Abstract
Post-translational farnesylation or geranylgeranylation at a C-terminal cysteine residue regulates localization and function of over 100 proteins, including the Ras isoforms, and is a therapeutic target in diseases including cancer and infection. Here we report global and selective profiling of prenylated proteins in living cells enabled by development of isoprenoid analogues YnF and YnGG in combination with quantitative chemical proteomics. Eighty prenylated proteins were identified in a single human cell line, 64 for the first time at endogenous abundance without metabolic perturbation. We further demonstrate that YnF and YnGG enable direct identification of post-translationally processed prenylated peptides, proteome-wide quantitative analysis of prenylation dynamics and alternative prenylation in response to four different prenyltransferase inhibitors, and quantification of defective Rab prenylation in a model of the retinal degenerative disease Choroideremia.
Introduction
Protein prenylation is a class of irreversible post-translational modification predicted to affect hundreds of proteins in the human proteome1. Farnesyl transferase (FTase) and geranylgeranyl transferase type 1 (GGTase-1) catalyze attachment of a single farnesyl or geranylgeranyl isoprenoid to a canonical C-terminal CXXX-motif (C = Cys; X = any amino acid) on proteins including Ras and Rho GTPases, nuclear lamins and gamma subunits of heterotrimeric G-proteins (Figure 1a). RabGGTase (GGTase-2) attaches one or two geranylgeranyl chains to members of the Rab family at a variety of cysteine-containing C-terminal motifs, with the assistance of Rab Escort Proteins, REP1 or REP2. Prenylation plays an important role in all normal cells, and in numerous human pathologies including cancers2, cardiovascular3 and retinal4 diseases, viral infections5,6 and Hutchinson-Gilford progeria syndrome (HGPS)7.
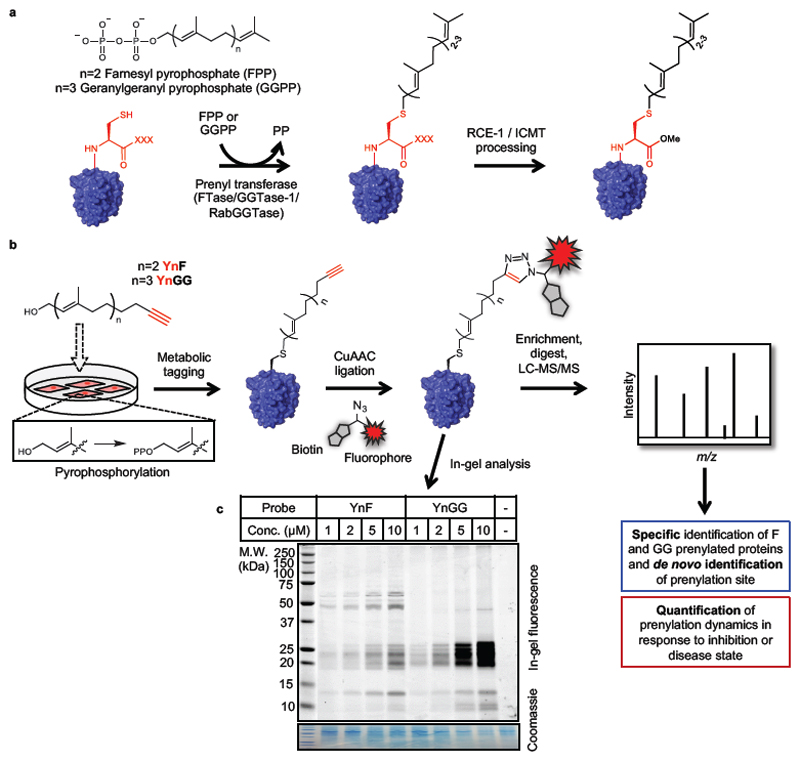
Figure 1. Global labeling of prenylated proteins using novel alkyne-tagged isoprenoid analogues.
(a) Prenylation, the covalent attachment of a farnesyl or geranylgeranyl isoprenoid to a cysteine near the protein C-terminus, is catalyzed by FTase or GGTase-1 on proteins bearing a C-terminal CXXX motif (C = Cys; X = any amino acid), or by RabGGTase at the C-terminus of Rab proteins. Some CXXX proteins undergo further processing by RCE-1 (cleavage of the C-terminal -XXX residues) and ICMT (carboxyl methylation). (b) Schematic of metabolic labeling workflow by novel alkyne-tagged prenyl probes YnF and YnGG. Probe-labeled proteins can be processed by Cu(I)-catalyzed click chemistry to attach capture reagents with various functionality such as affinity tags or fluorophores to enable enrichment for mass spectrometric analysis or (c) visualization by in-gel fluorescence in EA.hy926 cells.
Prenyl transferase inhibitors (PTIs) have been proposed as inhibitors of Ras signaling in cancer, but despite extensive clinical trials none of these agents has yet been approved for clinical use2. Further development is hampered by limited validation of the cellular targets of prenylation, and poor understanding of the dynamic interplay between prenyl transferases and their substrates in response to inhibition which can drive certain substrates to ‘switch’ between farnesylation and geranylgeranylation, making patient selection highly challenging1. Recently, farnesyl transferase inhibitors (FTIs) have shown positive clinical results in HGPS7 and chronic hepatitis delta virus (HDV) infection6, diseases in which FTI efficacy is thought to occur mainly through a specific prenylated protein. To explain past failures and set future directions there is a need to understand the prenylated proteome in specific cellular contexts, and to determine which proteins are targeted by inhibition of each prenyl transferase2.
Chemical proteomics is a powerful methodology to decipher substrates of post-translational modifications (PTMs), particularly for analytically challenging PTMs such as lipidation8–10. Several useful chemical probes for protein prenylation have been reported11–16, but currently-available methodologies provide limited coverage of the prenylated proteome and poor selectivity for farnesylation vs. geranylgeranylation pathways. There thus remains a need for chemical probes which can enable high-throughput quantification of specific changes in prenylation in disease models or in response to PTIs, at the whole proteome level.
Here we report a high-throughput approach for exploration of protein prenylation in living cells, exploiting a pair of novel alkyne-tagged prenyl probes coupled with multifunctional capture reagents and a quantitative chemical proteomics workflow (Figure 1b). We demonstrate that this methodology can be used to identify novel prenylated proteins at proteome-wide scale and at endogenous cellular abundance, and to quantify substrate-specific inhibition of prenylation upon treatment with different classes of PTIs, enabling system level validation and invalidation of widely-used tool inhibitors. We further show the broad utility of these probes to quantify “rescue” prenylation induced by treatment with a potent FTI, directly identify post-translational processing of prenylated C-termini, and quantify the complex effect on geranylgeranylation resulting from REP1 knock-out in a model of the X-linked retinal disease Choroideremia. Together, these probes and capture reagents significantly extend the existing tools available for metabolic prenyl labeling (Supplementary Table 1), enabling new applications in the exploration of protein prenylation in living cells.
