Abstract
A scalably manufacturable oral antibody technology that can interfere with gastrointestinal (GI) targets is needed. Contrary to the complex native secretory IgA, we achieve this using a single-gene encoded monomeric-IgA-like antibody, composed of camelid VHH fused to IgA Fc (mVHH-IgA). This can be produced in soybean seeds or secreted from Pichia pastoris yeast, freeze-or spray-dried, and when delivered in food prevents enterotoxigenic Escherichia coli (F4-ETEC) infection in piglets.
With the rapidly increasing knowledge of the role of the gut microbiome in diverse aspects of human and veterinary health, antibody-type drug-mediated methodology to specifically interfere with the microbiome or host factors in the gut is needed1–4. The challenge is that, unlike the systemically deliverable IgG1 scaffold-based antibodies, neither a comparable scaffold nor an integrated technology for orally deliverable antibodies is available.
The predominant antibody isoform at the GI mucosal surfaces is the secretory IgA (SIgA). SIgAs are complex tetravalent, abundantly glycosylated, heterodecameric antibodies, composed of four heavy chains, four light chains, a joining chain (J-chain) and a secretory component (SC) (Fig. 1a). SIgA is suitable for passive mucosal protection5–8, but recombinant SIgA production is challenging, as it requires expression and precise assembly of the 10 protein chains coded by four different genes6. There is also no industrially scaled affinity resin for downstream purification of SIgA9. Alternatively, expression and administration of antibodies in a food-grade matrix is attractive for GI-tract delivery, as it circumvents challenging purification and would allow for cost-effective scalable manufacturing.
Figure 1.
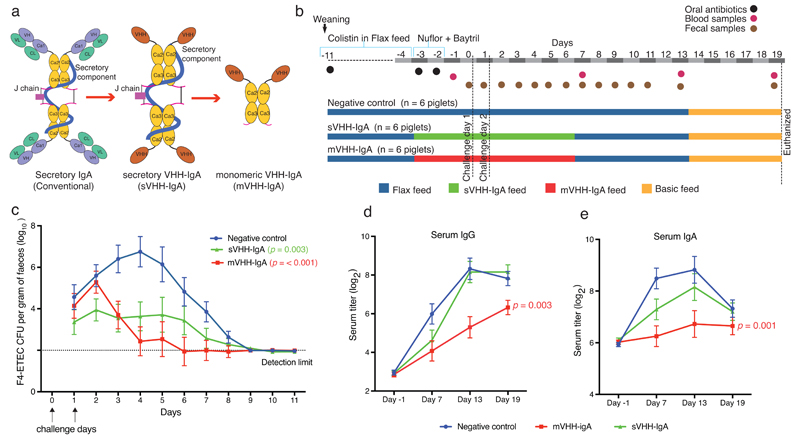
Monomeric IgA in a plant seed matrix prevents F4-ETEC infection in piglets. The conventional secretory IgA was deconstruction to VHH-IgA based secretory IgA (sVHH-IgA) and monomeric IgA (mVHH-IgA) (a), produced in Arabidopsis seeds and delivered feed-admixed to evaluate efficacy in F4-ETEC challenged piglets (b), which showed that the mVHH-IgA fed group rapidly cleared the bacteria (c), and had correspondingly low seroconversion of anti-F4-ETEC IgG (d) and IgA (e) serum titers. The line graphs depict group mean and error bars represent the standard error of the mean. Statistical significance (p values) of changes in feed effects over time compared with negative control were assessed by an approximate F-test following a repeated measurements analysis using the residual maximum likelihood.
We previously showed that replacing the Fab of IgA with a camelid VHH antibody fragment can reduce the number of required genes from four to three to produce a seed-bioencapsulated SIgA-analog (sVHH-IgA) in model plant Arabidopsis10. Feed containing seeds producing VHH-IgA formats protected, while monomeric VHH-IgG failed to protect piglets from F4 fimbriae-bearing enterotoxigenic E. coli (F4-ETEC) infection10. F4-ETEC is an important disease in pig rearing, causing economic losses due to post-weaning diarrhea, and is currently managed using antibiotics11. Mechanistically this swine infection is akin to cholera and ETEC-caused traveler’s diarrhea in humans12.
