Abstract
Motivation
Skeletal muscle dysfunction is a systemic effect in one-third of patients with chronic obstructive pulmonary disease (COPD), characterized by high reactive-oxygen-species (ROS) production and abnormal endurance training-induced adaptive changes. However, the role of ROS in COPD remains unclear, not least because of the lack of appropriate tools to study multifactorial diseases.
Results
We describe a discrete model-driven method combining mechanistic and probabilistic approaches to decipher the role of ROS on the activity state of skeletal muscle regulatory network, assessed before and after an 8-week endurance training program in COPD patients and healthy subjects. In COPD, our computational analysis indicates abnormal training-induced regulatory responses leading to defective tissue remodeling and abnormal energy metabolism. Moreover, we identified tnf, insr, inha and myc as key regulators of abnormal training-induced adaptations in COPD. The tnf-insr pair was identified as a promising target for therapeutic interventions. Our work sheds new light on skeletal muscle dysfunction in COPD, opening new avenues for cost-effective therapies. It overcomes limitations of previous computational approaches showing high potential for the study of other multi-factorial diseases such as diabetes or cancer.
1. Introduction
Systems biology aims to explain physiology, development, and pathology based on modular networks of expression, interaction, regulation, and metabolism (Long et al., 2008 and Wellmer et al., 2010). Gene regulatory networks (GRNs) (Barabasi and Oltvai, 2004) influence the adaptation to environmental perturbations via cell type-specific gene expression and interactions between transcription factors (TFs) and regulatory promoter regions (Tian et al., 2014). These mechanisms may affect the metabolic network by regulating the activity of key enzymes. One of the major challenges of the application of systems biology to the medical field is the study of crosstalks between metabolic and gene regulatory networks and their role in cellular response to environmental conditions and external perturbations (Coughlin et al., 2014). This is especially crucial in multi-factorial diseases (Gomez-Cabrero et al., 2014), in which multiple regulatory mechanisms lead the cell to an aberrant metabolism and abnormal adaptation to environmental conditions that explain heterogeneities of disease progress and clinical manifestations among patients.
Chronic obstructive pulmonary disease (COPD) is a complex chronic disease caused by inhalation of irritants in susceptible individuals. Pulmonary manifestations of the disease are characterized by alterations in lung function such as expiratory flow limitation (low forced expiratory volume during the first second, FEV1) and abnormal pulmonary gas exchange (arterial hypoxemia) (Vestbo et al., 2013). COPD imposes a high burden on health systems worldwide and is the third leading cause of death in the United States (Murray and Lopez, 2013). Skeletal muscle dysfunction is a characteristic systemic effect of the disease that affects approximately one third of patients (Maltais et al., 2014). It has a multi-factorial nature (Maltais et al., 2014), and it is acknowledged that high reactive oxygen species (ROS) generation in the mitochondrial electronic transport chain (ETC) is likely to play a relevant role in systemic mani-festations of the disease affecting the GRN (Rabinovich et al., 2007). Moreover, altered skeletal muscle redox status plays a central role in the abnormal physiological adaptations observed in COPD patients after endurance training (Rabinovich et al., 2001). It has been hypothesized that the systemic effects of COPD might share abnormal regulation of key metabolic pathways (Miralles et al., 2014). Thus, the state of the mitochondrial ETC depends on carbohydrate metabolism and transport of adenine nucleotides. All these factors, as well as altered cellular oxygenation (Cano et al., 2014), can have a functional impact on the respiratory chain and on mitochondrial ROS production through electron transport chain complex III (Selivanov et al., 2009).
Different data and model-driven approaches have been proposed to explore correlations between muscle transcriptomic or proteomic profiles and abnormal training-induced adaptive changes observed in COPD patients. One of these model approaches has been Bayesian networks (Morrow et al., 2015; Zhang et al., 2015). However, although Bayesian networks are able to assess causalities and may provide a rough idea of the processes associated with a given disease (Saraswati and Sitaraman, 2014), they are not suitable for deciphering the molecular mechanisms and complex dynamics of GRN involved in the abnormal training-induced physiological changes observed in COPD patients (Himes et al., 2009). On the other hand, mechanistic approaches such as kinetic models have limitations when dealing with large and highly inter-connected networks due to the paucity of precise knowledge on necessary parameters (Fisher and Henzinger, 2007). Thus, our knowledge of the underlying mechanisms of abnormal adaptation to training in COPD patients remains limited by the lack of suitable computational analyses.
In order to overcome the limitations of the current methods, we have developed a novel computational approach combining the benefits of probabilistic and model-driven methods to address the complex dynamics of energy-metabolism-associated GRN.
Our aim was to assess the importance of ROS-driven molecular mechanisms in explaining the defective muscle adaptation to training in COPD patients as a case of concept. Briefly, our approach is based on: i) the reconstruction of GRN using muscle-biopsy-transcriptomic data obtained in previous studies (Rodriguez et al., 2011; Turan et al., 2011; [GSE27536]) and manual curation based on bibliographic data (Supplementary material) (Figure 1B, step 5); and, ii) integration of the reconstructed GRN and constraints based on probabilistic approaches, into a discrete model-based analysis using Thomas formalism (Thomas and Kaufman, 2001) (Figure 1C, step 8). Our discrete GRN model unveiled alterations in TCA cycle, electron transport chain, creatine kinase and insulin receptor factor as key players in the abnormal training-induced responses in COPD patients. We suggest that these alterations result in anomalous GRN dynamics leading to less efficient energy metabolism, abnormal cytokine regulation and defective tissue remodeling. Importantly, the approach developed here showed superior accuracy and predictive ability compared to existing data-driven methods for analyzing the transcriptomic data (Supplementary. material 1).