Results
Novel alkyne-tagged isoprenoid analogues YnF and YnGG enable identification and quantification of prenylated proteins
We reasoned that structural similarity of chemical probes to the natural products farnesol (FOH) and geranylgeraniol (GGOH) should lead to high fidelity incorporation into cellular prenylated targets. Accordingly, we developed two prenyl probes designed to target farnesylation and geranylgeranylation selectively, termed YnF and YnGG, respectively (Figure 1b, synthetic methodology in Supplementary Information), in which an alkyne moiety enables post-incorporation coupling of functional ‘capture reagents’ bearing fluorophores and/or affinity tags through copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (so-called ‘click’) chemistry17,18. The corresponding pyrophosphates of each probe (YnFPP and YnGGPP, Supplementary Figure 1) were also synthesized, and their incorporation into peptides mimicking prenylated protein substrate C-termini was compared against FPP and GGPP, for FTase and GGTase-1 (Supplementary Figure 1), using a biochemical enzyme assay (Supplementary Information). In line with the biomimetic design of the probes, transfer of each probe was found to exhibit rates of incorporation and Km values aligned with those of the canonical native prenyl pyrophosphate for FTase (RHEB) and GGTase-1 (RHOA) peptide substrates, with no cross-reactivity with the non-substrate peptide/transferase pair (Supplementary Figure 2).
Probe incorporation in cells was first examined in an endothelial cell line (EA.hy92619), adding YnF or YnGG at a range of concentrations to cells in culture over 24 hours, followed by isolation of protein and ligation by CuAAC to AzTB (Supplementary Figure 8), an azide-tagged capture reagent bearing a TAMRA fluorophore and a biotin handle18. Analysis by in-gel fluorescence showed excellent incorporation of the probes at low micromolar concentration, with labeling evident at 1 µM probe, and increasing at 10 µM (Figure 1c). Previous studies using prenyl probes have typically pretreated cells with a HMG-CoA reductase inhibitor (statin) to deplete endogenous isoprenoids and enhance labeling11,15,16; however, statins exert widespread effects on metabolism, including changes in the levels of cholesterol metabolites and of prenylated proteins20. In contrast, incorporation of YnF and YnGG proceeds with high efficiency without statin treatment, enabling study of prenylation under unperturbed conditions. Both probes showed strong labeling in the 20-25 kDa region by in-gel fluorescence, consistent with labeling of small GTPases, but each probe also labeled an orthogonal set of bands, suggesting that selective farnesyl and geranylgeranyl probes may provide enhanced coverage of the whole complement of prenylated proteins. Consistent with selective labeling, probe incorporation was sensitive to competition by the respective natural isoprenoid substrate, farnesol (FOH) or geranylgeraniol (GGOH) (Supplementary Figure 3).
We next employed a quantitative chemical proteomics strategy to identify and validate protein targets labeled by the probes, using competition against natural isoprenoids. EA.hy926 cells were treated with 10 µM YnF or YnGG in the presence of increasing concentrations of FOH (0, 5, 25 µM) or GGOH (0, 2.5, 10 µM), and cell lysates spiked with SILAC lysate dual labeled with 13C615N4-Arg and 13C615N2-Lys (R10K8) and the relevant prenyl probe, providing a mass standard against which competition could be quantified21. Following CuAAC ligation, affinity enrichment and on-bead tryptic digest, samples were analyzed by nanoLC-MS/MS on a high resolution Orbitrap mass spectrometer (Supplementary Data 1 and 2). Although some overlap in protein identification between the two probes was evident, 75% of proteins which significantly responded to competition were uniquely detected by one probe only (Supplementary Figure 3), underlining the advantages of a dual farnesyl and geranylgeranyl probe system. All proteins sensitive to competition by natural isoprenoids carried a potential prenylation motif, with the exception of four proteins known to interact with Rabs (COPA, CD9, CD151, PODXL), enabling robust identification of prenylated proteins over background (Figure 2a and 2b, Supplementary Figure 4, Supplementary Data 1 and 2). The probes discriminate with high selectivity between known non-prenylated or prenylated proteins, with only the latter showing significant changes in the presence of competing canonical substrate (FOH or GGOH). YnF also identified 7 novel farnesylated proteins which showed identical competition behavior to the set of known farnesylated proteins (Figure 2a), including serine-threonine kinase ULK3, DDB1- and CUL4-associated factor 8 (DCAF8/WDR42A), centrosomal protein CEP85, leucine-rich repeat protein LRRFIP1, nuclear assembly protein NAP1L4, RhoBTB subfamily member RHOBTB3, and protein DPCD. Our approach also enables objective identification of CXXX motif proteins which are not substrates for probe incorporation (Figure 2a and 2b). For example, ubiquitin C-terminal hydrolase L1 (UCHL1), an abundant deubiquitinase previously reported to be farnesylated22, was not identified as a substrate for either probe in EA.hy926 cells. A UCHL1 C-terminal peptide was also not a substrate for prenylation in biochemical assays with any prenyl pyrophosphate (FPP, YnFPP, GGPP or YnGGPP; Supplementary Figure 2). Of the 80 prenylated proteins identified here with high confidence, 64 are shown to be prenylated at endogenous cellular abundance for the first time, without the use of disruptive protein overexpression or treatment with a statin (Supplementary Data 1), providing the most comprehensive set of prenylated proteins for a single cell type determined to date. A further putative 3 farnesylated and 6 geranylgeranylated proteins were identified for which competition resulted in non-detection of the protein even at the lowest concentration of natural isoprenoid (Supplementary Data 1, ‘low confidence’ substrates).
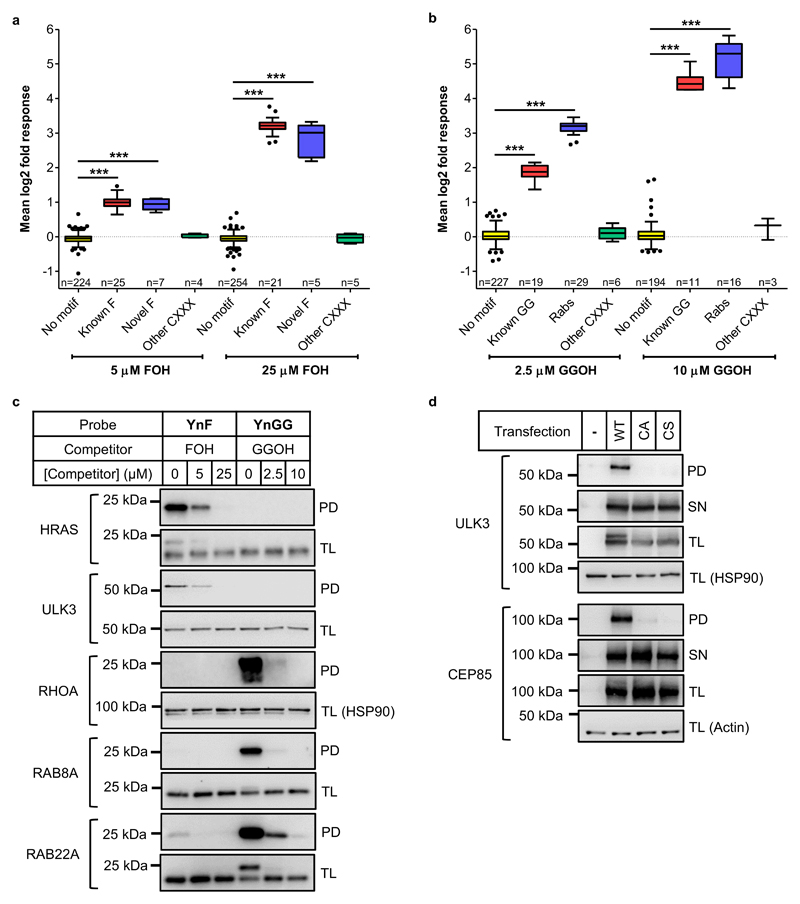
Figure 2. Prenylated protein discovery in EA.hy926 cells using prenyl probes YnF and YnGG.
