecule with the original C(CH3)2 groups here. Time snapshots shown in Figure 5 and Supporting Information movieS1 reveal that the dyes stay in close proximity throughout the simulation (2 nsec). While the Cy3 monomer preferably lies in the major groove of the DNA (see Cy3_monomer.pdb file in the Supporting Information), the dyes in the dimer configuration stay near the negatively charged backbone, as it can be expected for a positively charged molecule. For some time during the propagation, the attraction between the molecules is visible as one or both leave their planar form, which is normally the natural configuration for molecules with such a π-system. The edges are bent towards each other to maximize the interaction. Furthermore, as the bulky C(CH3)2 groups prevent the molecule from aligning perfectly, the dye molecules rotate slightly against each other, forming an average angle of 11.1°, confirming the results of the CD spectroscopy.
Figure 5.
Snapshots of MD simulations at the specified time frames. The Cy3 molecules are visible on the left and they are located, throughout the whole simulation, close to the DNA backbone. No intercalation is observed during the observation time of 2 ns. The molecules show a non-prefect parallel alignment and a fluctuating reciprocal position, which is consistent with the experimental observations of CD and the co-existence of monomer and dimer peaks.
Conclusion
In conclusion we demonstrated that with our relatively simple system of cyanine dyes covalently attached to DNA backbones we can artificially create molecular excitonic states by deterministically changing the reciprocal distance of the dyes and the rigidity of the scaffolding constructs. Our pre-programmable DNA-based approach demonstrates excellent control over dye assembly and designed dimer interaction strengths thus allowing optical tuning and theoretical modeling of dye aggregates. Furthermore, the use of DNA as the underlying scaffolding material opens the possibility for complex integrated nanostructures, thereby widening the toolbox of nano-optical components available for photonics and nano-optics.
Supplementary Material
DNA-Dye coupling scheme; free monomeric Cy3 absorption spectrum; samples images; absorption spectra of control experiments i.e. different construct concentration and ds-DNA/dye mixture; absorption spectrum of constructs with alternative DNA-Cy3 functionalization scheme; optimized molecular structures for theoretical modeling; calculated molecular orbitals; pdb file of MD simulations corresponding to the Cy3-DNA monomer construct; video of MD simulations corresponding to the Cy3-DNA dimer construct.
Acknowledgements
This work was supported by the European Commission through the ERC grant agreement n° 336440, ORCA and by the Deutsche Forschungsgemeinschaft through the SFB749. M.D.A is thankful to CRUK for program funding and E.A.H. acknowledges support from the Schweizerischer Nationalfonds (SNF).
We thank Prof Ulrich Keyser, Dr Alex Chin, Dr Celestino Creatore and Dr. Mauricio Pilo Pais for useful discussions. F.N acknowledges Timon Funk for the help with CD measurements and Luisa Kneer for the initial support in the project.
References
- (1).Scholes GD, Fleming GR, Olaya-Castro A, van Grondelle R. Lessons from Nature about Solar Light Harvesting. Nat Chem. 2011;3(10):763–774. doi: 10.1038/nchem.1145. [DOI] [PubMed] [Google Scholar]
- (2).Wasielewski MR. Self-Assembly Strategies for Integrating Light Photosynthetic Systems. Acc Chem Res. 2009;42(12):1910–1921. doi: 10.1021/ar9001735. [DOI] [PubMed] [Google Scholar]
- (3).Tame MS, McEnery KR, Özdemir ŞK, Lee J, Maier SA, Kim MS. Quantum Plasmonics. Nat Phys. 2013;9(6):329–340. [Google Scholar]
- (4).Saikin SK, Eisfeld A, Valleau S, Aspuru-Guzik A. Photonics Meets Excitonics: Natural and Artificial Molecular Aggregates. Nanophotonics. 2013;2(1):21–38. [Google Scholar]
- (5).Czikkely V, Försterling HD, Kuhn H. Light Absorption and Structure of Aggregates of Dye Molecules. Chem Phys Lett. 1970;6(1):11–14. [Google Scholar]
- (6).Reen M, Smith TW, Chens LB. J-Aggregate Formation of a Carbocyanine as a Quantitative Fluorescent Indicator of Membrane Potential. Biochemistry. 1991;30(5):4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- (7).Mateer DL, Ormerod AP, Harrison WJ, Edwards DJ. Highly Ordered Aggregates in Dilute Dye-Water Systems. Langmuir. 1995;11(26):390–393. [Google Scholar]