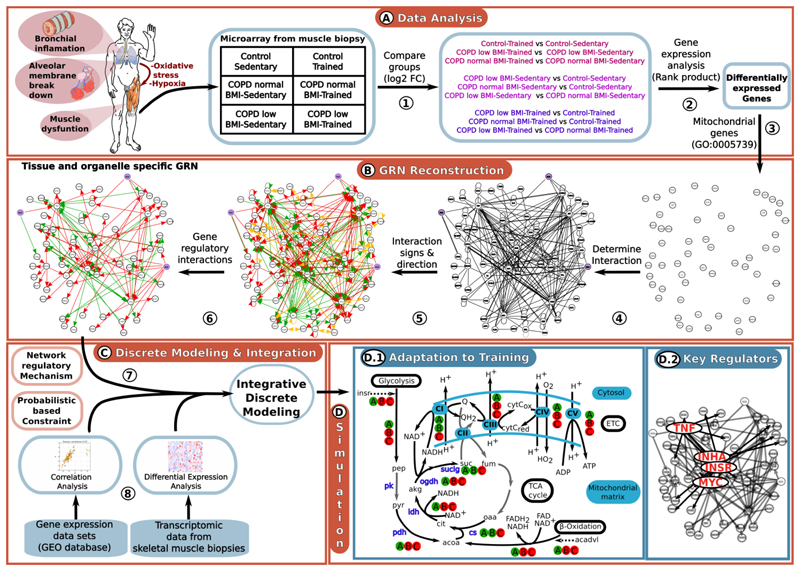
Fig. 1. Overall process of GRN reconstruction and its integration with probabilistic-based constraints into a discrete-model driven analysis.
Fig. 1A, step 1: Comparison of the trancriptomic data of the six study states Fig. 1A, step 2: Identify differentially expressed genes between conditions Fig. 1A, step 3: Since both energy metabolism and ROS production occur mainly in the mitochondria, the GRN reconstruction was focused on differentially expressed genes associated with mitochondrial processes using GO and Human Proteins Atlas databases. Fig. 1B, step 4: Automated interaction network reconstruction generated by IPA software (Krämer et al., 2014), DroID (Murali et al., 2011) and BioXM (Maier et al., 2011) databases based on the list of differentially expressed genes. This network includes gene regulatory and protein-protein interactions. Fig. 1B, step 5: Manual curation of the GRN reconstruction using a bibliomic data sources (Supplementary material 1.2) to identify and correct incomplete or erroneous annotations and identify the direction and sign of interactions. Fig. 1B, step 6: Since our analysis is based on transcriptomic data, we focused on gene regulatory interactions only. The nodes in the networks represent genes that are either differentialy expressed between groups or were included to construct a single GRN; violet nodes represent ROS and edges represent interactions between nodes, with green and red arrows indicating negative and positive interactions, respectively. Interactions with unknown direction and/or sign are in yellow. Fig. 1C step 7: Mathematical formalization of the reconstructed GRN based on R. Thomas formalism (Thomas and Kaufman, 2001). This approach describes mechanistically the interactions between differentially expressed mitochondrial-associated genes. Fig. 1C step 8: Impose constraints based on publicly available muscle-related gene expression data-sets Fig D.1: Applying the discrete model which integrates the gene-regulatory mechanisms (step 7) and the constraints (step 8) to determine the mechanisms associated with abnormal adaptation to training in COPD patients. Graphical representation of the predicted metabolic adaptation to endurance training program in healthy people (A), COPD patients with normal BMI (B) and COPD patients with low BMI (C). The figure represents a scheme of glycolysis, β-oxidation, TCA cycle and electric transport chain (ETC). Continuous arrows represent the metabolic reactions, while dashed arrows represent the activation of glycolysis and β-oxidation by insr and acadvl, respectively. Metabolic reactions/pathways associated with genes predicted to be up-regulated (down-regulated) after the training program are colored in green (red). White arrows represent metabolic reactions not directly associated with any gene in the reconstructed GRN. Gene abbreviations: insr: insulin receptor factor, acadvl: acyl-CoA dehydrogenase, very long chain. Metabolic reaction abbreviations: pk: pyruvare kinase, pdh: pyruvate dehydrogenase, cs: citrase synthase, idh: isocitrate dehydrogenase, ogdh: oxoglutarate dehydrogenase, suclg: succinate-CoA ligase, CI: NADH dehydrogenase, CII: succinate dehydrogenase, CIII: cytochrome c reductase, CIV: cytochrome c oxidase, CV: ATP synthase; the metabolites and cofactors represented in this figure are: pep: phosphoenol pyruvate, pyr: pyruvate, acoa: acetyl CoA, cit: citrate, akg: alpha-ketoglutarate, suc: succinate, fum: fumarate, oaa, oxalacetate, H+: proton, NAD+: nicotinamide adenine dinucleotide oxidized, NADH: nicotinamide adenine dinucleotide reduced, FAD: flavin adenine dinucleotide oxidized, FADH2: flavin adenine dinucleotide reduced, ATP: adenosine triphosphate, ADP: adenosine diphosphate, Q: ubiquinol, QH2: ubiquinone, cytCox: cytochrome c oxidized, cytCred: cytochrome c reduced. Fig D.2: Prediction of key regulators underlying the abnormal adaptation to training in COPD.
2. Methods
2.1. Experimental data
In this study, we explored abnormal muscle adaptation to training associated with COPD with the aim of examining the role of ROS in the activity state of the GRN in six different states: control group (12 individuals), COPD with normal BMI (13 patients), and COPD with low BMI (6 patients) (Rodriguez et al., 2011; Turan et al., 2011) before and after an 8-week training program (Sala et al., 1999).
2.2. Identifying differentially expressed genes
We performed a rank product analysis (Breitling et al., 2004) of transcriptomic data generated for the 3 study groups (control group, COPD with normal BMI, and COPD with low BMI) before and after an 8-week endurance training program (Supplementary material). Rank product is one of the most robust and widely used methods for analyzing gene expression data and provided a list of genes with significant differences between conditions (Figure 1A, step 2).
2.3. Gene regulatory network reconstruction
In order to reconstruct a GRN considering the effect of both training and the disease, we built a first draft using automated GRN reconstruction methods. To this end, we used Ingenuity Pathway Analysis (IPA) software and the IPA database (Ingenuity® Systems) (Krämer et al., 2014) to establish the relationships between those genes defined as differentially expressed. This software provides a platform that allows the integration of different layers of information such as genomics or proteomics and its graphical representation in a single interaction network. IPA Core Analysis can be used to link genotypes (molecular profiling data) to phenotypes and molecular events (signaling and metabolic pathways, molecular networks, etc), defining the reported interactions between genes, DNA, proteins, etc. (Krämer et al., 2014). In addition, we manually enriched our analysis by including information extracted from BioXM database (Maier et al., 2011).