(a) YnF and (b) YnGG labeling of prenylated CXXX substrates (Known F, Novel F and Known GG) and Rab proteins show dose-dependent sensitivity to competition with natural isoprenoid substrates farnesol (FOH) or geranylgeraniol (GGOH), quantified by spike-in SILAC. Proteins lacking a CXXX motif (No motif) and non-substrate CXXX proteins (Other CXXX) do not respond to isoprenoid competition. Kruskal-Wallis test with Dunn’s post-test (***p<0.01). Box and whisker plots represent median values (center lines) and 25th and 75th percentiles (box limits) with Tukey whiskers. (c) Immunoblot analysis validates selective labeling of known (HRAS) and novel (ULK3) farnesylated proteins and geranylgeranylated proteins (RHOA, RAB8A and RAB22A). (d) Transient expression of wild-type (WT) or cysteine mutant (CA and CS) proteins validate dependence of YnF-labeling on the C-terminal cysteine of novel farnesylated CXXX motif substrates ULK3 and CEP85. PD, pulldown; SN, supernatant; TL, total lysate. Note that addition of capture reagent AzTB alters the electrophoretic mobility of some probe-labeled substrates, accounting for the double band pattern apparent in some TL blots.
Competition experiments are widely-used in chemical proteomic analyses to differentiate between selective labeling and non-specific pull-down (e.g. due to interaction with the affinity resin)17,23,24; however these data do not differentiate between competition at the levels of pyrophosphate biosynthesis or transferase recognition. We therefore further probed concentration-dependent probe incorporation into farnesylated and geranylgeranylated substrates in cells in the absence of competing FOH or GGOH, comparing incorporation in cells treated with 1, 2, 5 or 10 µM YnF or YnGG using a 10-plex tandem mass tag (TMT10) methodology to enable relative quantification between all concentrations across both probes, and against a vehicle (DMSO) control (Supplementary Data 3). A clear probe- and concentration-dependent response was observed for known and novel farnesylated or geranylgeranylated substrates identified in competition experiments (Supplementary Figure 5), with strong selectivity for YnF or YnGG for FTase or GGTase-1 substrates, respectively (Supplementary Figure 5). These data mirror the biochemical selectivity of the probes, and further support transferase-selective incorporation of each probe in cells.
Validation of known and novel substrates was performed by affinity enrichment of labeled proteins followed by immunoblot analysis, and mirrored the results obtained in the proteomics analysis. Known farnesylated protein HRAS and novel target ULK3 were preferentially labeled by YnF, known geranylgeranylated proteins (RHOA, RAB8A and RAB22A) preferentially incorporated YnGG (Figure 2c), and labeling was sensitive to competition with the relevant natural isoprenoid. Furthermore, ULK3 and CEP85 mutants in which the CXXX cysteine was mutated to serine or alanine (CS and CA mutants) were not labeled by YnF, confirming that probe modification is dependent on the predicted site of prenylation (Figure 2d).
We next confirmed that the probes are effective in a range of different cell types, including cervical cancer (HeLa), breast cancer (MCF7) and monocytes (THP1). In each case and for each probe labeling could be readily observed, without addition of statin, at concentrations from 1 to 10 µM, although with some variations in apparent efficiency of incorporation (Supplementary Figure 6). Selectivity towards farnesylated (ULK3) and geranylgeranylated (RHOA) substrates was maintained across all cell types, as demonstrated by affinity enrichment and immunoblot analyses (Supplementary Figure 7).
YnF and YnGG enable direct detection of prenylation by mass spectrometry
Direct detection of the site of protein lipidation by mass spectrometry is notoriously challenging and a general proteome-wide approach has not been reported for protein prenylation, which for many substrates is further complicated by a cascade of post-translational proteolysis and methylation (Figure 1a). Building on our previous demonstration of direct identification of protein myristoylation using multifunctional capture reagents17,18,25, we employed AzRB, AzRTB and Az3MRB18 (Supplementary Figure 8) to the direct analysis of protein prenylation. Each of these reagents harbors an enzyme-cleavable sequence to enhance release and detection of modified peptides upon protein digestion with trypsin, or lysarginase, a thermophilic proteinase which can enhance identification of C-terminal peptides26 (Figure 3a). Analysis of the MS/MS spectra of identified YnF- and YnGG-modified peptides revealed characteristic C-S bond scission marker ions analogous to canonical prenylated peptides (Supplementary Figures 9 and 10)27, and cross-validation against orthogonal marker ions from each capture reagent enabled robust sequence assignment (Figure 3b).
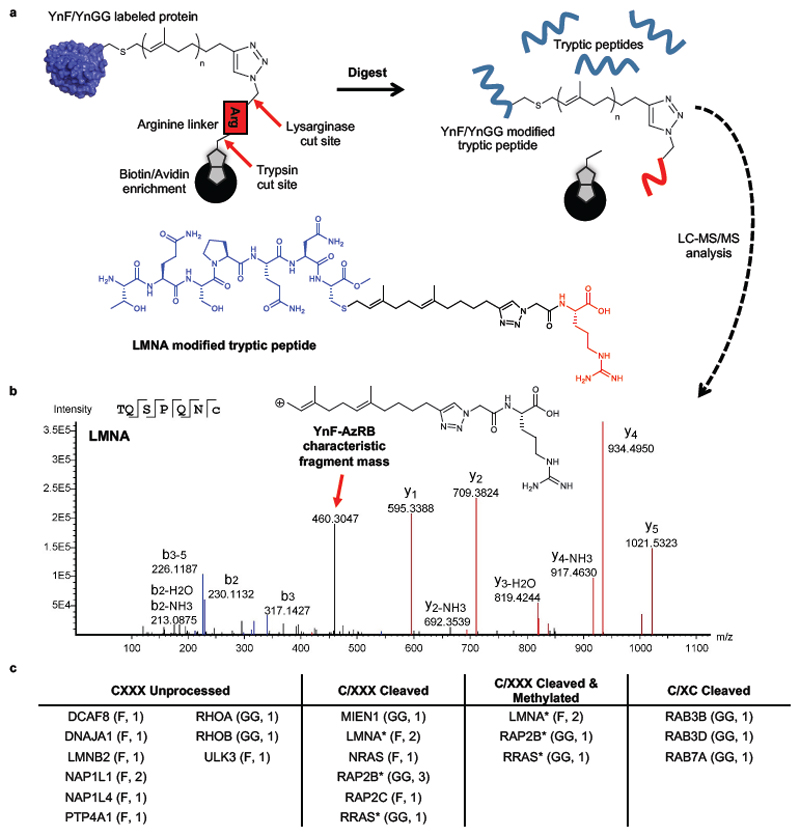
Figure 3. Direct detection of post-translational processing of prenylated peptides by LC-MS/MS.