- (8).Harrison WJ, Mateer DL, Tiddy GJT. Liquid-Crystalline J-Aggregates Formed by Aqueous Ionic Cyanine Dyes. J Phys Chem. 1996;100(6):2310–2321. [Google Scholar]
- (9).Würthner F, Kaiser TE, Saha-möller CR. J-Aggregates : From Serendipitous Discovery to Supra- Molecular Engineering of Functional Dye Materials. Angew Chem Int Ed. 2011;50:3376–3410. doi: 10.1002/anie.201002307. [DOI] [PubMed] [Google Scholar]
- (10).Eisfeld A, Briggs JS. The J- and H-Bands of Organic Dye Aggregates. Chem Phys. 2006;324:376–384. [Google Scholar]
- (11).Kopainsky B, Hallermeier JK, Kaiser W. The First Step of Aggregation of Pic: The Dimerization. Chem Phys Lett. 1981;83(3):498–502. [Google Scholar]
- (12).Roller E-M, Argyropoulos C, Högele A, Liedl T, Pilo-Pais M. Plasmon–Exciton Coupling Using DNA Templates. Nano Lett. 2016;16(9):5962–5966. doi: 10.1021/acs.nanolett.6b03015. [DOI] [PubMed] [Google Scholar]
- (13).Zengin G, Johansson G, Johansson P, Antosiewicz TJ, Käll M, Shegai T. Approaching the Strong Coupling Limit in Single Plasmonic Nanorods Interacting with J-Aggregates. Sci Rep. 2013;3 doi: 10.1038/srep03074. 3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Slavnova TD, Chibisov AK. Kinetics of Salt-Induced J-Aggregation of Cyanine Dyes. J Phys Chem. 2005;109:4758–4765. doi: 10.1021/jp058014k. [DOI] [PubMed] [Google Scholar]
- (15).Armitage B, Retterer J, Brien DFO. Dimerization of Cyanine Dyes in Water Driven by Association with Hydrophobic Borate Anions. J Am Chem Soc. 1993;115(20):10786–10790. [Google Scholar]
- (16).Renge I, Wild UP. Solvent, Temperature, and Excitonic Effects in the Optical Spectra of Pseudoisocyanine Monomer and J-Aggregates. J Phys Chem A. 1997;101(43):7977–7988. [Google Scholar]
- (17).Von Berlepsch H, Bo C, Ouart A, Burger C, Da S. Supramolecular Structures of J -Aggregates of Carbocyanine Dyes in Solution. J Phys Chem B. 2000;104(22):5255–5262. [Google Scholar]
- (18).Cheng Y-C, Fleming GR. Dynamics of Light Harvesting in Photosynthesis. Annu Rev Phys Chem. 2009;60:241–262. doi: 10.1146/annurev.physchem.040808.090259. [DOI] [PubMed] [Google Scholar]
- (19).Stein IH, Schüller V, Böhm P, Tinnefeld P, Liedl T. Single-Molecule FRET Ruler Based on Rigid DNA Origami Blocks. ChemPhysChem. 2011;12(3):689–695. doi: 10.1002/cphc.201000781. [DOI] [PubMed] [Google Scholar]
- (20).Mitchell GP, Mirkin CA, Letsinger RL. Programmed Assembly of DNA Functionalized Quantum Dots. J Am Chem Soc. 1999;121(10):8122–8123. [Google Scholar]
- (21).Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (22).Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Organization of “Nanocrystal Molecules” Using DNA. Nature. 1996:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- (23).Hannah KC, Armitage BA. DNA-Templated Assembly of Helical Cyanine Dye Aggregates : A Supramolecular Chain Polymerization. Acc Chem Res. 2004:845–853. doi: 10.1021/ar030257c. [DOI] [PubMed] [Google Scholar]
- (24).Asanuma H, Shirasuka K, Takarada T, Kashida H, Komiyama M. DNA - Dye Conjugates for Controllable H * Aggregation. J Am Chem Soc. 2003;(4):9042–9048. doi: 10.1021/ja021153k. [DOI] [PubMed] [Google Scholar]
- (25).Brotschi C, Leumann CJ. DNA with Hydrophobic Base Substitutes: A Stable, Zipperlike Recognition Motif Based On Interstrand-Stacking Interactions. Angew Chemie Int Ed. 2003;42(14):1655–1658. doi: 10.1002/anie.200250516. [DOI] [PubMed] [Google Scholar]
- (26).Bouamaied I, Nguyen T, Thomas R, Stulz E. Supramolecular Helical Porphyrin Arrays Using DNA as a Scaffold. Org Biomol Chem. 2008;6:3888–3891. doi: 10.1039/b813584c. [DOI] [PubMed] [Google Scholar]
- (27).Fendt L, Bouamaied I, Tho S, Amiot N, Stulz E. DNA as Supramolecular Scaffold for Porphyrin Arrays on the Nanometer Scale. J Am Chem Soc. 2007;129(14):1842–1844. doi: 10.1021/ja075711c. [DOI] [PubMed] [Google Scholar]
- (28).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, et al. Gaussian 09, Revision A. 1. Gaussian Inc.; Wallingford, CT: 2009. [Google Scholar]
- (29).Miertuš S, Scrocco E, Tomasi J. Electrostatic Interaction of a Solute with a Continuum. A Direct Utilizaion of {AB} Initio Molecular Potentials for the Prevision of Solvent Effects. Chem Phys. 1981;55:117–129. [Google Scholar]