Owing to the paucity of reported regulatory interactions in human skeletal muscle tissue, some genes remained unconnected in the draft reconstruction. To bridge the existing gaps, we imposed putative interactions by expanding our analysis, firstly to other muscular tissues in human (smooth and cardiac), secondly to other excitable tissues in humans (i.e. neurons) and finally to the Drosophila gene regulatory network (DroID, Murali et al., 2011). It was also extended to the mouse but this analysis did not add any new interactions to our network. For some of the new putative interactions it was necessary to include new genes into the network. We chose these new nodes (genes) while trying to use the minimum amount of new genes to connect the unconnected genes. Additionally, for the genes extracted from the Drosophila interaction network, we required that their human homologs were expressed in skeletal muscle based on the Atlas protein database (Uhlen et al., 2010).
Finally, we manually curated the GRN reconstruction using data from a large number of bibliographic sources (Supplementary material 1). The main objective of literature curation was to identify and correct incomplete or erroneous annotation and identify the direction and the sign of the interactions. This pipeline provided a highly curated mitochondrial muscle-specific GRN that integrated information of both disease effects (COPD) and adaptation to training.
2.4. Discrete modeling of the GRN
A discrete network model is a network abstracted in such a way that it can serve as a simple, efficient tool for the extraction of the very basic design principles of molecular regulatory networks (such as a GRN or a signaling pathway), without having to deal with all the biochemical details (de Jong et al., 2006). Typically, the mathematical description of GRN using discrete modeling approach is based on the asynchronous logical description proposed by R. Thomas (Thomas and Kaufman, 2001). This approach allows a semi-qualitative analysis of complex biological networks that describes the dynamics of the system (Corblin et al., 2010) (Figure 1C). A detailed description of the basis of discrete modeling is provided as on-line Supplementary Material 1. This approach was used to study the functional behavior of the reconstructed GRN.
2.5. Constraint integration into the model
To improve the predictive capabilities of the discrete model representing the reconstructed GRN, constraints must be integrated. This was performed in two different ways (Figure 1C, step 8).
Firstly, we introduced constraints based on differential gene expression analysis by analyzing the microarray data from skeletal muscle biopsies (Rodriguez et al., 2011) of control group and COPD patients with normal or low BMI before and after training (6 states). Here, we compared the same group before and after training and the three study groups at pre- and post-training state (9 comparisons, Figure 1A). This information was integrated into the discrete model in the form of inequalities. In this way, we formalized the situation in which the expression of gene “a” is significantly higher in “state 1” than in “state 2” (log2FC>0, p<0.01) as follows: Gastate1> Gastate2, which imposes a higher discrete value to gene “a” in “state 1” (Gastate1) than in “state 2” (Gastate2). Thus, we defined correlations in the discrete values of the nodes between states.
A second set of constraints, based on correlation analysis, was integrated as constraints in the discrete model in the form of inequalities. This analysis is based on the rationale that the discrete values of two genes that are significantly correlated cannot evolve in opposite directions (one increasing its expression and the other decreasing its expression), analogously two anti-correlated genes cannot evolve in the same direction.
Table 1 illustrates an example of how this approach is implemented. Here, we have two genes, “A” and “B”, with 4 and 3 possible discrete values, respectively, and no direct interactions between them. Assuming a significant correlation between gene “A” and “B” and on the basis of the discrete values depicted in table 1, there is only one possible discrete value of B in condition 2 (Bcon2), 2. This scenario excludes Bcon2=0 and 1. If instead, there is a significant anti-correlation, then Bcon2 only can be 0, excluding Bcond2=2 and 1.
Table 1.
Illustrative example of using expression correlations as constraints
| Genes/Prediction | Condition 1 | Condition 2 |
|---|---|---|
| A | 2 | 3 |
| B | 1 | Bcond2 |
Discrete values for genes A and B in conditions 1 and 2
We applied this approach to analyze a skeletal muscle gene expression dataset [GSE9405] in order to determine the existing correlations between the genes of different sample. Next, those correlations with a level of significance above a certain threshold are converted into constraints based on the criteria previously described and integrated into the discrete model in the form of inequalities. This approach is based on the Pearson correlation test and can be used to integrate information from probabilistic analysis into a deterministic model as constraints.
The probabilistic approaches, based on correlation and differential expression analysis, were integrated into our discrete model-based analysis to further reduce the number of feasible solutions.
2.6. Determine the level of significance of model predictions
To determine the accuracy of the predicted gene expression levels in response to an 8-week training program in healthy individuals and COPD patients by applying cross-validation. This method can be used to assess how the results of our analysis can be generalized to an independent data set. Here, we simulated the dynamics of the activity state of the GRN in response to endurance training by randomly removing a random number of constraints. Next, the results were compared with the results of the analysis considering all the constraints. This analysis is performed up to 1,000 times in order to build a matrix of contingencies. Finally, the p-value is calculated by χ2 test analysis on the table of contingencies.
2.7. Evaluate the effect of perturbation on key regulatory nodes aiming at retrieving post-training COPD to a healthy state
We simulated the effect of perturbations (e.g., a drug affecting the expression of a given gene) on one or a pair of genes that, along with 8-week endurance training program (Rodriguez et al., 2011), return the COPD patients to a healthy state (Figure 1 D.2). This is achieved in three main steps evaluating: i) the different adaptation to training between control group and COPD patients, ii) the effects of perturbations affecting key regulatory nodes in the adaptation to training in the COPD groups and iii) synergies between key nodes. To this end, we analyzed three aspects of the similarities observed between groups (number of nodes with the same response to endurance training): i) the degree of similarity between COPD and control groups, ii) the degree of improvement when a single gene is targeted in COPD groups compared with the wild type and iii) the degree of synergy when targeting a pair of nodes compared with the expected additive. The details of these analyses are explained in the Supplementary material 1.