(a) Enzyme-cleavable capture reagents allow release of YnF- and YnGG-labelled peptides after enrichment on NeutrAvidin beads, and subsequent identification by LC-MS/MS analysis. Note: Upon digest with lysarginase the arginine linker residue remains bound on the beads. (b) MS/MS spectrum of cleaved and methylated C-terminal tryptic peptide belonging to prelamin-A/C (LMNA) in EA.hy926 cells, modified by YnF and capture reagent AzRB. (c) List of proteins for which modified peptides were detected in EA.hy926 cells, grouped according to their apparent post-prenylation processing. Modification and number of unique peptides detected for each protein is indicated in brackets (Supplementary Data 3). *Peptides identified for proteins under more than one grouping.
The 26 distinct prenylated peptides identified in our analysis (Figure 3c, Supplementary Data 4) mapped to the C-terminal CXXX motif of one of 18 distinct proteins, including both well-established prenylated substrates (e.g. nuclear lamins, Rho, and Ras protein family members) and novel prenylated proteins ULK3, DCAF8 and NAP1L4. 12 peptides had undergone C-terminal hydrolytic cleavage generating a prenylated C-terminal cysteine, a further 4 peptides were cleaved and methylated, and the remaining 10 featured unprocessed C-termini (Supplementary Data 4). These findings demonstrate that YnF and YnGG labeling is compatible with endogenous post-prenylation processing by Ras-converting enzyme 1 (RCE1) and Isoprenylcysteine carboxyl methyltransferase (ICMT)28. Interestingly, we also found strong evidence for novel C-terminal processing of geranylgeranylated RAB3B, RAB3D, and RAB7A, in which the two terminal amino acids of the canonical XCXC motif have been removed (Supplementary Figure 10). Taken together, these data provide the first example of direct identification of numerous prenylated and processed prenylation sites at endogenous abundance, at the whole proteome level.
Chemical proteomics enables proteome-wide quantification of protein prenylation in response to prenyl transferase inhibition
As noted above, intense effort has been put into the development of inhibitors of prenyltransferases and enzymes involved in biosynthesis or processing steps up- or downstream of protein prenylation1,2. However, inconsistent efficacy in clinical trials2 suggests that the mode of action of PTIs is more complex than initially supposed, and the selectivity and impact of these inhibitors on prenylation has not been explored at the whole proteome level. We used YnF and YnGG to compare and quantify changes in prenylation in response to perturbation by four tool compounds widely used in cell and animal studies: FTI-277 (a FTase inhibitor)29 and GGTI-2133 (a GGTase-I inhibitor); the natural product Manumycin A (reported to be an inhibitor of FTase)30; and the drug Tipifarnib31, which is representative of sub-nM potency FTIs that have entered over 30 clinical trials2. EA.hy926 cells were labeled with probe in the presence of increasing concentrations of PTI, and analyzed and quantified using spike-in SILAC methodology.
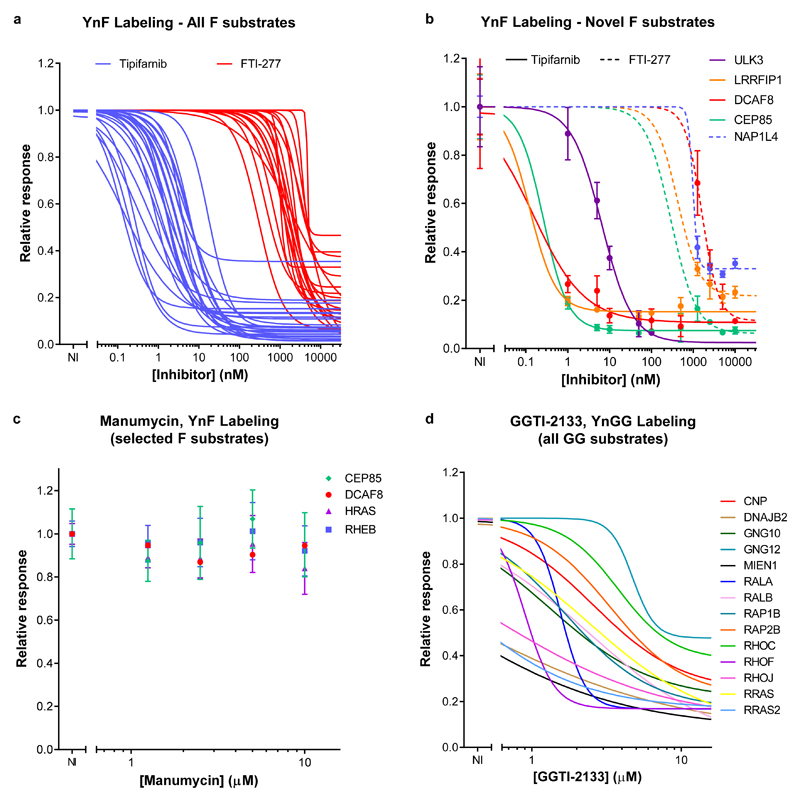
Consistent with their known potencies, the FTase inhibitors Tipifarnib and FTI-277 exhibited low nanomolar and micromolar mean in-cell IC50 responses, respectively (Figure 4a and 4b). Quantitative chemical proteomics enabled dose-response relationships to be determined for each individual FTase substrate in the cell, revealing a wide range in estimated IC50. Notably, all novel farnesylated proteins identified above responded to farnesyltransferase inhibitors, further validating them as bona fide FTase substrates (Figure 4b, Supplementary Data 1 and 5), and immunoblot analysis of HRAS, ULK3 and DCAF8 confirmed that YnF labeling of all three proteins is sensitive to Tipifarnib (Supplementary Figure 11). In contrast, there was no evidence that Manumycin A impacted farnesylation up to the highest concentration tested (10 µM, Figure 4c, Supplementary Data 5); acute cell toxicity was evident above this concentration (data not shown), strongly suggesting that phenotypes previously observed with this compound are unlikely to be directly related to prenylation and may be due to off-target effects and/or cytotoxicity. YnGG was similarly used to interrogate the effect of GGTase-1 inhibitor GGTI-2133 (Figure 4d, Supplementary Data 6), providing an in-cell dose-response for 14 known geranylgeranylated proteins.
Figure 4. Global in-cell characterization of prenyl transferase inhibitors by quantitative chemical proteomics in EA.hy926 cells.