Nonetheless, introducing three genes in homozygous condition still makes translation to a scalable seed crop species (such as soybean; Glycine max) tedious, especially with crop lifecycle of up to 6 months, successive crossing would require several years. Therefore, using the F4-ETEC piglet model, we first investigated whether a drastically simplified single-gene encoded monomeric VHH-IgA (mVHH-IgA) format would be efficacious in the gut (Fig. 1a). Arabidopsis seed stocks 10 were upscaled, containing four different anti-F4-ETEC VHH-IgAs (V1A, V2A, V3A and V4A), in either the sVHH-IgA or mVHH-IgA formats, wherein the VHH-IgA was about 0.2% of seed weight10. F4-ETEC-susceptible piglets receiving mVHH-IgA, sVHH-IgA or no antibodies (control group) in their feed were challenged with F4-ETEC (Fig. 1b, Supplementary Table 1a). Both the sVHH-IgA (p = 0.003) and mVHH-IgA (p = <0.001) fed groups had significantly lower shedding of the challenged strain vs. the control group (Fig. 1c). Although the mean shedding was low in the sVHH-IgA pen, a single piglet showed excessive shedding (>7 log10 colony forming units (CFU)) (Supplementary Table 2), which may have boosted the average seroconversion of this group (Fig.1d,e). The low anti-F4-ETEC IgG and IgA levels in the blood serum of the mVHH-IgA group (Fig. 1d,e) corroborated the immediate clearance of F4-ETEC by mVHH-IgA administered in feed. Our previous in vitro analysis showed that these antibody formats agglutinated F4-ETEC and prevent attachment to villous enterocytes10.
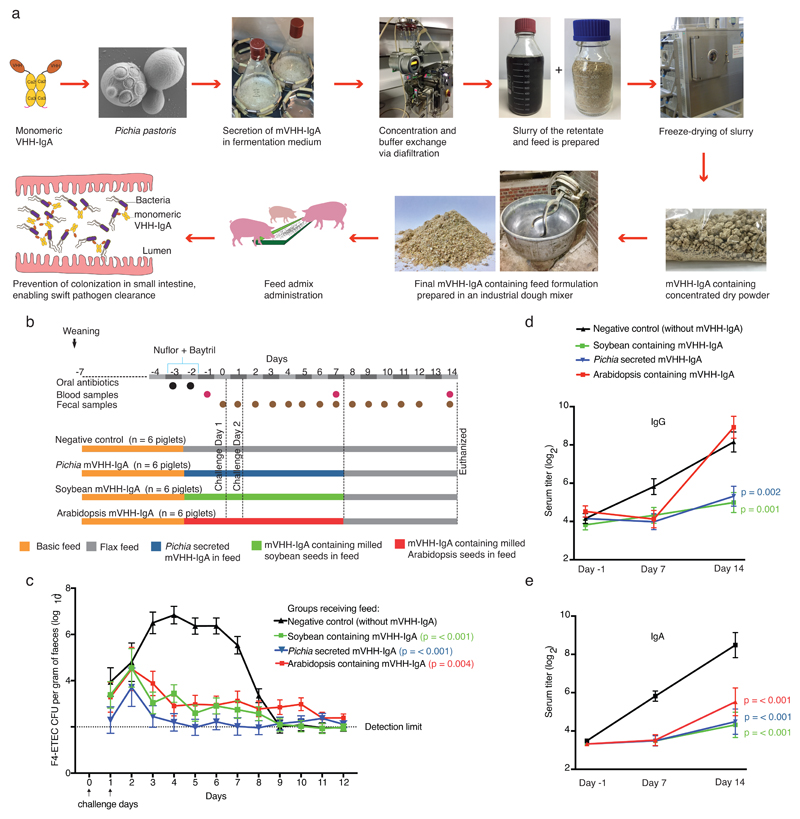
This finding is important for translation, as a single transgene-requiring mVHH-IgA (dimerization-free and SC-free) is much easier to express in diverse expression systems. We hence produced mVHH-IgA in soybean. Soybean seeds containing mVHH-IgA at about 0.2% of seed weight were generated in sufficient amounts for a piglet trial, in about 1.5 years (Supplementary Fig. 1a). However, seeking an alternative to the time (about 10 years) and capital-intensive (possibly beyond 100 million USD) GM-plant regulatory pathway13, we successfully secreted functional mVHH-IgA from the yeast Komagataella phaffii (i.e., Pichia pastoris) (Supplementary Fig. 1b). Yeast-based protein expression is scalable and utilized to cost-effectively express industrial and food-processing enzymes14,15. However, the mVHH-IgA produced through the yeast secretory pathway (required for proper protein folding), is secreted into the growth medium and is no longer bio-encapsulated, unlike when it is produced in the plant seed. To investigate whether such non-encapsulated Pichia mVHH-IgA would be efficacious in blocking F4-ETEC, we conducted another piglet challenge experiment (Fig. 2).