3. Results
3.1. Gene Regulatory Network reconstruction of mitochondrial muscle-specific adaptation to training in COPD patients
We reconstructed a mitochondrial skeletal muscle-specific GRN that integrates information on mechanisms of training-induced adaptations in health and in COPD. This was achieved in three steps: i) gene expression analysis, ii) automated regulatory network reconstruction and iii) manual curation of the network reconstruction.
First, we determined the genes showing significant differences between the study groups (Rodriguez et al., 2011; Turan et al., 2011) by normalizing gene expression (Irizarry et al., 2003) and applying the rank product method (Breitling et al., 2004). We focused our network reconstruction on genes associated with mitochondrial processes, using the Gene Ontology Database (GO) (Berardini et al., 2010), String (Franceschini et al., 2013), Uniprot (Apweiler et al., 2014) and Human Protein Atlas (Uhlen et al., 2010) to identify those genes related with mitochondrial processes (GO: 0005743) expressed in human skeletal muscle. Finally, we obtained a list of 105 genes that showed significant variations between two or more of the six states of study in the context of mitochondrial processes (Supplementary Material 1, Table S1).
Next, these genes were used to reconstruct a mitochondrial musclespecific GRN by using a variety of tools and databases (Methods and Supplementary material 1). Since this study is focused on conditions in which the machinery of ROS detoxification is saturated, this node was defined just as an input node. In other words, it is not directly affected by the network but rather its activity state can be inferred from the activity state of the network (Figure 1B, step 4 and Supplementary Figure S6). Figure 1B shows an interaction network where the nodes represent two types of genes: i) genes differentially expressed between at least two of the six states (control, COPD with normal and low BMI, before and after training) and ii) genes included to connect the above-mentioned genes in a single GRN. The arrows represent the interactions between the nodes (activations or inhibitions). Thus, the resulting GRN accounted for 59 nodes (58 genes + ROS) and 170 gene regulatory interactions (Supplementary material 2) (Figure 2B, step 6 and Supplementary Figure S17). Since the resulting GRN captures the key genes, interactions and mechanisms related with responses to endurance training and with COPD disease, it offers an excellent platform to integrate various sorts of omics data into a discrete model, allowing the identification of the dynamics governing abnormal adaptation to training in COPD patients. It was also useful for simulating physiological responses of the GRN to perturbations produced by training-induced ROS levels.
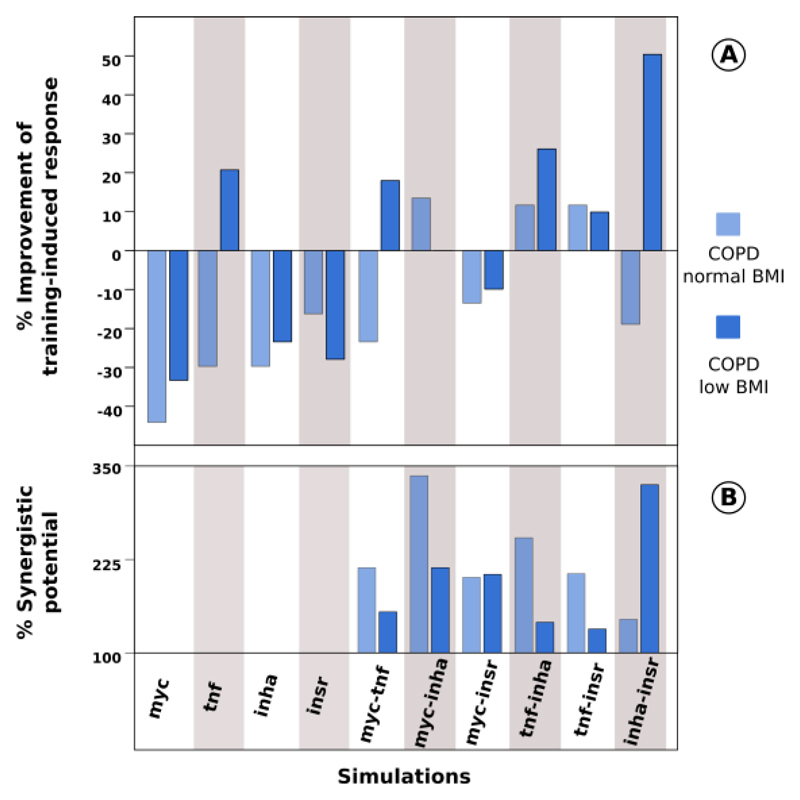
Fig. 2. Evaluation of potential therapeutic targets in COPD.
In the overall figure the first four columns represent the results of simulations on single target nodes and the six last correspond to simulations targeting pairs of nodes. Light and dark blue bars represent the results of the simulations of COPD patients with normal and low BMI, respectively Fig. 2 A: Evaluation of the % of improvement to training-induced response in both COPD groups compared with the results from table 2 and using as reference the response to training in control group. Fig. 1 B Evaluation of the % of synergistic potential compared with the expected additive effect.
We evaluated the predictive capability of our GRN reconstruction by integrating a 'validation data set' into a discrete model-driven analysis by using SysBiOX framework (Corblin et al., 2009; Thomas and Kaufman, 2001) (Methods and Supplementary material 1). The expression datasets corresponded to mouse muscle biopsies from three independent single gene knock-out experiments of myc, tp53 and insr and the gene expression information of these genes was omitted from the analysis. The resulting models correctly predicted the lack of activity of the corresponding knock-out genes in all three datasets, lending support to the GRN reconstruction (Supplementary material 3).
3.2. Building a multi-state discrete model of mitochondrial muscle-specific GRN on the basis of molecular mechanisms and probabilistic-based constraints
We performed a reduction transformation of the GRN while conserving the properties of the initial network (Kobayashi et al., 2003 and Naldi et al., 2011) (Methods). We also integrated constraints through various probabilistic approaches (Methods and Supplementary material 1).