Dose-response curves for YnF labeling of (a) all farnesylated substrates and (b) novel farnesylated substrates, in response to treatment with FTase inhibitors FTI-277 or Tipifarnib. (c) YnF-labeling of selected known and novel farnesylated substrates is insensitive to treatment with Manumycin A. (d) Dose-response curves of YnGG labeling in response to treatment with GGTase-1 inhibitor GGTI-2133. NI, no inhibitor. All data sets are presented as mean (n=3); error bars in (b) and (c) represent standard deviation. For visual clarity individual data points and error bars were omitted in (a) and (d); this data is available in Supplementary Data 5 and 6.
YnF and YnGG describe dynamic response of prenylation to inhibition in living cells
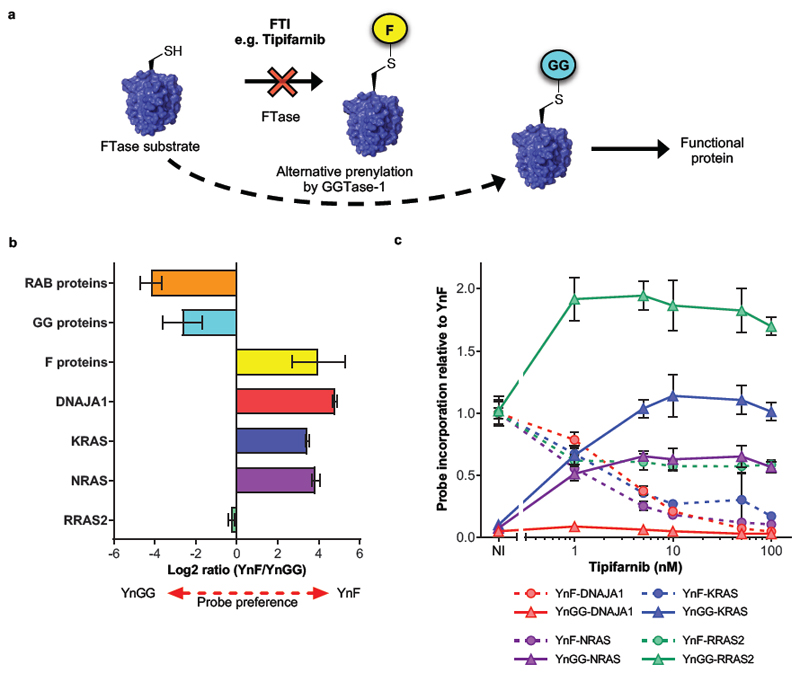
Some proteins are thought to be substrates for both FTase and GGTase-1 under physiological conditions, or to switch to GGTase-1 upon inhibition of FTase1 (Figure 5a). We used our two-probe system to explore the balance between farnesylation and geranylgeranylation in the unperturbed cellular environment, and in response to FTI treatment. Spiking lysates with a reference sample labeled with R10K8 and both prenyl probes enabled relative quantification of probe preference, confirming strong preferential incorporation of YnF or YnGG into farnesylated and geranylgeranylated substrates, respectively (Figure 5b, Supplementary Figure 12). Interestingly, RRAS2 was found to be prenylated by both probes, consistent with the observation that recombinant RRAS2 is a FTase and GGTase-1 substrate in vitro32.
Figure 5. Interrogation of alternative prenylation in response to PTI treatment in EA.hy926 cells.
(a) Upon pharmacological inhibition of FTase some farnesylated substrate proteins undergo alternative prenylation by GGTase-1 and are thought to retain biological function. (b) CXXX substrates that are farnesylated under physiological conditions, including DNAJA1, KRAS and NRAS show preferential incorporation of YnF, whereas geranylgeranylated CXXX substrates and Rab proteins preferentially incorporate YnGG. RRAS2, which is a substrate of both FTase and GGTase-1, shows a balanced probe preference. Data are presented mean (n=3) ± standard deviation. (c) Upon inhibition of FTase by Tipifarnib treatment, only KRAS, NRAS and RRAS2, but not DNAJA1, show a sustained dose-dependent increase in YnGG labeling, concomitant with loss of YnF labeling. Data points represent mean (n=3) ± standard deviation.
Having established a method to quantify relative levels of probe incorporation we interrogated the dynamics of prenylation in response to PTI treatment. The failure in the clinic of FTIs as anticancer agents has been attributed in part to “rescue prenylation” by GGTase-1 of substrates such as KRAS and NRAS1,2. However, the prevalence of alternative prenylation of proteins other than the Ras isoforms remains largely unexplored, despite its potentially important impact on the clinical efficacy and tolerability of FTIs. We compared proteome-wide changes in the ratio of YnF/YnGG incorporation to inhibition of YnF labeling in response to treatment with the clinically relevant FTI Tipifarnib over a range of inhibitor concentrations (Figure 5c, Supplementary Figure 12, Supplementary Data 7). KRAS, NRAS and RRAS2 displayed robust dose-dependent increases in YnGG labeling, with a maximum effect evident at 10, 5 and 1 nM Tipifarnib, respectively. The increase in YnGG incorporation mirrored the dose-dependent decrease in YnF labeling over the same concentration range (Figure 5c), indicating switch-like behavior for these three proteins on treatment with an FTI. The previously identified robust response of RRAS2 YnGG labeling to GGTI-2133 (Figure 4d) further suggests that this protein is prenylated by both FTase and GGTase-1 in cells, and it appears that inhibition of FTase can shift this balance towards geranylgeranylation. Remarkably, no other farnesylated protein showed a significant or robust response to inhibition (Supplementary Figure 12, Supplementary Data 7), showing for the first time that rescue prenylation is restricted to a small set of specific proteins at the whole proteome level.
YnGG quantifies reduced geranylgeranylation of a specific subset of Rab proteins in a REP-1 knock-out model of Choroideremia
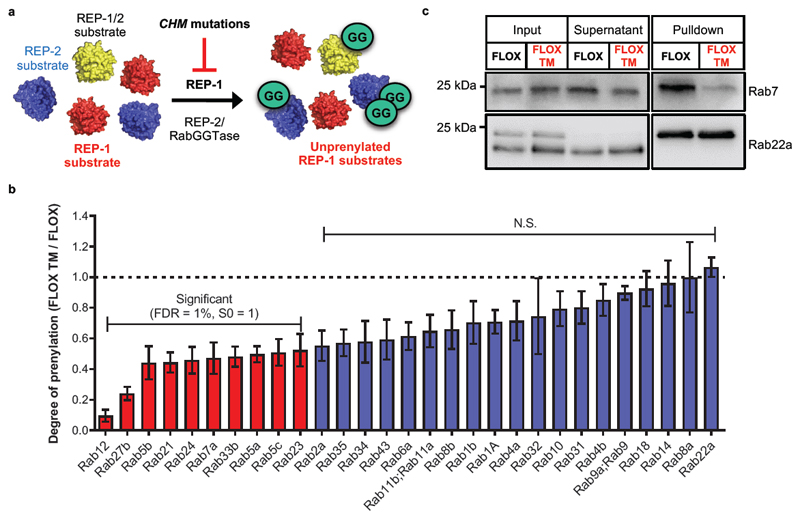
Choroideremia is an X-linked genetic disease caused by loss-of-function mutations in the CHM gene, which encodes Rab Escort Protein 1 (REP-1)4,33. Loss of REP-1 function affects approximately 1 in 50,000 people, and causes degeneration of the choroid, retinal pigment epithelium and photoreceptors, leading to gradual loss of vision in affected patients34. REP-2 is apparently unable to compensate for loss of REP-1 in the eye35, and ongoing gene therapy trials aim to prevent loss of vision by adding functional CHM cDNA36,37. REP proteins are required for prenylation by RabGGTase, and loss of REP-1 results in accumulation of non-prenylated Rab proteins (Figure 6a); however, the quantitative impact on Rab protein prenylation at the whole proteome level in a disease-relevant model remains unknown.