Figure 2.
Pichia-secreted mVHH-IgA prevents F4-ETEC infection in piglets. Pichia-secreted mVHH-IgAs were produced in a simple integrated fashion as food ingredient and formulated in feed (a) to evaluate efficacy alongside feed admixed milled soybean or Arabidopsis seeds producing mVHH-IgA in an F4-ETEC piglet challenge experiment (b). Swift clearance and low shedding of F4-ETEC in groups fed with Pichia, soybean- and Arabidopsis-produced mVHH-IgA demonstrated protection (c), corroborated with low anti-F4-ETEC titers of serum IgG (d) and IgA (e). The line graphs depict group mean and error bars represent standard error of the mean. Statistical significance (p values) of changes in feed effects over time compared with negative control were assessed by an approximate F-test following a repeated measurements analysis using the residual maximum likelihood.
For this experiment the antibody cocktail was simplified, using equal proportion of V2A and V3A, as VHHs V1 and V2 bind to the same epitope, whereas V3 binds to another conformational epitope16. The Pichia-secreted and soybean produced mVHH-IgAs differed in molecular weight due to differences in N-glycosylation (Supplementary Fig. 1). Based on average mVHH-IgA ELISA end-point titers of Pichia clones and the seed stocks, we observed that 1 ml of Pichia shake flash culture supernatant contained equivalent mVHH-IgA quantities as 5 mg of soybean or Arabidopsis seeds. Based on this ratio, experimental feeds were formulated aiming for piglet daily dose of 5 mg mVHH-IgA (V2A+V3A) produced in each system (a four-fold reduction in dose vs. the experiment in Figure 1).
A purification-devoid simple scalable manufacturing process was established for edible Pichia mVHH-IgA (Fig. 2a). The Pichia culture medium was clarified by centrifugation, concentrated and buffer exchanged via diafiltration and mixed with pig feed, generating a slurry, which was dried in a food-grade lyophilizer. The process yielded consistent mVHH-IgA levels in four production batches (Supplementary Fig. 2). The freeze-dried batches were mixed with pig feed to attain 18 kg of Pichia mVHH-IgA feed (Fig 2a, Supplementary Table 1B). We validated the antigen-binding efficacy of the mVHH-IgAs in this formulation stored at room temperature for 2 years (Supplementary Fig. 3). For soybean and Arabidopsis, we added milled V2A- and V3A-producing seeds to piglet feed (Supplementary Table 1B). The mVHH-IgA quantity and quality were assessed in each feed formulation via immunoblot densitometry analysis and antigen-binding ELISA (Supplementary Fig. 3).
On challenging with F4-ETEC (Fig. 2b), piglets receiving feed devoid of antibody showed successful colonization of the small intestine (Fig. 2c) but feed containing mVHH-IgA produced in Arabidopsis, soybean or Pichia prevented the F4-ETEC infection (Fig. 2c, Supplementary Table 3). In mVHH-IgA receiving groups, F4-ETEC appeared to be swiftly cleared from the GI tract, as the shedding declined from day 3 onwards and remained low, often below detectable levels (2 log10 CFU) thereafter (Fig. 2c, Supplementary Table 3). Rapid clearance and immune-exclusion of F4-ETEC was further supported by low anti-F4-ETEC seroconversion (Fig. 2d-e).
Given this success, we further explored whether Pichia-derived mVHH-IgA, which is quite heat-resistant (first melting transition at about 65°C, Supplementary Fig. 4a) could be spray-dried, a low-cost industrial alternative to freeze-drying, which requires mVHH-IgA to withstand short but intense heating (~170°C). After spray-drying, the mVHH-IgA remained intact and retaining its antigen binding capacity (Supplementary Fig. 4).