Next, we analyzed the system in order to infer the parameter ranges (and thus possible functional behaviors) of the observed activity state of the network in the six different conditions. To this end, we imposed an objective function that minimizes the overall discrete value of the nodes of the network, which maximizes the gene expression efficiency. As a result of this procedure, we obtained a vector of discrete values representing the activity state of the genes (nodes in the model) in each of the six conditions. Some of the values in these vectors were directly inferred from the gene expression data that were previously integrated as constraints (Methods), and the others were predicted. Next, we performed a cross-validation analysis to assess the predictive capability of our method. In this process we simulated the evolution of the node values in the GRN in response to endurance training by randomly removing a random number of constraints (extracted from gene expression analyses). This analysis was performed 1,000 times and showed that our computational approach was able to predict 85.37% of the node changes associated with the 8-week endurance training program, with an associated p-value lower than 10-5 (χ2 test) (Methods and Supplementary material 3).
3.3. Key adaptive differences associated with abnormal metabolic adaptations to training in COPD patients
COPD patients show abnormal physiological adaptations to training compared with the control group, as indicated in Table 2, wherein changes displayed are consistently predicted by all best-fit models in our analysis. Specifically, endurance training provokes more extensive transcriptional changes in the control group than in both COPD groups (Table 2 and Figure 1 D.1).We observed well-defined patterns of alterations in energy metabolism and muscle remodeling in COPD patients (Table 2). As compared to healthy individuals, the models predicted an abnormal low activity of ETC, TCA cycle and creatine kinase in response to an 8-week training program in COPD patients with low BMI. Interestingly, for COPD patients with normal BMI, the models predict up-regulation of complex I (as in healthy patients) and a slight increase in citrate synthase and succinyl-CoA ligase. However, the models also predict that complexes III, IV and V, other key enzymes of TCA cycle, and creatine kinase activities decrease after training in COPD patients with normal BMI, suggesting a defective energetic metabolism adaptation to training in this group. Thus, the physiological response to endurance training in the healthy individuals is consistent with a higher capacity for O2 utilization and energy production than in COPD patients. Interestingly glycerol-3-phosphate dehydrogenase cytosolic isoform is up-regulated in healthy individuals and down-regulated in the two COPD groups. We observed the opposite pattern in the mitochondrial isoform which enters electrons from NADH into the ETC that could partially compensate the reduction in ETC and TCA cycle activity. The model also shows differences in cdk4 and tnf in response to exercise in both COPD groups compared with healthy individuals. Both cdk4 and tnf have an important role promoting adaptive changes in myofiber cytoar-chitecture and protein composition in muscle (muscle remodeling) (Lee et al., 2009). The models also predicted differences in acadvl (very long-chain specific acyl-CoA dehydrogenase), insulin receptor factor (insr) and pyruvate dehydrogenase (pdha) that are up-regulated after training in healthy group and down-regulated in both COPD groups. While pdh and insr regulate glycolysis, advl is a key activator of beta-oxidation. These pathways fuel TCA cycle and ETC. Then, unlike control group, COPD patients reduce the expression of these enzymes that in turns deprives the energy metabolism. The predicted abnormal metabolic response to endurance training in COPD could explain the potential harm of short duration, high-intensity endurance training programs in severe COPD patients (Barreiro et al., 2009). The fact that abnormal adaptation to training affects genes and proteins related with muscle remodeling could explain the loss of muscle mass observed in the most severe group of COPD patients. In addition, cancer-associated genes such as tnf or tp53 also increase their activity in response to training in both COPD groups which could be related with the reported increase of COPD-lung cancer co-morbidity (Cohen et al., 2014). To determine the robustness of our computational method predicting specific gene changes associated with the endurance training, we evaluated the adaptation to training in the activity state of one specific node (value of a given gene at trained state compared with sedentary state) when the constraints related with that node were omitted. This analysis was performed with all the genes of the network (the node values supported by experimental measurements and the node values with no associated differential expression data, 58 analyses in total). We observed that the non-constrained analyses provided the same predictions for the unconstrained nodes as the analysis considering all the constraints in 74.01% of the cases, which is higher than expected by chance (P<10-5, χ2 test, Supplementary material 3).
Table 2.
Heatmap of the inferred activity changes following training. The three study groups are in rows: Control group, COPD patients with normal BMI (COPDBMI_N) and with low BMI (COPDBMI_L). Columns represent the genes considered in our analysis, grouped by pathways or cellular function: ETC (electronic transport chain), TCA cycle (tricarboxylic acid cycle), Glycolysis, ROS (reactive-oxygen species metabolism), Transc. Reg. (transcriptional regulation), Cancer Met. (cancer metabolism), Transport, Lipid Met (lipid metabolism). Cell Cycle and Prot. Deg. (protein degradation). Adaptation to training is represented using the following code: i) red: decrease in the discrete value of the node in response to training and ii) green: increment in the discrete value of the node in response to training. In both cases, the intensity of the colors indicates: i) predicted response to training supported by between 67 and 99% of the models are highlighted with the lightest colors, ii) predictions supported by the 100% of the models are highlighted with medium intensity color and iii) darkest color indicates nodes with no adaptation to training (supported by the 100% of the models). Finally, results supported by experi-mental evidences that were integrated as constraints in our analysis, are labeled with an asterisk (*).