Figure 6. Rep-1 knockout reduces geranylgeranylation of a subset of Rab proteins in mouse embryonic fibroblasts.
(a) Schematic of REP-1/2 mediated geranylgeranylation of Rab proteins by RabGGTase. Loss of REP-1 is only partially rescued by REP-2, resulting in accumulation of unprenylated Rab substrates. (b) Relative quantification of YnGG labeling in Rep-1 knockout mouse fibroblasts (FLOX TM) versus control (FLOX) shows that a sub-set of Rab proteins display reduced prenylation upon Rep-1 loss. Data are presented as mean (n=3) ± standard deviation. ANOVA analysis (Permutation-based FDR=1%, S0=1); N.S., not significant. (c) Immunoblot analysis confirms that loss of Rep 1 affects geranylgeranylation of Rab7, but not Rab22a, without significantly altering overall expression. Note that convention has been followed whereby mouse genes are given lower case names (e.g. Rep), whilst human proteins are in capitals (REP).
To interrogate which Rab substrates are most dependent on REP-1 for efficient prenylation, we applied YnGG to quantify differences in Rab geranylgeranylation levels in mouse embryonic fibroblasts (MEFs) derived from the conditional mouse Rep-1 knock-out model of Choroideremia38. Note that here as elsewhere, by convention gene/protein names are in lowercase for mouse (e.g. Rep-1), and uppercase for human (REP-1). Rep-1 knock-out (FLOX TM) or control (FLOX) MEFs were incubated with YnGG (10 µM, 24 hours), the resultant lysate was spiked with a heavy standard generated from MEF cell line NIH 3T3 labeled with both R10K8 and YnGG, and processed for quantitative proteomic analysis as described above to generate relative quantification of changes in prenylation across 29 Rab proteins in Rep-1 knockout cells vs. wild type control (Figure 6b, Supplementary Figure 13, Supplementary Data 8). The prenylation of 10 Rab proteins was significantly reduced upon Rep-1 knock-out, with prenylation of Rab12 and 27b reduced to 10% and 24%, respectively, of the level observed in Rep-1 functional cells. As expected, YnGG incorporation into GGTase-1 substrates was largely unaffected by Rep-1 knock-out (Supplementary Figure 13). Protein levels in whole cell lysates were further quantified by proteomics (Supplementary Figure 14), demonstrating no significant changes in the expression of Rabs or CXXX substrate proteins, including Rab7a, 5c and 21, each of which showed significantly reduced levels of prenylation in Rep-1 knockout cells. Immunoblot analysis of Rab22a, Rab7 and a GGTase-1 substrate (RhoA) recapitulated the results obtained through quantitative proteomics, confirming that only Rab7 YnGG labeling is affected by Rep-1 knockout, with no changes in overall expression (Figure 6c and Supplementary Figure 13).
Discussion
In this study we present a methodological approach to identify and quantify protein prenylation using a combination of alkyne-tagged prenylation probes and quantitative proteomics. Novel prenyl probes YnF and YnGG show preferential incorporation into farnesylated and geranylgeranylated proteins, respectively, maintaining selectivity for each transferase by mimicking overall chain length and hydrophobicity. Previous studies using prenyl analogues which depart significantly from the native lipid structure (for example by direct integration of a fluorophore or branching in the isoprenoid chain) have variously reported negligible13 or significant39 impact on the biochemical efficiency of transfer to specific peptide or protein substrates, although these studies did not explore incorporation in a cellular setting. Here we have presented multiple lines of evidence supporting transferase selectivity and substrate fidelity of YnF and YnGG, including enzyme kinetics, whole-proteome competition and concentration-dependent incorporation in cells for known and predicted farnesylated or geranylgeranylated proteins, cross-confirmation for known and novel substrates by immunoblot analyses in a range of cell lines, dose-dependent responses to selective FTase and GGTase-1 inhibitors including recapitulation of known prenyl switch responses, and direct mass spectrometric detection of specific probe incorporation into sites of prenylation, compatible with downstream processing. Taken together, these data strongly support YnF and YnGG as useful probes for exploring protein prenylation in cells.
The large proportion of substrates detected uniquely by either YnF or YnGG highlights the advantages of a two-probe system to maximize discovery of the prenylated proteome and provides substantially greater coverage than studies using previous probes, which vary probe chain length, introduce hydrophilic linkers, and rely on pretreatment with statins in order to achieve probe incorporation (see Supplementary Table 1 for a comparison with previous probes and studies). These probes cause no overt cytotoxicity and the degree of incorporation is probably modest as a proportion of total prenylated protein, which is likely to minimize phenotypic effects of probe incorporation; nevertheless, all metabolic labeling approaches involve a perturbation of the biological system. Several of the seven novel farnesylated proteins identified in this study are known to have important biological roles. CEP85 plays a role in controlling centriole duplication40, whilst ULK3 regulates cytokinetic abscission41, autophagy and Gli signaling42. LRRFIP1, a cytosolic nucleic acid binding protein, is a key mediator of Wnt/β-catenin signalling43. Prenylation data compare favorably with bioinformatic predictions using PrePS44, a widely used predictor of non-Rab prenylation motifs, with the majority of experimentally determined substrates predicted to be prenylated by at least one of the prenyl transferase enzymes (Supplementary Data 1). Conversely, CXXX proteins that did not respond to competition are not predicted substrates, nor do they respond to inhibition by any PTI tested, further suggesting that these proteins (including previously hypothesized substrates UCHL1 and annexin ANXA1) are not prenylated45.
The present study is the first proteome-scale analysis to identify a large number of prenylated and processed peptides from substrates at endogenous abundance (i.e. without substrate overexpression), and the first to demonstrate the compatibility of prenyl probes with the cascade of post-prenylation processing in cells. Furthermore, the novel discovery of C-terminal processing of Rab proteins which normally carry a double geranylgeranyl CXCX motif (RAB3B, RAB3D, and RAB7A) may indicate the existence of a previously unknown mechanism to control prenylation stoichiometry of Rab family members, a phenomenon which merits further investigation. Identification of prenylated peptides is technically challenging for multiple reasons; the high frequency of Arg/Lys-rich stretches proximal to the prenylation site coupled with post-prenylation processing results in short peptides which have sub-optimal chromatographic separation and can be difficult to assign, and this is further compounded by the generally poor ionization of C-terminal tryptic peptides46. Identification of an even greater range of modified peptides might be achieved in future through adaptation of the analytical approach to enhance detection of short C-terminal tryptic peptides, partially addressed here by the application of lysarginase. We note that discovery of intact CXXX peptides may be due to detection of substrates prior to cleavage or native retention of the XXX motif, since recent studies demonstrate that not all prenylated proteins are subject to quantitative processing47. Recent reports using protein overexpression and peptide biochemical assays suggest that CXXXX motif proteins may also be substrates for FTase48; we have undertaken careful and detailed inspection of our data in search of CXXXX motif proteins, including both proteomic analyses and direct site identification, but could not find evidence for this non-canonical prenylation motif in this mammalian cell line.