It is likely that a combination of several features enables the non-encapsulated Pichia mVHH-IgAs to work so efficiently in the GI tract. First, the VHH domain is small and robust17, facilitating drying, formulating in feed and intact gastric passage. Second, when secreted from yeast, the IgA Fc molecules get modified with very large N-glycans (Supplementary Fig. 1b), possibly enhancing stability of the entire molecule. Third, the mixing of the antibodies with the feed into a dry product may further provide stability.
Our future work will be directed toward translating this application for farm use. Subsequent farm-trials will enable mVHH-IgA impact assessment on F4-ETEC caused diarrhea and weight loss. As the porcine monogastric GI tract model has similarity to the human GI tract, future application development for human medicine appears warranted.
Online Methods
VHH-IgA-based antibody production in different systems
The anti-ETEC mVHH-IgA and sVHH-IgA producing Arabidopsis seeds were generated as described10. Briefly, the VHHs V1, V2, V3 and V4 were selected from a llama immune library, such that they recognized the three circulating serotypes of the F4 fimbrial tip adhesin FaeG. The sequences of the respective VHHs were fused to that of porcine IgAb Fc, and expressed in Arabidopsis seeds under the control of the β-phaseolin promoter and 3’ arcelin terminator. Within a similar expression cassette in separate T-DNAs, the porcine SC and J-chain were cloned. Starting from triple co-transformation of SC, J chain and one of the VHH-IgAs, homozygous seed stocks accumulating high amounts of each of the VHH-IgA-based bivalent monomeric IgA (mVHH-IgA) were identified and named mV1A, mV2A, mV3A and mV4A. Furthermore, plants co-expressing porcine J-chain and SC with one of the VHH-IgAs, enabled generation of homozygous lines accumulating VHH-IgA-based tetravalent secretory IgA (sVHH-IgA), named sV1A, sV2A, sV3A, sV4A10. The four mVHH-IgA and sVHH-IgA lines were upscaled in the greenhouse to raise 150 g seeds of each line to formulate the mVHH-IgA and sVHH-IgA feeds (daily dose 20 mg/day/pig) (Fig. 1 and Supplementary Table 1A). An additional 75 g of seeds containing mV2A and mV3A were raised to formulate a 5 mg daily dose for the Arabidopsis mVHH-IgA group in the second challenge experiment (Fig. 2 and Supplementary Table 1B).
For expression of V2A and V3A in soybean seeds, the genes V2A and V3A from entry plasmid pEV2A and pEV3A10, were recombined into the Gateway expression vector pGW4318 (Invitrogen). The resulting expression vectors were named pMXV2A and pMXV3A, and introduced into Agrobacterium strain EHA101 for transformation of the soybean plants (Glycine max cv. Williams 82) at the Plant Transformation Facility of Iowa State University, as described19. The VHH-IgA antibody accumulation was evaluated in the T2 seeds (first segregating seed stock) from extracts made by dissolving the seed powder obtained by drilling into one of the cotyledons without damaging the embryo axis, in the extraction buffer (50 mM Tris-HCl, 200 mM NaCl, 5 mM EDTA and 0.1% Tween 20 v/v, together with one cOmplete™ (Sigma) protease inhibitor cocktail tablet per 50 ml of buffer). Serial dilutions of seed extracts were evaluated in ELISA with FaeG (serotype ‘ac’) antigen-coated wells (concentration of 1 µg/ml in 0.1 M NaHCO3 pH 8.2) and detected with goat anti-pig IgA conjugated to horse-radish peroxidase (HRP) (BioRad), as described10. The same ELISA set up was used for evaluating VHH-IgA produced in Arabidopsis or Pichia, henceforth referred to as FaeG-ELISA. Ten to twenty seeds from transformants expressing high amounts of antibody were retained for growing T2 plants. To formulate the soybean-produced mVHH-IgA-bearing diet for the second piglet feed-challenge experiment, 75 g of each, V2A (T5 seeds) and V3A (T2 seeds) producing seeds were used (Supplementary Table 1B).