| ETC | TCA cycle | Glycolysis | ROS | Transc. Reg. | Cancer Mat. | Transport | Lipid Met. | Cell Cycle | Prot. Deg. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| ros | atp5d | cox6a2 | ndufsl | ndufv 1 | ndufs7 | ndufb7 | uqcrc1 | ckmt2 | cs | idh2 | ogdh | suclgl | pkm2 | pdha 1 | pdk4 | gpd2 | gpdl | sod2 | glrx | insr | mapk12 | nol3 | ppp2ca | srekl | hspb7 | mcd30 | crebl | parpl | flna | akpl | prkce | hsp9()ab | tnf | tp53 | rtn4ipl | lin9 | myb | myc | mll | stx4 | P2ry2 | trakl | tspo | pam 16 | acbd3 | col4a3bp | acadvl | fasn | gpam | PParg | inha | acsl5 | casp8 | cdk4 | app | fastk | eln | ubc | ||
| Adaptation of training | Control | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||||||||||||||||||||||||||||||||||||||
| COPDBMI_N | * | * | * | * | * | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| COPDBMI_L | * | * | * | * | * | * | * | * | * | |||||||||||||||||||||||||||||||||||||||||||||||||||
3.4. Anomalous ROS levels in low-BMI COPD patients in response to training
All the models predicted a reduction of ROS in healthy subjects after training, which is consistent with previously reported observations (Puente-Maestu et al., 2012; Rabinovich et al., 2001). A training-induced reduction of ROS levels in COPD group with normal BMI was also consistently predicted. Moreover, ROS levels in COPD group with low BMI were predicted to increase in response to the endurance training, as observed in independent studies (Rabinovich et al., 2001). Since the intracellular production of ROS may lead to oxidative stress and the damage of cellular components, the increment of ROS could explain the deficient adaptation to training observed in COPD patients with low BMI (Barreiro et al., 2009). To examine the robustness of these findings, we analyzed how ROS was predicted to change after training assuming that: i) all the sedentary groups had the same levels of ROS and ii) omitting all the constraints relating to ROS from experimental measurements (i.e. discrete value of ROS in the control group is equal or higher before than after training; see Methods for details of the analysis) (Puente-Maestu et al., 2012; Powers and Jackson, 2008). This non-constrained ROS analysis provided the same predictions as our original ROS analysis (Supplementary material 3). Additionally, the changes in ROS levels predicted in the control group in response to endurance training are consistent with experimental observations (Puente-Maestu et al., 2012; Powers and Jackson, 2008). These results validate the predictions on the variations in the level of ROS in response to endurance training in COPD patients.
3.5. Key players underlying abnormal metabolic adaptation to training in COPD patients
According to our model, the differences lie mainly in those genes that are directly related with the training-induced energy metabolism response (Table 2), which is activated in healthy people but is abnormally low in both COPD patients groups. We next identified key nodes regulating this abnormal adaptation to training. To this end, we explored the effects of training on the activity state of gene regulatory interactions in the three study groups (i.e. activation or inactivation in response to training). In our discrete model, if the effector node is below the threshold of a given interaction, the interaction is inactive and the target node is not perturbed through this interaction. The analysis identified different abnormal adaptations to training, at the level of gene regulatory interactions, in COPD individuals compared to healthy ones. Interestingly, these interactions were not randomly distributed in the network, but were concentrated around specific effectors: myc, tnf, inha or insr (Figure 1 D.2). Moreover, in the previous analyses tnf and insr presented clear and robust differences in their adaptations to training in healthy people compared with both COPD groups (Table 2). These results indicate that changes in activity state in tnf and insr in response to endurance training may be propagated through the GRN with significant effects on the state of the overall network, regulating the different adaptation to training observed in the three patient groups.
3.6. Potential therapeutic targets to modulate skeletal muscle dysfunction in COPD
We next evaluated the effects of in silico inhibition/activation of the key regulatory nodes: myc, tnf, inha and insr. These simulations were performed by perturbing key regulatory nodes, single or pairs of nodes, in order to explore their potential to retrieve post-training COPD patients to a state as close as possible to the healthy state. The aim of the simulations was threefold: i) to identify the functional impact of perturbing all single and pairwise combinations of nodes; ii) to assess the synergies between nodes; and, iii) to explore potential therapeutic strategies to modulate muscle dysfunction in COPD, either based on novel therapies or drug repositioning. To this end, we fixed the training-induced response in healthy individuals (Table 2) and eliminated all the constraints related with adaptation to training in both COPD groups (Methods). Thereafter, we analyzed the training response of the two COPD groups while imposing the same post-training value on the key target node(s) in both COPD groups as observed in post-training healthy subjects. Next, we evaluated if the training response was closer to healthy group adaptation compared with the results of each COPD group summarized in table 2. This approach was performed for all the key single nodes and their possible pairs (Methods and Supplementary Material 1).
Most of the perturbations targeting pairs of regulatory nodes yielded functional improvement in retrieving COPD patients to a state closer to healthy individuals. The combination of tnf and insr showed high potential to improve the training-induced response in both COPD groups (13% and 11% in COPD normal BMI and low BMI, respectively; Figure 2A). Moreover, the tnf-insr pair showed high synergistic potential. More specifically, our simulations predicted that the improvement in the training-induced response in COPD patients with normal BMI was 106% higher than expected by additive effects while it was 36% higher in COPD patients with low BMI (Figure 2B and Supplementary material 1). We note that while tnf is down-regulated in healthy and up-regulated in both COPD groups after training, insr has an opposite pattern. The predicted adverse effects of tnf on skeletal muscle bioenergetics is consistent with the reported correlation between COPD severity and tnf levels (Victor Pinto-Plata et al., 2012; Qi et al., 2014) and is consistent with our predictions that the impact of using tnf inhibitors as a drug is higher in COPD with low BMI than in COPD patients with normal BMI, compared with the wild types simulations (Table 2; 23% higher and 33% lower in COPD with normal and low BMI respectively; Figure 2A). The synergies between tnf and insr rely on bibliographic evidence indicating that COPD may directly increase insulin resistance through the effects of chronic inflammation on skeletal muscle insr (Lorenzo et al., 2008; Plomgaard et al., 2005). Overall, these reports support the results of our simulations predicting low effects of an insr-related drug alone (18% and 31% lower COPD with normal and low BMI respectively; Figure 2A). While most of the perturbations on target regulatory pairs of genes that included insr showed low effectiveness, interventions on the pair tnf-insr generated a significant improvement in both COPD groups. The effects predicted in our simulations were supported by a number of experimental results (Pinto-Plata et al., 2012; Qi et al., 2014; Lorenzo et al., 2008).
The current findings on potential therapeutic interventions seem to pave the way for novel drug repositioning for COPD patients. For example, it has been demonstrated that statins affect insulin receptors by decreasing skeletal muscle insulin resistance (Lalli et al., 2008; Saito et al., 2007). Moreover, compounds with statin activity, such as simvastatin, can inhibit tnf alpha activity (Lin et al., 2013) and are used, along with exercise, to reduce elevated lipid levels and prevent cardiovascular diseases (Pyörälä et al., 2004). Our predictions suggest the potential for drug repositioning (i.e statins) in COPD with important clinical implications.