Quantitative analysis of prenylation in response to PTIs demonstrates a range of sensitivity across substrates, which may correspond to differential efficiency of prenylation at each substrate in the complex environment of the cell. Similar nuances have been observed in vitro using peptide substrates (e.g. Tipifarnib IC50 of 0.86 nM and 7.9 nM for farnesylation of peptide substrates based on lamin B and KRasB, respectively31), and may influence the mode of action of these compounds in a given disease context. Probes selective towards each transferase enabled the first quantitative whole-proteome analysis of the phenomenon of dynamic switching of FTase substrates to prenylation by GGTase-1 in response to FTI treatment, uncovering the novel and intriguing finding that the large majority of substrates show no change in geranylgeranylation at concentrations of Tipifarnib sufficient to eliminate FTase activity. The switching propensity of a given protein cannot be reliably predicted based on sequence, and further studies will be required to understand the regulatory mechanisms leading to this surprising result. Whilst FTI-277 and particularly Tipifarnib were confirmed as potent tool inhibitors with excellent selectivity at the whole proteome level, our data provide compelling system-level evidence that any effects on prenylation observed with Manumycin A are likely to be due to off-target cytotoxic effects, leading to the conclusion that this compound is ineffective as a chemical probe. Although Manumycin A is a weak FTI (IC50 5 µM) and less widely applied than FTI-277 or Tipifarnib, it has been used as a tool inhibitor in over 100 previously published studies; we suggest that previous conclusions drawn using Manumycin A as an FTI should be treated with caution, unless cross-validated with complementary approaches or more selective tool compounds.
In summary, the novel probes YnF and YnGG provide a unique window on the prenylated proteome, providing enhanced coverage and selectivity in metabolically active model systems, and complementing in vitro ‘re-prenylation’ approaches13. In combination with optimized capture reagents and quantitative proteomic workflows, this method offers a new perspective on prenylation dynamics, prenylated protein processing, and inhibitor selectivity, and will be useful for identifying potential novel therapeutic uses for PTIs and for understanding the role and regulation of prenylation in health and disease.
Methods
Full materials and methods and details for the synthesis and characterization of all compounds are provided in Supplementary Information.
Supplementary Material
Table of content summary.
Chemical probes YnF and YnGG in combination with quantitative chemical proteomics technologies enable global analysis of protein prenylation, identification of prenylated peptides, interrogation of prenylation dynamics in response to pharmacological inhibition of prenyl transferase enzymes and quantification of defective Rab prenylation in a model of the retinal degenerative disease Choroideremia.
Acknowledgments
The authors would like to thank Lisa Haigh (Department of Chemistry Mass Spectrometry Facility, Imperial College London) for assistance in acquiring nanoLC-MS/MS and HRMS data, Sibyl Sheppard and Ben Chappell for their contributions to prenyl probe synthesis, Nicola O’Reilly (Francis Crick Institute) for peptide substrate synthesis, and Tony Magee (Imperial College London) for insightful comments on the manuscript. This study was supported by Cancer Research UK (Programme Foundation Award C29637/A20183 to E.W.T.), the British Heart Foundation (PhD studentship to E.M.S. and Project Grant PG/12/67/29773 to B.W.S. and E.W.T.), the European Union Framework Program 7 (Marie Curie Intra European Fellowship to J.Mo.S. and E.W.T.), Wellcome Trust (Programme grant 093445/Z/10/Z to M.C:S., and WT102871MA to J.Ma.S), and Royal Thai Government scholarship (PhD studentship to N.P.).
Footnotes
Data Availability Statement
All relevant data are available from the authors, and proteomics datasets have been deposited to public repositories, as detailed below.
Accession codes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository49 with the dataset identifier PXD009155.
Author contributions: E.M.S., J.Mo.S., R.A.S., N.P., B.W.S. and E.W.T designed the experiments. E.M.S. performed chemical synthesis of reagents, initial method validation and optimization, experiments related to isoprenoid competition and inhibitor treatment and analyzed data. J.Mo.S. performed experiments related to isoprenoid competition, inhibitor treatment, prenylation dynamics and Rep-1 knock-out, and analyzed data. R.A.S. performed chemical synthesis of reagents and performed data analysis of modified peptides. N.P. performed chemical synthesis of pyrophosphate derivatives, biochemical enzyme assays, experiments related to concentration-dependent probe incorporation, and analyzed data. T.L.-H. designed the biochemical enzyme assays and analyzed data. T.T. and M.C.S. provided mouse embryonic fibroblasts. L.N.V. and J.Ma.S. provided ULK3 and CEP85 protein constructs. E.W.T. conceived and directed the study. E.M.S., J.Mo.S., R.A.S. and E.W.T. wrote the manuscript, with input from all authors.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Wang M, Casey PJ. Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol. 2016;17:110–122. doi: 10.1038/nrm.2015.11. [DOI] [PubMed] [Google Scholar]
- 2.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oesterle A, Laufs U, Liao JK. Pleiotropic Effects of Statins on the Cardiovascular System. Circ Res. 2017;120:229–243. doi: 10.1161/CIRCRESAHA.116.308537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roosing S, Collin RW, den Hollander AI, Cremers FP, Siemiatkowska AM. Prenylation defects in inherited retinal diseases. J Med Genet. 2014;51:143–151. doi: 10.1136/jmedgenet-2013-102138. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Koh C, et al. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet Infect Dis. 2015;15:1167–1174. doi: 10.1016/S1473-3099(15)00074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon LB, et al. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grammel M, Hang HC. Chemical reporters for biological discovery. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tate EW, Kalesh KA, Lanyon-Hogg T, Storck EM, Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr Opin Chem Biol. 2015;24:48–57. doi: 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grocin AG, Serwa RA, Sanfrutos JM, Ritzefeld M, Tate EW. Whole Proteome Profiling of N-Myristoyltransferase Activity and Inhibition Using Sortase A. Mol Cell Proteomics. 2019;18:115–126. doi: 10.1074/mcp.RA118.001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kho Y, et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan LN, et al. A novel approach to tag and identify geranylgeranylated proteins. Electrophoresis. 2009;30:3598–3606. doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen UT, et al. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 14.Berry AF, et al. Rapid multilabel detection of geranylgeranylated proteins by using bioorthogonal ligation chemistry. Chembiochem. 2010;11:771–773. doi: 10.1002/cbic.201000087. [DOI] [PubMed] [Google Scholar]