- (30).Grimme S, Antony J, Ehrlich S, Krieg H. A Consistent and Accurate Ab Initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J Chem Phys. 2010;132(15):154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- (31).Santoro F, Improta R, Lami A, Bloino J, Barone V. Effective Method to Compute Franck-Condon Integrals for Optical Spectra of Large Molecules in Solution. J Chem Phys. 2007;126 doi: 10.1063/1.2437197. 84509. [DOI] [PubMed] [Google Scholar]
- (32).Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, et al. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- (34).Vlahovicek K, Pongor S. Model.it : Building Three Dimensional DNA Models from Sequence Data. Bioinformatics. 2000;16(11):1044–1045. doi: 10.1093/bioinformatics/16.11.1044. [DOI] [PubMed] [Google Scholar]
- (35).Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J Comput Chem. 2000;21(12):1049–1074. [Google Scholar]
- (36).Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of A/γ Conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and Testing of a General Amber Force Field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- (38).Graen T, Hoefling M, Grubmüller H. AMBER-DYES: Characterization of Charge Fluctuations and Force Field Parameterization of Fluorescent Dyes for Molecular Dynamics Simulations. J Chem Theory Comput. 2014;10(12):5505–5512. doi: 10.1021/ct500869p. [DOI] [PubMed] [Google Scholar]
- (39).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- (40).Scholes GD. Long-Range Resoncance Energy Transfer in Molecular Systems. Annu Rev Phys Chem. 2003;(18):57–87. doi: 10.1146/annurev.physchem.54.011002.103746. [DOI] [PubMed] [Google Scholar]
- (41).Beljonne D, Curutchet C, Scholes GD, Silbey RJ. Beyond Förster Resonance Energy Transfer in Biological and Nanoscale Systems. J Phys Chem B. 2009;113(19):6583–6599. doi: 10.1021/jp900708f. [DOI] [PubMed] [Google Scholar]
- (42).Czikklely V, Forsterling HD, Kuhn H. Extended Dipole Model for Aggregates of Dye Molecules. Chem Phys Lett. 1970;6(3):207–210. [Google Scholar]
- (43).Fulton RL, Gouterman M. Vibronic Coupling. II. Spectra of Dimers. J Chem Phys. 1964;41(8):2280–2286. [Google Scholar]
- (44).Tinland B, Pluen A, Sturm J, Weill G. Persistence Length of Single-Stranded DNA. Macromolecules. 1997;30(19):5763–5765. [Google Scholar]
- (45).Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys J. 2004;86(4):2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lin KTH, Silzel JW. Relation of Molecular Structure to Franck–Condon Bands in the Visible-Light Absorption Spectra of Symmetric Cationic Cyanine Dyes. Spectrochim Acta Part A Mol Biomol Spectrosc. 2015;142:210–219. doi: 10.1016/j.saa.2015.01.032. [DOI] [PubMed] [Google Scholar]
- (47).Bertolino CA, Ferrari AM, Barolo C, Viscardi G, Caputo G, Coluccia S. Solvent Effect on Indocyanine Dyes: A Computational Approach. Chem Phys. 2006;330:52–59. [Google Scholar]
- (48).Kasha M, Rawls HR, Ashraf El-Bayoumi M. The Exciton Model in Molecular Spectroscopy. Pure Appl Chem. 1965;11:371–392. [Google Scholar]
- (49).Guthmuller J, Zutterman F, Champagne B. Prediction of Vibronic Coupling and Absorption Spectra of Dimers from Time-Dependent Density Functional Theory: The Case of a Stacked Streptocyanine. J Chem Theory Comput. 2008;4(12):2094–2100. doi: 10.1021/ct8003047. [DOI] [PubMed] [Google Scholar]
- (50).Laurent AD, Jacquemin D. TD-DFT Benchmarks: A Review. Int J Quantum Chem. 2013;113(17):2019–2039. [Google Scholar]
- (51).Yamaoka K, Resnik RA. The Extrinsic Cotton Effect of Acridine Orange Bound to Native DNA and Helical Poly-a L-Glutamic Acid. J Phys Chem. 1966;70(14):4051–4066. [Google Scholar]
- (52).Stennett EMS, Ma N, Van Der Vaart A, Levitus M. Photophysical and Dynamical Properties of Doubly Linked Cy3-DNA Constructs. J Phys Chem B. 2014;118(1):152–163. doi: 10.1021/jp410976p. [DOI] [PubMed] [Google Scholar]
- (53).Seibt J, Engel V. On the Calculation of Circular Dichroism Spectra Using Quantum Wave-Packet Dynamics with an Application to Molecular Dimers. J Chem Phys. 2007;126(7) doi: 10.1063/1.2472365. 74110. [DOI] [PubMed] [Google Scholar]
- (54).Seibt J, Lohr A, Wurthner F, Engel V. Circular Dichroism and Absorption Spectroscopy of Merocyanine Dimer Aggregates: Molecular Properties and Exciton Transfer Dynamics from Time-Dependent Quantum Calculations. Phys Chem Chem Phys. 2007;9(47):6214–6218. doi: 10.1039/b712155e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.