4. Conclusion
In this study, we developed a novel strategy to reconstruct a highly curated mitochondrial muscle-specific GRN and the integration of mechanisms described by this network with information extracted from probabilistic approaches into a discrete model-driven analysis.
The novel computational method developed in this work combines probabilistic and model-driven approaches. Model-driven approaches can be used to analyze the dynamics of a system which cannot be carried out through classical gene expression data analysis. This is because these approaches are based only on the significant gene expression differences between conditions but do not take into account the interactions between the elements of the system. Thus, relatively small changes in a key gene may modify dramatically the behavior of a system but not be captured by significant changes in its gene expression value. However, model-driven methods are limited in size due to the lack of parameters that drive it to a large number of feasible solutions. On the other hand, probabilistic approaches allow the integration of constraints from a large number of sources which significantly reduces the number of feasible solutions allowing the expanded use of model-driven method to larger interaction networks. Our combined method has permitted us to overcome existing limitations in mechanistic approaches to deal with complex networks. More specifically, this approach allowed the data-driven integration of large-scale constraints into a model-driven analysis, enabling its application to large systems that cannot otherwise be studied due the lack of experimentally measured quantitative parameters. Additionally, it has enabled us to expand the use of probabilistic methods to the study of underlying molecular mechanisms. In addition, our approach can provide more information than other currently available data-driven methods. Thus, for instance, rank product analysis (R Breitling et al, 2004) depicted in table S1 (Supplementary material 1) shows few gene expression differences between conditions while our analysis was able to determine how training modulates the expression of all the genes in the three study groups. We also analyzed the skeletal muscle transcriptomic data using SamNet (Gosline SJ. et al, 2012), a tool which permits the integration of gene expression data into a case-specific interaction network reconstruction analysis. Importantly, in all the analyses our model-driven approach showed superior predictive performance compared to SamNet. More specifically, our approach, unlike SamNet, was able to integrate all the relevant genes into a single GRN adding fewer new nodes and a larger number of interactions per node. In addition, our approach could predict with more accuracy the activity state of each interaction in response to training and consequently define the key genes underlying abnormal metabolic adaptation to training in COPD patients (Supplementary material 1).
Our study has permitted us to unlock important mechanisms underlying the abnormal adaptation to training in COPD patients and the role of ROS in this disease. This new understanding can be used to design more efficient strategies combining optimal endurance training programs with drug treatments targeting specific key nodes of the energy metabolism.
This novel approach can be also extrapolated to other complex and multifactorial diseases with strong metabolic components, such as cancer, cardiovascular disorders or diabetes that often occur as comorbid conditions in COPD patients. In addition, our in silico analysis is suitable for integrating not only transcriptomic data, but also proteomic data. Further efforts to improve automated network reconstruction and probabilistic-based constraint integration methods that use patient-specific multi-omic data are expected to extend the applicability of this approach for personalized medicine. We anticipate that the approach presented here will open a new avenue in the study of COPD and other multifactorial and complex diseases, facilitating the development of more efficient treatments.
Supplementary Material
Supplementary data are available at Bioinformatics online.
Acknowledgements
This work was supported by the European Commission Seventh Framework Programme FP7 (Synergy-COPD project grant agreement n° 270086); the Spanish Government funds (SAF2014-56059-R (MINECO, Spain); the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR)-Generalitat de Catalunya (2014SGR1017); the 'Lendület' Programme of the Hungarian Academy of Sciences (BP) and The Wellcome Trust (BP). the "ICREA Academia" prize for excellence in research, ICREA foundation-Generalitat de Catalunya (MC). We also acknowledge the contribution of the reviewers to the improvement of the manuscript.
References
- Apweiler R, et al. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42(Database issue):D191–8. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A, Oltvai Z. Network biology: understanding the cells' functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barreiro E, et al. Chronic endurance exercise induces quadriceps nitrosative stress in patients with severe COPD. Thorax. 2009;64(1):13–9. doi: 10.1136/thx.2008.105163. [DOI] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, et al. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, et al. Biochemistry. 5th edition. New York: W H Freeman; 2002. sect 18.5. [Google Scholar]
- Breitling R, et al. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Cano I, et al. Oxygen pathway modeling estimates high reactive oxygen species production above the highest permanent human habitation. PloS One. 2014;9(11):e111068. doi: 10.1371/journal.pone.0111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, et al. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2014;14(1):58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- Corblin F, et al. A declarative constraint-based method for analyzing discrete genetic regulatory networks. Biosystems. 2009;98(2):91–104. doi: 10.1016/j.biosystems.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Corblin F, et al. Applications of a formal approach to decipher discrete genetic networks. BMC Bioinformatics. 2010;11:385. doi: 10.1186/1471-2105-11-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin SS. Toward a road map for global -omics: a primer on -omic technologies. Am J Epidemiol. 2014;180(12):1188–95. doi: 10.1093/aje/kwu262. [DOI] [PubMed] [Google Scholar]
- de Jong H, Ropers D. Qualitative approaches to the analysis of genetic regulatory networks. MIT Press; 2006. pp. 125–148. [Google Scholar]
- Fisher J, Henzinger TA. Executable cell biology. Nat Biotechnol. 2007;25(11):1239–49. doi: 10.1038/nbt1356. [DOI] [PubMed] [Google Scholar]
- Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cabrero D, et al. Synergy-COPD: a systems approach for under-standing and managing chronic diseases. J Transl Med. 2014;12(Suppl 2):S2. doi: 10.1186/1479-5876-12-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, et al. Prediction of chronic obstructive pulmonary disease (COPD) in asthma patients using electronic medical records. J Am Med Inform Assoc. 2009;16(3):371–9. doi: 10.1197/jamia.M2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, et al. Modeling genetic switches with positive feedback loops. J Theor Biol. 2003;221(3):379–99. doi: 10.1006/jtbi.2003.3190. [DOI] [PubMed] [Google Scholar]