- 15.Charron G, Li MM, MacDonald MR, Hang HC. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc Natl Acad Sci U S A. 2013;110:11085–11090. doi: 10.1073/pnas.1302564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palsuledesai CC, et al. Metabolic Labeling with an Alkyne-modified Isoprenoid Analog Facilitates Imaging and Quantification of the Prenylome in Cells. ACS Chem Biol. 2016;11:2820–2828. doi: 10.1021/acschembio.6b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thinon E, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5 doi: 10.1038/ncomms5919. 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broncel M, et al. Multifunctional reagents for quantitative proteome-wide analysis of protein modification in human cells and dynamic profiling of protein lipidation during vertebrate development. Angew Chem Int Ed Engl. 2015;54:5948–5951. doi: 10.1002/anie.201500342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Oost BA, Edgell CJ, Hay CW, MacGillivray RT. Isolation of a human von Willebrand factor cDNA from the hybrid endothelial cell line EA.hy926. Biochem Cell Biol. 1986;64:699–705. doi: 10.1139/o86-096. [DOI] [PubMed] [Google Scholar]
- 20.Holstein SA, Wohlford-Lenane CL, Hohl RJ. Consequences of mevalonate depletion. Differential transcriptional, translational, and post-translational up-regulation of Ras, Rap1a, RhoA, AND RhoB. J Biol Chem. 2002;277:10678–10682. doi: 10.1074/jbc.M111369200. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat Protoc. 2011;6:147–157. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, et al. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson's disease. Proc Natl Acad Sci U S A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clulow JA, et al. Competition-based, quantitative chemical proteomics in breast cancer cells identifies new target profiles for sulforaphane. Chem Commun (Camb) 2017;53:5182–5185. doi: 10.1039/c6cc08797c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalesh KA, Clulow JA, Tate EW. Target profiling of zerumbone using a novel cell-permeable clickable probe and quantitative chemical proteomics. Chem Commun (Camb) 2015;51:5497–5500. doi: 10.1039/c4cc09527h. [DOI] [PubMed] [Google Scholar]
- 25.Wright MH, et al. Global analysis of protein N-myristoylation and exploration of N-myristoyltransferase as a drug target in the neglected human pathogen Leishmania donovani. Chem Biol. 2015;22:342–354. doi: 10.1016/j.chembiol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huesgen PF, et al. LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat Methods. 2015;12:55–58. doi: 10.1038/nmeth.3177. [DOI] [PubMed] [Google Scholar]
- 27.Kassai H, Satomi Y, Fukada Y, Takao T. Top-down analysis of protein isoprenylation by electrospray ionization hybrid quadrupole time-of-flight tandem mass spectrometry; the mouse Tgamma protein. Rapid Commun Mass Spectrom. 2005;19:269–274. doi: 10.1002/rcm.1782. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Rodriguez V, et al. a-Factor analogues containing alkyne- and azide functionalized isoprenoids are efficiently enzymatically processed and retain wild type bioactivity. Bioconjug Chem. 2017;29:316–323. doi: 10.1021/acs.bioconjchem.7b00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuire TF, Qian Y, Vogt A, Hamilton AD, Sebti SM. Platelet-derived growth factor receptor tyrosine phosphorylation requires protein geranylgeranylation but not farnesylation. J Biol Chem. 1996;271:27402–27407. doi: 10.1074/jbc.271.44.27402. [DOI] [PubMed] [Google Scholar]
- 30.Hara M, et al. Identification of Ras farnesyltransferase inhibitors by microbial screening. Proc Natl Acad Sci U S A. 1993;90:2281–2285. doi: 10.1073/pnas.90.6.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.End DW, et al. Characterization of the antitumor effects of the selective farnesyl protein transferase inhibitor R115777 in vivo and in vitro. Cancer Res. 2001;61:131–137. [PubMed] [Google Scholar]
- 32.Carboni JM, et al. Farnesyltransferase inhibitors are inhibitors of Ras but not R-Ras2/TC21, transformation. Oncogene. 1995;10:1905–1913. [PubMed] [Google Scholar]
- 33.Coussa RG, Traboulsi EI. Choroideremia: a review of general findings and pathogenesis. Ophthalmic Genet. 2012;33:57–65. doi: 10.3109/13816810.2011.620056. [DOI] [PubMed] [Google Scholar]
- 34.Dimopoulos IS, Radziwon A, St Laurent CD, MacDonald IM. Choroideremia. Curr Opin Ophthalmol. 2017;28:410–415. doi: 10.1097/ICU.0000000000000392. [DOI] [PubMed] [Google Scholar]
- 35.Seabra MC, Brown MS, Goldstein JL. Retinal degeneration in choroideremia: deficiency of rab geranylgeranyl transferase. Science. 1993;259:377–381. doi: 10.1126/science.8380507. [DOI] [PubMed] [Google Scholar]
- 36.Edwards TL, et al. Visual Acuity after Retinal Gene Therapy for Choroideremia. N Engl J Med. 2016;374:1996–1998. doi: 10.1056/NEJMc1509501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue K, et al. Beneficial effects on vision in patients undergoing retinal gene therapy for choroideremia. Nat Med. 2018;24:1507–1512. doi: 10.1038/s41591-018-0185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolmachova T, et al. Independent degeneration of photoreceptors and retinal pigment epithelium in conditional knockout mouse models of choroideremia. J Clin Invest. 2006;116:386–394. doi: 10.1172/JCI26617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jennings BC, et al. Analogs of farnesyl diphosphate alter CaaX substrate specificity and reactions rates of protein farnesyltransferase. Bioorg Med Chem Lett. 2016;26:1333–1336. doi: 10.1016/j.bmcl.2015.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04122-x. 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caballe A, et al. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. Elife. 2015;4:e06547. doi: 10.7554/eLife.06547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goruppi S, et al. The ULK3 Kinase Is Critical for Convergent Control of Cancer-Associated Fibroblast Activation by CSL and GLI. Cell Rep. 2017;20:2468–2479. doi: 10.1016/j.celrep.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douchi D, et al. Silencing of LRRFIP1 reverses the epithelial-mesenchymal transition via inhibition of the Wnt/beta-catenin signaling pathway. Cancer Lett. 2015;365:132–140. doi: 10.1016/j.canlet.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 44.Maurer-Stroh S, et al. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onono FO, et al. A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol Cell Proteomics. 2010;9:742–751. doi: 10.1074/mcp.M900597-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilling O, Barre O, Huesgen PF, Overall CM. Proteome-wide analysis of protein carboxy termini: C terminomics. Nat Methods. 2010;7:508–511. doi: 10.1038/nmeth.1467. [DOI] [PubMed] [Google Scholar]
- 47.Hildebrandt ER, et al. A shunt pathway limits the CaaX processing of Hsp40 Ydj1p and regulates Ydj1p-dependent phenotypes. Elife. 2016;5 doi: 10.7554/eLife.15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanden MJ, et al. Efficient farnesylation of an extended C-terminal C(x)3X sequence motif expands the scope of the prenylated proteome. J Biol Chem. 2018;293:2770–2785. doi: 10.1074/jbc.M117.805770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.