For V2A and V3A expression in Pichia, the VHH-IgA fusion genes V2A and V3A were PCR-amplified from pEV2A and pEV3A10 using the primer set Alfa-V2 (FW: 5’-CTCTCTCGAGAAGAGAGAGGCCGAAGCTCAGGTGCAGCTGC-3’) and IgA-NotI (REV: 5’-CCTCTTGAGCGGCCGCCCTTTAGTAGCATATGCCTTCTG-3’), as described for VHH-IgG20, and cloned in frame with the Saccharomyces cerevisiae α–mating factor prepro sequence within the pPpT4_Alpha_S expression vector21, which was linearized using the enzyme PmeI (NEB) and introduced into Pichia pastoris strain NRRL Y-11430 via electroporation22. The positive Pichia colonies were selected on YPD agar plated with 100 µg/mL of Zeocin®. The expression of 20 individual colonies was analyzed in a 24-well system as described previously20. The medium containing the secreted VHH-IgA antibodies was typically harvested after 48 h of methanol induction. The expression level was evaluated via immunoblots and functional FaeG-ELISA, with and without Endoglycosidase T (Endo T) or Endoglycosidase H (Endo H)23 treatment for removal of N-glycosylations. Clones with high expression were identified and stored as glycerol-stock. As analytical standard, a carboxy-terminal histidine tag version of V2A was expressed in Pichia as described above, purified using IMAC column chromatography and quantified using spectrophotometry at 280 nm.
Seed-based antibody feed formulation
The soybean or Arabidopsis antibody-containing seeds were weighed (Supplementary Table 1), crushed in a chilled knife-mill (Retsch Grindomix GM200) and then mixed with pig feed (Voeders Van Haecke) in two steps to ensure thorough homogeneity. First as a premix of crushed seeds and pig feed with a hand-held electric balloon whisk (Braun), followed by mixing the premix with the pig feed in a fork dough mixer (150 L bowl capacity) for 30 min, to attain the daily antibody dose (20 mg or 5 mg, first and second challenge experiment, respectively) per 300 g of feed, which was the daily ration of feed accounted per piglet. To maintain equal nutrition in all groups throughout each experiment, flax seeds were used to replace Arabidopsis seeds. Additionally, in the second challenge experiment with soybean-produced antibodies, to account for the additional soybean proteins, wild-type soybean seeds were added to the Pichia, Arabidopsis and negative control groups at equal proportions (Supplementary Table 1B). All formulated feeds were bagged and stored at dry ambient temperature.
Pichia-based antibody feed formulation
Cultures for either mV2A or mV3A were grown in 2l baffled shake-flasks in two batches of 7.5 l (i.e. 30 flasks each with 250 ml culture, per batch) amounting to 15 l. Cultures were grown for 48 h in BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, 1.34% yeast nitrogen base, 1% glycerol) up to stationary phase. Cells were induced by replenishing the medium with BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate 1.34% yeast nitrogen base, 1% methanol) for 48 h. Induction was maintained by spiking the cultures with 1% (v/v) of methanol every 12 h. After 48 h of induction, the culture medium was harvested by centrifugation. The cell-free supernatant was concentrated via diafiltration to ~1.5 - 2 l and buffer-exchanged with a sodium-phosphate buffer (20 mM Na2HPO4, 18.75 mM NaCl, pH 6), using the Centramate™ 500 S tangential flow filtration system (Pall Life Science) fitted with a 5 kDa Omega™ Centramate filter cassette. To the resultant ~1.5-2 l protein solution containing Pichia-produced mVHH-IgA, an equal weight of commercial pig feed (Voeders Van Haecke) was added and mixed with a handheld paddle to avoid any foaming, and the slurry was lyophilized using a pilot-scale freeze dryer (Epsilon 2-10 D LSC- Martin-Christ, Germany) for 47 h. The resulting dried powder was then used as premix, which was further mixed in a fork dough mixer for 30 min together with the pig feed to result in 18 kg of final Pichia-produced VHH-IgA-bearing feed (Supplementary Table 1B). All the formulated feeds were bagged and stored at dry ambient temperature.
Evaluation of the mVHH-IgA in the final feed formulation
A 10% (w/v) suspension of the four final feed formulations (Pichia mVHH-IgA, soybean mVHH-IgA, Arabidopsis mVHH-IgA or negative control feed) was made in phosphate buffer saline. On brief centrifugation, the clarified fraction containing soluble proteins was analyzed via immunoblotting and ELISA assay. For immunoblot based analysis the samples were first treated with Endo H to remove N-glycan heterogeneity, resolved with SDS-PAGE under reducing conditions and probed with goat anti-pig IgA polyclonal antibodies conjugated to HRP. A volume extract corresponding to 1.2 mg of the original dry material was loaded per well. Purified poly-histidine tagged mVHH-IgA V2A secreted from Pichia, similarly treated with Endo H was used in a 2-fold dilution series on the same blot as reference standard. For ELISA based analysis the samples were 5-fold serially diluted and tested in the FaeG-ELISA as described above.