- Krämer A, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli CA, et al. Statin modulates insulin signaling and insulin resistance in liver and muscle of rats fed a high-fat diet. Metabolism. 2008;57(1):57–65. doi: 10.1016/j.metabol.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Lee SJ, et al. TNF-alpha regulates vascular smooth muscle cell responses in genetic hypertension. Int Immunopharmacol. 2009;9(7–8):837–43. doi: 10.1016/j.intimp.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Lin YC, et al. Simvastatin attenuates the additive effects of TNF-α and IL-18 on the connexin 43 up-regulation and over-proliferation of cultured aortic smooth muscle cells. Cytokine. 2013;62(3):341–51. doi: 10.1016/j.cyto.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Long TA, et al. Systems approaches to identifying gene regulatory networks in plants. Annu Rev Cell Dev Biol. 2008;24:81–10. doi: 10.1146/annurev.cellbio.24.110707.175408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M, et al. Insulin resistance induced by tumor necrosis factor-alpha in myocytes and brown adipocytes. J Anim Sci. 2008;86(14 Suppl):E94–104. doi: 10.2527/jas.2007-0462. [DOI] [PubMed] [Google Scholar]
- Maier D, et al. Knowledge management for systems biology a general and visually driven framework applied to translational medicine. BMC Syst Biol. 2011;5 doi: 10.1186/1752-0509-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltais F, et al. An official American Thoracic Society/European Respiratory Society statement: update on limb muscle dysfunction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;189:e15–62. doi: 10.1164/rccm.201402-0373ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, et al. Predictive medicine: outcomes, challenges and opportunities in the Synergy-COPD project. J Transl Med. 2014;12(Suppl 2):S12. doi: 10.1186/1479-5876-12-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JD, et al. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med. 2015;8(1):1. doi: 10.1186/s12920-014-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali T, et al. DroID 2011:a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39(Database issue):D736. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJ, Lopez AD. Measuring the global burden of disease. N Engl J Med. 2013;369(5):448–57. doi: 10.1056/NEJMra1201534. [DOI] [PubMed] [Google Scholar]
- Naldi A, et al. Dynamically consistent reduction of logical regulatory graphs. Theorl Comput Sci. 2011;412:2207–2218. [Google Scholar]
- Pinto-Plata V, et al. Inflammatory and repair serum biomarker pattern. Association to clinical outcomes in COPD. Respir Res. 2012;13:71. doi: 10.1186/1465-9921-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomgaard P, et al. Tumor necrosis factor-alpha induces skeletal muscle insulin resistance in healthy human subjects via inhibition of Akt substrate 160 phosphorylation. Diabetes. 2005;54(10):2939–45. doi: 10.2337/diabetes.54.10.2939. [DOI] [PubMed] [Google Scholar]
- Powers SK, Jackson MJ. Exercise-induced oxidative stress:cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–76. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente-Maestu L, et al. Site of Mitochondrial ROS Production in Skeletal Muscle of COPD and its Relationship with Exercise Oxidative Stress. Am J Respir Cell Mol Biol. 2012;47(3):358–62. doi: 10.1165/rcmb.2011-0382OC. [DOI] [PubMed] [Google Scholar]
- Pyörälä K, et al. Reduction of cardiovascular events by simvastatin in nondiabetic coronary heart disease patients with and without the metabolic syndrome: subgroup analyses of the Scandinavian Simvastatin Survival Study. Diabetes Care. 2004;27(7):1735–40. doi: 10.2337/diacare.27.7.1735. [DOI] [PubMed] [Google Scholar]
- Qi Y, et al. Inhibition of AMPK expression in skeletal muscle by systemic inflammation in COPD rats. Respir Res. 2014;15:156. doi: 10.1186/s12931-014-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich RA, et al. Reduced muscle redox capacity after endurance training in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;164(7):1114–8. doi: 10.1164/ajrccm.164.7.2103065. [DOI] [PubMed] [Google Scholar]
- Rabinovich RA, et al. Mitochondrial dysfunction in COPD patients with low body mass index. Eur Respir J. 2007;29(4):643–50. doi: 10.1183/09031936.00086306. [DOI] [PubMed] [Google Scholar]
- Rodriguez DA, et al. Muscle and blood redox status after exercise training in severe COPD patients. Free Radic Biol Med. 2011;52(1):88–94. doi: 10.1016/j.freeradbiomed.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Sala E, et al. Effects of endurance training on skeletal muscle bioenergetics in chronic obstructive pulmonary disease. AmJRespir Crit CareMed. 1999;159:1726–34. doi: 10.1164/ajrccm.159.6.9804136. [DOI] [PubMed] [Google Scholar]
- Saraswati S, Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Front Microbiol. 2014;5:764. doi: 10.3389/fmicb.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, et al. Statin reverses reduction of adiponectin receptor expression in infarcted heart and in TNF-alpha-treated cardiomyocytes in association with improved glucose uptake. Am J Physiol Heart Circ Physiol. 2007;293(6):H3490–7. doi: 10.1152/ajpheart.00310.2007. [DOI] [PubMed] [Google Scholar]
- Selivanov VA, et al. Bistability of mitochondrial respiration underlies paradoxical reactive oxygen species generation induced by anoxia. PLoS Comput Biol. 2009;5:e1000619. doi: 10.1371/journal.pcbi.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Kaufman M. Multistationarity, the basis of cell differentiation and memory. II. Logical analysis of regulatory networks in terms of feed-back circuits. Chaos. 2001;11:180–195. doi: 10.1063/1.1349893. [DOI] [PubMed] [Google Scholar]
- Tian C, et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol. 2014;10 doi: 10.15252/msb.20145470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan N, et al. A systems biology approach identifies molecular networks defining skeletal muscle abnormalities in chronic obstructive pulmonary disease. PLoS Comput Biol. 2011;7(9):e1002129. doi: 10.1371/journal.pcbi.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Vestbo J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187(4):347–65. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- Gosline SJ, Spencer SJ, Ursu O, Fraenkel E. SAMNet: a network-based approach to integrate multi-dimensional high throughput datasets. Integr Biol (Camb) 2012;4(11):1415–27. doi: 10.1039/c2ib20072d. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends Genet. 2010;26:519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, et al. Quantitative Equilibrium Signal Mapping: A Reliable Alternative to CT in the Assessment of Emphysema in Patients with Chronic Obstructive Pulmonary Disease. Radiology. 2015;7:132953. doi: 10.1148/radiol.14132953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.