Piglet feed-challenge experiment
The Animal Care and Ethics Committee, Faculty of Veterinary Medicine, Ghent University, Belgium approved both of the piglet challenge experiments (ethical dossier number EC2014/02 and EC2015/47). The first challenge experiment aimed at comparing the efficacy of administrating 20 mg daily dose of Arabidopsis seed-contained mVHH-IgA or sVHH-IgA compared with no antibody in feed (Fig. 1) and the second at evaluating a four-fold lower (5 mg) daily dose of mVHH-IgA secreted from Pichia, or contained in transgenic soybean or Arabidopsis seeds, administered in feed (Fig. 2). Suckling piglets (breed: Belgian Landrace) were screened from local farms, and those meeting the inclusion criteria of being seronegative for F4-ETEC and positive for the MUC13-genetic test24, which correlates with the presence of F4-ETEC receptors, were selected for the experiment. Eighteen piglets were sourced from a commercial farm for the first experiment, and 24 piglets from the farms of the ILVO institute for the second experiment. To prevent the contingency of F4-ETEC infection prior to the experiment, piglets from the commercial farm received 150,000 U/kg dose of colistin orally at weaning and feed mixed with colistin (2 g/kg feed) for a week after weaning, as in our previous experiment10. Similarly, based on an antibiogram analysis, the piglets from the ILVO farm were intramuscularly administered amoxicillin instead of oral colistin, in a regimen of 0.1 ml dose on the day of birth and every other day until 6 days after birth, which was increased to a 0.6 ml dose administered on the day before weaning and further on the 2nd and 4th day post-weaning. This protocol was approved by the ILVO institutional ethics board (dossier number- EC2016/267).
On weaning, the piglets were brought to the laboratory stables, given ear tags, and housed in groups of six piglets each. The challenge procedure was performed as described previously10. Briefly, the piglets were challenged on two consecutive days with 1010 F4-ETEC bacteria (strain- GIS26Rstrep)10, via intragastric intubation under sedation after neutralization of gastric pH with bicarbonate buffer for 30 min. The first day of challenge is considered as day 0 in the timeline. Prior to the challenge on day -3 and day -2, piglets were orally administered 1 ml of Nuflor (florfenicol) and 2.5 mg/kg body weight of Baytril (enrofloxacin), to increase F4-ETEC susceptibility by disturbing the gut microflora (dysbiosis). The specific modification, sample collection and manipulations with the animals, the group-specific feed regimen and day of euthanasia are schematically depicted in Fig. 1b and 2b, for the first and second challenge experiment, respectively. The antibody-containing feed was administered for 10 days, starting three (Fig. 1b) or two (Fig. 2b) days before the challenge. During these 10 days, the daily experimental feed diet of 300 g per piglet was provided in a common feeding vat per group. The negative control group always received feed without antibody-containing substrate. To monitor the shedding of the F4-ETEC challenge strain GIS26Rstrep, dilutions of daily fecal samples were immediately plated on blood agar plates with streptomycin (1 mg/mL) selection. Serum antibody against the F4-fimbriae were determined according to the ELISA setup as described previously10. The serum was 2-fold serially diluted and endpoint titers are denoted as the first serum dilution having an OD value below the cut off. The cut off was calculated equal to the OD of a negative reference serum plus two times the standard deviation.
An autopsy was performed post euthanasia, and the phenotypic expression of F4-ETEC receptors was confirmed on excised villous enterocytes in all piglets, as previously described10 and showed 41 to 85 bacteria bound per 250 µm of the cell surface. One piglet from the group receiving 5 mg of Arabidopsis seed produced mVHH-IgA (piglet 18) was excluded from the analysis and statistical calculations, due to umbilical hernia detected post-mortem leading to extreme strangulation of the small intestine, but the data for this piglet is reported in Supplementary Table 3. CFU counts and titers below the detection limit were imputed with values generated as a random sample from a skewed left tailed beta distribution Beta (5,1) for plotting the graphs and statistical calculations.
Melting curve determination
The purified histidine tagged version of V2A as described above, was used for determination of melting curve via the thermal shift assay using Sypro Orange, as described25. Melting curves were obtained for purified V2A within phosphate buffer saline or spray-drying buffer (20 mM Na2HPO4, 18.75 mM NaCl, pH 6 with 10% (w/v) maltodextrin).
Spray-dried Pichia produced VHH-IgA
Fifteen liters each of mVHH-IgA V2A and V3A containing Pichia medium were produced in shake flask, subsequently concentrated and buffer exchanged as described above to one liter of retentate. The retentate was transported on ice and diluted to 25 l of sodium- phosphate buffer (20 mM Na2HPO4, 18.75 mM NaCl, pH 6) with 10% maltodextrin. The liquid was mixed thoroughly using an industrial blender for 5-7 minutes and then fed into the spray-drier, with parameters set to 45°C preheating of the feeding liquid, and 170°C inlet air temperature. During the drying process, the average outlet air temperature was about 80°C, and a constant liquid pumping speed was maintained. Approximately 2.3 kg of dried V2A and V3A containing powder was recovered. A 10% (w/v) solution of the mVHH-IgA containing powder and of a control sample of the excipient used for spray-drying were evaluated for their functional antibody content in the FaeG-ELISA (Supplementary Fig. 4).
Statistical analysis
Log-transformed shedding and titer longitudinal data were analyzed (repeated measurements over time) using the residual maximum likelihood (REML) as implemented in Genstat v18 (VSN International). Briefly, a linear mixed model (random terms underlined) of the form log(y) = μ + feed + time +feed.time +pig.time was fitted to the longitudinal data. The term pig.time represents the residual error term with dependent errors because the repeated measurements are taken in the same individual, causing correlations among observations. Times of measurement were set as equally spaced, and the best model for correlation on the longitudinal data (e.g. uniform, autoregressive, antedependence, unstructured) was selected based on the likelihood ratio test (LRT) statistic and the Aikake Information Coefficient (AIC). Significance of feed effects over time (i.e. feed.time) and changes in differences between feed effects over time were assessed by an approximate F-test, of which the denominator degrees of freedom were calculated using algebraic derivatives as implemented in Genstat v18. Additional information is included in the Life Sciences Reporting Summary.
Reporting summary
Further details of the experimental design can be found in the Life Sciences Reporting Summary linked to this article
Data availability
The data of each piglet from the two challenge experiments plotted in Fig 1 and Fig 2 are reported herein; the shedding data in Supplementary Table 2 and Supplementary Table 3, and the serum titers in Supplementary Table 4 and Supplementary Table 5. Additional data is available from the corresponding authors on request.
Supplementary Material
Acknowledgments
We thank E. Van Lerberge and J. Nolf for overall technical support, particularly for upscaling seeds, antibody expression analysis, and protein analysis. We thank S. Brabant for help with blood sampling and analysis of the seroconversion, R. Cooman for animal caretaking and management of the stables and U. Van Nguyen for performing the villous adhesion assay. We thank K. Wang of Iowa State University for the soybean transformation, J. Haustraete of the Protein Service facility of VIB for large scale production of Pichia cultures, and D. De Paepe and K. Coudyser of ILVO food pilot for lyophilisation of Pichia spent medium. For access to weaned piglets we acknowledge the farms of K. Devolder and ILVO, and thank their resident animal husbandry team. We are grateful of S. Millet for feed consultation and access to large industrial mixers for feed formulation at ILVO, and T. Moravec of the Institute of Experimental Botany Prague, J. Mar Björnsson of ORF Genetics, and J. Van Huylenbroeck of ILVO for kindly leasing additional greenhouse space for soybean. The authors would finally like to thank M. Vuylsteke and V. Storme for help with the statistical analysis and A. Bleys for help with editing the manuscript. This work was supported by an IWT-innovation fellowship (IM-140851) awarded to V.V, co-sponsored by AVEVE Biochem, AVEVE Group. J.P. received a Ph.D. stipend from the Research Foundation Flanders (FWO project grant G0C9714N). B.L has been supported by the European Research Council’s consolidator grant awarded to N.C. (ERC-2013-CoG-616966). Overall we would like to acknowledge institutional funding and support from Ghent University and VIB.
Footnotes
Author Contributions
V.V., E.V., N.C, E.C., and A.D. designed research; V.V., J.P., B.L., S.R. performed research; V.V., J.P., B.L., E.C, A.D. and NC analyzed data; and V.V., A.D and N.C. wrote the paper.
Competing Financial Interests
E.V. is an employee of AVEVE Biochem. V.V., B.L., N.C. and A.D. are inventors on one or more patent applications related to the inventions reported in this publication. Research in the author’s laboratories has been sponsored in part by AVEVE Group.
References
- 1.Gagniere J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katsanos KH, Papadakis KA. Inflammatory bowel disease: updates on molecular targets for biologics. Gut Liver. 2017;11:455–463. doi: 10.5009/gnl16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Förster B, Chung PK, Crobach MJT, Kuijper EJ. Application of antibody-mediated therapy for treatment and prevention of Clostridium difficile infection. Front Microbiol. 2018;9:1382. doi: 10.3389/fmicb.2018.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly D, Yang L, Pei Z. Gut microbiota, Fusobacteria, and colorectal cancer. Diseases. 2018;6:109. doi: 10.3390/diseases6040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corthésy B. Recombinant secretory immunoglobulin A in passive immunotherapy: linking immunology and biotechnology. Curr Pharm Biotechnol. 2003;4:51–67. doi: 10.2174/1389201033378020. [DOI] [PubMed] [Google Scholar]
- 7.Moor K, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 2017;544:498–502. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- 8.Strugnell RA, Wijburg OLC. The role of secretory antibodies in infection immunity. Nat Rev Microbiol. 2010;8:656–667. doi: 10.1038/nrmicro2384. [DOI] [PubMed] [Google Scholar]
- 9.Reinhart D, Kunert R. Upstream and downstream processing of recombinant IgA. Biotechnol Lett. 2015;37:241–251. doi: 10.1007/s10529-014-1686-z. [DOI] [PubMed] [Google Scholar]
- 10.Virdi V, et al. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc Natl Acad Sci USA. 2013;110:11809–11814. doi: 10.1073/pnas.1301975110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairbrother JM, Nadeau É, Gyles CL. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev. 2005;6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- 12.Guerrant RL, Steiner TS, Lima AA, Bobak DA. How intestinal bacteria cause disease. J Infect Dis. 1999;179:S331–337. doi: 10.1086/513845. [DOI] [PubMed] [Google Scholar]
- 13.Smart RD, Blum M, Wesseler J. Trends in approval times for genetically engineered crops in the United States and the European Union. J Agric Econ. 2017;68:182–198. [Google Scholar]
- 14.Ciofalo V, Barton N, Kreps J, Coats I, Shanahan D. Safety evaluation of a lipase enzyme preparation, expressed in Pichia pastoris, intended for use in the degumming of edible vegetable oil. Regul Toxicol Pharmacol. 2006;45:1–8. doi: 10.1016/j.yrtph.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Raveendran S, et al. Applications of microbial enzymes in food industry. Food Technol Biotechnol. 2018;56:16–30. doi: 10.17113/ftb.56.01.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moonens K, et al. Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies. Vet Res. 2015;46:14. doi: 10.1186/s13567-015-0151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 18.Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 19.Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 2006;25:206–213. doi: 10.1007/s00299-005-0048-7. [DOI] [PubMed] [Google Scholar]
- 20.De Meyer T, et al. Comparison of VHH-Fc antibody production in Arabidopsis thaliana, Nicotiana benthamiana and Pichia pastoris. Plant Biotechnol J. 2015;13:938–947. doi: 10.1111/pbi.12330. [DOI] [PubMed] [Google Scholar]
- 21.Näätsaari L, et al. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs PP, Geysens S, Vervecken W, Contreras R, Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat Protoc. 2009;4:58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 23.Stals I, et al. Identification of a gene coding for a deglycosylating enzyme in Hypocrea jecorina. FEMS Microbiol Lett. 2010;303:9–17. doi: 10.1111/j.1574-6968.2009.01849.x. [DOI] [PubMed] [Google Scholar]
- 24.Goetstouwers T, et al. Refined candidate region for F4ab/ac enterotoxigenic Escherichia coli susceptibility situated proximal to MUC13 in pigs. PLoS One. 2014;9:e105013. doi: 10.1371/journal.pone.0105013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of each piglet from the two challenge experiments plotted in Fig 1 and Fig 2 are reported herein; the shedding data in Supplementary Table 2 and Supplementary Table 3, and the serum titers in Supplementary Table 4 and Supplementary Table 5. Additional data is available from the corresponding authors on request.